Abstract
Background: Cryptosporidium is an intestinal protozean parasite causing waterborne and foodborne outbreaks of diarrheal diseases. The present study was performed in order to find prevalence and subtypes of Cryptosporidium among children with diarrhea in Gonbad Kavoos City, Northern Iran.
Methods: Diarrheic samples were collected from 547 children. The initial parasitological diagnosis was made based on detection of oocysts using the modified Ziehl-Neelsen acid-fast staining method. The positive microscopically samples were selected for sequence analysis of partial 60 kDa glycoprotein (gp60) gene.
Results: Out of 547 collected samples, 27 (4.94%) were positive for Cryptosporidium oocysts. Fifteen from 27 positive samples successfully amplified in PCR. Sequences analysis of gp60 gene in 15 Cryptosporidium isolates revealed that all of them (100%) were C. parvum. The results showed three subtypes of IIa subtype family (7 cases) including IIaA16G2R1, IIaA17G1R1, IIaA22G3R1 and one subtype of IId subtype family (8 cases). The most common allele was IId A17G1d (53.3%).
Conclusion: The predominance of zoonotic subtype families of C. parvum species (IIa, IId) in the present study is in concordance with previous studies in Iran and emphasizes the significance of zoonotic transmission of cryptosporidiosis in the country.
Key Words: Cryptosporidium, Subtypes, Gp60 gene, Children, Iran
Introduction
Cryptosporidiosis is one of the most important zoonotic protozoan diseases caused by Cryptosporidium spp. The organism has a wide host range that includes humans and domestic animals throughout the world. Transmission of infection can be occurred by ingesting oocysts of the parasite thorough the fecal oral route. Many vertebrates, including human, are affected by pathologic changes created by this parasite (1).
Cryptosporidium spp. is a main pathogen causing acute diarrhea, nonspecific signs such as dehydration, anorexia, fever, and weakness. Diarrhea is generally self-limiting in immunocompetent persons. However, it may be major public health importance in children as well as in immunocompromised people (2). Molecular biology has established powerful new tools for categorizing Cryptosporidium and has revealed significant variation within the genus. Currently, the genus Cryptosporidium consists of 30 species. C. parvum and C. hominis are two species predominantly found in humans. However, other species such as C. meleagridis, C. muris, C. felis, C. canis, C. suis and C. andersoni have been occasionally detected in feces of immunocompetent and immunocompromised individuals. Recently, sequencing data of 60 KDa glycoprotein (gp60) gene have revealed substantial genetic heterogeneity among C. hominis and C. parvum isolates establishing different subtype families within both species including Ia, Ib, Id, Ie, If and Ig for C. hominis and IIa, IIb, IIc, IId, IIe, IIf , IIg, IIh, IIi, IIk, and IIl for C. parvum (3, 4).
As there is no genetic data about Cryptosporidium isolates in Gonbad Kavoos City, Northern Iran, this study aimed to find prevalence and identify the subtypes of the Cryptosporidium isolates from children with diarrhea using sequence analysis of the partial gp60 gene in this region.
Materials and Methods
Study population
The study was performed between November 2011 and October 2012 in two hospitals, namely Social Security and Taleghani hospitals in Gonbad kavoos City located in Golestan Province, Northern Iran, south eastern the Caspian Sea. A total of 547 children with diarrhea were examined for this study.
Collection of samples
Stool specimens were collected from each child. An informed consent was obtained from one of children's parents. The samples were concentrated by formalin-ethyl-acetate sedimentation method and stained using the modified Ziehl-Neelsen technique to detection of cryptosporidiosis. Aliquots of Cryptosporidium oocysts positive samples were preserved in 2.5% potassium dichromate and kept at 4 °C until DNA extraction.
