Isolated oceanic islands are characterized by patterns of biological diversity different from those on continents. Nucleotide sequences from chloroplast and nuclear genes were used to examine the origins and diversity of the cosmopolitan fern genus Pteridium on the Galapagos Islands. We found evidence for multiple origins of the widespread allotetraploid P. caudatum. We also show that the Galapagos Islands are home to P. caudatum as well as diploid P. esculentum subsp. arachnoideum and possible hybrids between the two. Haplotype diversity indicates that Pteridium has colonized the islands multiple times and probably from diverse mainland sources.
Keywords: Biogeography, bracken, ferns, Galapagos, hybridization, islands, nuclear genes, phylogeny, Pteridium
Abstract
Isolated oceanic islands are characterized by patterns of biological diversity different from that on nearby continental mainlands. Isolation can provide the opportunity for evolutionary divergence, but also set the stage for hybridization between related taxa arriving from different sources. Ferns disperse by haploid spores, which are produced in large numbers and can travel long distances in air currents, enabling these plants to become established on most oceanic islands. Here, we examine the origins and patterns of diversity of the cosmopolitan fern genus Pteridium (Dennstaedtiaceae; bracken) on the Galapagos Islands. We use nucleotide sequences from two plastid genes, and two nuclear gene markers, to examine phylogeography of Pteridium on the Galapagos Islands. We incorporate data from a previous study to provide a worldwide context. We also sampled new specimens from South and Central America. We used flow cytometry to estimate genome size of some accessions. We found that both plastid and nuclear haplotypes fall into two distinct clades, consistent with a two-diploid-species taxonomy of P. aquilinum and P. esculentum. As predicted, the allotetraploid P. caudatum possesses nuclear haplotypes from both diploid species. Samples from the Galapagos include P. esculentum subsp. arachnoideum, P. caudatum and possible hybrids between them. Multiple Pteridium taxa were also observed growing together at some sites. We find evidence for multiple origins of Pteridium on the Galapagos Islands and multiple origins of tetraploid P. caudatum throughout its range in Central and South America. We also posit that P. caudatum may include recent diploid hybrids, backcrosses to P. esculentum, as well as allotetraploid plants. The Galapagos Islands are positioned close to the equator where they can receive dispersing propagules from both hemispheres. This may partly explain the high levels of diversity found for this cosmopolitan fern on these islands.
Introduction
Oceanic islands provide an ideal biological setting for evolutionary change and thus for the study of evolutionary processes. On islands that are distantly isolated from continents, there can be an increased opportunity for organisms to diverge genetically from those in the original source populations. Furthermore, remote islands can act as a sink for individuals of the same species (or closely related species) arriving from more than one original source, thereby setting the stage for hybridization, increased genetic diversity, or both. Angiosperms and gymnosperms colonize islands most often via seeds, which contain diploid embryos. This is contrasted in ferns (monilophytes) and lycophytes, which are dispersed by haploid spores. In many cases, spore-bearing plants require two spores, and subsequently two gametophytes, for successful establishment (Soltis and Soltis 1987; Soltis et al. 1988). Moreover, spore-dispersed plants appear to have high levels of gene flow relative to seed plants (Soltis and Soltis 1987), presumably because spores are smaller and more easily transported in the air than most seeds. An increased dispersal potential can result in colonization of new areas from multiple spore sources, and may involve an increased opportunity for hybridization relative to seed plants. Here, we explore this possibility as we assess the origins and diversity of the cosmopolitan fern genus Pteridium on the Galapagos Islands.
The Galapagos Islands are a group of ∼14 main islands and ∼100 rocks or islets, ∼1000 km west of mainland Ecuador (Snell et al. 1996). The islands vary in age ranging from ∼0.7 to ∼4.2 million years since emergence above sea level (White et al. 1993). However, additional evidence points to a much older archipelago existing at the same spot prior to the current emergence (Christie et al. 1992). Rassmann (1997) cited this idea to explain why estimates of some lineage ages are more than 5 million years old. The relative isolation of the islands from the nearest mainland coasts (Ecuador and Costa Rica) has resulted in a high level of endemism. There are estimated to be 236 endemic plant species on the islands (Tye and Francisco-Ortega 2011). Origins of the Galapagos flora are now thought to be quite diverse and include northern and southern Andes and other parts of South America, as well as Central America and the Caribbean (Tye and Francisco-Ortega 2011). The wide range of elevations and rainfall patterns results in a diversity of ecological habitats (Itow 2003).
Pteridium is a worldwide genus that has been treated from as few as one species to >20. There are several taxonomic challenges in the genus, one of which is the high level of variability and phenotypic plasticity for morphological characters, including those that are used for taxonomic treatments. Furthermore, regional and local treatments often do not incorporate the context of variation that is seen at the global scale. A few authors have examined Pteridium in a worldwide context. For example, Ching (1940) considered the genus to comprise 6 species, whereas a year later, Tryon (1941) treated Pteridium as a single species with 2 subspecies and 12 varieties. Page (1976) reviewed information on geographic variation and concluded that there is probably more than one species, but he made no formal taxonomic changes. More recently, in a series of articles (Thomson 2000, 2012; Thomson et al. 2008), Thomson has recognized two main diploid species: P. aquilinum (corresponding to Tryon's subspecies aquilinum) from Europe, North America, Asia and Africa and P. esculentum (corresponding approximately to Tryon's subspecies caudatum). Pteridium esculentum is treated by some authors as two species: P. esculentum in Australia and New Zealand and P. arachnoideum in South America (see, for example, Schwartsburd et al. 2014), whereas others treat esculentum and arachnoideum as subspecies of P. esculentum (Thomson 2012; Zhou et al. 2014), a system we follow here. Regardless of rank assignment, evidence for two main clades of Pteridium includes analyses of plastid DNA variation (Der et al. 2009; Zhou et al. 2014). Further, several hybrids and allotetraploids have been examined (Thomson and Alonso-Amelot 2002; Zhou et al. 2014). Der et al. (2009) noted that development of nuclear genomic markers would be critical for establishing the origins of hybrid taxa and for other systematic studies of Pteridium.
South America is home to two main Pteridium taxa: diploid P. esculentum subsp. arachnoideum and allotetraploid P. caudatum, the latter a hybrid between P. esculentum from South America and P. aquilinum from North America (Thomson and Alonso-Amelot 2002). Tetraploidy was inferred on the basis of Feulgen cytometry (Tan and Thomson 1990) and spore size and guard cell length (Thomson and Alonso-Amelot 2002). The hybrid origin of P. caudatum is further supported by the additive pattern of DNA markers from P. aquilinum and P. esculentum (Thomson 2000; Thomson and Alonso-Amelot 2002). Additional characters that can be used to distinguish P. caudatum from P. esculentum subsp. arachnoideum in South America include the presence of gnarled trichomes between veins abaxially (Thomson and Martin 1996) and free laminar lobes on P. esculentum. An additional taxon was recently described from north eastern Brazil (Schwartsburd et al. 2014).
Most chromosome counts of Pteridium show 2n = 104 (Page 1976; Sheffield et al. 1989; Thomson 2000; Tindale and Roy 2002; Bainard et al. 2011), with other complements, such as triploidy (Sheffield et al. 1993), assumed to be rare. One count of 2n = 52 from Spain (Löve and Kjellqvist 1972) has not been corroborated despite resampling from the same area (Sheffield et al. 1989). Jarrett et al. (1968) reported the first observation of cytological variation in the genus, with a count of 2n = 208 (tetraploid) for one sporophyte of Pteridium from the Galapagos Islands. This report has been the motivation for previous as well as the current focus on Pteridium from these islands. Klekowski (1973) used gametophytes grown from spores collected from the islands to examine the ability to self-fertilize and cross with Pteridium from other sources. The results demonstrated that bracken from Hawaii (P. aquilinum) and samples from the Galapagos Islands were interfertile with Pteridium from Central and South America. However, Hawaiian and Galapagos Pteridium were intersterile with each other.
Recent examination of Pteridium collections from the Galapagos islands suggests that more than one taxon is present on Galapagos. We set out to examine Galapagos Pteridium with the following objectives:
To examine the origins of P. caudatum on both islands and mainland.
