SYNOPSIS
Glucocorticoids are primary stress hormones that regulate a variety of physiologic processes and are essential for life. The actions of glucocorticoids are predominantly mediated through the classic glucocorticoid receptor (GR). Glucocorticoid receptors are expressed throughout the body, but there is considerable heterogeneity in glucocorticoid sensitivity and biological responses across tissues. Ligand-activated GR induces or represses the transcription of thousands of genes through direct binding to DNA response elements, physically associating with other transcription factors, or both. The conventional belief that glucocorticoids act through a single GR protein has changed dramatically with the discovery of a diverse collection of receptor isoforms. These GR variants are derived from a single gene by alternative splicing and alternative translation initiation mechanisms. Moreover, posttranslational modifications of these GR isoforms further expand the heterogeneity of glucocorticoid signaling. In this chapter, we provide an overview of the molecular mechanisms that regulate glucocorticoid actions, highlight the dynamic nature of hormone signaling and discuss the molecular properties of the GR isoforms.
Keywords: glucocorticoid, glucocorticoid receptor, glucocorticoid signaling, hypothalamic-pituitary-adrenal axis, isoforms, phosphorylation and polymorphism
INTRODUCTION
Corticosteroids are a class of steroid hormones released by the adrenal cortex, which includes glucocorticoids and mineralocorticoids1. However, the term “corticosteroids” is generally used to refer to glucocorticoids. Named for their effect in carbohydrate metabolism, glucocorticoids regulate diverse cellular functions including development, homeostasis, metabolism, cognition and inflammation2. Due to their profound immune-modulatory actions, glucocorticoids are one of the most widely prescribed drugs in the world and the worldwide market for glucocorticoids is estimated to be worth more than USD 10 billion per year [3]. Glucocorticoids have become a clinical mainstay for the treatment of numerous inflammatory and autoimmune diseases, such as asthma, allergy, septic shock rheumatoid arthritis, inflammatory bowel disease, and multiple sclerosis. Unfortunately, the therapeutic benefits of glucocorticoids are limited by the adverse side effects that are associated with high dose (used in the treatment of systemic vasculitis and SLE) and long-term use. These side effects include osteoporosis, skin atrophy, diabetes, abdominal obesity, glaucoma, cataracts, avascular necrosis and infection, growth retardation, and hypertension3.
Furthermore, patients on long-term glucocorticoid therapy also develop tissue-specific glucocorticoid resistance4. Understanding the molecular mechanisms underlying the physiological and pharmacological actions of glucocorticoids is of great importance as it may aid in developing synthetic glucocorticoids with increased tissue selectivity, which can thereby minimize the side effects by dissociating the desired anti-inflammatory functions from undesirable adverse outcomes. Here, we summarize the recent advances and molecular processes involved in glucocorticoid action and function and discuss in detail the potential role of the glucocorticoid receptor (GR) in determining cellular responsiveness to glucocorticoids.
GLUCOCORTICOID SYNTHESIS, SECRETION AND BIOAVAILABILITY
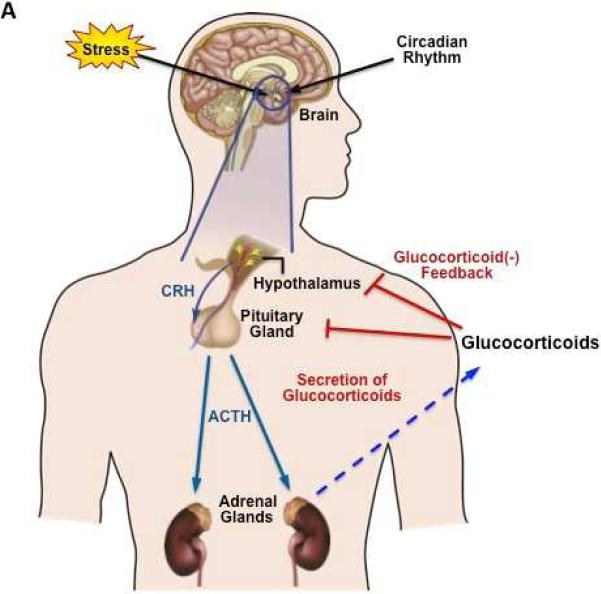
Glucocorticoids (cortisol in man and corticosterone in rodents) are steroid hormones synthesized and released by the adrenal glands in a circadian manner, in response to physiological cues and stress5. The circadian profile of glucocorticoid release from the adrenal glands is regulated by the hypothalamic-pituitary-adrenal (HPA) axis. Inputs from the suprachiasmatic nucleus (SCN) stimulate the para-ventricular nucleus (PVN) of the hypothalamus to release corticotrophin-releasing hormone (CRH) and arginine vasopressin (AVP). These hormones act on the anterior pituitary where they activate corticotroph cells to secrete adrenocorticotrophin hormone (ACTH) into the general circulation. Subsequently, ACTH acts on the adrenal cortex to stimulate the synthesis and release of glucocorticoids (Fig 1A)6. Once released from the adrenal glands into the blood circulation, glucocorticoids access target tissues to regulate a myriad of physiologic processes, including metabolism, immune function, skeletal growth, cardiovascular function, reproduction, and cognition. Due to its lipophilic nature, glucocorticoids cannot be pre-synthesized and stored in adrenal glands, but have to be rapidly synthesized (using a number of enzymatic reactions) upon ACTH stimulation. This feed-forward mechanism within the HPA system is balanced by negative feedback of glucocorticoids acting at both the anterior pituitary and within the hypothalamus to inhibit further release of ACTH and CRH, respectively (Fig 1A)7.
FIG 1.
A. Regulation of glucocorticoid secretion by the hypothalamic-pituitary-adrenal (HPA) axis. Stress induces the release of CRF from the hypothalamus, which is transported to the anterior pituitary, where it triggers the release of ACTH into the blood stream. ACTH stimulates the adrenal cortex to synthesize and release the glucocorticoids (cortisol in humans or corticosterone in rodents). Subsequently, the glucocorticoids act on the hypothalamus and pituitary to dampen excess activation of the HPA axis (“negative feedback system”). CRH, Corticotrophin-releasing hormone, ACTH, Adrenocorticotropic hormone. B. Domain structure of hGR-α. GR contains three major functional regions, the N-terminal transactivation domain (NTD), the central DBD and the C-terminal LBD. The region located between the DBD and LBD is known as the hinge region (H). Regions involved in transcriptional activation (AF1 and AF2), dimerization, nuclear localization and chaperone hsp90 binding are indicated.
Adapted from Ramamoorthy S, Cidlowski JA (2013) Exploring the molecular mechanisms of glucocorticoid receptor action from sensitivity to resistance. Endocr Dev 24:41–56. doi: 10.1159/000342502; with permission.
Biologically active glucocorticoids are synthesized from cholesterol through a multienzyme process termed steroidogenesis5, 7. ACTH increases adrenal gland activity via Protein Kinase A (PKA) activation leading to non-genomic regulation of steroidogenic proteins. This includes phosphorylation of hormone sensitive lipase (HSL), a protein that increases the levels of intracellular cholesterol, and phosphorylation of steroidogenic acute regulatory protein (StAR), which promotes the transport of cholesterol into the mitochondria, where cholesterol is converted into pregnenolone by the enzyme side-chain cleavage cytochrome P450 (P450scc). This process is followed by a number of enzymatic reactions within the mitochondria and the endoplasmic reticulum that ultimately leads to glucocorticoid synthesis within the cells, which, in turn, is released into the general blood circulation7.
