Abstract
Recent genome-wide association studies have identified variations in the recombination repair gene, RAD52, that are associated with increased lung cancer risk, and particularly with the development of lung squamous cell carcinomas (LUSC). LUSC development is strongly associated with smoking. DNA repair is increased in the lung tissues of smokers, presumably because of ongoing DNA damage from exposure to tobacco smoke. A key player in the DNA damage response, RAD52 plays a role in DNA strand exchange and annealing during homologous recombination (HR) in mammalian cells. In this study, we discovered two cis-expression quantitative trait loci (eQTL) SNPs in the RAD52 gene that are associated with its expression and are also associated with LUSC risk. In addition, we report that amplification of the genomic region 12p13.33, which contains the RAD52 gene, is significantly associated with the development of LUSC in the TCGA database and that somatic overexpression of RAD52 was confirmed to be significant in LUSC tumors from our own patient cohort. Consistent with these genetic findings, we demonstrate that blockade of Rad52 slows cell growth and induces senescence in mouse bronchial epithelial cells. In contrast, overexpression of Rad52 leads to an increased rate of cell proliferation. We show that depletion of Rad52 in mouse lung tumor cells alters cell cycle distribution and increases DNA damage accumulation associated with increased tumor cell death. Our genetic and functional data implicate RAD52 as a significant determinant of risk in the development of LUSC.
Keywords: eQTL, Non-small cell lung carcinoma, lung cancer, DNA damage, Cancer susceptibility gene, senescence, gene amplification
Introduction
Lung cancer currently ranks as the foremost cause of cancer deaths among men and women in the United States with more than one-quarter of all cancer deaths due to this disease [1]. Squamous cell lung cancer (LUSC), which is particularly associated with smoking, comprises about 25% to 30% of all lung cancers and begins in squamous cells which are flat cells that line the inside of the bronchial airways. Furthermore, while 80–90% of lung cancer patients have a history of smoking, only about 10% of heavy smokers develop lung cancer [2]. This suggests that although tobacco smoke undoubtedly increases one’s risk for lung cancer, genetic risk factors most likely predispose certain individuals to disease development as part of a complex interplay between carcinogens and genetic determinants. One would expect certain genes which influence exposure to, metabolism of, or response to carcinogens to play a role in lung cancer susceptibility, especially in LUSC.
Recent genome-wide association studies (GWAS) have identified a region on chromosome 12p13.33 as harboring a susceptibility locus for LUSC. This locus includes the recombination repair gene RAD52 which harbors a specific single nucleotide polymorphism (SNP) - rs6489769 that is associated with LUSC [3]. RAD52, originally described in yeast as playing a key role in recombination repair, is involved in strand exchange and annealing of strands during homologous recombination and is predominantly recruited for DNA repair during S phase of the cell-cycle [4,5].
Similarly, recent additions to the 1000 genomes project, including four genome-wide association studies of lung cancer in populations of European ancestry, identified large-effect genome-wide associations for squamous cell lung cancer with the rare variant of BRCA2-K3326X [6]. While BRCA2 is the more recognized factor in human HR, studies performed in BRCA2-deficient mammalian cells implicate RAD52 as an independent and alternate participant in HR [7]. Upon RAD52 depletion, both the frequency of double-strand break-induced HR and ionizing radiation-induced RAD51 foci decreased significantly. Likewise, RAD52-RAD51 foci were shown to form equally in the presence or absence of BRCA2 [8]. As well, inactivation of RAD52 in BRCA2 competent mouse ES cells did result in a modest reduction in HR [9].
In humans, RAD52 is a key player in the regulation of HR-related genomic instability that may lead to an increased LUSC risk. Furthermore, when combined with depletion of BRCA2 or PALB2, depletion of human RAD52 is synthetically lethal [10]. Thus, in human cells deficient in the PALB2 or BRCA2 genes, RAD52 depletion may decrease cell survival by reducing rates of homologous recombination and by increasing damage-induced chromosomal abnormalities [3]. However, the exact role of RAD52 in human lung cancer remains elusive and more effort is needed to fill this gap in the knowledge base.
In this study, we sought to investigate the role of RAD52 in lung cancer susceptibility. As mentioned previously, squamous cell lung carcinoma is particularly associated with smoking. Thus, it fits that variation in this DNA repair gene would impact one’s risk for lung cancer, suggestive of a potentially decreased ability to repair carcinogen-induced damage. Surprisingly, our copy number variation (CNV) analysis of LUSC tumors revealed a copy number gain in the region of RAD52, resulting in a statistically significant level of amplification for RAD52 in lung squamous cell carcinoma patients. We also identified two correlated cis-eQTL SNPs that exhibit cis-regulation of RAD52 expression and significant association with LUSC. Elevated RAD52 protein expression was also significant in LUSC tumors compared to that in matched normal lung tissues.
In line with the statistical evidence portraying a role for RAD52 in developing lung cancer, we functionally demonstrate that upon Rad52 depletion, the rate of cell proliferation was attenuated and senescence was induced in non-tumorigenic bronchial epithelial mouse cells. We also show that loss of RAD52 slows cell proliferation while overexpression of RAD52 enhances proliferation in mouse lung cancer cells. Additionally, in mouse tumor cell lines we demonstrate that depletion of RAD52 alters cell cycle distribution by decreasing the fraction of cells in G0/G1 and significantly increasing the fraction of cells in G2/M. Finally, we demonstrate that loss of Rad52 expression increases DNA damage accumulation in mouse normal bronchial and tumor cells.