To determine how many Pteridium taxa are on the Galapagos Islands.
To examine whether different Pteridium taxa occupy different islands, or elevations on the Galapagos Islands.
To examine the possible mainland origins of Galapagos Pteridium.
Methods
We sampled 17 Pteridium from three of the Galapagos Islands (Fig. 1; Table 1): Santa Cruz, Isabela and San Cristobal. We also scouted on Floriana, but were unable to locate any Pteridium on that island. At each site, we collected expanding frond segments onto silica gel, and collected intact fronds for herbarium specimens, deposited at the herbarium of the Charles Darwin Research Station (CDS). In addition, we included a selection of DNA samples from the plastid gene study of Der et al. (2009) to obtain nuclear gene sequences. We selected representatives of each of the major plastid clades of Der et al. (2009). We also included additional Pteridium samples from the mainland of Central and South America, and outgroups Histiopteris, Blotiella and Paesia (Table 1).
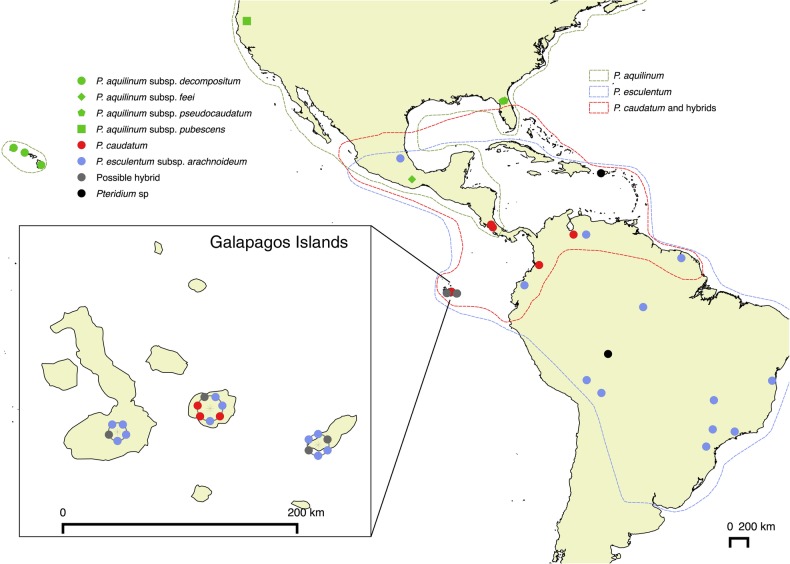
Figure 1.
Map of America showing locations of Pteridium sampled for this study. Inset shows details of Galapagos Islands. Approximate taxon boundaries are based on Tryon (1941), Tryon and Tryon (1982) and Mickel and Smith (2004).
Table 1.
Voucher and locality information for samples used in this study. Code (as used in tree figures) indicates collector and number, with full name in parentheses when abbreviated in code. ‘Possible hybrids’ are likely to be between P. esculentum subsp. arachnoideum and P. caudatum.
| Code | Herbarium | Taxon | Country | Island/province/state | Latitude (°) | Longitude (°) | Elevation (m) |
|---|---|---|---|---|---|---|---|
| Wolf 1001 | CDS | P. esculentum subsp. arachnoideum | Ecuador | Santa Cruz | −0.63 | −90.38 | 592 |
| Wolf 1002 | CDS | P. caudatum | Ecuador | Santa Cruz | −0.66 | −90.40 | 420 |
| Wolf 1003 | CDS | P. esculentum subsp. arachnoideum | Ecuador | Santa Cruz | −0.64 | −90.33 | 874 |
| Wolf 1004 | CDS | P. esculentum subsp. Arachnoideum | Ecuador | Santa Cruz | −0.65 | −90.33 | 732 |
| Wolf 1005a | CDS | Possible hybrid | Ecuador | Santa Cruz | −0.66 | −90.33 | 580 |
| Wolf 1005c | CDS | P. caudatum | Ecuador | Santa Cruz | −0.66 | −90.33 | 580 |
| Wolf 1006 | CDS | P. caudatum | Ecuador | Santa Cruz | −0.67 | −90.32 | 476 |
| Wolf 1007 | CDS | P. esculentum subsp. arachnoideum | Ecuador | Isabela | −0.81 | −91.09 | 1009 |
| Wolf 1008 | CDS | P. esculentum subsp. arachnoideum | Ecuador | Isabela | −0.83 | −91.09 | 1006 |
| Wolf 1009 | CDS | P. esculentum subsp. arachnoideum | Ecuador | Isabela | −0.84 | −91.09 | 822 |
| Wolf 1010 | CDS | P. esculentum subsp. arachnoideum | Ecuador | Isabela | −0.84 | −91.07 | 627 |
| Wolf 1011 | CDS | Possible hybrid | Ecuador | Isabela | −0.85 | −91.04 | 405 |
| Wolf 1012 | CDS | P. esculentum subsp. arachnoideum | Ecuador | San Cristobal | −0.91 | −89.55 | 381 |
| Wolf 1013 | CDS | P. esculentum subsp. arachnoideum | Ecuador | San Cristobal | −0.90 | −89.48 | 683 |
| Wolf 1014 | CDS | Possible hybrid | Ecuador | San Cristobal | −0.90 | −89.48 | 676 |
| Wolf 1015 | CDS | P. esculentum subsp. arachnoideum | Ecuador | San Cristobal | −0.90 | −89.52 | 739 |
| Wolf 1016 | CDS | P. esculentum subsp. arachnoideum | Ecuador | San Cristobal | −0.90 | −89.53 | 544 |
| Wolf 1017 | CDS | Possible hybrid | Ecuador | San Cristobal | −0.90 | −89.53 | 544 |
| Wolf 1018 | UTC | P. aquilinum subsp. decompositum | USA | Hawaii | 19.43 | −155.28 | 1247 |
| Wolf 1019 | UTC | P. aquilinum subsp. pseudocaudatum | USA | Florida | 29.63 | −81.92 | 42 |
| AL 147 (A. Larsson) | DUKE | P. aquilinum subsp. feei | Mexico | Oaxaca | 17.17 | −96.60 | 2660 |
| IJ 786 (Jiménez) | LPB, UC | P. esculentum subsp. arachnoideum | Bolivia | Franz Tamayo | −14.62 | −68.95 | 2350 |
| IJ 1245 (Jiménez) | LPB, UC | P. esculentum subsp. arachnoideum | Bolivia | Ayopaya | −16.65 | −66.62 | 2750 |
| IJ 2048 (Jiménez) | LPB, UC | Pteridium sp. | Bolivia | Federico Román | −10.48 | −65.57 | 140 |
| Wolf 1020 | UTC | P. caudatum | Costa Rica | San Jose | 9.56 | −83.80 | 2270 |
| Wood 15788 | HAW | P. aquilinum subsp. decompositum | USA | Hawaii | 22.15 | −159.65 | 1280 |
| Wolf 1023 | HAW | P. aquilinum subsp. decompositum | USA | Hawaii | 21.40 | −157.89 | 419 |
| Worthington 35231 | DUKE | Pteridium sp. | Puerto Rico | Ponce | 18.13 | −66.68 | 792 |
| JJdG 14388 (de Granville) | NSW 729390 | P. esculentum subsp. arachnoideum | French Guiana | Saint-Laurent-du-Maroni | 4.70 | −53.97 | 480 |
| Matos 231 | NY 01198119 | P. esculentum subsp. arachnoideum | Brazil | Bahia | −14.71 | −39.60 | 700 |
| Ortiz 497 | NY 00089157 | P. esculentum subsp. arachnoideum | Ecuador | Esmeraldas | 0.40 | −78.80 | 1925 |
| Delprete 10293 | NY 01019119 | P. esculentum subsp. arachnoideum | Brazil | Goias | −17.80 | −48.82 | 1150 |
| Prado 2351 | SP | P. esculentum subsp. arachnoideum | Brazil | Paranà | −25.14 | −50.03 | 1000 |
| Prado 2337 | SP | P. esculentum subsp. arachnoideum | Brazil | São Paulo | −22.77 | −45.53 | 1888 |
| Wolf 795 | UC 1622577 | H. incisa | |||||
| Wolf 387 | UTC | P. scaberula | |||||
| Wolf 376 | UTC | B. pubescens | La Réunion |
Morphology
Preliminary morphological analysis of samples was undertaken independently of molecular studies. Photographs of fresh unpressed pinnae supplemented by macrophotographs of the abaxial surface of individual ultimate segments were used for the examination of gross features of laminal dissection, presence/absence of free laminal lobes on pinna and pinnule axes, and abaxial laminal indumentum.
