Summary
The yeast homologues of the ATM and ATR DNA damage response kinases play key roles in telomerase-mediated telomere maintenance, but the role of ATM/ATR in the mammalian telomerase pathway has been less clear. Here we demonstrate the requirement for ATM and ATR in the localization of telomerase to telomeres and telomere elongation in immortal human cells. Stalled replication forks increased telomerase recruitment in an ATR-dependent manner. Furthermore, increased telomerase recruitment was observed upon phosphorylation of the shelterin component TRF1 at an ATM/ATR target site (S367). This phosphorylation leads to loss of TRF1 from telomeres and may therefore increase replication fork stalling. ATM and ATR depletion reduced assembly of the telomerase complex, and ATM was required for telomere elongation in cells expressing POT1ΔOB, an allele of POT1 that disrupts telomere length homeostasis. These data establish that human telomerase recruitment and telomere elongation are modulated by DNA damage-transducing kinases.
Graphical Abstract

Introduction
Vertebrate telomeres are repetitive TTAGGG DNA sequences located at the ends of chromosomes, which protect the coding regions of DNA. In mammalian germline cells and ~85% of cancers, telomere length is maintained by the dimeric ribonucleoprotein telomerase, which catalyzes the addition of TTAGGG repeats to counteract telomere shortening and cellular senescence (Shay and Bacchetti, 1997; Kim et al., 1994; Wenz et al., 2001). The minimal catalytic core of human telomerase consists of the telomerase reverse transcriptase protein (hTERT), telomerase RNA (hTR) and the protein dyskerin (Cohen et al., 2007).
The differentiation of telomeres from broken chromosome ends is conferred by a family of six telomere-specific binding proteins collectively termed ‘shelterin’ (de Lange, 2005). This complex consists of the double stranded binding proteins TRF1 and TRF2, the single stranded binding proteins POT1 and TPP1, the bridging protein TIN2 which links these two groups of proteins, and Rap1 (reviewed in Palm and de Lange, 2008). TRF1 protects the telomere and negatively regulates telomerase-mediated telomere lengthening (van Steensel and de Lange, 1997; Smogorzewska et al., 2000; Ancelin et al., 2002; Karlseder et al., 2002). TRF1 also facilitates the progression of the replication machinery; deletion of TRF1 increases replication fork stalling, leading to ATR kinase activation and a “fragile telomere” phenotype (Sfeir et al., 2009; Martinez et al., 2009). The TRF1-mediated repression of the ATR response requires recruitment of the shelterin components TIN2 and the TPP1/POT1 heterodimer (Zimmermann et al., 2014).
TPP1 and POT1 also have roles in mediating telomere length regulation. A surface on the N-terminal oligonucleotide/oligosaccharide-binding (OB) domain of TPP1 termed the TEL patch is important for activation of telomerase, by stimulating telomerase processivity and providing a direct binding site for telomerase recruitment to telomeres; mutation of the TEL patch can lead to telomere shortening syndromes characterized by bone marrow failure (Abreu et al., 2010; Nandakumar et al., 2012; Zhong et al., 2012; Kocak et al., 2014; Guo et al., 2014; Dalby et al., 2015). Additionally, mutation analyses at sites independent of the TEL patch have implicated TPP1 as part of a telomere length-dependent feedback loop that regulates telomere length homeostasis (Sexton et al., 2014). A mutant form of POT1 that abrogates binding to single stranded DNA (POT1ΔOB) deregulated telomere length control (Loayza and de Lange, 2003), indicating that the DNA binding capability of POT1 is vital as a negative regulator of telomere length. The impact of human POT1 on telomere length is complex, since both depletion and overexpression of POT1 lead to telomere lengthening (Ye et al., 2004; Veldman et al., 2004; Colgin et al., 2003; Armbruster et al., 2004). POT1 function as a positive or negative regulator of telomerase activity at the telomere depends on its position of binding relative to the DNA 3′ end, and is also modulated by its binding partner TPP1 (Zaug et al., 2005; Wang et al., 2007; Lei et al., 2005; Kelleher et al., 2005).
Telomerase action at the telomere is highly regulated; it preferentially elongates the shortest telomeres, and recruitment of the enzyme complex to the telomere occurs in mid-S phase of the cell cycle (Bianchi and Shore, 2007; Britt-Compton et al., 2009; Teixeira et al., 2004; Hemann et al., 2001; Tomlinson et al., 2006; Jady et al., 2006). In both budding and fission yeasts, the preference of telomerase to extend the shortest telomeres requires the activity of Tel1, the yeast homolog of human ATM (Sabourin et al., 2007; Hector et al., 2007; Arneric and Lingner, 2007). ATM and ATR are kinases within the phosphatidylinositol-3 kinase-related kinase (PIKK) family, which regulate cellular responses to DNA damage, mRNA decay and nutrient-dependent signalling (Lovejoy and Cortez, 2009). Activation of these DNA damage pathways is dampened at telomeres; in mammalian cells, TRF2 represses activation of ATM while POT1 represses ATR (Karlseder et al., 2004; Celli and de Lange, 2005; Denchi and de Lange, 2007; Guo et al., 2007; Okamoto et al., 2013). Nevertheless, there is a large amount of evidence that their yeast homologues play a positive role in facilitating telomere extension by telomerase (Moser et al., 2009; Moser et al., 2011; Yamazaki et al., 2012; Churikov et al., 2013).
It is not known whether the role of the ATM and ATR pathways in recruiting telomerase is conserved in mammals. Although ATM deficiency or ATR mutations can induce telomere shortening or instability in human and mouse cells (Metcalfe et al., 1996; Smilenov et al., 1997; Wong et al., 2003; Wu et al., 2007; Pennarun et al., 2010), these kinases were reported to be dispensable for elongation of the shortest telomeres in mouse models (Feldser et al., 2006; McNees et al., 2010). Also, immortalized cell lines from human patients with ATM mutations are able to maintain their telomeres with telomerase, albeit at short lengths (Sprung et al., 1997). Nonetheless, there is evidence that TRF1-mediated telomere length regulation in human cells involves ATM. Inhibition of human ATM resulted in increased TRF1 at the telomere, and phosphorylation of TRF1 on serine 367, an ATM/ATR target site, reduced the interaction of TRF1 with telomeres and abrogated its ability to inhibit telomere lengthening (McKerlie et al., 2012; Wu et al., 2007).
In this study, we report that both ATM and ATR are required for the recruitment of human telomerase to telomeres.
Results
ATM and ATR are both required for the presence of human telomerase at telomeres
We used fluorescence in situ hybridization (FISH) to detect human telomerase RNA (hTR) in combination with a probe for telomeres to visualize the presence of endogenous telomerase at telomeres in HEK 293T cells (Stern et al., 2012) (Figure 1A). Depletion of ATM and ATR using siRNA resulted in reduction of levels of each protein by ~95% and ~80% respectively (Figure 1B), without causing significant perturbations in progression of the cells through the cell cycle (Figure S1A). Depletion of either kinase resulted in a significant decrease in telomerase recruitment to telomeres in asynchronously-growing cells (Figures 1A and 1C). The roles of both ATM and ATR in telomerase recruitment were confirmed with a second siRNA against each protein (Figure 1D).
Figure 1. ATM and ATR are both required for the presence of human telomerase at telomeres.
(A) Representative images of hTR/telomere FISH in 293T cells treated with the indicated siRNAs or kinase inhibitors. Cells were synchronized to mid-S phase of the cell cycle and probed with hTR probes (green) or a telomere probe (red). Co-localizations are indicated by white arrows in the merge row. Scale bar = 10 μm.
(B) Immunoblot of 293T cells with either ATM (left panel) or ATR (right panel) siRNA-mediated knockdown, using the respective antibodies, with vinculin as a control.
