Abstract
Vascular smooth muscle cells (SMC), like all cells, acquire a cell-specific epigenetic signature during development that includes acquisition of a unique repertoire of histone and DNA modifications. These changes are postulated to induce an open chromatin state (referred to as euchromatin) on the repertoire of genes that are expressed in differentiated SMC including SMC-selective marker genes like Acta2 and Myh11, as well as housekeeping genes expressed by most cell types. In contrast, genes that are silenced in differentiated SMC acquire modifications associated with a closed chromatin state (i.e. heterochromatin) and transcriptional silencing. Herein we review mechanisms that regulate epigenetic control of the differentiated state of SMC. In addition, we identify some of the major limitations in the field and future challenges including development of innovative new tools and approaches for performing single-cell epigenetic assays and locus-selective editing of the epigenome that will allow direct studies of the functional role of specific epigenetic controls during development, injury-repair, and disease including major cardiovascular diseases such as atherosclerosis, hypertension, and microvascular disease associated with diabetes.
The vascular smooth muscle cell (SMC) is a highly specialized cell whose principal functions are contraction and regulation of blood pressure and blood flow distribution1. Mature differentiated SMC express a unique repertoire of proteins required for their contractile function2, 3. Importantly, a major subset of this SMC-specific repertoire, including the SMC differentiation marker genes smooth muscle myosin heavy chain (Myh11), smooth muscle alpha actin (Acta2) and SM22 alpha (Tagln) are transcriptionally regulated in a SMC-specific fashion by the CArG box/Serum Response Factor (SRF)/Myocardin (or myocardin-related transcription factor, MRTF) complex4–6. Myocardin (Myocd) is a potent SRF co-activator that is exclusively expressed in SMC and cardiomyocytes7–9 that binds to SRF on CArG regions of the SMC marker genes, but not to other SRF-dependent genes such as c-fos8, 10, 11. This highly specialized transcriptional regulation leads to a cell-specific and coordinate activation of most although not all of the repertoire of SMC marker genes4. There is considerable evidence that epigenetic mechanisms contribute to SMC-specific transcriptional regulation in mature and phenotypically modulated SMC12. Epigenetic regulation is classically defined by mechanisms controlling the heritability of traits or phenotypes from mother to daughter cells without modification of the DNA sequence but rather by modification of DNA bases (methylation, hydroxymethylation)13–15, post-translational modifications of the histone tails (methylation, acetylation, phosphorylation, ubiquitinylation)16, 17, histone subunit variants18 and non-coding RNA19. Due to space constraints and the recent publication of several excellent reviews on epigenetic regulation of SMC differentiation and phenotypic switching12, 20–23, we choose to focus this review on epigenetic programming associated with histone and DNA modifications that dynamically control the chromatin structure and thereby contribute to gene activation or repression in SMC during development, tissue repair/remodeling, and cardiovascular disease.
Vascular SMC acquire a unique cell-specific epigenetic signature during development
There is evidence suggesting that SMC-selective changes in chromatin remodeling contribute to the coordinate activation of CArG-dependent SMC marker genes during differentiation of these cells from embryonic stem cells (ESC)10, 23, 24. Indeed, studies by our lab have shown that binding of the SRF/Myocardin complex to CArG regions within the SMC marker genes is developmentally regulated and SMC selective. Key observations include the following. First, chromatin remodeling characterized by histone acetylation, a potent mechanism of gene activation, occurs on Myh11 and Acta2 early in the process of SMC differentiation from SMC precursors in response to treatment with retinoic acid (RA)24. Interestingly, despite being expressed in both undifferentiated and RA-differentiated SMC precursors, SRF binds only to the SMC marker genes that have been enriched in histone acetylation in RA-treated SMC precursor cells. Although correlative, these data suggest that changes in chromatin accessibility, mediated at least in part by histone modifications, render SMC marker genes permissive for subsequent activation by the SRF/Myocardin complex during SMC differentiation. Moreover, inhibition of Histone Acetyl Transferases (HATs) or expression of Histone Deacetylases (HDACs) leads to decreased SMC marker gene promoter activity in cultured SMC25, 26. Second, there is marked enrichment of the histone modification H3K4me2 on SMC marker genes including Myh11, Acta2, and Tagln in mature SMC as well as SMC progenitor cells committed to differentiate into SMC. In contrast, this enrichment was not seen in ESC and non-SMC somatic cells10. Third, Chromatin immunoprecipitation (ChIP) assays in SMC lines stably transfected with either Acta2 CArG-wild type or CArG-mutant promoters demonstrated that H3K4me2 enrichment occurs in absence of binding of SRF to CArG elements suggesting it is regulated through an upstream CArG-SRF independent process10. Fourth, there is selective tethering of Myocardin to H3K4me2-modified histone tails. We postulate that this helps stabilize binding of the SRF-Myocardin complex to the CArG regions which in the case of many SMC promoters are degenerate and exhibit reduced SRF binding affinity10.
These histone modifications are believed to alter the higher order chromatin structure through attraction or repelling of charged histone tails thereby regulating nucleosome density and the “accessibility” of various cis promoter-enhancer control elements. However, covalent modifications of DNA nucleotides have also been implicated in gene regulation. For example, methylation of cytosine residues has long been recognized as a stable mechanism of gene repression and chromatin compaction, and for many years was incorrectly thought to be relatively irreversible. Indeed, identification of mechanisms that mediate demethylation of DNA at discrete gene loci with associated gene de-repression is currently an area of intense interest in the epigenetic field27–29. For example, it has recently been shown that conversion of 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC) by Ten-Eleven Translocation (TET) enzyme-mediated oxidation appears to be an important mechanism of reactivation of genes14, 30. The most notable example of the latter occurs during reprogramming of somatic cells into induced pluripotency stem (iPS) cells31 for which Yamanaka received the 2012 Nobel Prize32 albeit this particular example is induced by artificial hyper-physiological over-expression of the iPS cell reprogramming factors Oct4, Sox2, c-myc, and Klf4. However, it has also been shown that 5hmC plays a major role in maintenance of pluripotency, self-renewal, and proper differentiation of embryonic stem cells (ESC) during early development33, 34. The role of 5hmC in somatic cells is poorly understood. However, several studies suggest that 5hmC promotes cell-specific differentiation programs in somatic cells via the enzyme TET235–37. Recent innovative studies by Liu et al.35 provide evidence that 5hmC also plays a key role in regulating SMC selective gene expression during development and phenotypic switching. TET2 expression is higher in mature SMC compared with ESC and is upregulated following treatment with rapamycin in vitro. TET2 is also highly expressed in medial SMC from healthy aortas. Moreover, knock down of TET2 by shRNA induces repression of SMC-specific genes including Myh11, Acta2, Myocd, and SRF and a correlated decrease in enrichment in 5hmC on these genes in cultured SMC.
