Abstract
Background: Viscerotropic leishmaniasis caused by Leishmania tropica poses a significant problem in the diagnosis and treatment management. Since differential gene expression is more important in outcome of the infection, we employed proteomic approach to identify potential proteins involved in visceralization of L. tropica.
Methods: The proteomes profiling of L. tropica isolated from cutaneous and visceral tissues of one host were compared by 2-DE/MS proteomics study. Moreover, the transcript level of some identified proteins was confirmed using real-time RT-PCR.
Results: Of the 700 protein spots that were detected reproducibly on each gel, 135 were found to be differentially expressed (P≤ 0.05). Most of responsive proteins in visceral isolate changed in less abundant compared to cutaneous isolate. Among differentially expressed proteins, 56 proteins were confidently identified and classified according to the biological process. The largest groups consist of proteins involved in carbohydrate metabolism and protein synthesis. Most of the identified proteins, which implicated in energy metabolism, cell signaling and virulence were down-regulated, whereas some proteins that have a role in protein folding, antioxidant defense and proteolysis were up-regulated in visceral form. Moreover, the transcript level of some identified proteins such as co-chaperon was confirmed using real-time RT-PCR.
Conclusion: L. tropica probably uses different mechanisms for survival and multiplication in viscera to establish viscerotropic leishmaniasis. The current study provides some clues into the mechanisms underlying the dissemination of L. tropica.
Key Words: L. tropica, Viscerotropic leishmaniasis, Proteome, 2-DE, LC mass spectrometry
Introduction
The order Kinetoplastidae includes a large number of pathogenic parasite species, which could infect a wide range of hosts, including humans, canine, rodents etc. There are approximately 21 species of the genus Leishmania cause leishmaniasis. Depending on the involved species, infection could consist of a spectrum of disease ranging from simple self-limiting cutaneous forms, namely cutaneous leishmaniasis (CL), mucocutaneous leishmaniasis (MCL) to a rather fatal if untreated, visceral leishmaniasis (VL) (1, 2). These clinical forms are primarily attributed to the species of parasites involved and the host immune system, although the mechanisms of species tissue tropism are largely unknown (3).
However, the species of Leishmania is the main factor that determines the clinical presentation. For example, in the old world and south Asia including Iran, L. tropica and L. major are the causative agents of CL and L. infantum is responsible for VL (4, 5). In recent decades, exceptional cases have been observed, such as visceral outcomes in infected individuals with L. tropica and cutaneous outcomes in cases infected with L. infantum (6- 8). Moreover, other confirmed cases were reported in human patients and animal reservoirs such as canine (dogs) which referred to as viscerotropic leishmaniasis (VTL) (9, 4). In fact, viscerotropic leishmaniasis is a concomitant form of cutaneous and visceral disease caused by L. tropica. Unlike classical VL, the variable pathology, lacking hepato splenomegaly and lower serum titers of anti-leishmania antibodies were observed in infected VTL cases (9).
The availability of the genome sequences will serve in ongoing efforts to study the parallel expression of genes and protein contents by variety of proteomic approaches like 2-dimentional gel electrophoresis (2-DE) and mass- spectrometry (10,11). In this regard, proteomics will permit the determination of how parasites interact with their hosts, respond to anti parasitic drugs and develop mechanisms to escape from immune response (12).
In this study, we employed proteomic approach for the first time in order to identify potential proteins implicated in disseminating of L. tropica from cutaneous to the viscera. Two cutaneous and visceral L. tropica isolates were subjected to 2-DE analysis by using pH [4-7] followed by liquid chromatography mass spectrometry (LC/MS) for protein identification. Here the goal was to identify differential protein abundance that may possible represent in the process of viscerotropism in L. tropica.
Materials and Methods
Leishmania tropica isolates
We used the Leishmania isolates obtained from a 5-month old owner dog with multiple cutaneous lesions from Karaj, central Iran was referred to Faculty of Veterinary Medicine and School of Public Health, Tehran university of Medical Sciences (13). The animal had no systemic clinical signs. Biopsy specimens were collected aseptically from cutaneous and visceral tissues including spleen and liver (was later named cutaneous isolate (CI) and viscerotropic isolate (VTI)).
Parasites were grown in RPMI 1640 (Gibco, Germany) with 20% FBS (Gibco) at 23 ºC. The identities of the isolates were obtained by DNA extraction from all the obtained tissues, skin, spleen, and liver followed by PCR amplification of NAGT gene and RFLP and sequencing (14). The nucleotide sequence data were submitted to the GenBank database and registered with accession numbers HM234011 and HM234012 for visceral and cutaneous tissues, respectively.
Protein extraction
Proteomics analysis was performed on triplicate cultured promastigotes obtained from both cutaneous and visceral tissues. Promastigotes were harvested by centrifugation at 3000 rpm for 20 min at 4 ºC and washed in sterile PBS, pH: 7.2-7.4 for 10 min three times. The cells were resuspended in 5 mM Tris–HCl, pH 7.8, containing 1 mM PMSF (Merck, Germany) as a protease inhibitor. The samples were sonicated at 40 Hz 3 times for 10 s with 50 s intervals on ice bath. Proteins were precipitated by 10% (w/v) TCA (Merck, Germany) in acetone (Merck) with 0.07 % (w/v) DTT (Merck) for 1 hour at -20 °C. The samples were then centrifuged at 17500 g (Hettich, Germany) for 15 minutes at 4 °C the pellets were washed with ice-cold acetone containing 0.07% DTT, incubated at -20 °C for 1 h and centrifuged at 4 °C. Washing and sedimentation of the pellets repeated three times and then residual acetone was allowed to air-dry. The samples powders were then solubilized in lysis buffer [9.5 M urea (Merck), 2% (w/v) CHAPS (Merck), 0.8% (w/v) Ampholyte (Bio-Rad, USA) pH 3-10, 1% (w/v) DTT]. The concentration of protein was measured by the Bradford assay (Bio-Rad, USA) with BSA (Merck) as the standard.
2-Dimentional Electrophoresis
The isoelectric focusing and the second dimension were performed as previously described with some modifications (15). For analytical and preparative gels, 120 μg and 1.2 mg of extracted promastigotes proteins were loaded respectively. Isoelectric focusing was carried out on the 18 cm immobilized pH gradient (IPG) strips (pH 4-7) (Bio-Rad, USA). IPG strips were rehydrated overnight by loading the samples diluted with rehydration buffer containing 8 M urea, 4% CHAPS, 2% ampholyte, 50 mM DTT, and traces of bromophenol blue (Merck). Isoelectric focusing (IEF) was conducted at 20 °C with Mutiphor II and a DryStrip kit (GE Healthcare, Germany). The running condition was as follows: 300 V for 90 minute, followed by 500 V for 90 min, 1000 V for 3 hour and finally 3500 V for 16 h. The focused strips were equilibrated twice for 15 min in 10 ml equilibration solution. The first equilibration was performed in a solution containing 6 M urea, 20% (w/v) glycerol, 2% (w/v) SDS (Merck), 1% (w/v) DTT, and 50 mM Tris-HCl (Merck) buffer, pH 8.8. The second equilibration was performed in a solution with 2.5% (w/v) iodoacetamide (Merck). Separation in the second dimension was performed by SDS-PAGE in a vertical slab of acrylamide (Merck) (12% total monomer, with 2.6% cross-linker) using a PROTEAN II Multi Cell (BioRad). The protein spots in analytical and preparative gels were visualized by silver nitrate (Merck, Germany) and CBB/ G-250 (Sigma, Germany) respectively (16, 17).
Staining and gel image analysis
GS-800 densitometer (Bio-Rad) at a resolution of 600 dots per square inch (dpi) was used for scanning of silver stain gels. The scanned gels saved as TIF images for subsequent analysis. Gels were analyzed using the Melanie 6 software (GeneBio, Geneva, Switzerland). Spot detection, protein quantification, and spot pairing were carried out based on software settings. The molecular masses of protein on gels were determined by co electrophoresis of standard protein markers (GE Heathcare) and pI of the proteins were determined by migration of the protein spots on 18 cm IPG (pH 4-7, linear) strips. The percent volume of each spot was estimated and analyzed to protein abundance determination. 2-dimensional gel per sample was run for three biologically independent replicates. Spots were determined to be significantly up- or down-regulated when P< 0.05. The induction factor (IF) was calculated by dividing the percent volume of spots in viscerotropic isolate (VTI) to the percent volume of spots in cutaneous isolate (CI). Statistical analysis of protein variations was carried out using the Student t-test with a confidence level of 95% on relative volume of matched spots.
