Abstract
Tooth formation is tightly regulated by epithelial-mesenchymal interactions via hierarchic cascades of signaling molecules. The glycosaminoglycan (GAG) chains covalently attached to the core protein of proteoglycans (PGs) provide docking sites for signaling molecules and their receptors during the morphogenesis of tissues and organs. While PGs are believed to play important roles in tooth formation, little is known about their exact functions in this developmental process and the relevant molecular basis. Family with sequence similarity member 20-B (FAM20B) is a newly identified kinase phosphorylating the xylose in the common linkage region connecting the GAG with the protein core of PGs. The phosphorylation of xylose is essential to the common linkage elongation and the subsequent GAG assembly. In this study, we generated Fam20B-floxed allele in mice and found that inactivating Fam20B in the dental epithelium leads to supernumerary maxillary and mandibular incisors. This finding highlights the pivotal role of PGs in tooth morphogenesis and opens a new window for understanding the regulatory mechanism of PG-mediated signaling cascades during tooth formation.
Keywords: glycosaminoglycan; kinase; proteoglycan; tooth development, supernumerary teeth
Proteoglycans (PGs) are large molecules that have one or more glycosaminoglycan (GAG) side chains attached to the protein core via an O-linkage through serine residue(s) (1). The assembly of GAG chains is initiated by the synthesis of the GAG-protein linkage region (GlcAβ1–3Galβ1–3Galβ1–4Xylβ1-O-Ser), which is formed through a sequential addition of monosaccharide residues catalyzed by respective glycosyltransferases (2). The repeating disaccharide of heparan sulfate, heparin, chondroitin sulfate and dermatan sulfate are synthesized on the linkage region (3), followed by numerous modifications including sulfation, epimerization and desulfation.
Mature PGs are present on the cell surface and in the extracellular matrix (ECM) of various tissues. The GAG chains are involved in complex biological functions such as assembling and activating the complexes of growth factors or growth factor receptors, as well as regulating pathways in the coagulation process and complement cascades (4). Heparan sulfate (HS) is known to be important modulators of WNT, BMP, FGF, and SHH signaling pathways (5). Chondroitin sulfate (CS) and dermatan sulfate (DS) have also been recognized as co-receptors and signal modulators in multiple signaling pathways (6).
The development of teeth is regulated by complex reciprocal interactions between the dental epithelium and dental mesenchyme. The proper patterning and morphogenesis of the tooth organ depend on a delicate balance of various signaling pathways such as those of WNT, BMP, FGF, and SHH, which are reiteratively used in the determination of tooth position, number, type, and shape (7). The response of the cells to the changes in the extracellular matrix involves integral cell surface molecules to recognize their matrix ligand and initiate intracellular signaling.
While PGs have been thought to be important for tooth formation, little is known about their specific roles in this developmental process. The current knowledge gained from previous studies represents a significant gap in understanding the role of PGs in tooth morphogenesis and the relevant molecular basis. Several studies reported the expression patterns of several PGs in embryonic teeth (8–14) and binding activity between mesenchymal syndecan and tenascin (15). A line of transgenic mice overexpressing heparan sulfate proteoglycan 2 (HSPG2) in the enamel organ display dull-ended crowns and divergent tooth roots (16, 17), indicating the need for an appropriate amount of HSPG2 during tooth morphogenesis. The oligodontia and split crowns in zebrafish lacking heparan sulfate proteoglycans (HSPGs) are partially attributed to an attenuated FGF8 signaling (18). The inactivation of endosulfatases in mice leads to mild dentin defects and impaired WNT signaling in odontoblasts, suggesting an essential role of HSPG desulfation in dentinogenesis (19). A more recent study utilizing an in vitro tooth culture system found that the differential expression of syndecan 1 and N-deacetylase/N-sulfotransferases (NDSTs), as well as their mediation on FGF retention on the cell surface, are responsible for the differential odontogenic potency between the reaggregated incisor and molar dental mesenchymal cells (20).
Family with sequence similarity 20 (FAM20) is a group of evolutionarily conserved molecules, which include FAM20A, FAM20B and FAM20C. FAM20C is a Golgi-enriched kinase responsible for the phosphorylation of secretory calcium-binding phospho-protein (SCPP) family, which includes “small-integrin-binding ligand, N-linked glycoproteins” (SIBLINGs) and several enamel-matrix proteins (21, 22). Fam20C-knockout mice develop severe hypophosphatemic rickets due to an increased renal phosphate wasting that is likely attributed to the remarkable elevation of serum FGF23 (23), while their dentin and enamel defects are largely independent from the hypophosphatemia and appear to be local effects of phosphorylation failure in the SCPP proteins (24, 25, 26). FAM20A is believed to be a pseudokinase that forms a functional complex with FAM20C, and this complex enhances extracellular protein phosphorylation within the secretory pathway (27). Loss of function in FAM20A leads to amelogenesis imperfecta with gingival fibromatosis syndrome (28) and enamel renal syndrome (29) in humans and mice (30). FAM20B is a newly identified kinase phosphorylating the xylose in tetrasaccharide linkage region that connects glycosaminoglycan (GAG) chains with the core protein of proteoglycans (31). The phosphorylation of this xylose is essential for galactosyltransferase II (GalT-II)-mediated elongation of the tetrasaccharide bridge and subsequent assembly of GAGs (32, 33). Genetic studies showed that loss-of-function mutations in the Fam20B in zebra fish decrease the amount of cellular GAG chains and cause cartilage and skeleton defects, while constitutive deletion of Fam20B in mice results in embryonic lethality at E13.5 (30, 34).
In this study, we generated a Fam20B-floxed allele and crossbred the Fam20B-floxed mice with Keratin 14 promoter-driven Cre (K14-Cre) mice to generate K14-Cre;Fam20Bflox/flox mice, in which Fam20B was inactivated in the oral and dental epithelium. The K14-Cre;Fam20Bflox/flox mice exhibit supernumerary incisors.
