Abstract
Background:
Bovine ephemeral fever (BEFV) is an arthropod-borne disease of cattle and water buffaloes. BEFV occurs seasonally in tropical, subtropical and temperate regions of Africa, Asia and Australia. It has been known for the past decades in Iran based on clinical signs but lack of an accurate diagnosis has made the real feature of disease obscured. This is the first scientific report on isolation and identification of the agent in which molecular diagnosis of BEFV was also set up with high sensitivity and specificity.
Methods:
The viral agent was successfully isolated through serial passages in brain of suckling mice and cell culture. In addition, the circulating virus during the autumn 2012 in Iran was molecularly characterized based on partial G gene.
Results:
Alignment of 3 virus sequences from different parts of Iran revealed that they are identical suggesting that the circulating viruses were most likely the same in this period. Phylogenetic analysis of the Iranian sequences with 17 sequences in the GenBank from the world showed that it is identical to the virus circulated in Turkey during the same period suggesting that the virus was circulated in a large geographic region.
Conclusion:
These results offer primary information about BEFV in Iran. To better understanding the epidemiology of the virus, further studies based on seroepidemiology, molecular epidemiology, entomology and meteorology together with finding the model of animal transportation in the region are necessary.
Keywords: Bovine ephemeral fever, Iran, Diagnosis, Isolation, Phylogenetic analysis
Introduction
Bovine ephemeral fever (BEFV) is a non-contagious, arthropod-borne disease of cattle and water buffaloes proposed to be transmitted by Culicoides biting midges and mosquitoes. BEFV has been isolated from Culicoides brevitarsis, Culicoides coarctatus, and Anopheles bancroftii, however the arthropod vectors are yet to be defined (Walker et al. 2012). The viral agent is classified as the type species of the genus Ephemerovirus within the Rhabdoviridae family. BEFV is bullet shaped and its genome consists of five structural protein genes; N, P, M, G and L (Walker et al. 2012). The G gene encodes glycoprotein G which is responsible for cell attachment and entry, and also contains type-specific and neutralizing antigenic sites (Cybinski et al. 1990, Walker et al. 1991).
BEFV occurs seasonally in tropical, subtropical and temperate regions of Africa, Asia, Australia and the Middle-East and is a disabling disease with considerable economic impact due to loss of milk production, loss of condition in beef herds, infertility and abortion. The morbidity rate is high (up to 100%), however the mortality rate is generally low (rarely exceeds 1%). Typical clinical signs include a sudden onset of fever as high as 41 °C, a sudden and severe drop in milk production, inappetence, lethargy, salivation, nasal discharge, depression, stiffness, ruminal stasis and dyspnea (Walker et al. 2012). Treatment is based on remission of clinical signs using non-steroidal anti-inflammatory drugs and administration of calcium borogluconate to hypocalcemic animals. Vaccination is considered as the only effective approach to control the disease. Live-attenuated, inactivated, subunit and recombinant vaccines have been applied experimentally and commercially while live vaccines generally induce more-prolonged immunity (Radostitis et al. 2007).
Because of the short course of viremia in affected animals and proper shipping requirements of blood samples to the laboratory, detection and isolation of BEFV is not usually successful. Bovine ephemeral fever has been reported for the past decades in Iran and some efforts have been taken to isolate the virus (Hazrati et al. 1975). According to clinical observations, the economic losses due to the virus seem to be significantly high, however due to the lack of an accurate laboratory diagnosis the real feature of the disease remained unknown.
The aim of this study was isolation and adaptation of the viral agent on cell culture, and molecular characterization of the circulating virus in Iran during the autumn 2012 while accurate diagnosis of BEFV was also set up. The results are useful for any epidemiological study and are prerequisites for further research to manufacture a potent vaccine against the virus in Iran.
Materials and Methods
Sampling
Blood samples with anticoagulant were taken from clinically suspected animals during febrile phase and shipped on ice to the Animal Virology Department, Research and Diagnosis of Razi Vaccine and Serum Research Institute.
