Abstract
Olfactory tissue undergoes lifelong renewal, due to the presence of basal neural stem cells. Multiple categories of globose basal stem cells have been identified, expressing markers such as Lgr5, Ascl1, GBC-2, and c-Kit. The differentiation potential of individual globose cells has remained unclear. Here, we utilized Cre/loxP lineage tracing with a multicolor reporter system to define c-Kit+ cell contributions at clonal resolution. We determined that reporter expression permitted identification of c-Kit derived progeny with fine cellular detail, and that clones were found to be comprised by neurons only, microvillar cells only, microvillar cells and neurons, or gland/duct cells. Quantification of reporter-labeled cells indicated that c-Kit+ cells behave as transit amplifying or immediate precursors, although we also found evidence for longer-term c-Kit+ cell contributions. Our results from the application of multicolor fate mapping delineate the clonal contributions of c-Kit+ cells to olfactory epithelial renewal, and provide novel insight into tissue maintenance of an adult neuroepithelium.
Keywords: Olfaction, stem cells, neurogenesis, regeneration, receptor tyrosine kinase
INTRODUCTION
The olfactory epithelium lines portions of the nasal cavity and contains chemosensory neurons, which are replaced continually in adult mammals (Graziadei and Graziadei, 1979; Schwob, 2002). Moreover, experimental injury induces a coordinated rapid reconstitution of the neuroepithelium (Schwob et al., 1995; Bergman et al., 2002; Jia and Hegg, 2012). Olfactory tissue self-renewal is supported by the presence of stem and progenitor cells situated in the basal germinal zone of the neuroepithelium (Goldstein and Schwob, 1996; Huard et al., 1998; Calof et al., 2002; Fletcher et al., 2011). Several of these basal cell populations, identified by genetic fate mapping studies, contribute to olfactory tissue maintenance, including keratin5+, Ascl1+, Lgr5+, or c-Kit+ basal cells (Leung et al., 2007; Gokoffski et al., 2011; Chen et al., 2014; Goldstein et al., 2015). Ongoing interest in determining the identity and functional potential of marker-defined olfactory neural stem cells arises for several reasons, including: (a) efforts to understand the mechanisms involved in normal olfactory maintenance, or in failures of this process due to injury or disease; (b) use of the olfactory epithelium as an experimental model for adult neurogenesis in general; and (c) the possibility of utilizing the nose as a source of autologous neural stem cells with therapeutic potential (Goldstein et al., 1998; Leung et al., 2007; Jang et al., 2008).
The c-Kit+ olfactory basal cell is of particular interest. Using single-color genetic fate mapping, as well as a conditional diphtheria toxin cell ablation model, we reported previously that c-Kit+ globose basal cells are required for olfactory neuron maintenance (Goldstein et al., 2015). Also, we found that Bowman’s glands can arise from c-Kit+ cells. However, aspects of the functional potential of individual c-Kit+ progenitors have remained unclear. For instance, c-Kit+ cells might function as immediate precursors, which undergo a terminal mitosis as they produce differentiated progeny. Alternatively, they may function as transit amplifying progenitors, or as more upstream stem cells that give rise to immediate neural precursors. An additional question is whether c-Kit+ cells are lineage committed or multipotential. Accordingly, here we utilized the R26R-Confetti Cre reporter system (Snippert et al., 2010) to determine directly the functional behavior of c-Kit+ olfactory progenitors with inducible c-KitCreERT2/+ mice (Klein et al., 2013). To address the clonality of c-kit cell contribution to neuroepithelium physiologically and in the case of injury, we studied unlesioned normal olfactory development as well as experimentally-induced neuroepithelial reconstitution in adult mice. The application of the multicolor Cre reporter technique (Livet et al., 2007; Snippert et al., 2010) to olfactory renewal, to discern greater detail of progenitor cell function and clonal relationships among reporter-labeled progeny, has not been reported previously.
METHODS
Animals
All experimental procedures were approved by the University of Miami Institutional Animal Care and Use Committee, and were performed in full compliance with the NIH Guidelines for the Care and Use of Laboratory Animals. The c-KitCreERT2/+ mouse line was provided by Dr. Dieter Saur (Klein et al., 2013). The multicolor Cre reporter, Gt(ROSA)26Sortm1(CAG-Brainbow2.1)Cle/J (Stock Number: 013731), abbreviated here as R26R-Confetti, was obtained from the Jackson Laboratory (Bar Harbor, ME). For conditional labeling of c-Kit+-derived cells, c-KitCreERT2/+ mice were crossed with R26R-Confetti mice. PCR genotyping for c-KitCreERT2/+ was performed as described (Klein et al., 2013); R26R-Confetti mice were bred as homozygotes. In initial experiments, tamoxifen (Sigma, St. Louis, MO) 20 mg/mL in peanut oil (Sigma) was given at 2 mg intraperitoneally at designated times to c-KitCreERT2/+; R26R-Confetti adults, or 0.2 mg to postnatal mice. For clonal analysis of the c-Kit+ olfactory lineage, animals were given a single low dose of tamoxifen, determined empirically to yield sufficiently sparse labeling: 1 mg for adult mice, and 0.0125 mg for neonates.
Tissue Processing
Adult mice were euthanized by exsanguination from perfusion with saline followed by fixative, under deep ketamine–xylazine anesthesia. After perfusion with phosphate buffered saline (PBS) followed by 4% paraformaldehyde in phosphate buffer, adult nasal tissue was dissected from surrounding muscle and bone, postfixed for 1–2 hours, rinsed in PBS, and then treated with 30% sucrose/250 mM EDTA in PBS for 3–4 days. Specimens were then embedded in O.C.T. compound (VWR, Radnor, PA) and frozen in liquid nitrogen. Tissue was cryosectioned at 60 μm, collected on Superfrost Plus slides (VWR), and stored at −20°C.