Materials and methods
All animal procedures were approved by the Institutional Animal Care and Use Committee of Texas A&M University Baylor College of Dentistry (Dallas, TX, USA) and performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Generation of Fam20B floxed allele and K14-Cre;Fam20Bflox/flox Mice
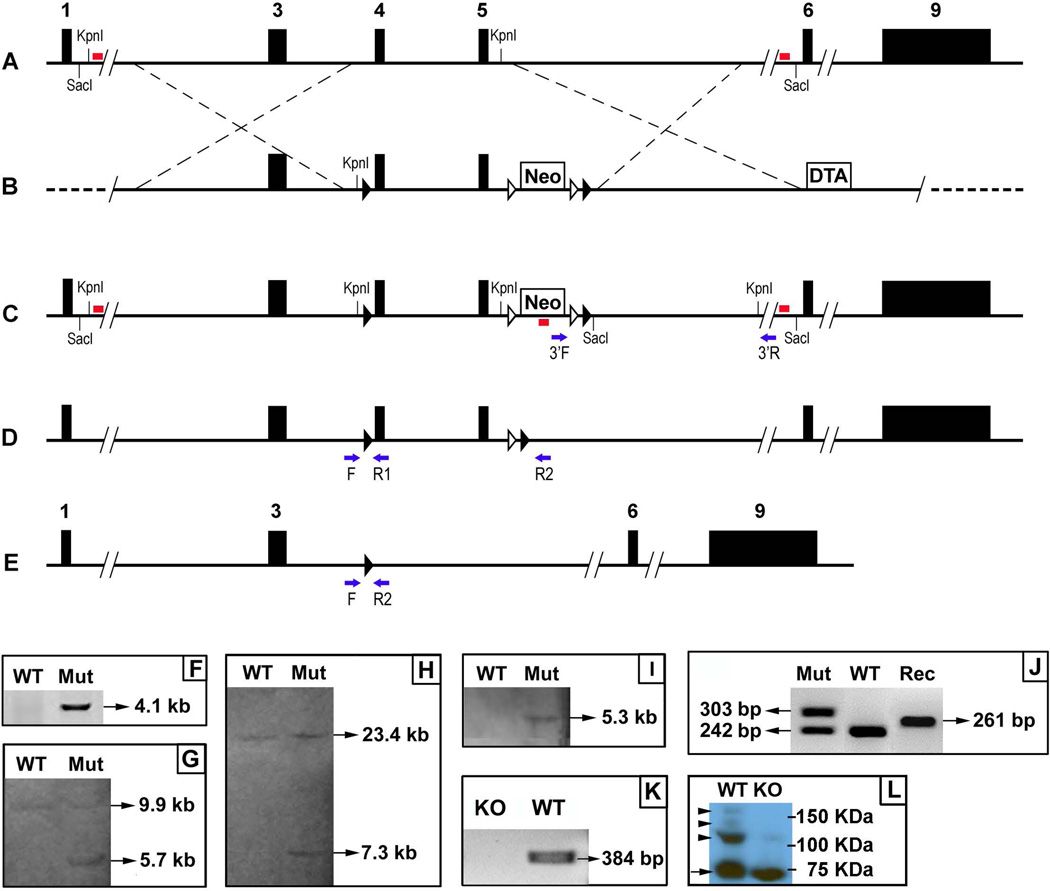
Murine Fam20B gene (NM_145413.4) contains nine exons. We chose exons 4 and 5 as the floxed region. To engineer the targeting vector, a 4.6 kb 5’ homology arm, a 2.5 kb 3’ homology arm, and a 4.6 kb fragment containing the exons 4 and 5 were amplified by polymerase chain reaction (PCR) using the template DNA extracted from a BAC clone RP24-262C9 (CHORI, Oakland, CA, USA). The 4.6 kb fragment that contains the exons 4 and 5 was subcloned between two loxP sequences in the targeting vector. The 5’ homologous arm was subcloned into the vector upstream from the 5’ loxP site, and the 3’ homologous arm was subcloned into the downstream of the 3’ loxP site. A PGK-DTA (diphtheria toxin A) cassette was located downstream from the 3’ homologous arm and used as negative selection for the targeting construct. The final construct was linearized by NotI restriction enzyme and electroporated into PluriStem C57BL/6N Murine ES cells (EMD Millipore, Darmstadt, Germany) (Cyagen Biosciences, Santa Clara, CA, USA). After positive (G418) and negative (DTA) selection, genomic DNA was extracted from the ES clones and a 3’ PCR screen was performed to identify the targeted clones. A positive clone was identified by PCR screening and confirmed by Southern blotting using the 5’, 3’, and Neo probes against the genomic DNA digested by KpnI or SacI restriction enzymes, respectively. The presence of 5’ and 3’ loxP sites in the targeted ES cells was confirmed by DNA sequencing (Fig. 1) (Supporting Table S1).
Figure 1. Generation of Fam20B conditional knockout (KO) allele.
A. Fam20B wild type (WT) allele. The black boxes indicated exons. The red bars represented 5' and 3' probes for Southern blotting. B. Fam20B targeting vector. The black triangles indicated loxP sequences. The white triangles represented Frt sequences. The crossed dashed lines indicated the position of 5' and 3' homologous arms. Neo, neomycin resistance. DTA, diphtheria toxin A. C. Targeted allele. The red bars indicated 5', 3', and Neo probes for Southern blotting. The blue arrows represented primers for 3' PCR screening for the targeted allele. D. Targeted allele without Neo cassete. The Neo cassete was removed by Flp-Frt mediated recombination. The blue arrows indicate PCR primers used to detect the flox allele (F and R1) and Cre-loxP mediated recombination event (F and R2). E. Conditional knockout allele. The exons 4 and 5 were removed by Cre-loxP mediated recombination. The blue arrows indicate the PCR primers for detecting the recombination event. F. PCR 3' screening for the targeted ES clone. The targeted allele gave rise to a 4.1 kb fragment, while the WT allele produced no band. Mut, Fam20B-floxed allele. G. Southern blotting of KpnI-digested gDNA with 5' probe. The probe identified a 9.9 kb fragment in the WT allele, and a 5.7 kb fragment in the targeted allele. H. Southern blotting of SacI-digested gDNA with 3' probe. The probe identified a 23.4 kb fragment in the WT allele, and a 7.3 kb fragment in the targeted allele. I. Southern blotting of KpnI-digested gDNA with Neo probe. The probe did not detected any fragment in the WT allele, but identified a 5.3 kb fragment in the targeted allele. J. PCR genotyping. The WT allele gave rise to a 242 bp fragment. The floxed allele produced a 303 bp fragment. The Cre-mediated recombination generated a 261 bp fragment. Rec, loxP-Cre mediated recombination in the Fam20B allele. K. RT-PCR using primers spanning exons 4 and 5 identified a 384 bp product in WT but no band was generated from the K14-Cre;Fam20Bflox/flox (KO) mice. L. Western immunoblotting using the antibody against the core protein of syndecan-1 (arrow) in the dental epithelium of WT and KO embryos. The epitheliun from the KO mice had a lower level of proteoglycan form (arrowheads), compared with that from the WT.