Viral RNA isolation and PCR amplification
Viral RNA was extracted from whole blood, cell culture and tissues using RNTX™ solution (Cinagen), according to manufacturer’s protocol. The purified RNA was subjected to reverse transcription (RT).
Reverse transcription (RT) was carried out using RevertAid™ First Strand cDNA synthesis Kit (Fermentas) as follows: 6 μl RNA and 1 μl Oligo (dT) primer with 5 μl DEPC-treated water heated at 65 °C for 10 minutes and cooled on ice. Then, 4 μl 5X Reaction buffer, 1 μl Ribolock™ Rnase inhibitor (20u/μl), 2 μl of 10 mM dNTP Mix, 1 μl RevertAid™ M-MulV Reverse Transcriptase (200 u/μl) were added to the solution to a final volume of 20 μl.
The resulting cDNA was amplified by polymerase chain reaction (PCR) using AccuPower® PCR PreMix (BIONEER), in which 2 μl of cDNA was mixed with 10 pmol of each forward: 5′-ATGTTCAAGGTCCTCATAATTACC-3′ (nt 1–24) and reverse: 5′-AATGATCAAAGAACCTATCATCAC-3′ (nt 1874–1871) (Wang et al. 2001) and dionized water to a final volume of 20 μl to amplify full length of G gene. Thermal cycling condition was initial denaturation at 95 °C for 5 min then 35 cycles of 95 °C for 40 s, 52 °C for 40 s, 72 °C for 2 min and 30 s and final extension of 72 °C for 10 min. The PCR product (1μl) was diluted (1/100) and 1 μl of the dilution was subjected for Nested-PCR using AccuPower® PCR PreMix (BIONEER) as described above. Nested-PCR amplified an 809 bp PCR product (nt 346–1155) using forward: 5′- TATTACCCTCCTGCCGGATGTTT-3′ and reverse: 5′-AGGTCTGTATTCGCACCAAGCTCT-3′ primers (Aziz-Boaron et al. 2012). Thermal cycling program was carried out as initial denaturation at 95 °C for 4 min then 35 cycles of 95 °C for 30 s, 56 °C for 30 s, 72 °C for 40 s and final extension of 72 °C for 5 min. All PCR products were run on agarose gel and stained with ethidium bromide and amplicons were observed in all positive samples.
In order to minimize the risk of nucleotide miscorporation for phylogenetic analysis of the 809 bp amplicon, additional PCR reaction was carried out on cDNAs using Pfu enzyme with the same thermal cycling condition. The PCR reaction mixture (50 μl total volume) comprised 5 μl 10× Pfu buffer, 2 μl dNTP mix (5mM), 1 μl (10 pmol) of each primer, 5 μl cDNA, 3.2 μl MgSo4 (25 mM) and 0.8 μl Pfu polymerase (2.5 u/μl, Fermentas).
Virus isolation
BEFV was successfully grown and adapted on cell culture by inoculating the virus intracerebrally into suckling mice primarily (Van der Westhuizen 1967). Using this approach, viral isolation was achieved as explained briefly. The blood samples were centrifuged at 2000 ×g for 5 min. Buffy coats of blood in anticoagulant were washed with phosphate-buffered saline (PBS). The buffy coats were resuspended in PBS and were frozen and thawed 3 times and 20 μl was inoculated into suckling mice’ brains. The suckling mice were examined carefully every day for clinical signs. After 6 days, their brain were extracted and were grinded with PBS, the homogenates were frozen and thawed 3 times, centrifuged at 2000 × g for 5 min and the supernatant was inoculated into suckling mice’s brains again until the 3rd passages when clinical signs were observed. The controlled mice were injected with 20 μl PBS repeatedly in all 3 passages. Viral RNA extraction and RT-PCR were performed on all inoculated and controlled mice. The homogenates from the 3rd passage were used for inoculation of cell culture.