Immunohistochemistry
Slides were rinsed in PBS, and blocking was performed using a solution of PBS, 10% normal serum (Jackson ImmunoResearch, West Grove, PA), 4% bovine serum albumin (BSA, Sigma), 5% nonfat dry milk, and 0.1% Triton X-100 (Sigma) for 30–60 minutes, followed by primary antibody diluted in the same solution overnight at 4°C. Primary antibodies used here include: goat anti-olfactory marker protein (OMP), 1:1000 (WAKO #019-22291, Richmond, VA), rat anti-CD73, 1:1000 (eBioscience #16-0731, San Diego, CA), rabbit anti-GAP43, 1:800 (Abcam #ab75810, Cambridge, MA), chick anti-GFP, 1:500 (Life Technologies #A10262, Carlsbad, CA), and rabbit anti-Trpm5, 1:100 (Alomone Labs, Jerusalem, Israel, #ACC-045). Note, heat-mediated antigen retrieval was performed using Tris pH 8.0 for anti-Trpm5. The antigen retrieval destroys XFP fluorescence, so anti-GFP, which cross-reacts with the other XFPs, was used to co-visualize Cre reporter and anti-Trpm5. Slides were rinsed in PBS and incubated with either fluorescent-conjugated secondary antibody or biotinylated secondary (Jackson ImmunoResearch) for 30–45 minutes in the same blocking solution. Fluorescent tertiary reagent was applied for 30 min, for visualization of biotinylated secondary. Slides were then rinsed and coverslipped with Vectashield containing 4,6-diamidino-2- phenylindole (DAPI; Vector).
Imaging and cell counting
Tissue sections were analyzed on an Olympus IX81 epifluorescent microscope or a Zeiss LSM-710 confocal microscope for detection of cytoplasmic red fluorescent protein (RFP), nuclear-localized green fluorescent protein (GFP), cytoplasmic yellow fluorescent protein (YFP) and membrane-tethered cyan fluorescent protein (CFP). We refer to these fluorescent reporters generically as XFPs. DAPI nuclear stain was excited at 405 nm, CFP was excited at 458 nm, GFP at 488 nm, YFP at 514 nm and RFP at 561 nm. The laser power was minimized and detection wavelength range was gated narrowly to avoid overlap among channels, especially between GFP and YFP. The nuclear localization of GFP and cytoplasmic localization of YFP also clearly discriminated between these reporters. Images were acquired with Zeiss Zen 2010 software and analyzed using FIJI ImageJ. Pseudocoloration and brightness/contrast adjustment were performed using either ImageJ or Adobe Photoshop CS3. For image capture of reporter-labeled clones, confocal Z-stacks were assembled from consecutive optical sections from 60 μm cryosections, using ImageJ. Labeled cells were counted for entire 60 μm coronal sections through similar antero-posterior regions of the nasal fossa, chosen to ensure that the superior turbinate (I), branching middle turbinate (IId and IIv), and inferior turbinates (III and IV) were included on each section. After titration of tamoxifen dosing, low-dose tissue was used for analyzing labeled clones, defined as non-overlapping single color clusters from these sparsely labeled tissues. At least three sections per animal from n=3–6 animals were analyzed for each condition.
Statistical Analysis
Significance testing was performed using either two-tailed Student’s t-test (α = 0.05) or one-way analysis of variance (ANOVA) with Tukey post-hoc analysis to compare means, or Kruskall–Wallis one-way analysis of variance by ranks and Dunn’s post-hoc analysis to compare medians and observation distributions.
RESULTS
Multicolor Cre fate mapping permits clonal analysis of c-Kit-derived progeny during injury-induced olfactory epithelium reconstitution
To determine the functional capacity of individual stem or progenitor cells via genetic fate mapping, it is necessary to produce separately distinguishable reporter-labeled clusters, which can be considered to be single clones when labeling is sufficiently sparse. Utilizing inducible c-KitCreERT2/+ mice crossed to the R26R-Confetti Cre reporter, we reasoned that low-dose tamoxifen induction would produce sparse single-color reporter-labeled clusters (Fig. 1). The R26R-Confetti mouse utilizes the Brainbow 2.1 construct, four fluorescent protein reporters (XFPs) downstream of a floxed neomycin roadblock aligned in tandem with multiple loxP sites, such that Cre activation will randomly yield expression of one of the four possible XFPs (Livet et al., 2007). This construct was inserted into the Rosa26 locus, which can drive expression in all tissues (Snippert et al., 2010). Thus, crossing the R26R-Confetti reporter to a Cre-driver of interest provides a stochastic multicolor Cre-reporter, which has been applied to other organs, including the heart (Fioret et al., 2014) and intestinal crypt (Snippert et al., 2010). In initial experiments, we confirmed that the 4 fluorescent reporter colors are identifiable and can be discriminated following high-dose tamoxifen (Fig. 2A). We subsequently titrated a single dose of intraperitoneal tamoxifen and determined that, in adult mice, a 1 mg dose is optimal during lesion-induced epithelial renewal (Fig. 2B, C). With this approach, labeled cell clusters identifiable in cryosections examined by confocal microscopy were found to be non-overlapping and comprised by only one of the four possible fluorescent reporters. Therefore, we consider these clusters to be clones that arose from Cre-mediated recombination in a single c-Kit+ founder cell.
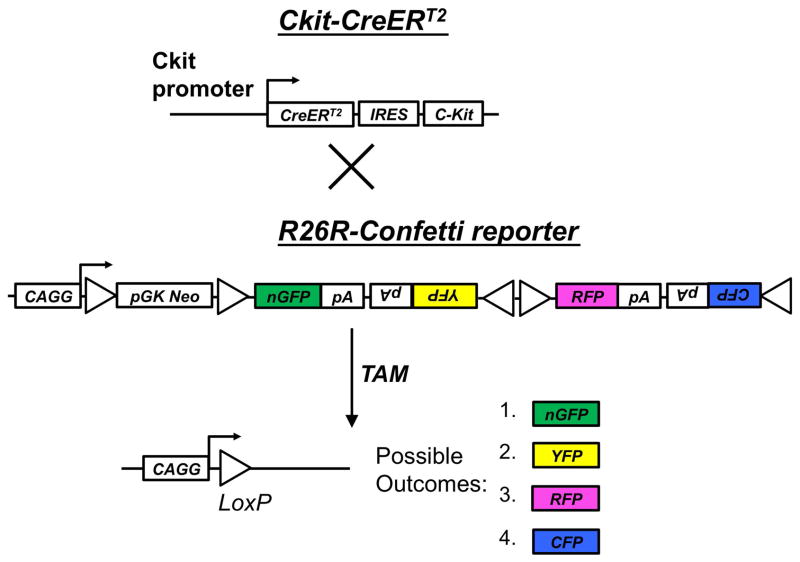
Figure 1.