(C) Average co-localizations between telomerase and telomeres in unsynchronized 293T cells treated with control siRNA (gray), ATM siRNA (red) (*p = 0.012) or ATR siRNA (purple) (**p = 0.0095).
(D) Quantitation of decrease in telomerase recruitment in S-phase synchronized 293T cells following treatment with two independent ATM and ATR siRNAs; **p < 0.01. Cells were synchronized with a thymidine/aphidicolin block (ATM) or sorted into cell cycle phases by FACS based on DNA content (ATR).
(E) Average telomerase co-localization with telomeres in S-phase synchronized 293T cells after treatment with DMSO vehicle (gray), 1.5 μM KU-55933 (light yellow) or 500 nM VE-822 (dark yellow); *p < 0.05.
(F) Telomerase co-localization with telomeres in 293T cells at the indicated number of hours after release from a thymidine/aphidicolin block, treated with control (gray) or ATM (red) siRNA.
(G) Telomerase co-localization with telomeres in 293T cells treated with control (gray) or ATM (red) siRNA. Cells were stained with the DNA dye VyBrant DyeCycle Violet and isolated into cell cycle phases with FACS. Enrichment of cells in the indicated phases was confirmed by flow cytometry of sorted cells (Figure S1A).
(H) Telomerase co-localization with telomeres in 293T cells treated with control (gray) or ATR (purple) siRNA and sorted by FACS as in Figure 1G.
In all panels, data is presented as the mean of three independent experiments ± SD. See also Figure S1.
The involvement of the kinase activities of both ATM and ATR were demonstrated by using the kinase inhibitors KU-55933 and VE-822 (Figures 1A and 1E), at concentrations sufficient to inhibit ATM autophosphorylation and ATR activation, respectively (Figure S1B), without inhibiting related kinases (1.5 μM for KU-55933 and 0.5 μM for VE-822; Hickson et al., 2004; Fokas et al., 2014).
Telomerase recruitment across the cell cycle was examined by synchronizing cells at the G1/S boundary with overnight thymidine and aphidicolin treatments, followed by release from the cell cycle block and harvest at hourly intervals (Figure S1A). Telomerase presence at the telomere peaks in mid-S phase (Figure 1F), as previously described (Tomlinson et al., 2006; Jady et al., 2006). Depletion of ATM abolished recruitment of telomerase to the telomere across all phases of the cell cycle (Figures 1F and S1C). Since this synchronization procedure is likely to cause DNA damage, we confirmed these observations by sorting ATM- and ATR-depleted cells by flow-activated cell sorting (FACS) into different cell cycle phases based on DNA content (Figure S1A). Despite reduced numbers of hTR-telomere foci in control cells across the cell cycle (discussed further below), lack of either kinase again resulted in a large decrease in telomerase recruitment, particularly in S phase (Figures 1G and 1H). Reduction of hTR-telomere colocalizations across the cell cycle after ATM depletion was also demonstrated in a second human cell line, HeLa (Figure S1D–G). These results provide direct evidence that telomere shortening in telomerase-positive human cells defective in ATM and ATR activity is at least partially the result of diminished recruitment of telomerase to telomeres.
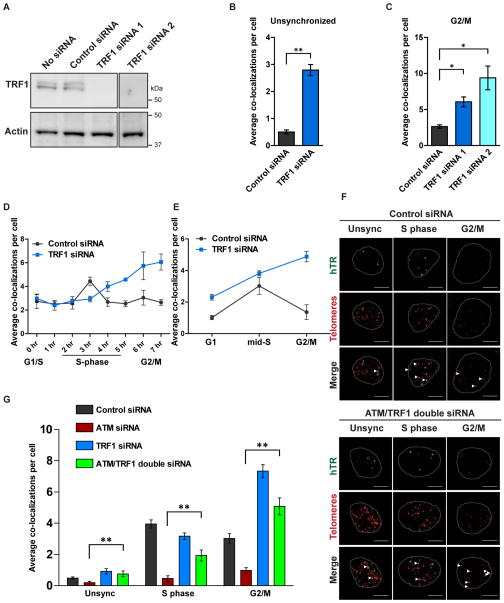
ATM effect on telomerase recruitment is partially mediated by TRF1
It is known that TRF1 is a substrate of ATM and/or other PIKK kinases, and such phosphorylation events lead to changes in TRF1 localization and telomere length (Kishi et al., 2001; Wu et al., 2007; McKerlie et al., 2012). To investigate whether these changes in telomere length are due to changes in the presence of telomerase at telomeres, we knocked down TRF1 by ~70–80% with two independent siRNAs (Figures 2A and S2A–B) and examined hTR-telomere colocalization. Knockdown of TRF1 resulted in large increases in telomerase recruitment in asynchronous cells or in cells synchronized to G2/M (Figures 2B, 2C and S2C). Analysis of telomerase recruitment over the cell cycle revealed a deregulation of recruitment, particularly in G2/M phase, in both thymidine/aphidicolin synchronized and FACS sorted 293T and HeLa cells (Figures 2D, 2E and S2F–H). TRF1 knockdown also resulted in increased hTR/telomere co-localizations in G1 phase in FACS-sorted cells, demonstrating that TRF1 is involved in the signalling pathway that restricts telomerase presence at the telomere to S phase.
Figure 2. ATM effect on telomerase recruitment is partially mediated by TRF1.
(A) Immunoblot of 293T cells treated with control or two different TRF1 siRNAs. All lanes are from the same immunoblot, which was cropped to remove intervening irrelevant lanes. Actin was probed as a control.
(B) Average recruitment of telomerase to telomeres in asynchronous 293T cells treated with control (gray) or TRF1 (blue) siRNA; **p = 0.0011.
(C) Average co-localization of telomerase with telomeres in 293T cells synchronized to G2/M of the cell cycle by release from a thymidine/aphidicolin block; cells were treated with control (gray) or two different TRF1 siRNAs (blue); *p < 0.05.
(D) Telomerase co-localization with telomeres in thymidine/aphidicolin synchronized 293T cells, treated with control (gray) or TRF1 (blue) siRNA. The values along the x-axis represent the number of hours since release of cells from G1/S boundary. The control data are the same as those in Figure 1F since these experiments were performed simultaneously.
(E) Telomerase co-localization with telomeres in 293T cells, treated with control (gray) or TRF1 (blue) siRNA; cells were stained with the DNA dye VyBrant DyeCycle Violet and isolated into the cell cycle phases with FACS. The control data are the same as those in Figure 1G since these experiments were performed simultaneously.
(F) FISH for hTR (green) and telomeres (red) in 293T cells treated with control or combined ATM and TRF1 siRNAs. Cells were either asynchronous, or synchronized with thymidine and aphidicolin and harvested 3 – 4 h (S phase) or 7 h (G2/M) after release from the G1/S boundary. Co-localizations are indicated by the white arrows in the merge row. Scale bar = 10 μm.
(G) Quantitation of Figure 2F; average telomerase-telomere co-localizations after control (gray), ATM only (red), TRF1 only (blue) and ATM/TRF1 (green) siRNAs, in asynchronous, S phase or G2/M phase cell populations; n = 4; **p < 0.01.
Data is presented as the mean of three independent experiments except where indicated otherwise, ± SD. See also Figure S2.
To confirm that ATM and TRF1 function in the same pathway, we performed a double knockdown of ATM and TRF1 (Figure S2D–E). If ATM involvement in telomerase recruitment is mediated by the phosphorylation and removal from the telomere of TRF1 (Wu et al., 2007), we predict that depletion of TRF1 will render telomerase insensitive to the absence of ATM. Simultaneous knockdown of ATM and TRF1 significantly elevated the levels of telomerase recruitment in S and G2/M phases compared to depletion of ATM alone (p = 0.0018 and p = 0.0097, respectively) (Figures 2F and 2G). A full recovery of recruitment was not observed in S phase cells; this may be partially due to residual TRF1 remaining after knockdown, but also indicates that other TRF1-independent ATM substrates may be involved in telomerase recruitment (discussed further below).
