Abstract
The mosquitocidal crystal protein, Cry19Aa, from Bacillus thuringiensis ssp. jegathesan, has high toxicity to Anopheles stephensi and Culex pipiens but is less toxic to Aedes aegypti. To study the functional role of putative domain II and surface residues in mosquito toxicity, 16 alanine substitution mutations were introduced into Cry19Aa. All mutant constructs were expressed as 65-kDa protoxins and subsequently digested by trypsin to produce further fragmented polypeptides of 40 and 25 kDa. With chymotrypsin, however, most protoxins were digested to 60 kDa and minor bands. The circular dichroism spectra of the chymotrypsin-activated toxins of Cry19Aa and muteins, Y324A, W357A, Y412A, Y414A, W416A, D418A and F485A indicated that there was no significant variation in their structure. In mosquito bioassays, Y324A, W357A, Y410A, W416A, D418A and F485A muteins showed substantial reductions in mosquitocidal activity toward A. aegypti and C. pipiens. These muteins also showed reduced competition with wild-type fluorescein 5-isothiocyanate-labeled Cry19Aa for binding to C. pipiens brush border membrane vesicles. These data suggest that the reduction of toxicity was a result of the reduced binding affinity. From these studies we have identified loop residues of domain II that are important in toxicity and receptor binding to Culex larval midgut.
Keywords: Bacillus thuringiensis, Cry19Aa, mutagenic analysis, competition binding
Introduction
Biopesticides from Bacillus thuringiensis ssp. israelensis and Bacillus sphaericus have been successfully used worldwide as biological control agents for mosquito and black fly (Becker & Margalit, 1993; Porter et al., 1993). Bacillus thuringiensis ssp. israelensis crystals are composed of four major polypeptides: Cry4Aa, Cry4Ba, Cry11Aa and Cyt1Aa (125, 135, 68 and 28 kDa in molecular mass, respectively) (Crickmore et al., 1998). Bacillus sphaericus has a binary toxin consisting of two proteins of 51 and 42 kDa (Charles et al., 1996). These crystal proteins are very highly toxic to important human disease vectors such as Anopheles, Aedes and Culex species. However, field resistance to B. sphaericus has been reported in Culex species (Silva-Filha & Regis, 1997). Although there are no reports of field resistance B. thuringiensis ssp. israelensis, laboratory selections using cloned israelensis toxins have shown that resistance can develop to individual Cry toxins (Georghiou & Wirth, 1997).
To date there have been several reports of B. thuringiensis strains of various serotypes with mosquito activity, such as ssp. morrisoni, canadiensis thompsoni, medellin, malaysiensis and jegathesan. Among them, B. thuringiensis ssp. jegathesan serotype H28a28c has been the focus of attention as an alternative to B. thuringiensis ssp. israelensis against Anopheles. Unfortunately it is less active against Aedes and Culex (Seleena et al., 1995). Bacillus thuringiensis ssp. jegathesan crystal is composed of seven major proteins of 80, 70, 65, 37, 26 and 16 kDa, which are not related to proteins of israelensis (Ragni et al., 1996). The genes encoding the 80 and 65 kDa have been cloned and referred to as Cry11Ba and Cry19Aa (Delécluse et al., 1995; Rosso & Delecluse, 1997). Cry11B was highly toxic for mosquito larvae of the species Aedes aegypti, Culex pipiens and Anopheles stephensi compared with Cry11A. Cry19Aa is highly toxic to C. pipiens, Culex quinquefasciatus and A. stephensi but has low activity against A. aegypti (Rosso & Delecluse, 1997). Cry19Aa, about 65 kDa in size, and Orf2 are coexpressed as one operon in B. thuringiensis cells. Orf2 might be important to stabilize expression of Cry19Aa (Rosso & Delecluse, 1997). Homolog scanning and site-directed mutagenesis techniques have been conducted extensively within each domain to localize regions responsible for toxicity, receptor binding and pore formation in B. thuringiensis Cry toxins. In this study, we demonstrate that alanine substitution of loop regions in domain II of Cry19Aa significantly affect larvicidal potency for Culex larvae.
