Abstract
Parkinsonism is an umbrella term for a group of disorders characterized by the clinical signs of tremor, bradykinesia, rigidity, and postural instability. On neuropathologic examination parkinsonism can display alternate protein pathologies (e.g. α-synucleinopathy or tauopathy) but the degeneration of nigral neurons is consistent. The main forms of parkinsonism are, Parkinson's disease (PD), Dementia with Lewy Bodies (DLB), Multiple System Atrophy (MSA), Progressive Supranuclear Palsy (PSP) and Corticobasal Degeneration (CBD). Genetic studies from candidate gene, to unbiased genome-wide approaches including association and next-generation sequencing have nominated a number of disease determinants. Within this review we will highlight the genetic loci that are associated with disease and discuss the implications and importance for a better understanding of the genes involved and thus the underlying pathophysiology of these disorders.
Keywords: Parkinson's disease, Dementia with Lewy Bodies, Multiple System Atrophy, Progressive Supranuclear Palsy, Corticobasal Degeneration, genes
Genetic findings over the last 25 years have altered our perception of what parkinsonism is; this was driven mostly by classical linkage methods in large familial aggregates with Mendelian patterns of disease inheritance [1]. Although for the most part these genetic variants account for a very small proportion of patients, they have provided crucial insights into the underlying pathophysiology of Parkinsonian disorders. The familial genes include SNCA, LRRK2, VPS35, PARKIN, PINK1 and DJ-1 for what is generally considered Parkinson's disease (PD), and MAPT for Frontotemporal dementia and parkinsonism linked to chromosome 17 with tau pathology (FTDP-17t), Progressive Supranuclear Palsy (PSP) and Corticobasal Degeneration (CBD). Notably the SNCA (α-synuclein) and MAPT (tau) genes encode the two major protein pathologies of parkinsonism labeled the α-synucleinopathies or tauopathies.
The vast majority of patients suffering with an age-related Parkinsonian disorder present clinical signs without a known family history of disease. In the case of PD, the most common form of parkinsonism, somewhere between only 15-20% of patients report another affected family member [2]. However other Parkinsonian disorders including PSP, CBD, and Multiple System Atrophy (MSA) report much less familial aggregation if any. In this scenario it is much less likely that the classical familial genetic approaches will be successful in identifying the genetic variation that influences the individual susceptibility to these disorders.
To address the underlying genetics of sporadic disease, population-based (i.e. unrelated case-control) studies were employed. For a longtime the only option was a candidate gene approach, whereby the most likely biologically relevant genes were selected for restricted genetic analysis. These studies had limited success, however, they did identify common variation in the SNCA and MAPT genes that associated with risk of PD, PSP and CBD. As genotyping technology advanced the first giant leap forward occurred with the evolution of the large-scale unbiased genome-wide association studies (GWAS). This methodology allowed the simultaneous genotyping of hundreds of thousands of single nucleotide polymorphisms (SNPs) dispersed across the genome and, by exploiting the phenomenon of linkage disequilibrium, measure the association of disease risk from common variation. Although early GWAS suffered from limited coverage and low sample numbers, the recent larger efforts have successfully identified a number of loci that alter the individual's susceptibility to parkinsonism.
This review will briefly describe the common variant loci that have been nominated through studies in the different Parkinsonian disorders and comment on the up-coming use of population-based Next-Generation Sequencing (NGS) approaches and the fundamental inherent complexities associated with polygenic disease.
Parkinson's disease (PD)
PD (OMIM168601) is the most frequent neurodegenerative movement disorder and second neurodegenerative disorder after Alzheimer's disease (AD). PD is characterized by the main clinical signs of resting tremor, rigidity, postural instability and bradykinesia, it is typically asymmetric at onset and the symptoms progressively worsen [3]. However, PD is recognized as a more systemic disease and non-motor signs include constipation, anosmia, sleep abnormalities, autonomic dysfunction, behavioral changes, depression and dementia. PD is a synucleinopathy with the pathologic presence of Lewy bodies and dopaminergic cell death in the substantia nigra confirming a definite diagnosis.