The targeted ES cells were injected into the blastocysts of C57BL/6 mice (Cyagen Biosciences). The chimeras were crossbred with B6.129S4-Gt(ROSA)26Sortm1(FLP1)Dym/RainJ mice (Jackson Laboratory, Bar Harbor, Maine, USA) to remove the Neo cassette from the F1 mice (Fig. 1).
To generate conditional knockout mice in which Fam20B is specifically inactivated in the epithelial tissues (including the oral and dental epithelium), we crossbred the mice following the strategy as previously described (24, 25). Briefly, the F1 heterozygous (Fam20Bflox/+) mice were crossbred with K14-Cre transgenic mice to produce K14-Cre;Fam20Bflox/+ mice, and the offspring were inbred to produce the K14-Cre;Fam20Bflox/flox mice. The K14-Cre transgenic mouse line used in this study was KRT14-Cre 43 transgenic mice (Jackson Laboratory), a K14-Cre transgene made by inserting a modified Cre cDNA fragment PCR-amplified from the pACN vector (35) into the BamHI site of a K14 expression vector (36). In addition, we generated Sox2-Cre;Fam20Bflox/flox mice by mating the Fam20Bflox/flox mice with Sox2-Cre transgenic mice, a line expressing Cre transgene at ~E6.5 and inactivating the floxed alleles in nearly all tissues.
Genotyping for detecting the floxed alleles, the Cre transgene and the Cre-loxP-mediated recombination events were performed by PCR analyses using specific primers (Table S) and the genomic DNA from tail lysates.
X-ray and micro-computed tomography
The lower and upper jaws dissected from 4-wk-old mice were analyzed with plain X-ray radiography (Faxitron Bioptics, Tucson, Arizona USA). Micro-CT analyses were performed using a Scanco micro-CT35 imaging system (Scanco Medical, Wayne, PA, USA) with a medium-resolution scan (7.0 µm slice increment) on the dissected tissues, as previously described (23, 26). The images were reconstructed with the EVS Beam software using a global threshold at 240 Hounsfield units. The samples from 6 K14-Cre;Fam20Bflox/flox, K14-Cre;Fam20Bflox/+, or WT mice (3 males and 3 females of each genotype) were examined by X-ray and micro-CT analyses, respectively.
Backscattered scanning electron microscopy
For the scanning electron microscopy (SEM) analyses, the lower and upper jaws from 4-wk-old mice were fixed in 4% paraformaldehyde overnight and dehydrated through a graded series of ethanol concentrations (70–100%), then embedded in methylmethacrylate without prior decalcification. Transectioning of the incisors and sagittal sections of the first molars were mounted, carbon coated and examined with field emission scanning electron microscopy (Philips XL30, FEI Company, Hillsboro, Oregon, USA). The samples from 6 K14-Cre;Fam20Bflox/flox, K14-Cre;Fam20Bflox/+, or WT mice (3 males and 3 females of each genotype) were examined in the SEM analyses.
Reverse transcription polymerase chain reaction
The dental epithelium was dissected from the first lower molars of 3-d-old mice. Total RNA was prepared using a Rneasy Mini Kit (Qiagen, Valencia, CA, USA) and converted into cDNA using a Reverse Transcription Kit (Qiagen), following the manufacturer’s instructions. RT-PCR was performed to examine the deletion of exons 4 and 5 in the Fam20B mRNA extracted from the K14-Cre;Fam20Bflox/flox mice (Table S).
Western immunoblotting
We dissected and pooled dental epithelium from the mandibles of E13.5 K14-Cre;Fam20Bflox/flox and WT mouse embryos after treating the mandibles with 1.8 U/ml dispase (Roch, Indianapolis, IN, USA) at room temperature for 15 min. The total protein lysates prepared from the dental epithelium were loaded onto 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western immunoblotting using the anti-syndecan-1 antibody (Santa Cruz Biotechnology, USA) at a concentration of 200 ng/ml.
Crossbreeding K14-Cre mice with Rosa26 reporter mice and X-galactosidase staining
The K14-Cre mice were crossbred with Rosa26 reporter mice (Jackson Lab), and the embryos were collected at E10.5, E11.5, E12.5 and E13.5, followed by X-Gal staining as previously described (20).
Statistical analysis
Data were expressed as mean ± SD. The Student’s t-test was used to compare the means between two groups. For analyses where comparisons were made among more than two groups, an analysis of variance (ANOVA) was used followed by Bonferroni method of multiple comparisons to determine which groups are significantly different from each other. A value of p < 0.05 was considered statistically significant.
Results
Validation of Fam20B inactivation
RT-PCR results confirmed the deletion of exons 4 and 5 in the Fam20B mRNA extracted from the dental epithelium of 3-d-old K14-Cre;Fam20Bflox/flox mice compared with their wild type (WT) littermates (Fig. 1). The Sox2-Cre;Fam20Bflox/flox mice showed embryonic lethality at E13.5 as the constitutive Fam20B-knockout mice did (34).
Reduction of Syndecan-1 in the dental epithelium
The results of Western immunoblotting showed remarkably less proteoglycans of syndecan-1 in the dental epithelium of K14-Cre;Fam20Bflox/flox mice compared with that from the WT mice, indicating a possible GAG deficiency in the K14-Cre; Fam20Bflox/flox mice (Fig. 1L).
Supernumerary teeth
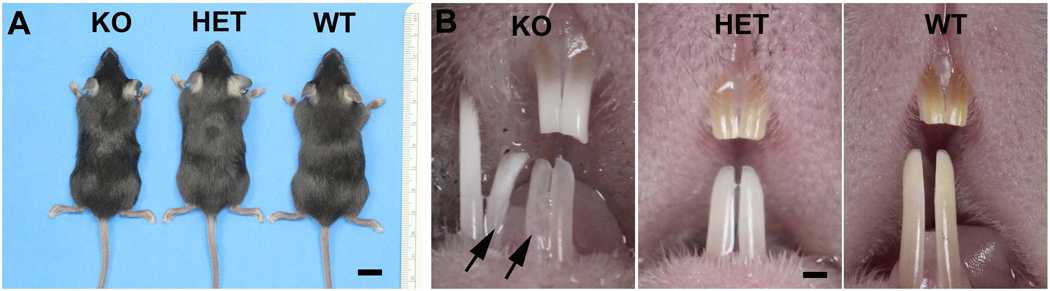
The Fam20Bflox/flox and K14-Cre;Fam20Bflox/+ mice were viable and fertile. We did not observe apparent differences in body size between these mice and WT mice (Fig. 2); the average body weight of 4-wk-old K14-Cre;Fam20Bflox/flox mice was 15.80 ± 1.92 g and that of WT mice was 15.69 ± 2.10 g (n = 12, P>0.05).
