Abstract
The role of epithelial to mesenchymal transition (EMT) in metastasis is a longstanding source of controversy, largely due to an inability to monitor transient and reversible EMT phenotypes in vivo. We established an EMT lineage tracing system to monitor this process, using a mesenchymal-specific Cre-mediated fluorescent marker switch system in spontaneous breast-to-lung metastasis models. We confirmed that within a predominantly epithelial primary tumor, a small portion of tumor cells undergo EMT. Strikingly, lung metastases mainly consisted of non-EMT tumor cells maintaining their epithelial phenotype. Inhibiting EMT by overexpressing miR-200 did not impact lung metastasis development. However, EMT cells significantly contribute to recurrent lung metastasis formation after chemotherapy. These cells survived cyclophosphamide treatment due to reduced proliferation, apoptotic tolerance, and elevated expression of chemoresistance-related genes. Overexpression of miR-200 abrogated this resistance. This study suggests the potential of an EMT-targeting strategy, in conjunction with conventional chemotherapies, for breast cancer treatment.
Keywords: Epithelial to mesenchymal transition, EMT, MET, breast cancer, lung metastasis, chemoresistance
Despite significant advances in diagnosing and treating cancer, metastasis persists as a barricade to successful therapy and the main cause of cancer-related death1. The epithelial to mesenchymal transition (EMT), wherein epithelial cells depolarize, lose their cell-cell contacts, and gain an elongated, fibroblast-like morphology, is a potential mechanism by which tumor cells gain metastatic features. Functional implications of EMT include enhanced mobility, invasion, and resistance to apoptotic stimuli2,3. Moreover, through EMT tumor cells acquire cancer stem cell, secondary tumor-initiating and chemoresistance properties4–6. However, the importance of EMT in vivo is fiercely debated due to significant challenges: mesenchymal tumor cells cannot easily be distinguished from neighboring stromal cells, and metastatic lesions mostly exhibit epithelial phenotypes7. The latter may be due to the hypothesized reverse process, mesenchymal to epithelial transition (MET), of the disseminated tumor cells. Studies have confirmed that mesenchymal cells are more capable of escaping the primary tumor, and of reaching distant sites, but it remains unproven that those same cells complete the full metastatic cascade in the form of a secondary nodule. Without evidence for the dissemination, colonization, and metastatic outgrowth of mesenchymal tumor cells, the role of EMT will remain contested. In this study, we employed multiple transgenic mouse models, establishing a cell lineage tracing approach together with characterization of epithelial and mesenchymal markers, to address the requirement of EMT in metastasis. The newly established transgenic model also provided us a unique opportunity to study the contribution of EMT to chemoresistance.
EMT lineage tracing during metastasis
To track EMT during metastasis in vivo, we generated a mesenchymal-specific, Cre-mediated fluorescent marker switch strategy and established a triple transgenic mouse model (Tri-PyMT, MMTV-PyMT/Rosa26-RFP-GFP/Fsp1-Cre, Fig. 1a). In these mice, spontaneous multifocal breast adenocarcinomas with distinct epithelial characteristics resembling the human luminal subtype develop in the mammary glands, and give rise to lung metastases with high penetrance8,9. The FSP1 (fibroblast specific protein 1) promoter drives expression of Cre recombinase in cells of mesenchymal lineage10. A Cre-switchable fluorescent marker (lox-RFP-stop-lox-GFP) is ubiquitously expressed under the control of the β-actin promoter in the Rosa26 locus11. FSP1 is the critical gate-keeping gene of EMT initiation12, and its early activation in this process13 allows for lineage tracing of tumor cells that have undergone EMT in vivo. Importantly, the color switch system is irreversible—even if the mesenchymal tumor cells undergo MET in the metastatic organs14, they would remain GFP+.
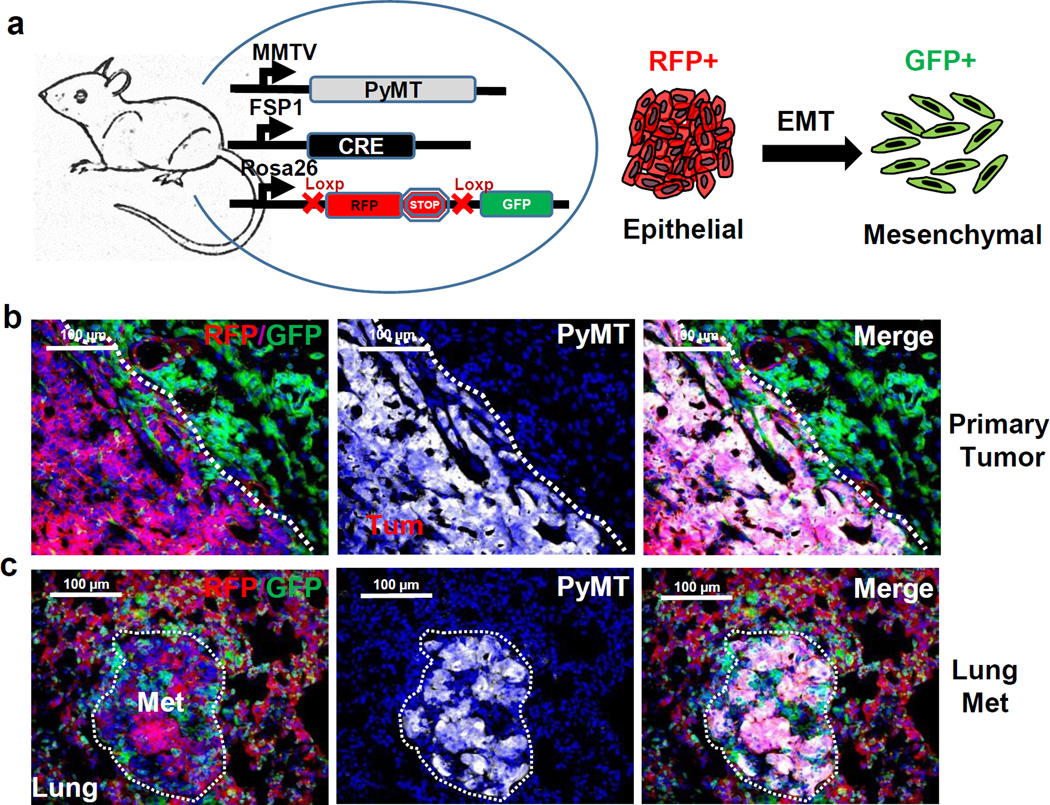
Figure 1. Establishing an EMT lineage tracing system in triple transgenic mice.
a, Schematic of triple transgenic mice carrying Polyoma Middle-T (PyMT) or Neu oncogenes driven by the MMTV promoter, Cre recombinase under the control of the FSP1 promoter, and floxed RFP-STOP followed by GFP under control of the β-actin promoter in the Rosa26 locus. RFP+ epithelial tumor cells undergoing EMT permanently convert into GFP+ cells following activation of Fsp-1 Cre.
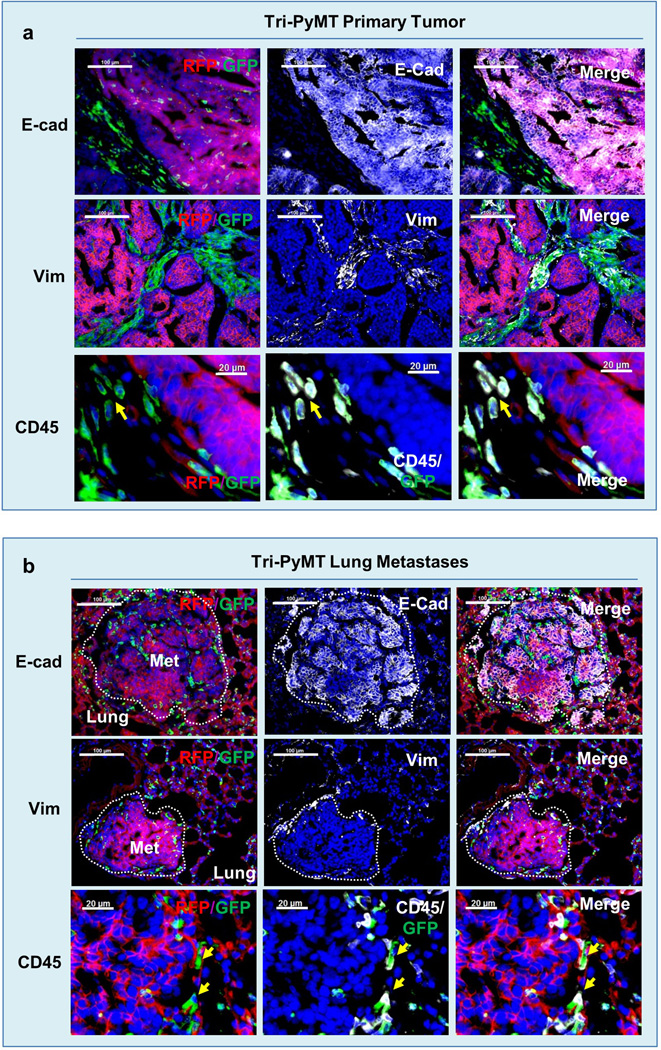
b and c, Immunofluorescent microscopy images of Tri-PyMT primary tumors (b) and lung metastases (c) (>10 sections from 3 mice), depicting RFP+ and GFP+ cells within the tumor bed, and staining (white, pseudo-colored) for PyMT.
Primary breast tumors developed in the Tri-PyMT mice at 8 weeks of age. Immunofluorescence revealed that the majority of tumor cells, identified by PyMT oncogene expression, were RFP positive (Fig. 1b). These cells expressed E-cadherin and lacked vimentin (Extended Data Fig.1a) indicating their epithelial phenotype. The GFP+ cells detected in the tumor bed were largely hematopoietic cells as they are PyMT negative and express CD45, a pan-hematopoietic marker (Extended Data Fig.1a), which is consistent with previous reports15. Altogether, this data suggests that tumor cells maintain their original RFP expression and epithelial phenotype in the primary tumor.
Lung metastasis developed spontaneously in Tri-PyMT lungs at 12 weeks of age. Surprisingly, the PyMT-positive metastatic lesions were RFP+ (Fig. 1c), and epithelial (E-cadherin+/vimentin-) (Extended Data Fig.1b), whereas only non-tumor cells expressed GFP. These results indicate that tumor cells did not activate the mesenchymal-specific FSP1 promoter, and retained their epithelial phenotype during metastasis. Thus, tumor cells may not undergo EMT to form metastatic lesions.