Candidate gene studies in PD identified common variation at SNCA, LRRK2, MAPT and GBA which were later confirmed in the large GWAS [4]. Common variation has been shown at SNCA, LRRK2 and MAPT to contain both protective and risk haplotypes which suggests the function of these genes could be modulated to offer a therapeutic hope [5]. Indeed for some of the reported families with SNCA multiplication mutations evidence exists that nature itself has evolved mechanisms to cope with pathogenic mutations [6]. Identifying those individuals who appear protected against pathogenic mutations in PD may offer alternate druggable targets for future development.
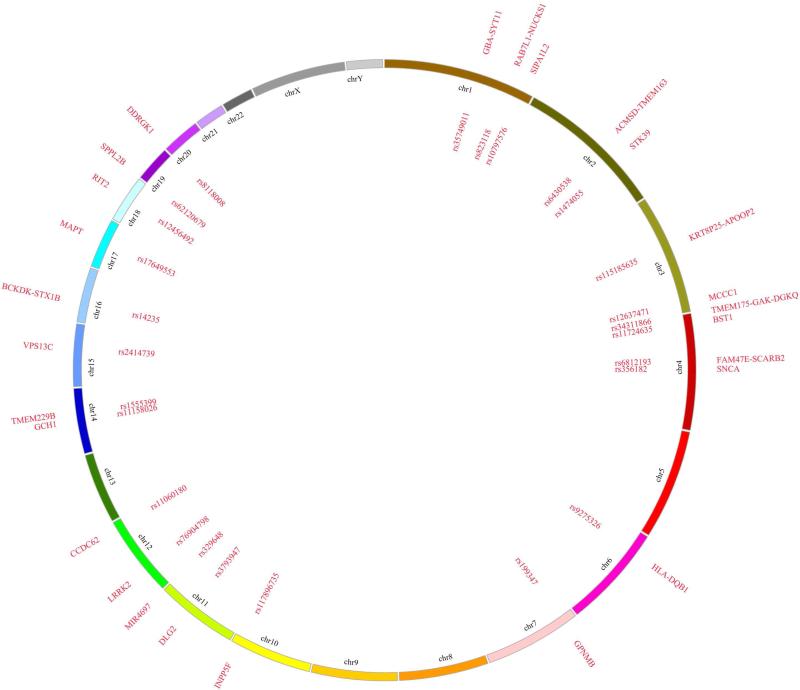
To date, 26 independent signals have been nominated in the most recent GWAS which was a meta-analysis of previous smaller studies (Figure 1) [7]. The caveat of GWAS is that although they nominate genomic loci they do not pinpoint the specific gene which allows for multiple biologically-relevant candidates. For some regions the best candidate is clear (e.g. SNCA and LRRK2) while for others it remains unclear and this might again be the most appropriate time to use the PARK loci nomenclature that was created for linkage regions.
Figure 1.
Genomic map of PD GWAS association signals. Here we show the 26 independent loci nominated as PD susceptibility loci in the Nalls et al. (2014) study. The circos plot represents the following features from the inside of the circle to outside: rs SNPID#, chromosome name, names of nearest genes or previously published proximal genes to the loci [30].