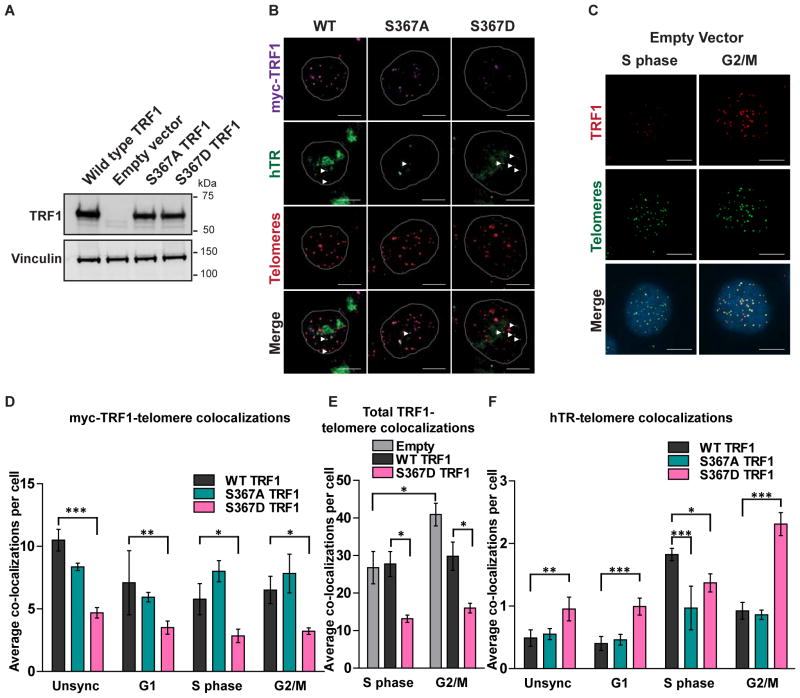
Phosphorylation of TRF1 at serine 367 controls TRF1 and telomerase localization to telomeres
We have demonstrated that phosphorylation of serine 367 of TRF1 results in localization of TRF1 to proteasome centres for degradation, resulting in telomere elongation (McKerlie et al., 2012). To demonstrate that such elongation events are due to changes in telomerase localization to telomeres, we transiently expressed in 293T cells similar levels of myc-tagged wild-type (WT) TRF1, or TRF1 containing mutations at S367 that either abolish (S367A) or mimic (S367D) phosphorylation at this residue (Figures 3A and S3A–B). Exogenous TRF1 was expressed at levels 10- to 15-fold greater than endogenous TRF1 (Figure 3A); this level of overexpression was sufficient for replacement of endogenous TRF1 with myc-tagged exogenous TRF1 at ~25% of telomeres (Figures 3B–E). S367D TRF1 demonstrated lower telomere association across all phases of the cell cycle than WT or S367A TRF1 (Figures 3B, 3D and S3C), consistent with its inability to bind telomeric DNA in vitro (McKerlie et al., 2012). Endogenous TRF1 was not depleted in this experiment, since we expected S367D TRF1 to behave in a dominant-negative manner to sequester endogenous TRF1 from the telomere, as has been observed for other DNA binding-defective TRF1 mutants (van Steensel and de Lange, 1997); we confirmed this using immunofluorescence with an antibody against TRF1 (Figures 3C and 3E). This observation coincided with an elevated level of telomerase recruitment in the presence of S367D TRF1 in G1 and G2/M phases (Figures 3B, 3F and S3C). Deregulation of telomerase recruitment in G1 and G2/M phases upon both TRF1 knockdown and S367D TRF1 expression was confirmed in a HeLa cell line stably expressing an shRNA against TRF1, together with shRNA-resistant S367D TRF1 (McKerlie et al., 2012; Figure S3D).
Figure 3. Phosphorylation of TRF1 at serine 367 regulates TRF1 and telomerase localization to telomeres.
(A) Immunoblot of over-expressed myc-tagged WT TRF1, empty vector, myc-S367A TRF1 or myc-S367D TRF1, probed with an anti-TRF1 antibody, and with vinculin probed as a control.
(B) Immunofluorescence with an anti-myc antibody (purple) and hTR/telomere FISH (green and red respectively) in asynchronous 293T cells over-expressing myc-tagged WT, S367A or S367D TRF1. Co-localizations between hTR and telomeres are indicated by white arrows. Scale bar = 10 μm.
(C) Immunofluorescence with an anti-TRF1 antibody (red) and telomere FISH (green) in 293T cells transfected with empty vector and sorted into S or G2/M phase based on DNA content. Scale bar = 10 μm.
(D) Quantitation of average co-localization of overexpressed myc-tagged TRF1 with telomeres across cell cycle stages in FACS sorted 293T cells; *p < 0.05, **p < 0.01, ***p < 0.005.
(E) Quantitation of total TRF1 at telomeres in S and G2/M phase sorted 293T cells over-expressing myc-tagged WT TRF1, empty vector, or myc-S367D TRF1; *p < 0.05.
(F) Quantitation of average co-localization of hTR and telomeres across cell cycle stages in FACS sorted 293T cells over-expressing myc-tagged WT TRF1, myc-S367A TRF1, or myc-S367D TRF1; *p < 0.05, **p < 0.01, ***p < 0.005.
In all panels, data is presented as the mean of three independent experiments ± SD. See also Figure S3.
Conversely, there was an ~50% decrease in telomerase recruitment at S phase in the presence of phosphodeficient S367A TRF1 compared to WT TRF1, in both 293T and HeLa cells (Figures 3F and S3D). These data suggest that phosphorylation of TRF1 at serine 367 results in dissociation of a portion of TRF1 from telomeres during S phase, and that this is necessary for full telomerase recruitment. This is supported by the slight increase in levels of endogenous TRF1 at telomeres in G2/M relative to S phase (Figures 3C and 3E). However, depletion of TRF1 levels at the telomere using siRNA, shRNA or expression of the dominant negative S367D TRF1 did not lead to substantially increased telomerase at the telomere during S phase (Figures 2D–G, S2H, 3E, 3F and S3D), suggesting that removal of TRF1 is necessary but not sufficient for full telomerase recruitment in S phase. Together, our data show that the major function of TRF1 is in the displacement of telomerase from the telomere outside S phase, for which removal of the phosphate group at S367 appears to be necessary.
Stalled replication forks trigger telomerase recruitment
The mechanism of telomerase regulation at the telomere by TRF1 may involve the known role of TRF1 in facilitating replication of telomeric DNA (Sfeir et al., 2009). Repetitive telomere repeats represent challenging templates for the canonical DNA replication machinery, often resulting in stalled replication forks and recruitment of single-stranded DNA binding protein RPA and activation of ATR (Ohki and Ishikawa, 2004; Sfeir et al., 2009; Fouche et al., 2006; Ivessa et al., 2002); in mammalian cells, this phenotype is exacerbated by depletion of TRF1 (Sfeir et al., 2009; Martinez et al., 2009). Our observation that treatment of cells with aphidicolin and thymidine results in an apparent increase in numbers of hTR-telomere foci (Figures 1F and 1G) suggests that DNA damage directly regulates telomerase recruitment. To directly examine whether this effect can be attributed to stalled replication forks, we subjected asynchronous 293T cells to a much shorter period of aphidicolin treatment. Telomerase recruitment increased ~2-fold after 30 min treatment with 0.5 μg/mL aphidicolin compared to controls, despite no significant change in cell cycle profile, and this increase was dependent upon ATR (Figures 4A, 4B and S4). Phosphorylation of the ATR target protein Chk1 demonstrated that a 30 min treatment with aphidicolin was sufficient to cause replication problems and activate the ATR pathway (Figure 4C). To ensure that this aphidicolin treatment did not result in damage signalling from DNA double strand breaks (DSBs) arising from collapsed replication forks, we examined activation of the ATM-Chk2 DSB response pathway (Figure 4C). Aphidicolin treatment caused a moderate activation of ATM and Chk2 phosphorylation (Figure 4C, lane 2, and Figure 4D), but this was abolished by knockdown of ATR (Figure 4C, lane 5, and Figure 4D), indicating that it results from the direct activation of ATM by ATR that is known to occur upon replication fork stalling (Stiff et al., 2006), rather than from DSB formation. In addition, induction of DSBs by γ-irradiation triggered robust ATR-independent ATM activation (Figure 4C, lanes 3 and 6) that did not result in increased telomerase recruitment to telomeres (Figures 4A and 4B). Together our data demonstrate that an increase in stalled replication forks and the resulting ATR signalling result in an increase in telomerase presence at the telomere. This is consistent with the telomere lengthening previously observed in telomerase-positive human cells after long-term treatment with aphidicolin (Sfeir et al., 2009).