DNA extraction
Genomic DNA was extracted from oosysts positive stool samples after washing three times with distilled water to removing the potassium dichromate. The QIAamp® DNA Stool Kit (Qiagen, Hilden, Germany) was employed for DNA extraction according to the manufacturer's instructions. The extracted DNA was stored at -20 °C until PCR analysis.
gp60 Nested Polymerase chain reaction and sequencing
A fragment of 400-500 bp within the gp60 gene (gp60) was amplified by nested PCR from genomic DNA samples as described previously (5). PCR was conducted in 20 µL volumes using 10 pmol of each primer, 200 µM of each dNTP, 2mM MgCl2, and 1U Taq DNA Polymerase (Cinnagen, Tehran, Iran) ,1–10 ng of template DNA using a Techne TC-412 Thermal Cycler (Bibby Scientific Limited, Staffordshire, UK). The following primers were used in the first round of PCR: 5′-ATA GTC TCC GCT GTA TTC-3′ and 5′-GCA GAG GAA CCA GCA TC-3′ employing the following cycling protocol: one cycle at 94 °C for 3 min (initial denaturation), followed by 32 cycles of 94° C for 30 s (denaturation), 42 °C for 30 s (annealing), and 72 °C for 1 min (extension), followed by a final extension at 72 °C for 7 min. In the second round, 1 µL of the primary amplicon was subjected to the PCR using primers 5′-TCC GCT GTA TTC TCA GCC-3′ and 5′-GAG ATATAT CTT GGT GCG -3′ employing the same cycling protocol. A known C. parvum and a sample without DNA were included in each set of PCR as positive and negative controls, respectively.
The secondary PCR products were sequenced using a BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) in a Genetic Analyzer PrismTM 3130x1 (Applied Biosystems, Foster City, CA). All sequences were analyzed and compared with each other and previously reported sequences for identification of the alleles and subtypes reference sequences using the Chromas software (v. 2.4).
Results
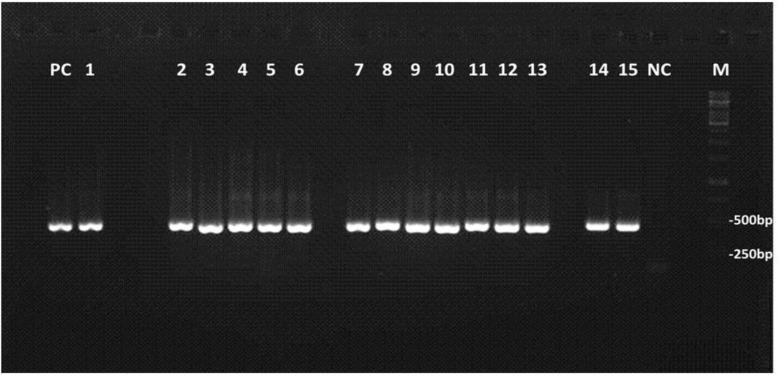
Out of 547 collected diarrheic samples from children, Cryptosporidium oocysts were found in 27 (4.9%) using the modified Ziehl-Neelsen technique. The PCR amplicons about 400 base pairs of gp60 gene were successfully obtained for 15 (of 27) Cryptosporidium positive cases on gel electrophoresis (Fig. 1). Sequence analyses of the partial gp60 sequence data, using well-defined reference sequences for comparison, allowed the genotypic and subgenotypic classification of isolates. Subtypes were identified according to the number of trinucleotide repeats (TCA or TCG) coding for the amino acid serine (6). All isolates were C. parvum species. Four subtypes within two subtype families were identified. Seven (of 15) isolates belonged to the subtype family IIa and remaining 8 isolates belonged to IId. Three subtypes were recognized within the subtype family IIa including IIaA16G2R1 (2/15), IIaA17G1R1 (1/15), IIaA22G3R1 (4/15) while IIdA17G1d (8/15) was the only subtype within IId subtype family (Table 1). Anthroponotic subtype family IIc was not observed among isolates. Four representative sequences of C. parvum subtypes obtained from 15 human isolate in this study submitted to the GenBank under accession numbers: KM114269 to KM114272.
Fig. 1.
PCR of Cryptosporidium isolates from Iranian children based on partial gp60 gene belonged to C. parvum. Lanes 1-15 Cryptospoidium isolates, PC: positive control, NC: negative control, M: DNA size marker
Table 1.