Genetic strategy for stochastic labeling of c-Kit lineage with one of four possible fluorescent reporters (XFPs). Tamoxifen (TAM)-inducible c-KitCreERT2/+ mice were crossed to the R26R-Confetti reporter, containing the Brainbow 2.1 construct at the R26R locus. Cre recombination leads to removal of the neo transcriptional roadblock, and random recombination leads to expression of one of four possible outcomes: nuclear localized GFP, cytoplasmic RFP or YFP, or membrane associated CFP.
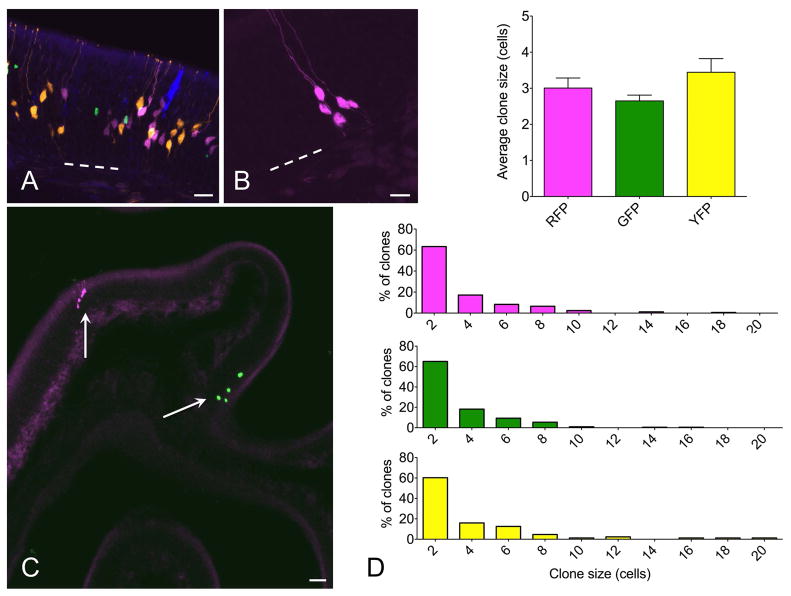
Figure 2.
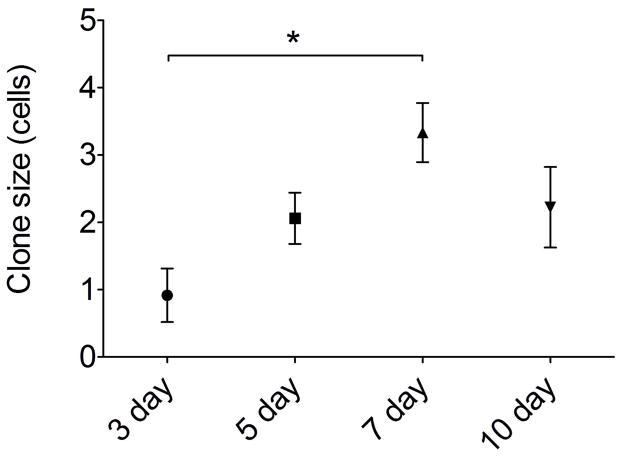
Multicolor fate mapping in olfactory neurogenesis. (A) In initial experiments, c-KitCreERT2/+; R26R-confetti mice were given 0.5 mg tamoxifen at p2 and euthanized at 2 weeks. All four XFPs were identified, with high density labeling precluding clonal assignment of labeled cells. (B, C) Adult mice were lesioned with methimazole and given 1 mg tamoxifen during epithelial reconstitution followed by euthanasia at 14 days. Individual single color non-overlapping XFP-labeled cell clusters were identifiable. At high magnification (B), fine cellular detail allowed definitive assessment of cell type. Here, 5 neurons are labeled by RFP. Low magnification overview demonstrates sparse labeling, permitting clonal assessment. Here, there is a single RFP+ and single nuclear-localized GFP+ clone well separated along a turbinate epithelium (arrows). (D) Quantification indicates that mean clone size and distribution is not different among different XFP-labeled clones. ANOVA for average clone size and Kruskall Wallis test for clone size distributions confirm no statistical differences, n=3 mice per time point. Bar = 25 μm in A, 10 μm in B, 50 μm in C; dashed line indicates basal lamina in A and B.
To stimulate olfactory neurogenesis, the methimazole lesion paradigm was used (Bergman et al., 2002). Following a single dose of methimazole (75 μg/g, IP), the olfactory epithelium degenerates and is then rapidly reconstituted from the activation of basal stem cells (Leung et al., 2007; Goldstein et al., 2015). The robust neuroepithelial renewal provides a useful model for the study of neural stem cells. To examine the activity of c-Kit+ cells in this setting, c-KitCreERT2/+; R26R-Confetti mice were treated with a single dose of tamoxifen (1 mg IP) at either day 3, 5, 7 or 10 following methimazole administration (n=3 mice per time point) and sacrificed at day 14. Analyzing 60 μm thick cryosections by confocal microscopy enabled us to visualize and quantify cell clusters in single sections, minimizing the chance that portions of clones would be distributed among adjacent thin sections (Fig 2B,C). In our hands, the membrane-tethered CFP reporter was often faint, ill defined, and difficult to reliably quantify in the olfactory epithelium. We, therefore, confined our quantitative analysis to RFP, YFP and GFP-labeled clones. Analysis revealed that the distribution of clones was not significantly different by XFP (Fig 2D), counting at least 3 sections per animal, n=3 animals per time point, 4 time points total. In addition, clone size did not differ by XFP color, with approximately 80% of clones comprised by ≤4 cells (Fig 2D).