The HPA axis has been shown to exhibit a circadian oscillation, thus coupling glucocorticoid synthesis to diurnal patterns. Consequently in humans, serum cortisol concentrations peak in the mornings and are lowest at night. HPA axis is the central stress response system responsible for the adaptation component of the stress response, which attempts to restore homeostasis8. Inappropriate regulation of the stress response has been linked to a wide array of pathologies including autoimmune disease, hypertension, affective disorders and major depression. Systemic serum glucocorticoid level is maintained by adrenal glucocorticoid synthesis, but glucocorticoid availability is further regulated at a tissue or cellular level. In humans, 80-90% of circulating glucocorticoids are bound to corticosteroid binding globulin (CBG) and 5-15% are bound to albumin to maintain the majority of glucocorticoids in an inactive form. Only 5% of systemic glucocorticoids are free and bioactive9. Hence, the accessibility of cortisol is regulated by CBG concentration.
Glucocorticoid availability at the cellular level is sustained by tissue-specific metabolic enzymes 11β-hydroxysteroid dehydrogenases (11β-HSDs)10. 11β-HSDs catalyze the interconversion of active glucocorticoids. 11β-HSD2, functions as a potent dehydrogenase that rapidly inactivates glucocorticoids (converts cortisol to cortisone), thus allowing aldosterone selective access to otherwise non-selective mineralocorticoid receptor in the kidney and the pancreas10. In contrast to endogenous glucocorticoids, most synthetic glucocorticoids do not bind CBG and are not metabolized by 11β-HSD2. However, 11β-HSD1 acts as a predominant 11β-reductase in all the glucocorticoid target tissues such as the liver, adipose tissue, brain and lung, and facilitates the conversion of inactive precursor cortisone to bioactive cortisol, thereby regenerating active glucocorticoids within tissues by exploiting the circulating high levels of inert cortisone11. Thus, the contrasting functions of the isoenzymes 11β-HSD1 and 11β-HSD2 maintain glucocorticoid availability and activity at the cellular level and inhibitors of 11β-HSD1 have been designed to limit the adverse metabolic side effects of increased endogenous glucocorticoids in conditions such as Cushing's syndrome. In fact, impaired hepatic 11b-HSD1 occurs in patients with polycystic ovary syndrome and primary obesity11.
GLUCOCORTICOID RECEPTOR
The physiological and pharmacological functions of glucocorticoids are mediated by the intracellular glucocorticoid receptor (GR), a member of the nuclear receptor family of ligand-activated transcription factors. The GR is a modular protein comprised of three functional domains: an N-terminal transactivation domain (NTD), a central DNA binding domain (DBD) and a C-terminal ligand-binding domain (LBD)12. Two nuclear localization signals are situated within the LBD and at the DBD-hinge region, respectively (Fig 1B). The NTD, covering amino acids 1–420 of the GR is least conserved and is thus the most variable domain among all the nuclear receptors. The NTD contains transcription activation function (AF1) that activates target genes in a ligand-independent fashion and is the primary site for all the posttranslational modifications. The central DBD is the most conserved domain across all the nuclear receptor proteins and harbors two zinc finger motifs that bind target DNA sequences called glucocorticoid response elements (GREs). The LBD houses the hydrophobic ligand-binding pocket formed by 12 α-helices and 4 β-sheets. A second activation function domain (AF-2) is located within this carboxy-terminal region along with sequences essential for ligand-dependent coregulator interactions13.
MECHANISMS OF GLUCOCORTICOID ACTION
GENOMIC ACTIONS OF GLUCOCORTICOIDS
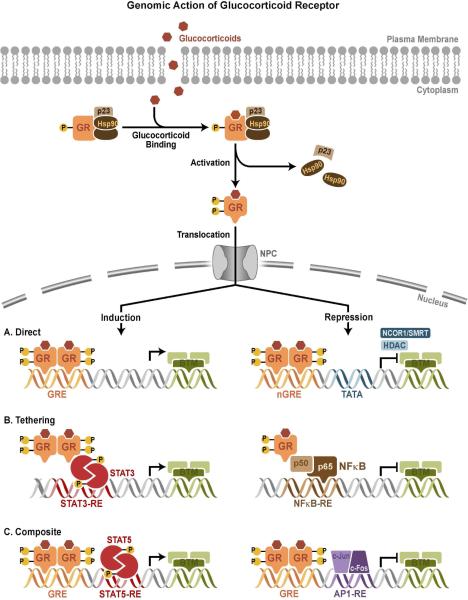
Glucocorticoids signal through genomic and non-genomic pathways. The classic, genomic actions of glucocorticoids are mediated through GR. In the absence of hormone, GR predominantly resides in the cytoplasm of cells as part of a large multi-protein complex that includes chaperone proteins (hsp90, hsp70, and p23) and immunophilins (FKBP51 and FKBP52)14. The multi-protein complex maintains GR in a conformation that favors high affinity ligand binding15. On binding ligand GR undergoes a conformational change, resulting in the dissociation of the multi-protein complex. This leads to a structural reorganization of the GR protein exposing the 2 nuclear localization signals, and the ligand bound GR is rapidly translocated into the nucleus through nuclear pores (Fig.2). Once inside the nucleus, GR binds directly to GREs and stimulates target gene expression. The consensus GRE is a palindromic sequence comprised of 2 half sites (GGAACAnnnTGTTCT) separated by a 3-nucleotide spacer16. GR binds GRE as a dimer and each half site is occupied by one receptor and thus the 3-nucleotide spacer between the 2 half sites is strictly required for GR:DNA interaction (Fig.2)17. Binding of GR to GRE induces conformational changes in GR leading to coordinated recruitment of coregulator and chromatin-remodeling complexes that influence the activity of RNA polymerase II and activates gene transcription and repression. A recent study has identified a negative glucocorticoid-responsive element (nGRE) that mediates glucocorticoid-dependent repression of target genes by recruiting corepressors (NCoR1 and SMRT) and histone deacetlyases (HDACs) (Fig.2)18. The consensus nGRE is palindromic (CTCC(n)0-2GGAGA), but differ from the classic GRE in having a variable spacer that ranges from 0-2 nucleotides and is occupied by 2 GR monomers19.
FIG 2. Genomic action of GR.
Upon binding glucocorticoids, cytoplasmic GR undergoes a conformation change (activation), becomes hyper-phosphorylated (P), dissociates from multi-protein complex, and translocates into the nucleus, where it regulates gene expression. GR activates or represses transcription of target genes by direct GRE binding, by tethering itself to other transcription factors apart from DNA binding, or in a composite manner by both direct GRE binding and interactions with transcription factors bound to neighboring sites. NPC = Nuclear pore complex; BTM = basal transcription machinery; TBP = TATA-binding protein; nGRE = negative GRE; RE = response element. Adated modified from88
Genome-wide GR recruitment studies have shown that only a small proportion of GREs are occupied by GR and specific GR binding sites vary between tissues due to differences in chromatin landscape which influences GRE accessibility20, 21. Comprehensive GR binding analyses revealed that many GR-binding sites identified are located far from the promoter proximal region of target genes and showed an unexpected difference between the activation and repressive functions of the GR [22]. For example, glucocorticoid induction of b-arrestin 1 and repression of b-arrestin 2 occurs through an intron 1 GRE and an intron 11 nGRE, respectively 23. Another example of a GR-binding site located a great distance from the transcription start site is the intragenic nGRE recently identified in exon 6 of the GR gene that mediates homologous down-regulation of GR expression24. A significant proportion of the GR-binding sites lack a consensus GRE element, which suggests that binding of GR to the chromatin may in many cases occur by tethering to other transcription factors. What remains to be established, however, is the functionality of these distant GR-binding sites in relation to the transcription of genes or other undiscovered functions encoded in the GR protein.