Materials and Methods
Study subjects
The cis-eQTL analysis of RAD52 was conducted on a cohort of 70 normal lung tissue samples provided by the Mayo Clinic [11]. The genetic association study conducted between RAD52 SNPs and lung cancer phenotypes was performed by analyzing a patient population of European ancestry (n=28,998) used in a recent large-scale GWAS study for lung cancer [6]. Gene expression analyses for RAD52 used the RNA-seq data for a cohort of tumor-normal paired RNA samples from 45 non-small cell lung cancer patients (36 lung adenocarcinoma (LUAD) and 9 lung squamous cell carcinoma (LUSC) recruited at the Medical College of Wisconsin.
eQTL and expression analysis of RAD52
The genotyping of the RAD52 SNPs for cis-eQTL analysis was part of a previous GWAS project using Illumina HumanHap 370k and 610k BeadChips [11] (Illumina, San Diego, CA, USA). CEPH DNA samples (a family trio) were included in each 96-well plate to monitor genotyping, and the concordance between the replicates was 99.5%. The RAD52 gene expression data in 70 normal lung tissues were part of the results of the genome microarray analysis done with the Illumina Human WG DASL array (Illumina, Inc, San Diego, CA, USA) as detailed previously [11]. The genetic association analysis between RAD52 and lung cancer was part of the recent large-scale lung cancer GWAS research detailed previously [6]. RNA sequencing for the tumor-normal paired RNA samples from 45 NSCLC patients followed the protocol from our previous publication [12]. Briefly, for data analysis, we used TopHat to conduct alignment of reads to the human reference genome (hg19). We used Cufflinks to compute expression levels of transcripts. Fragments per kilobase of exon per million fragments mapped (FPKM) values were summarized to the gene level by taking the sum of Cufflinks FPKMs estimated for the alternative transcripts of each Ensemble gene [12]. We specifically extracted expression data for RAD52 and conducted a pairwise comparison of the expression of this gene for the 45 lung tumor-normal pairs.
Statistical analysis
The linkage disequilibrium analysis for the RAD52 regions was conducted using Haploview software (http://www.broadinstitute.org/scientific-community/science/programs/medical-and-population-genetics/haploview/haploview) and the data from the Caucasian population of the 1000 Genomes Project. For the association testing between RAD52 SNPs and lung cancer risk, we derived joint odds ratios (ORs) and 95% confidence intervals (CIs) under a fixed-effect model for each SNP and the associated per-allele P values. To explore variability in associations according to tumor histology, we derived ORs for all lung cancer, LUAD and LUSC in the same way as in our previous study [6]. For the eQTL analysis, a linear regression model adjusted for age and sex was used to assess the correlation between genotypes (independent variable, coded as 0, 1, or 2) and transcript expression levels of RAD52 (eQTL, dependent variable) in 70 samples of normal lung tissue. The analysis was done with SAS version 9.0. Confounding factors that may be associated with lung cancer risk or gene expression were adjusted in the same way as in our previous publications (9,10). Therefore, pulling data from different cohorts did not introduce confounding information. In terms of RNA-seq based differential gene expression analysis of RAD52, a paired Student’s T-test was done to compare RAD52 expression between tumor samples and samples of adjacent normal tissue. We analyzed 36 LUAD (data not shown) and 9 LUSC tumor samples with matched normal tissue. The Wilcoxon signed rank test for the RAD52 gene showed differential expression using the tumor-normal pairs. We also downloaded somatic copy number alteration data in the 12p13.33 region containing RAD52 and somatic gene expression data of RAD52 generated by TCGA (The Cancer Genome Atlas, https://cghub.ucsc.edu/) to check the somatic copy number change and its association with somatic RAD52 expression changes in LUSC samples.
Cell culture
The immortalized, nontumorigenic mouse lung epithelial cell line, E10, as well as the mouse lung tumor cell lines obtained from spontaneous lung tumors in the A/J mouse (Spon2 and Spon6), as well as MC7 cells established from a methylene chloride-induced nonmetastatic lung tumor from B6C3F1 and the metastatic nicotine-derived nitrosamine ketone (NNK)-induced A/J mouse lung tumor (CL25M cell line) were cultured as previously described [13].
Lentivirus construction and transduction for short hairpin RNA (shRNA) expression
The pLKO.1 lentiviral system (GE Healthcare, PA, USA) was used to generate replication-deficient lentiviral particles for stable transduction of shRNA. The mature anti-sense strand of the shRNA sequences are as follows, Rad52-01: 5′-TAACTGCACCTTTACAAATGC -3′, Rad52-03: 5′-TAGTCCTCTAAAGAGTTCAGC -3′, Rad52-04: 5′-ATTCATCCGCTGTATACTGGC -3′, shSCRAM:5′-CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG -3′. Lentivirus stocks were prepared in 293T cells, infected into target cells with 8 ug/ml polybrene and selected with 2 ug/ml puromycin. Puromycin selected lines were used for experimental analysis from 4–25 days post-infection.
Overexpression of RAD52
The plasmids encoding empty vector pOPUR and pOPUR/Rad52 were a generous gift from Dr. Jac A. Nickoloff and were constructed according to his protocol [14]. Briefly, mammalian expression vector pOPUR was constructed to carry the RSV LTR promoter and triplicated lac operators from pOPI3CAT (Stratagene, La Jolla, CA) and the puromycin resistance gene from pPUR (Clontech, Palo Alto, CA). Human Rad52 cDNA was inserted downstream of the RSV LTR promoter in order to create a Rad52 overexpression plasmid that would retain its stability and not degrade post-transcription due to mRNA degradation signals in the 3′-untranslated region [14].
Immunoblot analysis of cellular lysates
Protein from E10, CL25M, Spon6 and Spon2 cells with shRNA targeting RAD52, constructs for overexpression of RAD52, scrambled vector control shRNA or empty pOPUR vector were extracted at 72–96 hours post transfection for transient transfections and in line with each experiment for stable stransfections. Cells were lysed with 100 μL of 1X RIPA lysis buffer containing proteinase inhibitor cocktails (Fisher Scientific, Pittsburg, PA), sonicated for 10 seconds, and spun at 13,000 RPM for 30 minutes at 4°C. Purified protein concentration was then calculated by the Bradford method, and boiled for 5 min. Normalized lysate was resolved by 4–12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Invitrogen, Carlsbad, CA) and transferred to PVDF membranes. Commercial antibodies used included: Rad52 (Cell Signaling, Danvers, MA, USA, 2762), Rad52 (Santa Cruz, Dallas, TX, USA, SC20044), Anti-phospho-Histone H2A.X (Ser139) (EMD Millipore, Billerica, MA, USA), p21 (F-5) (Santa Cruz, Dallas, TX, USA, SC20044), and beta-Actin (Santa Cruz, Dallas, TX, USA, SC20044). Immunoblots were developed using Clarity Western ECL Substrate (BioRad, Hercules, CA, USA) and quantitated on a ChemiDoc XRS+ Imaging System (BioRad, Hercules, CA, USA).