Materials and methods
Construction of alanine substitution mutants
The putative loop regions were predicted by modeling the structure of Cry19Aa (Abdullah & Dean, 2004) and aligning the amino acid sequences of Cry4Ba and Cry19Aa with CLUSTAL X ver. 1.83. The residues of putative loops 0, 1, 2 and 3 of Cry19Aa and two surface residues were mutated to alanines. Y324 and F325 in loop 0, Y356 and W357 in loop 1, Y410, E411, Y412, I413, Y414, W416, D418 and V420 in loop 2 and T484 and F485 in loop 3 were mutated. Additionally, surface residues, T390 and T610 were chosen for alanine substitution because they were predicted to be located in a receptor proximal position on the Cry19Aa structure. The template DNA, pJEG65.5 was a kind gift from A. Delécluse (Pasteur Institute, Paris, France). Alanine substitution was performed by the site-directed mutagenesis method based on QuikChange manufacturer’s manual (Stratagene). Mutations were confirmed by automated DNA sequencing (Plant–Microbe Genomics Facility, The Ohio State University).
Expression and activation of Cry19Aa muteins
Each mutated construct was transformed into B. thuringiensis 4Q7 strain by electroporation using a Gene Pulser (Bio-Rad) under the conditions of 2.5 kV, 25 μF and 400 Ω. Transformants were cultured in Schaeffer sporulation media (Schaeffer et al., 1965) with 25 μg erythromycin mL−1 and grown for 72 h at 30 °C in an incubator–shaker at 200 r.p.m. Cells were harvested and lysed, and crystal inclusions were washed as described previously (Lee et al., 1996) except for ultrasonication step (output level 5.0, 10 s on/10 s off for 5 min, two times) with Sonicator 3000 (Misonix). Crystal inclusion protein was solubilized in carbonate buffer (30 mM Na2CO3, 20 mM NaHCO3, 10 mM dithiothreitol, pH 11.5), and protoxin was digested with L-1-tosyl amino-2 phenylethyl-chloromethyl ketone (TPCK)-treated trypsin or N-α-tosyl-L-lysine chloromethyl ketone (TLCK)-treated α-chymotrypsin with a protease : protoxin ratio of 1 : 20 (w/w) for 2 h at 37 °C. Protoxin and toxins were analyzed by sodium dodecyl sulfate-polyacrylamide gel (10%) electrophoresis (SDS-PAGE) as described by Laemmli (1970). For further experiments, the chymotrypsinized toxin was purified by anion exchange chromatography with a Q-sepharose (Pharmacia) column using salt elution (0.2, 0.3, 0.4 and 0.6 M NaCl) and HPLC with a Superdex 200 (Pharmacia) column, sequentially.
Circular dichroism (CD) spectroscopy
CD spectra of wild-type and mutant toxins were collected with an Aviv Circular Dichroism spectrophotometer-model 62A DS (Lakewood, NJ) in a 21-Q-1 quartz cuvette at 25 °C using STAR STATIONARY 3.0 software. The purified toxins were at 2 μM in the carbonate buffer (30 mM Na2CO3, 20 mM NaHCO3, pH 10). Data shown were based on scanning from 300 to 200 nm at 1.0-nm steps and averaged from five scans.
Toxicity assays
Mosquitoes were reared in an environment-controlled room at 28 °C and 85% humidity, with a photoperiod of 14 h of light and 10 h of dark. The C. pipiens (recently isolated from nature in Ohio) were provided to us by R. Robish and W. Foster (Ohio State University) and A. aegypti cultures were originally obtained from A. Yousten (Virginia Polytechnic Institute). Fourth-instar larvae were used for Culex bioassays and second-instar larvae were used for Aedes. In Culex bioassays, a total of 30 larvae per 30 mL of distilled water with one replicate in a 24-well tray [one well is 5.5 (H) × 5.5 (W) × 5.5 (D) cm in size] were fed on a serial dilution of Cry toxins (as inclusions), and the number of mortalities were counted after a 24-h incubation at 28 °C. The 50% lethal concentration (LC50) was calculated by a Probit method using SOFTTOX version 1.1 (WINDOWCHEM). In the Aedes bioassay, a total of six larvae per 2.5 mL with triplicate in a 24-well Costar cell culture plate (Corning) were fed and mortality (%) was checked after 48 h.
Preparation of brush border membrane vesicles (BBMV)
Fourth-instar C. pipiens larvae were filtered and dried briefly on a filter paper (Fisher) under vacuum suction. Harvested larvae were frozen at −80 °C until needed. BBMV were prepared from whole C. pipiens larvae as described (Abdul-Rauf & Ellar, 1999). The BBMV pellet was resuspended in binding buffer (60 mM K2HPO4, 5 mM KH2PO4, 150 mM NaCl, 10 mM EGTA, pH 7) supplemented with complete protease inhibitor cocktail (Roche). The BBMV were used on the day of preparation. Leucine aminopeptidase and alkaline phosphatase activity were determined as described previously (Lowry et al., 1954; Erlanger et al., 1961). Culex pipiens BBMV was averagely enriched about fivefold in aminopeptidase activity and 4.5-fold in alkaline phosphatase over that in the homogenate.