Additionally, GR can physically interact with the members of the signal transducer and activator of transcription (STAT) family, either in conjunction with binding a GRE or apart, to enhance transcription of certain target genes (Fig.2)25. Most of the anti-inflammatory effects of glucocorticoids appear to result from an important negative regulatory mechanism called transrepression26, in which ligand-bound GR is recruited to chromatin by protein-protein interactions with DNA-bound transcription factors, particularly NF-κB and activator protein-1 (AP-1). GR directly binds the Jun subunit of AP1 and the p65 subunit of NF-kB and interferes with the transcriptional activation of these 2 proteins (Fig.2). For certain genes, transrepression is accomplished by the GR tethering itself to these DNA-bound transcription factors without itself directly interacting with the DNA27,28. However, for some genes, GR functions in a composite manner, binding directly to a GRE and physically associating with AP1 or NF-kB bound to a neighboring site on the DNA (Fig.2).
Glucocorticoid-induced gene expression is frequently cell type-specific and only a small proportion of genes are commonly activated between different tissues29. Tissue-specific target gene activation by glucocorticoids has been shown to be dependent on accessibility of the GR-binding site which in turn is determined by DNA methylation and higher order chromatin structures like long-range chromatin loops. Thus, tissue-specific target gene activation may be determined by the tissue-specific chromatin landscape, which influences binding of GR to the cognate DNA elements20, 30. Transcriptional regulation by GR is also modulated by recruitment of co-activators, which mediate posttranslational modifications of histones (acetylation and methylation)31, 32. This property aids in altering the chromatin structure and recruiting other cofactors, thus making the chromatin more accessible for the assembly of general transcription factors and the RNA polymerase complex at the target gene promoter33, 34. The identity of co-regulators that contribute to GR transactivation has grown in the recent years to numbers in the hundreds. Some of the well-studied GR co-regulators are the SRC family proteins, mediator complex and SWI/SNF complexes, NCoR1 and SMRT35.
NON-GENOMIC ACTIONS OF GLUCOCORTICOIDS
Rapid, non-genomic glucocorticoid actions are mediated through physiochemical interactions with cytosolic GR or membrane-bound GR. Unlike genomic effects, non-genomic effects of glucocorticoids do not require protein synthesis, and occur within seconds to minutes of GR activation36. A growing body of evidence suggests that the rapid non-genomic functions of GR utilize the activity of various kinases, like phosphoinositide 3-kinase, AKT, and mitogen-activated protein kinases (MAPKs)37. Binding of glucocorticoids to GR not only activates the receptor, but also liberates accessory proteins that participate in secondary signaling cascades. For example, when released from the inactive GR protein complex, c-Src activates signaling cascades that inhibit phospholipase A2 activity, phosphorylate annexin 1, and impair the release of arachidonic acid38, 39. In thymocytes, activated GR translocates to mitochondria and regulates apoptosis40. GR has also been reported to localize in caveolae and glucocorticoid mediated activation of this membrane-associated GR regulates gap junction intercellular communication and neural progenitor cell proliferation41, 42. Thus, rapid non-genomic GR signaling adds greater complexity and diversity to glucocorticoid dependent biological actions.
GLUCOCORTICOID RECEPTOR HETEROGENEITY
GR SPLICE VARIANTS
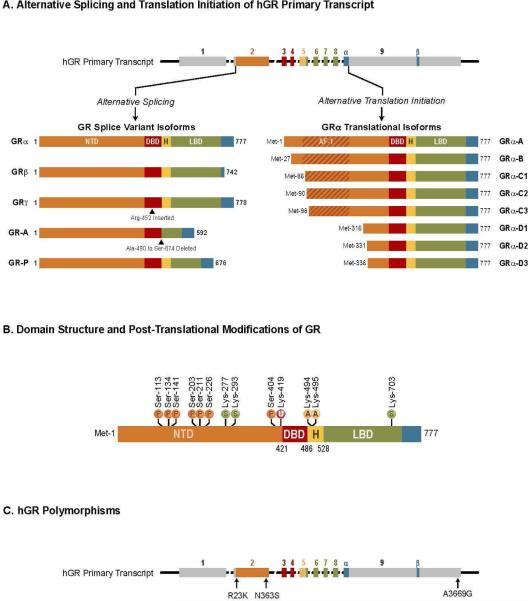
The GR protein is encoded by the Nr3c1 gene and consists of 9 exons; exon 1 forms the 5’-untranslated region, while exon 2–9 code for the GR protein. Exon 2 forms the N-terminal domain of GR, exon 3–4 constitute the central DBD, while exons 5–9 code for the hinge and ligand-binding domain. Alternative splicing at exon 9 of primary GR transcript generates two highly homologous mRNA transcripts that results in the production of two GR isoforms termed GRα and GRβ (Fig.3A)43, 44. The two isoforms are identical up to amino acid 727, but vary beyond this position. GRα, the predominant form of GR is composed of an additional 50 amino acids, while GRβ contains an additional 15 nonhomologous amino acids (Fig.3A)44. The distinct carboxy-terminal residues in GRβ confer unique properties to this GR isoform. GRβ does not bind ligand (due to the lack of helix 12), resides predominantly in the nucleus and is inactive on glucocorticoid-responsive reporter genes45, 46. Conversely, in the presence of GRα, GRβ functions as a dominant negative inhibitor and antagonizes GRα activity on many glucocorticoid-responsive target genes. Recent studies from multiple laboratories suggest that GRβ can function as a bona fide transcription factor by directly inducing and repressing a large number of genes independent of its dominant negative activity on GRα. Indeed, GRβ has been shown to recruit histone deacetylases and repress certain genes such as interleukin (IL)-5 and IL-1347, 48. Although GRβ does not bind glucocorticoids, it actively binds GR antagonist mifepristone (RU486),49 and the endogenous ligand for GRβ is currently unknown.
FIG 3.