Figure 4. Stalled replication forks trigger telomerase recruitment.
(A) FISH for hTR (green) and telomeres (red) in asynchronous 293T cells treated with control (3 left panels) or ATR (3 right panels) siRNA. DNA damage was induced by either 30 min of aphidicolin treatment to induce stalled replication forks or 2 Gy γ-irradiation to induce double strand breaks. Co-localizations between hTR and telomeres are indicated by the white arrows. Scale bar = 10 μm.
(B) Quantitation of telomerase localization to telomeres in cells from Figure 4A; *p < 0.05.
(C) Immunoblot of cells treated as in Figure 4A, probed for ATR, pS1981 ATM, ATM, pT68 Chk2, Chk2, pS345 Chk1, Chk1 and vinculin as a control.
(D) Quantitation of ATM activation (pS1981 ATM levels) from blot in Figure 4C; *p = 0.028.
In all panels, data is presented as the mean of three independent experiments ± SD. See also Figure S4.
Telomere elongation triggered by POT1 mutation is dependent on ATM
As a TRF1-independent mechanism of exposing single-stranded DNA at telomeres, we infected HeLa204 cells (Takai et al., 2010) with a retrovirus encoding the human POT1 construct myc-POTΔOB (Loayza and de Lange, 2003). Myc-POT1ΔOB contains a truncation of one of the two POT1 OB folds, allowing POT1 to localize to telomeres but abolishing its ability to bind single-stranded telomeric DNA, and abrogating POT1-mediated telomere length regulation (Loayza and de Lange, 2003; Zhong et al., 2012). As expected, myc-POTΔOB caused dramatic telomere length elongation over 35 – 37 population doublings (PD) (Figure 5A). Simultaneous expression of three independent shRNAs against ATM (Figure 5B) completely abrogated telomere lengthening (Figure 5A), demonstrating that telomerase-mediated telomere lengthening in this context is dependent on ATM.
Figure 5. Telomere elongation triggered by POT1 mutation is dependent on ATM.
(A) Terminal restriction fragment (TRF) assay to determine telomere length. HeLa204 cells were infected with retrovirus encoding luciferase shRNA (ctrl1), no shRNA (ctrl 2) or 3 different ATM shRNAs, together with myc-POT1ΔOB overexpression, and cultured for 32 – 37 population doublings (PD). All lanes are from the same blot, which was cropped to remove intervening irrelevant lanes.
(B) Immunoblot to determine ATM knockdown following shRNA treatment of cells from (A), with α-tubulin as a control.
ATM and ATR also affect telomerase complex assembly
The inability of TRF1 depletion to completely rescue telomerase recruitment after ATM silencing (Figure 2G) implies that ATM can affect other TRF1-independent pathways of telomerase regulation. We therefore investigated the levels of cellular telomerase components and their assembly capabilities. Measurement of total cellular hTR levels with a dot blot using an hTR specific probe revealed a slight inhibitory effect of TRF1 siRNA on hTR abundance (Figures 6A, 6B and S5A–B). The amount of hTR recovered after immunoprecipitation with an antibody against hTERT (Figures 6A and 6C) reflected the levels of hTR in crude lysates, indicating that TRF1 depletion did not impact assembly of hTERT and hTR into an active complex. Conversely, treatment with ATM siRNA did not affect cellular hTR abundance (Figures 6A and 6B), but reduced telomerase complex assembly by ~50% across all phases of the cell cycle (Figures 6A and 6C). Double ATM/TRF1 knockdown also resulted in a similar assembly defect (Figures 6D and S5C–D), indicating that TRF1 is unlikely to be the ATM target responsible for this effect. Telomerase activity, measured by a direct non-PCR assay (Cohen and Reddel, 2008) and normalized against the amount of hTR recovered in hTERT immunoprecipitates, revealed no significant changes in the specific activity of telomerase after depletion of either ATM or TRF1 (Figures 6E, 6F and S5E–G).
Figure 6. ATM affects telomerase assembly.
(A) Dot blot for hTR in 293T cells treated with control, ATM or TRF1 siRNA and synchronized with thymidine/aphidicolin. Crude cell lysates (top panel) contain total levels of cellular hTR while hTERT immunoprecipitated samples (IP) contain only hTR that has assembled with hTERT. Top row: in vitro-transcribed hTR standard.
(B) Quantitation of total hTR levels following control (gray), ATM (red) or TRF1 (blue) siRNA treatments (fmol of hTR in 1 × 106 cells) from Figure 6A; *p = 0.032.
(C) Quantitation of hTR assembled with hTERT (fmol of hTR in 2 × 106 cells) from Figure 6A, following control (gray), ATM (red) or TRF1 (blue) siRNA treatments; *p < 0.05, ***p < 0.005.
(D) Quantitation of hTR assembled with hTERT after treatment with combined ATM and TRF1 siRNAs (green); *p < 0.05, ***p < 0.005.
(E) Direct telomerase activity assay of immunoprecipitated telomerase from cells treated with control, ATM and TRF1 siRNA, synchronized to the indicated cell cycle stages. LC represents an 18 nt loading control.
(F) Telomerase specific activity, derived from total telomerase activity (Figure 6E) normalized to levels of hTR after hTERT immunoprecipitation (Figure 6C).
(G) Dot blot for hTR in 293T cells treated with control or two different ATR siRNAs and synchronized with thymidine/aphidicolin. The combination of ATR siRNA and thymidine/aphidicolin treatment perturbs progression of the cells through S phase, so this experiment could only be performed on unsynchronized or G1 cells. Crude cell lysates (top panel) contain total levels of cellular hTR while hTERT immunoprecipitated samples (IP) contain only hTR that has assembled with hTERT. Top row: in vitro-transcribed hTR standard.
(H) Quantitation of hTR assembled with hTERT after treatment with two different ATR siRNAs (purple); *p < 0.05.
(I) Telomerase specific activity, derived from total telomerase activity (Figure S5K) normalized to levels of hTR after hTERT immunoprecipitation (Figure 6H).
In all panels, data is presented as the mean of three independent experiments ± SD. See also Figure S5.
We also examined the effect of ATR silencing on levels of cellular telomerase RNA and telomerase assembly. Treatment with two different ATR siRNAs (Figure S5H) reduced telomerase complex assembly by up to ~50% in unsynchronized and G1 phase cells (Figures 6G and 6H) without affecting total hTR levels or telomerase specific activity (Figures 6I, S5I–K).
Discussion
In this study we have demonstrated that ATM and ATR are both necessary for full telomerase recruitment to telomeres in human cell lines. This provides an explanation, at least in part, for the long-standing observation of short telomeres in the ATM-deficient cells of ataxia telangiectasia (AT) patients (Metcalfe et al., 1996; Smilenov et al., 1997), and the telomere shortening observed upon inhibition of ATM in telomerase-positive immortal human cell lines (Wu et al., 2007). Consistently, while AT patient cells can become immortalized by activation of telomerase, most of these cell lines harbor very short telomeres (Sprung et al., 1997).