Fluorescein 5-isothiocyanate (FITC) labeling
Cry19Aa toxin (in 30 mM Na2CO3, 20 mM NaHCO3, pH 10) purified on a Superdex 200 gel filtration column using AKTA 1000 FPLC (GE Healthcare), was incubated with FITC (Anaspec Inc.) in a 1 : 100 M ratio for 4 h at room temperature. The labeled toxin was separated from the nonbound FITC using a PD-10 column (GE Healthcare) and verified on a UV transilluminator (312 nm) after SDS-PAGE separation. The fluorescence was measured on a Fluoromax-3 spectrofluorometer (JY Horiba) with excitation at 494 nm and emission at 518 nm using DATAMAX ver. 2.10 software.
Competition binding assay
Homologous and heterologous competition binding assays with FITC-labeled Cry19Aa were performed in triplicate in a total volume of 200 μL of binding buffer containing 0.1% (w/v) bovine serum albumin. Culex pipiens BBMV (10 μg mL−1) was incubated with 2 nM FITC-labeled toxin with increasing amounts of unlabeled toxin (0–2000 nM) and 100 nM unlabeled Cry19Aa as a blocker of productive binding sites for 1 h at room temperature. The reaction mixture was centrifuged at 30 000 g for 15 min to separate unbound labeled toxin from the BBMV. The fluorescence of bound labeled toxin was obtained by subtracting a fluorescence of the supernatant from the fluorescence of 2 nM FITC-labeled Cry19Aa toxin. To verify binding of Cry19Aa along with increasing unlabeled D418A, 100 nM unlabeled D418A was added as a blocker with or without C. pipiens BBMV. Data were plotted with SIGMAPLOT ver. 8.0 (SPSS Inc.) and the experiment was repeated with unlabeled muteins as competitors.
Results
Alignment and alanine substitutions
For mutation analysis, a homology model structure of Cry19Aa was obtained from a modeled structure of Cry4Ba as a template (Abdullah & Dean, 2004). Recently, the Cry4Ba structure was solved by Boonserm et al. (2005), but we could not acquire the Cry19Aa model from the coordinates of the solved Cry4Ba structure. Thus, the two sequences were aligned (Fig. 1) using CLUSTAL X (ver. 1.83) in order to compare loop regions of domain II. This alignment indicated that loops of the determined Cry4Ba structure were similar to our predicted Cry19Aa model. In the present study, loops of domain II and predicted surface residues in Cry19Aa were analyzed to localize regions and residues for toxicity by alanine substitution mutagenesis.
Fig. 1.
Amino acid sequence alignment of Cry19Aa with Cry4Ba. This was analyzed by CLUSTAL X (ver. 1.83). Upper- and underlines indicate loops in domain II of Cry4Ba and Cry19Aa, respectively. Circled T residues indicate predicted surface residues that were mutated. *, exact amino acid match; :, similar amino acid properties; ., different amino acid properties.
Mutations in loop 0 (319GLMTPYF325), in loop 1 (354ISYW357), in loop 3 (483ETFG486) and at two surface residues (T390 and T610) were made based on a residue’s exposed position and hydrophobicity. Eight alanine mutants were made in all of the loop 2 residues (410YEYIYPWGDV420), except for P415 and G417, because those residues could introduce structural modification in wild-type toxin. In addition, two protruding threonines on Cry19Aa structure were mutated.