Lineage tracing in additional models
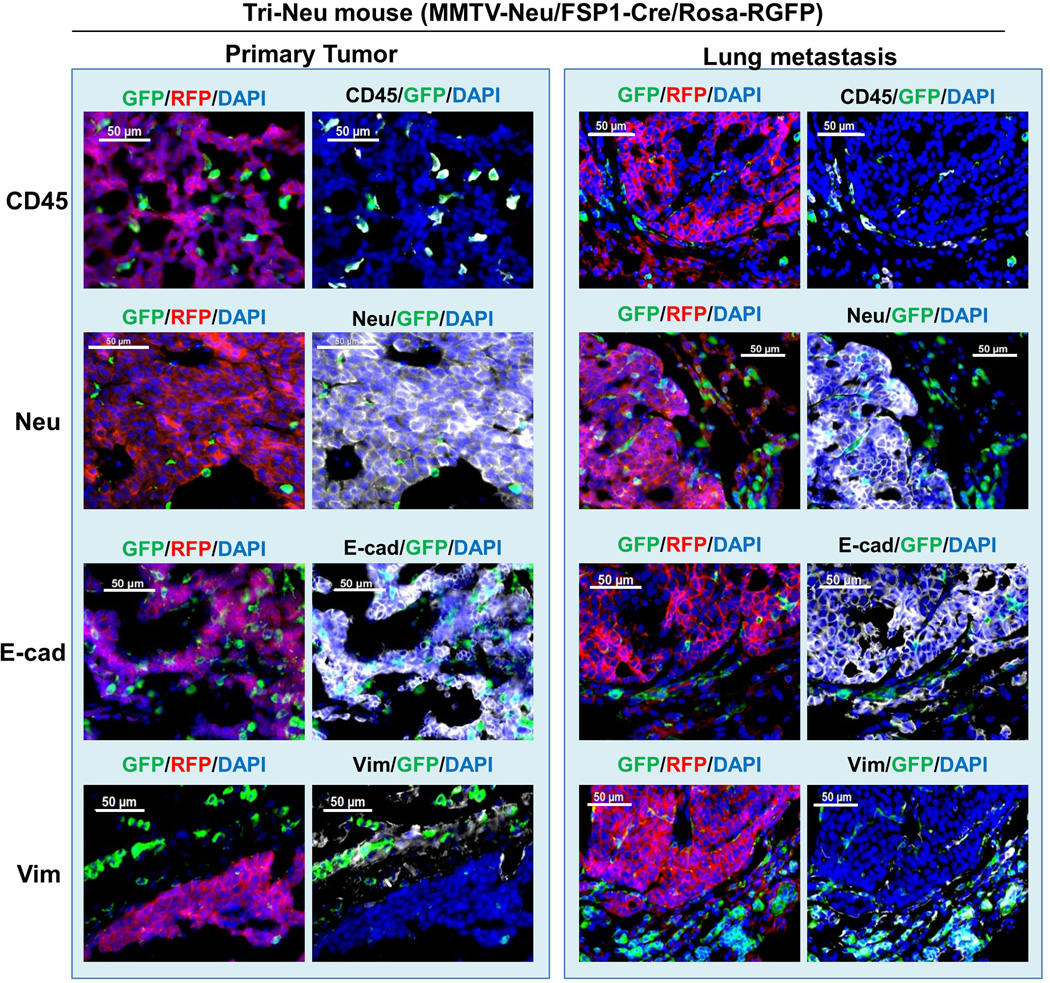
To exclude the possibility that the absence of EMT in metastasis may be unique to PyMT-driven breast tumors, we established EMT lineage tracing in the Neu oncogene-driven16 spontaneous breast cancer model (MMTV-neu/Rosa26-RFP-GFP/Fsp1-Cre, Tri-neu mouse). The Neu (ErbB-2) proto-oncogene is associated with 20–30% of human breast cancers, and MMTV-Neu transgenic mice spontaneously develop focal adenocarcinomas resembling human luminal phenotypes after an extended latency at 6–8 months of age. Lung metastases are frequently (72%) observed in these transgenic mice at 9–12 months of age. Mirroring the Tri-PyMT model, the Neu+ tumor cells in both primary and metastatic lesions in Tri-Neu mice were also RFP+ and epithelial (E-cad+/Vim-) (Extended Data Fig. 2). Therefore, the absence of EMT during metastasis formation is an oncogene-independent phenomenon, manifesting in both PyMT and Neu-driven tumors.
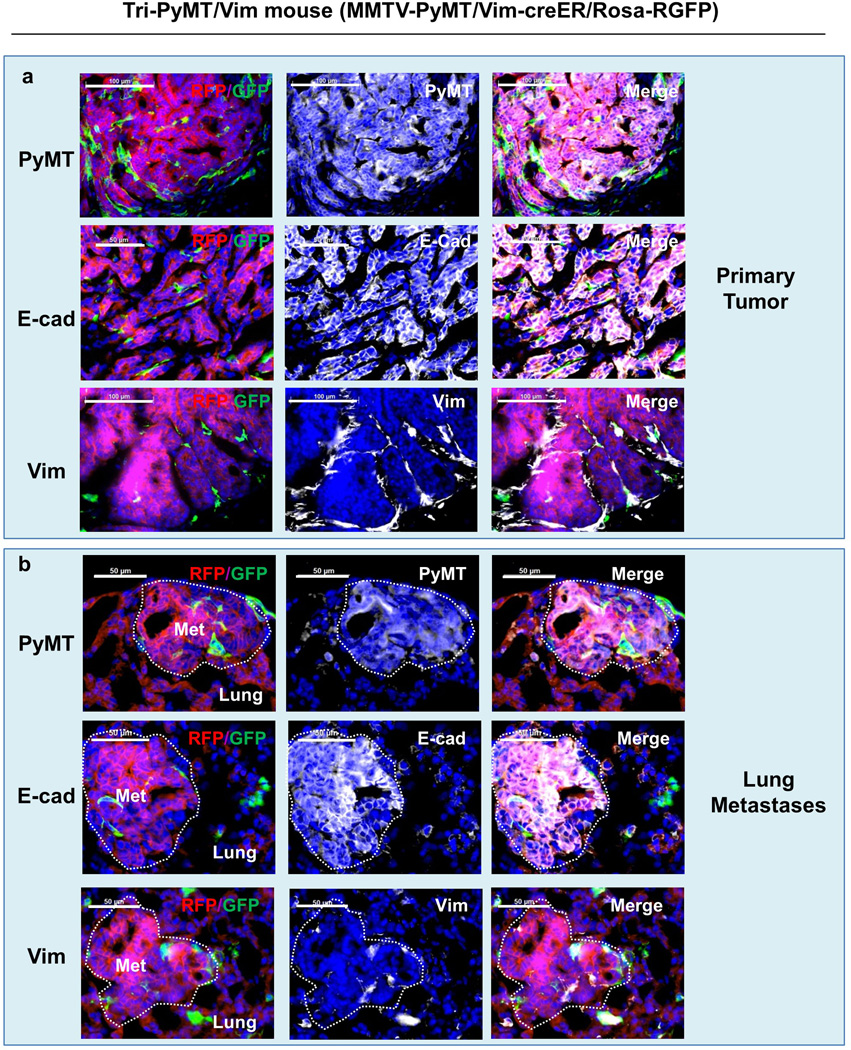
To overcome the limitation of using solely Fsp1-Cre to indicate EMT, we acquired the Vimentin-CreERT transgenic mouse, which successfully traced mesenchymal lineage cells during liver fibrosis17, and generated an additional EMT lineage tracing model (Tri-PyMT/Vim mice, MMTV-PyMT/Rosa26-RFP-GFP/Vimentin-CreERT). After continuous induction of Cre activity by Tamoxifen injection (2mg, i.p., 3 times per week starting when the primary tumors appear at 8 weeks of age) the majority of tumor cells in both the primary and metastatic lesions in Tri-PyMT/Vim mice were RFP+ (Extended Data Fig. 3)—suggesting an absence of vimentin promoter activation during lung metastasis formation. EMT marker staining also revealed the epithelial phenotype (E-cad+/Vim-) of the tumor cells in both primary and metastatic lesions (Extended Data Fig. 3).
Together, results from two oncogene-driven metastatic tumor models (MMTV-PyMT and MMTV-Neu) and two independent mesenchymal-specific reporters (Vim-Cre and Fsp1-Cre) suggest that EMT does not significantly contribute to the development of lung metastases.
Validating EMT lineage tracing
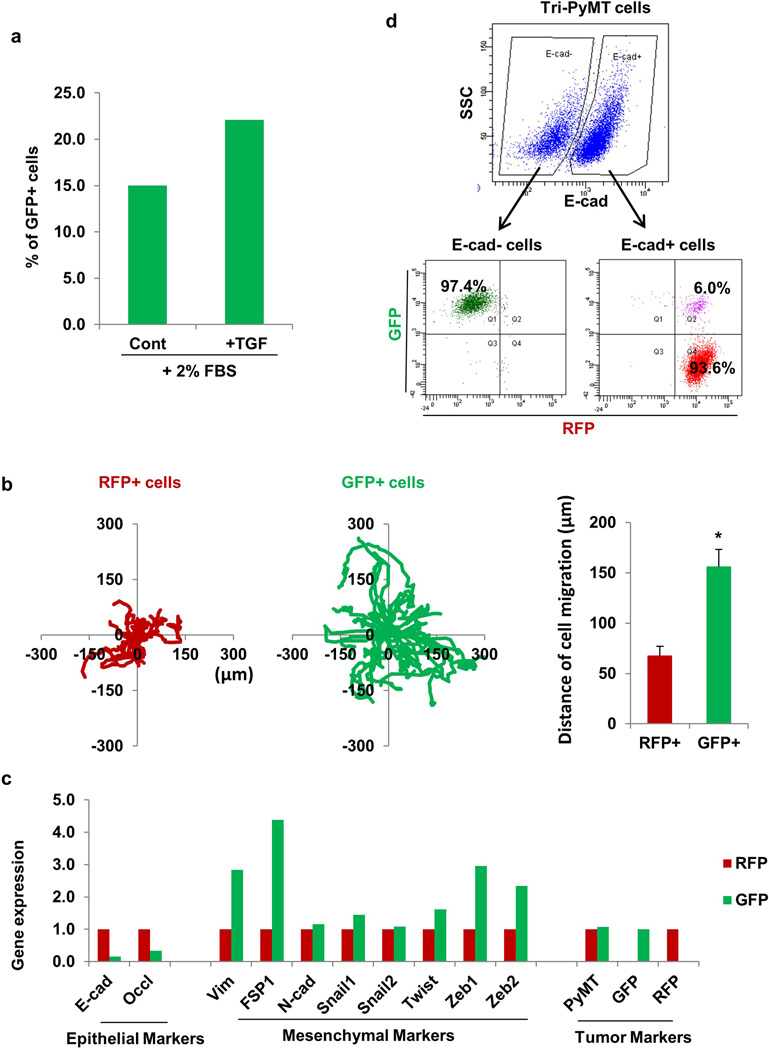
To evaluate the specificity and sensitivity of the EMT lineage tracing system, we established a cell line from the Tri-PyMT breast tumors. In culture, RFP+ Tri-PyMT cells switched their fluorescent marker expression to GFP, as indicated by the presence of a RFP+/GFP+ double-positive transitioning population (Fig. 2a). The cells were cultured in 10% FBS, and serum is known to be enriched for many EMT promoting factors including TGFs18. Moreover, addition of TGF-β1 in low-serum conditions (2% FBS), yielded an increase in GFP+ cells (Extended Data Fig. 4a). In concert with the fluorescent marker switch, Tri-PyMT cells changed their morphology from cobblestone-like clusters of epithelial cells to dispersed spindle-shaped mesenchymal cells (Fig. 2b). Reflecting the morphologic differences, the GFP+ cells were more motile than RFP+ cells (Extended Data Fig. 4b).
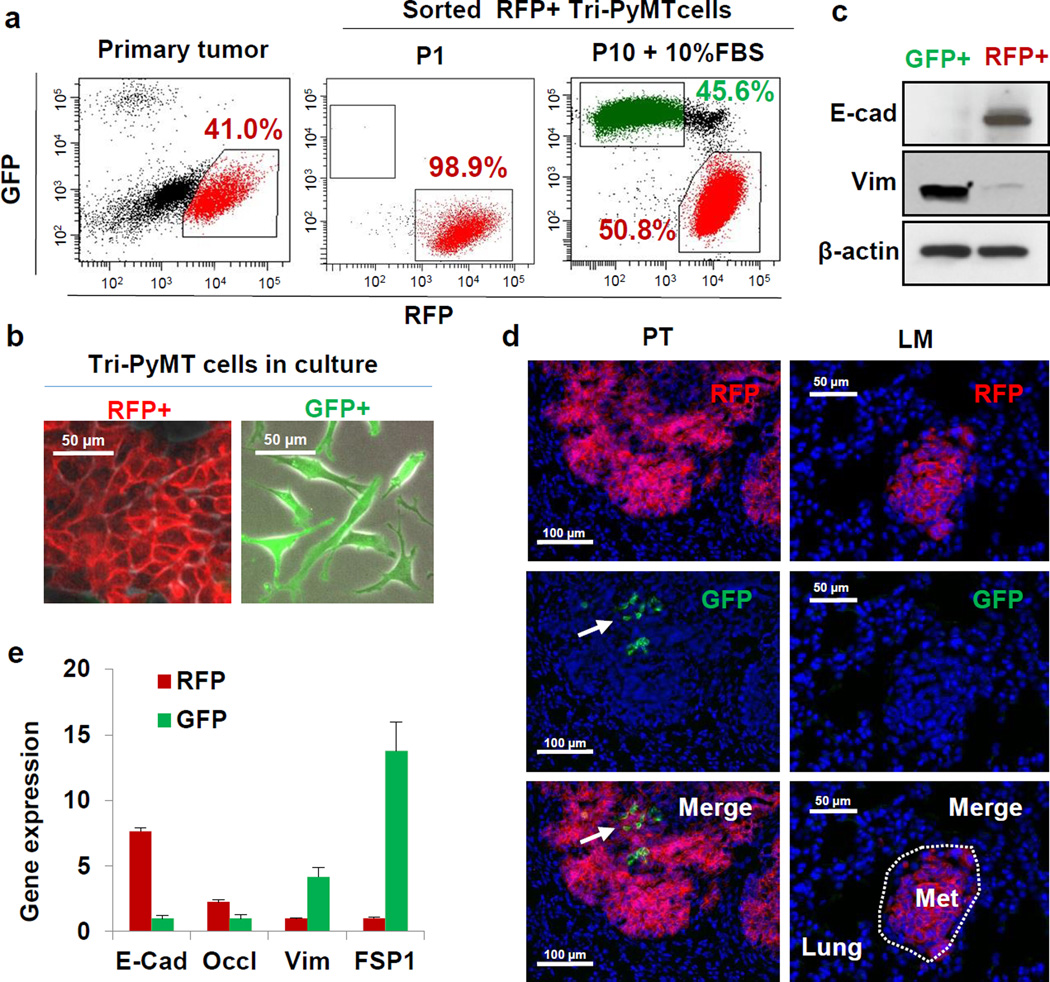
Figure 2. The EMT lineage tracing system reports EMT in tumor cells with high fidelity.
