Abstract
Background:
The aim of the study was to identify the prevalence of different species of Plasmodium and haplotypes of pfcrt in Plasmodium falciparum from the selected area.
Methods:
Overall, 10,372 blood films of suspected malarial patients were examined microscopically from rural health center Sinawan, district Muzaffargarh, Pakistan from November 2008 to November 2010. P. falciparum positive samples (both whole blood and FTA blood spotted cards) were used for DNA extraction. Nested PCR was used to amplify the pfcrt (codon 72–76) gene fragment. Sequencing was carried out to find the haplotypes in the amplified fragment of pfcrt gene.
Result:
Over all slide positivity rate (SPR), P. vivax and P. falciparum positivity rate was 21.40 %, 19.37 % and 2.03% respectively. FTA blood spotted cards were equally efficient in the blood storage for PCR and sequencing. Analysis of sequencing results of pfcrt showed only one type of haplotype SagtVMNT (AGTGTAATGAATACA) from codon 72–76 in all samples.
Conclusion:
The results show high prevalence of CQ resistance and AQ resistant genes. AQ is not recommended to be used as a partner drug in ACT in this locality, so as to ward off future catastrophes.
Keywords: Malaria, pfcrt, Resistance, Pakistan
Introduction
Globally 106 countries from tropical and subtropical are endemic for malaria (WHO 2010). These are in tropical or subtropical zones with are suitable breading places far anopheline mosquito. The regions included in this area are Asia, Africa, Islands of South, West and Central Pacific Ocean, Latin America, Turkey and certain Caribbean Islands (Lamar et al. 2007). In the areas of malaria transmission, epidemics of malaria have been leading cause of high morbidity and mortality. Complex epidemics of malaria have been observed in Kenya (May 1999–August 1999, Burundi (September 2000–May 2001), South Sudan (June 2003–November 2003), in two areas of Ethiopia (July 2003–February 2004) and India (January 2005–March 2005) (Checchi et al. 2006, Kumari 2009).
In 3rd world countries including Pakistan, malaria has always been major public health problem and it accounts for more than 95% of regional burden (Huda and Zamani 2009). The main reason of malariogenic potential in Pakistan is extensive agricultural practices, vast irrigation network, monsoon rains and poverty (WHO 2005). Annually, about 500000 episodes of malaria infection occur (Yasinzai and Kakharsulemankhel 2008) in rural areas, were associated with more people live below poverty line (38.65%) than urban areas (22.39%).
The main-stay in the control of malaria is the increasing frequency of drug resistant parasites especially P. falciparum (Gama et al. 2010).
First reports of resistance against CQ came from South America (Moore and Lanier 1961), South East Asia (Harinausta et al. 1965). Later, it reached to Africa. Across the world, CQR strain of P. falciparum originated from at least six locations independently: in Java, Philippines, Papua New Guinea, South East Asia and two locations in South America (Wootton et al. 2002, Chen et al. 2003). In Pakistan, the resistance of P. falciparum to CQ was first detected in 1981 from Sheikhupura and National Institute of Malaria Research and Training (NIMRT) revealed that R1 level of CQ resistance was widespread in Pakistan (frequency ranging from 30–84%) (Shah et al. 1997). Chloroquine was used as a first line treatment in Pakistan from 1950–2007 (Asif 2008). In the district Muzaffargarh, response of P. falciparum to chloroquine was found out during the time period of 2003–2005. RI was 29.36 %, and RII was 29.41 % (Rana 2009).
In 1990s, chloroquine resistant P. falciparum spread widely in Pakistan (Rab et al. 2001). Factors involved in the spread of drug resistance are drug use practices, drug half life, transmission intensity, clone multiplicity, parasite density, host immunity and genetic basis of drug resistance (Hasting and Watking 2006, Pongtavornpinyo et al. 2008). Resistance against many antimalarials has arisen in the same geographic areas. These areas have low transmission rate, low immunity level and higher parasites biomass (White and Pongtavornpinyo 2003). Use of substandard medicines can also be involved in the emergence of drug resistance (Leslie et al. 2009).
With the advancement in molecular biology and isolation of CQ resistance clone (Dd2) helped in identification of molecular markers in P. falciparum associated with drug resistance (Jiang et al. 2008). In 1990, Thomas Wellem along with his team performed a cross between HB3, a chloroquine sensitive (CQS) and Dd2, a chloroquine resistant (CQR) strain (Wellem et al. 1990). It was found that the main determinant of CQ resistance is a locus on chromosome 7 of P. falciparum (Wellem et al. 1991). It was further noted that it is a 38 Kb fragment (10 genes) (Su et al. 1997). Fidock et al. (2000) reported that a highly polymorphic 3.1 Kb gene (composed of 13 exons) is involved in resistance. Martin and Kirk (2004) reported that it is a 48.6 kDa protein of 424 amino acids long. It forms 10 transmembrane domains. The protein is located on digestive vacuole and it has transporter-like properties. It was called P. falciparum chloroquine resistant transport gene (pfcrt) and considered as a member of drug metabolite transporter super family (Tran and Saier 2004). In the past, SVMNT (ser-val-meth-asn-thr) allele carrying parasites had been reported twice, in 1996–1997 from Africa and in 2004 from Tanzania (Alifrangis et al. 2006). Three common haplotypes of pfcrt are CVMNK (CQS, present in all geographic regions), CVIET (CQR found in Africa and South East Asia), SVMNT (CQR, SagtVMNT is present in some countries of Asia and Stct VMNT is found in South America) (Wootton et al. 2002, Ursing et al. 2006). The high level of resistance to AQ metabolite desethylamodiaquine (DEAQ) is found to be associated with the presence of pfcrt codon 72–76 haplotype SVMNT. It replaces CVMNK found in CQS strain (Wootton et al. 2002, Warhurst et al. 2003, Sa et al. 2009, Beshir et al. 2010). In oocyte system, competition of drugs with CQ was checked by mutated pfcrt. Drug quinine and AQ at normal concentration and mefloquine and primaquine at higher concentration were able to compete with CQ for transport by mutated pfcrt while piperaquine and artemisinin did not interact with pfcrt (Martin et al. 2009).