Distribution of Cryptosporidium parvum subtypes in isolates from Iranian diarrheic children from Gonbad Kavoos City, Northern Iran
Discussion
Various prevalences have been described for cryptosporidiosis from different parts of Iran. In this study, a prevalence rate of 4.94% (27/547) was obtained for cryptosporidiosis among diarrheic children from Gonbad Kavoos City, northern Iran.
Similar prevalence has been described from Isfahan (4.6%) the country among diarrheic children (7). Lower prevalence have been reported from Mazandaran Province (0 and 2.3%) in the north, and central provinces of Tehran and Qazvin (1.1%, 2.5%) (8-11). However, higher prevalence has been stated from West Azerbayjan (7.66%) in the Northwest and Bandarabbas (7%) and Shiraz (25.6%) southern the country (12-14). An extensive range of prevalence of the disease have also been described in diarrheic children from other countries, including 0.9% in Malaysia, 1.9% in Philippine, 3.4% in Kuwait, 18.9% in Iraq, 25.3% in Uganda (15-18). The type of technique used for diagnosis and geographical area has been stated to affect the prevalence of cryptosporidiosis (19). Furthermore, dissemination of the parasite in each community and country depends on extent of contamination of water and food, animal contact, health measurements etc.
Previous studies have identified both C. parvum and C. hominis in human, with C. parvum as the predominant species responsible for human cryptosporidiosis in Iran (8, 20-23).
In this study, using sequence analysis of partial gp60 (pgp60) gene, all Cryptosporidium isolates (100%) from diarrheic children were identified as C. parvum species and none of them belonged to C. hominis. That emphasizes the importance of zoonotic transmission of cryptosporidiosis in the country. The predominance of C. parvum species in human cryptosporidiosis in Iran is consistent with studies from some developing and developed countries such as Malaysia, Kuwait, Yemen, Sweden, United Kingdom, Netherland, France, Portugal, Nicaragua (6, 24-31). In contrast, the predominance of C. hominis in human isolates have been reported from Australia (80.2%), India (75%), Egypt (60.5%), Mexico (83.33%) and Peru (70%) (32-36).
Other Cryptosporidium species reported to infect human such as C. meleagridis, C. muris, C. felis, C. canis, and C. andersoni were not found in the present study (37).
Some of C. parvum subtype families such as IIa and IId, are found in both human and livestock responsible for zoonotic transmission of cryptosporidiosis. IId is a major zoonotic subtype family reported in Europe, Asia, Egypt and Australia (38). Eight of fifteen (53.3%) isolates in our study belonged to this subtype family. IIdA17G1d was the most common subtype and the only subtype within IId subtype family. This subtype has been reported in calves from Sweden (39). IIa is the predominant subtype family in animals and human worldwide (38). Seven of fifteen (46.7%) isolates in the current study belonged to IIa subtype family. The second common subtype was IIaA22G3R1 (4/15). This subtype previously has been reported in human isolates from Australia (40, 41). Two isolates were identified as subtype IIaA16G2R1. This subtype previously has been identified in calves from Poland, Netherland, Belgium, France, Spain, Portugal, United States and Canada (42-45). In addition, this subtype has been found in a sample of UV treated water from Portugal (46). Subtype IIaA17G1R1 was identified in one isolate that have been described in human isolates from England and Sweden (30, 47). This subtype has been extensively reported in calves from European countries and Argentina (45, 48). Recently, this subtype has been found in Romanian newborn lambs (49).
The predominance of subtype families IId and IIa and lack of anthroponotic IIc subtype family in the current study is in a close agreement with few subtype analysis studies of cryptosporidiosis that have been performed using sequence analysis of pgp60 gene in Iran. In the study of Nazemalhosseini-Mojarad et al. 36.4% (8/22) and 63.6% (14/22) of human isolates belonged to IIa and IId subtype family, respectively and they did not report any other subtype family (21). In Tehran, 58% (11/19) human isolates were IId subtype family and 31.5% (6/19) were IIa subtype family and 10.5% (2/19) belonged to If subtype family (22). In the current study subtypes IIaA16G2R1, IIaA17G1R1, IIaA22G3R1 and IIdA17G1d are reported for the first time in Iran.