We used small subsamples of each accession comprising one or two dried pinnules for more detailed microscopic study following Thomson and Martin (1996). We examined the indument of abaxial pinnulet and segment midveins, determined presence versus absence of gnarled intervein trichomes, measured false-indusial width, estimated the number of cells per millimetre along the outer margin of the false indusium and measured stomatal guard cell length. Taxonomic designation was based on previous descriptions of the characters we used (Thomson 2000, 2012; Thomson and Alonso-Amelot 2002; Thomson et al. 2008).
DNA sequencing
DNA was extracted from fresh, desiccated or herbarium tissue using the DNeasy Plant Mini kit (QIAGEN, Valencia, CA, USA), following the manufacturer's protocol.
The plastid markers trnS–rpS4 (spacer + gene) and rpL16 intron were amplified in 25 μL polymerase chain reactions (PCRs) using the fern-specific primers published in Small et al. (2005). However, the complete plastid genome sequence of P. aquilinum (GenBank accession NC_014348) was used to redesign the rpl16 reverse and the trnS primers (names now with ptaq suffix; see Table 2).
Table 2.
Primer sequences for PCR and DNA sequencing. Suffix ‘ptaq’ denotes primers designed in this study.
| Primer name | Primer sequence, 5′–3′ |
|---|---|
| rpl16_r_ptaq | TCCTCTATGTTGCTTACGATAT |
| trns_gga_ptaq | CTACCGAGGGTTCAAATCCCTC |
| SQD_r2_ptaq | CCTTTGCCATAAACTGTAAGGGGGTG |
| EMSQD1E1F6 | GCAAGGGTACHAAGGTHATGATCATAGG |
| ApPEFP_f25_ptaq | AATGCTCTAAGTCATTGTTACCGATC |
| ApPEFP_C4218_r7 | TTGTAAATCTCTGTRTCRGATGYYGT |
| rps4_int_f1 | CAGATTACTGAAAAACTAGC |
| rps4_int_r1 | AGAAGAGCGAAAGGGTTC |
| rpl16_int_f1 | GCGAAGCTGAAAACGATGCC |
| rpl16_int_r1 | GTTCCATTTCTAAATAGCGG |
Nuclear primers were based on those of Rothfels et al. (2013). We chose two nuclear genes SQD1 (Region 1) and ApPEFP_C (Region 2), redesigning (now with suffix _ptaq) the forward ApPEFP_C and SQD1 reverse, based on Pteridium sequences from a study of the transcriptome (Der et al. 2011). SQD1 encodes sulfoquinovosyldiacylglyerol 1 involved in the biosynthesis of sulfolipids and ApPEFP_C encodes an appr-1-p processing enzyme family protein, ADP-ribose-1-monophosphatase (Appr-1-pase), a ubiquitous cellular processing enzyme. The PCR primer sequences (Table 2) used were as follows: SQD_r2_ptaq combined with EMSQD1E1F6, and ApPEFP_f25_ptaq combined with ApPEFP_C4218_r7. Polymerase chain reaction conditions followed Der et al. (2009), annealing at 56.5 °C for the plastid and nuclear genes. For sequencing, we used all PCR primers plus new internal primers (Table 2) for the two plastid genes: rps4_int_f1, rps4_int_r1, rpl16_int_f1 and rpl16_int_r1. In many samples, the two nuclear gene amplicons contained multiple haplotypes. To sequence each haplotype separately, we cloned the PCR products using the StrataClone PCR Cloning Kit (Agilent Technologies, Santa Clara, CA, USA). DNA sequences were assembled and edited using Sequencher 4.6 (Gene Codes Corporation, Ann Arbor, MI, USA).
Phylogenetic analysis
Sequences were aligned with MAFFT version 7.215 using the L-INS-i algorithm for accurate alignments. Newly generated sequences for the plastid genes rps4 and rpl16 were combined with data from Der et al. (2009; GenBank accession numbers FJ177158–FJ177206 for the trnS–rps4 spacer + gene and FJ177239–FJ177287 for the rpL16 intron) and concatenated for phylogenetic analysis. Maximum likelihood (ML) phylogenetic inference was performed separately for each nuclear gene and the plastid data with RAxML version 8.1.17 using 100 rapid bootstrap replicates followed by a ML search under the GTRGAMMA model of evolution. Trees were rooted with the three outgroups.
Flow cytometry
Genome size was determined using flow cytometry. Approximately 0.75 cm2 of fresh leaf tissue and 0.5 cm2 of standard, Vicia faba (26.9 pg; Doležel et al. 1998), were co-chopped on a chilled surface using a fresh razor blade in 500 μL of ice-cold extraction buffer (0.1 M citric acid, 0.5 % v/v Triton X-100) (Hanson et al. 2005), with 1 % w/v PVP-40 (Yokoya et al. 2000). Tissue was chopped into a semi-fine slurry, and the suspension was swirled by hand until the liquid reached a light green tinge. The suspension was poured through a cell strainer (BD Falcon; Becton, Dickinson and Company, Franklin Lakes, NJ, USA). RNaseA (1 mg mL−1) and 350 µL of propidium iodide staining solution (0.4 M NaPO4, 10 mM sodium citrate, 25 mM sodium sulfate, 50 µg mL−1 propidium iodide) were added to 140 µL of filtrate, incubated at 25 °C for 30 min, followed by up to 2 h on ice. The stained solutions were analysed with an Accuri C6 using a 488 nm laser, and 10 000 events were captured per sample. The relative genome size was calculated using the ratio of the mean fluorescent peak of the sample to the internal standard multiplied by the genome size of the standard.
Results
In general, we found Galapagos Pteridium to be highly variable for both morphological and molecular characters. We find evidence of two Pteridium taxa plus possible hybrids, and multiple colonization events from different mainland sources.
Morphology
Pteridium caudatum and P. esculentum can be distinguished morphologically by a combination of characters (Thomson 2000; Thomson and Alonso-Amelot 2002; Table 3). We inferred that 11 of our samples were clearly P. e. subsp. arachnoideum, 2 were clearly P. caudatum and 4 were difficult to determine and inferred to be possible hybrids. The two P. caudatum samples were found at the two lowest sites on Santa Cruz. The possible hybrids were found at the lowest sites on Isabela and San Cristobal and mid-elevation sites on Santa Cruz and San Cristobal. We found different Pteridium taxa growing within a kilometre of each other on Santa Cruz and San Cristobal. Samples from one site included one frond that was P. e. arachnoideum (Wolf 1005a) and another frond (collected within 1 m of the other) was P. caudatum (Wolf 1005c) or a hybrid.
Table 3.