a, Scatter plots from flow cytometry analysis of Tri-PyMT primary tumor cells, depicting GFP+ and RFP+ populations in the primary tumor immediately after sorting of RFP+ cells (P1), and after 10 passages in culture with 10% FBS (P10+10%FBS). Numbers indicate the percentage of RFP+ and GFP+ cells in the total population.
b, Phase contrast/fluorescent overlay image of Tri-PyMT cells in culture.
c, Western blot of sorted RFP+ and GFP+ Tri-PyMT cells for E-cadherin, vimentin and β-actin as a loading control. Representative of 2 individual experiments. For gel source data, see Supplementary Figure 1.
d, Representative imaging of GFP+ and RFP+ tumor cells in primary tumors (PT) and lung metastases (LM) in the orthotopic model (n=8 mice). Arrow indicates scattered GFP+EMT tumor cells in the primary tumor.
e, Q-RT-PCR analysis of relative expression of EMT markers in RFP+ and GFP+ cells sorted from orthotopic Tri-PyMT primary tumors. GAPDH served as the internal control. Data are reported as mean ± SEM, n=4 primary tumors.
The fidelity of the EMT lineage tracing system was confirmed by analysis of EMT marker expression in sorted RFP+ and GFP+ Tri-PyMT cells. RFP+ cells expressed elevated levels of epithelial markers including E-cadherin and Occludin, while GFP+ cells expressed several mesenchymal markers including vimentin, FSP1, Twist, Zeb1 and Zeb2 as determined by Q-RT-PCR (Extended Data Fig. 4c). Both RFP+ and GFP+ Tri-PyMT cells expressed the PyMT oncogene. Consistently, western blot analysis confirmed the differential expression of E-cadherin and vimentin in RFP+ and GFP+ cells (Fig. 2c). Flow cytometry for E-cadherin revealed that the majority of E-cadherin-negative cells were GFP+ (97.4%) (Extended Data Fig. 4d). Of note, the E-cadherin+ cells were either RFP+ (93.6%) or RFP+/GFP+ (6.0%), demonstrating that tumor cells switch their fluorescent marker expression prior to the loss of epithelial markers, and validating the early reporting of EMT in our system. These results confirm that the FSP1-Cre-mediated fluorescent marker switch in tumor cells reports EMT with high fidelity and efficiency.
Rare EMT events in tumor progression
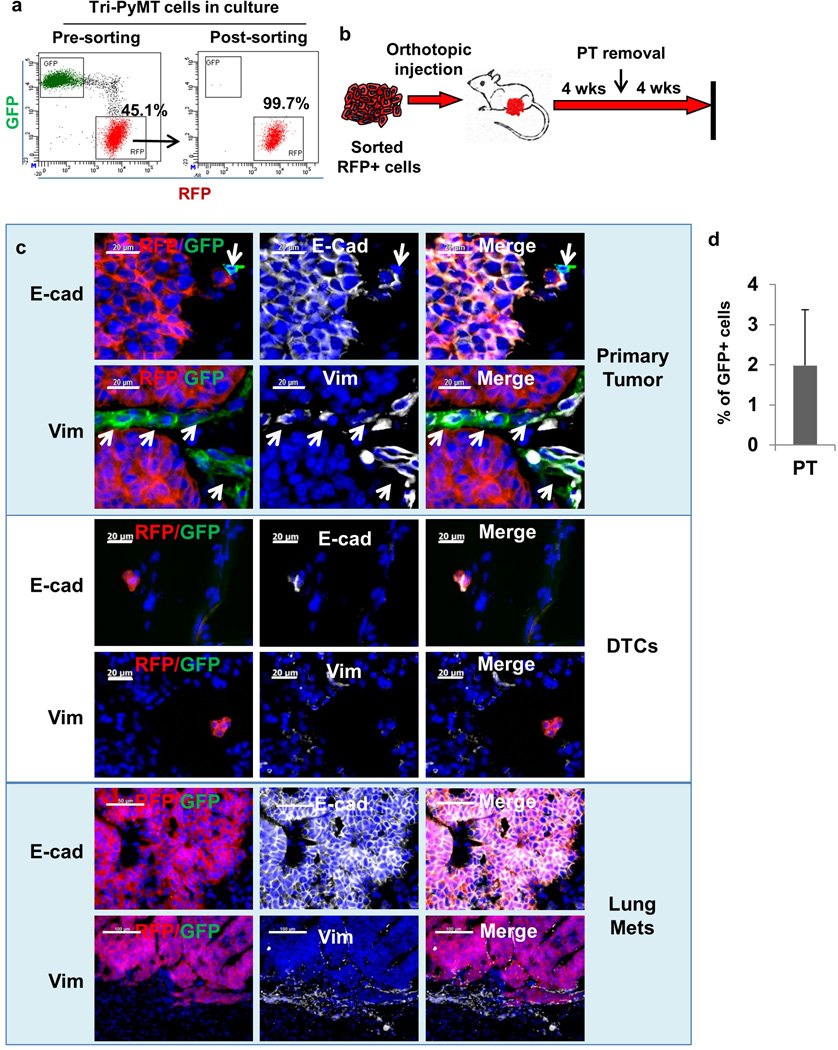
In the triple-transgenic models, ubiquitous expression of GFP in the tumor microevironment precluded detection of potentially rare GFP+ tumor cells. To confine the fluorescence to tumor cells, we established an orthotopic model by implanting purified RFP+ Tri-PyMT cells in the wild type mice (Extended Data Fig. 5a,b). Consistent with observations in the triple transgenic mice, primary tumors contained RFP+ epithelial cells (Fig. 2d). However, GFP+ cells were detected, indicating tumor EMT (Fig. 2d, Extended Data Fig. 5c, upper panel). These cells lacked E-cadherin (Extended Data Fig. 5c, upper panel) and made up 1.98 ± 1.40% (n=6) of the total tumor cells (Extended Data Fig. 5d). Q-RT-PCR analysis of EMT markers comparing sorted RFP+ and GFP+ cells from the same primary tumor confirmed the mesenchymal phenotype of the GFP+ cells (Fig. 2e). Importantly, these GFP+ EMT tumor cells did not contribute to lung metastasis. Early disseminated tumor cells detected in the lungs were epithelial and RFP+ (Extended Data Fig. 5c, middle panel), and 28 lung nodules detected in 8 mice maintained the epithelial phenotype (Fig. 2d, Extended Data Fig. 5c lower panel).
We also established an orthotopic Tri-PyMT/Vim model, wherein Tamoxifen was administered directly after orthotopic injection to ensure immediate tracing of EMT events. Consistently, the majority of tumor cells in both primary and metastatic tumors were RFP+ and epithelial (Extended Data Fig. 6). Again, GFP+ EMT events (4.46 ± 1.0% of total tumor cells, n=3) were detected in the primary tumors.
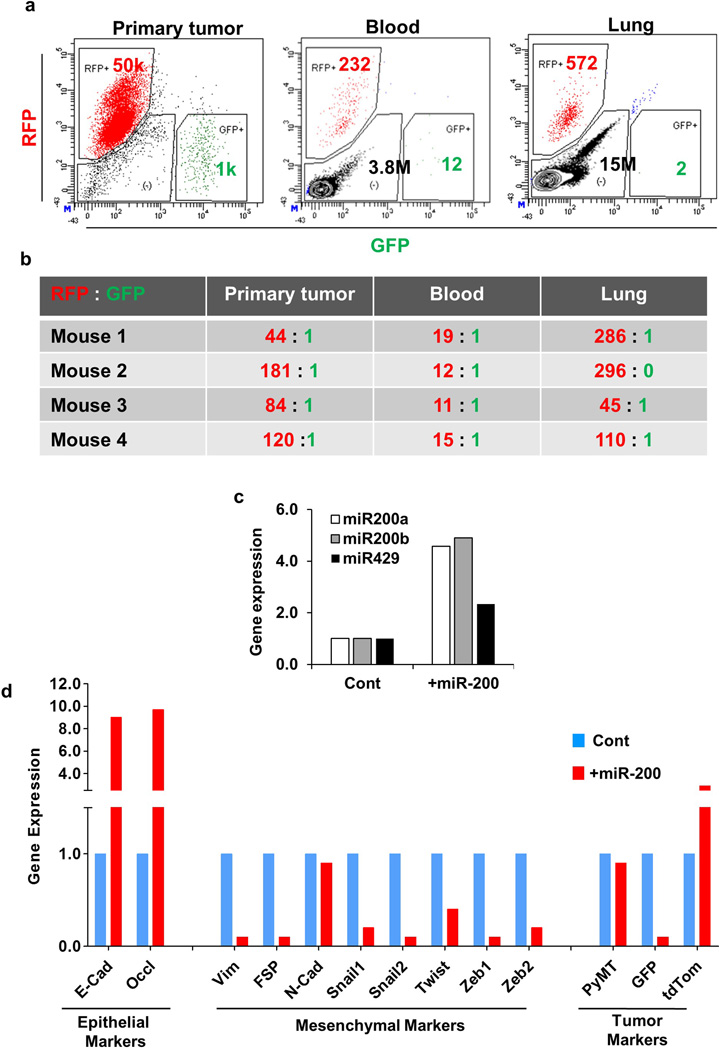
To further dissect the metastatic cascade, we quantified the relative numbers of RFP+ and GFP+ cells in the primary tumor, blood, and metastases of the Tri-PyMT orthotopic model by flow cytometry. An RFP to GFP ratio of ∼100:1 in the primary tumor and ∼15:1 in the blood was observed (Extended Data Fig. 7a,b). However, gain by GFP+ cells did not translate to an advantage in metastatic outgrowth, as the RFP:GFP ratio in the lung was ∼150:1. Altogether, these findings are consistent with our observations in the triple-transgenic models, suggesting that the majority of breast tumor cells persist in an epithelial state during primary tumor growth and lung metastasis formation.
EMT inhibition and metastasis formation
In spite of the extensive characterization of the EMT reporter system, there was still the distant possibility of our reporter failing to manifest all EMT events in vivo. Therefore, we sought to inhibit EMT and determine its impact on metastasis. We ectopically expressed miR-200, a well-known inhibitor of EMT that directly targets ZEB1 and ZEB2 – the transcriptional repressors of E-cadherin19,20. We posited that stably expressing miR-200 in Tri-PyMT cells would block EMT and trap tumor cells in a permanent epithelial state. Compared with control cells, miR-200 overexpressing cells (Extended Data Fig. 7c) showed elevated expression of epithelial cell markers and reduced expression of mesenchymal markers (Extended Data Fig. 7d). As expected, overexpression of miR-200 inhibited the RFP to GFP conversion (> 90% remaining RFP+, Fig. 3a). These results substantiate effective miR-200 suppression of EMT in the Tri-PyMT cells.