Protein digestion, peptide extraction and mass analysis
The proteins spots that showed significant statistically changes in VTI compare to CI were excised from the CBB, stained 2-DE gels and subjected to in-gel trypsin digest as described previously (18). Peptides were solubilized in 0.5% formic acid and fractionated on a nanoflow uHPLC system (Thermo RSLCnano) before online analysis by electrospray ionisation (ESI) MS on an Amazon ion trap MS/MS (Bruker Daltonics). Peptide separation was performed on a Pepmap C18 reversed phase column (LC Packings), using a 5 - 85% v/v acetonitrile gradient (in 0.5% v/v formic acid) run over 45 min. at a flow rate of 0.2 l / min. Mass spectrometric (MS) analysis was performed using a continuous duty cycle of survey MS scan followed by up to ten MS/MS analyses of the most abundant peptides, choosing the most intense multiply charged ions with dynamic exclusion for 120 s.MS data was processed using Data Analysis software (Bruker) and the automated Matrix Science Mascot Daemon server (v2.1.06). Protein identifications were assigned using the Mascot search engine to interrogate in house databases of protein sequences for L. major. In all identified proteins, the probability score was greater than the one fixed by MASCOT as being significant, that is, a p value < 0.05.
Real-Time RT-PCR analysis
RNA extraction and cDNA synthesis
Total RNA was extracted from 108 promastigotes of cutaneous and viscerotropic L. tropica isolates during the early stationary phase using Tripure reagent (Roch, Mannheim, Germany) according to the manufacture’s instruction. The quantity and quality of RNA were analyzed using nanodrop (ND-1000, Thermo Scientific Fisher, US) and gel electrophoresis, respectively. The RNAs were treated with RNase-free DNase I (Fermentas, Burlington, Canada) to avoid any genomic contamination. Complementary DNA (cDNA) was synthesized from 1 µg of total RNA using Transcriptor first strand cDNA synthesis Kit (Roch, Mannheim, Germany) following the manufacturer’s instructions.
Real-time RT-PCR analysis
Real-time reverse transcriptase-PCR (RT-PCR) was conducted to investigate the differences in gene expression of a number of dominant proteins between cutaneous and viscerotropic L. tropica isolates. Target gene primers were designed by primer 3 software version 0.4.0 (http://frodo.wi.mit.edu/) according to the identified proteins (Table 1). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene was included for normalization purposes, referred as internal control. RT-PCR was performed in 20 µl reactions containing 1 µl cDNA target, 100 nM forward and reverse primers and 1x SYBR® Premix Ex Taq TM II (Takara, Tokyo, Japan). Experiments were carried out in triplicate using a StepOne TM Real-Time PCR System (Applied Biosystems, Life Technologies, USA). The PCR condition was as follows: activation at 95 °C for 30 s, amplification at 95 °C for 5 s, 60 °C for 30 s for 40 cycles. The relative value of the expression level of each gene was determined based on the threshold cycle (CT) value of the target genes, normalized to that of reference genes (GAPDH) using the 2-∆∆ct method and the level of significance acceptable was 95% (P<0.05) (19).
Table 1.
Sequences of Primers used in Real-Time RT-PCR
| Primers name | Sequence (5' → 3') | Product size |
|---|---|---|
| Triosephosphate isomerase (TPI) | F acacaacatctcccatgacg R gatcggcattgacacttcac |
157 |
| Calmodulin-like protein (CLP) | F gctcgacgtggaacctctt R cagcttaatgaatgcgtcgt |
164 |
| Co-chaperone GrpE (Co-CHP) | F aggcgttttctgccttttc R tggggtcgaactttgtaccta |
159 |
| Elongation factor 1-alpha (EF-1 alpha) | F gatcgagaagttcgagaaggag R acttccacagcgcaatgtc |
125 |
| Small ubiquitin protein (SUP) | F gatatcgctgaaggtcgtcaa R ccctgcttcttgcagtacg |
105 |
| Glyceraldehydes-3-phosphate dehydrogenase (GAPDH) | F gaagtacacggtggaggctg R cgctgatcacgaccttcttc |
206 |
F: Forward, R: Reverse
Results
Proteomic response
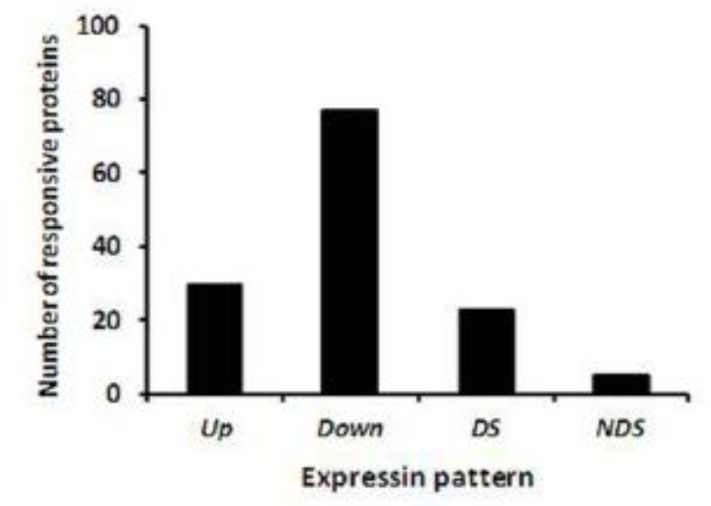
We analyzed the gels from three independent replicates of both cutaneous and visceral L. tropica isolates (6 gels in total). Of the 723 protein spots that were detected reproducibly on each gel, 135 spots showed statistically significant changes (P≤ 0.05) in VTI compared to CI (Fig. 1 and Table 2). Most of responsive proteins (107 proteins) in VI changed in abundance compared to CI, and only 28 proteins changed in gel position (present/absent) (Table 2). The majority of changes in VI proteins were seen as decreased in abundance; out of 135 proteins, only 30 proteins increased in abundance, while 77 proteins down-regulated in VI compared to CI. Some 23 proteins were present only in viscera isolate, while five proteins were only observed in CI and absent in VI (Fig. 2 and Table 2).
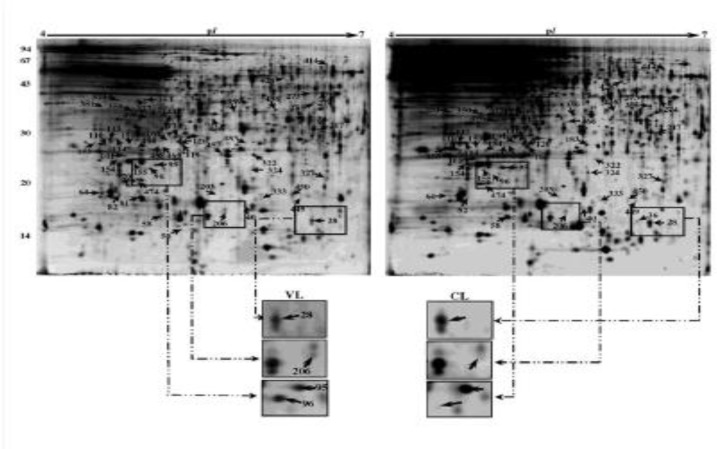
Fig. 1.
2-D gel analysis of proteins extracted from Leishmania tropica isolated from cutaneous and visceral (spleen) of a 5-month old dog. In first dimension (IEF), 120 µg was loaded on a 18-cm IPG strip with a linear gradient of pH 4-7. In the second dimension, 12% SDS-PAGE gels were used, with a well for molecular weight standards. Proteins were visualized by silver staining. Arrows represent spots identified by MS (Tables 2-3). Examples of changes in protein abundance between viscerotropic (VTI) and cutaneous (CI) samples have been presented
Table 2.