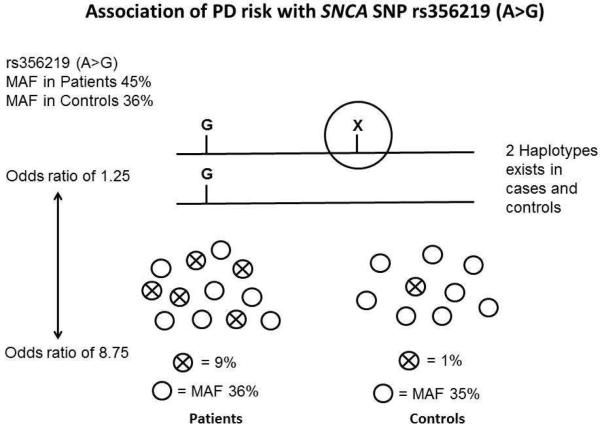
A small number of studies have been performed looking at the possible genetic interactions for some of the loci (i.e. SNCA, MAPT, LRRK2 and PARK16) [8-10]. The idea that a ‘genomic risk score’ can be created to predict the individual's likelihood to develop disease is an attractive goal however when using GWAS data to either measure the genetic contribution to disease (and what is accounted for) or to predict risk there is a fundamental caveat. The premise of the GWAS is that you do not need to genotype the functional variant to identify the risk at the loci due to the inherent linkage dysequilibrium with common variation. The caveat is that by not genotyping the functional variant, you do not have a true measure of the patient-control allele frequency and thus the true effect size (Figure 2). Large-scale targeted sequencing efforts are underway for each of the GWAS loci in an attempt to identify the functional variant/s that account for the association signal to address these concerns.
Figure 2.
Caveat interpreting GWAS association signal. The figure shows the variation in odds ratio that can be observed for an association when the functional variant (X) is not directly genotype but rather alleles in linkage disequilibrium are used as a measure of signal.
Dementia with Lewy bodies (DLB)
DLB (OMIM 127750) is one of the most common types of dementia in the elderly, and has clinical and neuropathological similarities with both AD and PD. Clinical signs of DLB include progressive dementia, parkinsonism, visual hallucinations, and fluctuations in cognition. At autopsy, DLB is classified as an α-synucleinopathy owing to the typical widespread presence of cortical Lewy bodies, and usually also the neurofibrillary tangles and senile plaques of AD. Only a handful of genetic studies have been performed in DLB, but candidate approaches nominated genes that are associated with PD or AD, including SNCA, GBA and APOE [4, 11-13].
The most recent study exploited the more mature fields of GWAS in PD and AD by examining the known GWAS loci from each of the diseases in a large series of DLB patients and controls [14]. Although the study had a smaller sample size than most of the recent PD and AD GWAS significant associations were observed for SNCA, SCARB2 (a PD GWAS locus) and APOE; a number of other loci reached nominal significance including MAPT. These findings suggest that there is risk overlap from both PD and AD within the disease etiology of DLB and may indicate a unique set of genetic risk variants are yet to be found. Presently, efforts are on-going to generate an unbiased GWAS dataset for DLB.
Multiple System Atrophy (MSA)
MSA (OMIM 146500) is a fulminant neurodegenerative disorder, clinically characterized by autonomic dysfunction, parkinsonism and cerebellar or pyramidal symptoms [15, 16]. A diagnosis of definite MSA requires a neuropathologic examination with striatonigral degeneration or olivopontocerebellar ataxia in the presence of α-synuclein-positive glial cytoplasmic inclusions (GCI). MSA is by nature a sporadic disorder with only a handful of small families ever described. This has led to the understanding that genetics has a lesser role to play in disease risk and thus has limited genetic studies.
To date, no GWAS or large-scale association studies have been published for MSA. Genomic multiplication of the SNCA locus has been observed to cause parkinsonism, dementia and autonomic dysfunction, characteristic of the MSA phenotype [17]. In 2009, Scholz and colleagues reported the recessive association of a SNP (rs111931074) in the 3’ untranslated region of the SNCA gene increases the risk for MSA [18]. Our group confirmed the association in a second pathologically confirmed series showing that the homozygous recessive genotype rs111931074-TT was increased in cases compared to controls [19]. Our genetic studies of MSA have also nominated common variation at other PD-loci; MAPT and LRRK2 [20, 21].