A. Alternative splicing and translation initiation of hGR primary transcript. The hGR primary transcript is composed of 9 exons, with exon 2 encoding most of the N- terminal domain (NTD), exons 3 and 4 encoding the DBD, and exons 5–9 encoding the hinge region (H) and LBD. GR splice variant isoform: The classic GRα protein results from splicing of exon 8 to the beginning of exon 9. GRβ is produced from an alternative splice acceptor site that links the end of exon 8 to downstream sequences in exon 9, encoding a variant with a unique 15 amino acid at C terminus (positions 728–742). GRγ is generated from an alternative splice donor site in the intronic sequence separating exons 3 and 4, resulting in a protein with an arginine insertion (Arg-452) between the two zinc fingers of the DBD. GR-A is produced from alternative splicing that joins exon 4 to exon 8, deleting the proximal 185 amino acids of the LBD (Ala-490-Ser-674) encoded by exons 5–7. GR-P is formed by a failure to splice exon 7 to exon 8. The retained intronic sequence introduces a stop codon, resulting in a truncated receptor mutant missing the distal half of the LBD. GRα translational isoforms: Domain organization of the GRα translational isoforms. Initiation of translation from eight different AUG start codons in a single GR-mRNA generates receptor isoforms with progressively shorter N-terminal domains. This generates the GRα translational isoforms GRα-A, B, C1, C2, C3, D1, D2 and D3. B. Domain structure and posttranslational modifications of hGR-α. Sites of posttranslational modifications like phosphorylation (P), sumoylation (S), ubiquitination (U) and acetylation (A) are indicated. C. hGR polymorphisms. Arrows indicate polymorphisms that result in amino acid changes and A3669G which leads to GR stability. Adopted and modified from88
GRβ is expressed in different tissues but generally at lower levels than GRα. However, GRβ is abundant in certain cell types, such as neutrophils and epithelial cells50. The molecular factors that control GRβ expression are poorly understood, but several studies have implicated the involvement of the splicing factor, SRp30c51, 52. Besides, the expression of GRβ can be increased by pro-inflammatory cytokines and other immune activators and lead to reduced GRα:GRβ ratio and glucocorticoid resistance. Reduced GRα:GRβ ratio has been associated with mood disorders such as schizophrenia, bipolar and major depressive disorders. Elevated GRβ levels have been associated with glucocorticoid resistance in several inflammatory diseases, including asthma, rheumatoid arthritis, ulcerative colitis, nasal polyposis, systemic lupus erythematosus, sepsis, acute lymphoblastic leukemia, and chronic lymphocytic leukemia46. Thus, manipulating GRα:GRβ expression ratios, may provide a means to modulate glucocorticoid sensitivity. Another significant finding has been the discovery of GRβ in mice, rat and zebra fish. These isoforms arise from a distinct splicing mechanism that employs alternative splice donor sites in the intron separating exons 8 and 9, resulting in a GRβ isoform similar in structure and function to human GRβ 53, 54, 55.
The additional splice variants of GR, GRγ, GR-A, and GR-P, were discovered in glucocorticoid-resistant cancer cells, and later in healthy tissues. Initially identified in cancer cells and blood mononuclear cells, GRγ is a splice variant in which exon 4 is alternatively spliced to exon 3, thereby including 3 bp of the intron region resulting in an additional arginine residue between the zinc fingers of the DBD (Fig.3A)56. GRγ exhibits ~50% of the activity of GRα for canonical glucocorticoid target genes, and GRγ expression in childhood acute lymphoblastic leukemia has been shown to correlate with resistance to glucocorticoid treatment57. Glucocorticoid resistance in small cell lung carcinoma and corticotroph adenomas is also associated with GRγ expression in these cancers58. The GR-A variant is generated by splicing of exon 4 to exon 8, resulting in a transcript lacking exon 5-7, which encoded the amino-terminal half of the LBD (Fig.3A). Failure to splice at exon 7/8 boundary yields GR-P isoform, which lacks the carboxy-terminal half of the LBD (Fig.3A). Due to defective LBD, both GR-A and GR-P do not bind glucocorticoids. Little is known about the GR-A's biological functions, however, GR-P has been shown to modulate the transcriptional activity of GRα in a cell type–specific manner59, 60. The GR-P variant is expressed in normal tissue and has been reported to be up-regulated in many glucocorticoid resistant hematological malignancies (Acute lymphoblastic leukemia, non-Hodgkin's lymphoma and multiple myeloma)61.
GR TRANSLATIONAL ISOFORMS
Apart from GR splice variants, alternative translation initiation from the single GRα mRNA produces an additional cohort of diverse GR proteins, adding to glucocorticoid receptor heterogeneity62, 63. Eight highly conserved AUG start codons in exon 2 of the GR transcript give rise to eight GRα variant with progressively shorter N-terminus. These receptor isoforms are designated GRα-A, -B, -C1, -C2, -C3, -D1, -D2 and -D3 (Fig.3A). The GRα-A isoform is the classical full-length receptor containing amino acids1–777. Ribosomal leaky scanning and ribosomal shunting mechanisms are involved in the generation of the GRα subtypes. Each of the GR splice variants (GRβ, GRγ, GR-A, and GR-P) would also be predicted to give rise to a similar complement of N-terminal isoforms. The GRα translational isoforms, distinguished only by the length of the NTD, have similar affinity for glucocorticoids and similar GRE binding capability following ligand-dependent activation. However, the subcellular localization of the isoforms differs, with GRα-D isoforms residing constitutively in the nucleus. In contrast, GRα-A, GRα-B, and GRα-C isoforms are localized in the cytoplasm of cells in the absence of hormone and translocate to the nucleus on glucocorticoid binding62. An interesting difference among the GRα isoforms, is that each GR variant possesses a distinct transcription profile. When individual isoforms are expressed in U2OS osteosarcoma or Jurkat T lymphoblastic leukemia cells, they each regulate a unique set of genes, with less than 10% being commonly regulated by all the isoforms64, 65. The isoform specific gene-regulatory profile produced functional differences in glucocorticoid-induced apoptosis in these cells. Cells expressing GRα-C3 exhibited highest sensitivity to glucocorticoid-induced apoptosis, while the GRα-D3 expressing cells were the most resistant66. The GRα-C3 is the most active and the heightened activity has been linked to an N-terminal motif (residues 98-115) that is sterically hindered in the larger GR isoforms. The unobstructed AF1 domain of GRα-C3 effectively recruits various co-regulators and enhances the transcriptional activity of GRα-C3 67. However, the lack of AF1 domain might be responsible for diminished transcriptional activity in GRα-D3. Unlike the other receptor isoforms, GRα-D3 does not repress the transcription of anti-apoptotic genes Bcl-xL, cellular inhibitor of apoptosis protein 1 and survivin. The inability of GRα-D to down-regulate the expression of these genes is associated with a weak interaction between GRα-D and NF-κB66.
The translational isoforms exhibit extensive tissue distribution, although their relative levels vary both between and within cells. In rodents, the GRα-A and GRα-B isoforms are the most abundant GR proteins in many tissues62. The highest GRα-C expression is found in pancreas, lung, and colon. The GRα-D variants are prevalent in the spleen and bladder but are expressed at low levels, while GRα-B is more abundant in thymus and colon. Recent studies have also demonstrated changes in the cellular complement of GRα translational isoforms in response to various cellular stimuli [68]. Moreover, the relative levels of the GRα subtypes expressed in the human brain were found to change during development and the aging process69, 70. The molecular mechanisms governing the expressed complement of translational isoforms are poorly understood. Therefore, genetic manipulation of the GR translational isoforms in animal models may shed new light on the biological importance of these intriguing GR variants, and it will be important to determine the contributions of a single GR isoform in a whole animal. Furthermore, it is critical to verify if glucocorticoid-resistant cells exhibit an altered pattern of GR isoform expression.
POST-TRANSLATIONAL MODIFICATIONS OF GR
Posttranslational modifications of GR further modulate the transcriptional landscape of the receptor. The most extensively studied covalent modification of GR is phosphorylation and at least seven serine residues (Ser-113, Ser-134, Ser-141, Ser-203, Ser-211, Ser-226 and Ser-404) that are phosphorylated in hGR, and all these sites are also conserved in mouse and rat (Fig.3B). Other phosphorylation sites include Ser-45 and 267. The receptor displays a basal level of phosphorylation and becomes hyperphosphorylated upon binding glucocorticoids, however, the structure of the ligand determines both the pattern and extent of GR phosphorylation. Different kinases are involved in the phosphorylation of GR, which includes MAPKs, cyclin-dependent kinases, casein kinase II, and glycogen synthase kinase 3β. Phosphorylation of GRα changes its transcriptional activity, often in a gene-specific manner71, 72. Ligand-dependent phosphorylation of GR at Ser-211 correlates with elevated transcriptional activity, while phosphorylation at Ser-226 decreases GR transcriptional activity. Impaired Ser-211 phosphorylation might lead to glucocorticoid resistance in malignant lymphoid cells, in contrast, hyperphosphorylation at Ser-226 might account for decreased GR signaling in the pathology of depression72, 73. Ligand-induced phosphorylation at Ser-404 has been shown to impact transcriptional activity of GR by impairing both activation and repression of target genes74. Ser-134 is exclusive in that it is phosphorylated in a glucocorticoid-independent manner by stress stimuli including glucose starvation, oxidative stress, UV irradiation and osmotic shock75.