Cellular Senescence by SA-beta-gal staining
Cellular senescence was assayed as described previously [15]. Briefly, equal numbers of E10 Immortalized, nontumorigenic mouse lung epithelial cells stably infected with either scrambled pLKO.1 vector or shRNA constructs targeting the Rad52 gene (shRad52-03,04) were plated in 6-well sterile tissue culture plates 48 hours prior to analysis and at a confluence of approximately 30%. Senescence was measured according to manufacturer’s protocol (Cell Biolabs, San Diego, CA). Briefly, the cells were washed with PBS. After washing, 1x Fixing Solution (25% Glutaraldehyde) was added to each well and the cells were incubated at room temperature for 5 minutes. Fixation solution was removed, the cells were washed in PBS and cell staining working solution was prepared fresh and added to each well. The cells were then incubated overnight at 37°C, protected from light. The development of blue color was detected visually under a light microscope. Data correspond to the percentage of SA-beta-Gal-positive cells counted for each cell population. Numbers on the graphs indicate a percentage of SA-beta-Gal-positive cells and were derived from counting 200 cells in triplicate plates.
Apparent Doubling Time
E10 cells were infected with either Scr. pLKO.1 or shRad52-03. Cells underwent puromycin selection for 48 hours until all cells in the mock infected plate were dead. Puromycin selected populations were then seeded at equal densities, while remaining in media with puromycin, and incubated under normal growth conditions for 48–72 hours. Cell number and viability were determined by the Countess Automated Cell Counter (Invitrogen, Carlsbad CA) and apparent doubling time was calculated. 2x = n final/n start for the number of doublings (x) and (h/x) for the apparent doubling time, where h is the varying periods of time.
Cell Proliferation Curves and Plating Efficiency
Cells were harvested with trypsin, counted on the Countess automated cell counter (Invitrogen, Carlsbad, CA) and plated at a density of 3,000 cells per well in multiple replicates on a 96-well tissue culture plate. Cell numbers were measured starting 4 days after infection and 2 days after completing selection in puromycin, although cells remained on puromycin throughout the length of the experiment to ensure measurement of the correct population. Cell confluence was measured every two hours on an Incucyte live cell imager (Essen Biosciences, Ann Arbor, MI), through photomicrographs. Cell culture confluence was measured using Incucyte software (Essen Biosciences, Ann Arbor, MI).
Analysis of cell cycle distribution by propidium iodide staining
Mouse tumor cell lines were cultured in RPMI with 10% serum over a period of 14 days. Flow cytometry was employed to analyze cell cycle distribution on triplicate samples of cells using ethanol fixing and propidium iodide staining with RNAse to assess relative DNA content. Sub-confluent cultures of cells were harvested by trypsinization. Post-harvest, cells were centrifuged in 15 ml conical tubes at 1000 rpm for 5 min, re-suspended in cold PBS, then centrifuged again in the 15 ml conical at 500 ×g for 20 minutes. Next, the supernatant was discarded and 1 ml cold PBS was added to wash the cells. The cells were then centrifuged again at 500 ×g for 20 minutes, the supernatant was discarded and the cells were thoroughly suspended in 500 ul cold PBS, avoiding formation of cell clumps. Next, using the Cell Countess, cells were measured to a concentration of 2×106 cells/ml in PBS. For fixation, 5 ml of ice cold 95%/Absolute ethanol was added to the cells in PBS while gently vortexing. Cells were then held on ice for 2 hours to complete fixation. For staining, cells were spun down at 1500 rpm for 5 minutes to pellet. The ethanol was aspirated and the cells were washed twice with PBS, leaving a small amount to re-suspend the cells in. One ml of PI staining solution (900 ul PBS, 50ul PI stock soln, 50 ul RNase stock soln) was then added to the cell pellet. Finally, the cells were incubated in the dark for 3 hours at 4 degrees Celsius to ensure the RNase had digested all the RNA. Flow cytometry was performed on the Accuri C6 cytometer (BD Biosciences, San Jose CA). Data were compiled and analyzed using CFlow Plus and FlowJo software.
Comet Assay
A Comet Assay kit (Trevigen, Helgermen CT) was used to quantify and analyze DNA damage in individual E10 and Spon2 cells. The image that is obtained resembles a “comet” with a distinct head and tail. The head is composed of intact DNA, while the tail consists of damaged DNA. Cells are scored based on their Olive Tail Moment (OTM). The OTM is defined as the product of the tail length and the fraction of total DNA in the tail. One group of shSCRAM cells in each cell line was treated with Hypochlorous acid (HOCL) at 4°C in suspension to serve as a positive control for DNA damage accumulation.
Results
eQTL containing RAD52 is associated with lung cancer development
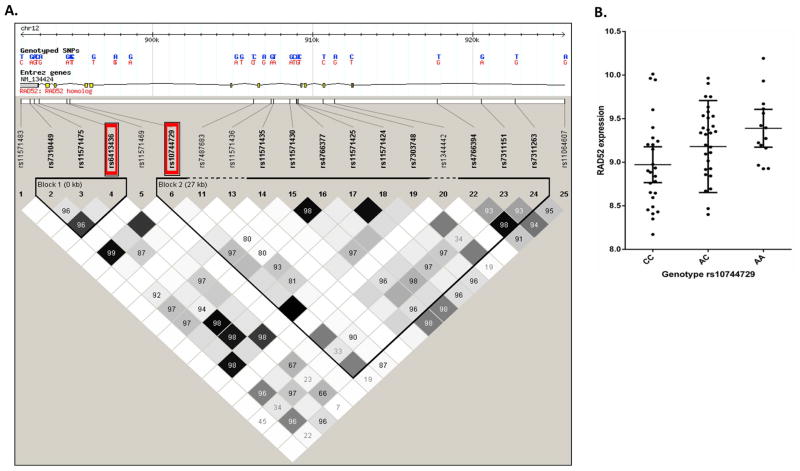
We found two SNPs whinin RAD52 that are associated with RAD52 gene expression in both lung tumor tissue as well as with non-small cell lung cancer phenotypes, especially LUSC. The first SNP, rs6413436, is significantly associated with LUSC in our GWAS cohort (p = 1.0×10−3 after multiple testing adjustment, odds ratio (OR) =1.06 for overall lung cancer (95% CIs: 1.024–1.10), n=28,998; p = 2.7×10−3, OR=1.09 for LUSC (95% CIs: 1.03–1.15), n=19,437). Although this SNP is not genotyped in our eQTL cohort of 70 lung tissues, it is in strong linkage disequilibrium (LD) (D′=1 and r2=0.82) with a nearby SNP, rs10744729, which we have genotyped in our eQTL cohort and found to be significantly associated with RAD52 gene expression (p = 0.01, n=70). Figure 1A shows the linkage disequilibrium plot of the tagged SNPs for the RAD52 region based on data from the Caucasian population of the 1000 Genomes Project. SNPs rs6413436 and rs10744729 are located in RAD52 intron 11 and intron 9, and the distance between them is relatively small, at about 1.9 kb. These are the two tagged SNP markers in Block 1 of RAD52 (marker #4 and #6 in Figure 1A red box). The SNP rs10744729 also showed a significant association with LUSC in our GWAS study (p = 0.02 after multiple testing adjustment). And the linkage disequilibrium between rs10744729 and rs6413436 in our dataset is close to complete LD (r2 = 0.82). There is complete LD between these 2 SNPs as revealed by HapMap data for whites (r2 = 1, Figure 1A). It is thus reasonable to conclude that rs10744729 is a proxy SNP for rs6413436. The significant association of both rs10744729 and rs6413436 with LUSC could be explained by the strong LD between these 2 SNPs. This plus that rs10744729 was significantly associated with RAD52 expression in lung tissues suggest that these 2 SNPs were eQTL SNPs that associate with LUSC risk by influencing RAD52 gene expression.