Figure 2. Gross defects in the K14-Cre;Fam20Bflox/flox mice.
A. Gross observation did not show apparent difference in the body size, hair, and skin among the 4-wk-old wild type (WT), K14-Cre;Fam20Bflox/+ heterozygous (Het) and K14-Cre;Fam20Bflox/flox (KO) mice. Scale bar, 10 mm. B. The K14-Cre;Fam20Bflox/flox mice showed supernumerary incisors. The incisors in the K14-Cre;Fam20Bflox/flox mice were opaque and showed a white color, while the incisors in the WT and Het mice were transparent and had a brown color. Scale bar, 1mm.
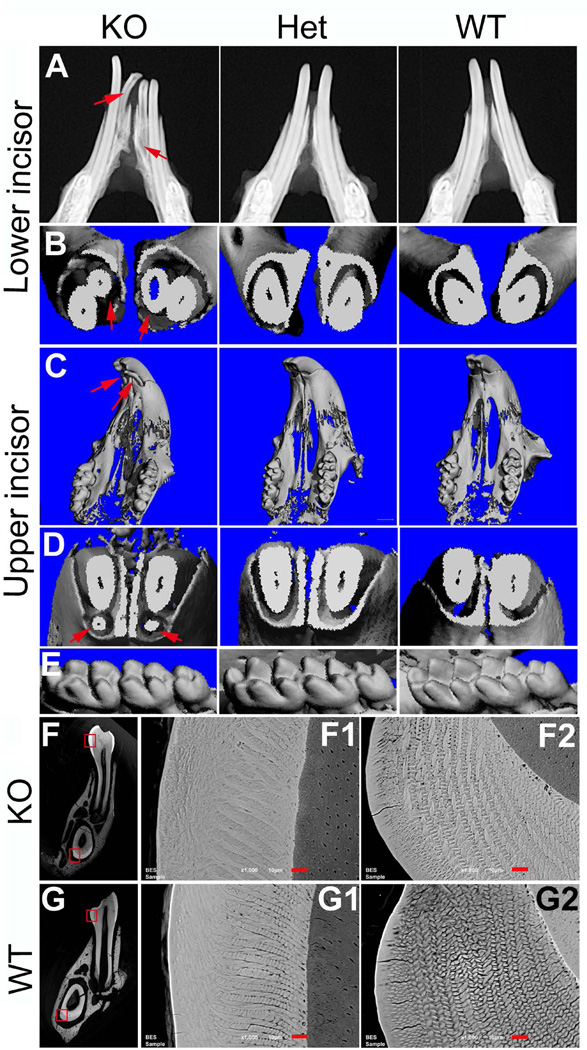
All of the K14-Cre;Fam20Bflox/flox mice displayed supernumerary incisors at the mesial sides of the lower incisors (i.e., 100% incidence). Approximately 23% of the K14-Cre;Fam20Bflox/flox mice had supernumerary incisors at the lingual side of the upper incisors (i.e., 23% incidence). There were no supernumerary molars in the K14-Cre;Fam20Bflox/flox mice (Fig. 3).
Figure 3. Supernumerary teeth and enamel phenotypes in the K14-Cre;Fam20Bflox/flox mice.
A. Plain X-ray. The supernumerary incisors (arrows) in the K14-Cre;Fam20Bflox/flox (KO) mice had smaller size and shorter length of roots than those in the normal mice. B. Micro-CT. Transection of lower incisors showed supernumerary incisors (arrows) in the KO mice. C. Micro-CT of upper jaws showed supernumerary incisors (arrows) in the KO mice. D. Micro-CT. Transection of upper incisors showed supernumerary teeth (arrows) in the KO mice. E. Micro-CT showed no morphology difference in the lower molars among the KO, heterozygous (Het) and WT mice. F and G. Backscatter SEM. The enamel of both molars (middle column) and incisors (right column) in the KO mice showed less clear structure of enamel prisms and seemingly more mineralization surrounding the prisms compared with those in the WT mice. F1 was the higher magnification view of the upper box area in F; F2 was the high magnification view of the lower box area in F. G1 was the high magnification view of the upper box area in G; G2 was the high magnification view of the lower box area in G. Scale bars, 10 µm.
Enamel phenotypes
Clinically, the tooth surface of the K14-Cre;Fam20Bflox/flox mice appeared opaque white compared to the brown-colored teeth in the WT mice (Fig. 2). SEM shows the enamel prisms of the K14-Cre;Fam20Bflox/flox mice to be less clearly defined in structure with more mineralization surrounding the prisms compared to similar sections from WT mice (Fig. 3), The enamel of the K14-Cre;Fam20Bflox/+ mice did not show apparent phenotypic differences from the WT mice (data not shown).