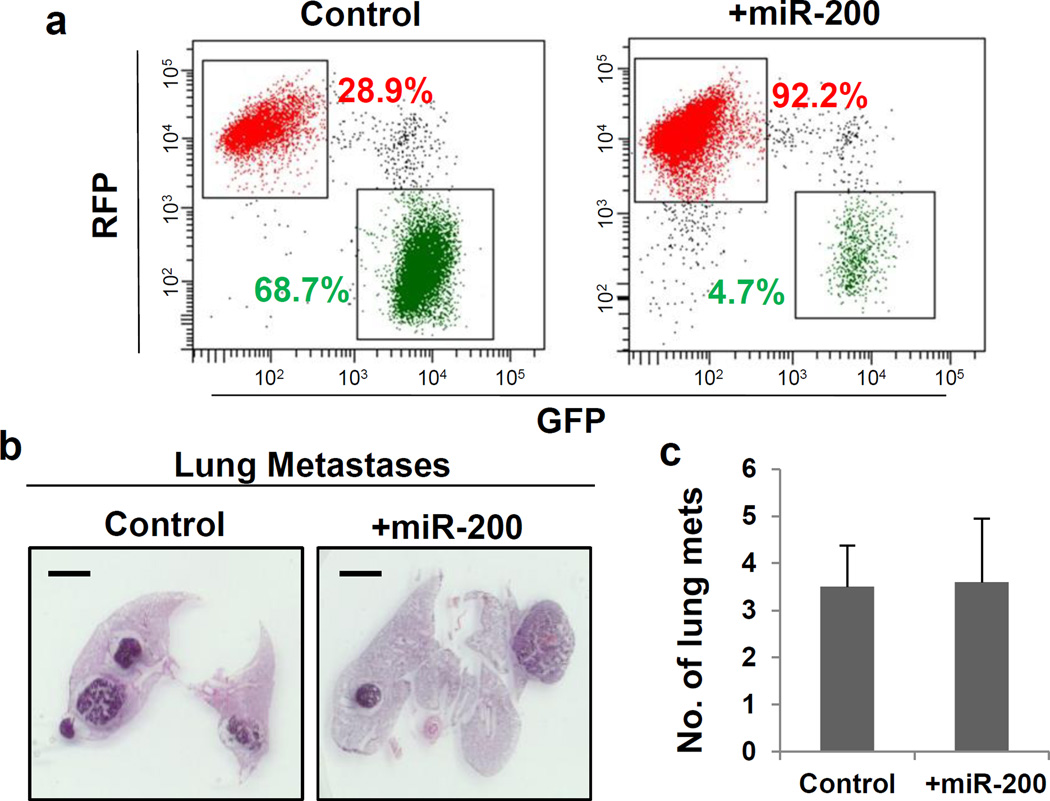
Figure 3. mir-200 inhibition of EMT in Tri-PyMT cells did not impact lung metastasis.
a, Flow cytometry analysis of Tri-PyMT control and +mir-200 cells, indicating percentage of RFP+ and GFP+ cells.
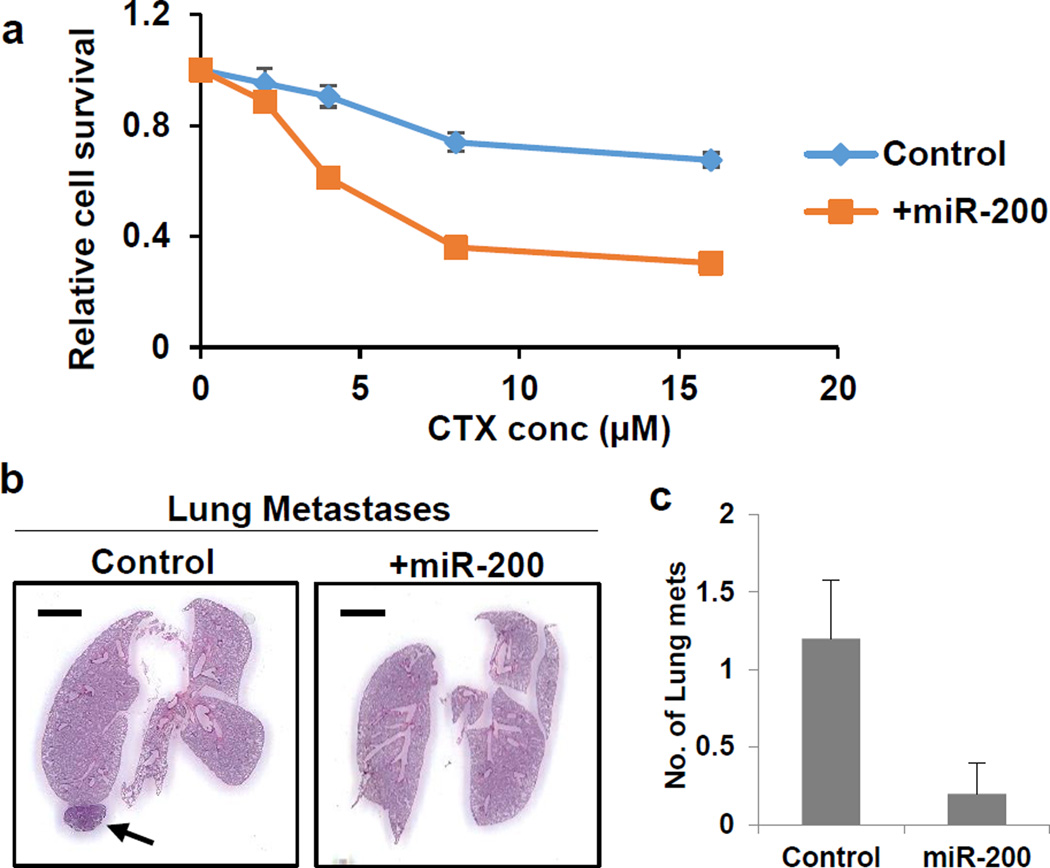
b, Representative histologic lung images in Tri-PyMT Cont and +mir-200 orthotopic mice (n=5).
c, Quantification of lung metastasis formation (number of individual nodules) in Tri-PyMT control and +mir-200 tumor-bearing mice (n=5).
To explore the impact of inhibiting EMT on metastasis formation in vivo, we orthotopically injected miR-200 overexpressing Tri-PyMT cells. We identified 18 metastases in 5 mice, a similar ratio to that observed in mice bearing control Tri-PyMT cells (28 metastases in 8 mice) (Fig. 3b,c). These results demonstrate that inhibition of EMT by miR-200 overexpression does not impair the ability of tumor cells to form distant lung metastases.
EMT is involved in chemoresistance
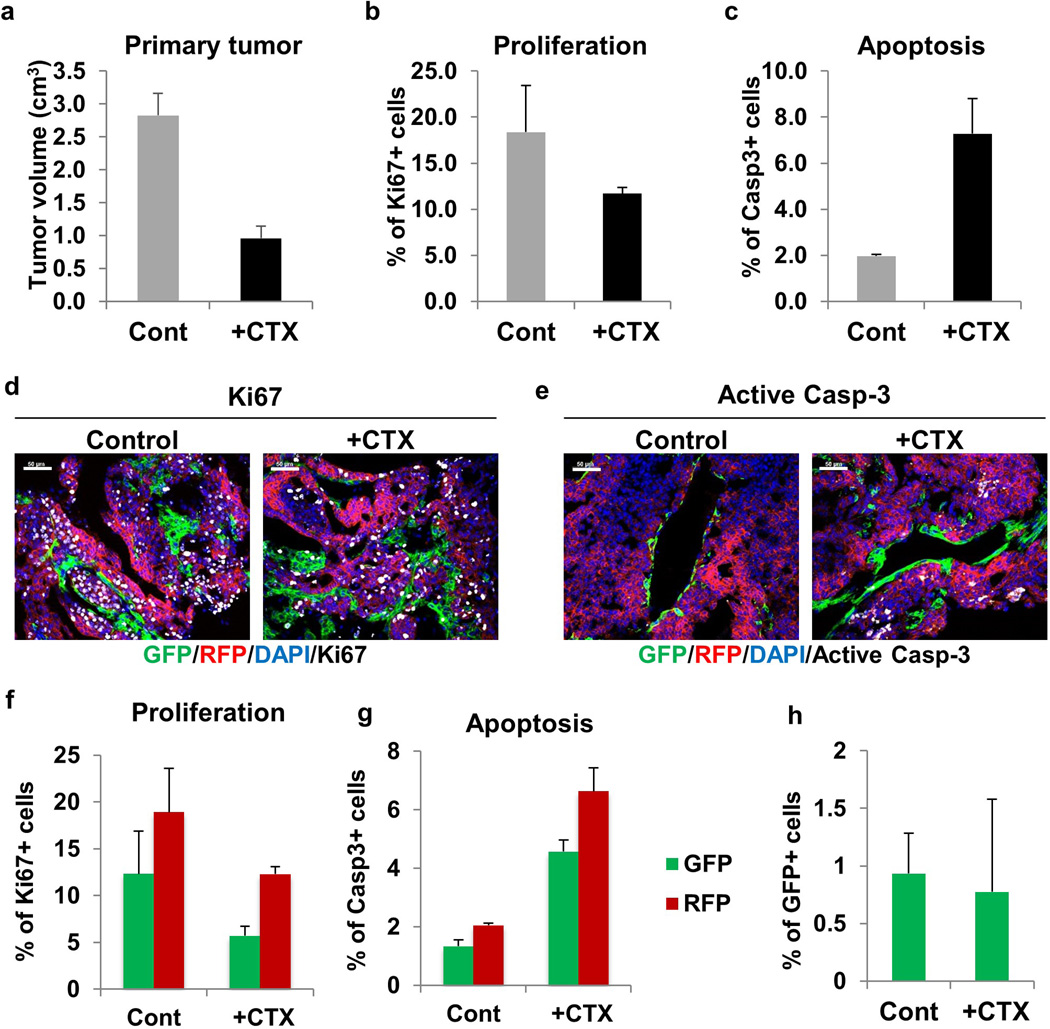
Emerging evidence suggests a molecular and phenotypic association between EMT and chemoresistance in several cancers21–23. Compellingly, residual breast cancers following chemotherapy display a mesenchymal phenotype and tumor initiating features23http://www.sciencedirect.com/science/article/pii/S0006291X1102170X - b0010. To determine if the acquisition of chemoresistance induces specific molecular changes consistent with EMT, we evaluated the orthotopic Tri-PyMT model under chemotherapy. Animals with established primary tumors were treated with cyclophosphamide, a commonly used drug in breast cancer treatment24 (CTX, 100mg/kg, once per week, for 2 weeks prior, and 2 weeks after surgery Fig. 4a). The tumors responded to chemotherapy, manifesting a 60% reduction in growth and markedly enhanced apoptotic activity (Extended Data Fig. 8a–c). Of note, the RFP+ cells were highly proliferative and apoptotic in comparison with GFP+ cells in CTX-treated mice (Extended Data Fig. 8d–g), suggesting that GFP+ cells have reduced susceptibility to chemotherapy. However, in the primary tumor, the GFP+ cell percentage remained static under CTX treatment (Extended Data Fig. 8h).