Microvillar cells arise from a c-Kit+ lineage
Despite intense interest in the regulatory and functional role of olfactory microvillar cells (Jia and Hegg, 2012; Pfister et al., 2012), direct evidence for the source of microvillar cells has been limited. Prior retroviral lineage tracing studies in rats, following methyl bromide-induced epithelial reconstitution, have shown that sustentacular cells can arise from basal cells, duct cells, or via their own mitotic self-renewal (Huard et al., 1998). However, evidence suggests that microvillar cells are likely post-mitotic, and it has been hypothesized that they might arise from basal cells or other cells such as duct progenitors (Pfister et al., 2012). Interestingly, a subset of microvillar cells that express the Trpm5 channel has been found to arise from a basal cell that expresses the Skn1a/Pou2f3 transcription factor (Yamaguchi et al., 2014). The fine cellular detail provided by sparse labeling with the Confetti reporters used here enabled us to clearly detect the presence of olfactory microvillar cells derived from c-Kit+ progenitors (Fig. 3). Microvillar cells, a non-neuronal cell type distinct from the apical sustentacular cells, have a broad cell body situated in the upper third of the epithelium, a wide apical projection, and a slender cytoplasmic process extending to the basement membrane. Biochemically, microvillar cells have been found to be the only cell type within the olfactory epithelium expressing the 5′-exonucleotidase CD73 (Pfister et al., 2012; Pfister et al., 2015). We found that reporter-labeled cells with microvillar morphology were co-labeled immunohistochemically by anti-CD73, confirming their phenotype (Fig. 3A). Neurons were occasionally present in the same clone as a microvillar cell (Fig 3A). Note that neurons are easily distinguished by a round soma with a dendritic knob that is not co-labeled by CD73. Thus, we provide here direct evidence that the c-Kit+ progenitor cell can give rise to microvillar cells. To determine if c-Kit+ cells may produce the Tprm5-expressing subset of microvillar cells, we also combined anti-TrpM5 staining with visualization of c-Kit derived XFP labeled cells (Fig. 3B–E). This co-staining required an antigen retrieval pre-treatment, which eliminated XFP fluorescence. Therefore, we visualized the XFP label using anti-XFP immunostaining. Although Trpm5 protein is not expressed by all microvillar cells (Fig 3B), we could identify examples of XFP+ cells in our c-Kit fate mapping specimens that were co-labeled by anti-Trpm5 (Fig 3C–E). We conclude that c-Kit+ progenitors produce microvillar cells, although these data do not exclude the possibility of other c-Kit-negative sources of microvillar cells. However, the low numbers of XFP microvillar cells identified in the present experiments reflect the low-dose tamoxifen approach utilized, to produce the sparse labeling required for clonal analyses.
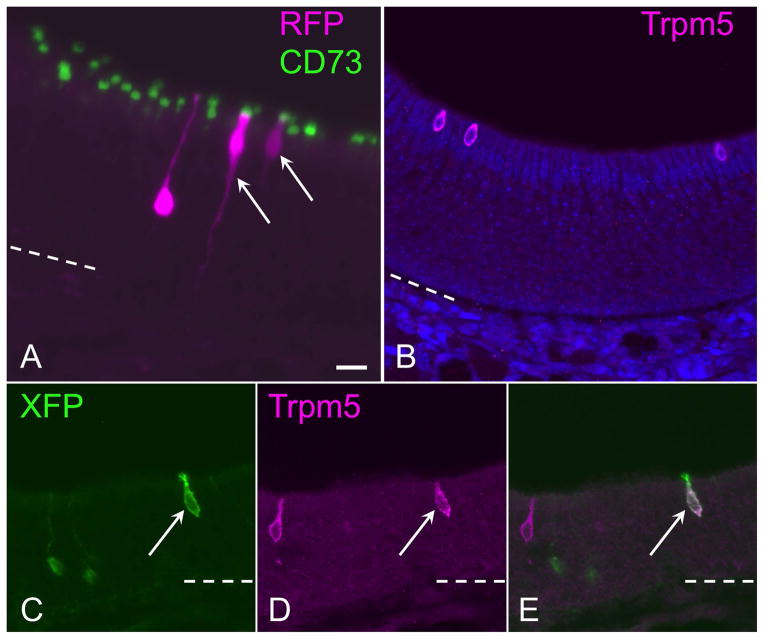
Figure 3.
Microvillar cells arise from c-Kit+ progenitors. (A) CD73, a general marker for olfactory microvillar cells, co-localizes to reporter-labeled c-Kit-derived microvillar cells. An RFP-labeled clone (magenta) contains an olfactory neuron and two microvillar cells (arrows). The neuronal dendrite is evident extending apically, ending in a dendritic knob unlabeled by CD73 (green). CD73 labeling localizes to the apical portion of 2 RFP+ microvillar cells (magenta) in the clone. In this image, a 60 μm thick 3-dimensional Z-stack was rotated 30 degrees along the Y-axis to visualize overlap versus co-localization of labeling. (B) The Trpm5 channel is expressed by only a subset of microvillar cells. Anti-Trpm5 (magenta) labels three microvillar cells in this high-power field of normal adult olfactory epithelium. (C–E) The Trpm5+ subset of microvillar cells can arise from c-Kit+ progenitors. Here, Trpm5 expression and Cre reporter was co-visualized with antibody to XFP (green), which labels any of the confetti reporters, and anti-Trpm5 (magenta). In this field, a c-Kit-derived membrane-tethered XFP+ microvillar cell is co-labeled by anti-Trpm5 (arrow); Trpm5-negative/reporter+ neurons are evident (green), as is a reporter-negative Trpm5+ microvillar cell (magenta). Bar = 20 μm in A; dashed line indicates basal lamina.
Clonality of neurons, microvillar cells and Bowman’s glands
We next analyzed in detail the cellular composition of individual clones (Table 1). Neurons were identifiable by their position and morphology, with their rounded soma, apical dendrite and dendritic knob. Additionally, co-staining with antibody to OMP was used to positively confirm neuronal identity (Fig. 4A–C). Characterization of microvillar cells was described above. Bowman’s glands were found to be easily identifiable by the localization of reporter label in tight groups of acinar cells in the lamina propria, often associated with a duct extending through the overlying neuroepithelium (Fig 4D, E). We previously reported that c-Kit-derived Bowman’s glands express cytokeratin 18, as expected (Goldstein et al., 2015).
Table 1.