Vascular SMC is not terminally differentiated and undergoes marked changes in phenotype during vascular injury-repair and in disease states
It has long been assumed that the SMC is not terminally differentiated, and that it can undergo rapid changes in phenotype in response to vascular injury, or in various cardiovascular diseases, including atherosclerosis [reviewed in12, 38–40]. However, until very recently, the evidence for this was nearly completely based on inferences from studies in cultured SMC including demonstration by our group41–44 and many others45–47 that treatment of cultured SMC with various growth factors such as PDGF (platelet derived growth factor) or bFGF (basic fibroblast growth factor), oxidized phospholipids, or pro-inflammatory cytokines resulted in coordinate reductions in expression of SMC marker genes, as well as increased migration, proliferation, and secretion of various extracellular matrix proteins and cytokines a process collectively referred to as “phenotypic modulation or switching”. That said, our understanding of phenotypic modulation and the fate of phenotypically modulated SMC in vivo are highly confounded by the fact that this process is characterized by down-regulation of the SMC marker genes like Acta2 and Myh11 making their identification in vivo unreliable without rigorous lineage tracing systems40. As such, the paradigm of SMC phenotypic switching during vascular injury and disease has been challenged by studies suggesting that a significant subset, if not the majority, of lesion and neointimal cells are of myeloid48, 49 or adventitial50 origin rather than SMC. The possibility that adventitial stem cells give rise to intimal lesion cells is intriguing but at present there is no direct evidence that they do so due to the lack of model systems with which to definitely lineage trace these cells in vivo. Moreover, initial claims that the majority of SMC-like cells within lesions are of myeloid origin by Nagai and co-workers48 were subsequently directly refuted in a series of bone marrow transfer (BMT) studies by Bentzon et al.51, 52 who claimed that myeloid cells fail to express Acta2. In direct contrast, a follow-up paper by the Nagai group53 using a combination of BMT studies and a Myh11 promoter reporter system, refuted their earlier studies48 by showing that the majority of SMC marker positive cells within lesions are not of myeloid origin, but that some myeloid cells can express early (Acta2, Tagln) but not late (Myh11 and Cnn1) SMC differentiation markers. These latter results are consistent with cross gender BMT human studies by Caplice et al.49 showing that approximately 10% of ACTA2+ cells in human lesions are of myeloid origin. Taken together, although there remains some controversy regarding the extent to which myeloid cells express SMC markers within lesions, our interpretation of the studies is that at least some do but that the majority of SMC marker positive cells within lesions are NOT of myeloid origin.
A much more profound claim that challenged the conventional dogma that SMC are the principal source of SMC marker positive cells within the neointima was made in a study by Tang et al54 who concluded that mature SMC are terminally differentiated and that the primary source of neointimal cells following vascular injury are derived from what they claimed was a novel previously uncharacterized medial stem cell population. However, the main conclusions of this paper were strongly refuted in an Editorial from a collective group of authors who are leaders in the field including ourselves55 on the basis of there being major technical and design flaws in those studies. More importantly, a number of subsequent rigorous lineage tracing studies by our group56, 57 and several others58–60 directly refute the major conclusions of Tang et al. and provide compelling evidence that SMC are plastic and exhibit phenotypic switching in vivo [for a more complete review of this critical topic please see recent reviews by Tabas et al.61 and Bennett et al.62].
We recently employed a novel SMC specific conditional lineage tracing mouse model [i.e. Myh11-CreERT2 ROSA floxed STOP eYFP mice, designated “SMC YFP+/+” mice] to show that >80% of SMC-derived cells within advanced ApoE−/− brachiocephalic artery (BCA) lesions lacked detectable expression of SMC markers such as Acta2 and could only be detected using a rigorous SMC lineage tracing system57. Moreover, we showed that approximately a third of these cells that were YFP+ but Acta2− expressed multiple markers of macrophages, and thus have likely been misidentified as being monocyte-derived macrophages in previous studies in the field. Most importantly, we showed that these phenotypic transitions of SMC were functionally important, in that we showed that SMC specific conditional KO of the pluripotency factor Klf4 did not prevent SMC phenotypic switching, but altered the nature of the phenotypic transitions including a marked reduction in SMC-derived macrophage-like cells. Consistent with these results, Feil et al.58 used a Tagln CreERT2 ROSA floxed STOP LacZ mouse model to show that a subset of SMC-derived cells express several macrophage markers, and SMC-derived cells within lesions underwent clonal expansion during atherosclerosis using a R26R-Confetti multicolor Cre reporter mice. Additional evidence that SMC are not terminally differentiated and undergo phenotypic transitions during vascular injury-repair can be found in studies by Nemenoff et al.59 who used a conditional Myh11 creERT2 ROSA26 LacZ reporter mouse to provide compelling evidence that the vast majority of neointimal cells following wire-induced injury of the femoral artery are derived from pre-existing differentiated (i.e. Myh11+) SMC59. These latter observations have been confirmed and extended in very elegant recent studies by Herring et al.60 which used conditional Myh11 and Acta2 creERT2 mTmG mice to show that previously differentiated SMC de-differentiate and constitute the main source of SMC within the neointima following carotid ligation injury.
Taken together, results indicate that typical methods for detecting SMC based on immunostaining for SMC marker genes not only greatly under-estimate the number of SMC derived cells within lesions, but also misidentify many of these cells since they lack SMC markers and have activated markers of other cell types including macrophages. Likewise, there is also clear evidence that cells other than SMC activate at least some SMC markers in the context of atherosclerotic lesions49, 53 [reviewed in40]. As such, studies establish that reliable identification of SMC derived cells within intimal lesions following injury or within atherosclerotic lesions must rely on use of rigorous SMC lineage tracing methods which is absolutely dependent on: 1) efficient and SMC-specific conditional activation of a sensitive lineage tracing gene; and 2) high resolution imaging to ensure the lineage tracing gene and markers of cell phenotype are present within the same cell (for a thorough review of requirements for definitive cell lineage tracing see this excellent review63.