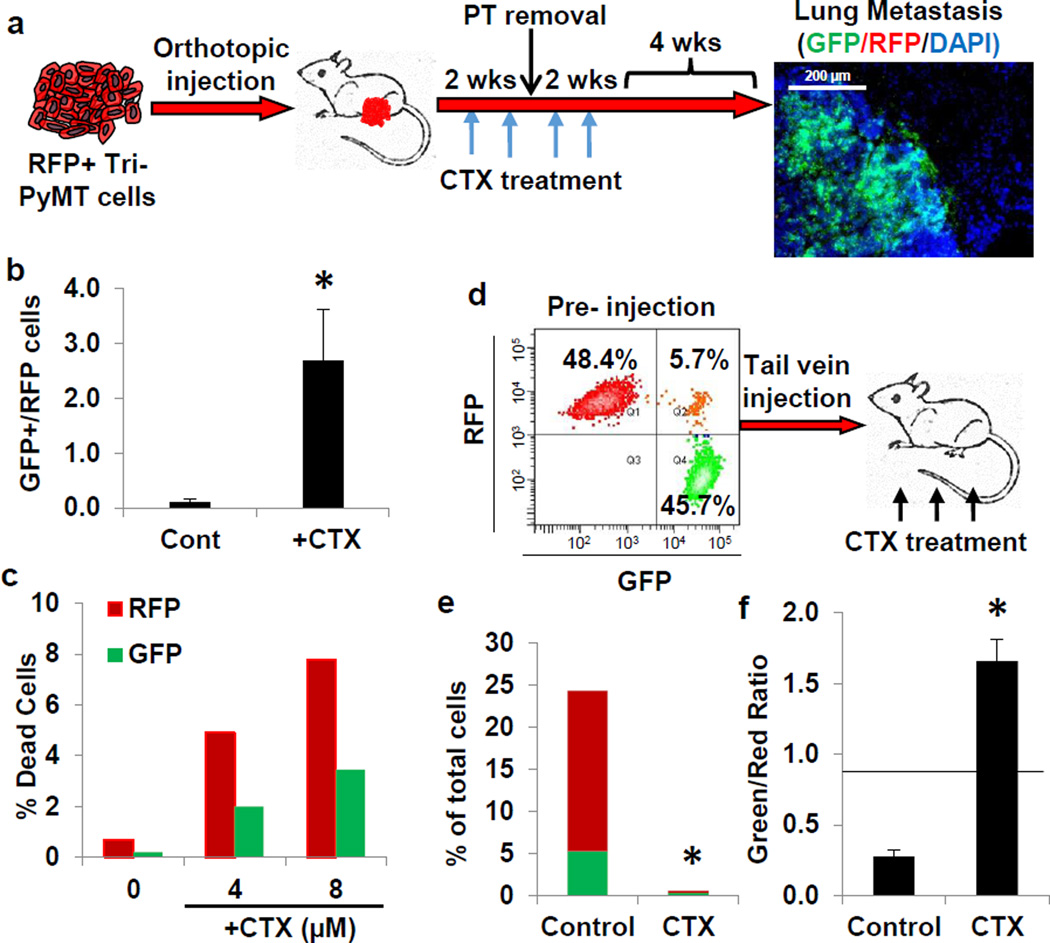
Figure 4. EMT tumor cells are resistant to chemotherapy.
a, Schema of cyclophosphamide (CTX) treatment in Tri-PyMT orthotopic model. Mice bearing an RFP+ primary tumor were treated with CTX (100mg/kg, once per week, for 4 weeks, as indicated by blue arrows). After two weeks of treatment, PT was removed (black arrow). Lung metastasis growth was permitted for 4 weeks post CTX treatment. Fluorescent imaging of lungs revealed the contribution of GFP+ tumor cells to lung metastases (n=9 mice).
b, Ratio of GFP+ to RFP+ cells in early metastatic lungs (4 weeks post orthotopic injection) of untreated (Cont) and CTX-treated (+CTX) mice as quantified by flow cytometry (n=4, *p<0.05).
c, Apoptosis (as measured by Annexin binding) of RFP+ and GFP+ Tri-PyMT cells treated with CTX (n=2 biological replicates).
d, Flow cytometry scatter plot showing the proportions of RFP+ and GFP+ Tri-PyMT cells prior to intravenous injection. Mice were treated with CTX (100mg/kg per week for 3 weeks, n=5 mice per group).
e, Quantification of flow cytometry data showing the percentage of RFP+ and GFP+ tumor cells (red and green bars, respectively) of total cells in the lung of the Control and CTX treated mice (CTX) (n=5 mice per group, *p<0.05).
f, Quantification of flow cytometry data showing the ratio of GFP+ to RFP+ cells in lungs of Control and CTX-treated mice. Black line represents the starting ratio of GFP+ to RFP+ cells prior to injection as derived from the data in Fig. 4d (*p<0.05).
Remarkably, in the early metastatic lungs (4 weeks after tumor inoculation), flow cytometry analysis revealed a 2.7:1 ratio of GFP+ to RFP+ cells in CTX-treated mice (Fig. 4b). Subsequently at 4 weeks after cessation of treatment, a significant contribution of GFP+ tumor cells was detected in 5 out of 17 metastatic lesions (Fig. 4a). This is in contrast to untreated mice, where all metastatic lesions were derived from RFP+ cells (Fig. 2d), suggesting that the EMT process may be involved in metastatic outgrowth in the context of chemotherapy.
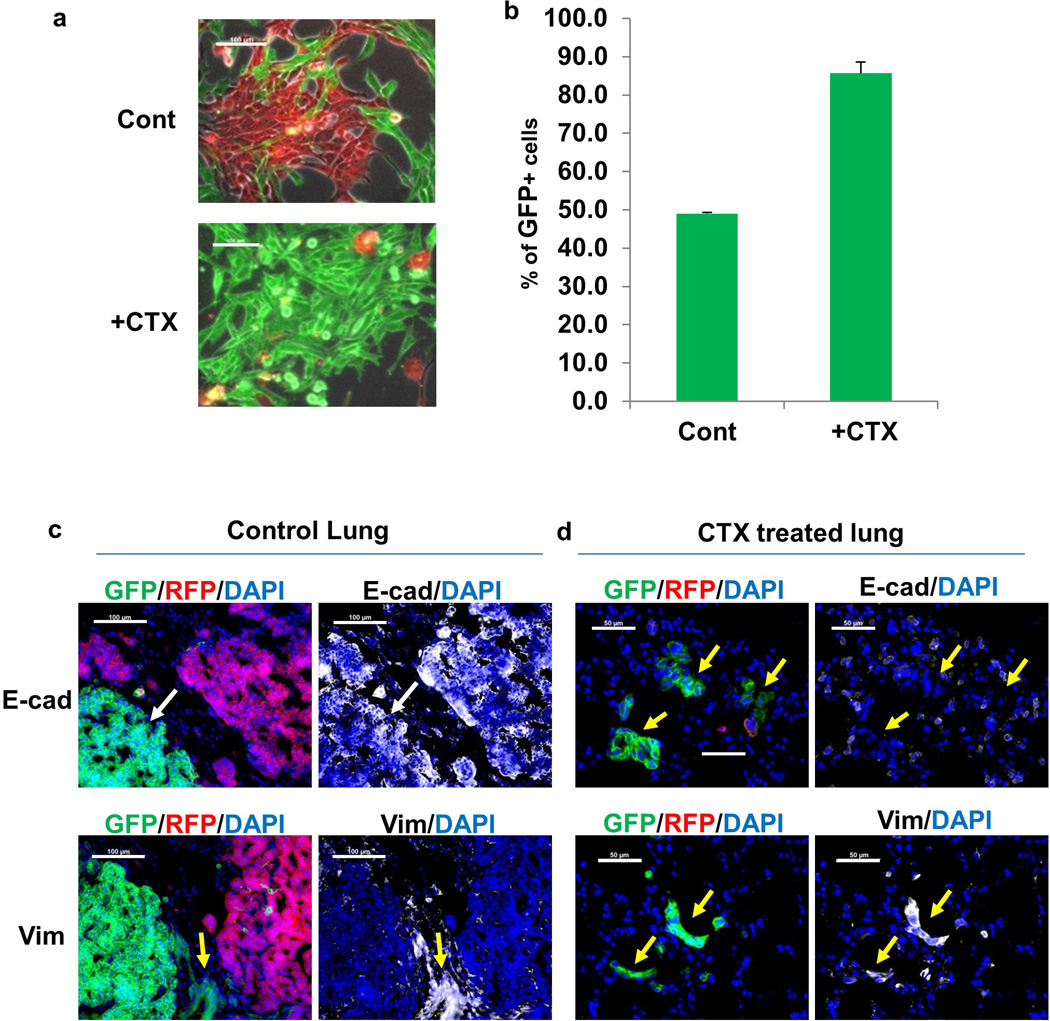
To evaluate the effects of CTX on the EMT and non-EMT cell populations, sorted GFP+ and RFP+ cells were incubated with CTX in vitro— the GFP+ cells were markedly more resistant to both short and long-term treatment (Fig. 4c, Extended Data Fig. 9a,b). The selective advantage of mesenchymal tumor cells in the context of chemotherapy was then corroborated by a competitive survival assay in vivo (Fig. 4d). Mice were injected intravenously with an equivalent number of RFP+ and GFP+ cells, and immediately received CTX (100mg/kg, once per week). After three weeks, lungs were harvested and the ratio of RFP+ and GFP+ cells was assessed by flow cytometry. CTX significantly inhibited outgrowth of lung metastasis from both RFP+ and GFP+ cells (Fig. 4e). The untreated lungs were morbidly overwhelmed with tumors, with nearly 80% of the tumor cells detected as RFP+. Conversely, in CTX-treated mice, more than 60% of the surviving tumor cells were GFP+, producing a significantly higher ratio of GFP:RFP cells in these mice (Fig. 4f). These results indicate that GFP+ EMT cells are more resistant to chemotherapy both in vitro and in vivo.
Immunostaining revealed that in the untreated mice, both RFP+ and GFP+ cells formed epithelial metastatic lesions (E-cad+/Vim-) (Extended Data Fig. 9c). Given the initial mesenchymal phenotypes of GFP+ cells prior to injection, this suggests that the GFP+ tumor cells have undergone MET in the metastatic organ. On the other hand, in CTX-treated mice the majority of surviving tumor cells were scattered mesenchymal GFP+ cells (E-cad-/Vim+) (Extended Data Fig. 9d). Together, these observations suggest that EMT tumor cells that sustain a mesenchymal phenotype are resistant to chemotherapy.
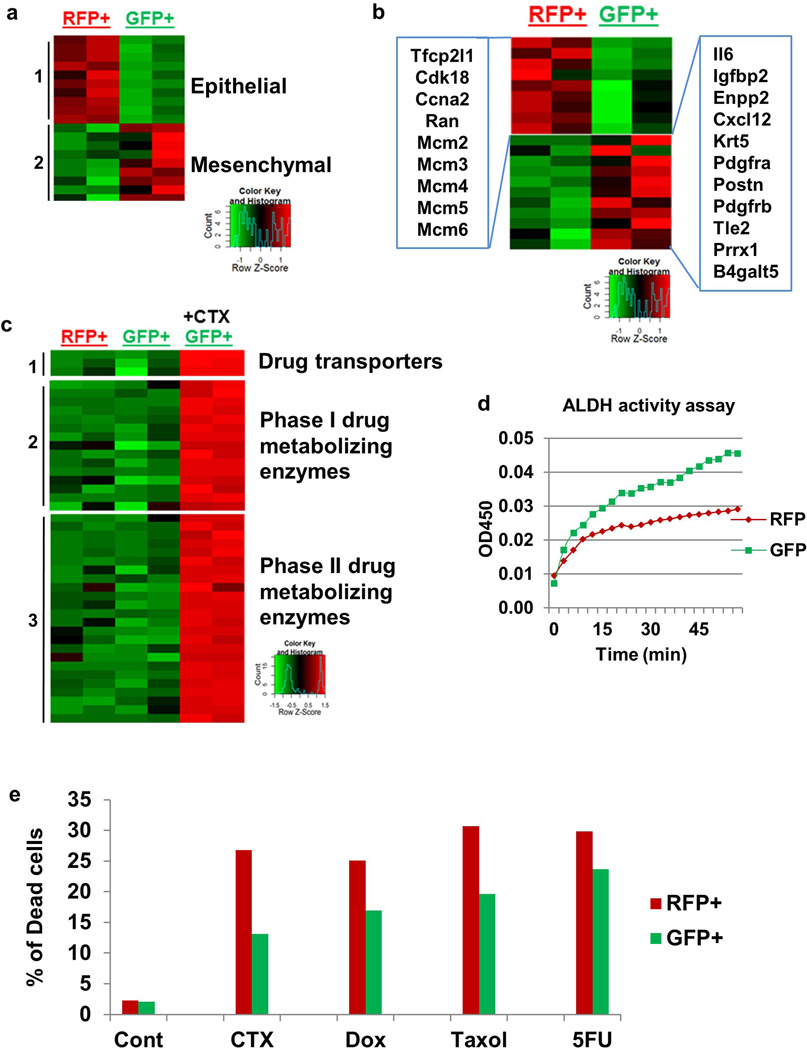
To begin to investigate the molecular underpinnings of mesenchymal tumor cell resistance, we analyzed the transcriptomic changes of EMT tumor cells. We sorted RFP+ and GFP+ cells and performed RNA-sequencing analysis (Supplementary Information Table 1). In addition to the expected changes in EMT marker expression (Extended Data Fig. 10a), the expression of many cell proliferation-related genes was reduced in GFP+ cells (Extended Data Fig. 10b), mirroring their phenotype of reduced proliferation in vivo. The GFP+ cells also showed increased expression of proven chemoresistance-related genes including IL6, Prrx1, Periostin, Enpp2, and Pdgfr25–29. Additionally, the CTX-treated GFP+ cells elevated their expression of many drug-metabolizing enzymes including drug transporters (Abcb1a, Abcb1b, and Abcc1), aldehyde dehydrogenases (ALDHs), cytochrome P450s, and glutathione metabolism related enzymes (Extended Data Fig. 10c). The main toxicity of CTX is due to its metabolite phosphoramide mustard, which is only formed in cells with low levels of ALDHs. ALDH converts the CTX-metabolite aldophosphamide into the non-toxic carboxyphosphamide30. In accordance with the transcriptomic data, GFP+ cells had significantly higher ALDH activity compared with RFP+ cells (Extended Data Fig. 10d). These properties of reduced proliferation, increased apoptotic resistance, and upregulation of chemoresistance and drug metabolizing genes in GFP+ EMT tumor cells may contribute to their insensitivity to CTX. Notably, GFP+ cells were also refractory to other commonly used chemotherapies including doxorubicin, paclitaxel, and 5-FU treatment (Extended Data Fig. 10e).