Proteins identified using LC/MS analysis
|
Expression pattern.
f
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
Cutaneous isolate (CI)
|
Viscerotropic isolate (VTI)
|
|||||||||
|
Spot
No. a |
Protein name |
Accession
No. b |
(PI/MW)
Exp. c |
(PI/MW)
Theo. d |
Protein Score. e | Ave %vol | Sd | Ave %vol | Sd | Induction factor (V/C). g |
| 28 | hypothetical protein | Q4QFW1 | 6.38/16 | 6.1/15 | 162 | 0.224 | 0.019 | 0.114 | 0.032 | 0.51 |
| 36 | hypothetical protein | Q4QFW1 | 6.3/16 | 6.1/15 | 79 | 0.037 | 0.007 | 0.000 | 0.000 | NDS |
| 44 | ADF/Cofilin | E9ADQ2 | 5.77/15 | 5.6/16 | 219 | 0.526 | 0.097 | 0.109 | 0.189 | 0.21 |
| 50 | 60S ribosomal protein L30 | E9AEK1 | 5.29/15 | 9.7/11 | 43 | 0.000 | 0.000 | 0.027 | 0.009 | DS |
| 58 | hypothetical protein, conserved (Alba superfamily) * | Q4QGA9 | 5.12/16 | 5.2/13 | 409 | 0.238 | 0.014 | 0.089 | 0.043 | 0.37 |
| 64 | 60S ribosomal protein L23a, putative | Q4QJ20 | 6.64/19 | 10.5/16 | 172 | 0.039 | 0.010 | 0.117 | 0.017 | 3.02 |
| 70 | hypothetical protein, conserved (Sfi1 spindle body protein) * | Q4Q6E8 | 4.86/20 | 10.6/121 | 72 | 0.000 | 0.000 | 0.107 | 0.021 | DS |
| 81 | elongation factor 1-alpha | Q4QEI9 | 4.96/18 | 9.0/49 | 167 | 0.000 | 0.000 | 0.222 | 0.031 | DS |
| 82 | 40S ribosomal protein S12, putative | Q4QG97 | 4.79/19 | 4.8/16 | 106 | 0.038 | 0.010 | 0.079 | 0.016 | 2.08 |
| 95 | p21 antigen protein | Q9U8C2 | 5.1/24 | 5.2/21 | 108 | 0.171 | 0.006 | 0.061 | 0.006 | 0.36 |
| 96 | hypothetical protein, unknown function | E9ACC9 | 5.04/23 | 5.3/18 | 145 | 0.007 | 0.002 | 0.057 | 0.006 | 8.38 |
| 101 | calmodulin-like protein | Q4QF91 | 4.8/25 | 4.9/21 | 264 | 0.090 | 0.008 | 0.033 | 0.005 | 0.37 |
| 110 | peroxidoxin | Q4QBH2 | 4.67/27 | 6.4/25 | 205 | 0.084 | 0.010 | 0.031 | 0.003 | 0.37 |
| 113 | proteasome beta 6 subunit, putative | Q4QJ65 | 4.71/29 | 6.5/28 | 174 | 0.163 | 0.037 | 0.068 | 0.007 | 0.42 |
| 114 | triosephosphate isomerase | Q4QAP8 | 4.77/29 | 8.6/27 | 216 | 0.113 | 0.009 | 0.038 | 0.008 | 0.34 |
| 119 | vacuolar sorting-like protein | Q4Q5H7 | 5.3/28 | 5.1/22 | 88 | 0.075 | 0.014 | 0.040 | 0.002 | 0.54 |
| 120 | hs1vu complex proteolytic subunit-like | Q4Q116 | 5.28/28 | 5.7/23 | 227 | 0.017 | 0.005 | 0.035 | 0.003 | 2.03 |
| 131 | hypothetical protein, conserved | Q4Q079 | 5.25/30 | 5.5/29 | 61 | 0.079 | 0.007 | 0.038 | 0.010 | 0.47 |
| 137 | hypothetical protein, conserved (SH3 domain protein) * | Q4QJ54 | 5.38/32 | 5.3/31 | 372 | 0.014 | 0.002 | 0.000 | 0.000 | NDS |
| 144 | glycosomal malate dehydrogenase | Q4QDF0 | 5.1/33 | 9.0/34 | 261 | 0.031 | 0.001 | 0.013 | 0.003 | 0.42 |
| 151 | hypothetical protein, conserved | Q4QED8 | 4.87/28 | 5.2/24 | 196 | 0.019 | 0.004 | 0.004 | 0.001 | 0.19 |
| 153 | peroxidoxin | Q4QBH2 | 4.58/28 | 4.6/25 | 228 | 0.102 | 0.016 | 0.036 | 0.013 | 0.35 |
| 154 | Qc-SNARE protein, putative | Q4QIG6 | 5.78/24 | 8.5/24 | 157 | 0.110 | 0.011 | 0.019 | 0.010 | 0.17 |
| 155 | eukaryotic translation initiation factor 1A, putative | Q4QF06 | 4.92/23 | 4.7/19 | 426 | 0.043 | 0.008 | 0.013 | 0.010 | 0.30 |
| 161 | hypothetical protein, conserved (Alba superfamily) * | Q4Q2T7 | 5.2/29 | 9.8/23 | 224 | 0.024 | 0.001 | 0.004 | 0.001 | 0.18 |
| 183 | hypothetical protein, conserved (Alba superfamily) * | Q4Q2T7 | 5.83/28 | 9.8/23 | 176 | 0.061 | 0.018 | 0.012 | 0.003 | 0.20 |
| Spot No. a |
Protein name | Accession No. b |
(PI/MW) Exp. c |
(PI/MW) Theo. d |
Protein Score. e | Ave %vol | Sd | Ave %vol | Sd | Induction factor (V/C). g |
| 203 | alpha tubulin | Q4QGC5 | 5.61/19 | 4.9/50 | 202 | 0.016 | 0.002 | 0.006 | 0.001 | 0.39 |
| 206 | Tryparedoxin | E9ADX3 | 5.64/16 | 6.6/17 | 40 | 0.018 | 0.001 | 0.037 | 0.007 | 2.11 |
| 217 | proteasome alpha 1 subunit, putative | E9AFW0 | 6.44/31 | 6.8/27 | 202 | 0.058 | 0.008 | 0.030 | 0.006 | 0.52 |
| 255 | cytosolic malate dehydrogenase | Q4Q7X6 | 6.4/37 | 5.8/34 | 493 | 0.061 | 0.011 | 0.032 | 0.013 | 0.53 |
| 273 | NADP-dependent alcohol dehydrogenase, putative | Q4QBD8 | 6.37/40 | 5.8/38 | 249 | 0.095 | 0.004 | 0.031 | 0.004 | 0.33 |
| 274 | NADP-dependent alcohol dehydrogenase, putative | Q4QBD8 | 6.48/40 | 5.8/38 | 366 | 0.287 | 0.031 | 0.146 | 0.019 | 0.51 |
| 322 | co-chaperone GrpE, putative | Q4Q7N4 | 5.89/26 | 7.7/24 | 242 | 0.019 | 0.002 | 0.083 | 0.011 | 4.28 |
| 324 | ADP-ribosylation factor-like protein | Q4Q756 | 5.86/23 | 6.0/20 | 169 | 0.053 | 0.015 | 0.017 | 0.001 | 0.32 |
| 327 | hypothetical protein, unknown function | Q4QEA4 | 6.5/21 | 9.3/17 | 46 | 0.000 | 0.000 | 0.027 | 0.007 | DS |
| 333 | endoribonuclease L-PSP (pb5), putative | Q4QBF5 | 5.96/18 | 5.5/17 | 228 | 0.007 | 0.003 | 0.016 | 0.004 | 2.38 |
| 350 | heat shock protein-like protein, putative | Q4Q584 | 4.9/35 | 5.0/36 | 442 | 0.039 | 0.008 | 0.020 | 0.007 | 0.52 |
| 353 | adenylate kinase, putative | Q4QC71 | 4.5/39 | 5.7/30 | 198 | 0.069 | 0.017 | 0.022 | 0.004 | 0.31 |