Figure 1.
(a) Linkage disequilibrium (LD) analysis of tagging SNPs based on the data of the Caucasian population used in 1000 Genome Project. The two cis-eQTL SNPs for RAD52 expression in lung tissue were rs6413436 and rs10744729, which were in strong linkage disequilibrium (D′=1 and r2=0.82 in our own sample and r2=1 in HapMap Caucasian sample). SNPs rs6413436 and rs10744729 are located in RAD52 intron 11 and intron 9, and the distance between them is about 1.9 kb. They are the two tagging SNP markers in Block 1 of RAD52 (marker # 4 and #6 in red box). Therefore, rs10744729 is a ‘proxy’ SNP for rs6413436. (b) The cis-eQTL SNP rs10744729 was significantly associated with the RAD52 gene expression (p = 0.01, n=70).
Figure 1B demonstrates that the risk allele ‘A’ of rs10744729 (in LD with the lung cancer risk allele ‘C’ of rs6413436) is associated with increased expression of RAD52 in a dose-dependent manner (homozygous reference alleles ‘CC’ < heterozygous ‘AC’ < homozygous mutants ‘AA’). ANOVA analysis showed that this SNP explains 9.2% of the expression variance of RAD52. Thus, the eQTL for rs10744729 and its associated SNP, rs6413436, accounts for a small proportion of the variance in RAD52 expression in lung tissues, an observation consistent with the relatively modest genetic risk observed with these two variants for LUSC (OR=1.09). By contrast, no significant association with lung adenocarcinoma (LUAD) was found for these two SNPs (p = 0.24, n=19,589), suggesting that these two eQTL SNPs were not related to the LUAD etiology.
Somatic copy number analysis in the RAD52 gene region using TCGA data
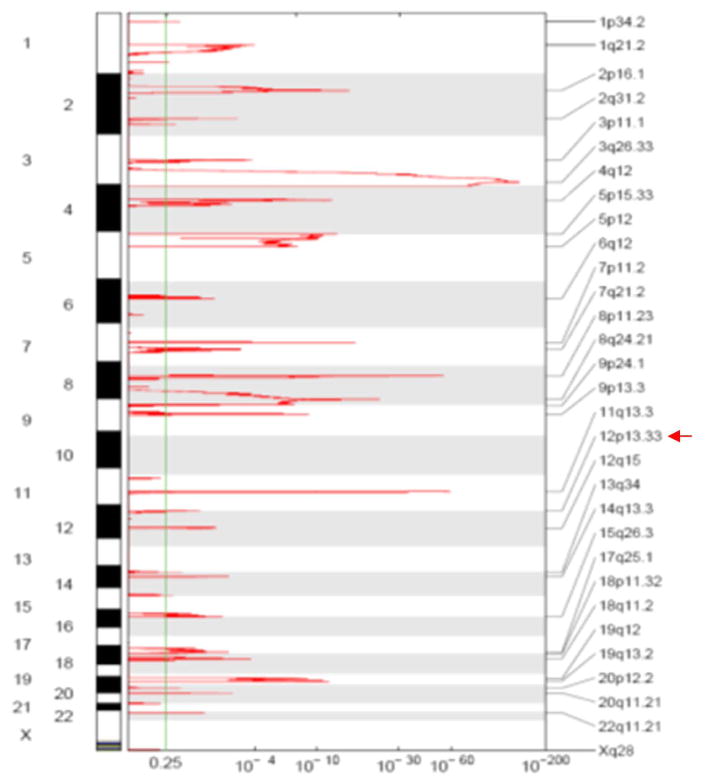
Using GISTIC software, somatic copy number analysis in 358 TCGA lung squamous cell carcinoma patients showed that the genomic region of 12p13.33 containing RAD52 reached a statistically significant level for focal amplification (q-value = 2.56×10−3, much less than the genome wide cutoff q = 0.25, Figure 2). Thirty-eight of 358 lung SCC tumors (11%) showed copy number gains in the RAD52 region (segments were reported as amplified if the corresponding estimated copy number ratio was greater than 1.25, i.e., log2 ratio > 0.32). These 38 identified TCGA LUSC tumors and their copy number amplification at the RAD52 gene region are listed in Supplemental Table 1. Gene expression levels of RAD52 are also positively correlated with somatic copy number gains in the RAD52 region in the TCGA LUSC samples (Supplementary Figure 1), suggesting that the somatic copy number gains in the LUSC tumors led to the corresponding increase in RAD52 expression.
Figure 2.
Somatic copy number analysis using the GISTIC program on 358 TCGA lung squamous cell carcinoma patients showed that the genomic region of 12p13.33 containing RAD52 reached a statistically significant level for focal amplification (q-value = 2.56×10−3, much less than the genome-wide cutoff q = 0.25).
Other significant RAD52 SNPs influencing LUSC
We identified a number of RAD52 SNPs which showed more significant P-values (P-values < 10−6 (Supplemental Table 2) for the associations with squamous cell lung cancer phenotype. However, these SNPs were not associated with RAD52 gene expression in our samples, suggesting that other mechanisms may underlie their associations with LUSC. The RAD52 SNPs we presented here are the ones associated with both RAD52 gene expression and LUSC risk. As well, these are cis-eQTL SNPs that may influence LUSC phenotype by affecting the expression level of RAD52. We replicated the significant association between RAD52 SNP rs6489769 and the risk of squamous cell carcinoma P = 9.4×10−7, (Supplemental Table 2) which was originally reported by Shi et al [3]; however, this SNP does not show significant association with our RAD52 gene expression data, suggesting it may not be a cis-eQTL SNP in RAD52. In fact, Shi et al. [3] did not find that the SNP rs6489769 was associated with RAD52 gene expression. Current evidence suggests that rs6489769 may influence one’s risk of developing LUSC via mechanisms other than RAD52 expression.