Typical morphologies for P. caudatum, P. esculentum subsp. arachnoideum, and possible hybrids (or introgressants) between them. Information based on Tryon (1941) and Thomson and Alonso-Amelot (2002).
| Determination | Wolf ID # | Free lobes on segment axes | False indusium: width (mm) | False indusium: cells/mm length along margin | Stomatal guard cell length (µm) | Abaxial surface between veins: gnarled trichomes | Abaxial surface: vein indumentum |
|---|---|---|---|---|---|---|---|
| P. caudatum | 1002, 1006 | Absent | 0.3–0.5 | ∼31 | >40 | Absent | Glabrous |
| P. esculentum subsp. arachnoideum | 1001, 1003, 1004, 1007, 1008, 1009, 1010, 1012, 1013, 1015, 1016 | Present | 0.1–0.3 | ∼48 | <40 | Present | Dense fine acicular white hairs, some twisted; fine white arachnoid hairs |
| Indeterminate: possibly introgressant | 1005a | Absent | 0.2 | 64 | 32.3 | Present | Vein hairs less dense than for typical subsp. arachnoideum |
| 1011 | Absent | 0.4 | 56 | 39.1 | Present | As for 1005a | |
| 1014 | Absent | 0.15 | 48 | 39.3 | Present | As for 1005a | |
| 1017 | Absent | 0.15–0.2 | 40 | 34.3 | Absent | As for 1005a |
Most of our Galapagos samples of Pteridium fell into one of the two distinct categories for stomatal guard cell length: those with a mean below 40 µm and those above 40 µm. Wolf 1002 and Wolf 1006 fall within the range expected for tetraploid P. caudatum (Thomson and Alonso-Amelot 2002) and close to the guard cell length (46.5 µm, Thomson 2000) for the Galapagos plant showing 4n = 208 (K Sheet H2146/97/1, Jarrett et al. 1968), corroborating our morphology-based determination of these samples (Table 3). Ploidy level of the other Galapagos samples studied cannot be determined due to the extended, apparently continuous, series of guard cell lengths represented (Table 3). The wide range of lengths observed suggests that both diploid and triploid levels might be represented. On the basis of morphology, 11 of our samples are P. esculentum subsp. arachnoideum and 4 may be hybrids carrying genomic elements from outside arachnoideum, and may be triploid (Table 3).
DNA
Overall, nucleotide data from the four genes contained 3365 characters of which 100 were phylogenetically informative, and 55 distinguished the P. aquilinum clade from the P. esculentum clade (Table 4). Phylogenetic analyses of the two plastid genes were congruent, as found previously (Der et al. 2009). Thus, alignments of the two plastid genes were concatenated for a combined analysis (Fig. 2). In the three analyses (two plastid genes concatenated, SQD1 and ApPEFP_C), the aquilinum and esculentum clades of Pteridium were sister to each other. Table 4 provides the ranges of GenBank accession numbers for each gene and Supporting Information—File S1 lists the GenBank accession number for each sequence. All trees and associated nucleotide alignments are deposited in TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S18018). We include outgroups for phylogenetic analysis of the nuclear genes. However, inclusion of outgroups in the plastid trees resulted in very short ingroup branch lengths, and is therefore not shown. The tree topology without outgroups is the same as that with outgroups.
Table 4.
Gene statistics and GenBank accession information.
| Gene | Number of characters | Variable characters | Parsimony-informative characters | Differences between aquilinum and esculentum haplotypes | GenBank accession numbers |
|---|---|---|---|---|---|
| ApPEFP_C | 785 | 51 | 28 | 17 | KT345729–KT345821 |
| SQD1 | 752 | 36 | 26 | 15 | KT345856–KT345898 |
| rps4 | 1036 | 33 | 25 | 11 + 1 indel | KT345822–KT345855 |
| rpl16 | 792 | 27 | 21 | 9 + 2 indels | KT345899–KT345934 |
| Total | 3365 | 147 | 100 | 55 |
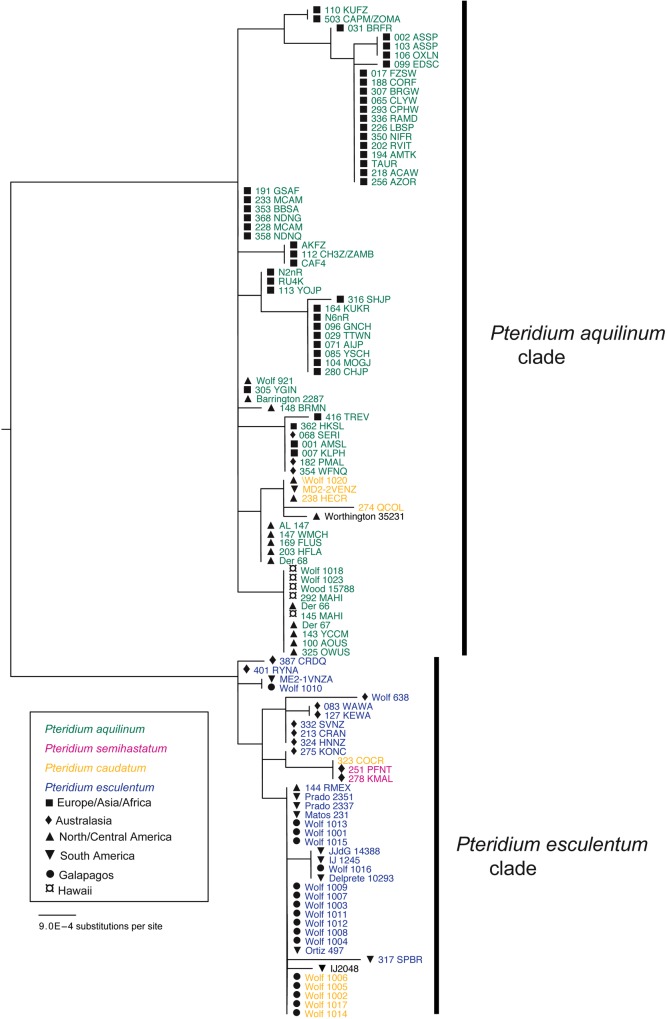
Figure 2.
Phylogenetic tree based on the combined plastid gene data set. The tree was rooted with Blotiella pubescens, Paesia scaberula and Histiopteris incisa.
We detected a total of 19 aquilinum and 31 esculentum plastid haplotypes. In samples from the Galapagos Islands, we detected 12 plastid haplotypes, one of which has been sampled previously in P. e. arachnoideum from Venezuela and Mexico (Der et al. 2009). The remaining Galapagos plastid haplotypes were nested within a clade that included haplotypes from Mexico and South America (Fig. 2). All samples of P. aquilinum had the expected plastid haplotype, and all new samples from South American Pteridium, including those from the Galapagos Islands, had esculentum haplotypes. A specimen from Costa Rica (Wolf 1020), which appears to be P. caudatum, had an aquilinum plastid haplotype, as did the single Mexican sample of P. a. feei (A. Larsson 147), a taxon not previously included in molecular studies. Previous studies (Thomson et al. 2008; Der et al. 2009) have noted two 5-bp polymorphic repeats in the trnS–rps4 spacer. Together, these polymorphisms account for three haplotypes: haplotype C in outgroups and the P. esculentum clade, haplotype A in P. aquilinum and haplotype B in only European and African P. aquilinum.