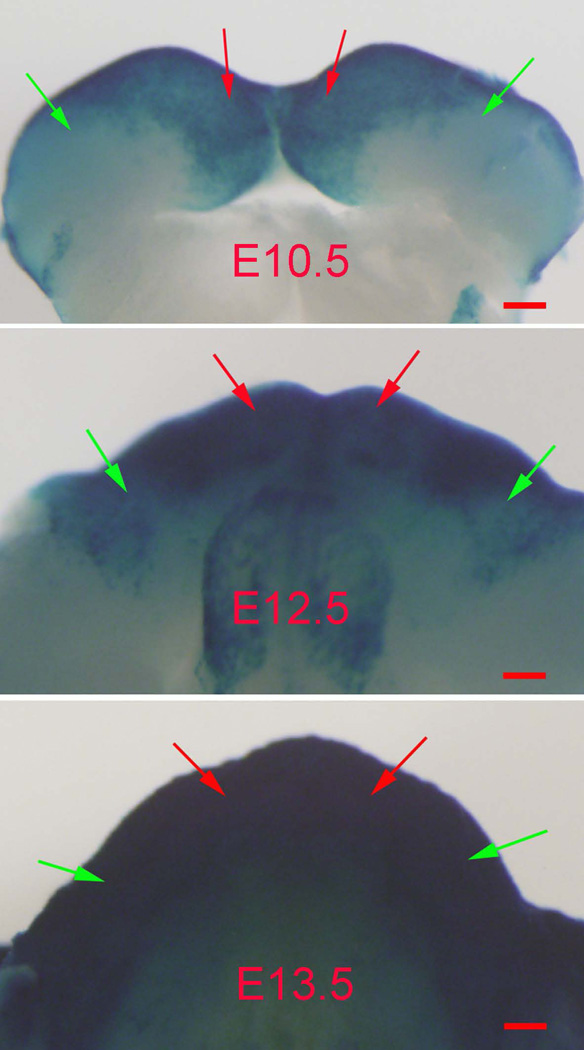
K14-Cre showed a lower level of expression in the incisor regions than in the molar regions at E10.5–E12.5
The X-Gal staining clearly showed that the molar regions had a lower level of K14-Cre expression than in the incisor regions at E10.5–E12.5 (Fig. 4). The intensity of K14-Cre increased in the molar regions over time and reached a level similar to that in the incisor regions at E13.5 (Fig. 4).
Figure 4. The molar and incisor regions in E10.5–E12.5 embryos showed different levels of K14-Cre activity.
X-Gal staining showed lower level of K14-Cre in the molar regions (green arrows) than in the incisor regions (red arrows) in the mouse embryos at E10.5 and E12.5. The K14-Cre activity in the molar regions increased to the similar level as the incisor regions at E13.5. Scale bars, 100 µm.
Discussion
FAM20B is a xylose kinase essential to the elongation of proteoglycan tetrasaccharide linkage region and the subsequent GAG assembly. The early embryonic lethality of Fam20B constitutive knockout mice makes it difficult to examine the tooth development in these mice (30). In this study, we generated Fam20B-floxed allele in which exons 4 and 5 were flanked by the loxP sequences. The deletion of exons 4 and 5 resulted in a frameshift from downstream exons that contain the highly conserved kinase domain, thus leading to Fam20B inactivation. We crossbred the Fam20B-floxed mice with K14-Cre transgenic mice to generate the K14-Cre;Fam20Bflox/flox mice in which Fam20B was specifically inactivated in the epithelial tissues. RT-PCR analyses confirmed the inactivation of Fam20B in the dental epithelium of the K14-Cre;Fam20Bflox/flox mice.
Inactivation of Fam20B impairs the GAG assembly of HS, CS and DS. It is intriguing that inactivating Fam20B in the dental epithelium leads to supernumerary incisors. This finding highlights the pivotal role of proteoglycans in the control of tooth number at the early stage of tooth development. Supernumerary tooth formation is closely related to upregulation or hypersensitivity of WNT, BMP, FGF, or SHH signaling. Gain-of-function in the WNT pathway by constitutive stimulation of β-catenin (37, 38) and overexpression of Lef1 (39) in the oral epithelium, or by inactivation of WNT negative regulators such as low density lipoprotein receptor-related protein 4 (Lrp4) (40), adenomatous polyposis coli (Apc) (41), epiprofin (42), and ectodin (43–49), led to supernumerary tooth formation. The supernumerary teeth in the mice overexpressing ectodysplasin A (Eda) (50, 51), ectodysplasin receptor (Edar) (52, 53), IκB kinase β (IKKβ) (54), or nuclear factor kB (NF-kB) (55) were accompanied by upregulation of WNT signaling. In addition to WNT signaling, gain-of-function in BMP signaling shown in ectodin- (43), CCAAT enhancer-binding protein beta- (CEBPB−) (56), and Osr2-KO mice (57), led to supernumerary teeth as part of a phenotype. Inactivation of genes encoding FGF antagonists, Sprouty2 and 4, led to supernumerary teeth due to hypersensitivity in FGF signaling (58, 59). Notably, the activity changes in these signaling pathways ultimately need an ectopic SHH expression to induce the supernumerary tooth formation (43, 60, 61). Given the regulatory roles of GAGs in mediating the WNT, BMP, FGF and SHH signaling pathways, and the correlation between the activity changes of these pathways and the supernumerary teeth, we speculate that the supernumerary teeth in the K14-Cre;Fam20Bflox/flox mice may be associated with the activity changes in one or several preceding pathways that are tightly regulated by the GAGs.
The exclusive location of supernumerary teeth in the incisor regions of the K14-Cre;Fam20Bflox/flox mice may be attributed to a differential expression of Fam20B, proteoglycans, or their downstream signaling molecules in the dental epithelium between the incisor and molar regions. It may also arise from a different deletion efficiency of Fam20B by the K14-Cre between the incisor and molar regions, seeing that the K14-Cre activity in the molar regions was remarkably lower than that in the incisor regions at E10.5–E12.5 (Figure 3), a time window critical for dental epithelium thickening, invaginating, and budding.
The enamel of K14-Cre;Fam20Bflox/flox mice showed an opaque-white color in contrast to a brown color in the normal mice, while the enamel thickness was not affected. The enamel prisms in K14-Cre;Fam20Bflox/flox mice showed a less clear structure compared with WT, and seemingly had more mineralization surrounding the enamel prisms. It remains to be determined whether the reduction of proteoglycans in enamel matrix had facilitated the degradation of matrix proteins and accelerated the mineralization process of enamel matrix. The FAM20 family is composed of three members — FAM20A, FAM20B, and FAM20C. The enamel phenogypes in the Fam20C- and Fam20A-KO mice are remarkablly different from the enamel appearance in Fam20B-KO mice. The former two are very similar in the morphology alteration in ameloblasts and the lack of enamel formation, which is likely attributed to the fact that FAM20C phosphorylates SCPP proteins that are essential for the assembly and mineralization of enamel matirx, and that FAM20A is a pseudokinase essential for the kinase activity of FAM20C in ameloblasts by forming a complex with the latter.
In summary, this study revealed previously unknown functions of FAM20B and proteoglycans in tooth development, hereby opens a new window for exploring the regulatory role of proteoglycans in the signaling cascades governing tooth formation.