Modifications of the SMC epigenetic programming are associated with SMC phenotype modulation
There is evidence suggesting that epigenetic mechanisms play an important role in regulating phenotypic switching of SMC. For example, we showed that PDGF BB-induced phenotypic transition of cultured SMC was associated with: 1) decreased histone acetylation mediated through Klf4-dependent recruitment of HDACs 2, 4, and 5 to multiple CArG-dependent SMC marker genes10, 42; and 2) enrichment of the silencing modification H3K9me3 on SMC promoters10. Consistent with these findings, subsequent studies by our lab64 showed that silencing of the Tagln gene following ligation-induced carotid injury in vivo also appeared to be mediated through binding of HDAC2 to the CArG region as well as pELK and reduced H4 acetylation, mediated through Klf4 dependent binding to a G/C repressor element in the Tagln promoter that is also found in most other SMC CArG-dependent marker genes. That is, Klf4 is upregulated following injury, binds to G/C repressor elements and recruits both HDACs and pElk that contribute to silencing of SMC marker genes. Of major interest, we have recently provided evidence indicating that similar mechanisms contribute to silencing of SMC marker genes within phenotypically modulated SMC within atherosclerotic lesions, in that we showed marked enrichment of Klf4 on the promoters of Acta2, Myh11, and Tagln based on in vivo ChIP assays on chromatin extracted from advanced ApoE2212;/2212; BCA lesions57.
Although likely to be of critical importance, unfortunately very little is currently know with respect to the contribution of DNA methylation in SMC phenotypic switching. Several lines of evidence suggest that changes in the DNA methylation pattern might play a role in major vascular diseases such as atherosclerosis and SMC phenotypic switching65. First, a process of global hypomethylation and decrease in DNA methyltransferases expression have been observed in human and mouse atherosclerotic lesions66. Nevertheless, this study does not provide direct evidence that the hypomethylation phenomenon occurs in SMC and the full spectrum of genes impacted by the hypomethylation is unknown. Second, SMC marker genes including Tagln and Myh11 promoters are hypomethylated in SMC compared with 10T1/2 cells67. In addition, the Myh11 promoter in cultured SMC contains significantly more 5mC as compared to freshly isolated aortic SMC67. These latter observations suggest that dynamic modifications of DNA methylation patterns of SMC marker genes may contribute to changes in expression of these genes during phenotypic transitions, at least during adaptation of cells to culture. Third, modulation of DNA demethylase TET2 expression and activity and the concomitant changes in hydroxymethylation of the SMC marker genes have been associated with SMC phenotypic switching within atherosclerotic lesions and following femoral artery injury35. TET2 expression is reduced in injured and atherosclerotic vessels. Moreover, TET2 knockdown induced increased neointimal proliferation post-injury, whereas overexpression of TET2 had the opposite effect35. Taken together, results suggest that changes in DNA methylation are likely to contribute to changes in gene expression during SMC phenotypic transitions but much further work is needed in this critical area.
Of major importance, we have observed that at least a part of the SMC epigenetic programming established during SMC differentiation is retained during phenotypic switching. Indeed, H3K4me2 is stably enriched on the Myh11 and Acta2 promoters in SMC treated with PDGF-BB35, or oxidized phospholipids56 compared with untreated SMC. Rigorous investigation of epigenetic modifications within SMC in vivo is particularly challenging considering the fact that normal and diseased tissues are made up of a large number of heterogeneous cell types. Indeed, this is a major general limitation in the epigenetic field that has greatly confounded efforts attempting to elucidate how any given epigenetic modification might alter the function of a particular cell type within a complex tissue. To circumvent this limitation, we developed a method which we refer to as ISH-PLA (in situ hybridization-proximity ligation assay), to visualize histone modifications at single genomic loci within individual cells in tissue sections56. Importantly, since there was no precedent for doing these types of analyses within intact tissue sections, it was critical to rigorously validate the methodology including doing the following. First, we employed our SMC specific Myh11 YFP lineage tracing mice to demonstrate that the presence of H3K4me2 on the Myh11 promoter (Myh11 H3K4me2) was completely restricted to SMC within all mouse and human tissues examined including numerous arteries and arterioles. Second, an endothelial cell (EC) selective epigenetic modification, H3K4me2 of Cdh5 (vascular endothelial cadherin) is restricted to endothelial cells in all blood vessels examined in both mouse and human paraformaldehyde fixed tissue specimens. We subsequently used our ISH-PLA method in conjunction with our SMC lineage-tracing mice to show that H3K4me2 is retained in phenotypically modulated SMC within atherosclerotic lesions that lack detectable expression of SMC marker genes (i.e. YFP+Acta2− lesion cells)56. Moreover, Myh11 H3K4me2 was maintained in SMC that had undergone transition to a macrophage-like state within mouse and human lesions57. Remarkably, the combination of ISH-PLA and rigorous Myh11 YFP lineage tracing showed that >30% of Lgals3+ cells within advanced mouse lesions were of SMC and not myeloid origin as has been assumed in previous studies in the field. Moreover, using ISH-PLA we provided evidence that nearly 20% of macrophage marker positive cells within advanced human coronary artery lesions were of SMC- not myeloid origin, and >35% of cells dual positive for Lgals3 and Acta2 were of SMC origin. The latter is of major significance in that a recent paper from Gordon Francis68 showed that 50% of foam cells within advanced human coronary artery lesions are dual positive for these markers. Taken together, these results resolve the decade long debate as to whether SMC are a major source of foam cells within human lesions. Importantly, our findings were confirmed by performing a combination of ISH-PLA detection of the SMC specific epigenetic mark, Myh11 H3K4me2, and Y-chromosome tracking in advanced coronary artery lesions of a male subject who had received a female heart transplant57. That is, we observed Y-chromosome negative CD68+ cells in lesions from the female donor that had a SMC Myh11 H3K4me2 epigenetic signature. Importantly, in no case Y-chromosome+ cells (i.e. of myeloid origin from the male recipient) that had the SMC epigenetic signature were observed indicating that myeloid cells do not acquire this mark in the context of atherosclerotic lesions thus further validating these method and findings. These observations lead us to the conclusion that H3K4me2 on the SMC marker genes is a restricted and stable epigenetic signature of the SMC lineage distinguishing SMC from other cell types independently of their state of differentiation. The next section will consider the possible functional significance of this observation.