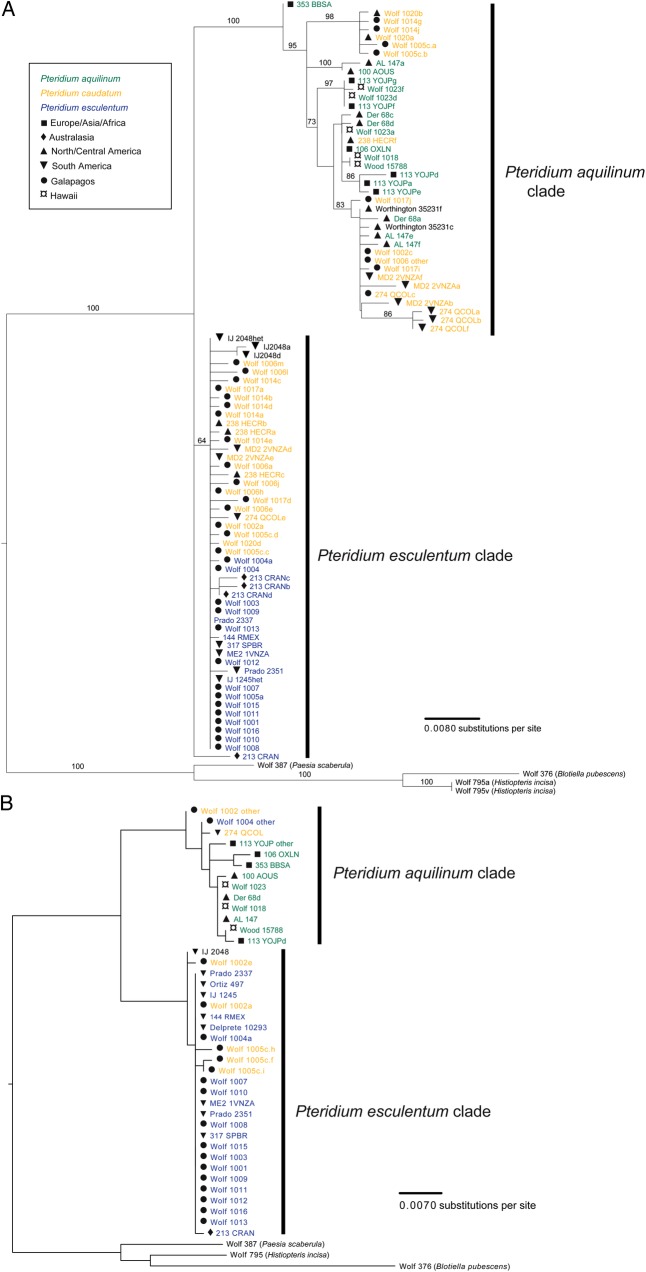
The two nuclear genes showed a similar pattern of differentiation as the plastid genes: a set of distinct nucleotide differences (15 for SQD1 and 17 for ApPEFP_C) distinguished aquilinum from esculentum haplotypes. For SQD1, samples of P. caudatum were heterozygous for the above nucleotide positions indicating that they were additive for P. aquilinum and P. esculentum haplotypes. However, although we were able to sequence a few distinct haplotypes from heterozygous plants, this was largely unsuccessful. Bacterial cells carrying the SQD1 PCR product appeared to be clumping so that single colonies were usually not single clones and therefore remained heterozygous, despite re-streaking of colonies. We suspect that clumping was a function of partial expression of the gene. Conversely, we were able to clone several haplotypes of ApPEFP_C from heterozygous individuals and we found that P. caudatum plants indeed possessed both aquilinum and esculentum haplotypes. We detected a total of 12 aquilinum and 13 esculentum haplotypes for SQD1 (Fig. 3), and 32 aquilinum and 30 esculentum haplotypes for ApPEFP_C (Fig. 3). In the samples from the Galapagos Islands, we detected 7 aquilinum and 14 esculentum ApPEFP_C haplotypes, and 1 esculentum and 9 aquilinum haplotypes for SQD1. Of the six samples that were heterozygous for SQD1, we were able to sequence five haplotypes from three individuals. Of the 17 samples that were heterozygous for ApPEFP_C, 8 had both aquilinum and esculentum haplotypes, 5 heterozygous samples had only aquilinum haplotypes and 4 samples had only esculentum haplotypes. All Galapagos specimens have esculentum nuclear haplotypes, 11 with an esculentum haplotype only, 10 of which were P. e. arachnoideum. Five Galapagos samples have aquilinum and esculentum haplotypes, two of which were P. caudatum, two appeared to be hybrids and one was P. e. arachnoideum. One specimen (Wolf 1004) appeared to have aquilinum and esculentum haplotypes for SQD1, but only aquilinum haplotypes for ApPEFP_C. All mainland samples of P. caudatum have both aquilinum and esculentum haplotypes. Diploid individuals should have no more than two haplotypes at a locus, whereas tetraploids are expected to have no more than four haplotypes. However, 8 of the 13 heterozygous individuals had extra haplotypes: 3 P. caudatum samples with 5, 5 and 7 haplotypes, 1 P. esculentum with 6 haplotypes and 4 P. aquilinum samples with 3, 3, 3 and 5 haplotypes. In most cases, extra haplotypes differed from others from the same individual by one nucleotide, and at the most three nucleotides. All extra haplotypes appear to be the result of single nucleotide autapomorphies, and we cannot account for extra haplotypes by recombination, whether in plant cells, PCR tubes or during cloning.
Figure 3.
Phylogenetic tree based on the nuclear genes ApPEFP_C (A) and SQD1 (B). Trees were rooted with B. pubescens, P. scaberula and H. incisa.
Flow cytometry
We were able to estimate c-values for fresh Pteridium fronds on a consistent basis, but we were unable to do so for any dry samples, including those in silica gel and herbarium specimens. We estimate haploid genome size (mean ± coefficient of variation) of 15.88 (±0.67) pg for P. a. decompositum from Hawaii (Wolf 1018), 16.13 (±0.67) pg for P. a. pseudocaudatum from Florida (Wolf 1019) and 29.2 (±2.1) pg for P. caudatum from Costa Rica (Wolf 1020). These are consistent with previous estimates for diploid and tetraploid Pteridium, respectively (Tan and Thomson 1990; Bainard et al. 2011).
Discussion
In this article, we examined morphological and molecular variation in Pteridium from Galapagos Islands. To make inferences about taxonomic variation and possible origins on the islands, we first provided context with other mainland and worldwide samples, including sequences from a previous study (Der et al. 2009). We first discuss variation for morphological and molecular characters, followed by the implications for the origins of Galapagos Pteridium.
Molecular data
A growing body of research has examined variation for nuclear-encoded genes within species of ferns (for example see Grusz et al. 2009; Nitta et al. 2011; Sigel et al. 2014). Many studies use nucleotide information from the plastid genome, which appears to be effectively haploid and non-recombining in most plants. Nuclear genes, however, are subject to different processes as a result of chromosomal behaviour at a range of genomic scales. For a single-copy gene, a diploid individual should carry one or two haplotypes (alleles), and a tetraploid can have up to four. Eight of our samples carried more than the expected number based on our estimate of ploidy. Several explanations can account for these results. Polymerase chain reaction and sequencing error could manifest as extra haplotypes within an individual. It is also possible that SQD1 and ApPEFP_C are not strictly single copy in Pteridium. Extra haplotypes can occur through several processes including segmental duplication of the chromosomal region carrying the gene, aneuploidy and polyploidy. All measures of genome size and chromosome number in Pteridium point to diploid and tetraploid being the most common arrangement. But samples sizes are small and we would benefit from population-level estimates of genome size, especially in areas with multiple species such as Galapagos Islands and areas of Central America. Regardless of the causes of extra haplotypes, they should be explored further. Meanwhile, because the extra haplotypes possessed only a few autapomorphic differences from others, they do not affect the phylogenetic inferences or estimates of origin numbers in this study.
Our only sample from the Caribbean was from a specimen represented by a young frond, and therefore, difficult to identify morphologically. Most descriptions indicate that P. e. arachnoideum is throughout the Caribbean, but this sample has aquilinum haplotypes. Future studies would benefit from increase sampling in the Caribbean.
Morphological variation
Pteridium is notorious for its phenotypic plasticity, including morphological variation among fronds within an individual clone and between pinnae on a frond (Tryon 1941; Sheffield et al. 1989; Ashcroft and Sheffield 1999; Thomson 2000). Wide morphological variability in P. caudatum led Tryon (1941) to recognize several ‘phases’. Ortega (1990) found in Venezuela both typical P. e. arachnoideum and a second more compact form lacking free lobes between ultimate segments, while Schwartsburd et al. (2014) recognized three morphotypes of P. e. arachnoideum from Brazil. The significance and genetic basis of these character suites is yet to be established, but their variation has led to many reports of apparent intermediates between P. caudatum, P. e. arachnoideum and other taxa (Tryon 1941; Mickel and Beitel 1988; Ortega 1990; Mickel and Smith 2004; Schwartsburd et al. 2014).
The relationship between stomatal guard cell length and ploidy level was clearly documented for ferns by Barrington et al. (1986) and later established for Pteridium (Tan and Thomson 1990; Sheffield et al. 1993; Thomson 2000; Thomson and Alonso-Amelot 2002). Guard cell length is quite variable within and between Pteridium taxa at the subspecies level (Thomson 2000; Thomson and Alonso-Amelot 2002), and its relationship with ploidy, therefore, requires calibration and validation for each particular comparison, which we followed here.