Activation of protoxins and structural analysis
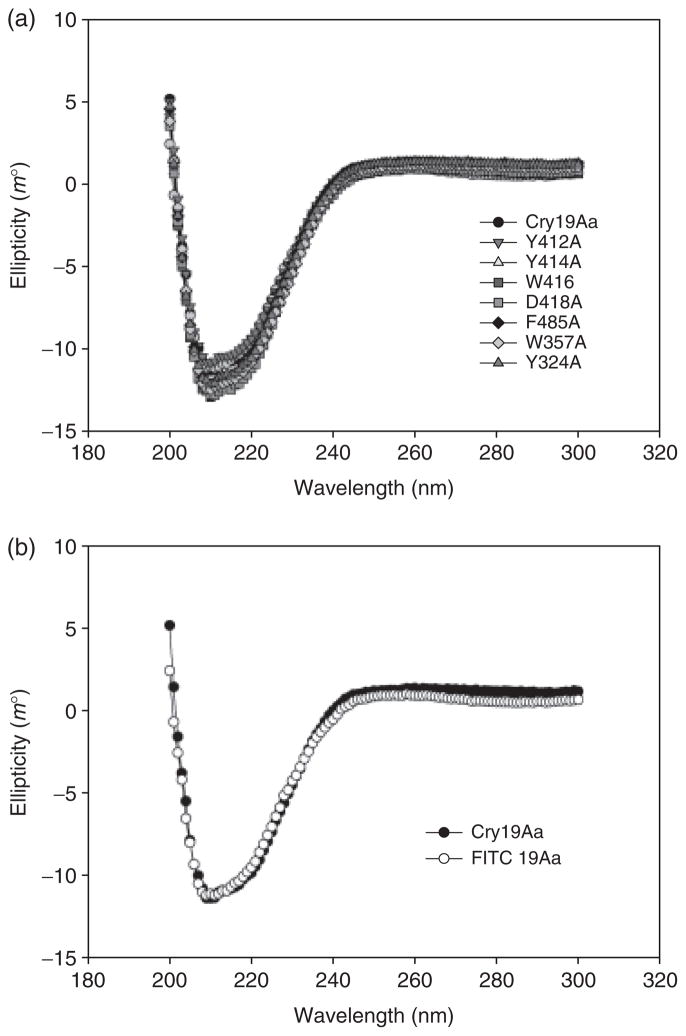
All the muteins were expressed in amounts comparable to wild type (Fig. 2a). The predominant bands of expressed proteins visualized on SDS-PAGE were 65 kDa. Subsequent digestion by protease and CD spectrum analysis were performed as presumptive experiments of global folding fidelity (Almond & Dean, 1993). The protoxins of all mutants were digested beyond the normal 60-kDa toxic core to 40 and 25 kDa if treated with trypsin (Fig. 2b). Chymotrypsin digested the protoxins of most mutants and wild type of Cry19Aa to 60 kDa and minor polypeptides of 32 kDa (Fig. 2c). Although all mutants produced stable protoxins, F325A, Y410A, V420A and T484A muteins were more sensitive to chymotrypsin than wild type, resulting in degradation of 60-kDa toxin. To prepare active toxins for further experiments such as CD analysis and binding assay, protoxins were treated by chymotrypsin because it produced dominant 60-kDa toxin. CD spectra were also obtained to investigate whether the mutations introduced significant structural changes. The CD spectra of the activated toxins of Cry19Aa and muteins, Y324A, W357A, Y412A, Y414A, W416A, D418A and F485A indicated that there was no global or significant variation in the secondary and tertiary structure of the toxins due to the alanine substitutions (Fig. 3).
Fig. 2.
Coomassie blue-stained SDS-10% PAGE of protoxins (a) and toxins after digestion with trypsin (b) and chymotrypsin (c) of Cry19Aa and muteins expressed in Bacillus thuringiensis 4Q7 cells. Lane 19, Cry19Aa; lane 1, Y410A; lane 2, E411A; lane 3, Y412A; lane 4, I413A; lane 5, Y414A; lane 6, W416A; lane 7, D418A; lane 8, V420A; lane 9, T484A; lane 10, F485A; lane 11, Y356A; lane 12, W357A; lane 13, Y324A; lane 14, F325A; lane 15, T390A; lane 16, T610A; lane M, Precision plus protein™ standards (Bio-Rad). Molecular mass marker positions (in kDa) are indicated to the left.
Fig. 3.
CD spectra of muteins (a) and FITC-labeled Cry19Aa (b) compared with wild-type Cry19Aa. The spectra of the purified toxins (2 μM) within the range from 200 to 300 nm were measured at 25 °C.
Mosquito bioassay of Cry19Aa and its muteins against C. pipiens and A. aegypti larvae
To determine whether mutations on the loop regions affected the toxicity of Cry19Aa, wild type and muteins were tested against A. aegypti and C. pipiens (Table 1). The LC50 for Cry19Aa toxin against second-instar A. aegypti larvae was 129 μg mL−1. For the A. aegypti bioassay, the concentration of the muteins was set as 100 μg mL−1 and mortality (%) was checked at 48 h because the LC50 value of Cry19Aa is high and does not yield reliable results at 24 h. All alanine substitutions in loop 1, 2 and 3 showed reduction in toxicity to Aedes except for F325A. However, T390A and T610A did not lose Aedes toxicity. In the Culex bioassay, Y356A, E411A, I413A and surface residue muteins have not lost toxicity. Specifically, Y324A (in loop 0), W357A (in loop 1), Y410A, W416A, D418A (in loop 2) and F485A (in loop 3) muteins showed significant loss of toxicity in mosquitocidal activities toward both Aedes (> 4 times) and Culex (> 130 times). Also, F325A, Y412A, Y414A, V420A and T484A were moderately reduced in Culex toxicity (8–20 times). These data suggest that domain II of Cry19Aa has an important role for toxicity as shown in the general mode of action of B. thuringiensis toxins. All four loops are involved in toxicity and particular residue positions, as opposed to a class of residue, are important for toxicity.