Confluent Green Monkey Kidney (Vero) cell monolayers in 25 cm2 flask were inoculated with 0.5 ml of homogenate from the 3rd passages of virus in brains of suckling mice and incubated at 37 °C for 1 h, then cell maintenance medium containing: DMEM (Gibco), Glucose, Calf serum (5%), NaHCO3, Penecillin and Streptomycin, was added into the monolayers. The cultures were maintained at 37 °C for 8 days and observed every 24 h intervals. The flasks were frozen-thawed 3 times, they were centrifuged and the supernatant were inoculated into new monolayers. A monolayer flask was also considered as control which was inoculated with PBS in serial passages. Viral RNA extraction and RT-PCR were also carried out on all inoculated and controlled monolayers.
Sequence and phylogenetic analysis
The 809 PCR products were purified by High pure PCR product purification kit (Roche) and directly sequenced in both orientations with PCR primers mentioned above. The obtained sequences were assembled by Genious (4.8.3) software. The partial G gene sequences from Miandoab (36° N, 46° E), Shahriar (35° N, 51° E) and Mamasani (30° N, 51° E) located in North-West, center and South of Iran, respectively, were aligned using Clustal W program (Thampson et al. 1994). An Iranian sequence named SH (Shahriar) was deposited in GenBank with Accession number of KF 14868. This sequence was then aligned with 17 sequences from Australia, Taiwan, Japan, China, Egypt, Israel and Turkey (Table 1). The phylogenetic tree was constructed by the maximum likelihood method (Tamura et al. 2011) using Kimura’s two-parameter formula (Kimura 1980). The reliability of the branching was evaluated by the bootstrap test with 100 replicates (Felsenstein 1985).
Table 1.
Bovine ephemeral fever virus sequences used for the phylogenetic analysis
| Isolates | Country | Year of isolation | Accession number |
|---|---|---|---|
| BB7921 | AUSTRALIA | 1968 | AF234533 |
| CS1619 | AUSTRALIA | 1998 | AF058323 |
| CS1647 | AUSTRALIA | 1984 | AF058322 |
| YHL | JAPAN | 1966 | AB462028 |
| ON-04-1 | JAPAN | 2004 | AB462044 |
| 1984/TW/TN1 | TAIWAN | 1984 | AY935239 |
| TN-2004-124 | TAIWAN | 2004 | AY818194 |
| TRCP77 | TURKEY | 2008 | GQ229452 |
| TR-ET/IK-2 | TURKEY | 2012 | KC609426 |
| 2012/TR/CP3 | TURKEY | 2012 | KC470310 |
| 2012/TR/SKR | TURKEY | 2012 | KC337118 |
| EGY-2005 | EGYPT | 2005 | GQ227745 |
| IL-2008 | ISRAEL | 2008 | JN646090 |
| ISR10/3 | ISRAEL | 2012 | JN833635 |
| JB76H | CHINA | 1976 | JX566640 |
| SHANDONG | CHINA | 2011 | JX174661 |
| JT02L | CHINA | 2002 | JX564639 |
| SH | IRAN | 2012 | KF148685 |
Results
Polymerase chain reaction (PCR)
All PCR reactions were successfully carried out. The first reaction amplified an 1871 bp product spanning the whole G gene and the nested reaction amplified an 809 bp product.