Most of our samples from the Galapagos Islands (Table 3) had distinct morphological signatures of P. e. arachnoideum or P. caudatum. However, four samples did not fall clearly into either morphological category (Table 3). Therefore, we infer that the latter samples are possible hybrids between P. caudatum and P. e. arachnoideum, or the result of a ploidy level other than diploid or tetraploid.
The origins of P. caudatum in South America
Thomson (2000) and Thomson and Alonso-Amelot (2002) first outlined P. caudatum as one of the fertile allotetraploids between P. aquilinum and P. esculentum. Furthermore, these authors speculated that P. caudatum has had multiple origins in Central and South America. This hypothesis was supported by analysis of plastid DNA (Der et al. 2009), which showed that some P. caudatum samples had the P. aquilinum plastid DNA, whereas others had that of P. esculentum. Here, we provide additional evidence for the hybrid origin of P. caudatum; all samples had the additive pattern with both P. aquilinum and P. esculentum nuclear gene haplotypes. As for many allotetraploids, multiple origins can be inferred (Soltis and Soltis 1991; Ranker et al. 1994; Meimberg et al. 2009). We detected three P. aquilinum plastid haplotypes and seven P. esculentum plastid haplotypes among P. caudatum accessions. Even more haplotypes are seen in the nuclear DNA, but that is expected because two haplotypes can be transferred in a single origin involving a heterozygous plant. We detected 7 P. aquilinum and 12 P. esculentum nuclear DNA haplotypes across our P. caudatum accessions. Inferring the minimum number of origins is difficult because we do not know how much nucleotide change has occurred since the origin. But given the range of variation found in P. caudatum, we can infer at least 8 separate origins among the 11 accessions sampled here. From examination of the phylogenetic trees, it seems that the P. aquilinum parent could be P. a. pseudocaudatum (Florida and Caribbean), P. a. latiusculum (eastern North America) or P. a. feei (Mexico). All have similar plastid and nuclear haplotypes so that distinguishing the P. aquilinum parent further is challenging. The esculentum parent of P. caudatum includes only P. e. arachnoideum. However, this taxon is highly variable and probably includes several taxa (Schwartsburd et al. 2014). Future sampling should aim to include more samples of P. e. arachnoideum from western South America as well as Brazil.
We sampled P. caudatum more densely on the Galapagos Islands than the mainland, so it is difficult to determine whether Pteridium on the Galapagos Islands is more variable than for an equivalent area on the mainland. However, the variation that we detected indicates that P. caudatum, P. e. arachnoideum and possible backcrosses can be found in close proximity. Thus, it is possible that plants referred to P. caudatum include stable fertile allotetraploids, recently formed allotetraploids, homoploid hybrids between P. aquilinum and P. e. arachnoideum, and even possible hexaploid hybrids between P. caudatum and P. e. arachnoideum. Given this level of possible hybridization, we suggest that treating New World Pteridium as three species—the diploids P. esculentum and P. aquilinum, and hybrid P. caudatum (in all its manifestations)—represents best the biological situation in the genus (Thomson and Alonso-Amelot 2002; Zhou et al. 2014).
Pteridium on Galapagos Islands
We found evidence of P. e. arachnoideum, P. caudatum and their possible hybrids inhabiting three Galapagos Islands (Fig. 1), often with more than one taxon in close proximity. There was a tendency for P. caudatum to be found in lower elevation agricultural areas, but it is not clear if this is because of a habitat preference of P. caudatum, or if P. caudatum has been introduced with agricultural material. Long distance dispersal via spores is the most likely explanation for colonization of Pteridium. In fact, there is evidence of Pteridium hybrids in Scotland involving a parent from North America, suggesting transatlantic dispersal of Pteridium spores (Rumsey et al. 1991). Pteridium is highly variable within a relatively small area on the Galapagos Islands, a pattern that has not been observed, to our knowledge, to this extent on the mainland. However, this could be because collections have not been made at the scale used here, or collectors tend to favour specimens that key more easily to one taxon or another, rather than hybrids. If the high variability on the Galapagos Islands is not an artefact, then it could be explained by the location of the Islands. Because the closest mainland areas include both South and Central America, if spores are continually being introduced, then they could easily be coming from multiple sources. This is certainly consistent with the high number of haplotypes on the islands. Introduction from multiple sources has also been inferred for other plant species on the Galapagos Islands (Andrus et al. 2009) and for ferns on other island systems (Shepherd et al. 2009).
Earlier descriptions of the origin of Galapagos flora attributed the majority of the flora to have a Caribbean origin (Duncan and Hargraves 1984) as the result of ancient vicariance events. Conversely, Porter (1979) hypothesized that the Galapagos flora as a whole was mostly of South American origin. More recently, Tye and Francisco-Ortega (2011) compiled phylogenetic data to infer origins and showed that whereas the largest source (45 % of documented colonization events) was South American, other significant sources included Central America and the Caribbean (12 %), and North America (5 %). Pteridium adds an interesting twist to the data because all the above regions appear to be involved, although we do not yet have conclusive evidence for exact sources from North America; they could be from anywhere from Mexico to Florida.
It is unfortunate that we were unable to determine c-values for any of our Galapagos samples because this could have been used to test the prediction that putative hybrids between P. caudatum and P. e. arachnoideum are triploid. Future efforts will be made to sample appropriately for flow cytometry.
Conclusions
The most striking pattern across worldwide Pteridium is the morphological and molecular distinction between the P. aquilinum and P. esculentum clades. About half of the parsimony-informative molecular characters account for this difference between these two diploid species. How might speciation have occurred in the face of gene flow? One possibility is that initial divergence coincided with the separation of the southern landmasses, which was initiated about 180 million years ago (Scotese 2001) and continued until about 30 million years ago (McLoughlin 2001). However, additional factors would be required to maintain such a pattern of separation between species. One factor evident today is that mostly easterly and westerly prevailing winds operate at the equatorial regions (Oort and Yienger 1996). This would explain the similarities within P. aquilinum and within P. esculentum (Thomson 2012). However, northerly or southerly wind patterns crossing the equator, such as the inter-tropical convergence zone (Oort and Yienger 1996; Wright et al. 2001), are relatively rare. Yet this phenomenon could provide the means necessary for gene flow across the equator to enable hybridization between P. aquilinum and P. esculentum, thus forming the hybrids P. caudatum in South America, and P. semihastatum in Australia and tropical Asia (Thomson and Alonso-Amelot 2002).
Our new evidence for P. caudatum as a hybrid between diverged diploid species adds to previous examples from studies on ferns. Nitta et al. (2011) found evidence of hybrids between geographically distinct clades in the filmy fern genus Crepidomanes. Sessa et al. (2012) found evidence of rampant hybridization in Dryopteris. Furthermore, Rothfels et al. (2015) reported hybridization between fern taxa diverged for approximately 60 million years ago. This ability to form hybrids has been attributed to a slower ‘speciation clock’ in plants that lack pre-mating isolation mechanisms that involve biotically mediated fertilization (Rothfels et al. 2015). Such patterns are consistent with our findings for Pteridium in South America, and particularly on the Galapagos Islands.
In order to gain more resolution on origins of Pteridium and its hybrids on the Galapagos Islands, we would need additional genetic resolution. This could best be achieved by sampling at a finer geographic scale, and with many plants per site. In addition to using phylogenetic analysis of nucleotide sequences, it would be useful to include microsatellite loci or single nucleotide polymorphisms (Miller et al. 2007; Hohenlohe et al. 2010). Given our current data, we found evidence for multiple taxa, multiple origins and likely hybridization on an oceanic archipelago. The results from Pteridium add to a growing body of work on the origins of ferns on oceanic islands (Geiger et al. 2007) as well as the origins of the general flora of the Galapagos Islands (Tye and Francisco-Ortega 2011). More studies are needed to test whether these results for Pteridium extend to spore-bearing plants in general.
Accession Numbers
All nucleotide sequences used in this manuscript study have been deposited in (and released by) GenBank. GenBank accession numbers are provided in Table 4 and in more detail in Supporting Information—File S1. All phylogenetic trees and associated nucleotide alignments are deposited in TreeBASE. These data, including the actual nucleotide sequences, can be accessed by reviewers at http://purl.org/phylo/treebase/phylows/study/TB2:S18018?x-access-code=e0885899a1b8bff199f59defe19bb535&format=html.