Conclusion
C. parvum was the only species found in Iranian children suffering from diarrhea and other Cryptosporidium species reported to infect the human were not found here. Detecting the subtype families IIa and IId of C. parvum in children suggest that zoonotic transmission play a more important role in human Cryptosporidiosis in Iran. Larger scale studies on subtype analysis of Cryptosporidium isolates from human and domestic animals in other regions of Iran is needed to improve our knowledge of cryptosporidiosis transmission in the country.
Acknowledgments
The authors would like to thank Zohreh Lasjerdi for her technical assistance. This work was financially supported by Golestan University of Medical sciences, project code: 930611115. The authors declare that there is no conflict of interests.
References
- 1.Fayer R. Cryptosporidium: a water-borne zoonotic parasite. Vet Parasitol. 2004;126(1-2):37–56. doi: 10.1016/j.vetpar.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Fayer R, Morgan U, Upton SJ. Epidemiology of Cryptosporidium: transmission, detection and identification. Int J Parasitol. 2000;30(12-13):1305–22. doi: 10.1016/s0020-7519(00)00135-1. [DOI] [PubMed] [Google Scholar]
- 3.Nazemalhosseini-Mojarad E, Feng Y, Xiao L. The importance of subtype analysis of Cryptosporidium spp. in epidemiological investigati-ons of human cryptosporidiosis in Iran and other Mideast countries. Gastroenterol Hepatol Bed Bench. 2012;5(2):67–70. [PMC free article] [PubMed] [Google Scholar]
- 4.Šlapeta J. Cryptosporidiosis and Cryptosporidium species in animals and humans: A thirty colour rainbow? Int J Parasitol. 2013;43(12–13):957–70. doi: 10.1016/j.ijpara.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Abe N, Matsubayashi M, Kimata I, Iseki M. Subgenotype analysis of Cryptosporidium parvum isolates from humans and animals in Japan using the 60-kDa glycoprotein gene sequences. Parasitol Res. 2006;99(3):303–05. doi: 10.1007/s00436-006-0140-0. [DOI] [PubMed] [Google Scholar]
- 6.Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, Iqbal J, Khalid N, Xiao L. Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol. 2005;43(6):2805–09. doi: 10.1128/JCM.43.6.2805-2809.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saneian H, Yaghini O, Yaghini A, Modarresi MR, Soroshnia M. Infection rate of Cryptosporidium parvum among diarrheic children in Isfahan. Iran J Pediatr. 2010;20(3):343–47. [PMC free article] [PubMed] [Google Scholar]
- 8.Keshavarz A, Athari A, Haghighi A, Kazami B, Abadi A, Nazemalhoseini Mojarad E, Kashi L. Genetic characterization of Cryptosporidium spp. among children with diarrhea in Tehran and Qazvin provinces, Iran. Iran J Parasitol. 2008;3(3):30–36. [Google Scholar]
- 9.Gholami S, Khanmohammadi M, Ahmadpour E, Paghe AS, Khadem Nakhjiri S, Ramazannip-our H, Shahbazi A. Cryptosporidium infection in patients with gastroenteritis in Sari, Iran. Iran J Parasitol. 2014;9(2):226–32. [PMC free article] [PubMed] [Google Scholar]
- 10.Nahrevanian H, Azarinoosh SA, Esfandiari B, Ziapoor SP, Shadifar M, Amirbozorgy G, Hayati E, Davoodi J. Current situation of Cryptosporidium and other enteroparasites among patients with gastroenteritis from western cities of Mazandaran province, Iran, during 2007-2008. Gastroenterol Hepatol Bed Bench. 2010;3(3):120–25. [Google Scholar]