Phylogenetic analysis of the partial G gene sequences
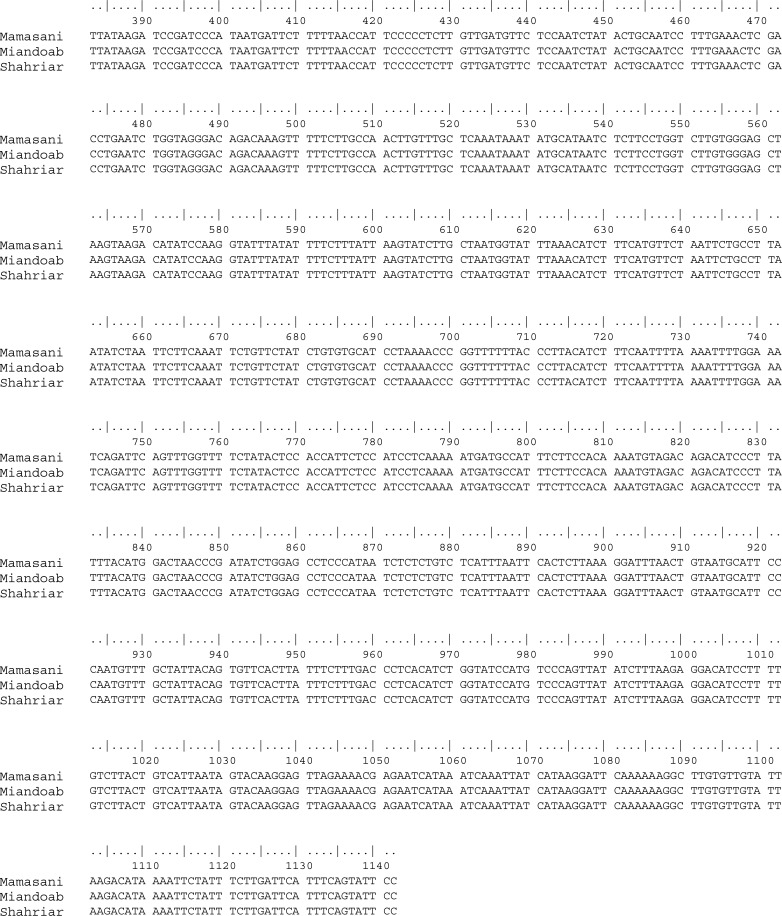
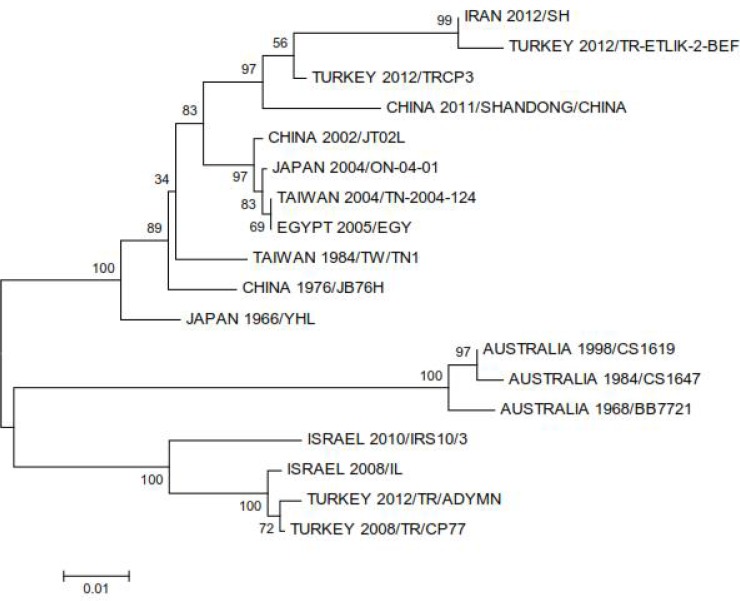
Using Clustal W program, the 3 sequences obtained from different parts of Iran including Shahriar, Miandoab and Mamasani were absolutely identical as depicted in Fig. 1. Additionally, alignment of Iranian sequence with 17 sequences from the world revealed that the Iranian sequence is 99.9 % identical with the Turkish isolate TRETLIK-2-BEF which was isolated in 2012. According to the phylogenetic tree, all sequences grouped into three distinct lineages including Australian, Middle- East and East Asian. The Iranian 2012/SH with the Turkish 2012/ TR-ETLIK-2-BEF and TRCP3 were placed with East Asian in one cluster (Fig. 2).
Fig. 1.
Alignment of partial G gene nucleotides (at residues 383–1142) of three bovine ephemeral fever isolates collected from different parts of Iran during autumn 2012
Fig. 2.
Phylogenetic tree of partial G gene sequences of the Iranian Shahriar isolate with 17 isolates from Turkey, Australia, Taiwan, Egypt, Israel, China and Japan during the years 1966–2012. The scale represents 1% sequence divergence
Virus isolation
From the third passage on suckling mice, clinical signs including paralysis, stiffness in hind legs and death were observed on fourth day after inoculation, while no clinical sign and death were observed in the control mice which were inoculated with PBS (Fig. 3). Moreover, the monolayer Vero cells showed CPE (Cytopathic effect) including rounding of cells, chromatin condensation followed by detachment from flasks on fifth day after inoculation from the second passage (Fig. 4). The presence of BEFV genome was confirmed in brains of suckling mice and cells inoculated with the virus by RT-PCR while the control mice and cells were negative.