Table 1.
Bioassays of Cry19Aa alanine muteins to the larvae of Culex pipiens and Aedes aegypti
| Toxin |
C. pipiens* |
A. aegypti† |
|---|---|---|
| LC50 (ng mL−1) | Mortality (%) at 100 μg mL−1 | |
| Cry19Aa | 149 (103–205) | 61.1 |
| Loop 0 | ||
| Y324A | > 20 000‡ | 11.1 |
| F325A | 2790 (1730–4900) | 33.3 |
| Loop 1 | ||
| Y356A | 198 (81–701) | 16.7 |
| W357A | > 20 000§ | 0 |
| Loop 2 | ||
| Y410A | > 20 000¶ | 0 |
| E411A | 189 (148–243) | 18.8 |
| Y412A | 2440 (2310–2580) | 0 |
| I413A | 289 (225–389) | 5.6 |
| Y414A | 1210 (1150–1260) | 0 |
| W416A | > 20 000|| | 0 |
| D418A | > 20 000** | 0 |
| V420A | 1760 (1680–1830) | 5.3 |
| Loop 3 | ||
| T484A | 2920 (2740–3110) | 5.0 |
| F485A | > 20 000†† | 16.7 |
| Surface | ||
| T390A | 224 (162–317) | 50.0 |
| T610A | 105 (45–168) | 38.9 |
Bioassays used inclusion crystal proteins and spore purified from Bacillus thuringiensis transformants.
Larval stages and mortality check times were 4th instar and 24 h for C. pipiens, and 2nd instar and 24 h for A. aegypti.
43.3% mortality was observed at this dose.
16.1% mortality was observed.
38.7% mortality was observed.
25.8% mortality was observed.
32.3% mortality was observed.
9.7% mortality was observed.
Competition binding studies to mosquito brush border membranes
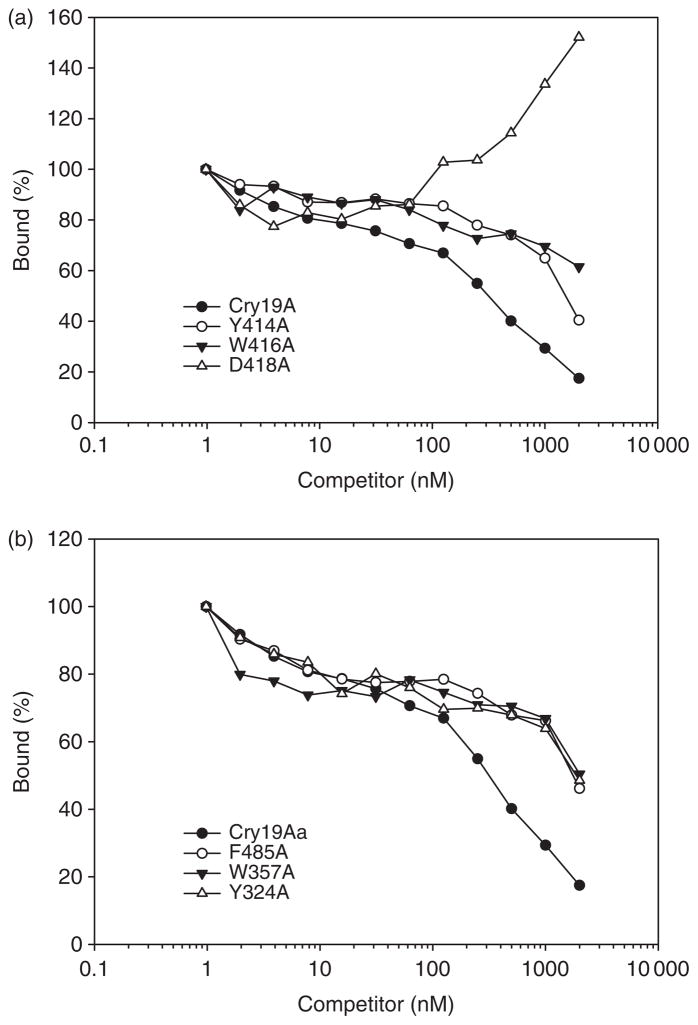
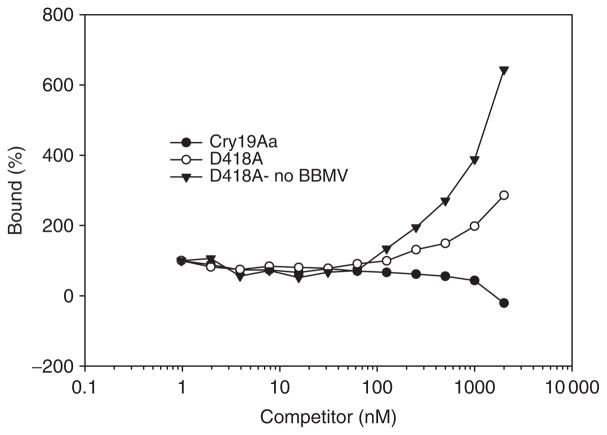
To evaluate the binding properties of muteins to C. pipiens BBMV, homologous and heterologous competition binding of FITC-labeled Cry19Aa toxin to BBMV were performed in the presence of increasing concentration of unlabeled wild type and muteins (Fig. 4). CD spectra of FITC-labeled toxin indicated no global perturbation in structure (Fig. 3b). In the absence of competitor, about 64% of labeled Cry19Aa toxin bound to the C. pipiens BBMV. Muteins Y324A, W357A, W416A and F485A did not compete with wild-type toxin as efficiently as homologous competition. This suggests that their reduction of toxicity resulted from reduced binding affinity. Also, Y414A (eight times less toxic mutein) did not significantly compete with wild-type toxin. This competition binding study demonstrated that a positive correlation between toxicity and binding affinity in domain II loops of Cry19Aa. Unexpectedly, in the competition with