Clone analysis; c-KitCreERT2/+; R26R-Confetti adults given low-dose tamoxifen at either 7 or 10 days post methimazole lesion; n=3 mice, 26 coronal sections for each group.
| Clone type | 7 days (clone number) | 7 days (clone size, mean±s.d, range) | 10 days (clone number) | 10 days (clone size, mean±s.d, range) |
|---|---|---|---|---|
| Neuron only | 399 | 2.7±2.3 (1–17) | 85 | 3.1±2.9 (1–14) |
| Microvillar cells only | 34 | 1.4± 0.9 (1–6) | 9 | 1.2±0.7 (1–3) |
| Mixed neuron and microvillar | 23 | 8.8± 7.5 (2–34) | 10 | 6.1±4.4 (2–15) |
| Gland/duct | 5 | (range 6–50) | 0 | n/a |
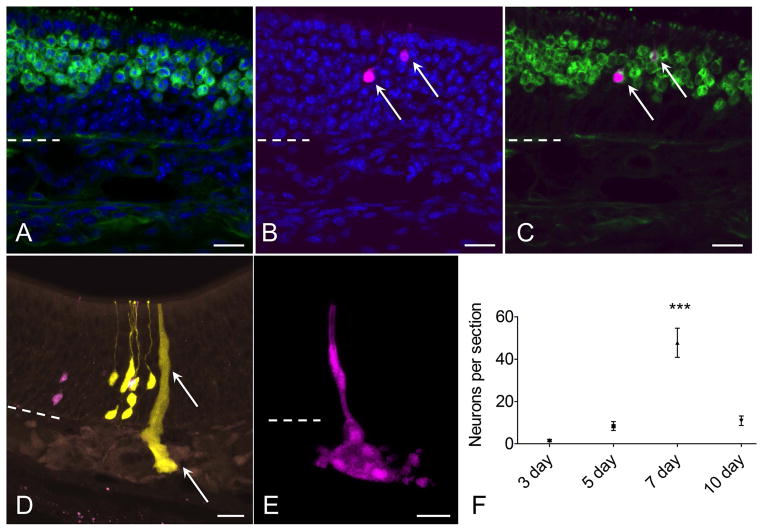
Figure 4.
Characterization of neurons and glands derived from c-Kit+ cells. (A–C) RFP+ (magenta) c-Kit-derived neuron phenotype is confirmed by co-labeling with antibody to olfactory marker protein (OMP, green, arrows). (D, E) Bowman’s glands and ducts also arise from c-Kit+ cells. In a section from a mouse given high-dose tamoxifen, RFP+ and YFP+ neurons are evident, along with a YFP+ gland and duct (arrow). Although these overlapping cells cannot be considered clonally related, the image demonstrates XFP reporter expression in glands, ducts and neurons. (E) From a low-dose TAM-treated mouse, an entire gland and duct RFP+ clone can be visualized from a 60 μm thick z-stack image. (F) Quantification of neurons per section indicates that neuron production from c-Kit+ globose basal cells is highest in mice given TAM at day 7 post lesion (P<0.001, ANOVA, n=3 mice per time point). Dashed line marks basal lamina. Bar = 20 μm in (A–E).
Quantification of clone size and cellular composition across the time points sampled during olfactory reconstitution was performed. These data indicated that c-Kit+ cells are most active at 7 days following lesion, with 50 ± 10 (SEM) XFP labeled neurons per section (Fig 4F). This represents approximately a 10-fold increase versus labeling in mice given tamoxifen at 5 days post lesion. There was a broad distribution of clone sizes, with the majority of clones being ≤4 cells (Fig 5). However, a number of larger neuron-containing clones were also identified (e.g. Fig 2D). Mean clone size was also largest in mice given tamoxifen at day 7 post lesion (Fig 5), with overall mean number of cells per clone of 3.33 ± 0.76 (SEM, n=3 mice). We interpret these results as evidence that c-Kit+ basal cells may behave as immediate precursors or as amplifying progenitors. Immediate precursors would produce only one or two-cell clones, whereas amplifying progenitors would produce larger clones.
Figure 5.
Neuron and microvillar clone size is greatest in mice given tamoxifen at day 7 following experimental lesion. XFP+ cells in the olfactory epithelium were quantified per clone; gland clones were excluded as they were generally very large clones (10–30 cells per gland). By ANOVA, overall clone size at day 7 (3.33±0.44 cells; SEM) was significantly greater compared to day3 (0.92±0.39 cells; P=0.015); other comparisons were not significantly different, n=3 mice per time point.
Interestingly, clones comprised by neurons only, microvillar cells only, or both neurons and microvillar cells were identified (e.g. Fig. 3). Thus, c-Kit+ basal cells do not appear to be strictly lineage-committed. Clone composition details are summarized in Table 1, for 7 and 10 day post-methimazole animals. Although mean clone size peaked at 7 days, the presence of mixed clones, comprised by microvillar cells and neurons, was greatest in mice that received tamoxifen induction at 10 days post-methimazole. At 7 days, 5% of epithelial clones were mixed, and this increased to 9.6% of clones at 10 days. However, Bowman’s gland and duct clones were always separate from microvillar cells or neurons, indicating that the c-Kit+ gland/duct progenitor is a distinct committed lineage. Gland clones were often very large, containing 20 or more cells (Fig 4E), always single-color clones, and were most also prominent in tissue from mice treated with tamoxifen at day 7 after methimazole.