Increased RAD52 gene expression in LUSC samples from NSCLC cohort
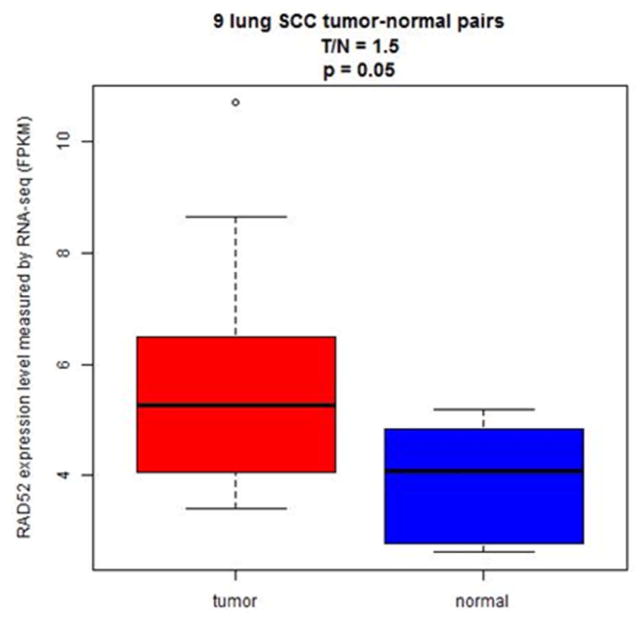
Based on our independent non-small cell lung cancer (NSCLC) patient dataset, we found that RAD52 was significantly overexpressed in the tumors of LUSC patients (p=0.05, n=9, Figure 3). Specifically, there was an average of a 50% increase in RAD52 expression in LUSC tumors compared to that in normal tissue samples (Figure 3). However, there was no significant difference in RAD52 expression between the tumor and normal lung tissues of LUAD patients (p=0.29, n = 36). This data suggests that RAD52 overexpression could contribute to LUSC etiology but not to LUAD carcinogenesis, which is consistent with our eQTL study showing that the identified eQTL SNPs in RAD52 are significantly associated with LUSC but not LUAD.
Figure 3.
Overexpression of RAD52 in lung squamous cell carcinoma tumors observed in our independent NSCLC patient dataset. RAD52 was significantly overexpressed in the tumors of LUSC patients (p=0.05, n=9). There was an average of a 50% increase in RAD52 gene expression in LUSC tumors compared to normal tissues.
Loss of Rad52 induces senescence in mouse immortalized lung epithelial cells
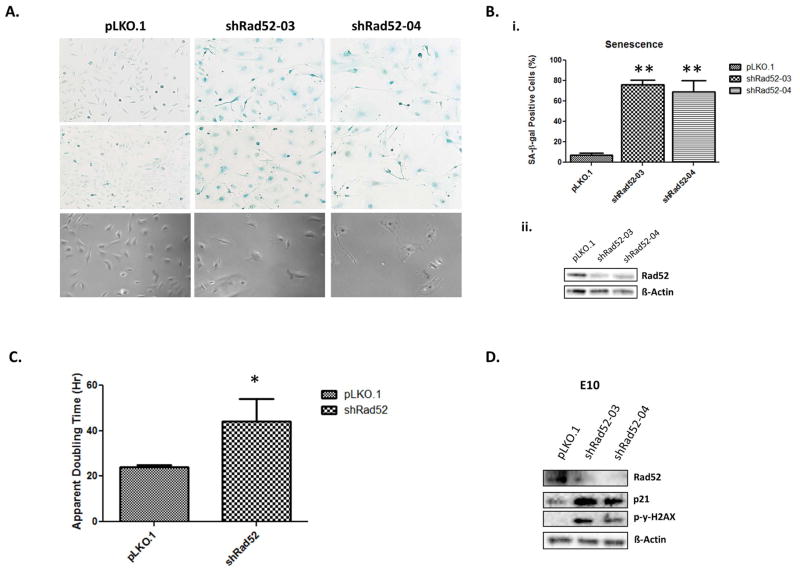
Although Rad52 acts in a parallel fashion to the known tumor suppressor BRCA2, depletion of Rad52 has actually been shown to slow cell proliferation in MCF7 breast cancer cells [8]. This suggests that although the inherent function of Rad52 is to repair damaged DNA, in a pro-tumorigenic setting, it may actually function to enhance cell proliferation or to protect tumor cells from apoptosis. Given Rad52’s recent identification as a susceptibility candidate associated with squamous cell lung carcinoma, we hypothesized that depletion of Rad52 might be a factor in the survival and proliferation of lung and lung tumor cells. To determine the effect of Rad52 depletion in mouse bronchial cells, we treated immortalized, non-tumorigenic mouse bronchial epithelial cells with either shRNA targeting RAD52 or a scrambled vector. RAD52 was depleted at the protein level from the E10 cells by 2.63 fold and 1.89 fold using shRNA constructs 03 and 04, respectively. The cells were cultured for 7–10 days post-infection before they stopped dividing, became morphologically round and flattened, and stained positive for senescence associated beta-galactosidase activity, indicating that loss of RAD52 resulted in senescence (Figure 4).
Figure 4.