D418A, increasing unlabeled D418A seemed to increase binding of Cry19Aa (Fig. 4a). However, further binding experiments with D418A as a competitor indicated that there is coprecipitation of labeled Cry19Aa with D418A even in the absence of C. pipiens BBMV (Fig. 5). This result indicated that D418A mutein did not have a direct effect on the binding property of Cry19Aa to C. pipiens BBMV.
Fig. 4.
Binding of loop 2 (a) and other loops (b) Cry19Aa muteins to Culex pipiens BBMV. FITC-labeled Cry19Aa was incubated with 10 μg mL−1 of BBMV and 100 nM unlabeled Cry19Aa as a blocker in the presence of increasing concentrations of unlabeled Cry19Aa or muteins. Binding is expressed as percentage of the amount bound upon incubation with labeled toxin alone. Data shown are obtained from the average of three experiments.
Fig. 5.
Binding of FITC-labeled Cry19Aa to Culex pipiens BBMV competing with Cry19Aa and D418A. FITC-labeled Cry19Aa was incubated with or without 10 μg mL−1 of BBMV and 100 nM unlabeled D418A as a blocker in the presence of increasing concentrations of unlabeled Cry19Aa or D418A. Binding is expressed as percentage of the amount bound upon incubation with labeled toxin alone.
Discussion
Site-directed mutagenesis has been extensively used to elucidate the functional role of the residues in each domain of Cry proteins (Wu & Dean, 1996; Lee et al., 1999). Alanine substitutions are employed because alanine residues eliminate the side-chain usually without disrupting the overall structure (Cunningham & Wells, 1989). The receptor binding region of coleopteran- and lepidopteran-specific Cry proteins has been located primarily in domains II and III (Schnepf et al., 1990; Wu & Dean, 1996). Recently, the three-dimensional structure of Cry4Ba was solved (Boonserm et al., 2005). The domain II loop regions of Cry4Ba were explored for receptor-binding epitopes by exchanging the predicted loop residues with sequences from the closely related Cry4Aa (Abdullah et al., 2003). Mutations in the predicted loop 3 induced a significant toxicity increase toward Culex species, against which Cry4Ba shows no natural activity, while mutations in the predicted loops 1 and 2 abolished the natural toxicity toward both Aedes and Anopheles, but did not introduce toxicity against Culex.