Recently, the first whole-genome sequencing study in a proband from a unique Japanese family with MSA nominated recessive mutations in the COQ2 gene to be the cause of the disease phenotype [22]. A number of other cases were identified and a common substitution (p.V393A) was associated with an increased risk of disease. The COQ2 mutations are predicted to result in a loss-of-function which subsequently leads to a COQ10 deficiency. Our follow-up studies suggested specific variants were increased in patients but we did not observe any common association as proposed in Asian populations [23]. Presently, efforts are on-going to publish an unbiased GWAS dataset for MSA.
Progressive Supranuclear Palsy (PSP)
PSP (OMIM 601104) is a rare neurodegenerative movement disorder clinically characterized by falls, axial rigidity, vertical supranuclear gaze palsy, bradykinesia and cognitive decline [24]. PSP is the second most common extrapyramidal disorder after PD. Histopathologically, PSP is classified as a tauopathy, due to the presence of neurofibrillary tangles (the major component of tangle pathology is abnormally hyperphosphorylated tau) and glial tau inclusions, with neuronal loss and gliosis in the globus pallidus, subthalamic nucleus and substantia nigra.
Given the presence of tau pathology it was established early that common genetic variability within the MAPT gene, denoted by the extended, non-recombining MAPT haplotype H1, was associated with disease risk [25]. However the identification of other susceptibility locus proved elusive. PSP-like signs and pathology were observed in some carriers of pathogenic familial LRRK2 mutations but there is no evidence of common variant association. In 2011, the first large PSP GWAS was published and confirmed MAPT as the most significant association by far (P=2.1×10−51), but also nominated a small number of additional loci (MOBP P=1.0×10−9; STX6 P=1.8×10−9 and EIF2AK3 P=7.4×10−7) [26]. However, given the rarity of the disease no follow-up studies have yet confirmed these hits or nominated additional loci.
Corticobasal degeneration (CBD)
CBD is a very rare, progressive neurodegenerative disorder with patients presenting motor, sensory, behavioral and cognitive signs [27]. The clinical phenotype, termed corticobasal syndrome (CBS), is a sporadic disorder comprising of asymmetric progressive rigidity and apraxia with limb dystonia and myoclonus. CBD pathology is characterized by circumscribed cortical atrophy with spongiosis and ballooned neurons with extensive neuronal and glial tau pathology. Abnormal tau accumulation within astrocytes forms pathognomonic astrocytic plaques.
Given the population frequency of CBD, large-scale genetic efforts have been limited. This year a global consortium published the first GWAS, with a relatively small series of just over 150 pathologically-confirmed CBD cases [28]. This study confirmed the established association with MAPT (P=1.42×10−12) and nominated two novel associations at KIF13B (P=7.1×10−7) and SOS1 (P=2.0×10−6). Interestingly, when the signals from the larger PSP GWAS were tested nominal association was observed with the MOBP locus (P=3.8×10−5; with same direction and effect size) but not STX6 or EIF2AK3. These findings may suggest that similar to DLB with PD and AD, there may be some overlap in the genetic risk determinants of PSP and CBD however some specific distinct associations exist.
Perspectives
The nature of parkinsonism is characterized by a late-onset sporadic phenotype that is influenced by a gradient of environmental and genetic determinants. Identification of the environmental factors, given their likely quantitative effect, will be difficult without a clear understanding of the genetic background. The causative spectrum of disease phenotypic risk from 99% genetic to 99% environmental covers the alternate hypothesizes underlying these disorders. From genetic examples such as SNCA triplications to environmental causes (such as post-encephalitic parkinsonism, MPTP-induced parkinsonism), most patients probably fall somewhere in-between. Within the current review we have tried to outline some of the genetic variants and loci that influence those patients who may not have one major disease determinant. The identification of these factors and the elucidation of their clinical relevance remains one of the greatest challenges within the field of parkinsonism genetics.