Fig. 3.

Suckling mice inoculated with bovine ephemeral fever presented paralysis, stiffness and death while control mouse marked with blue dot on the head (left) showed no clinical signs
Fig. 4.
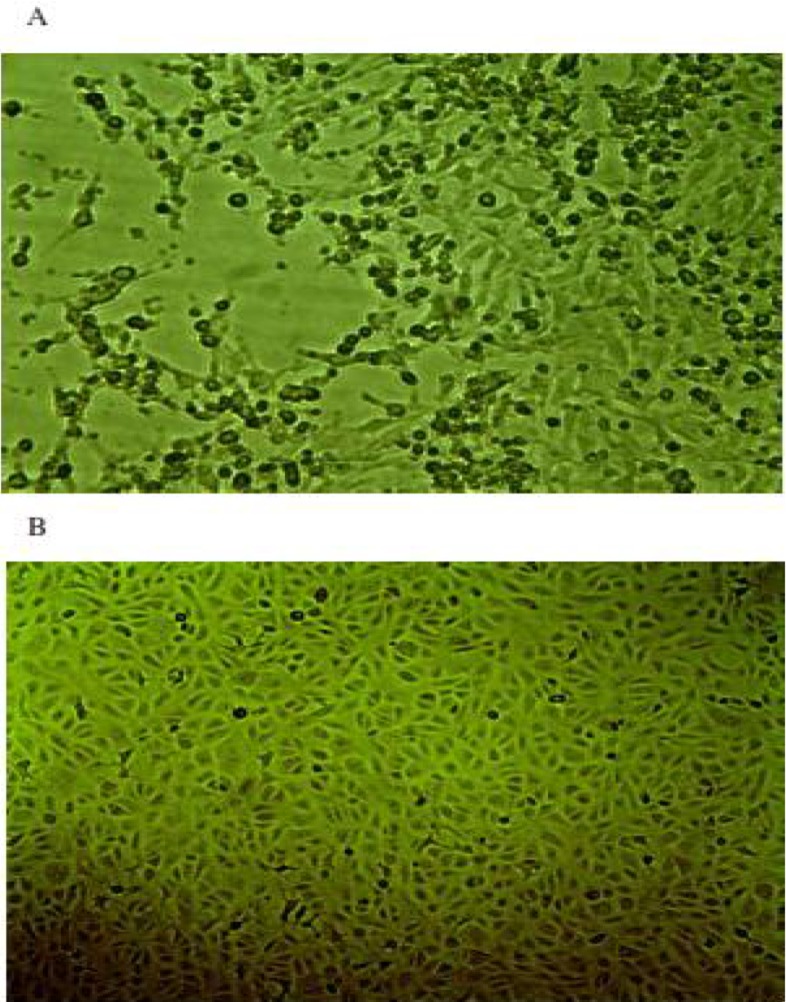
A) Cytopathic effect in Vero cell inoculated with bovine ephemeral fever. B) Control Vero cell line showed no Cytopathic effect
Discussion
Bovine ephemeral fever is one of the most important infectious diseases of livestock in Iran. The disease occurs usually in southern regions but sometimes observed as epidemic in other parts. Differential diagnosis of BEF based on clinical observation is sometimes difficult and complicated. Therefore, tentative diagnosis is essential and necessary to implement urgent and proper prophylactic action, evaluation of the extent of the disease and further epidemiological studies. This study represents an accurate and reliable diagnosis using molecular approach for detection of the widely spread viral agent in Iran. In future, application of a real time PCR as introduced by Stram et al. (2005) will definitely increase the sensitivity of BEFV diagnosis and will also help to reveal the real extent of the virus.