Our data show that TRF1 is involved in the signalling pathway restricting telomerase access to the telomere to S phase; loss of TRF1 results in an increase in telomerase at the telomere in both G1 and G2/M phases. Evidence from this and previous studies suggests that phosphorylation of TRF1 at serine 367 results in partial TRF1 dissociation from telomeres and its degradation; removal of this phosphate is necessary for correct cell cycle control of telomerase presence at the telomere. Constitutive expression of a phosphomimetic of S367 TRF1 also leads to inappropriate retention of telomerase at the telomere outside S phase, leading to a telomere length increase (Wu et al., 2007; McKerlie et al., 2012 and this study, Figure 3). The mechanism for this regulatory function of TRF1 may include its known role in facilitating telomere replication; TRF1 dissociation from telomeres induces replication fork stalling that activates ATR (Sfeir et al., 2009), and we provide evidence that replication fork stalling leads to an ATR-dependent increase in telomerase recruitment (Figure 4). The control of telomere replication and telomerase presence at the telomere by human TRF1 (Figure 7A) appears analogous to the situation in Schizosaccharomyces pombe, in which deletion of the double-stranded telomeric binding protein Taz1 results in stalled telomeric replication forks (Miller et al., 2006), and leads to deregulation of the cell cycle control of telomerase at the telomere (Dehe et al., 2012; Chang et al., 2013). It is possible that in the absence of TRF1, aberrant products of stalled replication forks may persist into the subsequent G2/M and G1 phases, forming substrates for telomerase, as has been postulated in the case of Taz1 in S. pombe (Dehe et al., 2012). It has been proposed that it is the tendency of the replication machinery to stall in repetitive DNA that forms the signal for telomerase recruitment specifically in S phase (Rog and Cooper, 2008; Verdun and Karlseder, 2006; Wu et al., 2007; Stern and Bryan, 2008; Dehe et al., 2012; Chang et al., 2013). In this report we provide direct evidence for this concept.
Figure 7. Model for ATM and ATR involvement in human telomerase recruitment to telomeres.
(A) During S phase, ATM and/or other PIKKs phosphorylates TRF1 at S367, which leads to partial dissociation of TRF1 from telomeres (McKerlie et al., 2012; Wu et al., 2007). Depletion of TRF1, together with its protein partners Tin2, TPP1 and Pot1, causes telomeric replication forks to stall, leading to recruitment of RPA and ATR (Sfeir et al., 2009; Martinez et al., 2009; Zimmermann et al., 2014). ATR phosphorylates an unknown substrate to mediate telomerase recruitment. Replication fork stalling caused by aphidicolin treatment also leads to telomerase recruitment (this study) and telomere elongation (Sfeir et al., 2009).
(B) An independent role of ATM and ATR involves stimulation of telomerase assembly, which is a prerequisite for telomerase localization to telomeres. This model does not preclude involvement of other unidentified substrates of ATM, ATR and other PIKKs.
The downstream targets of ATR involved in mediating human telomerase recruitment have not yet been identified. Under stalled fork conditions, activated ATR is able to phosphorylate and activate ATM (Stiff et al., 2006, and Figure 4C and D); whether this phosphorylation of ATM participates in a positive feedback loop in removing TRF1 to accelerate ATR-mediated telomere length regulation remains to be investigated.
We also provide evidence that exposing telomeric single-stranded DNA in a different way, by overexpression of POT1 lacking its DNA-binding domain (Loayza and de Lange, 2003), provides a signal for rapid telomere lengthening that is also ATM-dependent (Figure 5). Overexpression of POTΔOB does not perturb binding of TRF1 to the telomere (Loayza and de Lange, 2003); the ATM-dependence of telomere lengthening in this context reinforces the notion that in addition to mediating telomerase recruitment through a TRF1-dependent pathway, there must be other TRF1-independent mechanisms by which ATM mediates telomerase localization at telomeres. This is consistent with the observation that lack of TRF1 does not completely rescue the telomerase recruitment defect caused by ATM depletion (Figure 2G).
We demonstrated that one TRF1-independent function of ATM is its impact upon the ability of hTR and hTERT to assemble into a functional enzyme complex (Figure 6), which is a prerequisite for localization of hTR to telomeres (Tomlinson et al., 2008). ATR also plays a role in assembly of human telomerase; we do not know if the substrates of these two kinases in this process are the same. This role is reflected in a substantial decrease in the amount of hTR recovered after hTERT IP, and in the total immunoprecipitated telomerase activity following ATM and ATR knockdown. Telomerase specific activity remains unchanged, demonstrating that both ATM and ATR have no effect on telomerase catalytic activity, consistent with results in yeast (Chan et al., 2001). No consensus PIKK phosphorylation motifs exist in the RNA binding domain of hTERT, implying either that ATM or ATR can mediate telomerase assembly by targeting regions not in the RNA binding domain, or that they can regulate telomerase assembly by phosphorylating unknown substrates (Figure 7B).
Our data support a model incorporating multiple roles for ATM and ATR in the presence of human telomerase at telomeres (Figure 7). One pathway involving both ATM and ATR is mediated by phosphorylation of TRF1 and its removal from the telomere, leading to replication fork stalling in telomeric DNA, which acts as a trigger for telomerase recruitment. A second pathway involves the role of ATM and ATR in facilitating telomerase assembly; additional phosphorylation targets of ATM, ATR and other PIKKs in the telomerase recruitment process may remain to be identified. These data reveal that, although it is important for telomeres to repress DNA damage signalling in order to avoid deleterious fusions, telomeres have also evolved the ability to carefully exploit aspects of DNA damage signalling pathways to regulate telomerase presence at the telomere. Increased understanding of regulation of telomerase assembly and access to the telomere may provide valuable insight in the process of developing highly specific cancer therapeutics.
Experimental Procedures
Plasmids
A TRF1 expression plasmid containing myc-tagged TRF1 was constructed by subcloning the TRF1 open reading frame from pCMV-GFP.hTRF1 plasmid into a pcDNA3.1/myc-HisB plasmid by In-Fusion® cloning (Clontech Laboratories, Inc.) according to the manufacturer’s protocol. The mutation of serine 367 of TRF1 into alanine or aspartic acid was achieved using the Stratagene® QuikChange® II XL Site-Directed Mutagenesis kit. POT1 lacking the OB fold domain (POT1ΔOB) was expressed from the pLPC retroviral vector as described (Loayza and de Lange, 2003), followed by infection with retrovirus vectors encoding shRNAs against ATM.
Cell culture and transfections
HEK 293T cells (from Dr. T. Adams, CSIRO), HeLa cells (American Type Culture Collection), HeLa204 cells (Takai et al., 2010) or HeLa cells stably overexpressing TRF1 (McKerlie et al., 2012) were cultured in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) in a humidified 37°C incubator with 5% CO2. Cells were transfected with 90 pmol of siRNA, and 5.5 μL of lipofectamine® RNAiMAX (Life Technologies) in 310 μL of opti-MEM® (Life Technologies). siRNA sequences and catalogue numbers are provided in Table S1 (Supplemental Data). Cells overexpressing myc-TRF1 were transfected with 2.5 μg of WT TRF1 or 1.0 μg of S367A/D TRF1 expression plasmid to ensure even protein expression levels, in DMEM for 34 h. HEK 293T cells were then synchronized at G1/S phase by adding 2 mM thymidine (Sigma-Aldrich) for 15 h, releasing for 9 h, and adding 0.5 μg/mL aphidicolin (Sigma-Aldrich) for 16 h. Cells were harvested 3 h after release and confirmed to be in mid-S phase by analysis of DNA content by flow cytometry on a FACSDiva (Becton Dickinson). HeLa cells grown in DMEM media (+ 10% FBS) were synchronized with medium containing 2 mM thymidine (Sigma-Aldrich) for 18 h, released into thymidine-free DMEM (+ 10% FBS) for 9 h and blocked with a second round of 2 mM thymidine for 15 h. For kinase inhibitor treatment, 1.5 μM KU-55933 (Merck Millipore), 500 mM VE-822 (Selleckchem) or DMSO was added to cells released from synchronization for 3 h before harvesting in mid S phase.