To demonstrate that the EMT is required for the generation of CTX resistance, we first tested in vitro the effect of treatment on control and miR-200 overexpressing Tri-PyMT cells. With increasing concentrations of CTX, the miR-200 cells were significantly more susceptible to therapy (Fig. 5a). We then expanded upon this finding in vivo, establishing orthotopic control and miR-200 primary tumors, and applying the pre-and-post surgery CTX regimen. We found that by blocking EMT in tumor cells, we effectively ablated metastatic growth (Fig. 5b,c). Thus, EMT contributes to the development of chemoresistant metastasis.
Figure 5. miR-200 overexpression abrogates CTX resistance.
a, Sensitivity of Control and +miR-200 Tri-PyMT tumor cells to CTX treatment as measured by CellTiter-Glo®. n=4 biological replicates per condition
b, Representative histologic lung images in Tri-PyMT Cont and +mir-200 tumor-bearing mice treated with CTX, (n=5).
c, Quantification of lung metastasis formation (number of individual nodules) in Tri-PyMT control and +mir-200 tumor-bearing mice CTX-treated mice (n=5).
Discussion
Using two independent EMT lineage tracing strategies in two disparate oncogene-driven autochthonous models of breast cancer, we demonstrated that lung metastases are derived from non-EMT tumor cells, contradicting the original EMT/MET hypothesis2,31. In a tracing system similar to our own, EMT was identified in primary tumors, but the mesenchymal lineage status of the metastatic nodules was not pursued32. Ultimately in our models we found that tumor cells disseminate and form metastases while persisting in their epithelial phenotype, in accordance with a recent study33. To underline that EMT is not required for metastasis, overexpression of miR-200—a microRNA that is incongruously associated with both reduced invasion19,20 and increased metastasis34—resulted in combined suppression of the EMT-promoting transcription factors Snail1/2, Twist, Zeb1 and Zeb2, but had no effect on metastasis. Given that both epithelial and mesenchymal tumor cells have the potential to disseminate, it is plausible that the larger fraction of highly proliferative epithelial cells out-compete the minor EMT tumor cell population in generating macrometastatic lesions.
Until now, the majority of data connecting EMT with chemoresistance was largely derived from in vitro studies, or clinical prognostic data. We demonstrated that highly proliferative non-EMT cells were sensitive to chemotherapy, and the emergence of recurrent EMT-derived metastases was observed after treatment. There is a great emphasis towards developing EMT-targeting therapies35,36, and our studies suggest that while EMT blockade may not affect metastasis formation, specifically targeting EMT tumor cells will be synergistic with conventional chemotherapy. Thus, our EMT lineage tracing system provides a unique preclinical platform to develop combination therapies that will eliminate both populations, and combat chemoresistance.
Methods
Animals
Wild type C57BL/6 and FVB/n mice, and transgenic mice with ACTB-tdTomato-EGFP (Stock# 007676), FSP1-Cre (Stock# 012641), MMTV-PyMT (Stock# 002374), and MMTV-Neu (Stock# 002376) were obtained from The Jackson Laboratory (Bar Harbor, Maine). The Vimentin-Cre mouse was a kind gift from the laboratory of Dr. Schwabe at Columbia University. CB-17 SCID mice were obtained from Charles River (Wilmington, MA). All mouse strains obtained were bred in the animal facility at WCMC. All animal work was conducted in accordance with a protocol approved by the Institutional Animal Care and Use Committee at Weill Cornell Medical College.
The ACTB-tdTomato-EGFP and FSP1-Cre mice were bred together to obtain double transgenic mice and then bred with MMTV-PyMT or MMTV-Neu mice to obtain the Tri-PyMT and Tri-Neu triple transgenic mice, respectively. Double transgenic male mice carrying ACTB-tdTomato-EGFP and MMTV-PyMT alle were crossed with the Vimentin-Cre mice to obtain the Tri-PyMT-Vim triple transgenic mice. Genotyping for each transgenic line was performed following the standardized protocols as described in the website of The Jackson Laboratory. Genotyping for Vimentin-Cre was done using forward primer 5′-CCCCTTCCTCACTTCTTTCC and reverse primer 5′-ATGTTTAGCTGGCCCAAATG.
Tamoxifen Injection
To induce Vim-Cre activities in the Tri-PyMT/Vim mice, Tamoxifen (Sigma-Aldrich, 2 mg/mouse dissolved in corn oil) was administered through intraperitoneal injections, 3 times per week starting when the primary tumors appear (at 8 weeks of age) and continuing for 6 weeks until metastasis developed in the lung.
Establishing Tri-PyMT cell line
The primary tumor of the Tri-PyMT mouse (12 weeks old female) was surgically removed under sterile conditions. Tumor tissue was sliced into ∼1 mm3 blocks and implanted into the fat pad (#4 on the right side) of CB-17 SCID mice. The secondary tumor was used to establish the Tri-PyMT cell line, eliminating the contamination of fluorescent positive stromal cells in the tumor tissue from Tri-PyMT transgenic mice.
Tumor tissue was minced and digested with an enzyme cocktail (Collagenase A, elastase, and DNase I, Roche Applied Science) in HBSS buffer at 37 °C for 30 min. The cell suspension was strained through a 40µm cell strainer (BD Biosciences). Cells were washed with PBS 3 times and uploaded in the Aria III cell sorter (BD Biosciences). The sorted RFP+ cells were cultured in DMEM supplemented with 10% fetal bovine serum. The PyMT oncogene expression in the established cell line was confirmed by RT-PCR (Extended Data Fig. 4c). The tumorigenic ability of these cells was confirmed throughout the study.
To determine EMT induction by TGF-β, cells were cultured for one week in DMEM with 2% FBS and 2ng/ml TGF-β1 (R&D Systems). The GFP+ cell ratio was quantified by flow cytometry.
To generate the miR-200 overexpressing cell line, a pLenti 4.1 Ex miR200b-200a-429 construct20, was obtained from Addgene. To eliminate the contamination of fluorescent marker expression in targeted cells, the GFP gene in this construct was removed by BstBI/XbaI digestion followed by blunted self-ligation. Lentivirus was packaged by co-transfection of the pLenti-miR200s construct and packaging plasmids into HEK293T cells. Tri-PyMT cells (passage 2) were infected with the lentivirus. Infected cells (Tri-PyMT miR-200) were selected by culturing with puromycin (2µg/mL) for 14 days. A control Tri-PyMT cell line was generated by infecting cells with lentivirus carrying the puromycin resistance gene, following the same procedure in parallel.
Orthotopic breast tumor model
To establish an orthotopic breast tumor model, we first purified RFP+ cells from passages 10–15 of Tri-PyMT cell culture by FACS. The purified RFP+ Tri-PyMT cells (1×106 cells with purity >99%, Extended Data Fig. 5a) were injected into the mammary fat pad of 8-week old female CB-17 SCID mice. The growth of the primary tumor was monitored by external caliper measurement once a week. In approximately 4 weeks, the primary tumor was surgically removed and the incision was closed with wound clips. The tumor size did not exceed 5% of total body weight as permitted in the IACUC protocol. Animals were sacrificed 4 weeks after primary tumor removal to analyze the development of pulmonary metastasis. For animals subjected to chemotherapy, Cyclophosphamide (CTX, Sigma-Aldrich, 100mg/kg) was administered once per week, for 2 weeks prior and 2 weeks after surgery.
Tissue processing, Immunofluorescence and Microscopy
The harvested primary tumors and PBS-perfused lungs bearing metastases were fixed in 4% paraformaldehyde overnight, followed by 30% sucrose for 2 days, and then embedded in Tissue-tek O.C.T. embedding compound (Electron Microscopy Sciences). Serial sections (10µm, at least 10 sections) were prepared for histological analysis by Hematoxylin & Eosin (H&E) staining, and immunofluorescent staining following standardized protocols.
Primary antibodies used in this study include CD45 (30-F11, BioLegend), E-cadherin (DECMA-1, BioLegend), vimentin (sc-7557, Santa Cruz), PyMT (ab15085, Abcam), Neu (sc-284, Santa Cruz), Ki67 (ab15580, Abcam), and Active Caspase-3 (C92–605, BD Pharmingen). Primary antibodies were directly conjugated to Alexa Fluor 647 using an antibody labeling kit (Invitrogen) performed as per manufacturer’s instructions and purified over BioSpin P30 columns (Bio-Rad). GFP+ and RFP+ cells were detected by inherent fluorescence.
Fluorescent images were obtained using a computerized Zeiss fluorescent microscope (Axiovert 200M), fitted with an apotome and an HRM camera. Images were analyzed using Axiovision 4.6 software (Carl Zeiss Inc.).
Flow cytometry and cell sorting
For the metastatic lungs and primary tumors, cell suspensions were prepared by digesting tissues with an enzyme cocktail (Collagenase A, elastase, and DNase I, Roche Applied Science) in HBSS buffer at 37°C for 30 min. For cultured cells, cells were collected through trypsinization. A single cell suspension was prepared by filtering through a 30-µm cell strainer (BD Biosciences). Then cells were stained following a standard immunostaining protocol. Briefly, cells were pre-blocked with 2% FBS plus Fc block (CD16/CD32, 1:30, BD Biosciences) and then incubated with the primary antibody against E-cadherin (DECMA-1, BioLegend). SYTOX Blue (Invitrogen) was added to the staining tube in the last 5 minutes to facilitate the elimination of dead cells. GFP+ and RFP+ cells were detected by their intrinsic signals. The stained samples were analyzed using the LSRII flow cytometer coupled with FACS Diva software (BD Biosciences). Flow cytometry analysis was performed using a variety of controls including isotype antibodies, unstained and single color stained samples for determining appropriate gates, voltages, and compensations required in multivariate flow cytometry.
For sorting live cells back for further culturing or injection into animals, we used the Aria II cell sorter coupled with FACS Diva software (BD Biosciences). The preparation of cells for sorting was performed under sterile conditions. The purity of subpopulations after sorting was confirmed by analyzing post-sort samples in the sorter again.