Phosphorylation of GR also modifies other properties of GR that affect GR signaling76. The cellular compartmentalization of GR is altered by phosphorylation, GR phosphorylation at Ser-203, Ser-226 or Ser-404 enhances cytoplasmic retention thus reducing transcriptional activity. Degradation of the GR protein is enhanced by glucocorticoid-dependent phosphorylation at Ser-404 because phosphorylation-deficient mutants are stabilized in the presence of glucocorticoids77.
GR protein is also subject to a variety of other posttranslational modifications that regulate the function of the receptor. Ubiquitin is a 76-amino-acid protein that, when attached to specific lysine residues, marks proteins for proteasomal degradation. GR is ubiquitinated at a conserved lysine residue located at position 419 (Lys-419), and this modification targets the receptor for degradation by the 26S proteasome (Fig.3B)78,79. Mutation of this conserved Lys residue enhances the glucocorticoid-induced transcriptional activity of GR and blocks ligand-dependent down-regulation of GR80. Another important posttranslational modification of GR is the covalent addition of a small ubiquitin-related modifier-1 (SUMO-1) termed sumoylation. GR is sumoylated at residues Lys-277, Lys-293 and Lys-703 (Fig.3B). Sumoylation of GRα has been shown to promote its degradation and inhibits the transcriptional activity of GR in a promoter-specific manner by recruiting corepressors81. It has been suggested that GR can be acetylated at lysine residues -494 and -495 and acetylation of GR alters the inhibitory actions of glucocorticoids on NF-κB (Fig.3B). Recent studies have shown that acetylation of GR by clock transcription factor reduces GR transcriptional activity82, 83. In summary, posttranslational modification of GR regulates multiple aspects of GR function and adds to receptor heterogeneity.
GLUCOCORTICOID RECEPTOR POLYMORPHISMS
A polymorphism is defined as an inheritable genetic germ line variant of a single locus (most frequently a single nucleotide variation) that is present in at least 1% of the population. Polymorphisms in the GR gene that alter the amino acid sequence are linked to impaired GR function as a transcriptional activator or repressor. The N363S polymorphism, located within exon 2 (Fig.3C), occurs in ~4% of the population, results in modest increases in GR transcriptional activity, and is associated with generalized increases in glucocorticoid sensitivity. N363S carriers have been reported to have an increased body mass index, coronary artery disease and decreased bone mineral density84. The ER22/23EK polymorphism that occurs in ~3% of the individuals results in an arginine (R) to lysine (K) change at position 23 (R23K) within the N terminus (Fig.3C). ER22/23EK is associated with decreased GR transcriptional activity. The ER22/23EK polymorphism has been shown to increase the ratio of GRα-A to GRα-B and the carriers of ER22/23EK polymorphism have a lower tendency to develop impaired glucose tolerance, type-2 diabetes and cardiovascular disease85.
The A3669G polymorphism in GRβ 3’ untranslated region results in an increase of both GRβ mRNA and protein (Fig.3C). Moreover, carriers of A3669G polymorphism have a higher incidence of rheumatoid arthritis and cardiovascular disease. Individuals homozygous for A3669G polymorphism were associated with a proinflammatory phenotype with an increased risk for myocardial infarction and coronary heart disease86.
SUMMARY AND FUTURE CONSIDERATIONS
Glucocorticoids are primary stress hormones that regulate a vast array of physiological processes, and synthetic derivatives of these molecules are widely used in the clinic for treating inflammatory disorders, autoimmune diseases and hematological cancers. Despite the efficacy of glucocorticoids in the treatment of inflammatory and immune disorders, their utility is limited by harmful side effects of chronic and/or high dose treatment. These side effects include diabetes, impaired wound healing, skin atrophy, muscle atrophy, HPA dysfunction, cataracts, peptic ulcers, hypertension, metabolic syndrome, osteoporosis, and water/electrolyte imbalance. Considerable effort has been dedicated over the last several decades to enhance glucocorticoid potency while minimizing adverse side effects by modifying the chemical structure of the natural glucocorticoids87. The discovery that multiple GR isoforms with unique expression, gene-regulatory, and functional profiles are generated by alternative splicing, alternative translation initiation of the mature mRNA, and posttranslational modifications have advanced our understanding of molecular basis for the diversity in glucocorticoid sensitivity (hyposensitivity or hypersensitivity). Genome wide GR recruitment studies have shown that tissue specific chromatin landscape also exhibits profound differences in glucocorticoid sensitivity.
An important challenge in the clinical application of glucocorticoids is the heterogeneity in glucocorticoid responsiveness among individuals, with a significant portion of the population (up to 30%) exhibiting some degree of glucocorticoid resistance. Progress in our understanding of glucocorticoid expression patterns have uncovered a variety of mechanisms that contribute to reduced glucocorticoid responsiveness, including increased expression of the GRβ and GRα-D isoforms, changes in GR phosphorylation and homologous down-regulation of GR. Dissecting the molecular mechanisms of resistance permits not only the prediction of patient responsiveness to glucocorticoids but also the design of novel therapeutic strategies for combating glucocorticoid insensitivity. In summary, understanding the heterogeneity of GR signaling in both health and disease will aid in the development of safer and more effective glucocorticoid therapies with improved benefit/risk ratios for patients.
KEY POINTS.
1. An important challenge in the clinical application of glucocorticoids is the heterogeneity in glucocorticoid responsiveness among individuals with a significant portion of the population exhibiting some degree of glucocorticoid resistance. 2. Glucocorticoid sensitivity and specificity is influenced by GR isoform expression profile. Inflammatory and pathological processes modulate cellular GR isoform profiles. 3. Assessing glucocorticoid sensitivity in individual patients is important for an optimal glucocorticoid treatment plan in the clinic. 4. Understanding the heterogeneity of GR signaling in both health and disease will aid in the development of safer and more effective glucocorticoid therapies with improved benefit/risk ratios for patients.
ACKNOWLEDGMENTS
Support provided by the Intramural Research Program of the National Institutes of Health/National Institute of Environmental Health Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURE: None
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no relevant conflicts of interest.