For cell sorting, cells were harvested and resuspended in Hank’s balanced salt solution (Life Technologies) at a concentration of 1 × 106 cells/mL. The cells were stained with VyBrant DyeCycle Violet (Life Technologies) at a concentration of 1 μL per 1 × 106 cells at 37°C for 30 min before cell sorting using a BD FACSAriaIII Cell Sorter (BD Bioscience, US). Confirmation of cell cycle phases was performed using the FACSDiva (Becton Dickinson).
Gamma (γ) irradiation
Gamma irradiation was performed using the Gammacell® 1000 Elite irradiator (Best Theratronics) with a 137Caesium source. The irradiated cells were returned to incubate at 37°C for an hour before harvesting for downstream analysis.
FISH of hTR and telomeric DNA, and immunofluorescence
Simultaneous FISH against hTR and telomeric DNA, with or without immunofluorescence for TRF1, was performed as previously described (Stern et al., 2012), with the exception that cells were fixed in 2% paraformaldehyde. Antibodies used for immunofluorescence were Myc-tag antibody (Cell Signaling, cat. no 2276; diluted 1:1,000) and polyclonal rabbit anti-TRF1 antibody (#370, de Lange lab; diluted 1:2,000). Staining was visualized on a Zeiss Axio Imager M1 microscope, with a Plan-Apochromat 63× oil objective (numerical aperture, 1.4), and an AxioCam MR digital camera (Carl Zeiss). Exposure times between treatments were consistent. For presentation purposes, pixel intensity histograms were adjusted in ZEN (Carl Zeiss), equally across all figure panels, and images were cropped in Adobe Photoshop. Colocalizing hTR and telomere foci were manually counted in >100 cells per treatment, and data were analyzed by Student t test for pairwise comparison.
Automated quantification of 3D co-localisations between TRF1 and telomeres
Immunofluorescence with anti-TRF1 antibody (Figure 3C) had sufficiently low background that quantitation of colocalizing foci was able to be automated using Metafer4 software (Metasystems, GmbH, Altlussheim, Germany) on a Zeiss Axioplan 2 microscope (Germany), with a 63× NA (1.4 Plan-Apochromat) oil objective, and appropriate filter cubes. DAPI stained nuclei were identified and background subtraction, image sharpening and TopHat transformation applied to the TRF1 immunofluorescence and telomere FISH channels. TRF1 immunofluorescence and telomere FISH foci were defined as foci of > 0.1 μm2, > 25% intensity over background, separated by a minimum distance of 0.5 μm. Co-localizations were events where the centre of a TRF1 and a telomere FISH focus were ≤ 0.3 μm apart in three dimensions. Colocalizing TRF1 and telomere foci were counted in 200 – 300 cells per treatment, and data were analyzed by Student t test for pairwise comparison.
Cell lysis and Western blot
The procedure for cell lysis and Western blot was performed as previously described (Guo et al., 2014), with the exception that the large proteins ATR and ATM were electrophoresed on 3–8% gradient Tris-Acetate gels (Life Technologies) at 150 V for 90 min. The following antibodies were used: ATM (Abcam rabbit polyclonal, diluted 1:2,000, cat. no AB32420), ATR (Cell Signaling rabbit polyclonal, diluted 1:1,000, cat. no 2790), anti-pS1981 ATM (Abcam rabbit polyclonal, diluted 1:1,000, cat. no AB81292), pT68 Chk2 (Cell Signaling rabbit polyclonal, diluted 1:1,000, cat. no 2661), Chk2 (Merck-Millipore mouse monoclonal, diluted 1:1,000, cat. no 05-649), pS345 Chk1 (Cell Signaling rabbit polyclonal, diluted 1:1,000, cat. no 23485), Chk1 (Cell Signaling mouse monoclonal, diluted 1:1,000, cat. no 23605), TRF1 (rabbit polyclonal, #370, de Lange lab, diluted 1:2,000), vinculin (Sigma-Aldrich mouse monoclonal, diluted 1:10,000, cat. no V9131), and actin (Sigma-Aldrich rabbit polyclonal, diluted 1:10,000, cat. no A2103).
Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)
RNA extraction was performed using the RNeasy Mini Kit (Qiagen) according to manufacturer’s protocol and RNA quantitated with a Nanodrop spectrophotometer (ThermoFisher ND-1000). RNA (1 μg) was digested with 1 U of DNase (Amp grade; Sigma-Aldrich) at room temperature for 15 min, before the addition of EDTA to 2.5 mM and heat inactivation at 65°C. cDNA synthesis was performed using the ‘Superscript III First-Strand Synthesis System for RT-PCR’ (Life Technologies) according to the manufacturer’s protocol. qPCR was performed in triplicate on each cDNA with corresponding GAPDH controls, using the Roche real-time PCR LightCycler® 96 system with 45 cycles of 95 °C for 15 s and 60 °C for 60 s, in Applied Biosystems SYBR green mix, with 0.5 μM forward and reverse primers (TRF1 forward: 5′-CGAGCTAGAAAAAGACAGGC, TRF1 reverse: 5′-AGTTTTAGTTTCTTCATGGT, GAPDH forward: 5′-ACCCACTCCTCCACCTTTG, GAPDH reverse: 5′-CTCTTGTGCTCTTGCTGGG). Analysis was carried out using the ΔΔCt method with the Roche LightCycler® 96 system software.
hTR dot blot
Harvested cells were lysed with telomerase buffer (20 mM HEPES-KOH pH 7.9, 300 mM KCl, 2 mM MgCl2, 10% v/v glycerol, 0.1% v/v Triton X-100, 5 mM dithiothreitol (DTT), 1 mM phenylmethanesulfonylfluoride (PMSF)) at a concentration of 105cel ls/μL, rotating at 4 C for 1 hr before centrifugation at 16,200 × g at 4 C for 20 min. 10 μL of the lysate was applied to a GE Healthcare Life Sciences Amersham™ Hybond™-XL membrane, and probed with a 32P-labeled oligonucleotide complementary to hTR as described (Jurczyluk et al., 2011).
Telomerase purification and activity assay
Immunopurification of telomerase was performed as described (Cohen and Reddel, 2008), using a polyclonal anti-hTERT antibody and elution with a competing peptide (both available from Abbexa Ltd., Cambridge, UK). Activity of the eluted telomerase was measured in a 20 μL solution phase extension reaction containing 20 mM HEPES-KOH pH 7.9, 2 mM MgCl2, 5 mM DTT, 1 mM spermidine, 0.1% Triton X-100, 0.5 mM dTTP, 0.5 mM dATP, 5 μM α-32P-dGTP (198 Ci/mmol) and 1 μM of DNA primer Bio-L-18GGG (5′-Biotin-CTAGACCTGTCATCA(TTAGGG)3) for 60 min at 37°C. The reaction was terminated with EDTA, and products isolated on Dynabead M280 streptavidin beads (Life Technologies), together with a 12 nt biotinylated control DNA, as described (Tomlinson et al., 2015). Products were separated on a 10% acrylamide/7M urea sequencing gel as described (Tomlinson et al., 2015).
Telomere Restriction Fragment (TRF) southern blot
Telomere length was measured by HinfI/RsaI digestion of genomic DNA, separation by agarose gel electrophoresis, transfer to nylon membrane and probing with a 32P-labelled telomeric probe, using standard techniques (de Lange, 1992).
The hTERT antibody, CMRI 276–294, and antigenic release peptide are available to researchers from Abbexa Ltd (Cambridge, UK), under exclusive license from the authors’ institution, Children’s Medical Research Institute.