Quantitative RT-PCR analysis
Total RNA was extracted by using the RNeasy Kit (Qiagen), and miRNA via the mirVana miRNA isolation kit (Life Technologies), and converted to cDNA using qScript™ cDNA SuperMix (Quanta Biosciences) and RT-PCR. Q-PCR was performed with the appropriate primers (sequences shown in the table) and iQ™ SYBER Green master mix (Bio-Rad). PCR protocol: initial denaturing at 95°C for 3 min, 40 cycles of 95°C for 20 sec, 60°C for 30 sec, and 72°C for 30 sec, followed by final extension at 72°C for 5 min and melt curve analysis was applied on a Bio-Rad CFX96 Real Time System (Bio-Rad) coupled with Bio-Rad-CFX Manager software.
| Target gene | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| GAPDH | GGTCCTCAGTGTAGCCCAAG | AATGTGTCCGTCGTGGATCT |
| E-cadherin | ACACCGATGGTGAGGGTACACAGG | GCCGCCACACACAGCATAGTCTC |
| Occludin | TGCTAAGGCAGTTTTGGCTAAGTCT | AAAAACAGTGGTGGGGAACGTG |
| Vimentin | TGACCTCTCTGAGGCTGCCAACC | TTCCATCTCACGCATCTGGCGCTC |
| N-cadherin | AAAGAGCGCCAAGCCAAGCAGC | TGCGGATCGGACTGGGTACTGTG |
| FSP-1 | CCTGTCCTGCATTGCCATGAT | CCCACTGGCAAACTACACCC |
| Snail1 | ACTGGTGAGAAGCCATTCTCCT | CTGGCACTGGTATCTCTTCACA |
| Snail2 | TTGCAGACAGATCAAACCTGAG | TGTTTATGCAGAAGCGACATTC |
| Twist1 | AGCTACGCCTTCTCCGTCTG | CTCCTTCTCTGGAAACAATGACA |
| Zeb-1 | GATTCCCCAAGTGGCATATACA | TGGAGACTCCTTCTGAGCTAGTG |
| Zeb-2 | TGGATCAGATGAGCTTCCTACC | AGCAAGTCTCCCTGAAATCCTT |
| PyMT | ACTGCTACTGCACCCAGACA | CTGGAAGCCGGTTCCTCCTA |
| GFP | CCACATGAAGCAGCACGACT | GGGTCTTGTAGTTGCCGTCG |
| RFP | AGCGCGTGATGAACTTCGAG | CCGCGCATCTTCACCTTGTA |
RNA-sequencing analysis
Total RNA was extracted from sorted RFP+ and GFP+ Tri-PyMT cells with the RNeasy Kit (Qiagen). RNA-Seq libraries was constructed and sequenced following standard protocols (Illumina). Single-end RNA-seq reads were mapped to UCSC mouse genome (GRCm38/mm10) using Tophat2. FPKM values for each gene were estimated by Cufflinks and statistical analysis was done using Cuffdiff2. Heatmaps for differentially expressed genes with adjusted p value <0.05 were drawn using gplots R package.
Western blot analysis
Cells were homogenized in 1x RIPA lysis buffer (Millipore) with protease inhibitors (Roche Applied Science). Samples were boiled in 1x Laemmli buffer and 10% β-mercaptoethanol, and loaded onto 12% gradient Tris-Glycine gels (Bio-Rad). Western blotting was performed using antibodies specific for E-cadherin (clone DECMA-1), vimentin (clone RV202, BD Pharmingen), and β-actin (clone AC-15, Sigma-Aldrich).
Cell apoptosis & viability assays
To determine apoptosis of RFP+ and GFP+ cells, Tri-PyMT cells (Passage 10) were seeded on adherent 6-well plates (1×106 cells), and treated with 4-Hydroperoxy Cyclophosphamide (Santa Cruz) for 48 hours. After treatment, cells were trypsinized and stained with APC conjugated Annexin V (BD Biosciences) and SYTOX Blue (Invitrogen) for apoptotic cells labeling. The stained cells were analyzed in the LSRII flow cytometer to quantify the percentage of apoptotic, dead, and live RFP+ and GFP+ cells by FACS Diva software. To determine the viability of Tri-PyMT Control and +miR-200 cells treated with CTX, cells were plated in 96-well adherent black-walled plates (1×104 cells), and treated with 4-Hydroperoxy Cyclophosphamide for 48 hours. After treatment, cell viability was measured with the CellTiter-Glo® Luminescent Cell Viability Assay (Promega).
Cell migration assay
1×105 Tri-PyMT cells were seeded in a 6-well plate. Real time images of cells (including phase, GFP and RFP channels) were taken under a computerized Zeiss microscope (Axiovert observation) every 10 minutes for 10 hours. Movement of individual cells (>10 RFP+ and >10 GFP+ cells in each field, >2 fields were analyzed) were tracked with ImageJ software, and the distance that was traveled during that time was measured as indicated.
ALDH activity assay
RFP+ and GFP+ Tri-PyMT cells (1×106 cells each) were freshly sorted from culture by FACS and then homogenized in cold ALDH Assay buffer provided in the ALDH Activity Colorimetric Assay Kit (Biovision Inc.) Following the protocol, ALDH substrate and acetaldehyde were added. ALDH activities in samples were measured by OD at 450 nm in a kinetic mode (every 3 minute for 60 min).
Statistical Analysis
To determine the sample size of animal experiments, we used power analysis assuming the (difference in means)/(standard deviation) is >2.5. Therefore, all animal experiments were conducted with ≥ 5 mice per group to ensure adequate power between groups by two-sample t-test comparison. Animals were randomized within each experimental group. No blinding was applied in performing experiments. Results were expressed as mean ± SEM. Data distribution in groups and significance between different treatment groups was analyzed by using the Mann-Whitney T test in GraphPad Prism software. P values < 0.05 were considered significant. Error bars depict SEM, except where indicated otherwise.
Extended Data
Extended Data Figure 1. Characterization of the primary tumor and lung metastasis of Tri-PyMT mice.
Sections of primary tumors (a) and lungs (b) from Tri-PyMT mice were immunostained for E-cadherin (E-cad, top), vimentin (Vim, middle) and CD45 (bottom) in white psuedocolor. Representative images are shown (n > 5 mice). Note the co-localization of PyMT with RFP, and CD45 with GFP (as indicated by arrows) in both primary tumors and lung metastases.
Extended Data Figure 2. Characterization of the primary tumor and lung metastasis of Tri-Neu mice.
Sections of primary tumors (left panel) and lungs (right panel) from Tri-Neu mice were immunostained for CD45, Neu, E-cadherin, and Vimentin (in white psuedocolor). Representative images are shown (n>5 mice). Note that both primary tumors and lung metastases are largely composed of epithelial RFP+ tumor cells.
Extended Data Figure 3. Characterization of the primary tumor and lung metastasis of Tri-PyMT/Vim mice.
Tri-PyMT/Vim mice were obtained by crossing MMTV-PyMT, Vimentin-Cre and Rosa26-GRFP transgenic mice. Sections of primary tumors (a) and lungs (b) were immunostained for PyMT, E-cadherin, and Vimentin (in white psuedocolor). Representative images are shown (n> 5 mice). Note that both primary tumors and lung metastases are largely composed of epithelial RFP+ tumor cells.
Extended Data Figure 4. Characterization of Tri-PyMT cells.
a, EMT of Tri-PyMT cells with TGF. RFP+ Tri-PyMT cells were sorted by flow cytometry and cultured in medium containing 2% FBS with or without TGF-β1 (2ng/mL) for 3 days. Plot shows quantification of the percentage of GFP+ cells analyzed by flow cytometry (n=2 biological replicates).
b, Cell migration assay of Tri-PyMT cells. The tracing plots show the movement of individual RFP+ and GFP+ cells in 10 hours of live imaging. Quantification plot (right panel) showed the average distance that RFP+ and GFP+ cells have moved during the time frame (n>20, *p<0.01).
c, Relative expression of epithelial, mesenchymal, and tumor markers in sorted RFP+ and GFP+ Tri-PyMT cells as determined by Q-RT-PCR with GAPDH as the internal control. n=2 individual experiments.
d, EMT of Tri-PyMT cells is reported by fluorescent marker switch. Flow cytometry plot shows E-cadherin- (E-Cad-) and E-cadherin+ (E-Cad+) subpopulations of Tri-PyMT cells (upper panel). Of the E-Cad- and E-Cad+ subsets, the populations were further dissected according to innate fluorescence (lower panel). Numbers indicate the percentage of GFP+, RFP+, or transitioning (Q2) cells in the parental E-Cad- or E-Cad+ subsets, respectively.
Extended Data Figure 5. Establishing an orthotopic model with sorted RFP+ Tri-PyMT cells.
a, Flow cytometry plots show Tri-PyMT cells before and after sorting for RFP+ cells. Numbers indicate the percentage and purity of RFP+ cells used for establishing orthotopic breast tumors in mice.
b, Schematic of the orthotopic breast tumor model with sorted RFP+ Tri-PyMT cells. Cells are injected into the mammary gland of wild type mice to generate primary breast tumors, resection of primary tumor at 4 weeks and lung metastases evaluation in another 4 weeks.
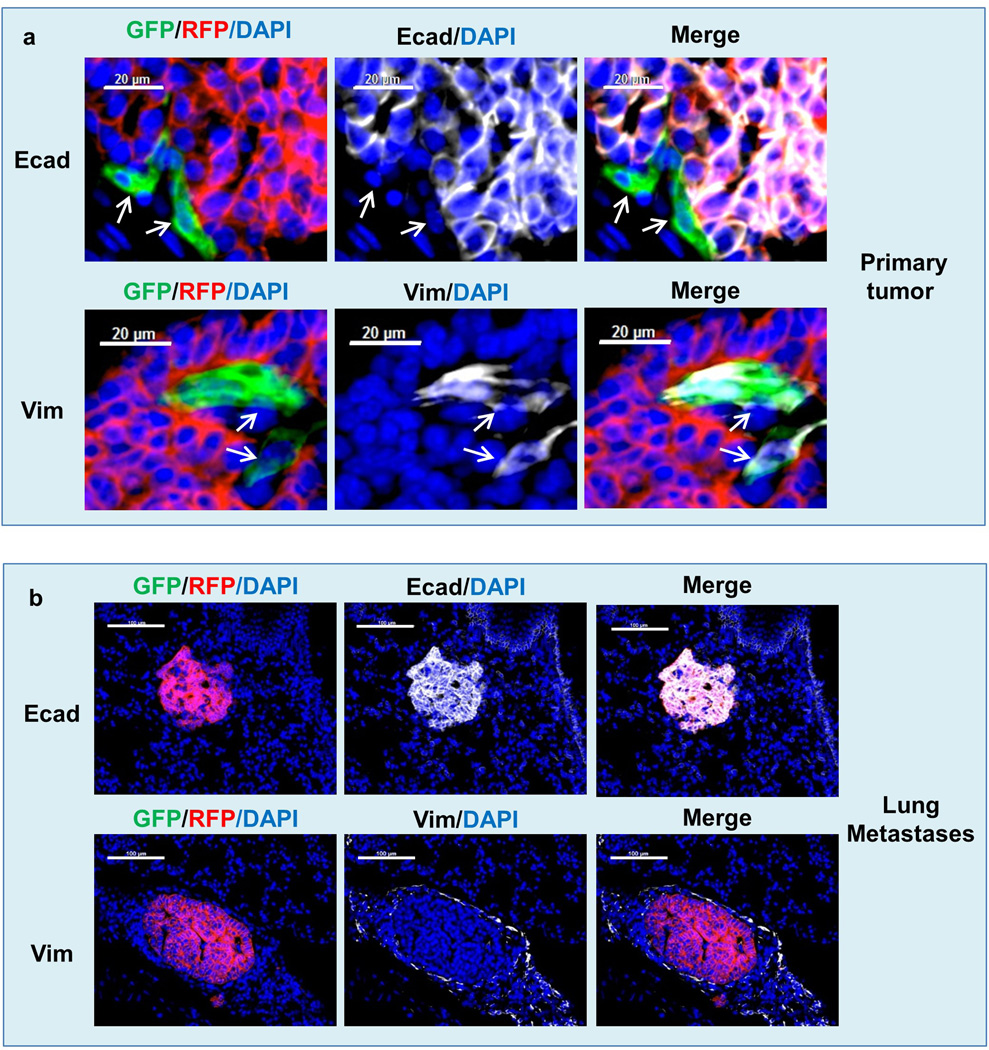
c, Characterization of tumor cells in the primary tumor, disseminated tumor cells (DTCs) and tumor cells in the lung metastasis of the Tri-PyMT orthotopic model. Sections of primary tumors and lungs from Tri-PyMT orthotopic mice were immunostained for E-cadherin and Vimentin (in white pseudocolor). Essentially all RFP+ tumor cells are detected as E-cad+/Vim−, while the scattered GFP+ tumor cells in the primary tumor are E-Cad−/Vim+ (as indicated by arrows in the top panel). Representative images are shown (n=8). d, Plot shows the percentage of GFP+ cells out of total tumor cells (GFP+ plus RFP+, n=6).
Extended Data Figure 6. Characterization of EMT status of orthotopic Tri-Vim-PyMT primary tumor.
Sections of Tri-Vim-PyMT orthotopic primary tumors (a) and metastatic lung (b) were immunostained for E-cadherin and vimentin (in white psuedocolor). As expected, RFP+ tumor cells are entirely E-cadherin positive and vimentin negative, GFP+ tumor cells are vimentin positive and E-cadherin negative, and lung metastases are epithelial and RFP+.