The use of NGS approaches (exome, whole-genome and transcriptome) in parkinsonism may help resolve the underlying genetic basis of what appears to be a complex multigenic disorder [29]. GWAS have nominated a number of loci through common variant association; NGS will hopefully provide further insight into rare variants and help resolve the pathogenic mechanisms of each gene/protein. In addition, exome and genome sequencing approaches will facilitate pathway analysis whereby the additive effects of multiple variants within the same pathway can be assessed. The concept of polygenic disease, with susceptibility determined by the additive or interactive effects of many low penetrant loci, is an attractive answer to late-onset sporadic disorders.
As observed for LRRK2 variants in PD it is clear that the frequency of specific variants will depend on the ethnic heritage of the patient group [5]. To date most studies have been performed in Caucasian populations, with only one Asian GWAS for PD. Interestingly, the MAPT H2 is almost absent in Asian populations and studies in PSP and CBD have been limited. As stated earlier the identification of environmental determinants will be difficult until we understand the genomic influence, however there the interaction of the two may be even more complex. Assuming specific variants only have an effect when a specific environmental agent is encountered (e.g. smoking) creates a scenario whereby straight-forward patient-control studies will not be helpful. Similar to polygenic effects, we will need to rely even more heavily on bioinformatics and computing power to analyze the big data sets for these types of gene-gene or gene-environment interactions.
Applying the genetic knowledge which has been gleaned from these large-scale efforts to direct therapeutic intervention strategies is critical. Identifying the functional/actionable variants at each locus and within each candidate gene will be crucial to understanding the disease mechanisms related to each specific protein/ locus. These genetic insights can drive the functional research and develop the biological tools and model systems that will be required for drug development. To date, little work has been successful in identifying genetic factors that modulate the age-at-onset which is an excellent phenotype to target for therapeutic intervention given the late-onset nature of parkinsonism.
We are entering an exciting time for genomic studies of parkinsonism. The technological revolution that has occurred over the last decade, from the growth of GWAS to the latest NGS studies has moved us closer to an answer. This period of precision/individualized medicine will be driven by our understanding of the genetic variation and how this will inform diagnostic/prognostic protocols. As targeted therapeutics are developed, genomics will inform criteria and selection for informed clinical trials. It is likely combination approaches will be used in parkinsonism, for example, some patients will benefit from α-synuclein knockdown approaches, others may require a LRRK2 inhibitor (if gain of function is the true toxic mechanism) and others will require a combination of both. Genomic information will be an indispensable part of the neurologist's armamentarium as we move forward to our ultimate goal of parkinsonism prevention.
Table 1.
The genes/loci associated with the alternate forms of parkinsonism.
| Disease | Gene | Locus | OMIM Gene Number |
|---|---|---|---|
| DLB | GBA | 1q22 | 606463 |
| APOE | 19q13 | 107741 | |
| SNCA | 4q22 | 163890 | |
| SCARB2 | 4q21 | 602257 | |
| MSA | SNCA | 4q22 | 163890 |
| MAPT | 17q21 | 157140 | |
| COQ2 | 4q21 | 609825 | |
| PSP | MAPT | 17q21 | 157140 |
| MOBP | 3p22 | 600948 | |
| EIF2AK3 | 2p11 | 604032 | |
| STX6 | 1q25 | 603944 | |