Keeping in view the situation of malaria, this study was designed to identify the chloroquine resistant genes of P. falciparum which convert a chloroquine sensitive parasite (CQS) to chloroquine resistant (CQR) type parasite.
Materials and Methods
Selection of Area for Sampling
Muzaffargarh is located at latitude 30.06° longitudes 71.02°, between the two rivers, Chenab in the East and Indus in the West. For present study, the area of Muzaffargarh was selected on the basis of high endemicity. Data obtained by the curtsy of Directorate General Health Punjab, Pakistan (through personal communication) reported “1388 laboratory confirmed cases” in Punjab were with 15 % in district Muzaffargarh in 2006. In 2007, total number of positive malarial cases was 1903, again highest positive cases (7.5%) were observed in Muzaffargarh.
Sample Collection
Blood samples were collected from individuals presented with fever, persistent headache and malaria like symptoms. Patient’s consent was obtained prior to sampling, finger prick blood was used for slide preparation. Thick and thin blood smears were prepared and stained by Giemsa stain (3% solution in 7.2 pH phosphate buffer) (WHO 1991). Two ml blood of P. falciparum positive patients was collected in EDTA vacutainers and spotted on Flinders Technical Associates (FTA) classic cards (Whatman) by micropipette according to manufacturer’s instruction. Vacutainers were stored at 4 °C and cards were stored at room temperature till use for DNA extraction. Sampling was done at Rural Health Centers Muzaffargarh from November, 2008 to November, 2010 (25 months) as shown in Fig. 1.
Fig. 1.
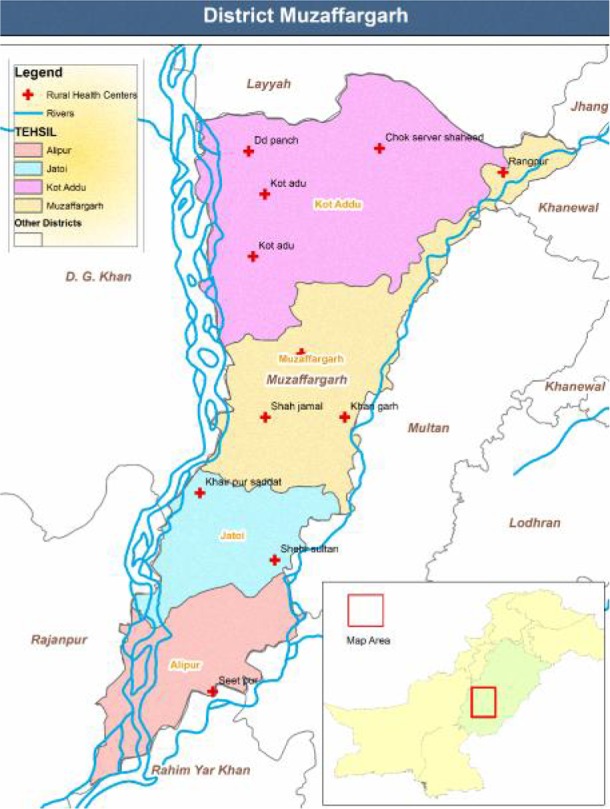
Map of district Muzaffargarh showing RHCs
DNA Extraction
For amplification of pfcrt gene, DNA was eluted both from FTA cards and whole blood. P. falciparum positive samples (by microscopic review and RDTs method). FTA Cards contain chemicals that lyses (removes) blood cells, denature proteins and protect nucleic acids from nucleases, oxidation and UV damage. Captured nucleic acid is ready for downstream applications in less than 30 min and is stable for years at room temperature. DNA was eluted according to manufacturer’s instructions (Whatman). While from whole blood, 200 μl of sample was used for DNA extraction with the QIA amp midi kit, as described by the manufacturer (Qiagen). Extracted DNA was analyzed by using 1 % agarose gel.
Amplification of pfcrt Gene Fragment
Two-step nested PCR was done to amplify pfcrt gene fragment of 145 base pairs. Primers were designed, using DNA sequence of P. falciparum chloroquine resistant strain Dd2/Indochina (by Pub Med accession number AF030694). The sequences of primers used in round 1 and round 2 are given in Table 1.