Do epigenetic mechanisms regulate cell lineage memory?
Biologists have been challenged for centuries trying to elucidate the mechanisms that contribute to maintenance of cellular lineage identify and integration of environmental cues across cell generations including during development of complex multicellular organisms from a single fertilized egg. Whereas it has been assumed that this critical cell property must be regulated through “epigenetic” mechanisms, and this might be the case, there are a number of major limitations in the experimental evidence in support of this concept including the following.
First, the vast majority of the functional epigenetic studies have been based on genetic deletion of various histone modifying enzymes and DNA methyltransferases that result in global and massive changes across the entire genome and which not surprisingly are often embryonic or postnatal lethal69. Similarly, pharmacological inhibition of histone modifying enzymes (e.g. HDAC inhibitors) leads to global epigenetic reprogramming of the genome. Whereas these sorts of studies establish that epigenetic controls are critical, by no means can one ascertain that a particular change at a specific locus is functional.
Second, tremendous emphasis has been made on the identification of cell-specific epigenetic programming by high-throughput sequencing of the whole genome and recent studies compiled and compared epigenetic profiles of somatic cells in culture or extracted from different tissues70 and ESC undergoing differentiation into primordial germ lines70–73. These data showed that lineages acquire specific epigenetic programming “signatures”. However, these studies are correlative and they fail to ascertain if acquisition of a cell specific epigenome is the cause of cell differentiation, or the consequence.
Third, by far the majority of epigenomic studies have been done on cultured cells systems that poorly replicate epigenetic controls in vivo. Indeed, cultured SMC are poorly differentiated and exhibit a phenotype very different than mature differentiated SMC in vivo because cell culture systems do not recapitulate complex environmental cues that regulate SMC differentiation in vivo under normal and pathological conditions1, 12. As a relevant illustration of this bias, studies by Zhu et al.70 involving genome wide mapping of the epigenome of various human tissues and cultured cells provide a striking confirmation of just how different cultured cells can be from their in vivo counterparts and how somatic cells from different lineages tend to undergo similar epigenetic reprogramming once they are cultured in vitro, becoming more and more like one another, while less and less like their in vivo brethren. Indeed, these latter studies showed that variety of cultured cells from different origins acquired a very similar epigenome during adaptation to growth in culture, and which was very distinct from that exhibited in vivo in the rare cases where this could be ascertained.
Fourth, studies including ChIP-seq performed on chromatin derived from multi-cellular tissue samples have attempted to differentiate which epigenetic signals come from each respective cell type using bioinformatics approaches74 that utilize epigenomes identified from studies of cultured cells. Although these studies have the advantage that assays were done on a homogenous cell population, as noted above, it is extremely unlikely the epigenome displayed accurately replicates that displayed by their in vivo counterparts due to the many differences in environmental cues present in the two systems. Ironically, the latter includes the absence of heterotypic cell interactions. As a relevant example, ChIP and ChIP-seq cannot be used to rigorously investigate SMC-specific epigenetic regulation in vivo because chromatin obtained from SMC-rich tissues is not exclusively derived from SMCs but is rather “contaminated” by chromatin derived from multiple cell types. Consequently, ChIP analyses on an atherosclerotic lesion specimen represent a composite signal derived from many different cell types (e.g. endothelial cells, monocytes, macrophages, adventitial fibroblasts) that are all susceptible to change their phenotype and their epigenetic programming during development of the disease40. Importantly, this issue has also not been adequately addressed in nearly all studies involving analyses of genome-wide DNA methylation data75, 76 and comparative studies using either ChIP-seq or DNA methylation datasets almost always rely on biased bioinformatics approaches. Importantly, we have identified a simple, albeit expensive and time-consuming means to at least partially resolve these limitations as exemplified in our recent studies examining the effect of SMC-specific conditional knockout of the stem cell pluripotency factor Klf4 within ApoE−/− mice fed a Western diet for 18 weeks to induce formation of advanced atherosclerotic lesions57. In brief, we performed differential Klf4 ChIP-seq analyses of chromatin samples obtained from brachiocephalic lesions of our Klf4 wild-type SMC lineage tracing atherosclerotic mice (i.e. SMC Klf4+/+ YFP+/+ Apoe−/−) versus SMC-specific Klf4 KO atherosclerotic mice (i.e. SMC Klf4Δ/Δ YFP+/+ Apoe−/−) thus enabling us to identify putative Klf4 target genes selective for SMC.
The last, and perhaps the most striking limitation of epigenetic studies in vivo is the failure to provide direct loss of function evidence that a given epigenetic modification has a specific functional effect in a given cell. Let us finish this review by addressing this considerable challenge.
Development of new methods for locus specific epigenetic editing and studying functional consequences
As noted previously functional epigenetic studies require the combination of locus-specific epigenomic editing with single cell epigenetic and functional analyses. While at one time this would have seemed impossible, it appears that all the pieces may finally be available, although as far as we are aware no one has yet successfully combined them. Here are the pieces.
First, several methods are now available for doing single-cell epigenetics [reviewed in77] including single-cell RNA-seq78 and single-cell genome-wide DNA methylation analysis79. Our ISH-PLA method allows investigation of epigenetic modification in single cells in vivo in fixed histological sections and is powerful for validation purposes, including in both normal and diseased human tissue sections, but by no means is this method conducive for high throughput screens or initially defining the epigenome of a given cell type in vivo. That is, you must first define what you want to look for using other methods and approaches.
Second, several recent papers have described innovative new methods and tools that permit reliable cell-specific/locus-specific editing of the epigenome80–83, which has become the Holy Grail for the epigenetic field and are essential for determining the functional role of specific epigenetic changes and reprogramming in major human diseases including cardiovascular pathologies. The use of technologies like Zinc fingers, TAL effector proteins and CRISPR-Cas9 initially designed for genome editing84–86 have recently been used to modify the epigenome by altering DNA methylation80 or histone modification81, 82 patterns at single genomic loci. Importantly, a recent Nature Biotechnology article from Bradley Bernstein’s lab81 described the first method to achieve locus specific editing of the epigenome. This study targets histone marks H3K4me2 and H3K27ac characteristic of active enhancers by the use of TAL effector proteins fused with the histone de-methylase LSD1 (enzyme responsible for removal of H3K4me2) to aim specific gene enhancer regions. With the 40 enhancers targeted, the authors observed a locus-specific decrease in H3K4me2 and H3K27ac in 65% of these enhancers. For nine of the TALE-LSD1 constructs, a comparison between ChIP-seq and RNA-seq demonstrated a correlation between H3K4me2 removal on enhancers and down-regulation of genes regulated by these enhancers.