REFERENCES
- 1.Van Staa TP, Leufkens HG, Abenhaim L, et al. Use of oral corticosteroids in the United Kingdom. QJM : monthly journal of the Association of Physicians. 2000;93:105–111. doi: 10.1093/qjmed/93.2.105. [DOI] [PubMed] [Google Scholar]
- 2.Rhen T, Cidlowski JA. Antiinflammatory action of glucocorticoids –new mechanisms for old drugs. N Engl J Med. 2005;353:1711–1723. doi: 10.1056/NEJMra050541. [DOI] [PubMed] [Google Scholar]
- 3.Schacke H, Docke WD, Asadullah K. Mechanisms involved in the side effects of glucocorticoids. Pharmacology & therapeutics. 2002;96:23–4. doi: 10.1016/s0163-7258(02)00297-8. [DOI] [PubMed] [Google Scholar]
- 4.Barnes PJ. Mechanisms and resistance in glucocorticoid control of inflammation. J Steroid Biochem Mol Biol. 2010;120:76–85. doi: 10.1016/j.jsbmb.2010.02.018. [DOI] [PubMed] [Google Scholar]
- 5.Miller WL, Auchus RJ. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocrine reviews. 2011;32:81–151. doi: 10.1210/er.2010-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Webster JI, Tonelli L, Sternberg EM. Neuroendocrine regulation of immunity. Annual review of immunology. 2002;20:125–163. doi: 10.1146/annurev.immunol.20.082401.104914. [DOI] [PubMed] [Google Scholar]
- 7.John CD, Buckingham JC. Cytokines: regulation of the hypothalamo-pituitary-adrenocortical axis. Current opinion in pharmacology. 2003;3:78–84. doi: 10.1016/s1471-4892(02)00009-7. [DOI] [PubMed] [Google Scholar]
- 8.Walker JJ, Spiga F, Gupta R, Zhao Z, Lightman SL, Terry JR. Rapid intra-adrenal feedback regulation of glucocorticoid synthesis. J. R. Soc. Interface. 2015;12:20140875. doi: 10.1098/rsif.2014.0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breuner CW, Orchinik M. Plasma binding proteins as mediators of corticosteroid action invertebrates. The Journal of endocrinology. 2002;175:99–11221. doi: 10.1677/joe.0.1750099. [DOI] [PubMed] [Google Scholar]
- 10.Seckl JR. 11beta-hydroxysteroid dehydrogenases: changing glucocorticoid action. Curr Opin Pharmacol. 2004;4:597–602. doi: 10.1016/j.coph.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Cooper MS, Stewart PM. 11Beta-hydroxysteroid dehydrogenase type 1 and its role in the hypothalamus-pituitary-adrenal axis, metabolic syndrome, and inflammation. The Journal of clinical endocrinology and metabolism. 2009;94:4645–4654. doi: 10.1210/jc.2009-1412. [DOI] [PubMed] [Google Scholar]
- 12.Kumar R, Thompson EB. Gene regulation by the glucocorticoid receptor: structure:function relationship. J Steroid Biochem Mol Biol. 2005;94:383–94. doi: 10.1016/j.jsbmb.2004.12.046. [DOI] [PubMed] [Google Scholar]
- 13.Bledsoe RK, Montana VG, Stanley TB, Delves CJ, Apolito CJ, McKee DD, et al. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell. 2002;110:93–105. doi: 10.1016/s0092-8674(02)00817-6. [DOI] [PubMed] [Google Scholar]
- 14.Grad I, Picard D. The glucocorticoid responses are shaped by molecular chaperones. Mol Cell Endocrinol. 2007;275:2–12. doi: 10.1016/j.mce.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Pratt WB, Toft DO. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–60. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 16.Beato M. Gene regulation by steroid hormones. Cell. 1989;56:335–44. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- 17.Freedman LP. Anatomy of the steroid receptor zinc finger region. Endocr Rev. 1992;13:129–45. doi: 10.1210/edrv-13-2-129. [DOI] [PubMed] [Google Scholar]
- 18.Surjit M, Ganti KP, Mukherji A, Ye T, Hua G, Metzger D, et al. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145:224–41. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 19.Hudson WH, Youn C, Ortlund EA. The structural basis of direct glucocorticoid-mediated transrepression. Nat Struct Mol Biol. 2013;20:53–8. doi: 10.1038/nsmb.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–8. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burd CJ, Archer TK. Chromatin architecture defines the glucocorticoid response. Mol Cell Endocrinol. 2013 doi: 10.1016/j.mce.2013.03.020. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, Myers RM. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oakley RH, Revollo J, Cidlowski JA. Glucocorticoids regulate arrestin gene expression and redirect the signaling profile of G protein-coupled receptors. Proc Natl Acad Sci U S A. 2012;109:17591–6. doi: 10.1073/pnas.1209411109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramamoorthy S, Cidlowski JA. Ligand-induced repression of the glucocorticoid receptor gene is mediated by an NCoR1 repression complex formed by long-range chromatin interactions with intragenic glucocorticoid response elements. Mol Cell Biol. 2013;33:1711–22. doi: 10.1128/MCB.01151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uhlenhaut NH, Barish GD, Yu RT, Downes M, Karunasiri M, Liddle C, et al. In-sights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol Cell. 2013;49:158–71. doi: 10.1016/j.molcel.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogatsky I, Ivashkiv LB. Glucocorticoid modulation of cytokine signaling. Tissue Antigens. 2006;68:1–12. doi: 10.1111/j.1399-0039.2006.00599.x. [DOI] [PubMed] [Google Scholar]
- 27.Nissen RM, Yamamoto KR. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxyterminal domain. Genes Dev. 2000;14:2314–29. doi: 10.1101/gad.827900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang-Yen HF, Chambard JC, Sun YL, Smeal T, Schmidt TJ, Drouin J, et al. Transcriptional interference between c-Jun and the glucocorticoid receptor: mutual inhibition of DNA binding due to direct protein-protein interaction. Cell. 1990;62:1205–15. doi: 10.1016/0092-8674(90)90396-v. [DOI] [PubMed] [Google Scholar]
- 29.Lamberts SW, Huizenga AT, de Lange P, de Jong FH, Koper JW. Clinical aspects of glucocorticoid sensitivity. Steroids. 1996;61:157–60. doi: 10.1016/0039-128x(96)00005-0. [DOI] [PubMed] [Google Scholar]
- 30.Reddy TE, Gertz J, Crawford GE, Garabedian MJ, Myers RM. The hypersensitive glucocorticoid response specifically regulates period 1 and expression of circadian genes. Mol Cell Biol. 2012;32:3756–67. doi: 10.1128/MCB.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenkins BD, Pullen CB, Darimont BD. Novel glucocorticoid receptor coactivator effector mechanisms. Trends Endocrinol Metab. 2001;12:122–6. doi: 10.1016/s1043-2760(00)00357-x. [DOI] [PubMed] [Google Scholar]
- 32.Lonard DM, O'Malley BW. Expanding functional diversity of the coactivators. Trends Biochem Sci. 2005;30:126–32. doi: 10.1016/j.tibs.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Rosenfeld MG, Glass CK. Coregulator codes of transcriptional regulation by nuclear receptors. J Biol Chem. 2001;276:36865–8. doi: 10.1074/jbc.R100041200. [DOI] [PubMed] [Google Scholar]