Supplementary Material
Acknowledgements
We thank Dr. Jianquan Feng and Jingya Wang for their help with the SEM analysis. We are grateful to Jeanne Santa Cruz for her assistance with the editing of this article. This research was supported by NIH Grants DE23873-01 (to XW) and DE022549 (to CQ). We thank China Scholarship Council (Grant 201406240152) for supporting the Visiting Scholar who was involved in this study.
Footnotes
Conflicts of interest
The authors state that they have no conflicts of interest.
Additional Supporting Information may be found in the online version of this article:
Table S1: Primers for PCR screen, South blotting probes, genotyping and RT-PCR.
References
- 1.Kjellén L, Lindahl U. Proteoglycans: structures and interactions. Annu Rev Biochem. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]
- 2.Sugahara K, Kitagawa H. Recent advances in the study of the biosynthesis and functions of sulfated glycosaminoglycans. Curr Opin Struct Biol. 2000;10:518–527. doi: 10.1016/s0959-440x(00)00125-1. [DOI] [PubMed] [Google Scholar]
- 3.Mizumoto S, Yamada S, Sugahara K. Human genetic disorders and knockout mice deficient in glycosaminoglycan. Biomed Res Int. 2014;2014:495764. doi: 10.1155/2014/495764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linhardt RJ, Toida T. Role of glycosaminoglycans in cellular communication. Acc Chem Res. 2004;37:431–438. doi: 10.1021/ar030138x. [DOI] [PubMed] [Google Scholar]
- 5.Sarrazin S, Lamanna WC, Esko JD. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol. 2011;3:a004952. doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mikami T, Kitagawa H. Biosynthesis and function of chondroitin sulfate. Biochim Biophys Acta. 2013;1830:4719–4733. doi: 10.1016/j.bbagen.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Jernvall J, Thesleff I. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech Dev. 2000;92:19–29. doi: 10.1016/s0925-4773(99)00322-6. [DOI] [PubMed] [Google Scholar]
- 8.Thesleff I, Jalkanen M, Vainio S, Bernfield M. Cell surface proteoglycan expression correlates with epithelial-mesenchymal interaction during tooth morphogenesis. Dev Biol. 1988;129:565–572. doi: 10.1016/0012-1606(88)90401-0. [DOI] [PubMed] [Google Scholar]
- 9.Vainio S, Jalkanen M, Thesleff I. Syndecan and tenascin expression is induced by epithelial-mesenchymal interactions in embryonic tooth mesenchyme. J Cell Biol. 1989;108:1945–1953. doi: 10.1083/jcb.108.5.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vainio S, Thesleff I. Sequential induction of syndecan, tenascin and cell proliferation associated with mesenchymal cell condensation during early tooth development. Differentiation. 1992;50:97–105. doi: 10.1111/j.1432-0436.1992.tb00490.x. [DOI] [PubMed] [Google Scholar]
- 11.Bai XM, Van der Schueren B, Cassiman JJ, Van den Berghe H, David G. Differential expression of multiple cell-surface heparan sulfate proteoglycans during embryonic tooth development. J Histochem Cytochem. 1994;42:1043–1054. doi: 10.1177/42.8.8027524. [DOI] [PubMed] [Google Scholar]
- 12.Hikake T, Mori T, Iseki K, Hagino S, Zhang Y, Takagi H, Yokoya S, Wanaka A. Comparison of expression patterns between CREB family transcription factor OASIS and proteoglycan core protein genes during murine tooth development. Anat Embryol (Berl) 2003;206:373–380. doi: 10.1007/s00429-003-0311-z. [DOI] [PubMed] [Google Scholar]
- 13.Jiang BZ, Yokohama-Tamaki T, Wang ZL, Obara N, Shibata S. Expression, localisation and synthesis of versican by the enamel organ of developing mouse molar tooth germ: an in vivo and in vitro study. Arch Oral Biol. 2010;55:995–1006. doi: 10.1016/j.archoralbio.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 14.Bernfield M, Sanderson RD. Syndecan, a developmentally regulated cell surface proteoglycan that binds extracellular matrix and growth factors. Philos Trans R Soc Lond B Biol Sci. 1990;327:171–186. doi: 10.1098/rstb.1990.0052. [DOI] [PubMed] [Google Scholar]
- 15.Salmivirta M, Elenius K, Vainio S, Hofer U, Chiquet-Ehrismann R, Thesleff I, Jalkanen M. Syndecan from embryonic tooth mesenchyme binds tenascin. J Biol Chem. 1991;266:7733–7739. [PubMed] [Google Scholar]
- 16.Yonemochi H, Saku T. Perlecan, a heparan sulfate proteoglycan, is a major constituent of the intraepithelial stroma functioning in tooth morphogenesis. J Oral Biosci. 2006;48:233–243. [Google Scholar]
- 17.Ida-Yonemochi H, Satokata I, Ohshima H, Sato T, Yokoyama M, Yamada Y, Saku T. Morphogenetic roles of perlecan in the tooth enamel organ: an analysis of overexpression using transgenic mice. Matrix Biol. 2011;30:379–388. doi: 10.1016/j.matbio.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Wiweger MI, Zhao Z, Van Merkesteyn RJ, Roehl HH, Hogendoorn PC. HSPG-deficient zebrafish uncovers dental aspect of multiple osteochondromas. PLoS One. 2012;7:e29734. doi: 10.1371/journal.pone.0029734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayano S, Kurosaka H, Yanagita T, Kalus I, Milz F, Ishihara Y, Islam MN, Kawanabe N, Saito M, Kamioka H, Adachi T, Dierks T, Yamashiro T. Roles of heparan sulfate sulfation in dentinogenesis. J Biol Chem. 2012;287:12217–12229. doi: 10.1074/jbc.M111.332924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C, Gu S, Sun C, Ye W, Song Z, Zhang Y, Chen Y. FGF signaling sustains the odontogenic fate of dental mesenchyme by suppressing β-catenin signaling. Development. 2013;140:4375–4385. doi: 10.1242/dev.097733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishikawa HO, Xu A, Ogura E, Manning G, Irvine KD. The Raine syndrome protein FAM20C is a golgi kinase that phosphorylates bio-mineralization proteins. PLoS ONE. 2012;7:e42988. doi: 10.1371/journal.pone.0042988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tagliabracci VS1, Engel JL, Wen J, Wiley SE, Worby CA, Kinch LN, Xiao J, Grishin NV, Dixon JE. Secreted kinase phosphorylates extracellular proteins that regulate biomineralization. Science. 