Taken together, these results provide a compelling proof of principle that locus-selective editing of the epigenome is possible although surprisingly this initial paper did not go onto examine how these site specific epigenomic modifications impacted cell function, and importantly studies were done entirely in vitro. However, the field appears to be poised to finally be able to combine single cell epigenetic assays with single cell locus-specific epigenetic editing to directly and rigorously begin to assess the exact functional roles of cell-specific epigenetic controls in regulation of cell differentiation, cell lineage memory, and phenotypic transitions during tissue injury, repair, and adaptation, as well as how these critical processes are altered in disease states, including major cardiovascular diseases. Indeed, it is an exciting time for the field.
Summary and New Perspective
Observations of a SMC-specific epigenetic signature including SMC-specific distribution of histone modification and DNA hydroxymethylation are consistent with the long-standing dogma in the epigenetic field that differences in chromatin organization and distribution of histone marks are crucial events modulating the “reading” of the unique genomic sequence and allowing generation of multiple somatic cell types from ESC. Importantly, there is evidence in vitro and in vivo that SMC retain their SMC-specific epigenetic signature (i.e. enrichment of H3K4me2 on the SMC marker genes) during phenotypic switching and their transition to alternative phenotypes. Nevertheless, a major challenge for the field is to determine if such epigenetic signatures play a critical functional role in controlling SMC lineage identity and functional phenotypes in vivo. Novel methodologies including rigorous lineage tracing, single-cell epigenetic assay and locus-specific epigenome editing tools provide unique new opportunities to investigate the functional relevance of epigenetic programming in controlling SMC identity and plasticity in the context of major cardiovascular diseases such as atherosclerosis.
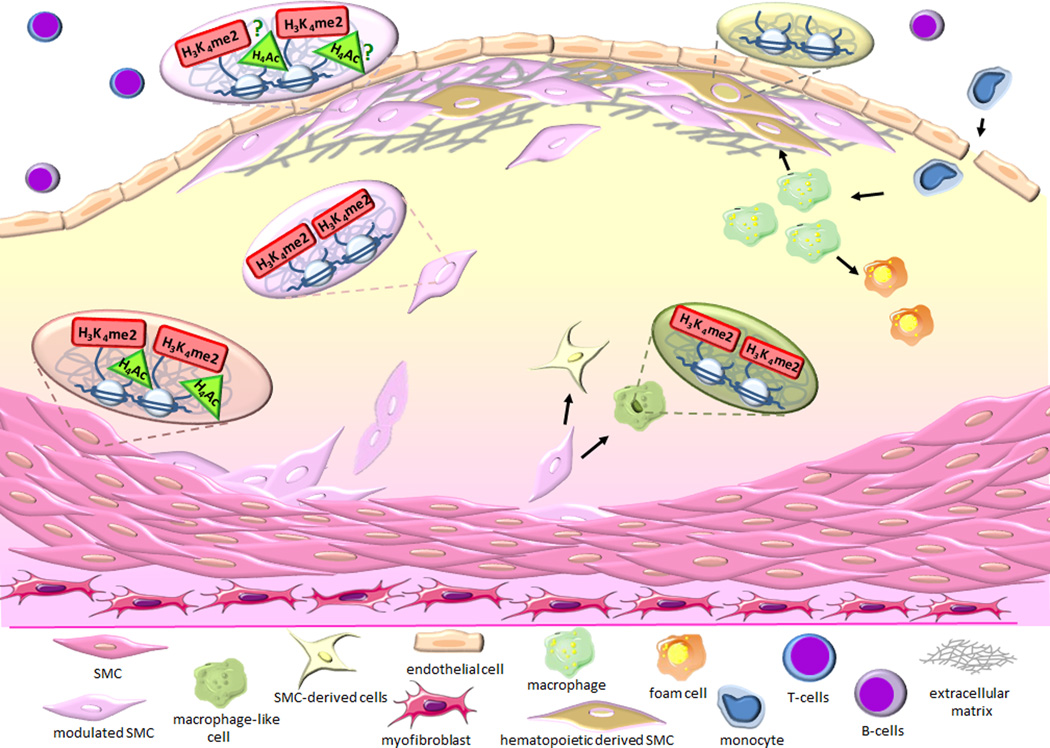
Figure 1. In vivo epigenetic signature in differentiated and phenotypically modulated SMC within atherosclerotic lesion.
Differentiated vascular smooth muscle cells (SMC) display a lineage restricted epigenetic signature characterized by the enrichment of histone modifications H3K4me2 and H4ac on the SMC marker genes including Acta2 and Myh1110, 56. Using SMC lineage tracing systems, tracking of SMC which have lost expression of the SMC marker repertoire allowed the rigorous identification of dedifferentiated SMC and of SMC transitions into macrophage-like, myofibroblast-like, and mesenchymal stem cell-like cells within advanced atherosclerotic lesions58. Development and utilization of in vivo single cell epigenetic assays such as ISH-PLA demonstrated that H3K4me2 on the Myh11 gene was retained during phenotypic switching and transition of SMC into macrophage-like cells56. In contrast, cells from myeloid origin do not acquire this SMC restricted signature. These observations lead to the hypothesis that appearance and retention of H3K4me2 on the SMC repertoire might play a critical role controlling SMC differentiation, identity and cell lineage memory.
ACKNOWLEDGMENTS
None.
Sources of Funding. This work was supported by US National Institutes of Health R01 grants HL057353, HL098538 and HL087867 to G.K.O., and American Heart Association grant 15SDG25860021 to D.G.
Footnotes
Disclosure. The authors declare no competing financial interests.