| 359 | hypothetical protein, conserved (P-loop_NTPase super family)* | Q4QFN8 | 5.72/36 | 5.6/32 | 85 | 0.074 | 0.013 | 0.030 | 0.012 | 0.40 |
| 374 | succinyl-CoA synthetase alpha subunit, putative | Q4Q9M4 | 4.77/38 | 9.2/31 | 313 | 0.000 | 0.000 | 0.023 | 0.010 | DS |
| 377 | dihydroorotate dehydrogenase | Q4QEW7 | 4.93/39 | 5.7/35 | 83 | 0.030 | 0.007 | 0.006 | 0.003 | 0.21 |
| 381 | hypothetical protein, conserved (Enkurin superfamily) * | Q4QAX0 | 4.65/36 | 9.0/31 | 152 | 0.015 | 0.004 | 0.001 | 0.000 | 0.04 |
| 389 | phosphoglycerate kinase B, cytosolic | Q4QD33 | 4.8/42 | 8.0/45 | 425 | 0.000 | 0.000 | 0.002 | 0.000 | DS |
| 414 | Structure-specific endonuclease subunit SLX1 homolog | Q4Q9W0 | 6.52/57 | 8.7/75 | 29 | 0.061 | 0.012 | 0.029 | 0.007 | 0.48 |
| 428 | elongation factor 1-alpha | Q4QEI9 | 6.14/47 | 9.0/49 | 202 | 0.058 | 0.005 | 0.009 | 0.002 | 0.16 |
| 436 | succinyl-CoA ligase [GDP-forming] beta-chain, putative | Q4Q1C4 | 5.56/33 | 6.5/44 | 345 | 0.000 | 0.000 | 0.047 | 0.012 | DS |
| 438 | hypothetical protein, conserved | O97202 | 4.1/36 | 9.6/46 | 95 | 0.015 | 0.001 | 0.060 | 0.012 | 3.97 |
| 449 | hypothetical protein, conserved | Q4QHN3 | 6.28/18 | 5.9/16 | 194 | 0.017 | 0.004 | 0.007 | 0.002 | 0.42 |
| 450 | hypothetical protein, conserved | Q4QHN3 | 6.22/19 | 5.9/16 | 152 | 0.015 | 0.005 | 0.103 | 0.025 | 6.75 |
| 457 | peroxidoxin | Q4QBH2 | 5.62/26 | 4.6/25 | 205 | 0.000 | 0.000 | 0.039 | 0.000 | DS |
| 466 | glycosomal malate dehydrogenase | Q4QDF0 | 5.66/33 | 9.0/34 | 217 | 0.024 | 0.004 | 0.000 | 0.000 | NDS |
| Spot No. a |
Protein name | Accession No. b |
(PI/MW) Exp. c |
(PI/MW) Theo. d |
Protein Score. e | Ave %vol | Sd | Ave %vol | Sd | Induction factor (V/C). g |
| 468 | heat shock protein 90 | E9ADS8 | 5.1/28 | 5.0/87 | 182 | 0.014 | 0.002 | 0.006 | 0.001 | 0.40 |
| 469 | hs1vu complex proteolytic subunit-like | Q4Q116 | 5.13/27 | 5.7/23 | 194 | 0.018 | 0.002 | 0.008 | 0.002 | 0.42 |
| 474 | small ubiquitin protein, putative | Q4QIC2 | 4.94/20 | 5.0/13 | 126 | 0.009 | 0.002 | 0.019 | 0.004 | 2.16 |
| 484 | peroxidoxin | Q4QBH2 | 4.94/27 | 4.6/25 | 226 | 0.044 | 0.007 | 0.022 | 0.008 | 0.50 |
| 496 | hypothetical protein, conserved (SNF-7-like protein) * | Q4Q130 | 5.02/34 | 4.8/25 | 234 | 0.034 | 0.005 | 0.006 | 0.003 | 0.18 |
The numbering corresponds to the 2-DE gel in Fig. 1./
Accession number in Swiss-Prot. /
Experimental pI and molecular weight.
Theoretical pI and molecular weight./
Mascot score greater than 26 were significant at p=0.05./
Expression pattern of spots that showed a statistically change in cutaneous and visceral leishmaniasis./
The induction factor calculated by dividing the percent volume of spots in gels corresponding to visceral leishmaniasis to the percent volume of spots in cutaneous leishmaniasis samples./ Average percent volume (Ave %vol) and standard deviation (sd) of spots from two samples (cutaneous and visceral) and Three replication have been presented. Spots were concluded to be significantly up- or down-regulated when p was <0.05./
Hypothetical proteins with special domains /The data-base search was run against the NCBI non-redundant protein data-base and Uniprot's Swiss-Prot.
Fig. 2.
Number of proteins differing significantly in abundance in Leishmania tropica isolated from visceral (spleen) (VTI) to cutaneous lesions (CI) of a dog infected to co-cutaneous/viscerotropic leishmaniasis compared.
Up: proteins more abundant in VTI.
Down: proteins less abundant in VTI.
DS: detected spots only in VTI (in fact not detected spots in CI(
NDS: not detected spots in VTI (in fact detected spots only in CI)
Protein identification
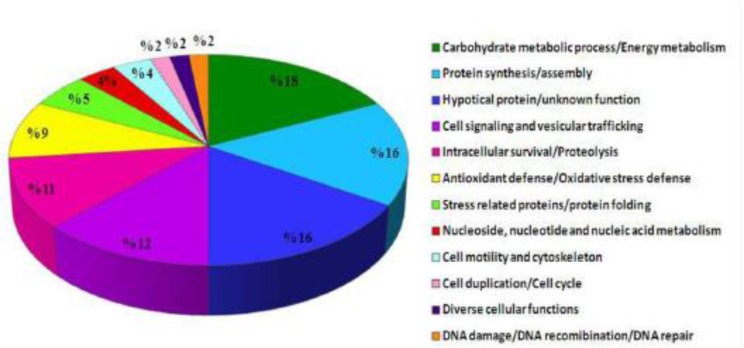
Of 135 differentially expressed proteins detected on the analytical gels, 61 proteins were detected reliably on CBB-stained preparative gels; analyzed using LC/MS/MS after excitation from CBB stained gels. Of these, 56 proteins were identified (Table 2 and Fig. 1). These proteins according to their functions and biological processes were classified in twelve categories: carbohydrate metabolism process, protein synthesis and assembly, cell signaling and vesicular trafficking, intracellular survival/proteolysis, antioxidant defense, stress related proteins/protein folding, cell motility and cytoskeleton, nucleoside, nucleotide and nucleic acid metabolism, cell duplication/cell cycle, diverse cellular functions, DNA damage/DNA recombination/DNA repair (Table 3 and Fig. 3). In addition, some of these proteins are hypothetical proteins, which their functions in Leishmania remain to be elucidated. Most proteins identified in this study were assigned for the first time to a proteome map of L. tropica. The transcript level of some identified proteins was confirmed using real-time RT-PCR.
Table 3.