Long term repopulating ability of c-Kit+-derived cells
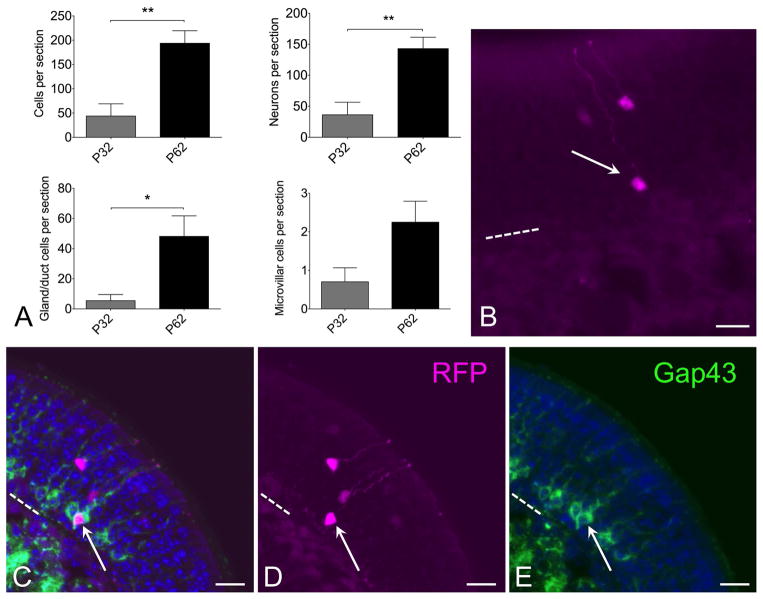
To determine whether progeny of c-Kit+ basal cells might function as reserve cells or contribute to ongoing neuroepithelial renewal, c-KitCreERT2/+; R26R-confetti mice were given a single low dose of tamoxifen (0.0125 mg IP) at postnatal day 2, and were euthanized at either day 32 or day 62 (Fig 6). Quantification of total labeled c-Kit+ progenitor-derived neurons per section revealed that there are a significantly greater number of labeled neurons in mice killed at p62 compared to p32 (Fig 6). There is potential variation in amount of reporter labeling among animals, despite identical tamoxifen dosing. Therefore, we treated two entire litters at p2 with tamoxifen, and randomly euthanized half of each litter at either p32 or p62, yielding n=5 mice at p32 and 6 mice at p62. Quantification revealed a mean of 36±20 (SEM) labeled neurons per section at p32 and 143±18 labeled neurons per section at p62, indicating a significant increase by t-test (p=0.0036; unpaired, two-tailed). In addition, total labeled cells per section, including glands, microvillar cells or neurons, was increased at p62 (Fig 6A). Counting the gland or duct labeled cell increase separately achieved significance. Counting labeled microvillar cells separately trended towards an increase but was not significant (Fig 6A). Taken together, we interpret this finding as evidence that progeny of the c-Kit+ cells present at p2, when tamoxifen was given, continue to produce new neural precursor cells beyond 1 month. Thus, at least a subset of c-Kit+ cells function as either a reserve cell or as ongoing amplifying progenitors.
Figure 6.
Progeny of c-Kit+ cells labeled at p2 continue to contribute to neuron production long term. (A) Quantification of XFP labeled cells from mice given a single dose of tamoxifen at p2 and killed at either p32 or p62 reveals increased c-Kit-derived cells at p62. Graphs show comparisons by total cells or by individual cell types. Note that neuronal counts indicate a significant increase at p62 versus p32 (t-test, P<0.01, n=5–6 mice per time point). Increased c-Kit-derived neuron label at longer survivals is consistent with ongoing c-Kit+ progenitor activity. (B) Both mature and new c-Kit-derived neurons are evident in olfactory epithelium from C-KitCreERT2/+; R26R-confetti mice treated with tamoxifen at p2 and killed at p62. RFP labeled neurons are evident in apical (mature) layers as well as basal (immature) layer (arrow). (C–E) Gap43 staining co-localizes with RFP+ immature neuron (arrow) in sections from c-KitCreERT2/+; R26R-confetti mice treated with tamoxifen on p2 and sacrificed p62. Older mature neurons are located more apically, in the Gap43 (−) layers. The presence of Gap43 (+)/XFP(+) neurons in this tissue reflects recent c-Kit+ neurogenesis. Nuclei are labeled with DAPI (blue). Dashed line marks basal lamina in (B–E); bar = 20 μm in (B–E).
Consistent with this interpretation, we observe that many of the labeled neurons identifiable in mice killed at p62 are situated deep in the neuroepithelium (Fig 6B). The olfactory epithelium is pseudostratified such that new, immature neurons are situated closer to the basement membrane, just above the globose basal cells. More mature, fully differentiated neurons are localized higher in the epithelium, in the OMP+ zone (e.g. Fig 4A–C). Tissue from mice killed at p62 would not contain labeled neurons situated in the deep layers if they had been produced over a month earlier; rather all labeled neurons would be situated much higher in the epithelium. To confirm that some of the XFP labeled neurons present in tissue from mice treated with tamoxifen day p2 and killed day 62 are new immature cells, sections were probed with antibody to Gap43, which specifically localizes only to new neurons situated just apical to the basal cell layers (Fig 6C–E). As expected, some XFP labeled neurons located basally were co-labeled by anti-Gap43. This finding provides strong evidence that these neurons were produced quite recently from progeny of c-Kit+ cells that underwent Cre-mediated recombination on postnatal day 2, i.e. that at least some c-Kit+ cells behave as stem cells.
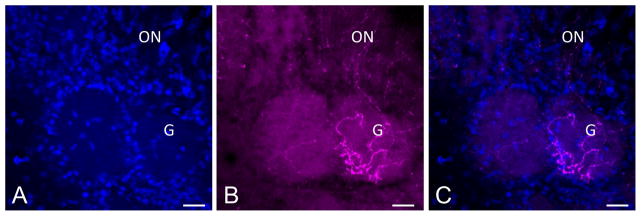
Finally, the excellent cytoplasmic distribution of RFP and YFP reporter protein enabled us to confirm that c-Kit-derived neurons do indeed project their axons to the glomerular layer of the olfactory bulb (Fig 7). Labeled axons can be visualized in the olfactory nerve layer of the bulb, with terminations present within glomeruli, the site of synapse with mitral and tufted cells, providing added confirmation that many c-Kit-derived cells are fully mature neurons.
Figure 7.
(A–C) Mature olfactory receptor neurons arising from c-Kit+ cells innervate the olfactory bulb. C-KitCreERT2/+; R26R-confetti mouse was treated with tamoxifen on p2 and sacrificed p62. RFP+ axons are evident innervating a glomerulus (G) in the olfactory bulb. Olfactory nerve layer is marked (ON). The excellent cytoplasmic distribution of RFP in this model may be useful for studies assessing glomerulus innervation. Nuclei are labeled with DAPI (blue). Bar = 50 μm.