Sources of Funding
This research project was supported by the Mary Gunson Memorial Bequest. C.J.V. was supported by the National Science Foundation Graduate research fellowship programme: DGE-1315138.
Contributions by the Authors
P.G.W. and J.T. conceived the project, planned the sampling and wrote the manuscript. J.T. conducted morphological and anatomical analyses. M.P.S. led the fieldwork part of the project. C.A.R. conducted all lab work and compiled the data. J.P.D. conducted all phylogenetic analyses and archiving of data. All authors contributed to and approved the manuscript.
Conflict of Interest Statement
None declared.
Supporting Information
The following additional information is available in the online version of this article –
File S1. List of all haplotypes from all four genes, with corresponding GenBank accession numbers.
Acknowledgements
We thank Tom Ranker, Mark Ellis, Alan Smith, Kent Perkins (FLAS), Robbin Moran (NYBG), Michael Kessler, Jefferson Prado and Layne Huiet (DUKE) for help obtaining and identifying specimens. We also thank the staff at CDF (Lenyn Betancourt, Frank Bungartz, Patricia Jaramillo, Sonia Cisneros and Solanda Rea) and the Park Nationale Galapagos for assistance in obtaining permits. We thank Margaret Mayger and Erick Yucailla for field assistance in the Galapagos. Two reviewers made helpful suggestions that improved the manuscript. The lab part of this research was performed while P.G.W. was on sabbatical in the labs of Kathleen Pryer (Duke University) and Pamela and Douglas Soltis (University of Florida). We thank all respective lab members for their hospitality.
Literature Cited
- Andrus N, Tye A, Nesom G, Bogler D, Lewis C, Noyes R, Jaramillo P, Francisco-Ortega J. 2009. Phylogenetics of Darwiniothamnus (Asteraceae: Astereae)—molecular evidence for multiple origins in the endemic flora of the Galápagos Islands. Journal of Biogeography 36:1055–1069. 10.1111/j.1365-2699.2008.02064.x [DOI] [Google Scholar]
- Ashcroft CJ, Sheffield E. 1999. Rejection of Pteridium aquilinum subspecies atlanticum (C. N. Page). Botanical Journal of the Linnean Society 130:157–170. [Google Scholar]
- Bainard JD, Henry TA, Bainard LD, Newmaster SG. 2011. DNA content variation in monilophytes and lycophytes: large genomes that are not endopolyploid. Chromosome Research 19:763–775. 10.1007/s10577-011-9228-1 [DOI] [PubMed] [Google Scholar]
- Barrington DS, Paris CA, Ranker TA. 1986. Systematic inferences from spore and stomate size in the ferns. American Fern Journal 76:149–159. 10.2307/1547723 [DOI] [Google Scholar]
- Ching RC. 1940. On natural classification of the family “Polypodiaceae”. Sunyatsenia 5:201–268. [Google Scholar]
- Christie DM, Duncan RA, Mcbirney AR, Richards MA, White WM, Harpp KS, Fox CG. 1992. Drowned islands downstream from the Galapagos hotspot imply extended speciation times. Nature 355:246–248. 10.1038/355246a0 [DOI] [Google Scholar]
- Der JP, Thomson JA, Stratford JK, Wolf PG. 2009. Global chloroplast phylogeny and biogeography of bracken (Pteridium; Dennstaedtiaceae). American Journal of Botany 96:1041–1049. 10.3732/ajb.0800333 [DOI] [PubMed] [Google Scholar]
- Der JP, Barker MS, Wickett NJ, Depamphilis CW, Wolf PG. 2011. De novo characterization of the gametophyte transcriptome in bracken fern, Pteridium aquilinum. BMC Genomics 12:99 10.1186/1471-2164-12-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J, Greilhuber J, Lucretti S, Meister A, Lysák MA, Nardi L, Obermayer R. 1998. Plant genome size estimation by flow cytometry: inter-laboratory comparison. Annals of Botany 82(Suppl A):17–26. 10.1006/anbo.1998.0730 [DOI] [Google Scholar]
- Duncan RA, Hargraves RB. 1984. Plate tectonic evolution of the Caribbean region in the mantle reference frame. Memoirs of the Geological Society of America 162:81–94. 10.1130/MEM162-p81 [DOI] [Google Scholar]
- Geiger JMO, Ranker TA, Neale JMR, Klimas ST. 2007. Molecular biogeography and origins of the Hawaiian fern flora. Brittonia 59:142–158. 10.1663/0007-196X(2007)59[142:MBAOOT]2.0.CO;2 [DOI] [Google Scholar]
- Grusz AL, Windham MD, Pryer KM. 2009. Deciphering the origins of apomictic polyploids in the Cheilanthes yavapensis complex (Pteridaceae). American Journal of Botany 96:1636–1645. 10.3732/ajb.0900019 [DOI] [PubMed] [Google Scholar]
- Hanson L, Boyd A, Johnson MAT, Bennett MD. 2005. First nuclear DNA C-values for 18 eudicot families. Annals of Botany 96:1315–1320. 10.1093/aob/mci283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA. 2010. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genetics 6:e1000862 10.1371/journal.pgen.1000862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itow S. 2003. Zonation pattern, succession process and invasion by aliens in species-poor insular vegetation of the Galapagos Islands. Global Environmental Research 7:39–58. [Google Scholar]
- Jarrett FM, Manton I, Roy SK. 1968. Cytological and taxonomic notes on a small collection of living ferns from Galapagos. Kew Bulletin 22:475–480. 10.2307/4108355 [DOI] [Google Scholar]
- Klekowski EJ., Jr 1973. Genetic endemism of Galapagos Pteridium. Biological Journal of the Linnean Society 66:181–188. 10.1111/j.1095-8339.1973.tb02168.x [DOI] [Google Scholar]
- Löve A, Kjellqvist E. 1972. Cytotaxonomy of Spanish plants. Lagascalia 2:23–35. [Google Scholar]
- Mcloughlin S. 2001. The breakup history of Gondwana and its impact on pre-Cenozoic floristic provincialism. Australian Journal of Botany 49:271–300. 10.1071/BT00023 [DOI] [Google Scholar]
- Meimberg H, Rice KJ, Milan NF, Njoku CC, Mckay JK. 2009. Multiple origins promote the ecological amplitude of allopolyploid Aegilops (Poaceae). American Journal of Botany 96:1262–1273. 10.3732/ajb.0800345 [DOI] [PubMed] [Google Scholar]
- Mickel JT, Beitel JM. 1988. Pteridophyte flora of Oaxaca, Mexico. Memoirs of the New York Botanical Garden 46:1–568. [Google Scholar]
- Mickel JT, Smith AR. 2004. The pteridophytes of Mexico. Memoirs of the New York Botanical Garden 88:1–1055. [Google Scholar]
- Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA. 2007. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Research 17:240–248. 10.1101/gr.5681207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta JH, Ebihara A, Ito M. 2011. Reticulate evolution in the Crepidomanes minutum species complex (Hymenophyllaceae). American Journal of Botany 98:1782–1800. 10.3732/ajb.1000484 [DOI] [PubMed] [Google Scholar]
- Oort AH, Yienger JJ. 1996. Observed interannual variability in the Hadley circulation and its connection to ENSO. Journal of Climate 9:2751–2767. [DOI] [Google Scholar]
- Ortega FJ. 1990. El genero Pteridium en Venezuela: taxonomia y distribucion geografica. Biollania 7:45–54. [Google Scholar]
- Page CN. 1976. The taxonomy and phytogeography of bracken—a review. Botanical Journal of the Linnean Society 73:1–34. 10.1111/j.1095-8339.1976.tb02010.x [DOI] [Google Scholar]