Functional and Biological process categories of the identified proteins
| Spot No a | Protein name | Accession No b | Induction factor (V/C) c | ||
|---|---|---|---|---|---|
| Carbohydrate metabolic process/Energy metabolism | |||||
| 114 | triosephosphate isomerase | Q4QAP8 | 0.34 | ||
| 131 | hypothetical protein, conserved (ATP synthase B super family) * | Q4Q079 | 0.47 | ||
| 144 | glycosomal malate dehydrogenase | Q4QDF0 | 0.42 | ||
| 255 | cytosolic malate dehydrogenase | Q4Q7X6 | 0.53 | ||
| 273 | NADP-dependent alcohol dehydrogenase, putative | Q4QBD8 | 0.33 | ||
| 274 | NADP-dependent alcohol dehydrogenase, putative | Q4QBD8 | 0.51 | ||
| Protein synthesis/assembly | |||||
| 58 | hypothetical protein, conserved (Alba superfamily) * | Q4QGA9 | 0.37 | ||
| 155 | eukaryotic translation initiation factor 1A, putative | Q4QF06 | 0.30 | ||
| 161 | hypothetical protein, conserved (Alba superfamily) * | Q4Q2T7 | 0.18 | ||
| 183 | hypothetical protein, conserved (Alba superfamily) * | Q4Q2T7 | 0.20 | ||
| 428 | elongation factor 1-alpha | Q4QEI9 | 0.16 | ||
| Cell signaling and vesicular trafficking | |||||
| 101 | calmodulin-like protein | Q4QF91 | 0.37 | ||
| 119 | vacuolar sorting-like protein | Q4Q5H7 | 0.54 | ||
| 154 | Qc-SNARE protein, putative | Q4QIG6 | 0.17 | ||
| 324 | ADP-ribosylation factor-like protein | Q4Q756 | 0.32 | ||
| 381 | hypothetical protein, conserved (Enkurin superfamily) * | Q4QAX0 | 0.04 | ||
| 496 | hypothetical protein, conserved (SNF-7-like protein) * | Q4Q130 | 0.18 | ||
| Intracellular survival/Proteolysis | |||||
| 120 | hs1vu complex proteolytic subunit-like | Q4Q116 | 2.03 | ||
| 333 | endoribonuclease L-PSP (pb5), putative | Q4QBF5 | 2.38 | ||
| 474 | small ubiquitin protein, putative | Q4QIC2 | 2.16 | ||
| Antioxidant defense/Oxidative stress defense | |||||
| 206 | Tryparedoxin | E9ADX3 | 2.11 | ||
| 457 | peroxidoxin | Q4QBH2 | DS | ||
| Stress related proteins/protein folding | |||||
| 322 | co-chaperone GrpE, putative | Q4Q7N4 | 4.28 | ||
| Cell motility and cytoskeleton | |||||
| 44 | ADF/Cofilin | E9ADQ2 | 0.21 | ||
| 203 | alpha tubulin | Q4QGC5 | 0.39 | ||
| Nucleoside, nucleotide and nucleic acid metabolism | |||||
| 353 | adenylate kinase, putative | Q4QC71 | 0.31 | ||
| 377 | dihydroorotate dehydrogenase | Q4QEW7 | 0.21 | ||
| Cell duplication/Cell cycle | |||||
| 70 | hypothetical protein, conserved (Sfi1 spindle body protein) * | Q4Q6E8 | DS | ||
| Table 3. (Continued). | |||||
| Spot No a | Protein name | Accession No b | Induction factor (V/C) c | ||
| Diverse cellular functions | |||||
| 359 | hypothetical protein, conserved (P-loop_NTPase super family) * | Q4QFN8 | 0.40 | ||
| DNA damage/DNA recomibination/DNA repair | |||||
| 414 | Structure-specific endonuclease subunit SLX1 homolog | Q4Q9W0 | 0.48 | ||
| Hypotical protein/unknown function | |||||
| 28 | hypothetical protein | Q4QFW1 | 0.51 | ||
| 36 | hypothetical protein | Q4QFW1 | NDS | ||
| 96 | hypothetical protein, unknown function | E9ACC9 | 8.38 | ||
| 151 | hypothetical protein, conserved | Q4QED8 | 0.19 | ||
| 327 | hypothetical protein, unknown function | Q4QEA4 | DS | ||
| 438 | hypothetical protein, conserved | O97202 | 3.97 | ||
| 449 | hypothetical protein, conserved | Q4QHN3 | 0.42 | ||
| 450 | hypothetical protein, conserved | Q4QHN3 | 6.75 | ||
The numbering corresponds to the 2-DE gel in Fig. 1./
Accession number in Swiss-Prot./
The induction factor calculated by dividing the percent volume of spots in gels corresponding to visceral leishmaniasis to the percent volume of spots in cutaneous leishmaniasis samples./ DS: detected spots only in visceral leishmaniasis, NDS: not detected spots in visceraleishmaniasis./ Significantly up-regulated proteins in visceral leishmaniasis compared to cutaneous leishmaniasis./ Significantly down-regulated proteins in visceral leishmaniasis compared to cutaneous leishmaniasis.
Hypothetical proteins with special domains
Fig. 3.
Functional annotation of the identified proteins classified by biological function and processes described in Table 3.
Discussion
Biological process of proteins identified
Proteins with carbohydrate metabolism activity comprised the largest category (18%) found in this analysis. Six proteins in this group including triosephosphate isomerase (spot 114), hypothetical protein, conserved (ATP synthase B super family) (spot 131), two species of glycosomal and cytosolic malate dehydrogenase (spot 144 and 255) and two species of NADP-dependent alcohol dehydrogenase, putative (spot 273 and 274) showed down-regulation in the viscerotropic isolate (Table 3 and Fig. 3).
The roles of these proteins have been implicated in glycolysis and the tricarboxylic acid cycle pathways that involved in the catabolism of glucose and energy production (20). Suppression of these proteins may suggest that the parasite might have reduced energy production and vital metabolism leading to decrease in multiplication of the parasite of immune system in viscera.
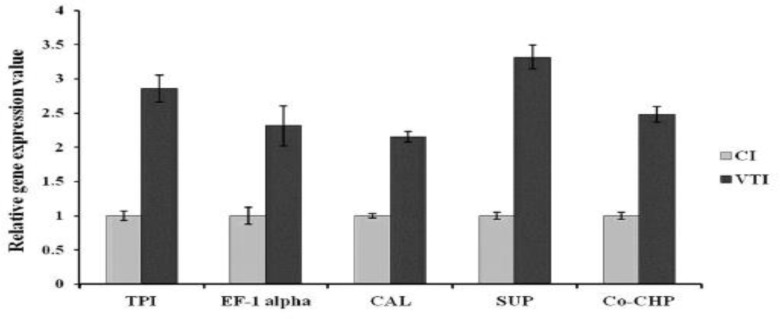
Among this cluster TPI is one of the critical and important glycolytic enzymes, which was identified as a T- cell stimulatory protein and considered as a potential vaccine candidate against VL (21). Down-regulation of this protein in visceral form, might contribute to diminish elicitation of immune response and symptoms in disseminated L. tropica. Moreover, in order to address the relationship of protein expression levels with transcript levels, the mRNA expressions of TPI in cutaneous and viscerotropic L. tropica isolates were analyzed using real-time RT-PCR. Fig. 4 shows a significant up-regulation of TPI (2.855 Fold) in visceral isolate compared to cutaneous one (P<0.05) while it was down regulated at protein level (Table 3). The discrepancy between the expression of TPI at the level of protein and transcriptome indicating that post-transcriptional and post-translational regulation plays an important role in regulating of gene expression (22).
Fig. 4.
Relative gene expression pattern of target genes in cutaneous and visceral L. tropica isolates by real time RT-PCR. The expression of GAPDH was used to normalize the data. The values are the mean ± SD of three independent experiments (P<0.05).
TPI: Triosephosphate isomerase, EF-1 alpha: Triosephosphate isomerase, CAL: Calmodulin-like protein, SUP: Small ubiquitin protein, Co-CHP: Co-chaperone GrpE
Protein synthesis and assembly
The second largest cluster of identified proteins (16%) consists of five down-regulated proteins (Table 3). These include three species of Alba super family (spots 58, 161 and 183), eukaryotic translation initiation factor 1A (TIF-1A) (spot 155) and Elongation factor-1alpha (EF-1alpha) (spot 428) which play a role in protein synthesis and assembly. Down-regulation of this cluster proposes that protein synthesis might be reduced in visceral form to reduce stimulation of immune response. Among this group, EF-1alpha deserves particular attention. It is a highly conserved GTP-binding protein involved in protein translation. In addition, it is an actin/microtubule-binding protein, which interacts with the cytoskeleton and recently in Leishmania, was identified as a virulence factor (23). This protein could diffuse into the cytosol of infected macrophages, where it is able to activate tyrosine phosphatase-1 leading to macrophage deactivation (24). Alternatively, it has been described as a potent antibody inducer involved in the humoral immune response during Mediterranean visceral leishmaniasis (25). Suppression of EF-1 alpha in visceral form could probably contribute to reduce elicitation of immune response and symptoms in viscerotropic form. Additionally, transcript level of EF-1 alpha was detected using real-time RT-PCR. Fig. 4 shows a significant up-regulation of EF-1 alpha (2.315 fold) in viscerotropic isolate in comparison with cutaneous one, which is in contrast with proteomics result, in which EF-1 alpha was down-regulated (Table 3). The inconsistency of results might be due to different post-translational regulation pathways such as changes in RNA stability (22).