2012;336:1150–1153. doi: 10.1126/science.1217817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Wang S, Li C, Gao T, Liu Y, Rangiani A, Sun Y, Hao J, George A, Lu Y, Groppe J, Yuan B, Feng J, Qin C. Inactivation of a novel FGF23 regulator, FAM20C, leads to hypophosphatemic rickets in mice. PLoS Genetics. 2012;8:e1002708. doi: 10.1371/journal.pgen.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Jung J, Liu Y, Yuan B, Lu Y, Feng JQ, Qin C. The specific role of FAM20C in amelogenesis. J Dent Res. 2013;92:995–999. doi: 10.1177/0022034513504588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Wang J, Liu Y, Yuan B, Ruest B, Feng JQ, Qin C. The Specific Role of FAM20C in Dentinogenesis. J Dent Res. 2015;94:330–336. doi: 10.1177/0022034514563334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Wang S, Lu Y, Gibson MP, Liu Y, Yuan B, Feng JQ, Qin C. FAM20C plays an essential role in the formation of murine teeth. J Biol Chem. 2012;287:35934–35942. doi: 10.1074/jbc.M112.386862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui J, Xiao J, Tagliabracci VS, Wen J, Rahdar M, Dixon JE. A secretory kinase complex regulates extracellular protein phosphorylation. Elife. 2015;4:e06120. doi: 10.7554/eLife.06120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Sullivan J, Bitu CC, Daly SB, Urquhart JE, Barron MJ, Bhaskar SS, et al. Whole-exome sequencing identifies FAM20A mutations as a cause of Amelogenesis Imperfecta and Gingival Hyperplasia Syndrome. Am J Hum Genet. 2011;88:616–620. doi: 10.1016/j.ajhg.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaureguiberry G, De La Dure-Molla M, Parry D, Quentric M, Himmerkus N, Koike T, et al. Nephrocalcinosis (enamel renal syndrome) caused by autosomal recessive FAM20A mutations. Nephron Physiol. 2013;122:1–6. doi: 10.1159/000349989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogel P, Hansen GM, Read RW, Vance RB, Thiel M, Liu J, Wronski TJ, Smith DD, Jeter-Jones S, Brommage R. Amelogenesis imperfecta and other biomineralization defects in Fam20a and Fam20c null mice. Vet Pathol. 2012;49:998–1017. doi: 10.1177/0300985812453177. [DOI] [PubMed] [Google Scholar]
- 31.Fujimori S, Novak H, Weissenböck M, Jussila M, Gonçalves A, Zeller R, Galloway J, Thesleff I, Hartmann C. Wnt/β-catenin signaling in the dental mesenchyme regulates incisor development by regulating Bmp4. Dev Biol. 2010;348:97–106. doi: 10.1016/j.ydbio.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koike T, Izumikawa T, Tamura J, Kitagawa H. FAM20B is a kinase that phosphorylates xylose in the glycosaminoglycan-protein linkage region. Biochem J. 2009;421:157–162. doi: 10.1042/BJ20090474. [DOI] [PubMed] [Google Scholar]
- 33.Wen J, Xiao J, Rahdar M, Choudhury BP, Cui J, Taylor GS, Esko JD, Dixon JE. Xylose phosphorylation functions as a molecular switch to regulate proteoglycan biosynthesis. Proc Natl Acad Sci U S A. 2014;111:15723–15728. doi: 10.1073/pnas.1417993111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eames BF, Yan YL, Swartz ME, Levic DS, Knapik EW, Postlethwait JH, Kimmel CB. Mutations in fam20b and xylt1 reveal that cartilage matrixcontrols timing of endochondral ossification by inhibiting chondrocyte maturation. PLoS Genet. 2011;7:e1002246. doi: 10.1371/journal.pgen.1002246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bunting M, Bernstein KE, Greer JM, Capecchi MR, Thomas KR. Targeting genes for self-excision in the germ line. Genes Dev. 1999;13:1524–1528. doi: 10.1101/gad.13.12.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andl T, Reddy ST, Gaddapara T, Millar SE. WNT signals are required for the initiation of hair follicle development. Dev Cell. 2002;2:643–653. doi: 10.1016/s1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- 37.Järvinen E, Salazar-Ciudad I, Birchmeier W, Taketo MM, Jernvall J, Thesleff I. Continuous tooth generation in mouse is induced by activated epithelial Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A. 2006;103:18627–18632. doi: 10.1073/pnas.0607289103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu F, Chu EY, Watt B, Zhang Y, Gallant NM, Andl T, Yang SH, Lu MM, Piccolo S, Schmidt-Ullrich R, Taketo MM, Morrisey EE, Atit R, Dlugosz AA, Millar SE. Wnt/β-catenin signaling directs multiple stages of tooth morphogenesis. Dev Biol. 2008;313:210–224. doi: 10.1016/j.ydbio.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou P, Byrne C, Jacobs J, Fuchs E. Lymphoid enhancer factor1 directs hair follicle patterning and epithelial cell fate. Genes Dev. 1995;9:700–713. doi: 10.1101/gad.9.6.700. [DOI] [PubMed] [Google Scholar]
- 40.Ohazama A, Johnson EB, Ota MS, Choi HY, Porntaveetus T, Oommen S, Itoh N, Eto K, Gritli-Linde A, Herz J, Sharpe PT. Lrp4 modulates extracellular integration of cell signaling pathways in development. PLoS One. 2008;3:e4092. doi: 10.1371/journal.pone.0004092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang XP, O'Connell DJ, Lund JJ, Saadi I, Kuraguchi M, Turbe-Doan A, Cavallesco R, Kim H, Park PJ, Harada H, Kucherlapati R, Maas RL. Apc inhibition of Wnt signaling regulates supernumerary tooth formation during embryogenesis and throughout adulthood. Development. 2009;136:1939–1949. doi: 10.1242/dev.033803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakamura T, De Vega S, Fukumoto S, Jimenez L, Unda F, Yamada Y. Transcription factor epiprofin is essential for tooth morphogenesis by regulating epithelial cell fate and tooth number. J Biol Chem. 2008;283:4825–4833. doi: 10.1074/jbc.M708388200. [DOI] [PubMed] [Google Scholar]