DISCUSSION
Neurogenesis occurs in the adult olfactory area to maintain the self-renewing epithelium, including the population of primary olfactory receptor neurons (Graziadei and Graziadei, 1979). Because of ongoing neuron production and rapid lesion-induced reconstitution, the olfactory tissue provides a widely used model system to study adult neural stem and progenitor cells (Schwob, 2002; Leung et al., 2007). Of the various markers identifying stem and/or progenitor olfactory basal cells, we reported recently that c-Kit, a cell-surface receptor, defines a population of neurogenic globose basal cells (Goldstein et al., 2015). The c-Kit+ population is of particular interest, because, using targeted diphtheria toxin-mediated cell ablation, we found that adult c-Kit+ cells are necessary for olfactory maintenance. Several lines of evidence have indicated that the globose basal cell population is functionally heterogeneous, with multipotent or lineage committed cells, immediate precursors, transit amplifying progenitors, and label-retaining reserve stem cells (Gordon et al., 1995; Goldstein et al., 1998; Huard et al., 1998; Chen et al., 2014; Jang et al., 2014). Given this heterogeneity, we sought here to determine the functional potential of individual c-Kit+ basal cells. The recent development of a multicolor Cre reporter system placed at the R26R locus provides a tool for genetic fate mapping at clonal resolution (Snippert et al., 2010). Here, we report the application of this system to the study of an adult neural progenitor population.
We reasoned that inducible c-KitCreERT2/+; R26R-confetti mice treated with low-dose tamoxifen induction would yield sparse, non-overlapping single color clusters that could be considered clonal (Fioret et al., 2014). Hypothetically, several outcomes might occur: (1) all clones could be comprised by one or two cells, indicating that the c-Kit+ cell is an immediate precursor undergoing a terminal mitosis; (2) clones could be distributed widely in size, consistent with c-Kit expression in transit amplifying progenitors, some of which re-enter the cell cycle repeatedly; (3) all clones could consist of a single cell type, if c-Kit+ cells are strictly lineage-committed; or (4) clones could consist of neurons and non-neuronal cells, if single c-Kit+ cells are multipotent.
Our results show that the c-Kit+ population functions as transit amplifying and immediate precursor cells. Although the majority of clones were comprised by one cell type, a subset of c-Kit+ cells are multipotent, producing both neurons and microvillar cells. That microvillar cells arise from c-Kit+ globose basal cells is also a novel finding of importance, since the microvillar cells are key regulators of basal stem cells (Jia and Hegg, 2012). In addition, experiments in which a single pulse of tamoxifen was used to induce recombination followed by long survival times demonstrated that at least a subset of c-Kit+-derived progeny continue to produce neurons long term. Finally, Bowman’s glands arise from lineage committed c-Kit+ progenitors that can produce entire acinar structures and ducts as single large clones.
Collectively, the data provided here regarding the role of c-Kit+ progenitors in olfactory tissue homeostasis, combined with previous data on globose basal cell properties, indicate the remarkable potential of adult nasal epithelium. There are obvious therapeutic implications for understanding the biology of defined olfactory progenitors. For instance, important clinical problems lacking treatment options include acquired hyposmia or anosmia conditions, such as post-viral olfactory disorder or post-head trauma olfactory loss (Goldstein and Lane, 2004; Holbrook and Leopold, 2006). The rational development of novel therapies requires an understanding of the cell types and regulatory mechanisms of olfactory tissue maintenance; the c-Kit+ population has a key role in this process. Additionally, the possibility of utilizing the nose as a source of autologous adult neural stem or progenitor cells has gained attention as the field of regenerative medicine advances rapidly (Tabakow et al., 2014), with particular interest from the lay press recently (Guest and Dietrich, 2014). The importance of c-Kit+ basal cells described here raises the possibility of therapeutic manipulation of this population, as c-Kit is expressed on the cell surface and has been a useful marker for stem cell purification in other tissue models (Rangel et al., 2013).
In summary, we have applied multicolor fate mapping using R26R-confetti mice to delineate the differentiation potential of olfactory c-Kit+ progenitors. Our findings indicate that the c-Kit+ population contains transit amplifying progenitors, multipotent cells capable of producing neurons and microvillar cells, and also gland-committed progenitors. These data provide new perspectives on the process of adult neurogenesis and olfactory maintenance.
Acknowledgments
This work was supported by NIH grant K08DC013556 to BJG.
Footnotes
The authors report no conflicts of interest.
References
- Bergman U, Ostergren A, Gustafson AL, Brittebo B. Differential effects of olfactory toxicants on olfactory regeneration. Arch Toxicol. 2002;76:104–112. doi: 10.1007/s00204-002-0321-2. [DOI] [PubMed] [Google Scholar]
- Calof AL, Bonnin A, Crocker C, Kawauchi S, Murray RC, Shou J, Wu HH. Progenitor cells of the olfactory receptor neuron lineage. Microsc Res Tech. 2002;58:176–188. doi: 10.1002/jemt.10147. [DOI] [PubMed] [Google Scholar]
- Chen M, Tian S, Yang X, Lane AP, Reed RR, Liu H. Wnt-responsive Lgr5(+) globose basal cells function as multipotent olfactory epithelium progenitor cells. J Neurosci. 2014;34:8268–8276. doi: 10.1523/JNEUROSCI.0240-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fioret BA, Heimfeld JD, Paik DT, Hatzopoulos AK. Endothelial cells contribute to generation of adult ventricular myocytes during cardiac homeostasis. Cell Rep. 2014;8:229–241. doi: 10.1016/j.celrep.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher RB, Prasol MS, Estrada J, Baudhuin A, Vranizan K, Choi YG, Ngai J. p63 regulates olfactory stem cell self-renewal and differentiation. Neuron. 2011;72:748–759. doi: 10.1016/j.neuron.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokoffski KK, Wu HH, Beites CL, Kim J, Kim EJ, Matzuk MM, Johnson JE, Lander AD, Calof AL. Activin and GDF11 collaborate in feedback control of neuroepithelial stem cell proliferation and fate. Development. 2011;138:4131–4142. doi: 10.1242/dev.065870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein BJ, Fang H, Youngentob SL, Schwob JE. Transplantation of multipotent progenitors from the adult olfactory epithelium. Neuroreport. 1998;9:1611–1617. doi: 10.1097/00001756-199805110-00065. [DOI] [PubMed] [Google Scholar]