Cry19Aa shows a comparable higher toxicity than Cry4Aa or Cry11Aa against C. pipiens (Liu & Dean, 2006) and also had less cross-resistance in Culex strains resistant to Cry4Aa, Cry4Ba and Cry11Aa (Wirth et al., 2001). These properties indicate that Cry19Aa is a valuable agent in mosquito control and resistance management as part of a synergistic set of mosquitocidal genes. The putative structure of Cry19Aa suggests that the apex of domain II is composed of four possible loops comprising residues 319–325 (designated as loop 0), 354–357 (loop 1), 410–420 (loop 2) and 483–486 (loop 3). Based on our knowledge, we investigated the role of domain II loops in mosquitocidal activity and receptor binding. In our previous Cry4Ba mutagenic study (Abdullah et al., 2003), loops 1 and 2 seem to play a role in the mechanism of action of Aedes and Anopheles while mutations of loop 3 were not critical for these insects (Abdullah et al., 2003). From the above results we expected one or two loops of Cry19Aa would be mainly involved with Culex toxicity. However, bioassay data from the present study show that Y324A in loop 0, W357A in loop 1, Y410A, W416A, D418A in loop 2 and F485A in loop 3 have substantially reduced toxicity to C. pipiens (>130 times) and A. aegypti (four times). Our results suggest that all loop regions of domain II are involved with Culex toxicity. Among six mutants, only Y410A had sensitivity to chymotrypsin showing degradation to small peptides instead of 60 kDa, suggesting that Y410A might be slightly more susceptible to mosquito gut proteases than wild type. Protease degradation is correlated with the loss of toxicity of Y410A. However, protein degradation was also observed in other muteins, F325A, V420A and T484A, which exhibited less toxin loss than Y410A. Further experiments on a cause of the loss of toxicity of Y410A will be needed. Competition binding assays revealed that four muteins (Y324A, W357A, W416A and F485A) that had substantially reduced toxicity to Culex showed no significant competition with wild-type toxin. The locations of the altered residues of these mutants reveal binding epitopes for Cry19Aa to its receptor. D418A is an exception. It shows the interesting attribute of aggregation with wild-type Cry19Aa. This deserves further study.
In summary, we have demonstrated that the loop regions of domain II are important in toxicity toward Culex and Aedes by playing a role in receptor binding and the receptor-binding residues of Cry19Aa are physically scattered among the loops of domain II, rather than clustered in a confined region of toxin. As a result, the loop regions are an excellent target for protein engineering to enhance toxicity toward Culex or Aedes as well as to understand the molecular mechanism of Cry19Aa mosquitocidal toxin.
Acknowledgments
We thank Dr Dan Zeigler for comments on the manuscript and helpful discussions. This work was supported by NIH grant to D.H.D. and M.J. Adang (Grant #R01 AI 29092).
References
- Abdul-Rauf M, Ellar DJ. Isolation and characterization of brush border membrane vesicles from whole Aedes aegypti larvae. J Invertebr Pathol. 1999;73:45–51. doi: 10.1006/jipa.1998.4792. [DOI] [PubMed] [Google Scholar]
- Abdullah MAF, Dean DH. Enhancement of Cry19Aa mosquitocidal activity against Aedes aegypti by mutations in the putative loop regions of domain II. Appl Environ Microb. 2004;70:3769–3771. doi: 10.1128/AEM.70.6.3769-3771.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdullah MAF, Alzate OM, Mohammad RJ, McNall AMJ, Dean DH. Introduction of Culex toxicity into Bacillus thuringiensis Cry4Ba by protein engineering. Appl Environ Microb. 2003;69:5343–5353. doi: 10.1128/AEM.69.9.5343-5353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almond BD, Dean DH. Structural stability of Bacillus thuringiensis δ-endotoxin homolog-scanning mutants determined by susceptibility to proteases. Appl Environ Microb. 1993;59:2442–2448. doi: 10.1128/aem.59.8.2442-2448.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker N, Margalit J. Use of Bacillus thuringiensis israelensis against mosquitoes and blackflies. In: Entwistle PF, Cory PF, Bailey MJ, Higgs S, editors. Bacillus thuringiensis an Environmental Biopesticide: Theory and Practice. John Wiley & Sons; New York: 1993. pp. 145–170. [Google Scholar]