Cell signaling and vesicular trafficking
Six of identified proteins were grouped in this cluster (spots 101, 119, 154, 324, 381 and 496) were down-regulated in VI (Table 3 and Fig. 3).One of the cell signaling proteins was calmodulin-like preotein (spot 101). Calmodulin is a kind of calcium binding protein, which expresses in all eukaryotic cells including members of the genus Leishmania; it participates in calcium signaling pathways that regulate multiple critical processes such as growth and proliferation (26). Moreover, it plays a vital role in virulence of Leishmania during invasion of macrophage (27). On the other hand, vacuolar sorting-like protein as a vesicular trafficking protein involved in sorting and delivering of vacuolar proteins to each intracellular compartment (28). In addition, this protein as a member of endosome sorting and authophagy pathways is essential for differentiation and virulence of Leishmania (29). Regarding down-regulation of this group, it is assumed that the parasite limited its central function in terms of cell signaling and vesicular trafficking to diminish induction of immune system. Furthermore, transcript level of CLP was up regulated about 2.156 fold in viscerotropic isolate compared to cutaneous (Fig. 4) which demonstrating of post-translational regulation.
Intracellular survival/Proteolysis
We identified three proteins (spots 120, 333 and 474) were involved in intracellular survival/proteolysis category and were up regulated in VI (Table 3). This cluster consists of hs1vu complex proteolytic subunit-like (spot 120), endoribonuclease L-PSP (pb5), putative (spot 333) and small ubiquitin protein putative (spot 474). Of particular interest is ubiquitin, which plays a vital role in the protein degradation mechanism through the ubiquitin-proteasome pathway (UPP). This pathway is an important protein quality control mechanism for the selective proteolysis of oxidized and misfolded proteins, which prevents the cell from toxic accumulation of abnormal proteins (30). Over expression of ubiquitin might contribute to resistance to oxidative stress, heath shock and antimonial toxicity through degradation of oxidatively damaged proteins (31, 32). Therefore, our results suggest that the up-regulation of proteolysis proteins such as ubiquitin in viscerotropic form could promote degradation of oxidized proteins and thereby protect cell from oxidative stress and increased temperature the parasite encountered in viscera.
Real-time RT-PCR result confirmed the expression pattern of small ubiquitin protein (SUP) in transcript level. As shown in Fig. 4, SUP was up-regulated (3.316 fold) in viscerotropic isolate compared to cutaneous one, which is in agreement with proteomics results (Table 3).
Antioxidant defense /Oxidative stress defense
We identified two proteins involved in antioxidant defense or detoxification of reactive oxygen species (ROS). One protein, tryparedoxin (spot 206) upregulated up to 2.1 fold in visceral isolate and the other one (spot 484) identified as peroxidoxin was detected only in VI (Table 3). Tryparedoxins are special thiol disulfide oxidoreductases related to thioredoxins, which play a crucial role in hydroperoxide detoxification cascades of Kinetoplastida. They mediate electron transfer between trypanothione and a peroxiredoxin leading to reduce and detoxify hydroperoxides and possibly peroxynitrite (33). There is solid evidence that increased levels of tryparedoxins and possibly other components of the parasite hydroperoxide elimination apparatus in the cytosol could provide resistance to host-derived radicals leading to facilitate infection (34). Peroxidoxin as the other identified protein is a tryparedoxin peroxidase, which participates in antioxidant defense by decomposing ROS and reactive nitrogen species (RNS) with cooperation of tryparedoxins (35). Over-expression of peroxidoxin in Leishmania has enhanced resistance to oxidative stress induced by macrophage (36). Hence, enhancement in expression of tryparedoxin and peroxidoxin in visceral form might contribute to protection against macrophage released toxic oxidants and parasite survival in viscera.
Stress related proteins/protein folding
Co-chaperone GrpE (spot 322) was classified in the stress proteins category and was over-expressed up to 4.28 fold in visceral form (Table 3). In addition, RNA expression level of Co-chaperone GrpE was analyzed using Real-time RT-PCR. As shown in Fig. 4 Co-chaperone GrpE was up-regulated (2.479 fold) in viscerotropic isolate compared to cutaneous one, which is well consistent with proteomics result.
Co chaperones are nonclient-binding partners of chaperones and are essential in the function of chaperones such as heat shock proteins (HSP). Co-chaperone GrpE are HSP70 co-chaperones regulating HSP70 action in protein folding (37). HSP 70 and its co-chaperones involved in different cellular processes such as assembly of newly synthesized proteins, refolding of misfolded proteins and regulation of the heat-shock and stress responses (38). Up-regulation of HSP70 protects cells from toxic effects of heat and oxidative stress due to preventing accumulation of misfolded proteins and cell death (39, 40). L. tropica increased co-chaperon expression to shield from increasing of temperature and oxidants, which encountered in dissemination to the viscera.
Cell motility and cytoskeleton
This cluster consists of two down-regulated proteins in visceral isolate identified as ADF/Cofilin (spot 44) and alpha tubulin (spot 203) (Table 3 and Fig. 3). ADF/cofilin exists in all eukaryotic organisms and is involved in multiple actin-based cellular activities, such as cell motility and cytokinesis (41). Leishmania parasites express only one species of ADF/cofilin, which is essentially needed in flagellar assembly and motility (42).
Alpha tubulin is known as one of the members of distinct microtubule networks in Leishmania, which is implicated in locomotion, cell shape and division (43). Alpha tubulin has also been identified as a vaccine candidate antigen from a phage expression library using sera of VL patients (44).
Nucleoside, nucleotide and nucleic acid metabolism
Two proteins recognized in this study belonged to nucleoside, nucleotide and nucleic acid metabolism. These include adenylate kinase putative (spot 353) and dihydroorotate dehydrogenase (spot 377), which both of them down regulated in VI (Table 3 and Fig. 3). There are different isoenzymic forms of adenylate kinase that catalyze the reversible transfer of the terminal phosphate group between ATP and AMP. This enzyme is involved in energy metabolism and nucleic acid synthesis, and is necessary for maintenance and cell growth (45). Earlier studies demonstrated that adenylate kinase may play a role in maintenance of ADP/ATP levels in L. donovani and it could be a potential target antigen for diagnosis or vaccination of leishmaniasis (46). Dihydroorotate dehydrogenas was another recognized protein classified in this category, which is the fourth enzyme in the pyrimidine biosynthetic pathway (47). This protein has been recognized as a potential virulence factors and drug targets in protozoan parasites (48). Down-regulation of aforementioned proteins in visceral form could conceivably reduce nucleotide acid metabolism.
Cell duplication/Cell cycle
In this group a hypothetical protein belonged to Sfi spindle body proteins (spot 70) was detected only in spleen (Table 3 and Fig. 3). In particular, Sfi1 is a centrin binding partner, which localizes to the half-bridge of the spindle pole body (SPB; the microtubule-organizing center) and has an essential function in SPB duplication and cell cycle (49). Moreover, depletion of this protein causes cell arrest with failure to form a mitotic spindle (50).
Diverse cellular functions
We identified a hypothetical protein associated with P-loop-NTPase superfamily (spot 359) which was down regulated in visceral form (Table 3 and Fig. 3). P-loop-NTPases represent a large protein super family and each subgroup member is involved in different cellular processes such as translation, signal transduction, metal insertion and protein transportation (51).
DNA damage/DNA recombination/DNA repair
A structure-specific endonulease subunit SLX homolog (spot 414) was classified in this group, which was down regulated in VI (Table 3 and Fig. 3). Structure-specific endonucleases are responsible for several kinds of DNA repair processes such as nucleotide excision repair and DNA inter strand crosslink repair (52).
We recognized eight hypothetical proteins with unknown function. Additionally, homologous domains were not found through data analysis (Tables 2, 3 and Fig. 3).