REFERENCES
- 1.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiological reviews. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 2.Owens GK. Regulation of differentiation of vascular smooth muscle cells. Physiological reviews. 1995;75:487–517. doi: 10.1152/physrev.1995.75.3.487. [DOI] [PubMed] [Google Scholar]
- 3.Somlyo AP, Somlyo AV. Ca2+ sensitivity of smooth muscle and nonmuscle myosin II: modulated by G proteins, kinases, and myosin phosphatase. Physiological reviews. 2003;83:1325–1358. doi: 10.1152/physrev.00023.2003. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida T, Sinha S, Dandre F, Wamhoff BR, Hoofnagle MH, Kremer BE, Wang DZ, Olson EN, Owens GK. Myocardin is a key regulator of CArG-dependent transcription of multiple smooth muscle marker genes. Circulation research. 2003;92:856–864. doi: 10.1161/01.RES.0000068405.49081.09. [DOI] [PubMed] [Google Scholar]
- 5.Du KL, Ip HS, Li J, Chen M, Dandre F, Yu W, Lu MM, Owens GK, Parmacek MS. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Molecular and cellular biology. 2003;23:2425–2437. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Kitchen CM, Streb JW, Miano JM. Myocardin: a component of a molecular switch for smooth muscle differentiation. Journal of molecular and cellular cardiology. 2002;34:1345–1356. doi: 10.1006/jmcc.2002.2086. [DOI] [PubMed] [Google Scholar]
- 7.Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z, Wang DZ, Pipes GC, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long X, Creemers EE, Wang DZ, Olson EN, Miano JM. Myocardin is a bifunctional switch for smooth versus skeletal muscle differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16570–16575. doi: 10.1073/pnas.0708253104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK. Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. The Journal of clinical investigation. 2006;116:36–48. doi: 10.1172/JCI26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miano JM. Serum response factor: toggling between disparate programs of gene expression. Journal of molecular and cellular cardiology. 2003;35:577–593. doi: 10.1016/s0022-2828(03)00110-x. [DOI] [PubMed] [Google Scholar]
- 12.Alexander MR, Owens GK. Epigenetic control of smooth muscle cell differentiation and phenotypic switching in vascular development and disease. Annual review of physiology. 2012;74:13–40. doi: 10.1146/annurev-physiol-012110-142315. [DOI] [PubMed] [Google Scholar]
- 13.Bird A. DNA methylation patterns and epigenetic memory. Genes & development. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 14.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 16.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 17.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 18.Henikoff S, Furuyama T, Ahmad K. Histone variants, nucleosome assembly and epigenetic inheritance. Trends in genetics : TIG. 2004;20:320–326. doi: 10.1016/j.tig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovascular research. 2011;90:430–440. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu R, Leslie KL, Martin KA. Epigenetic regulation of smooth muscle cell plasticity. Biochimica et biophysica acta. 2015;1849:448–453. doi: 10.1016/j.bbagrm.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spin JM, Maegdefessel L, Tsao PS. Vascular smooth muscle cell phenotypic plasticity: focus on chromatin remodelling. Cardiovascular research. 2012;95:147–155. doi: 10.1093/cvr/cvs098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Findeisen HM, Kahles FK, Bruemmer D. Epigenetic regulation of vascular smooth muscle cell function in atherosclerosis. Current atherosclerosis reports. 2013;15:319. [PubMed] [Google Scholar]
- 23.McDonald OG, Owens GK. Programming smooth muscle plasticity with chromatin dynamics. Circulation research. 2007;100:1428–1441. doi: 10.1161/01.RES.0000266448.30370.a0. [DOI] [PubMed] [Google Scholar]
- 24.Manabe I, Owens GK. Recruitment of serum response factor and hyperacetylation of histones at smooth muscle-specific regulatory regions during differentiation of a novel P19-derived in vitro smooth muscle differentiation system. Circulation research. 2001;88:1127–1134. doi: 10.1161/hh1101.091339. [DOI] [PubMed] [Google Scholar]
- 25.Qiu P, Li L. Histone acetylation and recruitment of serum responsive factor and CREB-binding protein onto SM22 promoter during SM22 gene expression. Circulation research. 2002;90:858–865. doi: 10.1161/01.res.0000016504.08608.b9. [DOI] [PubMed] [Google Scholar]
- 26.Cao D, Wang Z, Zhang CL, Oh J, Xing W, Li S, Richardson JA, Wang DZ, Olson EN. Modulation of smooth muscle gene expression by association of histone acetyltransferases and deacetylases with myocardin. Molecular and cellular biology. 2005;25:364–376. doi: 10.1128/MCB.25.1.364-376.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, Abramowitz LK, Bartolomei MS, Rambow F, Bassi MR, Bruno T, Fanciulli M, Renner C, Klein-Szanto AJ, Matsumoto Y, Kobi D, Davidson I, Alberti C, Larue L, Bellacosa A. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Y, Chen J, Li K, Wu T, Huang B, Liu W, Kou X, Zhang Y, Huang H, Jiang Y, Yao C, Liu X, Lu Z, Xu Z, Kang L, Chen J, Wang H, Cai T, Gao S. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell stem cell. 2013;12:453–469. doi: 10.1016/j.stem.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 33.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 34.Dawlaty MM, Breiling A, Le T, Barrasa MI, Raddatz G, Gao Q, Powell BE, Cheng AW, Faull KF, Lyko F, Jaenisch R. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Developmental cell. 2014;29:102–111. doi: 10.1016/j.devcel.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu R, Jin Y, Tang WH, Qin L, Zhang X, Tellides G, Hwa J, Yu J, Martin KA. Ten-eleven translocation-2 (TET2) is a master regulator of smooth muscle cell plasticity. Circulation. 2013;128:2047–2057. doi: 10.1161/CIRCULATIONAHA.113.002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin SG, Jiang Y, Pfeifer GP, Lu Q. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell reports. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Owens GK. Molecular control of vascular smooth muscle cell differentiation and phenotypic plasticity. Novartis Foundation symposium. 2007;283:174–191. doi: 10.1002/9780470319413.ch14. discussion 191-173, 238-141. [DOI] [PubMed] [Google Scholar]