Table 1.
Primers used for amplification of pfcrt gene fragment of 145 base pairs
| PCR Round | Primer | Primer Sequence (5′–3′) | Tm |
|---|---|---|---|
| 1st Round | Crtd-A (forward primer) | 5′-GGTGGAGGTTCTTGTCTTGGTA-3′ | 57.5 °C |
| Crtd-B (reverse primer) | 5′-GACCTATGAAGGCCAAAATGACTG-3′ | 56.7 °C | |
| 2nd Round | Crtd-C (forward primer) | 5′-TGTGCTCATGTGTTTAAACTT-3′ | 50.06 °C |
| Crtd –D (reverse primer) | 5′-CAAAACTATAGTTACCAATTTT-3′ | 46.10 °C |
Preparation of PCR Reaction Mix for Round 1 (pfcrt)
The PCR mixture contained 1X PCR buffer (Promega), 2.0 mM MgCl2 (Promega), 0.2 mM dNTPs, 0.25 μM each primer, 1.25 units, 0.5 units Taq polymerase (Promega) and 5 μl template DNA. The total reaction volume was 50 μl.
Cycling conditions for PCR were: one cycle of 95 °C for 5 minutes followed by 30 cycles of 95 °C for 10 seconds, 57 °C for 30 seconds and 72 °C for 30 seconds, followed by a final extension at 72 °C for 10 minutes.
PCR Reaction Mix Preparation for Round 2 (pfcrt)
The PCR mixture contained 1X PCR buffer (Promega), 2.0mM MgCl2 (Promega), 0.2 mM dNTPs, 0.25 μM each primer, 0.5 units Taq polymerase (Promega) and 5 μl template DNA. The total reaction volume was 50 μl.
Cycling conditions for PCR were: one cycle of 95 °C for 5 minutes followed by 35 cycles of 95 °C for 30 seconds, 48 °C for 30 seconds and 65 °C for 30 seconds, followed by a final extension at 65 °C for 3 minutes.
Amplified PCR products of each gene fragment were electrophoresed using 3 % agarose gel and visualized on UV. PCR products were cleaned using Exonuclease I (BioLabs Inc) and sequenced in both directions using Big Dye® Terminator system.
Statistical Analysis
Results of sequencing were analyzed by the programme of Applied Bio-system (sequence scanner V, 0.1).
Results
Sampling was carried out in Muzaffargarh, Pakistan from November 2008 to November 2010 (25 Months), in order to find out the malaria frequency. During this period, 10372 blood films of suspected malarial patients were prepared and examined. Their positivity rates for the infection of Plasmodium (both falciparum and vivax), P. vivax and P. falciparum were 2220, 2009 and 211 respectively. In this duration, slide positivity rate (SPR), P. vivax positivity rate % (VPR) and P. falciparum positivity rate (FPR) were 21.40 %, 19.37 % and 2.03 % respectively. The difference in P. vivax (90.49%) and P. falciparum (9.51%) infection was found highly significant (χ2= 1456, P< 0.001).
Extracted DNA
Samples positive for P. falciparum (211) were used for DNA extraction. Genomic DNA is shown in Fig. 2, 3.
Fig. 2.
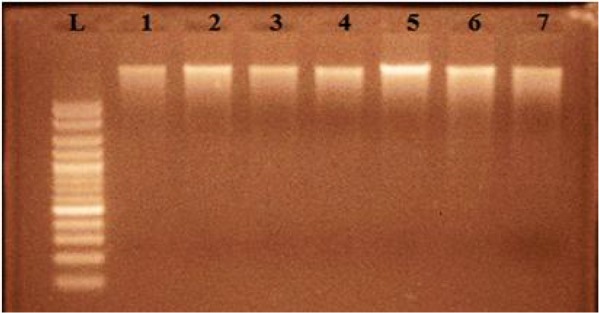
Gel electrophoresis picture showing extracted genomic DNA of falciparum positive malarial patients (Lane L shows 100bp ladder, lane 1–7 show genomic DNA)
Fig. 3.
Gel electrophoresis picture showing amplified pfcrt gene fragment (145bp) (by using DNA extracted from the whole blood). Lane 1 and 2 show negative and positive controls respectively, lanes 3 to 12 show patient samples, lane 13 shows 100bp DNA Marker
Amplification of pfcrt (codon 72–76)
All the 211 P. falciparum positive samples were amplified using polymerase chain reaction. Amplification was carried out of the extracted DNA from whole blood and from FTA blood spotted cards.
pfcrt (codon 72–76) Amplification (by using disc of blood spotted FTA cards)
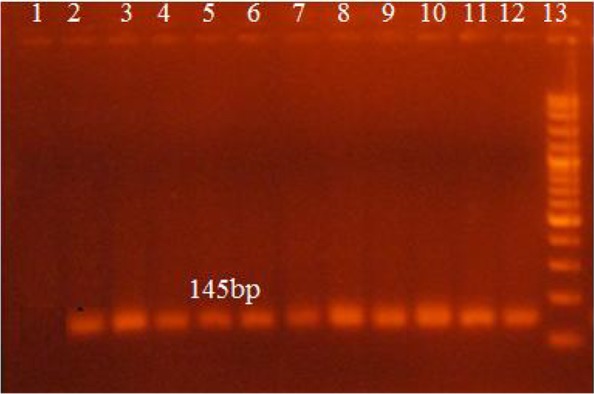
Amplification by using the disc (2mm) was also conducted by using punched disc from FTA blood spotted cards. Product of (145bp) was visible as show in Fig. 4.