Conclusion
L. tropica probably reduces the essential functions such as energy metabolisms, protein synthesis and cell signaling to lower its replication and induction of immune system contributing to establishment of infection in viscera. Moreover, down-regulation of some virulence factors such as elf1-alpha in visceral isolate could help parasite escape from immune system contributing to reduce host immune response and symptoms in viscerotropic form. Enhanced expression of co-chaperon, tryparedoxin and ubiquitin could assist parasite to shield from oxidants induced by immune system as well as higher temperature parasite exposed in viscera.
Further investigations are required to explore precise function of these proteins in this scenario.
Acknowledgments
The authors would like to express their gratitude to Prof. Ghasem Hosseini Salekdeh from Agricultural Biotechnology Research Institute of Iran, (ABRI), for their kind cooperation in proteomics laboratory. We are grateful to Elham Sarhadi from ABRI for valuable technical assistant. Also special thanks from Prof. Burchmore from University of Glasgow, United Kingdom and his colleague Suzzane Eideman for Mass spectrometry analysis. We thank Dr. Malmasi and Dr. Jamshidi from Faculty of Veterinary Medicine, University of Tehran, for providing the samples. This study received financial support from Vice Chancellor for Research, School of Advanced Technologies in Medicine, Tehran University of Medical Sciences (Project No: 10394-87-01-89).The authors declare no competing financial interest.
Abbreviations
ACL, anthroponotic cutaneous leishmaniasis; CI, cutaneous isolate; VTI (VI), Viscerotropic(visceral) isolate; 2 DE, two-dimensional electrophoresis; LC-MS, liquid chromatography/mass Spectrometry; Real-Time RT-PCR, Real-time reverse transcription-polymerase chain reaction; FBS, fetal bovine serum; NAGT, N- acetylglucosamine-1- phosphate transferase; PMSF, phenylmethylsulfonyl fluoride; DTT, dithiothreitol; TCA, trichloroacetic acid; BSA, bovine serum albumin; CHAPS, (3-cholamidopropyl) dimethylammonio
References
- 1.Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J, et al. Leishmaniasis worldwide and global estimates of its incidence. PloS One. 2012:7. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reithinger R, Dujardin JC, Louzir H, Pirmez C, Alexander B, Brooker S, et al. Cutaneous leishmaniasis. Lancet Infect Dis. 2007;7:581–96. doi: 10.1016/S1473-3099(07)70209-8. [DOI] [PubMed] [Google Scholar]
- 3.Foucher AL, Papadopoulou B, Ouellette M. Prefractionation by Digitonin Extraction Increases Representation of the Cytosolic and Intracellular Proteome of Leishmania infantum. J Proteome Res. 2006;5:1741–50. doi: 10.1021/pr060081j. [DOI] [PubMed] [Google Scholar]
- 4.Hajjaran H, Mohebali M, Mamishi S, Vasighe F, Oshaghi MA, Naddaf SR, et al. Molecular Identification and Polymorphism Determina-tion of Cutaneous and Visceral Leishmaniasis Agents Isolated from Human and Animal Hosts in Iran. BioMed Research International. 2013:ID 789326. doi: 10.1155/2013/789326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Control of the leishmaniases: report of a meeting of the WHO Expert Committee on the Control of Leishmaniases; WHO Technical Report No. 949; 22-26 March 2010; Geneva: [Google Scholar]
- 6.Badirzadeh A, Mohebali M, Ghasemian M, Amini H, Zarei Z, Akhoundi B, et al. Cutaneous and post kala-azar dermal leishmaniasis caused by Leishmaniainfantum in endemic areas of visceral leishmaniasis, northwestern Iran 2002‐2011: a case series. Pathog Glob Health. 2013;107:194–7. doi: 10.1179/2047773213Y.0000000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magill A J, Grogl M, Gasser Jr R A, Sun W, Oster CN. Visceral infection caused by Leishmaniatropica in veterans of Operation Desert Storm. N Engl J Med. 1993;328:1383–87. doi: 10.1056/NEJM199305133281904. [DOI] [PubMed] [Google Scholar]
- 8.Zijlstra EE, Musa AM, Khalil EA, el- Hassan IM, el-Hassan AM. Post-kala-azar dermal leishmaniasis. Lancet Infect Dis. 2003;3:87–98. doi: 10.1016/s1473-3099(03)00517-6. [DOI] [PubMed] [Google Scholar]
- 9.Dillon DC, Day CH, Whittle JA, Magill AJ, Reed SG. Characterization of a Leishmaniatropica antigen that detects immune responses in Desert Storm viscerotropic leishmaniasis patients. Proc Natl Acad Sci USA. 1995:7981–85. doi: 10.1073/pnas.92.17.7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fasel N, Acestor N, Fadili-Kundig A, Gonzalez IS. Leishmania After the Genome. Caiser Academic Press; 2008. The Leishmania proteom; pp. 55–65. [Google Scholar]
- 11.Kazemi-Rad E, Mohebali M, Khadem Erfan MB, Saffari M, Raoofian R, Hajjaran H, et al. Identification of antimony resistance markers in Leishmaniatropica field isolates through a cDNA-AFLP approach. Exp Parasitol. 2013;135:344–49. doi: 10.1016/j.exppara.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 12.Paape D, Aebischer T. Contribution of proteomics of Leishmania spp. to the understanding of differentiation, drug resistance mechanisms, vaccine and drug development. J Proteomics. 2011;74:1614–24. doi: 10.1016/j.jprot.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Mohebali M, Malmasi A, Hajjaran H, Jamshidi S, Akhoundi B, Rezaei M, et al. Disseminated leishmaniasis caused by Leishmaniatropica in a puppy from Karaj, Central Iran. Iran J Parasitol. 2011;6:69–73. [PMC free article] [PubMed] [Google Scholar]
- 14.Hajjaran H, Mohebali M, Teimouri A, Oshaghi M A, Mirjalali H, et al. Identification and phylogenetic relationship of Iranian strains of various Leishmania species isolated from cutaneous and visceral cases of leishmaniasis based on N-acetyl glucosamine-1-phosphate transferase gene. Infect Genet Evol. 2014;26:203–12. doi: 10.1016/j.meegid.2014.05.026. [DOI] [PubMed] [Google Scholar]
- 15.Bazargani MM, Sarhadi E, Bushehri AA, Matros A, Mock HP, Naghavi MR, et al. A proteomics view on the role of drought-induced senescence and oxidative stress defense in enhanced stem reserves remobiliza-tion in wheat. J Proteomics. 2011;74:1959–73. doi: 10.1016/j.jprot.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 16.Blum H, Beier H, Gross H J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–9. [Google Scholar]
- 17.Neuhoff V, Arold N, Taube D, Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 1988;9:255–62. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- 18.Daneshvar H, Wyllie S, Phillips S, Hagan P, Burchmore R. Comparative proteomics profiling of a gentamicin-attenuated Leishmaniainfantum cell line identifies key changes in parasite thiol-redox metabolism. J Proteomics. 2012;75:1463–71. doi: 10.1016/j.jprot.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 19.Livak K J, Schmittgen TD. Analysis of relative gene expression data using real- time quantitative PCR and the 2 (Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Opperdoes FR, Coombs GH. Metabolism of Leishmania: proven and predicted. Trends Parasitol. 2007;23:149–58. doi: 10.1016/j.pt.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Kushawaha PK, Gupta R, Tripathi CDP, Khare P, Jaiswal AK, Sundar S, et al. Leishmaniadonovani Triose Phosphate Isomerase: A Potential Vaccine Target against Visceral Leishmaniasis. PLoS One. 2012;7:e45766. doi: 10.1371/journal.pone.0045766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kramer S. Developmental regulation of gene expression in the absence of transcriptional control: the case of kinetoplastids. Mol Biochem Parasitol. 2012;181:61–72. doi: 10.1016/j.molbiopara.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Condeelis J. Elongation factor 1 alpha, translation and the cytoskeleton. Trends Biochem Sci. 1995;20:169–70. doi: 10.1016/s0968-0004(00)88998-7. [DOI] [PubMed] [Google Scholar]