| CBD | MAPT | 17q21 | 157140 |
| KIF13B | 8p12 | 607350 | |
| SOS1 | 2p22 | 182530 | |
| MOBP | 3p22 | 600948 |
Acknowledgements
We would like to thank all those who have contributed to our research, particularly the patients and families who donated DNA samples for this work. We acknowledge the assistance of Krutika Satish Gaonkar of Mayo Clinic Health Sciences Research (Biomedical Statistics and Informatics). The Ross lab is supported by The Little Family Foundation, the Mangurian Foundation for Lewy body research, The Beaches Parkinson's Support Group Jacksonville, the Michael J. Fox Foundation, a Morris K. Udall Parkinson's Disease Research Center of Excellence (NINDS P50NS072187), and NINDS R01NS078086.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Trinh J, Farrer M. Advances in the genetics of Parkinson disease. Nat Rev Neurol. 2013;9:445–54. doi: 10.1038/nrneurol.2013.132. [DOI] [PubMed] [Google Scholar]
- 2.Fujioka S, Ogaki K, Tacik PM, Uitti RJ, Ross OA, Wszolek ZK. Update on novel familial forms of Parkinson's disease and multiple system atrophy. Parkinsonism Relat Disord. 2014;20(Suppl 1):S29–34. doi: 10.1016/S1353-8020(13)70010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lees AJ, Hardy J, Revesz T. Parkinson's disease. Lancet. 2009;373:2055–66. doi: 10.1016/S0140-6736(09)60492-X. [DOI] [PubMed] [Google Scholar]
- 4.Sidransky E, Lopez G. The link between the GBA gene and parkinsonism. Lancet Neurol. 2012;11:986–98. doi: 10.1016/S1474-4422(12)70190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross OA, Soto-Ortolaza AI, Heckman MG, Aasly JO, Abahuni N, Annesi G, et al. Association of LRRK2 exonic variants with susceptibility to Parkinson's disease: a case-control study. Lancet Neurol. 2011;10:898–908. doi: 10.1016/S1474-4422(11)70175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishioka K, Ross OA, Ishii K, Kachergus JM, Ishiwata K, Kitagawa M, et al. Expanding the clinical phenotype of SNCA duplication carriers. Mov Disord. 2009;24:1811–9. doi: 10.1002/mds.22682. [DOI] [PubMed] [Google Scholar]
- 7.Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat Genet. 2014;46:989–93. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heckman MG, Elbaz A, Soto-Ortolaza AI, Serie DJ, Aasly JO, Annesi G, et al. Protective effect of LRRK2 p.R1398H on risk of Parkinson's disease is independent of MAPT and SNCA variants. Neurobiol Aging. 2014;35:266, e5–14. doi: 10.1016/j.neurobiolaging.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacLeod DA, Rhinn H, Kuwahara T, Zolin A, Di Paolo G, McCabe BD, et al. RAB7L1 interacts with LRRK2 to modify intraneuronal protein sorting and Parkinson's disease risk. Neuron. 2013;77:425–39. doi: 10.1016/j.neuron.2012.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soto-Ortolaza AI, Heckman MG, Labbe C, Serie DJ, Puschmann A, Rayaprolu S, et al. GWAS risk factors in Parkinson's disease: LRRK2 coding variation and genetic interaction with PARK16. Am J Neurodegener Dis. 2013;2:287–99. [PMC free article] [PubMed] [Google Scholar]
- 11.Nishioka K, Ross OA, Vilarino-Guell C, Cobb SA, Kachergus JM, Mann DM, et al. Glucocerebrosidase mutations in diffuse Lewy body disease. Parkinsonism Relat Disord. 2011;17:55–7. doi: 10.1016/j.parkreldis.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishioka K, Wider C, Vilarino-Guell C, Soto-Ortolaza AI, Lincoln SJ, Kachergus JM, et al. Association of alpha-, beta-, and gamma-Synuclein with diffuse lewy body disease. Arch Neurol. 2010;67:970–5. doi: 10.1001/archneurol.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuang D, Leverenz JB, Lopez OL, Hamilton RL, Bennett DA, Schneider JA, et al. APOE epsilon4 increases risk for dementia in pure synucleinopathies. JAMA Neurol. 2013;70:223–8. doi: 10.1001/jamaneurol.2013.