In this research, scientific procedure for the isolation of BEFV on cell culture was also successfully set up in the country. Complete characterization of the isolated viruses will certainly raise our knowledge about the epidemiology of the virus in the Middle-East and the world. These viruses can also be applied for manufacturing an appropriate domestic vaccine against the virus and are also efficient for seroepidemiologic studies using viral neutralization (VN) test.
As previously reported, the sequences from the world are clustered into three distinct lineages (Kato et al. 2009, Aziz-Boaro et al. 2012, Lin and Qiu 2012, Zheng and Qiu 2012, Walker et al. 2012). In the present study, Australian lineage is also a completely distinct lineage, Israeli and some Turkish isolates are grouped into a separate lineage and the East Asian isolates together with Iranian, Egyptian and the other Turkish sequences construct the third lineage. This geographic distinction explained to be resulted from circulation of the virus in a vast distance by vectors and winds in Australia and East Asia (Murray 1970, Ogawa 1992, Shirakawa et al. 1994, Finlaison et al. 2010). However, the circulation of BEFV in the Middle East has been proposed to occur by both winds and animal transport, this hypothesis was supported by close genetic relationship between some Middle East (Egyptian-2005) and East Asian sequences, and supportive documents indicating animal transport from China to Jordan in 2004 (Aziz-Baron et al. 2012). The Iranian 2012/SH was placed with the Turkish 2012/TR-ETLIK isolate in one branch (99.9% nucleotide identity), this branch with Chinese 2011/SHANDONG and Turkish 2012/TRCP3 constructed a sublineage within the East Asian Lineage, the high similarity of some Turkish and the Iranian sequences with that of Chinese again provide supportive evidence for the possible two routes of transmission in the Middle East.
Alignment of partial G gene sequence from 3 different regions (North-West, Centre and South) of Iran during the autumn of 2012 revealed that they are 100 % identical implying that the circulating virus was most likely the same in this period. However, this finding is not consistent with the sequences submitted in the GenBank from Turkey in the same period which are even separated into different lineages. These divergent sequences may also support the role of animal transport beside the wind and insect in transmission of the virus in the region. Turkey faced a severe BEF in 2012 with mortality rate of 15–20 % (Tonbak et al. 2012), this significant increase in mortality with acute form of BEF was also reported in China in 2011 (Zheng and Qiu 2012), but it is not clear whether the highly similar (96.4% nucleotide identity) Turkish (2012/TR-ETLIK) and Chinese (2011/SHANDONG) isolates or environmental factors (Walker 2013) are responsible for the change in virulence in these two far regions. It is also remain to be elucidated how a number of Turkish viruses were co-circulated in a specific period and whether this co-circulation affected the feature of the disease.
Conclusion
The geographic situation of Iran which is located between the East Asian and Middle-East countries places the country at high risk of BEFV transmission by both winds and animal transport. Characterization of more viruses together with analysis of meteorological and entomological data and finding the model of animal transport in years will help in evaluating the risk of the virus transmission in Iran and the Middle-East. In particular, further research is essentially required to define the vector species and their ecology in Iran. Due to the high risk of BEFV transmission and its consequences in the region, implementation of a specific surveillance system and large scale vaccination are recommended.
Acknowledgments
The author gratefully thanks to Razi Vaccine and Serum Research Institute for financial support of this study. The authors declare that there is no conflict of interests.