Fig. 4.
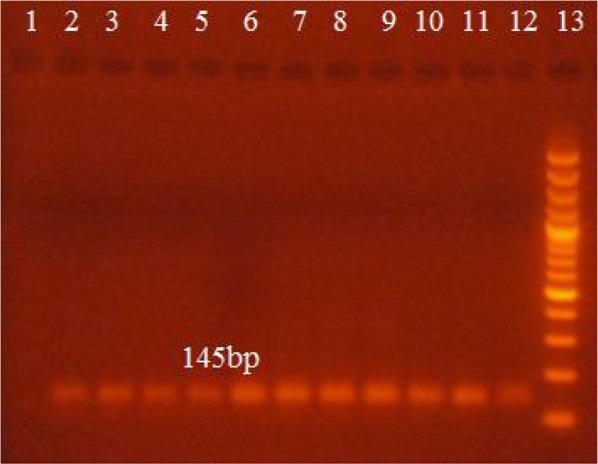
Picture of gel electrophoresis showing amplified pfcrt gene fragment (145bp) (by using disc of blood spotted card). Lane 1 and 2 show negative and positive controls respectively, lanes 3 to 12 show patient samples, lane 13 shows 100bp DNA Marker
Similar PCR results were obtained using DNA extracted from whole blood and FTA blood spotted cards which showed that FTA cards were found equally efficient for storage of blood for PCR reaction.
Sequencing Result
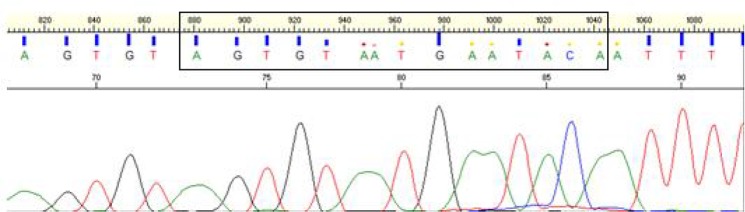
Sequencing results were analyzed by using Applied-Biosystem VI software. It showed only one type of haplotype sequence Sagt VMNT (AGTGTAATGAATACA)from codon 72–76 in all samples. No wild type CVMNT or other drug resistant (mutant) haplotype was found. Fig. 5 is depicting sequencing results of pfcrt (codon 72–76) with forward primer.
Fig. 5.
Sequencing results of pfcrt (codon 72–76)
Discussion
Present findings showed that overall SPR observed in Muzaffargarh, Pakistan was 21.40 %. According to the report of Ministry of Health, (2006) malaria was the second most prevalent disease in Pakistan (Murtaza et al. 2009). Akin figures of SPR (19.7% and 19.40%) were reported from Punjab and Khyber Pakhtunkhwa (Idris et al. 2007, Mohammad et al. 2010), which were respectively 1.64 % and 2.00 % less than present study. SPR reported from India has shown the percentages of 25.2 % and 24.54 % (Dhiman et al. 2010, Panigahi et al. 2011), which were 3.80% and 3.14% higher than in current study.
Although malaria is rife worldwide but the maximum numbers of devastations caused by it are in Sub-Saharan Africa (WHO 2010). In Africa 30.20 %, 27.07 % and 22.60 % SPR was reported from Uganda, Ethiopia and Tanzania (Jensen et al. 2009, Yohannes and Haile 2010, Mubi et al. 2011) and these figures indicated an enhancement of 8.80 %, 5.67 % and 1.20 % respectively than the figures of the current study. These differences may be due to geographical, climatic, economic and cultural variations.
In the current study, P. vivax, was the predominant specie of this region. The overall infection caused by P. vivax and P. falciparum was 90.49 % and 9.51 % respectively, and the difference between the infection of P. vivax and P. falciparum was highly significant (χ2= 1456, P< 0.001). Similar figures were collected from Muzaffarabad, Azad Kashmir and Khyber Pakhtunkhwa, occurrence of P. vivax infection (90.40% 92.20%, 92.20%) was far more than that caused by P. falciparum infection (9.6%, 7.8% and 7.8%) (Jan and Kiani 2001, Jamal et al. 2005, Jalalul-din and Ally 2006). Similarly higher incidence of P. vivax infection (72.4%) than P. falciparum infection (24.1%) was reported from Baluchistan province (Idris et al. 2007). Some other figures from the same province showed higher incidence of P. vivax infection (58.9%, 51.8%, 88.6%, 72.3%) than P. falciparum infection (41%, 48.1%, 11.3%, 27.6%) (Yasinzai and Kakarsulemankhel 2008). Infections caused by P. vivax were also reported more than those of P. falciparum from some areas of Sind (Mahmood et al. 2005, Atif et al. 2009). P. vivax infection (11.70%, 75%) was less than P. falciparum infection (88.29%, 85%) in some areas or Indian (Dhiman et al. 2010, Panigrahi et al. 2011). Similarly, P. falciparum (90%, 70%, 90.7% 68.53%) was predominant specie in some African regions (Pinto et al. 2000, Yohannes and Haile 2010, Lemma et al. 2011).