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bras J, Guerreiro R, Darwent L, Parkkinen L, Ansorge O, Escott-Price V, et al. Genetic analysis implicates APOE, SNCA and suggests lysosomal dysfunction in the etiology of dementia with Lewy bodies. Hum Mol Genet. 2014;23:6139–46. doi: 10.1093/hmg/ddu334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Low PA, Reich SG, Jankovic J, Shults CW, Stern MB, Novak P, et al. Natural history of multiple system atrophy in the USA: a prospective cohort study. Lancet Neurol. 2015;14:710–9. doi: 10.1016/S1474-4422(15)00058-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fanciulli A, Wenning GK. Multiple-system atrophy. N Engl J Med. 2015;372:1375–6. doi: 10.1056/NEJMc1501657. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs J, Nilsson C, Kachergus J, Munz M, Larsson EM, Schule B, et al. Phenotypic variation in a large Swedish pedigree due to SNCA duplication and triplication. Neurology. 2007;68:916–22. doi: 10.1212/01.wnl.0000254458.17630.c5. [DOI] [PubMed] [Google Scholar]
- 18.Scholz SW, Houlden H, Schulte C, Sharma M, Li A, Berg D, et al. SNCA variants are associated with increased risk for multiple system atrophy. Ann Neurol. 2009;65:610–4. doi: 10.1002/ana.21685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross OA, Vilarino-Guell C, Wszolek ZK, Farrer MJ, Dickson DW. Reply to: SNCA variants are associated with increased risk of multiple system atrophy. Ann Neurol. 2010;67:414–5. doi: 10.1002/ana.21786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vilarino-Guell C, Soto-Ortolaza AI, Rajput A, Mash DC, Papapetropoulos S, Pahwa R, et al. MAPT H1 haplotype is a risk factor for essential tremor and multiple system atrophy. Neurology. 2011;76:670–2. doi: 10.1212/WNL.0b013e31820c30c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heckman MG, Schottlaender L, Soto-Ortolaza AI, Diehl NN, Rayaprolu S, Ogaki K, et al. LRRK2 exonic variants and risk of multiple system atrophy. Neurology. 2014;83:2256–61. doi: 10.1212/WNL.0000000000001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mutations in COQ2 in familial and sporadic multiple-system atrophy. N Engl J Med. 2013;369:233–44. doi: 10.1056/NEJMoa1212115. [DOI] [PubMed] [Google Scholar]
- 23.Ogaki K, Fujioka S, Heckman MG, Rayaprolu S, Soto-Ortolaza AI, Labbe C, et al. Analysis of COQ2 gene in multiple system atrophy. Mol Neurodegener. 2014;9:44. doi: 10.1186/1750-1326-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Litvan I. Update on progressive supranuclear palsy. Curr Neurol Neurosci Rep. 2004;4:296–302. doi: 10.1007/s11910-004-0055-z. [DOI] [PubMed] [Google Scholar]
- 25.Conrad C, Andreadis A, Trojanowski JQ, Dickson DW, Kang D, Chen X, et al. Genetic evidence for the involvement of tau in progressive supranuclear palsy. Ann Neurol. 1997;41:277–81. doi: 10.1002/ana.410410222. [DOI] [PubMed] [Google Scholar]
- 26.Hoglinger GU, Melhem NM, Dickson DW, Sleiman PM, Wang LS, Klei L, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet. 2011;43:699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kouri N, Whitwell JL, Josephs KA, Rademakers R, Dickson DW. Corticobasal degeneration: a pathologically distinct 4R tauopathy. Nat Rev Neurol. 2011;7:263–72. doi: 10.1038/nrneurol.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kouri N, Ross OA, Dombroski B, Younkin CS, Serie DJ, Soto-Ortolaza A, et al. Genome-wide association study of corticobasal degeneration identifies risk variants shared with progressive supranuclear palsy. Nat Commun. 2015;6:7247. doi: 10.1038/ncomms8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonifati V. Genetics of Parkinson's disease--state of the art, 2013. Parkinsonism Relat Disord. 2014;20(Suppl 1):S23–8. doi: 10.1016/S1353-8020(13)70009-9. [DOI] [PubMed] [Google Scholar]
- 30.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]