Immortalized E10 mouse lung epithelial cells underwent spontaneous senescence upon Rad52 depletion. (a, top, middle) Senescence is demonstrated through senescence-associated β-galactosidase activity by staining cells with the chromogenic substrate 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal). (a, bottom) Representative phase-contrast images documenting the observed cells are shown for each population. (b, top) Indicated bronchial epithelial cells were infected with control shRNA or Rad52-shRNAs (03 or 04). Cells were fixed and stained for senescence-associated beta-galactosidase (SA-beta-Gal) activity 10 days after the infection. Numbers on the graphs indicate percentage of SA-beta-Gal-positive cells compared to total cell count and were derived from counting 200 cells in triplicate plates. (b, bottom) Densitometry using ImageJ software demonstrated 2.63 and 1.89 fold decreases in expression levels of Rad52 in E10 cells infected with shRad52-03 and shRad52-04 compared to scrambled control, respectively. (c) E10 cells infected with either Scr. pLKO.1 or shRad52-03 were seeded at equal densities and incubated under normal growth conditions for 48–72 hours. Apparent doubling time was calculated. (d) E10 Scr. pLKO.1 and shRad52-03,04 populations were cultured under normal growth conditions and cell lysates were prepared and subjected to immunoblot analysis. (ns, no significance; * p<0.05; **p<0.005)
Phosphorylation of gamma H2AX is an early sign of DNA damage induced by replication stalling [16]. The role of γ-H2AX foci in the DNA damage signaling pathway induced by double-strand breaks is to act as docking areas for the recruitment of repair factors and to bring the broken DNA ends closer so that DNA repair is accomplished [17]. Beard et al. found that upon induction of DNA damage, y-H2AX-proficient cells, as well as H2AX-complemented H2AX(−/−), cells reacted by increasing p21 levels and arresting the cell cycle. As our results also suggest, (Figure 4D), H2AX is required for p21-induced cell cycle arrest after replication stalling. In addition, γ-H2AX has been shown to be essential for concentrating repair proteins and maintaining the integrity of the DNA damage response foci [18]. Finally, the significance of H2AX in cell cycle arrest was previously demonstrated by Fernandez-Capetillo et al. [19]. This serves to confirm the observed senescence in our E10 bronchial epithelial cells and to suggest a further role for the DNA damage response in the aforementioned growth arrest discovered upon Rad52 depletion.
Expression of RAD52 affects growth in mouse lung epithelial and tumor cells
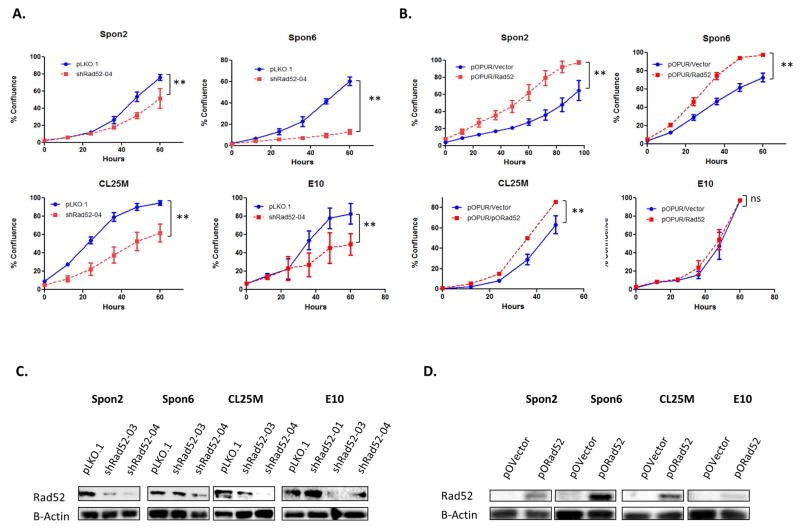
Utilizing distinct viral transfections, equal numbers of viable Spon2, Spon6, CL25M and E10 wild-type and RAD52-depleted cells were seeded in the Incucyte live cell imager, and the growth rate was observed over a period of 3 days. Upon depletion of RAD52 in mouse lung epithelial cells, we observed the rate of cell proliferation begin to decrease around 96 hours post infection and 48 hours post completion of puromycin selection, prior to the appearance of a senescent phenotype. Cellular proliferation in lung tumor cell lines (Spon2, Spon6 and CL25M) also decreased with depletion of RAD52 (Figure 4A). Admittedly, we recognize that percent confluence is not a pure measure of cell number; however, doubling time assays in E10 cells (Figure 4) along with MTS measurements of cell proliferation confirm that cell morphology did not affect proliferation rates. In effect, any error caused by differences in cell morphology would likely function to enlarge cells depleted of RAD52, enhancing their rate of confluence.
In comparing the apparent doubling time of E10 cells, we determined that depletion of RAD52 led to a significant increase in apparent doubling time (*P<0.05), increasing 1.83 fold from those infected with a scrambled control shRNA (Figure 4C). Moreover, although E10 cells overexpressing RAD52 did not show a growth difference, all tumor cell lines overexpressing RAD52 showed a significant increase in growth rate around 5 days post-infection.
Depletion of RAD52 alters cell-cycle distribution in lung tumor cells and induces accumulation of cells in G2/M
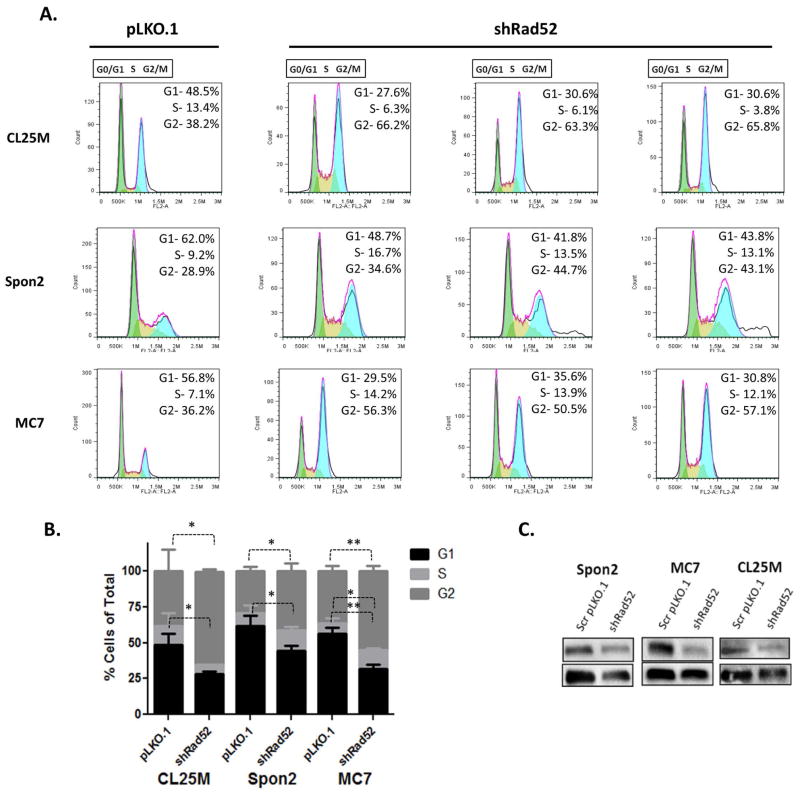
To further investigate whether changes in cell-cycle regulation correlate with the observed changes in cell growth, we compared cell-cycle profiles of different mouse lung tumor cell lines through flow cytometry with propidium iodide (PI) staining. The E10 mouse bronchial epithelial cell line became senescent post-RAD52 knockdown, which prevented the acquisition of enough cells for flow cytometry. Results from these studies demonstrate accumulation of tumor cell populations in the G2/M fraction and a decrease in cell populations in the G0/G1 fraction consistent with deficient DNA repair-induced senescence (Figure 6). These data strongly support a role for RAD52 in G2/M progression and/or passage through the G2/M checkpoint in mouse lung tumor cells. This G2 delay is likely to be the major determinant of the slower growth rate seen for the RAD52-depleted mouse cells (Figure 5). A less severe profile was seen in the Spon2 cell line (Figure 6A). This indicates that other factors besides Rad52 expression may be rate-limiting in the passage of Spon2 cells beyond the G2 phase.