Supplementary Material
Acknowledgments
Cell sorting was performed in the Flow Cytometry Core Facility that is supported by Westmead Research Hub, Cancer Institute New South Wales (CINSW) and National Health and Medical Research Council (NHMRC). Microscopy was performed in the ACRF Telomere Analysis Centre, supported by Australian Cancer Research Foundation and the Ian Potter Foundation, and with the assistance of Scott Page. Phosphoimaging analysis was performed on a Typhoon Biomolecular Imager purchased with support from the Ramaciotti Foundation. We thank Sonja Frölich and Anthony Cesare for development of the algorithm used for automated foci counting. We are grateful to Anthony Cesare, Scott Cohen and Christopher Tomlinson (Children’s Medical Research Institute) for helpful discussions and providing protocols. We thank Angus Ho in the Zhu laboratory for the generation of TRF1-depleted HeLa cells complemented with wild type and mutant TRF1 alleles. Work in the Bryan laboratory was supported by project grants awarded by Cancer Council NSW (RG10/02 and RG12/02) and NHMRC (571073). Work in the Zhu laboratory was supported by Canadian Institutes of Health Research (MOP-86620). The work on telomere length regulation was supported by an RO1 from the National Institutes of Health (to TdL). AST was supported by a Kids Cancer Alliance PhD Scholarship, MK by an Australian Awards Scholarship, and TMB by a Career Development Fellowship from CINSW (11/CDF/3-05). JLS was supported by an NHMRC Biomedical Research scholarship, the CINSW Research Scholar Award, and the Judith Hyam Memorial Trust.
Footnotes
Author Contributions
JLS, AST and TMB designed the project and conceived hypotheses; AST performed the majority of the experiments, with other experiments contributed by MK and JLS; AS and TdL contributed experiments related to Pot1ΔOB; all authors analyzed data and interpreted experimental results; AST and TMB wrote the manuscript, with input from all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol. 2010;30:2971–2982. doi: 10.1128/MCB.00240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancelin K, Brunori M, Bauwens S, Koering CE, Brun C, Ricoul M, Pommier JP, Sabatier L, Gilson E. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol Cell Biol. 2002;22:3474–3487. doi: 10.1128/MCB.22.10.3474-3487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster BN, Linardic CM, Veldman T, Bansal NP, Downie DL, Counter CM. Rescue of an hTERT mutant defective in telomere elongation by fusion with hPot1. Mol Cell Biol. 2004;24:3552–3561. doi: 10.1128/MCB.24.8.3552-3561.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arneric M, Lingner J. Tel1 kinase and subtelomere-bound Tbf1 mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep. 2007;8:1080–1085. doi: 10.1038/sj.embor.7401082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Shore D. Increased association of telomerase with short telomeres in yeast. Genes Dev. 2007;21:1726–1730. doi: 10.1101/gad.438907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt-Compton B, Capper R, Rowson J, Baird DM. Short telomeres are preferentially elongated by telomerase in human cells. FEBS Lett. 2009;583:3076–3080. doi: 10.1016/j.febslet.2009.08.029. [DOI] [PubMed] [Google Scholar]
- Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- Chan SW, Chang J, Prescott J, Blackburn EH. Altering telomere structure allows telomerase to act in yeast lacking ATM kinases. Curr Biol. 2001;11:1240–1250. doi: 10.1016/s0960-9822(01)00391-8. [DOI] [PubMed] [Google Scholar]
- Chang YT, Moser BA, Nakamura TM. Fission yeast shelterin regulates DNA polymerases and Rad3ATR kinase to limit telomere extension. PLoS Genet. 2013;9:e1003936. doi: 10.1371/journal.pgen.1003936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churikov D, Corda Y, Luciano P, Geli V. Cdc13 at a crossroads of telomerase action. Front Oncol. 2013;3:39. doi: 10.3389/fonc.2013.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, Reddel RR. Protein composition of catalytically active human telomerase from immortal cells. Science. 2007;315:1850–1853. doi: 10.1126/science.1138596. [DOI] [PubMed] [Google Scholar]
- Cohen SB, Reddel RR. A sensitive direct human telomerase activity assay. Nat Methods. 2008;5:355–360. doi: 10.1038/nmeth.f.209. [DOI] [PubMed] [Google Scholar]
- Colgin LM, Baran K, Baumann P, Cech TR, Reddel RR. Human POT1 facilitates telomere elongation by telomerase. Curr Biol. 2003;13:942–946. doi: 10.1016/s0960-9822(03)00339-7. [DOI] [PubMed] [Google Scholar]
- Dalby AB, Hofr C, Cech TR. Contributions of the TEL-patch amino acid cluster on TPP1 to telomeric DNA synthesis by human telomerase. J Mol Biol. 2015;427:1291–1303. doi: 10.1016/j.jmb.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. Human telomeres are attached to the nuclear matrix. EMBO J. 1992;11:717–724. doi: 10.1002/j.1460-2075.1992.tb05104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Dehe PM, Rog O, Ferreira MG, Greenwood J, Cooper JP. Taz1 enforces cell-cycle regulation of telomere synthesis. Mol Cell. 2012;46:797–808. doi: 10.1016/j.molcel.2012.04.022. [DOI] [PubMed] [Google Scholar]
- Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- Feldser D, Strong MA, Greider CW. Ataxia telangiectasia mutated (Atm) is not required for telomerase-mediated elongation of short telomeres. Proc Natl Acad Sci U S A. 2006;103:2249–2251. doi: 10.1073/pnas.0511143103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokas E, Prevo R, Hammond EM, Brunner TB, McKenna WG, Muschel RJ. Targeting ATR in DNA damage response and cancer therapeutics. Cancer Treat Rev. 2014;40:109–117. doi: 10.1016/j.ctrv.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Fouche N, Ozgur S, Roy D, Griffith JD. Replication fork regression in repetitive DNAs. Nucleic Acids Res. 2006;34:6044–6050. doi: 10.1093/nar/gkl757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Deng Y, Lin Y, Cosme-Blanco W, Chan S, He H, Yuan G, Brown EJ, Chang S. Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J. 2007;26:4709–4719. doi: 10.1038/sj.emboj.7601893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Kartawinata M, Li J, Pickett HA, Teo J, Kilo T, Barbaro PM, Keating B, Chen Y, Tian L, et al. Inherited bone marrow failure associated with germline mutation of ACD, the gene encoding telomere protein TPP1. Blood. 2014;124:2767–2774. doi: 10.1182/blood-2014-08-596445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector RE, Shtofman RL, Ray A, Chen BR, Nyun T, Berkner KL, Runge KW. Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol Cell. 2007;27:851–858. doi: 10.1016/j.molcel.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Hemann MT, Strong MA, Hao LY, Greider CW. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the Ataxia-Telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- Ivessa AS, Zhou JQ, Schulz VP, Monson EK, Zakian VA. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Richard P, Bertrand E, Kiss T. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell. 2006;17:944–954. doi: 10.1091/mbc.E05-09-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurczyluk J, Nouwens AS, Holien JK, Adams TE, Lovrecz GO, Parker MW, Cohen SB, Bryan TM. Direct involvement of the TEN domain at the active site of human telomerase. Nucleic Acids Res. 2011;39:1774–1788. doi: 10.1093/nar/gkq1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, de Lange T. The telomeric protein TRF2 binds the ATM kinase and can inhibit the ATM-dependent DNA damage response. PLoS Biol. 2004;2:E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlseder J, Smogorzewska A, de Lange T. Senescence induced by altered telomere state, not telomere loss. Science. 2002;295:2446–2449. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