- 39.Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473:317–325. doi: 10.1038/nature10146. [DOI] [PubMed] [Google Scholar]
- 40.Gomez D, Owens GK. Smooth muscle cell phenotypic switching in atherosclerosis. Cardiovascular research. 2012;95:156–164. doi: 10.1093/cvr/cvs115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holycross BJ, Blank RS, Thompson MM, Peach MJ, Owens GK. Platelet-derived growth factor-BB-induced suppression of smooth muscle cell differentiation. Circulation research. 1992;71:1525–1532. doi: 10.1161/01.res.71.6.1525. [DOI] [PubMed] [Google Scholar]
- 42.Yoshida T, Gan Q, Shang Y, Owens GK. Platelet-derived growth factor-BB represses smooth muscle cell marker genes via changes in binding of MKL factors and histone deacetylases to their promoters. American journal of physiology Cell physiology. 2007;292:C886–C895. doi: 10.1152/ajpcell.00449.2006. [DOI] [PubMed] [Google Scholar]
- 43.Pidkovka NA, Cherepanova OA, Yoshida T, Alexander MR, Deaton RA, Thomas JA, Leitinger N, Owens GK. Oxidized phospholipids induce phenotypic switching of vascular smooth muscle cells in vivo and in vitro. Circulation research. 2007;101:792–801. doi: 10.1161/CIRCRESAHA.107.152736. [DOI] [PubMed] [Google Scholar]
- 44.Alexander MR, Murgai M, Moehle CW, Owens GK. Interleukin-1beta modulates smooth muscle cell phenotype to a distinct inflammatory state relative to PDGF-DD via NF-kappaB-dependent mechanisms. Physiological genomics. 2012;44:417–429. doi: 10.1152/physiolgenomics.00160.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bornfeldt KE, Raines EW, Nakano T, Graves LM, Krebs EG, Ross R. Insulin-like growth factor-I and platelet-derived growth factor-BB induce directed migration of human arterial smooth muscle cells via signaling pathways that are distinct from those of proliferation. The Journal of clinical investigation. 1994;93:1266–1274. doi: 10.1172/JCI117081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaplan-Albuquerque N, Van Putten V, Weiser-Evans MC, Nemenoff RA. Depletion of serum response factor by RNA interference mimics the mitogenic effects of platelet derived growth factor-BB in vascular smooth muscle cells. Circulation research. 2005;97:427–433. doi: 10.1161/01.RES.0000179776.40216.a9. [DOI] [PubMed] [Google Scholar]
- 47.Millette E, Rauch BH, Defawe O, Kenagy RD, Daum G, Clowes AW. Platelet-derived growth factor-BB-induced human smooth muscle cell proliferation depends on basic FGF release and FGFR-1 activation. Circulation research. 2005;96:172–179. doi: 10.1161/01.RES.0000154595.87608.db. [DOI] [PubMed] [Google Scholar]
- 48.Sata M, Saiura A, Kunisato A, Tojo A, Okada S, Tokuhisa T, Hirai H, Makuuchi M, Hirata Y, Nagai R. Hematopoietic stem cells differentiate into vascular cells that participate in the pathogenesis of atherosclerosis. Nat Med. 2002;8:403–409. doi: 10.1038/nm0402-403. [DOI] [PubMed] [Google Scholar]
- 49.Caplice NM, Bunch TJ, Stalboerger PG, Wang S, Simper D, Miller DV, Russell SJ, Litzow MR, Edwards WD. Smooth muscle cells in human coronary atherosclerosis can originate from cells administered at marrow transplantation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4754–4759. doi: 10.1073/pnas.0730743100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu Y, Zhang Z, Torsney E, Afzal AR, Davison F, Metzler B, Xu Q. Abundant progenitor cells in the adventitia contribute to atherosclerosis of vein grafts in ApoE-deficient mice. The Journal of clinical investigation. 2004;113:1258–1265. doi: 10.1172/JCI19628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bentzon JF, Sondergaard CS, Kassem M, Falk E. Smooth muscle cells healing atherosclerotic plaque disruptions are of local, not blood, origin in apolipoprotein E knockout mice. Circulation. 2007;116:2053–2061. doi: 10.1161/CIRCULATIONAHA.107.722355. [DOI] [PubMed] [Google Scholar]
- 52.Bentzon JF, Weile C, Sondergaard CS, Hindkjaer J, Kassem M, Falk E. Smooth muscle cells in atherosclerosis originate from the local vessel wall and not circulating progenitor cells in ApoE knockout mice. Arteriosclerosis, thrombosis, and vascular biology. 2006;26:2696–2702. doi: 10.1161/01.ATV.0000247243.48542.9d. [DOI] [PubMed] [Google Scholar]
- 53.Iwata H, Manabe I, Fujiu K, Yamamoto T, Takeda N, Eguchi K, Furuya A, Kuro-o M, Sata M, Nagai R. Bone marrow-derived cells contribute to vascular inflammation but do not differentiate into smooth muscle cell lineages. Circulation. 2010;122:2048–2057. doi: 10.1161/CIRCULATIONAHA.110.965202. [DOI] [PubMed] [Google Scholar]
- 54.Tang Z, Wang A, Yuan F, Yan Z, Liu B, Chu JS, Helms JA, Li S. Differentiation of multipotent vascular stem cells contributes to vascular diseases. Nat Commun. 2012;3:875. doi: 10.1038/ncomms1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen AT, Gomez D, Bell RD, Campbell JH, Clowes AW, Gabbiani G, Giachelli CM, Parmacek MS, Raines EW, Rusch NJ, Speer MY, Sturek M, Thyberg J, Towler DA, Weiser-Evans MC, Yan C, Miano JM, Owens GK. Smooth muscle cell plasticity: fact or fiction? Circulation research. 2013;112:17–22. doi: 10.1161/CIRCRESAHA.112.281048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gomez D, Shankman LS, Nguyen AT, Owens GK. Detection of histone modifications at specific gene loci in single cells in histological sections. Nature methods. 2013;10:171–177. doi: 10.1038/nmeth.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shankman LS, Gomez D, Cherepanova OA, Salmon M, Alencar GF, Haskins RM, Swiatlowska P, Newman AA, Greene ES, Straub AC, Isakson B, Randolph GJ, Owens GK. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat Med. 2015;21:628–637. doi: 10.1038/nm.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feil S, Fehrenbacher B, Lukowski R, Essmann F, Schulze-Osthoff K, Schaller M, Feil R. Transdifferentiation of vascular smooth muscle cells to macrophage-like cells during atherogenesis. Circulation research. 2014;115:662–667. doi: 10.1161/CIRCRESAHA.115.304634. [DOI] [PubMed] [Google Scholar]