- 24.Nandan D, Yi T, Lopez M, Lai C, Reiner NE. Leishmania EF-1alpha activates the Src homology 2 domain containing tyrosine phosphatase SHP-1 leading to macrophage deactivation. J Biol Chem. 2002;277:50190–7. doi: 10.1074/jbc.M209210200. [DOI] [PubMed] [Google Scholar]
- 25.Kamoun-Essghaier S, Guizani I, Strub J. M., Van Dorsselaer A, Mabrouk K, Ouelhazi L, Dellagi K. Proteomic approach for characterization of immunodominant membrane-associated 30-to 36-kilodalton fraction antigens of Leishmaniainfantum promastigotes, reacting with sera from mediterranean visceral leishmaniasis patients. Clin Diagn Lab Immunol. 2005;12:310–320. doi: 10.1128/CDLI.12.2.310-320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chin D, Means A R. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–8. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 27.Lu HG, Zhong L, Chang KP, Docampo R. Intracellular Ca2+ pool content and signaling and expression of a calcium pump are linked to virulence in Leishmaniamexicana amazonesis amastigotes. J Biol Chem. 1997;272:9464–73. doi: 10.1074/jbc.272.14.9464. [DOI] [PubMed] [Google Scholar]
- 28.Seaman MN, Marcusson EG, Cereghino JL, Emr S D. Endosome to Golgi retrieval of the vacuolar protein sorting receptor, Vps10p, requires the function of the VPS29, VPS30, and VPS35 gene products. J Cell Biol. 1997;137:79–92. doi: 10.1083/jcb.137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Besteiro S, Williams RA, Morrison LS, Coombs GH, Mottram JC. Endosome sorting and autophagy are essential for differentiation and virulence of Leishmania major. J Biol Chem. 2006;281:11384–96. doi: 10.1074/jbc.M512307200. [DOI] [PubMed] [Google Scholar]
- 30.Shang F, Taylor A. Ubiquitin–proteasome pathway and cellular responses to oxidative stress. Free Radic Biol Med. 2011;51:5–16. doi: 10.1016/j.freeradbiomed.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng L, Watt R, Piper P. Polyubiquitin gene expression contributes to oxidative stress resistance in respiratory yeast (Saccharomyces cerevisiae) Mol Gen Genet. 1994;243:358–62. doi: 10.1007/BF00301072. [DOI] [PubMed] [Google Scholar]
- 32.Kazemi-Rad E, Mohebali M, Khadem-Erfan MB, Hajjaran H, Hadighi R, Khamesipour A, et al. Overexpression of Ubiquitin and Amino Acid Permease Genes in Association with Antimony Resistance in Leishmania tropica Field Isolates. Korean J Parasitol. 2013;51:413–9. doi: 10.3347/kjp.2013.51.4.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nogoceke E, Gommel DU, Kieß M, Kalisz HM, Flohé L. A unique cascade of oxidoreductases catalyses trypanothione-mediated peroxide metabolism in Crithidia fasciculata. Biol Chem. 1997;378:827–36. doi: 10.1515/bchm.1997.378.8.827. [DOI] [PubMed] [Google Scholar]
- 34.Castro H, Sousa C, Novais M, Santos M, Budde H, Cordeiro-da-Silva A, et al. Two linked genes of Leishmania infantum encode tryparedoxins localised to cytosol and mitochondrion. Mol Biochem Parasitol. 2004;136:137–47. doi: 10.1016/j.molbiopara.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 35.Rhee SG, Woo HA. Multiple functions of peroxiredoxins: peroxidases, sensors and regulators of the intracellular messenger H2O2, and protein chaperones. Antioxid. Redox Signal. 2011;15:781–94. doi: 10.1089/ars.2010.3393. [DOI] [PubMed] [Google Scholar]
- 36.Lin YC, Hsu JY, Chiang SC, Lee ST. Distinct overexpression of cytosolic and mitochondrial tryparedoxin peroxidases results in preferential detoxification of different oxidants in arsenite-resistant Leishmania amazonensis with and without DNA amplification. Mol Biochem Parasitol. 2005;142:66–75. doi: 10.1016/j.molbiopara.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 37.Caplan AJ. What is a co-chaperone? Cell Stress Chaperones. 2003;8:105–7. doi: 10.1379/1466-1268(2003)008<0105:wiac>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–84. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bellmann K, Jäättelä M, Wissing D, Burkart V, Kolb, H. Heat shock protein hsp70 overexpression confers resistance against nitric oxide. FEBS Lett. 1996;391:185–88. doi: 10.1016/0014-5793(96)00730-2. [DOI] [PubMed] [Google Scholar]
- 40.Chong KY, Lai CC, Lille S, Chang C, Su C Y. Stable overexpression of the constitutive form of heat shock protein 70 confers oxidative protection. J Mol Cell Cardiol. 1998;30:599–608. doi: 10.1006/jmcc.1997.0623. [DOI] [PubMed] [Google Scholar]
- 41.Ono S. Mechanism of Depolymerization and Severing of Actin Filaments and Its Significance in Cytoskeletal Dynamics. Int Rev Cytol. 2007;258:1–82. doi: 10.1016/S0074-7696(07)58001-0. [DOI] [PubMed] [Google Scholar]
- 42.Tammana TVS, Sahasrabuddhe A A, Mitra K, Bajpai VK, Gupta CM. Actin-depolymerizing factor, ADF/cofilin, is essentially required in assembly of Leishmania flagellum. Mol Microbiol. 2008;70:837–52. doi: 10.1111/j.1365-2958.2008.06448.x. [DOI] [PubMed] [Google Scholar]
- 43.Hiam A, Sebastien D, George B, Arlette F, Kalil J, Le Pape P. Microtubule target for new antileishmanial drugs based on ethyl 3-haloacetamidobenzoates. J Enzyme Inhib Med Chem. 2006;21:305–12. doi: 10.1080/14756360600700699. [DOI] [PubMed] [Google Scholar]
- 44.Theinert SM, Basu R, Forgber M, Roy S, Sundar S, Walden P. Identification of New Antigens in Visceral Leishmaniasis by Expression Cloning and Immunoblotting with Sera of Kala-Azar Patients from Bihar, India. Infect Immun. 2005;73:7018–21. doi: 10.1128/IAI.73.10.7018-7021.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan H, Tsai M. Nucleoside monophosphate kinases: structure, mechanism, and substrate specificity. Adv Enzymol Relat Areas Mol Biol. 1999;73:103–34. doi: 10.1002/9780470123195.ch4. [DOI] [PubMed] [Google Scholar]
- 46.Villa H, Pérez-Pertejo Y, García-Estrada C, Reguera RM, Requena JM, Tekwani BL, et al. Molecular and functional characterization of adenylate kinase 2 gene from Leishmania donovani. Eur J Biochem. 2003;270:4339–47. doi: 10.1046/j.1432-1033.2003.03826.x. [DOI] [PubMed] [Google Scholar]
- 47.Feliciano PR, Cordeiro AT, Costa-Filho AJ, Nonato MC. Cloning, expression, purification, and characterization of Leishmania major dihydroorotate dehydrogenase. Protein Expr Purif. 2006;48:98–103. doi: 10.1016/j.pep.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 48.Cuervo P, de Jesus JB, Junqueira M, Mendonca-Lima L, Gonzalez LJ, Betancourt L, et al. Proteome analysis of Leishmania(Viannia)braziliensis by two-dimensional gel ele-ctrophoresis and mass spectrometry. Mol Biochem Parasitol. 2007;154:6–21. doi: 10.1016/j.molbiopara.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 49.Kilmartin JV. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J Cell Biol. 2003;162:1211–21. doi: 10.1083/jcb.200307064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selvapandiyan A, Debrabant A, Duncan R, Muller J, Salotra P, Sreenivas G, et al. Centrin gene disruption impairs stage-specific basal body duplication and cell cycle progression in Leishmania. J Biol Chem. 2004;279:25703–10. doi: 10.1074/jbc.M402794200. [DOI] [PubMed] [Google Scholar]
- 51.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 52.Saito TT, Lui DY, Kim HM, Meyer K, Colaiácovo MP. Interplay between structure-specific endonucleases for crossover control during Caenorhabditis elegans meiosis. PLoS Genet. 2013:9–e1003586. doi: 10.1371/journal.pgen.1003586. [DOI] [PMC free article] [PubMed] [Google Scholar]