Currently, parasites strains are exhibiting resistance to all major classes of drug present in the field and some of them have been withdrawn from use because of their ineffectiveness. Unfortunately we are unaware that where the next generation of antimalarial will come from (Grimberg and Mehlotra 2011). Core objective of the study was the scrutiny of mutant and wild types of pfcrt (codon 72–76). pfcrt gene fragment of 145bp was amplified in all positive P. falciparum (211) samples. Results showed 100 % Sagt VMNT (ser-val-meth-asn-thr). No StctVMNT haplotype or wild type (CVMNK) was observed. Origin of SagtVMNT had been hypothesized from Mato Grasso, Brazil (Vieira et al. 2004) and was reported from Guyana, Peru, Suriname, Venezuela (Contreras et al. 2002, Cortese et al. 2002, Peek et al. 2005, Mehlotra et al. 2008, Griffing et al. 2010). Clinical resistance to chloroquine (CQ) and amodiaquine (AQ) was found associated with mutations in pfcrt gene (Picot et al. 2009, Beshir et al. 2010). Sa et al. (2009) correlated high level resistance to AQ metabolite desethylamodiaquine (DEAQ) in tests (in vitro) with the presence of SVMNT haplotype of pfcrt. Mehlotra et al. (2008), Zakeri et al. (2008), and Beshir et al. (2010) detected high prevalence of this haplotype in the parasitic population of Brazil, Papua New Guinea, Laos, Afghanistan and Iran. Esmaeili et al. (2008) also reported very high prevalence of K76T mutation in an endemic district of Iran. These results were also supported by the study carried out (in vitro) on natural P. falciparum isolate, laboratory reference strain, parasites transfected with allelic replacement and progeny of genetic crosses (Sidu et al. 2002, Sa et al. 2009). In the past, SVMNT (ser-val-meth-asn-thr) allele carrying parasites had been reported twice, in 1996–1997 from Africa and in 2004 from Tanzania (Alifrangis et al. 2006). Three common haplotypes of pfcrt are CVMNK (CQS, present in all geographic regions), CVIET (CQR found in Africa and South East Asia), SVMNT (CQR, Sagt VMNT is present in some countries of Asia and Stct VMNT is found in South America) (Wootton et al. 2002, Ursing et al. 2006). In the area of Muzaffargarh P. vivax was predominant specie so CQ exposure was higher. It may be the main reason of high prevalence of spreading CQR parasite (having pfcrt “SVMNT”) in this area. Similar findings were reported from India, mutant pfcrt “SVMNT” haplotype was endemic in those areas, where P. vivax was dominant specie (Mallick et al. 2012).
Conclusion
Prior to everything else, appropriate diagnosis of Plasmodium and its species is highly necessary. A treatment initiated without proper diagnosis not only increases the probabilities of drug resistance, but also invites complications. Malaria controlling policies should be decentralized: made and implemented at province and district levels. Proper understanding and careful evaluation of local epidemiological pattern and the other aspects of malaria can produce base-line data in planning task for devising control strategies. The drug resistance issue is likely to pose long term challenges for us. Hybrid of conventional and neo drugs can help to conserve the efficiency of valuable anti malarial medicines, but choosing the right combination partners will continue to require the study of presently known and emerging mechanism.
The results show high prevalence of CQ resistance and AQ resistant genes. Excessive presence of CQ and AQ resistant genes in plasmodium population isolated from the people of Muzaffargarh suggest establishing the base line for monitoring of mutations. This is evident now that in this area, CQ or AQ will not be effective for the victims of P. falciparum infection. AQ is not recommended to be used as a partner drug in ACT in this locality, so as to ward off future catastrophes.
Acknowledgments
Financial assistance was provided by University of the Punjab Research Committee 2010–2011 and Higher Education Commission Pakistan (Pin# 074-0551-(BM-4-033) is gratefully acknowledged. Thanks to Dr Hanan El Mohammady Ismail Head, Immunology Section Clinical Trials Research Programme US Naval Medical Research Unit #3 (NAMRU-3) Cairo, Egypt for valuable suggestions to improve the manuscript. This paper is produced from PhD project conducted by the principle author. The authors declare that there is no conflict of interests.