- 11.Tahvildar-Biderouni F, Salehi N. Detection of Cryptosporidium infection by modified ziehl-neelsen and PCR methods in children with diarrheal samples in pediatric hospitals in Tehran. Gastroenterol Hepatol Bed Bench. 2014;7(2):125–30. [PMC free article] [PubMed] [Google Scholar]
- 12.Hamedi Y, Safa O, Haidari M. Cryptosporidium infection in diarrheic children in southeastern Iran. Pediatr Infect Dis J. 2005;24(1):86–88. doi: 10.1097/01.inf.0000148932.68982.ec. [DOI] [PubMed] [Google Scholar]
- 13.Mirzaei M. Prevalence of Cryptosporidium sp. infection in diarrheic and non-diarrheic humans in Iran. Korean J Parasitol. 2007;45(2):133–37. doi: 10.3347/kjp.2007.45.2.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nouri M, Moghadam A, Haghighatnia H. Cryptosporidium infection in human diarrhea patients in West Azerbaijan, Iran. Med J Islamic Rep Iran. 1991;5(1,2):35–38. [Google Scholar]
- 15.Menon BS, Shukri Abdullah MD, Mahamud F, Morgan UM, Malik AS, Choo KE, Singh B. Low prevalence of Cryptosporidium parvum in hospitalized children in Kota bharu, Malaysia. Southeast Asian J Trop Med Public Health. 2001;32(2):319–22. [PubMed] [Google Scholar]
- 16.Natividad FF, Buerano CC, Lago CB, Mapua CA, de Guzman BB, Seraspe EB, Samentar LP, Endo T. Prevalence rates of Giardia and Cryptosporidium among diarrheic patients in the Philippines. Southeast Asian J Trop Med Public Health. 2008;39(6):991–9. [PubMed] [Google Scholar]
- 17.Tumwine JK, Kekitiinwa A, Nabukeera N, Akiyoshi DE, Rich SM, Widmer G, Feng X, Tzipori S. Cryptosporidium parvum in children with diarrhea in Mulago Hospital, Kampala, Uganda. Am J Trop Med Hyg. 2003;68(6):710–15. [PubMed] [Google Scholar]
- 18.Al-alousi TI, Mahmood OI. Detection of Cryptosporidium oocysts in calves and children in Mosul, Iraq. 2012;Proc 11th Vet Sci Conf:280–85. [Google Scholar]
- 19.Akinbo FO, Okaka CE, Omoregie R, Dearen T, Leon ET, Xiao L. Molecular characterization of Cryptosporidium spp. in HIV-infected persons in Benin city, Edo State, Nigeria. Fooyin J Health sci. 2010;2(3):85–89. [Google Scholar]
- 20.Meamar AR, Rezaian M, Rezaie s, Mohraz M, Mohebali M, Mohammad K, Golestan B, Guyot K, Dei-Cas E. SSU- rRNA gene analysis of Cryptosporidium spp. in HIV positive and negative patients. Iran J Public Health. 2006;35(4):1–7. [Google Scholar]
- 21.Nazemalhosseini-Mojarad E, Haghighi A, Taghipour N, Keshavarz A, Mohebi SR, Zali MR, Xiao L. Subtype analysis of Cryptosporidium parvum and Cryptosporidium hominis isolates from humans and cattle in Iran. Vet Parasitol. 2011;179(1-3):250–52. doi: 10.1016/j.vetpar.2011.01.051. [DOI] [PubMed] [Google Scholar]
- 22.Taghipour N, Nazemalhosseini- Mojarad E, Haghighi A, Rostami- Nejad M, Romani S, Keshavarz A, Alebouyeh M, Zali M. Molecular epidemiology of Cryptosporidiosis in Iranian children Tehran, Iran. Iran J Parasitol. 2011;6(4):41–45. [PMC free article] [PubMed] [Google Scholar]
- 23.Pirestani M, Sadraei J, Dalimi Asl A, Zavvar M, Vaeznia H. Molecular characterization of Cryptosporidium isolates from human and bovine using 18s rRNA gene in Shahriar county of Tehran, Iran. Parasitol Res. 2008;103(2):467–72. doi: 10.1007/s00436-008-1008-2. [DOI] [PubMed] [Google Scholar]
- 24.Alyousefi NA, Mahdy MA, Lim YA, Xiao L, Mahmud R. First molecular characterization of Cryptosporidium in Yemen. Parasitology. 2013;140(06):729–34. doi: 10.1017/S0031182012001953. [DOI] [PubMed] [Google Scholar]