- 59.Nemenoff RA, Horita H, Ostriker AC, Furgeson SB, Simpson PA, VanPutten V, Crossno J, Offermanns S, Weiser-Evans MC. SDF-1alpha induction in mature smooth muscle cells by inactivation of PTEN is a critical mediator of exacerbated injury-induced neointima formation. Arteriosclerosis, thrombosis, and vascular biology. 2011;31:1300–1308. doi: 10.1161/ATVBAHA.111.223701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herring BP, Hoggatt AM, Burlak C, Offermanns S. Previously differentiated medial vascular smooth muscle cells contribute to neointima formation following vascular injury. Vascular cell. 2014;6:21. doi: 10.1186/2045-824X-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tabas I, Garcia-Cardena G, Owens GK. Recent insights into the cellular biology of atherosclerosis. J Cell Biol. 2015;209:13–22. doi: 10.1083/jcb.201412052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bennett MR, Sinha S, Owens GK. Vascular smooth muscle cell in atherosclerosis. Circulation research. 2015 doi: 10.1161/CIRCRESAHA.115.306361. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buckingham ME, Meilhac SM. Tracing cells for tracking cell lineage and clonal behavior. Developmental cell. 2011;21:394–409. doi: 10.1016/j.devcel.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 64.Salmon M, Gomez D, Greene E, Shankman L, Owens GK. Cooperative binding of KLF4, pELK-1, and HDAC2 to a G/C repressor element in the SM22alpha promoter mediates transcriptional silencing during SMC phenotypic switching in vivo. Circulation research. 2012;111:685–696. doi: 10.1161/CIRCRESAHA.112.269811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hiltunen MO, Yla-Herttuala S. DNA methylation, smooth muscle cells, and atherogenesis. Arteriosclerosis, thrombosis, and vascular biology. 2003;23:1750–1753. doi: 10.1161/01.ATV.0000092871.30563.41. [DOI] [PubMed] [Google Scholar]
- 66.Hiltunen MO, Turunen MP, Hakkinen TP, Rutanen J, Hedman M, Makinen K, Turunen AM, Aalto-Setala K, Yla-Herttuala S. DNA hypomethylation and methyltransferase expression in atherosclerotic lesions. Vasc Med. 2002;7:5–11. doi: 10.1191/1358863x02vm418oa. [DOI] [PubMed] [Google Scholar]
- 67.Rozenberg JM, Tesfu DB, Musunuri S, Taylor JM, Mack CP. DNA methylation of a GC repressor element in the smooth muscle myosin heavy chain promoter facilitates binding of the Notch-associated transcription factor, RBPJ/CSL1. Arteriosclerosis, thrombosis, and vascular biology. 2014;34:2624–2631. doi: 10.1161/ATVBAHA.114.304634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Allahverdian S, Chehroudi AC, McManus BM, Abraham T, Francis GA. Contribution of intimal smooth muscle cells to cholesterol accumulation and macrophage-like cells in human atherosclerosis. Circulation. 2014;129:1551–1559. doi: 10.1161/CIRCULATIONAHA.113.005015. [DOI] [PubMed] [Google Scholar]
- 69.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nature reviews Genetics. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 70.Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, Bennett DA, Houmard JA, Muoio DM, Onder TT, Camahort R, Cowan CA, Meissner A, Epstein CB, Shoresh N, Bernstein BE. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, Yang H, Wang T, Lee AY, Swanson SA, Zhang J, Zhu Y, Kim A, Nery JR, Urich MA, Kuan S, Yen CA, Klugman S, Yu P, Suknuntha K, Propson NE, Chen H, Edsall LE, Wagner U, Li Y, Ye Z, Kulkarni A, Xuan Z, Chung WY, Chi NC, Antosiewicz-Bourget JE, Slukvin I, Stewart R, Zhang MQ, Wang W, Thomson JA, Ecker JR, Ren B. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J, Schier AF. Chromatin signature of embryonic pluripotency is established during genome activation. Nature. 2010;464:922–926. doi: 10.1038/nature08866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A, Shalek AK, Kelley DR, Shishkin AA, Issner R, Zhang X, Coyne M, Fostel JL, Holmes L, Meldrim J, Guttman M, Epstein C, Park H, Kohlbacher O, Rinn J, Gnirke A, Lander ES, Bernstein BE, Meissner A. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153:1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ernst J, Kellis M. Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nature biotechnology. 2015;33:364–376. doi: 10.1038/nbt.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bock C. Analysing and interpreting DNA methylation data. Nature reviews Genetics. 2012;13:705–719. doi: 10.1038/nrg3273. [DOI] [PubMed] [Google Scholar]
- 76.Michels KB, Binder AM, Dedeurwaerder S, Epstein CB, Greally JM, Gut I, Houseman EA, Izzi B, Kelsey KT, Meissner A, Milosavljevic A, Siegmund KD, Bock C, Irizarry RA. Recommendations for the design and analysis of epigenome-wide association studies. Nature methods. 2013;10:949–955. doi: 10.1038/nmeth.2632. [DOI] [PubMed] [Google Scholar]
- 77.Bheda P, Schneider R. Epigenetics reloaded: the single-cell revolution. Trends in cell biology. 2014;24:712–723. doi: 10.1016/j.tcb.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 78.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. mRNA-Seq whole-transcriptome analysis of a single cell. Nature methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 79.Smallwood SA, Lee HJ, Angermueller C, Krueger F, Saadeh H, Peat J, Andrews SR, Stegle O, Reik W, Kelsey G. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nature methods. 2014;11:817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maeder ML, Angstman JF, Richardson ME, Linder SJ, Cascio VM, Tsai SQ, Ho QH, Sander JD, Reyon D, Bernstein BE, Costello JF, Wilkinson MF, Joung JK. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nature biotechnology. 2013;31:1137–1142. doi: 10.1038/nbt.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mendenhall EM, Williamson KE, Reyon D, Zou JY, Ram O, Joung JK, Bernstein BE. Locus-specific editing of histone modifications at endogenous enhancers. Nature biotechnology. 2013;31:1133–1136. doi: 10.1038/nbt.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nature biotechnology. 2015 doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kabadi AM, Ousterout DG, Hilton IB, Gersbach CA. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic acids research. 2014;42:e147. doi: 10.1093/nar/gku749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, Holmes MC, Naldini L. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nature biotechnology. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 85.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nature biotechnology. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 86.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]