Figure 6.
Depletion of Rad52 alters the cell-cycle distribution of lung tumor cells inducing a decrease in cells in G0/G1 and an accumulation of cells in G2/M. (a) Cell cycle progression was assessed by propidium iodide staining as a measure of DNA content. Flow cytometry was performed on CL25M, Spon2 and MC7 Rad52-depleted and scrambled vector control cells. Fixed cells were acquired using the Accuri C6 Cytometer. (b) Cells were then analyzed using FlowJo software and the Watson-Pragmatic cell cycle method and percentage values for G0/G1 (2 NDNA content), S (intermediate DNA content) and G2/M (4 N DNA content) cells. (c) Confirmation of knockdown of Rad52 in mouse tumor cell lines using the shRad52-03 construct and scrambled control shRNA by western blot. (ns, no significance; * p<0.05; **p<0.005)
Figure 5.
Cellular proliferation of lung tumor and bronchial epithelial cells with altered RAD52 expression. (a) Cellular proliferation in lung tumor cells (Spon2, Spon6, CL25M) and bronchial epithelial cells (E10) transfected with shRNA against Rad52 compared to scrambled control. (b) Cellular proliferation in lung tumor cells (Spon2, Spon6, CL25M) and bronchial epithelial cells (E10) transfected with the pOPUR Rad52 overexpression plasmid compared to vector. (c–d) Transfected populations from all cell lines were cultured under normal growth conditions for 7–10 days, Cell lysates were prepared and subjected to immunoblot analysis. (ns, no significance; * p<0.05; **p<0.005)
Loss of RAD52 induces spontaneous DNA damage in immortalized bronchial epithelial and lung tumor cells
Highly efficient and accurate DNA repair throughout the replication cycle of a cell is critical in maintaining genome stability and thwarting disease-causing polymorphisms. When DNA damage accumulates, a cell may either die because of excessive damage, or the cell may mechanistically sidestep the damage through modulation of other checkpoints or tumor suppressive proteins, leading to replication of the damaged cell and disease.
In order to determine if the observed changes in cell growth and cell cycle distribution may be due to a deficiency in the DNA damage response, E10 and Spon2 cells were depleted using multiple shRNA constructs and subjected to the Comet assay to quantify DNA damage in individual cells (Supplementary figure 2).
Analysis of olive tail moment (OTM) showed that both wild type E10 and Spon2 cells displayed little endogenous DNA damage. However, Rad52 depleted cells cultured for 6–11 days contained relatively high levels of DNA damage accumulation (Supplementary figure 2). These results demonstrate that under cell-culture conditions where the repair system is impaired, RAD52 deficiency is associated with the accumulation of DNA damage.
Discussion
In between cycles of replication, eukaryotic cells will undergo damage to their genome by both endogenous and exogenous means. In order to survive, cells have developed different means of damage control and repair systems. As the main mechanism in the eukaryotic DNA damage response, homologous recombination serves as a safeguard in cell cycle progression, ensuring proper signaling of chromosome damage and accurate chromosome segregation during mitosis [20]. If unsuccessful in a critical repair process such as the DNA damage response, a cell may either undergo apoptosis or accumulate mutations that will ultimately evolve into genomic instability and cancer [7]. The complementary evidence determined by our genetic and functional studies suggests a potential role for the DNA repair gene, RAD52, in lung tumorigenesis. Our research into the role of RAD52 in susceptibility to lung carcinogenesis emerged from prior findings implicating variation in RAD52 as a factor in LUSC development [3]. Here, we have identified two candidate regions in RAD52 where variations in the gene are associated with increased copy number as well as increased RAD52 expression in lung tumor tissue compared to normal.
A study performed by the Barlow group demonstrated that by reducing homologous recombination in ATM-deficient mice through genetic deletion of the Rad52 gene, not only did the development of T-cell lymphomas decrease, but mice depleted of Rad52 also lived longer [21]. This report suggests that excessive homologous recombination, termed hyperrecombination, by a member of the normal DNA damage response may contribute to tumorigenesis. This hyperrecombinogenic, phenotype is known to be toxic to the cell, leading to chromosomal instability and disease through an accumulation of lethal recombination variants [21,22]. In A-T, disease phenotypes are believed to arise from a loss of ATM kinase activity as well as disrupted signaling to proteins such as p53, chk2 and DNA-repair proteins, such as Rad52, resulting in cell cycle checkpoint defects and impaired DNA repair [21]. Barlow’s group tested the hypothesis that excessive recombination might be responsible for the overlapping phenotypes in A-T and Blooms in vivo by crossing Atm−/− mice with mice deficient in the HR protein Rad52 [21]. Her group clearly demonstrated that by reducing homologous recombination through genetic deletion of the Rad52 gene, they were able to significantly decrease and delay the incidence of thymic tumors in Atm−/− mice: more than half of the Atm−/− mice developed tumors by 4 months of age compared to less than 20% of the Atm−/− Rad52−/− mice [21].
Therefore, when considering current GWAS studies, our eQTL analysis, and recent experimental studies, it is reasonable to suggest that although chromosomal instability is not rescued in the absence of Rad52, genetic deletion of Rad52 may result in a reduction of abnormally high levels of intrachromosomal recombination and that this reduction by Rad52 reduces tumorigenesis by protecting genome integrity. Given the recent genomic data linking Rad52 to lung cancer along with our data demonstrating Rad52 amplification in LUSC cases, along with the known role of Rad52 in recombination, it is rational to suggest that overexpression of Rad52 would aid in the development of a lung cancer phenotype.