References
- Alifrangis M, Dalgaard MB, Lusingu JP, Vestergaard LS, Staalsoe T, Jensen ATR, Enevold A, Rønn AM, Khalil IF, Warhurst DC. ( 2006) Occurrence of the Southeast Asian/ South American SVMNT haplotype of the chloroquine-resistance transporter gene in Plasmodium falciparum in Tanzania. J Infect Dis. 193( 12): 1738– 1741. [DOI] [PubMed] [Google Scholar]
- Asif SA. ( 2008) Departmental audit of malaria control programme 2001–2005 North West Frontier Province (NWFP). J Ayub Med Coll Abbottabad. 20( 1): 98– 102. [PubMed] [Google Scholar]
- Atif S, Farzana M, Naila S, Abdul F. ( 2009) Incidence and pattern of malarial infection at a tertiary care Hospital of Hyderabad. World J Med Sci. 4( 1): 9– 12. [Google Scholar]
- Beshir K, Sutherland CJ, Merinopoulos I, Durrani N, Leslie T, Rowland M, Hallett RL. ( 2010) Amodiaquine resistance in Plasmodium falciparum malaria in Afghanistan is associated with the Pfcrt SVMNT allele at codons 72 to 76. Antimicrob Agents Chemother. 54( 9): 3714– 3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checchi F, Cox J, Balkan S, Tamrat A, Priotto G, Alberti KP, Zurovac D, Guthmann JP. ( 2006) Malaria epidemics and interventions, Kenya, Burundi, southern Sudan, and Ethiopia, 1999– 2004. Emerg Infect Dis. 12: 1477– 1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Kyle DE, Pasay C, Fowler EV, Baker J, Peters JM, Cheng Q. ( 2003) pfcrt allelic types with two novel amino acid mutations in chloroquine-resistant Plasmodium falciparum isolates from the Philippines. Antimicrob Agents Chemother. 47( 11): 3500– 3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras CE, Cortese JF, Caraballo A, Plowe CV. ( 2002) Genetics of drug-resistant Plasmodium falciparum malaria in the Venezuelan state of Bolivar. Am J Trop Med Hyg. 67( 4): 400– 405. [DOI] [PubMed] [Google Scholar]
- Cortese JF, Caraballo A, Contreras CE, Plowe CV. ( 2002) Origin and dissemination of Plasmodium falciparum drug-resistance mutations in South America. J Infect Dis. 186( 7): 999– 1006. [DOI] [PubMed] [Google Scholar]
- Dhiman RC, Chavan L, Pant M, Pahwa S. ( 2010) National and regional impacts of climate change on malaria by 2030. Curr Sci. 101( 3): 372– 383. [Google Scholar]
- Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos L. ( 2002) Mutations in the Plasmodium falciparum digestive vacuole transmembrane protein Pfcrt and evidence for their role in chloroquine resistance. Mol Cell. 6( 4): 861– 871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama B, de Oliveira N, de Souza J, Santos F, de Carvalho L, Melo Y, Rosenthal P, Daniel-Ribeiro C, Ferreira-da-Cruz M. ( 2010) Brazilian Plasmodium falciparum isolates: investigation of candidate polymorphisms for artemisinin resistance before introduction of artemisinin-based combination therapy. Malar J. 9( 1): 355– 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffing S, Syphard L, Sridaran S, McCollum AM, Mixson-Hayden T, Vinayak S, Villegas L, Barnwell JW, Escalante AA, Udhayakumar V. ( 2010) pfmdr1 amplification and fixation of pfcrt chloroquine resistance alleles in Plasmodium falciparum in Venezuela. Antimicrob Agents Chemother. 54( 4): 1572– 1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimberg BT, Mehlotra RK. ( 2011) Expanding the Antimalarial Drug Arsenal-Now, But How? Pharmaceuticals. 4( 5): 681– 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings IM, Watkins WM. ( 2006) Tolerance is the key to understanding antimalarial drug resistance. Trends Parasitol. 22( 2): 71– 77. [DOI] [PubMed] [Google Scholar]
- Huda A, Zamani G. ( 2009) The progress of roll back malaria in Eastern Mediterranean region over the past decade. East Mediterr Health J. 14: 82– 9. [PubMed] [Google Scholar]
- Idris M, Sarwar J, Fareed J. ( 2007) Pattern of malaria infection diagnosed at Ayub Teaching Hospital Abbottabad. J Ayub Med Coll Abbottabad. 19( 2): 35– 36. [PubMed] [Google Scholar]
- Jalal-ud-din SAK, Ally SH. ( 2006) Malaria in children: study of 160 cases at a private clinic in Mansehra. J Ayub Med Coll Abbottabad. 18( 3): 44– 45. [PubMed] [Google Scholar]
- Jan A, Kiani T. ( 2001) Haematozoan parasites in Kashmiri refugees. Pak J Med Res. 40( 1): 10– 12. [Google Scholar]
- Jensen T, Bukirwa H, Njama-Meya D, Francis D, Kamya M, Rosenthal P, Dorsey G. ( 2009) Use of the slide positivity rate to estimate changes in malaria incidence in a cohort of Ugandan children. Malar J. 8( 1): 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari R, Joshi P, Lal S, Shah W. ( 2009) Management of malaria threat following tsunami in Andaman and Nicobar Islands, India and impact of altered environment created by tsunami on malaria situation of the islands. Acta Trop. 112( 2): 204– 211. [DOI] [PubMed] [Google Scholar]