- 25.Guyot K, Follet-Dumoulin A, Lelievre E, Sarfati C, Rabodonirina M, Nevez G, Cailliez JC, Camus D, Dei-Cas E. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J Clin Microbiol. 2001;39(10):3472–80. doi: 10.1128/JCM.39.10.3472-3480.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim YA, Iqbal A, Surin J, Sim BL, Jex AR, Nolan MJ, Smith HV, Gasser RB. First genetic classification of Cryptosporidium and Giardia from HIV/AIDS patients in Malaysia. Infect Genet Evol. 2011;11(5):968–74. doi: 10.1016/j.meegid.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 27.Alves M, Xiao L, Sulaiman I, Lal AA, Matos O, Antunes F. Subgenotype analysis of Cryptosporid-ium isolates from humans, cattle, and zoo ruminants in Portugal. J Clin Microbiol. 2003;41(6):2744–47. doi: 10.1128/JCM.41.6.2744-2747.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muñoz-Antoli C, Pavón A, Marcilla A, Toledo R, Esteban JG. Prevalence and molecular characterization of Cryptosporidium in schoolchild-ren from department of Rio San Juan (Nicaragua) Trop Biomed. 2011;28(1):40–47. [PubMed] [Google Scholar]
- 29.Wielinga PR, de Vries A, van der Goot TH, Mank T, Mars MH, Kortbeek LM, van der Giessen WB. Molecular epidemiology of Cryptosporidium in humans and cattle in The Netherlands. Int J Parasitol. 2008;38(7):809–17. doi: 10.1016/j.ijpara.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 30.Insulander M, Silverlås C, Lebbad M, Karlsson L, Mattsson JG, Svenungsson B. Molecular epidemiology and clinical manifestations of human cryptosporidiosis in Sweden. Epidemiol Infect. 2013;141(5):1009–20. doi: 10.1017/S0950268812001665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLauchlin J, Amar C, Pedraza-Díaz S, Nichols GL. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1,705 fecal samples from humans and 105 fecal samples from livestock animals. J Clin Microbiol. 2000;38(11):3984–90. doi: 10.1128/jcm.38.11.3984-3990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cama VA, Bern C, Roberts J, Cabrera L, Sterling CR, Ortega Y, Gilman RH, Xiao L. Cryptosporidium species and subtypes and clinical manifestations in children. Peru Emerg Infect Dis. 2008;14(10):1567–74. doi: 10.3201/eid1410.071273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valenzuela O, González-Díaz M, Garibay-Escobar A, Burgara-Estrella A, Cano M, Durazo M, Bernal RM, Hernandez J, Xiao L. Molecular characterization of Cryptosporidium spp. in children from Mexico. PLoS ONE. 2014;9(4):e96128. doi: 10.1371/journal.pone.0096128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma P, Sharma A, Sehgal R, Malla N, Khurana S. Genetic diversity of Cryptosporidium isolates from patients in North India. Int J Infect Dis. 2013;17(8):e601–e05. doi: 10.1016/j.ijid.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Helmy YA, Krücken J, Nöckler K, von Samson-Himmelstjerna G, Zessin KH. Molecular epidemiology of Cryptosporidium in livestock animals and humans in the Ismailia province of Egypt. Vet Parasitol. 2013;193(1–3):15–24. doi: 10.1016/j.vetpar.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 36.O’Brien E, McInnes L, Ryan U. Cryptosporidium GP60 genotypes from humans and domesticated animals in Australia, North America and Europe. Exp Parasitol. 2008;118(1):118–21. doi: 10.1016/j.exppara.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 37.Fayer R, Santín M, Macarisin D. Cryptosporidium ubiquitum n. sp. in animals and humans. Vet Parasitol. 