As with that study, we found that not only did lung tumor tissue samples express higher levels of RAD52 compared to normal lung tissue, but depleting RAD52 in functional in vitro studies led to phenotypes associated with decreased tumorigenesis. As our functional studies show, overexpression of Rad52 leads to an increased rate of cell growth, a prominent tumorigenic phenotype, further supporting a role for Rad52 in the development of lung cancer.
As mentioned above, in this study we sought to determine the effect of loss and overexpression of RAD52 on such phenotypes associated with lung tumorigenesis including induction of senescence, rates of cell proliferation, altered cell cycle distribution and accumulation of DNA damage. Current literature implicates a connection between a high frequency of structural variation in the genome with phenotypic diversity and predisposition to disease [23]. Hence, it would be reasonable to believe that DNA repair genes such as RAD52 may modulate oncogenic phenotypes related to DNA damage and genomic variation. We observed that E10 immortalized mouse lung epithelial cells went into a state of senescence 7–10 days post infection with shRNA constructs targeting Rad52. E10 cells express high levels of the known oncogene MDM2 and are ARF/p53-inactivated [13]. Thus, it makes sense that depletion of Rad52 in E10 cells lead to upregulation of p21 and phospho-gamma H2AX expression, similar to observations made by the MacLaren group upon knockdown of c-Jun [24]. This group found a potential role for c-Jun in the DNA damage response, linking its depletion to premature senescence, an accumulation of cells in G2/M phase of the cell cycle, and a persistence of DNA damage [24].
Furthermore, in mouse lung tumor cells, knockdown of Rad52 curbed proliferation and led to an accumulation of cells in the G2/M phase of the cell cycle (Figures 5,6). Alteration of cell-cycle regulation by RAD52 has not been demonstrated previously. The observed G2/M-accumulation upon RAD52 depletion may be due to variation in expression of other proteins or increased sensitivity to culture conditions.
Little is known of the effects of variation in RAD52 in regards to susceptibility to lung cancer and tumor development. However, genetic variations that alter homologous recombination, coupled with a carcinogenic environment, may plausibly contribute to tumorigenesis. Further understanding of the role that RAD52 plays in the development of NSCLC will ultimately yield clinically significant results through determination of prediction markers and prognostic factors in regard to LUSC therapeutics.
Supplementary Material
Acknowledgments
Grant Support: NIH, National Cancer Institute grants, U19CA148127 (Amos, You)
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Halmos B, Boiselle PM, Karp DD. Lung cancer. Primary Care Update for OB/GYNS. 2003;10(3):87–94. [Google Scholar]
- 3.Shi J, Chatterjee N, Rotunno M, et al. Inherited variation at chromosome 12p13. 33, including RAD52, influences the risk of squamous cell lung carcinoma. Cancer Discov. 2012;2(2):131–139. doi: 10.1158/2159-8290.CD-11-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lisby M, Antunez de Mayolo A, Mortensen UH, Rothstein R. Cell cycle-regulated centers of DNA double-strand break repair. Cell Cycle. 2003;2(5):479–483. [PubMed] [Google Scholar]
- 5.Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci U S A. 2001;98(15):8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, McKay JD, Rafnar T, et al. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat Genet. 2014;46(7):736–741. doi: 10.1038/ng.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lok BH, Powell SN. Molecular pathways: understanding the role of Rad52 in homologous recombination for therapeutic advancement. Clin Cancer Res. 2012;18(23):6400–6406. doi: 10.1158/1078-0432.CCR-11-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Z, Scott SP, Bussen W, et al. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc Natl Acad Sci U S A. 2011;108(2):686–691. doi: 10.1073/pnas.1010959107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rijkers T, Van Den Ouweland J, Morolli B, et al. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol Cell Biol. 1998;18(11):6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lok BH, Carley AC, Tchang B, Powell SN. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene. 2013;32(30):3552–3558. doi: 10.1038/onc.2012.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Sheu CC, Ye Y, et al. Genetic variants and risk of lung cancer in never smokers: a genome-wide association study. Lancet Oncol. 2010;11(4):321–330. doi: 10.1016/S1470-2045(10)70042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Govindan R, Ding L, Griffith M, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150(6):1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDoniels-Silvers AL, Herzog CR, Tyson FL, Malkinson AM, You M. Inactivation of both Rb and p53 pathways in mouse lung epithelial cell lines. Exp Lung Res. 2001;27(3):297–318. doi: 10.1080/019021401300054064. [DOI] [PubMed] [Google Scholar]
- 14.Kim PM, Allen C, Wagener BM, Shen Z, Nickoloff JA. Overexpression of human RAD51 and RAD52 reduces double-strand break-induced homologous recombination in mammalian cells. Nucleic Acids Res. 2001;29(21):4352–4360. doi: 10.1093/nar/29.21.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhuang D, Mannava S, Grachtchouk V, et al. C-MYC overexpression is required for continuous suppression of oncogene-induced senescence in melanoma cells. Oncogene. 2008;27(52):6623–6634. doi: 10.1038/onc.2008.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fragkos M, Jurvansuu J, Beard P. H2AX is required for cell cycle arrest via the p53/p21 pathway. Mol Cell Biol. 2009;29(10):2828–2840. doi: 10.1128/MCB.01830-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franco S, Gostissa M, Zha S, et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol Cell. 2006;21(2):201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5(7):675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez-Capetillo O, Chen HT, Celeste A, et al. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat Cell Biol. 2002;4(12):993–997. doi: 10.1038/ncb884. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Heyer WD. Who’s who in human recombination: BRCA2 and RAD52. Proc Natl Acad Sci U S A. 2011;108(2):441–442. doi: 10.1073/pnas.1016614108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treuner K, Helton R, Barlow C. Loss of Rad52 partially rescues tumorigenesis and T-cell maturation in Atm-deficient mice. Oncogene. 2004;23(27):4655–4661. doi: 10.1038/sj.onc.1207604. [DOI] [PubMed] [Google Scholar]
- 22.Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7(2):85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 24.MacLaren A, Black EJ, Clark W, Gillespie DA. c-Jun-deficient cells undergo premature senescence as a result of spontaneous DNA damage accumulation. Mol Cell Biol. 2004;24(20):9006–9018. doi: 10.1128/MCB.24.20.9006-9018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.