- Goldstein BJ, Goss GM, Hatzistergos KE, Rangel EB, Seidler B, Saur D, Hare JM. Adult c-Kit(+) progenitor cells are necessary for maintenance and regeneration of olfactory neurons. J Comp Neurol. 2015;523:15–31. doi: 10.1002/cne.23653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein BJ, Lane AP. Future directions in chemosensory research. Otolaryngol Clin North Am. 2004;37:1281–1293. doi: 10.1016/j.otc.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Goldstein BJ, Schwob JE. Analysis of the globose basal cell compartment in rat olfactory epithelium using GBC-1, a new monoclonal antibody against globose basal cells. J Neurosci. 1996;16:4005–4016. doi: 10.1523/JNEUROSCI.16-12-04005.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MK, Mumm JS, Davis RA, Holcomb JD, Calof AL. Dynamics of MASH1 expression in vitro and in vivo suggest a non-stem cell site of MASH1 action in the olfactory receptor neuron lineage. Mol Cell Neurosci. 1995;6:363–379. doi: 10.1006/mcne.1995.1028. [DOI] [PubMed] [Google Scholar]
- Graziadei GA, Graziadei PP. Neurogenesis and neuron regeneration in the olfactory system of mammals. II. Degeneration and reconstitution of the olfactory sensory neurons after axotomy. J Neurocytol. 1979;8:197–213. doi: 10.1007/BF01175561. [DOI] [PubMed] [Google Scholar]
- Guest J, Dietrich DW. Letter to the editor. Commentary regarding the recent publication by Tabarow et al, Functional regeneration of supraspinal connections in a patient with transected spinal cord following transplantation of bulbar olfactory ensheathing cells with peripheral nerve bridging. J Neurotrauma. 2014 doi: 10.1089/neu.2014.3790. [DOI] [PubMed] [Google Scholar]
- Holbrook EH, Leopold DA. An updated review of clinical olfaction. Curr Opin Otolaryngol Head Neck Surg. 2006;14:23–28. doi: 10.1097/01.moo.0000193174.77321.39. [DOI] [PubMed] [Google Scholar]
- Huard JM, Youngentob SL, Goldstein BJ, Luskin MB, Schwob JE. Adult olfactory epithelium contains multipotent progenitors that give rise to neurons and non-neural cells. J Comp Neurol. 1998;400:469–486. [PubMed] [Google Scholar]
- Jang W, Chen X, Flis D, Harris M, Schwob JE. Label-retaining, quiescent globose basal cells are found in the olfactory epithelium. J Comp Neurol. 2014;522:731–749. doi: 10.1002/cne.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang W, Lambropoulos J, Woo JK, Peluso CE, Schwob JE. Maintaining epitheliopoietic potency when culturing olfactory progenitors. Exp Neurol. 2008;214:25–36. doi: 10.1016/j.expneurol.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia C, Hegg CC. Neuropeptide Y and extracellular signal-regulated kinase mediate injury-induced neuroregeneration in mouse olfactory epithelium. Mol Cell Neurosci. 2012;49:158–170. doi: 10.1016/j.mcn.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S, Seidler B, Kettenberger A, Sibaev A, Rohn M, Feil R, Allescher HD, Vanderwinden JM, Hofmann F, Schemann M, Rad R, Storr MA, Schmid RM, Schneider G, Saur D. Interstitial cells of Cajal integrate excitatory and inhibitory neurotransmission with intestinal slow-wave activity. Nat Commun. 2013;4:1630. doi: 10.1038/ncomms2626. [DOI] [PubMed] [Google Scholar]
- Leung CT, Coulombe PA, Reed RR. Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat Neurosci. 2007;10:720–726. doi: 10.1038/nn1882. [DOI] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Pfister S, Dietrich MG, Sidler C, Fritschy JM, Knuesel I, Elsaesser R. Characterization and turnover of CD73/IP(3)R3-positive microvillar cells in the adult mouse olfactory epithelium. Chem Senses. 2012;37:859–868. doi: 10.1093/chemse/bjs069. [DOI] [PubMed] [Google Scholar]
- Pfister S, Weber T, Hartig W, Schwerdel C, Elsaesser R, Knuesel I, Fritschy JM. Novel role of cystic fibrosis transmembrane conductance regulator in maintaining adult mouse olfactory neuronal homeostasis. J Comp Neurol. 2015;523:406–430. doi: 10.1002/cne.23686. [DOI] [PubMed] [Google Scholar]
- Rangel EB, Gomes SA, Dulce RA, Premer C, Rodrigues CO, Kanashiro-Takeuchi RM, Oskouei B, Carvalho DA, Ruiz P, Reiser J, Hare JM. C-kit(+) cells isolated from developing kidneys are a novel population of stem cells with regenerative potential. Stem Cells. 2013;31:1644–1656. doi: 10.1002/stem.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob JE. Neural regeneration and the peripheral olfactory system. Anat Rec. 2002;269:33–49. doi: 10.1002/ar.10047. [DOI] [PubMed] [Google Scholar]
- Schwob JE, Youngentob SL, Mezza RC. Reconstitution of the rat olfactory epithelium after methyl bromide-induced lesion. J Comp Neurol. 1995;359:15–37. doi: 10.1002/cne.903590103. [DOI] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, Clevers H. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Tabakow P, Raisman G, Fortuna W, Czyz M, Huber J, Li D, Szewczyk P, Okurowski S, Miedzybrodzki R, Czapiga B, Salomon B, Halon A, Li Y, Lipiec J, Kulczyk A, Jarmundowicz W. Functional regeneration of supraspinal connections in a patient with transected spinal cord following transplantation of bulbar olfactory ensheathing cells with peripheral nerve bridging. Cell Transplant. 2014;23:1631–1655. doi: 10.3727/096368914X685131. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Yamashita J, Ohmoto M, Aoude I, Ogura T, Luo W, Bachmanov AA, Lin W, Matsumoto I, Hirota J. Skn-1a/Pou2f3 is required for the generation of Trpm5-expressing microvillous cells in the mouse main olfactory epithelium. BMC Neurosci. 2014;15:13. doi: 10.1186/1471-2202-15-13. [DOI] [PMC free article] [PubMed] [Google Scholar]