References
- Aziz-Boaron O, Klausner Z, Hasoksuz M, Shenkar J, Gafni O, Gelman B, David D, Klement E. ( 2012) Circulation of bovine ephemeral fever in the Middle East-Strong evidence for transmission by winds and animal transport. Vet Microbiol. 158: 300– 307. [DOI] [PubMed] [Google Scholar]
- Cybinski DH, Walker PJ, Byrne KA, Zakrzewski H. ( 1990) Mapping of antigenic sites on the bovine ephemeral fever virus glycoprotein using monoclonal antibodies. J Gen Virol. 71: 2065– 2072. [DOI] [PubMed] [Google Scholar]
- Felsenstain J. ( 1985) Confidence limits on phylogenesis: an approach using the bootstrap. Evolution. 39: 783– 791. [DOI] [PubMed] [Google Scholar]
- Finlaison DS, Read AJ, Kirkland PD. ( 2010) An epizootic of bovine ephemeral fever in New South Wales in 2008 associated with long-distance dispersal of vectors. Aust Vet J. 88: 301– 305. [DOI] [PubMed] [Google Scholar]
- Hazrati A, Hessami M, Roustai M, Dayhim F. ( 1975) Isolation of Bovine Ephemeral Fever Virus in Iran. Arch Razi Inst. 27: 80– 81 [Google Scholar]
- Kato T, Aizawa M, Katsunori T, Kokuba T, Yanase T, Shirafuji H, Tsuda T, Yamakawa M. ( 2009) Phylogenetic relationships of the G gene sequence of bovine ephemeral fever virus isolated in Japan, Taiwan and Australia. Vet Microbiol. 137: 217– 223. [DOI] [PubMed] [Google Scholar]
- Kimura M. ( 1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 28: 2731– 2739. [DOI] [PubMed] [Google Scholar]
- Murray MD. ( 1970) The spread of ephemeral fever of cattle during the 1967–68 epizootic in Australia. Aust Vet J. 46: 77– 82. [DOI] [PubMed] [Google Scholar]
- Ogawa T. ( 1992) Epidemiological investigation of bovine ephemeral fever outbreaks in Kyusyu Island in Japan during the fall of 1988. Prev Vet Med. 14: 67– 69. [Google Scholar]
- Shirakawa H, Ishibashi K, Ogawa T. ( 1994) A comparison of the epidemiology of bovine ephemeral fever in South Korea and south-western Japan. Aust Vet J. 71: 50– 52. [DOI] [PubMed] [Google Scholar]
- Radostitis OM, Gay CG, Hinchcliff KW, Constable PD. ( 2007) Veterinary Medicine. 10th edition, Sounders/Elsevier, Philadelphia. [Google Scholar]
- Stram Y, Kuznetzova L, Levin A, Yadin H, Rubinstein-Giuni M. ( 2005) A real-time RT-quantative (q) PCR for the detection of bovine ephemeral fever virus. J Virol Methods. 130: 1– 6. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. ( 1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position–specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673– 4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonbak S, Berber E, Yoruk MD, Azkur AK, Pestil Z, Bulut H. ( 2013) A large-scale outbreak of bovine ephemeral fever in Turkey, 2012. J Vet Med Sci. 75: 1511– 1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Westhuizen B. ( 1967) Studies on bovine ephemeral fever. I Isolation and preliminary characterization of a virus from natural and experimentally produced cases of bovine ephemeral fever. Onderstepoort J Vet Res. 34: 29– 40. [PubMed] [Google Scholar]
- Walker PJ, Byrne KA, Cybinski DH, Doolan DL, Wang Y. ( 1991) Proteins of bovine ephemeral fever virus. J Gen Virol. 80: 1211– 1220. [DOI] [PubMed] [Google Scholar]
- Walker PJ, Blasdell KR, Joubert DA. ( 2012) Ephemeroviruses: arthropod-borne rhabdoviruses of ruminants, with large and complex genomes. In: Dietzgen RG, Kuzmin IV. (Eds) Rhabdoviruses: Molecular Taxonomy, Evolution, Genomics, Ecology, Cytopathology and Control. Caister Academic Press, Norfolk, pp. 59– 88. [Google Scholar]
- Walker PJ. ( 2013) Bovine ephemeral fever: cyclic resurgence of a climate-sensitive vector-borne disease. Microbiol Aust. 41– 41. [Google Scholar]
- Wang F-I, Hsu AM, Huang KJ. ( 2001) Bovine ephemeral fever in Taiwan. J Vet Diagn Investi. 13: 462– 467. [DOI] [PubMed] [Google Scholar]
- Zheng F, Qiu C. ( 2012) Phylogenetic relationships of the glycoprotein gene of bovine ephemeral fever virus isolated from mainland China, Taiwan, Japan, Turkey, Israel and Australia. Virol J. 9: 268. [DOI] [PMC free article] [PubMed] [Google Scholar]