Extended Data Figure 7. Dissemination of Tri-PyMT cells in vivo.
a, Disseminated tumor cells are RFP+ and epithelial. RFP+ Tri-PyMT cells were injected into the fat pad of mice. The fluorescence of the primary tumor, circulating tumor cells in the blood and disseminated tumor cells in the lung was analyzed by flow cytometry. The flow cytometry plots depicted are the enumeration of RFP+ and GFP+ cells.
b, The ratios of detected RFP+ versus GFP+ cells are shown in the chart (n=4 mice).
c, Relative expression of miR-200 family microRNAs in Tri-PyMT control and miR-200 expressing cells. n=2 individual experiments.
d, Relative expression of EMT markers and tumor markers in Tri-PyMT control and +mir-200 cells as determined by Q-RT-PCR with GAPDH as the internal control, n=2 individual experiments.
Extended Data Figure 8. Effects of CTX therapy on primary tumors.
a, Quantification of primary tumor growth after 2 weeks of CTX therapy. For tumor growth data see Source Data File. b, Proliferation status of primary tumor cells as detected by Ki67 staining in control mice and after 2 weeks of CTX therapy. c, Level of apoptosis in primary tumors as detected by active caspase-3 staining in control mice and after 2 weeks of CTX therapy. Representative images of Ki67 (d) and active caspase-3 staining (e) (white pseudocolor) of primary tumors in control mice and CTX-treated mice. f, Proliferation status of RFP+ and GFP+ primary tumor cells as detected by Ki67 staining in control and CTX-treated mice. g, Level of apoptosis in RFP+ and GFP+ primary tumors as detected by active caspase-3 staining in control and CTX-treated mice. h, Percentage of GFP+ tumor cells in control and CTX-treated primary tumors. n=3 mice for all figures described above. Quantification performed utilizing ImageJ software.
Extended Data Figure 9. EMT tumor cells are resistant to CTX treatment both in vitro and in vivo.
a, b, Long term CTX treatment in vitro results in GFP+ population. Tri-PyMT cells were subjected to 2 weeks cyclophosphamide (+CTX) treatment (4 µM). Fluorescent imaging (a) and flow cytometry (quantified, b, n=3) exhibit the percentage of GFP+ cells in the CTX-treated culture compared to control untreated cells
c, d, EMT status of lung nodules in competitive survival assay. Representative fluorescent images of Tri-PyMT lung metastases in untreated control lungs (c) and CTX-treated lungs (d), depicting RFP+ and GFP+ tumor cells. Immunostaining showing E-cadherin (E-cad), vimentin (Vim) in white pseudocolor. White arrow indicates GFP+ tumor cells with epithelial phenotypes (E-cad+/Vim−), while the yellow arrow indicates GFP+ cells with mesenchymal phenotypes (E-cad−/Vim+). Nuclei were counter stained with DAPI. n =5.
Extended Data Figure 10. Gene expression profile analysis of RFP+ and GFP+ Tri-PyMT cells.
RFP+ and GFP+ Tri-PyMT cells were sorted by flow cytometry and subjected to transcriptomic analysis by RNA-sequencing.
a, Heatmap of differentially expressed genes (adjusted p<0.05) from RNA-Seq of sorted RFP+ and GFP+ Tri-PyMT cells, biologically duplicated. Genes that are established epithelial markers (Group 1) include E-cad, Dsp, Epcam, Fgfbp1, Krt18, Krt19, Ocln, Tjp3, Krt14, Tjp2; the mesenchymal markers (Group 2) include N-cad, Col23a1, Col3a1, Col5a1, Col6a2, fsp1, Mmp3, Wnt5a, Zeb1.
b, Cell cycle (left panel) and chemoresistance-related (right panel) genes alternatively regulated in RFP+ and GFP+ cells.
c, GFP+ Tri-PyMT cells were also sorted from 4-OH-cyclophosphamide (CTX, 4 µM) treated samples. Interestingly, a branch of genes related to drug metabolism were significantly elevated in CTX treated GFP+ cells. Group 1 genes are drug transporters including Abcb1a, Abcb1b, Abcc1. Group 2 genes are Phase I drug metabolizing enzymes including Adh7, Aldh1a1, Aldh1a3, Aldh1l1, Aldh1l2, Aldh2, Aldh3a1, Aldh3a2, Aldh3b2, Aldh4a1, Cyp1a1, Cyp2f2, Cyp2j6, Ptgs1, Ptgs2. Group 3 genes are Phase II drug metabolizing enzymes including Aox1, Blvrb, Ces2e, Ces2f, Ces2g, Chst1, Ephx1, Fmo1, Gpx2, Gsta3, Gsta4, Gstm2, Gsto1, Gstp1, Gstt3, Maoa, Mgst1, Mgst2, Nat6, Nat9, Nqo1, Pon3, Ugt1a6a, Ugt1a7c.
d, Aldehyde dehydrogenase (ALDH) activity assay. Cell lysates were prepared from flow cytometry sorted RFP+ and GFP+ Tri-PyMT cells. ALDH activity in samples was measured by OD at 450 nm in a kinetic mode (every 3 minute for 60 min). Representative result from two independent experiments depicted.
e, EMT tumor cells (GFP+ cells) showed resistance to multiple commonly used chemotherapies. Tri-PyMT cells were subjected to treatment with 4-OH-cyclophosphamide (CTX, 8 µM), doxorubicin (Dox, 2µM), paclitaxel (Taxol, 10µM) and 5FU (1.6µM) for 3 days. Flow cytometry analysis of apoptotic cells was performed after Annexin staining. The percentage of dead cells (Annexin+) in RFP+ and GFP+ cells, respectively, was quantified. n=2 biological replicates.
Supplementary Material
Acknowledgements
This work was supported by a grant from the US Department of Defense CDMRP LCRP (LC110643). K. F. is supported by a fellowship from the NIH (1 F31 CA186510-01). V.M. was supported by the National Cancer Institute subaward (U54 CA149196-05) and WCMC Meyer Cancer Center Pilot Funding. This work was also supported by funds from The Neuberger Berman Foundation Lung Cancer Research Center; the Arthur and Myra Mahon Donor-Advised Fund; the Liz Claiborne and Art Ortenberg Foundation; the Douglas & Katherine McCormick Family Foundation, the R. & M. Goldberg Family Foundation; the P. & C. Collins Fund; the Eliot Stewart “Wren” Fund; the William and Shelby Modell Family Foundation Trust; and generous funds donated by patients in the Division of Thoracic Surgery to N.K.A. The funding organizations played no role in experimental design, data analysis or manuscript preparation.
Footnotes
Author Contributions: D.G., K.R.F., and V.M. designed the experiments. K.R.F. and D.G. performed the experiments. A.D., S.L., H.C., T. E. R and S.R., provided technical support with experiments and animal work. L.T.V. and N.K.A. made critical comments to improve the study design. J.S., F.L., and S.W. performed RNA-sequencing analysis. J.T., and R.F.S. generated the Vim-Cre transgenic mice. K.R.F., D.G. and V.M. wrote and edited the manuscript with input from the other authors. All authors discussed the results and conclusions drawn from them.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Bastid J. EMT in carcinoma progression and dissemination: facts, unanswered questions, and clinical considerations. Cancer Metastasis Rev. 2012;31:277–283. doi: 10.1007/s10555-011-9344-6. [DOI] [PubMed] [Google Scholar]
- 2.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheel C, Weinberg RA. Phenotypic plasticity and epithelial-mesenchymal transitions in cancer and normal stem cells? Int J Cancer. 2011;129:2310–2314. doi: 10.1002/ijc.26311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gal A, et al. Sustained TGF beta exposure suppresses Smad and non-Smad signalling in mammary epithelial cells, leading to EMT and inhibition of growth arrest and apoptosis. Oncogene. 2008;27:1218–1230. doi: 10.1038/sj.onc.1210741. [DOI] [PubMed] [Google Scholar]
- 6.Hennessy BT, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69:4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao D, et al. Myeloid progenitor cells in the premetastatic lung promote metastases by inducing mesenchymal to epithelial transition. Cancer Res. 2012;72:1384–1394. doi: 10.1158/0008-5472.CAN-11-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin EY, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163:2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhowmick NA, et al. TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303:848–851. doi: 10.1126/science.1090922. [DOI] [PubMed] [Google Scholar]
- 11.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 12.Xue C, Plieth D, Venkov C, Xu C, Neilson EG. The gatekeeper effect of epithelial-mesenchymal transition regulates the frequency of breast cancer metastasis. Cancer Res. 2003;63:3386–3394. [PubMed] [Google Scholar]
- 13.Okada H, Danoff TM, Kalluri R, Neilson EG. Early role of Fsp1 in epithelial-mesenchymal transformation. Am J Physiol. 1997;273:F563–F574. doi: 10.1152/ajprenal.1997.273.4.F563. [DOI] [PubMed] [Google Scholar]
- 14.Gunasinghe NP, Wells A, Thompson EW, Hugo HJ. Mesenchymal-epithelial transition (MET) as a mechanism for metastatic colonisation in breast cancer. Cancer Metastasis Rev. 2012;31:469–478. doi: 10.1007/s10555-012-9377-5. [DOI] [PubMed] [Google Scholar]
- 15.Cabezon T, et al. Expression of S100A4 by a variety of cell types present in the tumor microenvironment of human breast cancer. Int J Cancer. 2007;121:1433–1444. doi: 10.1002/ijc.22850. [DOI] [PubMed] [Google Scholar]
- 16.Guy CT, et al. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci U S A. 1992;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Troeger JS, et al. Deactivation of hepatic stellate cells during liver fibrosis resolution in mice. Gastroenterology. 2012;143:1073–1083. doi: 10.1053/j.gastro.2012.06.036. e1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dumont N, et al. Sustained induction of epithelial to mesenchymal transition activates DNA methylation of genes silenced in basal-like breast cancers. Proc Natl Acad Sci U S A. 2008;105:14867–14872. doi: 10.1073/pnas.0807146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 21.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Toy KA, Kleer CG. Metaplastic breast carcinomas are enriched in markers of tumor-initiating cells and epithelial to mesenchymal transition. Mod Pathol. 2012;25:178–184. doi: 10.1038/modpathol.2011.167. 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Creighton CJ, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Minckwitz G. Docetaxel/anthracycline combinations for breast cancer treatment. Expert Opin Pharmacother. 2007;8:485–495. doi: 10.1517/14656566.8.4.485. [DOI] [PubMed] [Google Scholar]
- 25.Lau CK, et al. An Akt/hypoxia-inducible factor-1alpha/platelet-derived growth factor-BB autocrine loop mediates hypoxia-induced chemoresistance in liver cancer cells and tumorigenic hepatic progenitor cells. Clin Cancer Res. 2009;15:3462–3471. doi: 10.1158/1078-0432.CCR-08-2127. [DOI] [PubMed] [Google Scholar]
- 26.Xiao ZM, Wang XY, Wang AM. Periostin induces chemoresistance in colon cancer cells through activation of the PI3K/Akt/survivin pathway. Biotechnol Appl Biochem. 2013 doi: 10.1002/bab.1193. [DOI] [PubMed] [Google Scholar]
- 27.Yamada D, et al. Role of crosstalk between interleukin-6 and transforming growth factor-beta 1 in epithelial-mesenchymal transition and chemoresistance in biliary tract cancer. Eur J Cancer. 2013;49:1725–1740. doi: 10.1016/j.ejca.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Yao Z, et al. TGF-beta IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc Natl Acad Sci U S A. 2010;107:15535–15540. doi: 10.1073/pnas.1009472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ocana OH, et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22:709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Russo JE, Hilton J. Characterization of cytosolic aldehyde dehydrogenase from cyclophosphamide resistant L1210 cells. Cancer Res. 1988;48:2963–2968. [PubMed] [Google Scholar]
- 31.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trimboli AJ, et al. Direct evidence for epithelial-mesenchymal transitions in breast cancer. Cancer Res. 2008;68:937–945. doi: 10.1158/0008-5472.CAN-07-2148. [DOI] [PubMed] [Google Scholar]
- 33.Yu M, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korpal M, et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta PB, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diessner J, et al. Targeting of preexisting and induced breast cancer stem cells with trastuzumab and trastuzumab emtansine (T-DM1) Cell Death Dis. 2014;5:e1149. doi: 10.1038/cddis.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.