- 34.Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–10. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ronacher K, Hadley K, Avenant C, Stubsrud E, Simons SS, Jr, Louw A, et al. Ligand-selective transactivation and transrepression via the glucocorticoid receptor: role of cofactor interaction. Mol Cell Endocrinol. 2009;299:219–31. doi: 10.1016/j.mce.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 36.Groeneweg FL, Karst H, de Kloet ER, Joels M. Mineralocorticoid and glucocorticoid receptors at the neuronal membrane, regulators of nongenomic corticosteroid signaling. Mol Cell Endocrinol. 2012;350:299–309. doi: 10.1016/j.mce.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 37.Samarasinghe RA, Witchell SF, DeFranco DB. Cooperativity and complemenarity: synergies in non-classical and classical glucocorticoid signaling. Cell Cycle. 2012;11:2819–27. doi: 10.4161/cc.21018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Croxtall JD, Choudhury Q, Flower RJ. Glucocorticoids act within minutes to inhibit recruitment of signaling factors to activated EGF receptors through a receptor-dependent, transcription-independent mechanism. Br J Pharmacol. 2000;130:289–98. doi: 10.1038/sj.bjp.0703272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Solito E, Mulla A, Morris JF, Christian HC, Flower RJ, Buckingham JC. Dexamethasone induces rapid serine-phosphorylation and membrane translocation of annexin 1 in a human folliculostellate cell line via a novel nongenomic mechanism involving the glucocorticoid receptor, protein kinase C, phosphatidylinositol 3-kinase, and mitogen-activated protein kinase. Endocrinology. 2003;144:1164–74. doi: 10.1210/en.2002-220592. [DOI] [PubMed] [Google Scholar]
- 40.Boldizsar F, Talaber G, Szabo M, et al. Emerging pathways of non-genomic glucocorticoid (GC) signalling in T cells. Immunobiology. 2010;215:521–526. doi: 10.1016/j.imbio.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Matthews L, Berry A, Ohanian V, Ohanian J, Garside H, Ray D. Caveolin mediates rapid glucocorticoid effects and couples glucocorticoid action to the antiproliferative program. Mol Endocrinol. 2008;22:1320–30. doi: 10.1210/me.2007-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samarasinghe RA, Di Maio R, Volonte D, Galbiati F, Lewis M, Romero G, et al. Nongenomic glucocorticoid receptor action regulates gap junction intercellular communication and neural progenitor cell proliferation. Proc Natl Acad Sci U S A. 2011;108:16657–62. doi: 10.1073/pnas.1102821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bamberger CM, Bamberger AM, de Castro M, Chrousos GP. Glucocorticoid receptor beta, a potential endogenous inhibitor of glucocorticoid action in humans. J Clin Invest. 1995;95:2435–41. doi: 10.1172/JCI117943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oakley RH, Sar M, Cidlowski JA. The human glucocorticoid receptor beta isoform. Expression, biochemical properties, and putative function. J Biol Chem. 1996;271:9550–9. doi: 10.1074/jbc.271.16.9550. [DOI] [PubMed] [Google Scholar]
- 45.Kino T, Su YA, Chrousos GP. Human glucocorticoid receptor isoform beta: recent understanding of its potential implications in physiology and pathophysiology. Cell Mol Life Sci. 2009;66:3435–48. doi: 10.1007/s00018-009-0098-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lewis-Tuffin LJ, Cidlowski JA. The physiology of human glucocorticoid receptor beta (hGRbeta) and glucocorticoid resistance. Ann N Y Acad Sci. 2006;1069:1–9. doi: 10.1196/annals.1351.001. [DOI] [PubMed] [Google Scholar]
- 47.Kelly A, Bowen H, Jee YK, Mahfiche N, Soh C, Lee T, et al. The glucocorticoid receptor beta isoform can mediate transcriptional repression by recruiting histone deacetylases. J Allergy Clin Immunol. 2008;121:203–8. e1. doi: 10.1016/j.jaci.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 48.Kim SH, Kim DH, Lavender P, Seo JH, Kim YS, Park JS, et al. Repression of TNF-alpha-induced IL-8 expression by the glucocorticoid receptor-beta involves inhibition of histone H4 acetylation. Exp Mol Med. 2009;41:297–306. doi: 10.3858/emm.2009.41.5.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewis-Tuffin LJ, Jewell CM, Bienstock RJ, Collins JB, Cidlowski JA. The Human glucocorticoid receptor b (hGRb) binds RU-486 and is transcriptionally active. Mol Cell Biol. 2007;27:2266–82. doi: 10.1128/MCB.01439-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hauk PJ, Hamid QA, Chrousos GP, Leung DY. Induction of corticosteroid insensitivity in human PBMCs by microbial superantigens. J Allergy Clin Immunol. 2000;105:782–7. doi: 10.1067/mai.2000.105807. [DOI] [PubMed] [Google Scholar]
- 51.Jain A, Wordinger RJ, Yorio T, Clark AF. Spliceosome protein (SRp) regulation of glucocorticoid receptor isoforms and glucocorticoid response in human trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2012;53:857–66. doi: 10.1167/iovs.11-8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu J, Gong JY, Goodman OB, Jr, Cartegni L, Nanus DM, Shen R. Bombesin attenuates pre-mRNA splicing of glucocorticoid receptor by regulating the expression of serine-arginine protein p30c (SRp30c) in prostate cancer cells. Biochim Biophys Acta. 2007;1773:1087–94. doi: 10.1016/j.bbamcr.2007.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DuBois DC, Sukumaran S, Jusko WJ, Almon RR. Evidence for a glucocorticoid receptor beta splice variant in the rat and its physiological regulation in liver. Steroids. 2013;78:312–20. doi: 10.1016/j.steroids.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hinds TD, Jr, Ramakrishnan S, Cash HA, Stechschulte LA, Heinrich G, Najjar SM, et al. Discovery of glucocorticoid receptor-beta in mice with a role in metabolism. Mol Endocrinol. 2010;24:1715–27. doi: 10.1210/me.2009-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schaaf MJ, Champagne D, van Laanen IH, van Wijk DC, Meijer AH, Meijer OC, et al. Discovery of a functional glucocorticoid receptor beta-isoform in zebrafish. Endocrinology. 2008;149:1591–9. doi: 10.1210/en.2007-1364. [DOI] [PubMed] [Google Scholar]
- 56.Ray DW, Davis JR, White A, Clark AJ. Glucocorticoid receptor structure and function in glucocorticoid-resistant small cell lung carcinoma cells. Cancer Res. 1996;56:3276–80. [PubMed] [Google Scholar]
- 57.Beger C, Gerdes K, Lauten M, Tissing WJ, Fernandez-Munoz I, Schrappe M, et al. Expression and structural analysis of glucocorticoid receptor isoform gamma in human leukaemia cells using an isoform-specific real-time polymerase chain reaction approach. Br J Haematol. 2003;122:245–52. doi: 10.1046/j.1365-2141.2003.04426.x. [DOI] [PubMed] [Google Scholar]
- 58.Rivers C, Levy A, Hancock J, Lightman S, Norman M. Insertion of an amino acid in the DNA-binding domain of the glucocorticoid receptor as a result of alternative splicing. J Clin Endocrinol Metab. 1999;84:4283–6. doi: 10.1210/jcem.84.11.6235. [DOI] [PubMed] [Google Scholar]
- 59.de Lange P, Segeren CM, Koper JW, Wiemer E, Sonneveld P, Brinkmann AO, et al. Expression in hematological malignancies of a glucocorticoid receptor splice variant that augments glucocorticoid receptor-mediated effects in transfected cells. Cancer Res. 2001;61:3937–41. [PubMed] [Google Scholar]
- 60.Gaitan D, DeBold CR, Turney MK, Zhou P, Orth DN, Kovacs WJ. Glucocorticoid receptor structure and function in an adrenocorticotropin-secreting small cell lung cancer. Mol Endocrinol. 1995;9:1193–201. doi: 10.1210/mend.9.9.7491111. [DOI] [PubMed] [Google Scholar]
- 61.Krett NL, Pillay S, Moalli PA, Greipp PR, Rosen ST. A variant glucocorticoid re-ceptor messenger RNA is expressed in multiple myeloma patients. Cancer Res. 1995;55:2727–9. [PubMed] [Google Scholar]