- 43.Laurikkala J, Kassai Y, Pakkasjärvi L, Thesleff I, Itoh N. Identification of a secreted BMP antagonist, ectodin, integrating BMP, FGF, and SHH signals from the tooth enamel knot. Dev Biol. 2003;264:91–105. doi: 10.1016/j.ydbio.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 44.Kassai Y, Munne P, Hotta Y, Penttilä E, Kavanagh K, Ohbayashi N, Takada S, Thesleff I, Jernvall J, Itoh N. Regulation of mammalian tooth cusp patterning by ectodin. Science. 2005;309:2067–2070. doi: 10.1126/science.1116848. [DOI] [PubMed] [Google Scholar]
- 45.Murashima-Suginami A, Takahashi K, Kawabata T, Sakata T, Tsukamoto H, Sugai M, Yanagita M, Shimizu A, Sakurai T, Slavkin HC, Bessho K. Rudiment incisors survive and erupt as supernumerary teeth as a result of USAG-1 abrogation. Biochem Biophys Res Commun. 2007;359:549–555. doi: 10.1016/j.bbrc.2007.05.148. [DOI] [PubMed] [Google Scholar]
- 46.Munne PM, Tummers M, Järvinen E, Thesleff I, Jernvall J. Tinkering with the inductive mesenchyme: Sostdc1 uncovers the role of dental mesenchyme in limiting tooth induction. Development. 2009;136:393–402. doi: 10.1242/dev.025064. [DOI] [PubMed] [Google Scholar]
- 47.Ahn Y, Sanderson BW, Klein OD, Krumlauf R. Inhibition of Wnt signaling by Wise (Sostdc1) and negative feedback from Shh controls tooth number and patterning. Development. 2010;137:3221–3231. doi: 10.1242/dev.054668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho SW, Kwak S, Woolley TE, Lee MJ, Kim EJ, Baker RE, Kim HJ, Shin JS, Tickle C, Maini PK, Jung HS. Interactions between Shh, Sostdc1 and Wnt signaling and a new feedback loop for spatial patterning of the teeth. Development. 2011;138:1807–1816. doi: 10.1242/dev.056051. [DOI] [PubMed] [Google Scholar]
- 49.Kiso H, Takahashi K, Saito K, Togo Y, Tsukamoto H, Huang B, Sugai M, Shimizu A, Tabata Y, Economides AN, Slavkin HC, Bessho K. Interactions between BMP-7 and USAG-1 (uterine sensitization-associated gene-1) regulate supernumerary organ formations. PLoS One. 2014;9:e96938. doi: 10.1371/journal.pone.0096938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mustonen T, Pispa J, Mikkola ML, Pummila M, Kangas AT, Pakkasjärvi L, Jaatinen R, Thesleff I. Stimulation of ectodermal organ development by ectodysplasin-A1. Dev Biol. 2003;259:123–136. doi: 10.1016/s0012-1606(03)00157-x. [DOI] [PubMed] [Google Scholar]
- 51.Kangas AT, Evans AR, Thesleff I, Jernvall J. Nonin-dependence of mammalian dental characters. Nature. 2004;432:211–214. doi: 10.1038/nature02927. [DOI] [PubMed] [Google Scholar]
- 52.Pispa J, Mustonen T, Mikkola ML, Kangas AT, Koppinen P, Lukinmaa PL, Jernvall J, Thesleff I. Tooth patterning and enamel formation can be manipulated by misexpression of TNF receptor Edar. Dev Dyn. 2004;231:432–440. doi: 10.1002/dvdy.20138. [DOI] [PubMed] [Google Scholar]
- 53.Tucker AS, Headon DJ, Courtney JM, Overbeek P, Sharpe PT. The activation level of the TNF family receptor. Edar, determines cusp number and tooth number during tooth development. Dev Biol. 2004;268:185–194. doi: 10.1016/j.ydbio.2003.12.019. [DOI] [PubMed] [Google Scholar]
- 54.Page A, Cascallana JL, Casanova ML, Navarro M, Alameda JP, Pérez P, Bravo A, Ramírez A. IKKβ overexpression leads to pathologic lesions in stratified epithelia and exocrine glands and to tumoral transformation of oral epithelia. Mol Cancer Res. 2011;9:1329–1338. doi: 10.1158/1541-7786.MCR-11-0168. [DOI] [PubMed] [Google Scholar]
- 55.Blackburn J, Kawasaki K, Porntaveetus T, Kawasaki M, Otsuka-Tanaka Y, Miake Y, Ota MS, Watanabe M, Hishinuma M, Nomoto T, Oommen S, Ghafoor S, Harada F, Nozawa-Inoue K, Maeda T, Peterková R, Lesot H, Inoue J, Akiyama T, Schmidt-Ullrich R, Liu B, Hu Y, Page A, Ramírez Á, Sharpe PT, Ohazama A. Excess NF-κB induces ectopic odontogenesis in embryonic incisor epithelium. J Dent Res. 2015;94:121–128. doi: 10.1177/0022034514556707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang B, Takahashi K, Sakata-goto T, Kiso H, Togo Y, Saito K, Tsukamoto H, Sugai M, Akira S, Shimizu A, Bessho K. Phenotypes of CCAAT/enhancer-binding protein beta deficiency: hyperdontia and elongated coronoid process. Oral Dis. 2013;19:144–150. doi: 10.1111/j.1601-0825.2012.01963.x. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Z, Lan Y, Chai Y, Jiang R. Antagonistic actions of Msx1 and Osr2 pattern mammalian teeth into a single row. Science. 2009;323:1232–1234. doi: 10.1126/science.1167418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klein OD, Minowada G, Peterkova R, Kangas A, Yu BD, Lesot H, Peterka M, Jernvall J, Martin GR. Sprouty genes control diastema tooth development via bidirectional antagonism of epithelial-mesenchymal FGF signaling. Dev Cell. 2006;11:181–190. doi: 10.1016/j.devcel.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peterkova R, Churava S, Lesot H, Rothova M, Prochazka J, Peterka M, Klein OD. Revitalization of a diastemal tooth primordium in Spry2 null mice results from increased proliferation and decreased apoptosis. J Exp Zool Part B Mol Dev Evol. 2009;312B:292–308. doi: 10.1002/jez.b.21266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cobourne MT, Sharpe PT. Making up the numbers: The molecular control of mammalian dental formula. Semin Cell Dev Biol. 2010;21:314–324. doi: 10.1016/j.semcdb.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 61.Mizumoto S, Yamada S, Sugahara K. Human genetic disorders and knockout mice deficient in glycosaminoglycan. Biomed Res Int. 2014;2014:495764. doi: 10.1155/2014/495764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.