- Lamar J, Martschinske R, Tetreault G, Doud C. ( 2007) Navy Medical Department Pocket Guide to Malaria Prevention and Control. Technical Manual NEHCTM PM 6250.1
- Lemma H, San-Sebastian M, Lofgren C, Barnabas G. ( 2011) Cost-effectiveness of three malaria treatment strategies in rural Tigray, Ethiopia where both Plasmodium falciparum and P. vivax co-dominate. Cost Eff Resour Alloc. 9: 2– 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie T, Kaur H, Mohammed N, Kolaczinski K, Ord RL, Rowland M. ( 2009) Epidemic of Plasmodium falciparum malaria involving substandard antimalarial drugs, Pakistan, 2003. Emerging Infect Dis. 15( 11): 1753– 1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood K. ( 2005) Malaria in Karachi and other areas in Sindh. Pak Armed Forces Med J. 55: 345– 348. [Google Scholar]
- Mallick P, Joshi H, Valecha N, Sharma S, Eapen A, Bhatt R, Srivastava H, Sutton P, Dash A, Bhasin V. ( 2012) Mutant pfcrt “SVMNT” haplotype and wild type pfmdr1 “N86” are endemic in Plasmodium vivax dominated areas of India under high chloroquine exposure. Malar J. 1( 11): 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RE, Kirk K. ( 2004) The malaria parasite's chloroquine resistance transporter is a member of the drug/metabolite transporter superfamily. Mol Biol Evol. 21( 10): 1938– 1949. [DOI] [PubMed] [Google Scholar]
- Martin RE, Marchetti RV, Cowan AI, Howitt SM, Bröer S, Kirk K. ( 2009) Chloroquine transport via the malaria parasite's chloroquine resistance transporter. Sci. 325( 5948): 1680– 1682. [DOI] [PubMed] [Google Scholar]
- Mehlotra RK, Mattera G, Bockarie MJ, Maguire JD, Baird JK, Sharma YD, Alifrangis M, Dorsey G, Rosenthal PJ, Fryauff DJ. ( 2008) Discordant patterns of genetic variation at two chloroquine resistance loci in worldwide populations of the malaria parasite Plasmodium falciparum. Antimicrob Agents Chemother. 52( 6): 2212– 2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DV, Lanier JE. ( 1961) Observations on two Plasmodium falciparum infections with an abnormal response to chloroquine. Am J Trop Med Hyg. 10( 1): 5– 9. [DOI] [PubMed] [Google Scholar]
- Mubi M, Janson A, Warsame M, Martensson A, Kallander K, Petzold MG, Ngasala B, Maganga G, Gustafsson LL, Massele A. ( 2011) Malaria rapid testing by community health workers is effective and safe for targeting malaria treatment: randomized cross-over trial in Tanzania. Plos one. 6( 7): e19753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad W, Khan A, Faiz-ur-Rehman Ali A, Irshad S. ( 2010) Slide positivity rate in clinically suspected malaria cases. J Med Sci. 18( 2): 101– 103. [Google Scholar]
- Murtaza G, Memon IA, Memon AR, Lal M, Kallar NA. ( 2009) Malaria morbidity in Sindh and the Plasmodium species distribution. Pak J Med Sci. 25( 4): 646– 649. [Google Scholar]
- Panigrahi B, Kerketta A, Mohapatra A, Hazra R, Parida S, Marai N, Kar S, Mahapatra N. ( 2011) Effect of construction of an irrigation canal on malaria situation in two primary health centers of Dhenkanal district of Orissa, India. Trop Biomed. 28( 1): 76– 84. [PubMed] [Google Scholar]
- Peek R, Van Gool T, Panchoe D, Greve S, Bus E, Resida L. ( 2005) Drug resistance and genetic diversity of Plasmodium falciparum parasites from Suriname. Am J Trop Med Hyg. 73( 5): 833– 838. [PubMed] [Google Scholar]
- Peter BB. ( 2001) Drug Resistance in Malaria. WHO/CDS/CSR/DRS/2001.4, World Health Organization. [Google Scholar]
- Picot S, Olliaro P, De Monbrison F, Bienvenu AL, Price RN, Ringwald P. ( 2009) A systematic review and meta-analysis of evidence for correlation between molecular markers of parasite resistance and treatment outcome in falciparum malaria. Malar J. 8: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto J, Sousa C, Gil V, Goncalves L, Lopes D, do Rosario V, Charlwood J. ( 2000) Mixed-species malaria infections in the human population of Sao Tome island, West Africa. Trans R Soc Trop Med Hyg. 94( 3): 256– 257. [DOI] [PubMed] [Google Scholar]
- Pongtavornpinyo W, Yeung S, Hastings I, Dondorp A, Day N, White N. ( 2008) Spread of anti-malarial drug resistance: mathematical model with implications for ACT drug policies. Malar J. 7( 1): 229– 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prashant KM, Hema J, Neena V, Surya KS, Alex E, Rajendra MB, Harish CS, Patrick LS, Aditya PD, Virendra KB. ( 2012) Mutant pfcrt “SVMNT” haplotype and wild type pfmdr1“N86” are endemic in Plasmodium vivax dominated areas of India under high chloroquine exposure. Malar J. 11: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjab Strategic Plan for Malaria Control ( 2005) National Malaria Control Programme, Ministry of Health, Government of Pakistan. Strategic Plan; 15– 21. [Google Scholar]