- Kelleher C, Kurth I, Lingner J. Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro. Mol Cell Biol. 2005;25:808–818. doi: 10.1128/MCB.25.2.808-818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Kishi S, Zhou XZ, Ziv Y, Khoo C, Hill DE, Shiloh Y, Lu KP. Telomeric protein Pin2/TRF1 as an important ATM target in response to double strand DNA breaks. J Biol Chem. 2001;276:29282–29291. doi: 10.1074/jbc.M011534200. [DOI] [PubMed] [Google Scholar]
- Kocak H, Ballew BJ, Bisht K, Eggebeen R, Hicks BD, Suman S, O’Neil A, Giri N, Maillard I, Alter BP, et al. Hoyeraal-Hreidarsson syndrome caused by a germline mutation in the TEL patch of the telomere protein TPP1. Genes Dev. 2014;28:2090–2102. doi: 10.1101/gad.248567.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Zaug AJ, Podell ER, Cech TR. Switching human telomerase on and off with hPOT1 protein in vitro. J Biol Chem. 2005;280:20449–20456. doi: 10.1074/jbc.M502212200. [DOI] [PubMed] [Google Scholar]
- Loayza D, de Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;424:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- Lovejoy CA, Cortez D. Common mechanisms of PIKK regulation. DNA Repair (Amst) 2009;8:1004–1008. doi: 10.1016/j.dnarep.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez P, Thanasoula M, Munoz P, Liao C, Tejera A, McNees C, Flores JM, Fernandez-Capetillo O, Tarsounas M, Blasco MA. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009;23:2060–2075. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKerlie MA, Lin S, Zhu XD. ATM regulates proteasome-dependent subnuclear localization of TRF1, which is important for telomere maintenance. Nucleic Acids Res. 2012;40:3975–3989. doi: 10.1093/nar/gks035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNees CJ, Tejera AM, Martinez P, Murga M, Mulero F, Fernandez-Capetillo O, Blasco MA. ATR suppresses telomere fragility and recombination but is dispensable for elongation of short telomeres by telomerase. J Cell Biol. 2010;188:639–652. doi: 10.1083/jcb.200908136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe JA, Parkhill J, Campbell L, Stacey M, Biggs P, Byrd PJ, Taylor AM. Accelerated telomere shortening in ataxia telangiectasia. Nat Genet. 1996;13:350–353. doi: 10.1038/ng0796-350. [DOI] [PubMed] [Google Scholar]
- Miller KM, Rog O, Promisel Cooper J. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- Moser BA, Chang YT, Kosti J, Nakamura TM. Tel1ATM and Rad3ATR kinases promote Ccq1-Est1 interaction to maintain telomeres in fission yeast. Nat Struct Mol Biol. 2011;18:1408–1413. doi: 10.1038/nsmb.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser BA, Subramanian L, Khair L, Chang YT, Nakamura TM. Fission yeast Tel1 and Rad3 promote telomere protection and telomerase recruitment. PLoS Genet. 2009;5:e1000622. doi: 10.1371/journal.pgen.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature. 2012;492:285–289. doi: 10.1038/nature11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki R, Ishikawa F. Telomere-bound TRF1 and TRF2 stall the replication fork at telomeric repeats. Nucleic Acids Res. 2004;32:1627–1637. doi: 10.1093/nar/gkh309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Bartocci C, Ouzounov I, Diedrich JK, Yates JR, III, Denchi EL. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature. 2013;494:502–505. doi: 10.1038/nature11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Pennarun G, Hoffschir F, Revaud D, Granotier C, Gauthier LR, Mailliet P, Biard DS, Boussin FD. ATR contributes to telomere maintenance in human cells. Nucleic Acids Res. 2010;38:2955–2963. doi: 10.1093/nar/gkp1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog O, Cooper JP. Telomeres in Drag: dressing as DNA damage to engage telomerase. Curr Opin Genet Dev. 2008;18:212–220. doi: 10.1016/j.gde.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Sabourin M, Tuzon CT, Zakian VA. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol Cell. 2007;27:550–561. doi: 10.1016/j.molcel.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton AN, Regalado SG, Lai CS, Cost GJ, O’Neil CM, Urnov FD, Gregory PD, Jaenisch R, Collins K, Hockemeyer D. Genetic and molecular identification of three human TPP1 functions in telomerase action: recruitment, activation, and homeostasis set point regulation. Genes Dev. 2014;28:1885–1899. doi: 10.1101/gad.246819.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- Smilenov LB, Morgan SE, Mellado W, Sawant SG, Kastan MB, Pandita TK. Influence of ATM function on telomere metabolism. Oncogene. 1997;15:2659–2665. doi: 10.1038/sj.onc.1201449. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. Control of human telomere length by TRF1 and TRF2. Mol Cell Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprung CN, Bryan TM, Reddel RR, Murnane JP. Normal telomere maintenance in immortal ataxia telangiectasia cell lines. Mutat Res. 1997;379:177–184. doi: 10.1016/s0027-5107(97)00119-x. [DOI] [PubMed] [Google Scholar]
- Stern JL, Bryan TM. Telomerase recruitment to telomeres. Cytogenet Genome Res. 2008;122:243–254. doi: 10.1159/000167810. [DOI] [PubMed] [Google Scholar]
- Stern JL, Zyner KG, Pickett HA, Cohen SB, Bryan TM. Telomerase recruitment requires both TCAB1 and Cajal bodies independently. Mol Cell Biol. 2012;32:2384–2395. doi: 10.1128/MCB.00379-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, O’Driscoll M, Jeggo PA. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 2006;25:5775–5782. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai KK, Hooper SM, Blackwood SL, Gandhi R, de Lange T. In vivo stoichiometry of shelterin components. J Biol Chem. 2010;285:1457–1467. doi: 10.1074/jbc.M109.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- Tomlinson CG, Moye AL, Holien JK, Parker MW, Cohen SB, Bryan TM. Two-step mechanism involving active-site conformational changes regulates human telomerase DNA binding. Biochem J. 2015;465:347–357. doi: 10.1042/BJ20140922. [DOI] [PubMed] [Google Scholar]
- Tomlinson RL, Abreu EB, Ziegler T, Ly H, Counter CM, Terns RM, Terns MP. Telomerase reverse transcriptase is required for the localization of telomerase RNA to Cajal bodies and telomerese in human cancer cells. Mol Biol Cell. 2008;19:3793–3800. doi: 10.1091/mbc.E08-02-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell. 2006;17:955–965. doi: 10.1091/mbc.E05-09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- Veldman T, Etheridge KT, Counter CM. Loss of hPot1 function leads to telomere instability and a cut-like phenotype. Curr Biol. 2004;14:2264–2270. doi: 10.1016/j.cub.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- Wenz C, Enenkel B, Amacker M, Kelleher C, Damm K, Lingner J. Human telomerase contains two cooperating telomerase RNA molecules. EMBO J. 2001;20:3526–3534. doi: 10.1093/emboj/20.13.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KK, Maser RS, Bachoo RM, Menon J, Carrasco DR, Gu Y, Alt FW, DePinho RA. Telomere dysfunction and Atm deficiency compromises organ homeostasis and accelerates ageing. Nature. 2003;421:643–648. doi: 10.1038/nature01385. [DOI] [PubMed] [Google Scholar]
- Wu Y, Xiao S, Zhu XD. MRE11-RAD50-NBS1 and ATM function as comediators of TRF1 in telomere length control. Nat Struct Mol Biol. 2007;14:832–840. doi: 10.1038/nsmb1286. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Tarumoto Y, Ishikawa F. Tel1ATM and Rad3ATR phosphorylate the telomere protein Ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev. 2012;26:241–246. doi: 10.1101/gad.177873.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci U S A. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong FL, Batista LF, Freund A, Pech MF, Venteicher AS, Artandi SE. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell. 2012;150:481–494. doi: 10.1016/j.cell.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, Kibe T, Kabir S, de Lange T. TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev. 2014;28:2477–2491. doi: 10.1101/gad.251611.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.