2010;172(1-2):23–32. doi: 10.1016/j.vetpar.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 38.Wang R, Zhang L, Axen C, Bjorkman C, Jian F, Amer S, Liu A, Feng Y, Li G, Lv C, Zhao Z, Qi M, Dong H, Wang H, Sun Y, Ning C, Xiao L. Cryptosporidium parvum IId family: clonal population and dispersal from Western Asia to other geographical regions. Sci Rep. 2014;4:4208. doi: 10.1038/srep04208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silverlås C, Bosaeus-Reineck H, Näslund K, Björkman C. Is there a need for improved Cryptosporidium diagnostics in Swedish calves? Int J Parasitol. 2013;43(2):155–61. doi: 10.1016/j.ijpara.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waldron LS, Ferrari BC, Cheung-Kwok-Sang C, Beggs PJ, Stephens N, Power ML. Molecular Epidemiology and Spatial Distribution of a Waterborne Cryptosporidiosis Outbreak in Australia. Appl Environ Microbiol. 2011;77(21):7766–71. doi: 10.1128/AEM.00616-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jex AR, Whipp M, Campbell BE, Cacciò SM, Stevens M, Hogg G, Gasser RB. A practical and cost-effective mutation scanning-based approach for investigating genetic variation in Cryptosporidium. Electrophoresis. 2007;28(21):3875–83. doi: 10.1002/elps.200700279. [DOI] [PubMed] [Google Scholar]
- 42.Chalmers R, Jackson C, Elwin K, Hadfield S, Hunter P. Investigation of genetic variation within Cryptosporidium hominis for epidemiological purposes. Final report to DWI. National Public Health Service for Wales; 2007. [Google Scholar]
- 43.Wang R, Wang H, Sun Y, Zhang L, Jian F, Qi M, Ning C, Xiao L. Characteristics of Cryptosporidium Transmission in Preweaned Dairy Cattle in Henan, China. J Clin Microbiol. 2011;49(3):1077–82. doi: 10.1128/JCM.02194-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Follet J, Guyot K, Leruste H, Follet-Dumoulin A, Hammouma-Ghelboun O, Certad G, Dei-Cas E, Halama P. Cryptosporidium infection in a veal calf cohort in France: molecular characterization of species in a longitudinal study. Vet Res. 2011;42:116. doi: 10.1186/1297-9716-42-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imre K, Dărăbus G. Distribution of Cryptosporidium species, genotypes and C. parvum subtypes in cattle in European countries. Revista Sci Parasitol. 2011;12(1):1–9. [Google Scholar]
- 46.Lobo ML, Xiao L, Antunes F, Matos O. Occurrence of Cryptosporidium and Giardia genotypes and subtypes in raw and treated water in Portugal. Lett Appl Microbiol. 2009;48(6):732–37. doi: 10.1111/j.1472-765X.2009.02605.x. [DOI] [PubMed] [Google Scholar]
- 47.Bouzid M, Tyler K, Christen R, Chalmers RM, Elwin K, Hunter PR. Multi-locus analysis of human infective Cryptosporidium species and subtypes using ten novel genetic loci. BMC Microbiol. 2010;10(1):213. doi: 10.1186/1471-2180-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomazic ML, Maidana J, Dominguez M, Uriarte EL, Galarza R, Garro C, Florin-Christensen M, Schnittger L. Molecular characterization of Cryptosporidium isolates from calves in Argentina. Vet Parasitol. 2013;198(3–4):382–86. doi: 10.1016/j.vetpar.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 49.Imre K, Luca C, Costache M, Sala C, Morar A, Morariu S, Ilie MS, Imre M, Dărăbus G. Zoonotic Cryptosporidium parvum in Romanian newborn lambs (Ovis aries) Vet Parasitol. 2013;191(1-2):119–22. doi: 10.1016/j.vetpar.2012.08.020. [DOI] [PubMed] [Google Scholar]