- Rab M, Freeman T, Durrani N, De Poerck D, Rowland M. ( 2001) Resistance of Plasmodium falciparum malaria to chloroquine is widespread in eastern Afghanistan. Ann Trop Med Parasitol. 95( 1): 41– 46. [DOI] [PubMed] [Google Scholar]
- Rana SM. ( 2009) Treatment responses of uncomplicated falciparum malaria to different antimalaria drugs by mono and combination therapy in Punjab, Pakistan. [PhD thesis]. Department of Zoology, University of the Punjab Lahore, Pakistan. [Google Scholar]
- Sá JM, Twu O, Hayton K, Reyes S, Fay MP, Ringwald P, Wellems TE. ( 2009) Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci. 106( 45): 18883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandie M, Isabelle M, Rachida T, Collins S, Pembe IM, Xavier I, Françoise B, Brigitte L, Jean-François M, Parfait A, Leonardo KB, Antoine B. ( 2012) Molecular monitoring of Plasmodium falciparum drug susceptibility at the time of the introduction of artemisinin-based combination therapy in Yaoundé, Cameroon: Implications for the future. Malar J. 11: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah I, Rowland M, Mehmood P, Mujahid C, Razique F, Hewitt S, Durrani N. ( 1997) Chloroquine resistance in Pakistan and the upsurge of falciparum malaria in Pakistani and Afghan refugee populations. Ann Trop Med Parasitol. 91( 6): 591– 602. [DOI] [PubMed] [Google Scholar]
- Sidhu ABS, Verdier-Pinard D, Fidock DA. ( 2002) Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Sci. 298( 5591): 210– 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Kirkman LA, Fujioka H, Wellems TE. ( 1997) Complex polymorphisms in an 330 kDa protein are linked to chloroquine-resistant Plasmodium falciparum in Southeast Asia and Africa Cell. 91( 5): 593– 603. [DOI] [PubMed] [Google Scholar]
- Tran CV, Saier MH. ( 2004) The principal chloroquine resistance protein of Plasmodium falciparum is a member of the drug/metabolite transporter superfamily. Microbiol. 150( 1): 1– 3. [DOI] [PubMed] [Google Scholar]
- Ursing J, Zakeri S, Gil J, Bjorkman A. ( 2006) Quinoline resistance associated polymorphisms in the pfcrt, pfmdr1 and pfmrp genes of Plasmodium falciparum in Iran. Acta Trop. 97( 3): 352– 356. [DOI] [PubMed] [Google Scholar]
- Vieira PP, Urbano FM, Alecrim MG, Alecrim WD, da Silva LHP, Sihuincha MM, Joy DA, Mu J, Su X, Zalis MG. ( 2004) pfcrt Polymorphism and the spread of chloroquine resistance in Plasmodium falciparum populations across the Amazon Basin. J Infect Dis. 190( 2): 417– 424. [DOI] [PubMed] [Google Scholar]
- Warhurst DC. ( 2003) Polymorphism in the Plasmodium falciparum chloroquine-resistance transporter protein links verapamil enhancement of chloroquine sensitivity with the clinical efficacy of amodiaquine. Malar J. 2: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellems TE, Panton LJ, Guzman IY, do Rosario VE, Gwadz RW, Walker-Jonah A, Krogstad DJ. ( 1990) Chloroquine resistance not linked to mdr-1 like genes in a Plasmodium falciparum cross. Nature. 345: 253– 255. [DOI] [PubMed] [Google Scholar]
- Wellems TE, Walker-Jonah A, Panton LJ. ( 1991) Genetic mapping of the chloroquine-resistance locus on Plasmodium falciparum chromosome 7. Proc Natl Acad Sci. 88( 8): 3382– 3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White N, Pongtavornpinyo W. ( 2003) The denovo selection of drug–resistant malaria parasites. Proc R Soc Lond B Biol Sci. 270( 1514): 545– 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, Magill AJ, Su X. ( 2002) Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 418( 6895): 320– 323. [DOI] [PubMed] [Google Scholar]
- World Health Organization ( 2005) World malaria report. Geneva. [Google Scholar]
- World Health Organization ( 2010) World malaria report. Geneva. [Google Scholar]
- Yasinazi MI, Kakarsulemank JM. ( 2008) Incidence of Human Malaria Infection in Northern Hilly Region of Balochistan Adjoining with NWFP, Pakistan: District Zhob. Pak J Biol Sci. 11( 12): 1620– 1624. [DOI] [PubMed] [Google Scholar]
- Yohannes M, Haile M. ( 2010) The potential of in situ rain water harvesting for water resources conservation on malaria transmission in Tigray, Northern Ethiopia. Momona Ethiopian J Sci. 2( 2): 49– 63. [Google Scholar]
- Zakeri S, Afsharpad M, Kazemzadeh T, Mehdizadeh K, Shabani A, Djadid ND. ( 2008) Association of pfcrt but not pfmdr1 alleles with chloroquine resistance in Iranian isolates of Plasmodium falciparum. Am J Trop Med Hyg. 78( 4): 633– 640. [PubMed] [Google Scholar]