- 62.Lu NZ, Cidlowski JA. Translational regulatory mechanisms generate N-terminal glucocorticoid receptor isoforms with unique transcriptional target genes. Mol Cell. 2005;18:331–42. doi: 10.1016/j.molcel.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 63.Oakley RH, Cidlowski JA. Cellular processing of the glucocorticoid receptor gene and protein: new mechanisms for generating tissue-specific actions of glucocorticoids. J Biol Chem. 2011;286:3177–84. doi: 10.1074/jbc.R110.179325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu NZ, Collins JB, Grissom SF, Cidlowski JA. Selective regulation of bone cell apoptosis by translational isoforms of the glucocorticoid receptor. Mol Cell Biol. 2007;27:7143–60. doi: 10.1128/MCB.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu I, Shin SC, Cao Y, Bender IK, Jafari N, Feng G, et al. Selective glucocorticoid receptor translational isoforms reveal glucocorticoid-induced apoptotic transcriptomes. Cell Death Dis. 2013;4:e453. doi: 10.1038/cddis.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gross KL, Oakley RH, Scoltock AB, Jewell CM, Cidlowski JA. Glucocorticoid receptor a isoform-selective regulation of antiapoptotic genes in osteosarcoma cells: a new mechanism for glucocorticoid resistance. Mol Endocrinol. 2011;25:1087–99. doi: 10.1210/me.2010-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bender IK, Cao Y, Lu NZ. Determinants of the heightened activity of glucocorticoid receptor translational isoforms. Mol Endocrinol. 2013;27:1577–87. doi: 10.1210/me.2013-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cao Y, Bender IK, Konstantinidis AK, Shin SC, Jewell CM, Cidlowski JA, et al. Glucocorticoid receptor translational isoforms underlie maturational stage-specific glucocorticoid sensitivities of dendritic cells in mice and humans. Blood. 2013;121:1553–62. doi: 10.1182/blood-2012-05-432336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sinclair D, Webster MJ, Wong J, Weickert CS. Dynamic molecular and anatomical changes in the glucocorticoid receptor in human cortical development. Mol Psychiatry. 2011;16:504–15. doi: 10.1038/mp.2010.28. [DOI] [PubMed] [Google Scholar]
- 70.Sinclair D, Webster MJ, Fullerton JM, Weickert CS. Glucocorticoid receptor mRNA and protein isoform alterations in the orbitofrontal cortex in schizophrenia and bipolar disorder. BMC Psychiatry. 2012;12:84. doi: 10.1186/1471-244X-12-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beck IM, Vanden Berghe W, Vermeulen L, Yamamoto KR, Haegeman G, De Bosscher K. Crosstalk in inflammation: the interplay of glucocorticoid receptor-based mechanisms and kinases and phosphatases. Endocr Rev. 2009;30:830–82. doi: 10.1210/er.2009-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Z, Frederick J, Garabedian MJ. Deciphering the phosphorylation “code” of the glucocorticoid receptor in vivo. J Biol Chem. 2002;277:26573–80. doi: 10.1074/jbc.M110530200. [DOI] [PubMed] [Google Scholar]
- 73.Avenant C, Ronacher K, Stubsrud E, Louw A, Hapgood JP. Role of ligand-dependent GR phosphorylation and half-life in determination of ligand-specific transcriptional activity. Mol Cell Endocrinol. 2010;327:72–88. doi: 10.1016/j.mce.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 74.Galliher-Beckley AJ, Williams JG, Collins JB, Cidlowski JA. Glycogen synthase kinase 3beta-mediated serine phosphorylation of the human glucocorticoid receptor redirects gene expression profiles. Mol Cell Biol. 2008;28:7309–22. doi: 10.1128/MCB.00808-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Galliher-Beckley AJ, Williams JG, Cidlowski JA. Ligand-independent phosphorylation of the glucocorticoid receptor integrates cellular stress pathways with nuclear receptor signaling. Mol Cell Biol. 2011;31:4663–75. doi: 10.1128/MCB.05866-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Webster JC, Jewell CM, Bodwell JE, Munck A, Sar M, Cidlowski JA. Mouse glucocorticoid receptor phosphorylation status influences multiple functions of the receptor protein. J Biol Chem. 1997;272:9287–93. doi: 10.1074/jbc.272.14.9287. [DOI] [PubMed] [Google Scholar]
- 77.Chen W, Dang T, Blind RD, Wang Z, Cavasotto CN, Hittelman AB, et al. Glucocorticoid receptor phosphorylation differentially affects target gene expression. Mol Endocrinol. 2008;22:1754–66. doi: 10.1210/me.2007-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Deroo BJ, Rentsch C, Sampath S, Young J, DeFranco DB, Archer TK. Proteasomal inhibition enhances glucocorticoid receptor transactivation and alters its sub-nuclear trafficking. Mol Cell Biol. 2002;22:4113–23. doi: 10.1128/MCB.22.12.4113-4123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wallace AD, Cidlowski JA. Proteasome-mediated glucocorticoid receptor degradation restricts transcriptional signaling by glucocorticoids. J Biol Chem. 2001;276:42714–21. doi: 10.1074/jbc.M106033200. [DOI] [PubMed] [Google Scholar]
- 80.Wang X, DeFranco DB. Alternative effects of the ubiquitin-proteasome mediated by CHIP, an E3 ligase. Mol Endocrinol. 2005;19:1474–82. doi: 10.1210/me.2004-0383. [DOI] [PubMed] [Google Scholar]
- 81.Le Drean Y, Mincheneau N, Le Goff P, Michel D. Potentiation of glucocorticoid receptor transcriptional activity by sumoylation. Endocrinology. 2002;143:3482–9. doi: 10.1210/en.2002-220135. [DOI] [PubMed] [Google Scholar]
- 82.Ito K, Yamamura S, Essilfie-Quaye S, Cosio B, Ito M, Barnes PJ, et al. Histone deacetylase 2-mediated deacetylation of the glucocorticoid receptor enables NF-kappaB suppression. J Exp Med. 2006;203:7–13. doi: 10.1084/jem.20050466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Charmandari E, Chrousos GP, Lambrou GI, Pavlaki A, Koide H, Ng SS, et al. Peripheral CLOCK regulates target-tissue glucocorticoid receptor transcriptional activity in a circadian fashion in man. PLoS One. 2011;6:e25612. doi: 10.1371/journal.pone.0025612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jewell CM, Cidlowski JA. Molecular evidence for a link between the N363S glucocorticoid receptor polymorphism and altered gene expression. J Clin Endocrinol Metab. 2007;92:3268–77. doi: 10.1210/jc.2007-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van Rossum EF, Lamberts SW. Polymorphisms in the glucocorticoid receptor gene and their associations with metabolic parameters and body composition. Recent Prog Horm Res. 2004;59:333–57. doi: 10.1210/rp.59.1.333. [DOI] [PubMed] [Google Scholar]
- 86.Derijk RH, Schaaf MJ, Turner G, Datson NA, Vreugdenhil E, Cidlowski J, et al. A human glucocorticoid receptor gene variant that increases the stability of the glucocorticoid receptor beta-isoform mRNA is associated with rheumatoid arthritis. J Rheumatol. 2001;28:2383–8. [PubMed] [Google Scholar]
- 87.Clark AR, Belvisi MG. Maps and legends: the quest for dissociated ligands of the glucocorticoid receptor. Pharmacol Ther. 2012;134:54–67. doi: 10.1016/j.pharmthera.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 88.Ramamoorthy S, Cidlowski JA. Exploring the molecular mechanisms of glucocorticoid receptor action from sensitivity to resistance. Endocr Dev. 2013;24:41–56. doi: 10.1159/000342502. doi: 10.1159/000342502. [DOI] [PMC free article] [PubMed] [Google Scholar]