- Boonserm P, Davis P, Ellar DJ, Li J. Crystal structure of the mosquito-larvicidal toxin Cry4Ba and its biological implications. J Mol Biol. 2005;348:363–382. doi: 10.1016/j.jmb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Charles J-F, Nielsen-LeRoux C, Delecluse A. Bacillus sphaericus toxins: molecular biology and mode of action. Annu Rev Entomol. 1996;41:451–472. doi: 10.1146/annurev.en.41.010196.002315. [DOI] [PubMed] [Google Scholar]
- Crickmore N, Zeigler DR, Feitelson J, et al. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol Mol Biol R. 1998;62:807–813. doi: 10.1128/mmbr.62.3.807-813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- Delécluse A, Rosso M-L, Ragni A. Cloning and expression of a novel toxin gene from Bacillus thuringiensis subsp. jegathesan encoding a highly mosquitocidal protein. Appl Environ Microb. 1995;61:4230–4235. doi: 10.1128/aem.61.12.4230-4235.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlanger BF, Kokowsky N, Cohen W. The preparation and properties of two new chromogenic substrates of trypsin. Arch Biochem Biophys. 1961;95:271–278. doi: 10.1016/0003-9861(61)90145-x. [DOI] [PubMed] [Google Scholar]
- Georghiou GP, Wirth MC. Influence of exposure to single versus multiple toxins of Bacillus thuringiensis subsp. israelensis on development of resistance in the mosquito Culex quinquefasciatus (Diptera: Culicidae) Appl Environ Microb. 1997;63:1095–1101. doi: 10.1128/aem.63.3.1095-1101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee MK, You TH, Young BA, Valaitis AP, Dean DH. Aminopeptidase N purified from gypsy moth BBMV is a specific receptor for Bacillus thuringiensis CryIAc toxin. Appl Environ Microb. 1996;62:2845–2849. doi: 10.1128/aem.62.8.2845-2849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MK, You TH, Gould FL, Dean DH. Identification of residues in domain III of Bacillus thuringiensis Cry1Ac toxin that affect binding and toxicity. Appl Environ Microb. 1999;65:4513–4520. doi: 10.1128/aem.65.10.4513-4520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Dean DH. Redesigning Bacillus thuringiensis Cry1Aa toxin into a mosquito toxin. Protein Eng Des Sel. 2006;19:107–111. doi: 10.1093/protein/gzj009. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Roberts NR, Wu ML, Hixon WS, Crawford EJ. The quantitative histochemistry of brain. II. Enzyme measurements. J Biol Chem. 1954;207:19–37. [PubMed] [Google Scholar]
- Porter AG, Davidson EW, Liu J-W. Mosquitocidal toxins of bacilli and their genetic manipulation for effective biological control of mosquitoes. Microbiol Rev. 1993;57:838–861. doi: 10.1128/mr.57.4.838-861.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni A, Thiéry I, Delécluse A. Characterization of six highly mosquitocidal Bacillus thuringiensis strains that do not belong to the H-14 serotype. Curr Microbiol. 1996;32:48–54. doi: 10.1007/s002849900009. [DOI] [PubMed] [Google Scholar]
- Rosso ML, Delecluse A. Contribution of the 65-kilodalton protein encoded by the cloned gene cry19Ato the mosquitocidal activity of Bacillus thuringiensis subsp. jegathesan. Appl Environ Microb. 1997;63:4449–4455. doi: 10.1128/aem.63.11.4449-4455.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer P, Millet J, Aubert JP. Catabolic repression of bacterial sporulation. P Natl Acad Sci USA. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnepf HE, Tomczak K, Ortega JP, Whiteley HR. Specificity-determining regions of a lepidopteran-specific insecticidal protein produced by Bacillus thuringiensis. J Biol Chem. 1990;265:20923–20930. [PubMed] [Google Scholar]
- Seleena P, Lee HL, Lecadet MM. A new serovar of Bacillus thuringiensis possessing 28a28c flagellar antigenic structure: Bacillus thuringiensis serovar jegathesan, selectively toxic against mosquito larvae. J Am Mosquito Contr. 1995;11:471–473. [PubMed] [Google Scholar]
- Silva-Filha MH, Regis L. Reversal of low-level resistance to Bacillus sphaericus in a field population of the southern house mosquito (Diptera: Culicidae) from an urban area of Recife, Brazil. J Econ Entomol. 1997;90:299–303. doi: 10.1093/jee/90.2.299. [DOI] [PubMed] [Google Scholar]
- Wirth MC, Delecluse A, Walton WE. Lack of cross-resistance to Cry19A from Bacillus thuringiensis subsp. jegathesan in Culex quinquefasciatus (Diptera: Culicidae) resistant to Cry toxins from Bacillus thuringiensis subsp. israelensis. Appl Environ Microb. 2001;67:1956–1958. doi: 10.1128/AEM.67.4.1956-1958.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S-J, Dean DH. Functional significance of loops in the receptor binding domain of Bacillus thuringiensis CryIIIA δ-endotoxin. J Mol Biol. 1996;255:628–640. doi: 10.1006/jmbi.1996.0052. [DOI] [PubMed] [Google Scholar]