- Porter DM. 1979. Endemism and evolution in Galapagos Islands vascular plants. In: Bramwell D, ed. Plants and islands. London: Academic Press. [Google Scholar]
- Ranker TA, Floyd SK, Trapp PG. 1994. Multiple colonizations of Asplenium adiantum-nigrum onto the Hawaiian archipelago. Evolution 48:1364–1370. 10.2307/2410392 [DOI] [PubMed] [Google Scholar]
- Rassmann K. 1997. Evolutionary age of the Galápagos iguanas predates the age of the present Galápagos Islands. Molecular Phylogenetics and Evolution 7:158–172. 10.1006/mpev.1996.0386 [DOI] [PubMed] [Google Scholar]
- Rothfels CJ, Larsson A, Li FW, Sigel EM, Huiet L, Burge DO, Ruhsam M, Graham SW, Stevenson DW, Wong GKS, Korall P, Pryer KM. 2013. Transcriptome-mining for single-copy nuclear markers in ferns. PLoS ONE 8:e76957 10.1371/journal.pone.0076957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfels CJ, Johnson AK, Hovenkamp PH, Swofford DL, Roskam HC, Fraser-Jenkins CR, Windham MD, Pryer KM. 2015. Natural hybridization between genera that diverged from each other approximately 60 million years ago. The American Naturalist 185:433–442. 10.1086/679662 [DOI] [PubMed] [Google Scholar]
- Rumsey FJ, Sheffield E, Haufler CH. 1991. A re-assessment of Pteridium aquilinum (L.) Kuhn in Britain. Watsonia 18:297–301. [Google Scholar]
- Schwartsburd PB, De Moraes PLR, Lopes-Mattos KLB. 2014. Recognition of two morpho-types in eastern South American brackens (Pteridium—Dennstaedtiaceae—Polypodiopsida). Phytotaxa 170:103–117. 10.11646/phytotaxa.170.2.3 [DOI] [Google Scholar]
- Scotese CR. 2001. Atlas of Earth history. Arlington, TX: PALEOMAP Project. [Google Scholar]
- Sessa EB, Zimmer EA, Givnish TJ. 2012. Reticulate evolution on a global scale: A nuclear phylogeny for New World Dryopteris (Dryopteridaceae). Molecular Phylogenetics and Evolution 64:563–581. [DOI] [PubMed] [Google Scholar]
- Sheffield E, Wolf PG, Haufler CH, Ranker TA, Jermy AC. 1989. A re-evaluation of plants referred to as Pteridium herediae (Colmeiro) Löve and Kjellqvist. Botanical Journal of the Linnean Society 99:377–386. 10.1111/j.1095-8339.1989.tb00409.x [DOI] [Google Scholar]
- Sheffield E, Wolf PG, Rumsey FJ, Robson DJ, Ranker TA, Challinor SM. 1993. Spatial distribution and reproductive behaviour of a triploid braken (Pteridium aquilinum) clone in Britain. Annals of Botany 72:231–237. 10.1006/anbo.1993.1103 [DOI] [Google Scholar]
- Shepherd LD, De Lange PJ, Perrie LR. 2009. Multiple colonizations of a remote oceanic archipelago by one species: how common is long-distance dispersal? Journal of Biogeography 36:1972–1977. 10.1111/j.1365-2699.2009.02120.x [DOI] [Google Scholar]
- Sigel EM, Windham MD, Pryer KM. 2014. Evidence for reciprocal origins in Polypodium hesperium (Polypodiaceae): a fern model system for investigating how multiple origins shape allopolyploid genomes. American Journal of Botany 101:1476–1485. 10.3732/ajb.1400190 [DOI] [PubMed] [Google Scholar]
- Small RL, Lickey EB, Shaw J, Hauk WD. 2005. Amplification of noncoding chloroplast DNA for phylogenetic studies in lycophytes and monilophytes with a comparative example of relative phylogenetic utility from Ophioglossaceae. Molecular Phylogenetics and Evolution 36:509–522. 10.1016/j.ympev.2005.04.018 [DOI] [PubMed] [Google Scholar]
- Snell HM, Stone PA, Snell HL. 1996. A summary of geographical characteristics of the Galapagos Islands. Journal of Biogeography 23:619–624. 10.1111/j.1365-2699.1996.tb00022.x [DOI] [Google Scholar]
- Soltis DE, Soltis PS. 1987. Polyploidy and breeding systems in homosporous pteridophyta: a reevaluation. The American Naturalist 130:219–232. 10.1086/284706 [DOI] [Google Scholar]
- Soltis PS, Soltis DE. 1991. Multiple origins of the allotetraploid Tragopogon mirus (Compositae): rDNA evidence. Systematic Botany 16:407–413. 10.2307/2419333 [DOI] [Google Scholar]
- Soltis PS, Soltis DE, Holsinger KE. 1988. Estimates of intragametophytic selfing and interpopulational gene flow in homosporous ferns. American Journal of Botany 75:1765–1770. 10.2307/2444691 [DOI] [Google Scholar]
- Tan MK, Thomson JA. 1990. Variation of genome size in Pteridium. In: Thomson JA, Smith RT, eds. Bracken 89: bracken biology and management. Sydney: The Australian Institute of Agricultural Science. [Google Scholar]
- Thomson JA. 2000. Morphological and genomic diversity in the genus Pteridium (Dennstaedtiaceae). Annals of Botany 85:77–99. 10.1006/anbo.1999.1101 [DOI] [Google Scholar]
- Thomson JA. 2012. Taxonomic status of diploid southern hemisphere brackens (Pteridium: Dennstaedtiaceae). Telopea 14:43–48. 10.7751/telopea2012007 [DOI] [Google Scholar]
- Thomson JA, Alonso-Amelot ME. 2002. Clarification of the taxonomic status and relationships of Pteridium caudatum (Dennstaedtiaceae) in Central and South America. Botanical Journal of the Linnean Society 140:237–248. 10.1046/j.1095-8339.2002.00089.x [DOI] [Google Scholar]
- Thomson JA, Martin AB. 1996. Gnarled trichomes: an understudied character in Pteridium. American Fern Journal 86:36–51. 10.2307/1547367 [DOI] [Google Scholar]
- Thomson JA, Mickel JT, Mehltreter K. 2008. Taxonomic status and relationships of bracken ferns (Pteridium: Dennstaedtiaceae) of Laurasian affinity in Central and North America. Botanical Journal of the Linnean Society 157:1–17. 10.1111/j.1095-8339.2008.00791.x [DOI] [Google Scholar]
- Tindale MD, Roy SK. 2002. A cytotaxonomic survey of the Pteridophyta of Australia. Australian Systematic Botany 15:839–937. 10.1071/SB00034 [DOI] [Google Scholar]
- Tryon RM. 1941. A revision of the genus Pteridium. Rhodora 43:1–67. [Google Scholar]
- Tryon RM, Tryon AF. 1982. Ferns and allied plants with special reference to Tropical America. New York: Springer. [Google Scholar]
- Tye A, Francisco-Ortega J. 2011. Origins and evolution of Galapagos endemic vascular plants. In: Bramwell D, Caujapé-Castells J, eds. The biology of island floras. Cambridge: Cambridge University Press. [Google Scholar]
- White WM, Mcbirney AR, Duncan RA. 1993. Petrology and geochemistry of the Galápagos islands: portrait of a pathological mantle plume. Journal of Geophysical Reserch 98:19533–19563. 10.1029/93JB02018 [DOI] [Google Scholar]
- Wright SD, Yong CG, Wichman SR, Dawson JW, Gardner RC. 2001. Stepping stones to Hawaii: a trans-equatorial dispersal pathway for Metrosideros (Myrtaceae) inferred from nrDNA (ITS+ETS). Journal of Biogeography 28:769–774. 10.1046/j.1365-2699.2001.00605.x [DOI] [Google Scholar]
- Yokoya K, Roberts AV, Mottley J, Lewis R, Brandham PE. 2000. Nuclear DNA amounts in roses. Annals of Botany 85:557–561. 10.1006/anbo.1999.1102 [DOI] [Google Scholar]
- Zhou S, Dong W, Chen X, Zhang X, Wen J, Schneider H. 2014. How many species of bracken (Pteridium) are there? Assessing the Chinese brackens using molecular evidence. Taxon 63:509–521. 10.12705/633.9 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.