Abstract
The ability of cells to adhere and sense differences in tissue stiffness is crucial for organ development and function. The central mechanisms by which adherent cells detect extracellular matrix compliance, however, are still unknown. Using two single-molecule–calibrated biosensors that allow the analysis of a previously inaccessible but physiologically highly relevant force regime in cells, we demonstrate that the integrin activator talin establishes mechanical linkages upon cell adhesion, which are indispensable for cells to probe tissue stiffness. Talin linkages are exposed to a range of piconewton (pN) forces and bear, on average, 7–10 pN during cell adhesion depending on their association with f-actin and vinculin. Disruption of talin’s mechanical engagement does not impair integrin activation and initial cell adhesion but prevents focal adhesion reinforcement and thus extracellular rigidity sensing. Intriguingly, talin mechanics are isoform-specific so that expression of either talin-1 or talin-2 modulates extracellular rigidity sensing.
Introduction
Tissue rigidity is an epigenetic factor that governs tissue patterning and organ development1–3, while altered tissue mechanics is associated with numerous disease states including cardiovascular disorders, spinal cord injury or tumour formation4, 5. To distinguish differences in tissue stiffness, cells constantly probe the mechanical properties of their environment by anchoring and pulling on the surrounding extracellular matrix (ECM)6–8. This anchorage-dependent rigidity sensing is mediated by focal adhesions (FAs), subcellular structures in which ECM-binding integrin receptors are connected through adaptor proteins with the intracellular actin cytoskeleton9, 10. Although the important role of individual integrin subunits and distinct FA molecules such as focal adhesion kinase (FAK), paxillin or vinculin has been appreciated7, 11, 12, the central mechanism that couples cell adhesion with mechanosensing remained unknown.
Among the implicated regulators of FA mechanosensing are talins, primarily known for their essential role during integrin activation13. Talins directly bind and thereby activate integrin receptors with an N-terminal head-domain and are thought to transduce mechanical information by simultaneously connecting to the actin cytoskeleton with their C-terminal rod-domain14–16. Due to the lack of suitable techniques to measure subcellular talin forces, however, quantitative evidence for mechanical tension across talin in cells was missing. We therefore embarked on the development of biosensors to examine the piconewton (pN) mechanics of talin linkages in living cells.
Results
Single-molecule calibration of two genetically encoded tension sensors
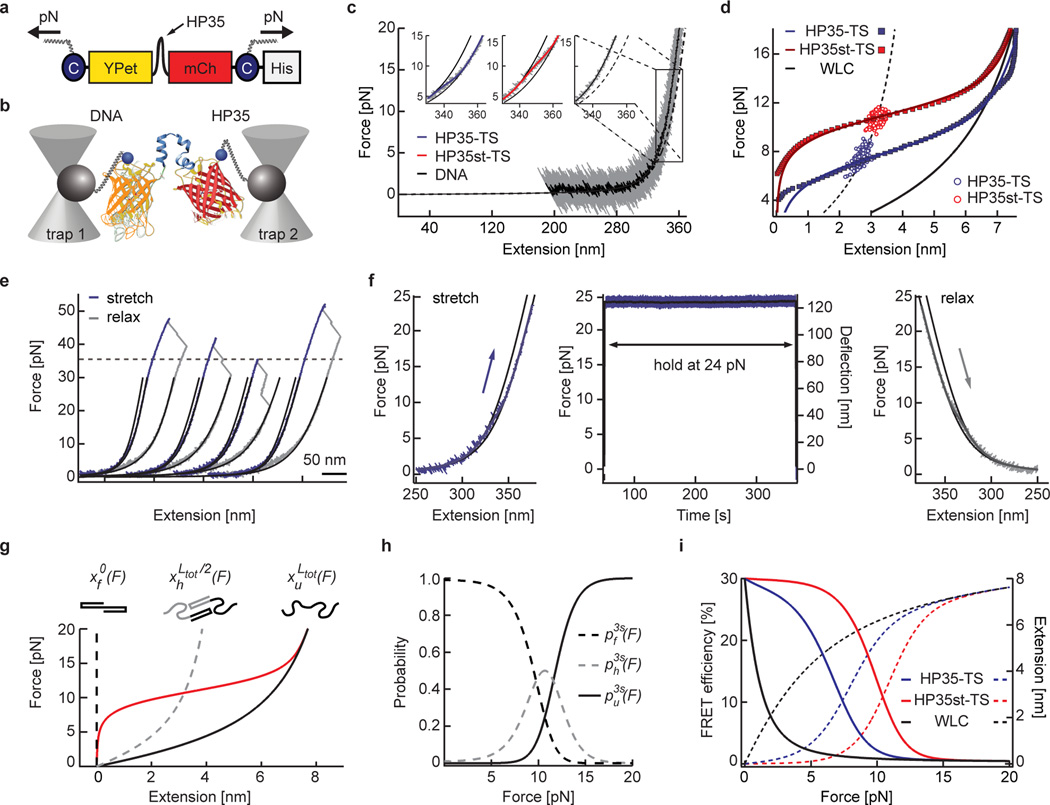
We have previously generated a probe (called TSMod), in which an elastic peptide is flanked by two fluorophores allowing the measurement of molecular forces between 1–6 pN using Förster resonance energy transfer (FRET)12, 17–19. Yet individual myosin motors can generate single pN forces20 and forces across distinct integrin receptors were recently shown to be significantly higher21, 22. This suggests that the proteins which directly connect adhesion receptors with actomyosin networks such as talin may experience higher mechanical forces as well. We therefore engineered two tension sensors using the 35 amino acid-long villin headpiece peptide (HP35) as a force-sensitive element flanked by an YPet/mCherry pair of fluorophores (Fig. 1a). HP35 is an ultrafast-folding peptide that undergoes an equilibrium unfolding/folding transition in response to mechanical forces of about 7 pN, whereas a stable HP35 mutant (HP35st) undergoes this transition at about 10 pN23, 24. To test whether HP35 unfolding/folding dynamics are affected by the presence of N- and C-terminally–fused fluorophores, we performed single-molecule calibrations using a custom-built optical tweezer setup (Fig. 1b, Supplementary Note and Online Methods). As expected, the average equilibrium transition mid-forces were at 7.4 pN (HP35-TS) and 10.6 pN (HP35st-TS), and both sensors quickly recovered their original conformation when forces were released (Fig. 1c, d and Supplementary Fig. 1a–e). Importantly, unfolding of fluorophores was not observed below 35 pN (Fig. 1e) and also did not occur when constructs were trapped at 24 pN for more than five minutes (Fig. 1f). The force-extension data of HP35-TS and HP35st-TS were well-fitted by a three-state model assuming HP35(st) to be either in a folded, half-folded/half-unfolded or unfolded state (Fig. 1g, Supplementary Note and Supplementary Fig. 1c, f–h). The resulting probabilities for HP35(st) to be in any of these conformations at a given force were used to calculate the biosensors’ force-FRET responses revealing highest sensitivity between 6–8 pN and 9–11 pN (Fig. 1i). Thus, HP35-TS and HP35st-TS are efficiently folding, rapidly responding and reversibly switching tension sensors with response thresholds at about 7 pN and 10 pN.
Figure 1.
Biosensor calibration using single-molecule force spectroscopy. (a) HP35-TS comprises two fluorophores, YPet and mCherry (mCh), which are linked by the villin headpiece peptide (HP35). Mechanical force across this biosensor leads to HP35 unfolding, increase in fluorophore separation distance and reduced FRET. For single-molecule (sm) calibration, DNA handles were attached using cysteines (C), a His-tag was used for purification. (b) Schematic illustration of the custom-built dual trap optical tweezer setup used for calibration. (c) 200 kHz resolution force-extension trace (FE) (gray) fitted with an extensible worm-like chain model (black). Inset: Zoom into representative FEs of individual HP35(st)-TS molecules as compared to DNA; the fit to HP35-TS data is shown in blue, HP35-st-TS in red and DNA in black. (d) Average FE of individual HP35-TS (blue) and HP35st-TS (red) molecules. Experimental data are shown as filled squares, solid lines are fits to the data, empty circles represent transition mid-point forces (HP35-TS: n=344 single pulls pooled from 15 independent repeats, i.e. different molecules; HP35st-TS: n=338 single pulls pooled from 10 independent repeats). (e) FE of four representative HP35-TS molecules showing fluorophore unfolding at high (>35 pN) forces. (f) After stretching to 24 pN, HP35-TS was exposed to high force for more than five min before relaxation; no indications of fluorophore unfolding were observed. (g) Average force-extension fit for HP35st-TS using a three-state model. The dashed black line represents the folded state, the gray dashed line the half-folded/half-unfolded state with contour length and the black solid line the completely unfolded state with contour length ; the red line indicates the average protein extension (h) Probability plot for the folded, half-folded/half-unfolded and unfolded state. (i) Modelled FRET-force (solid lines) and extension-force (dashed lines) correlations of HP35(st)-TS.
Talin tension sensor evaluation
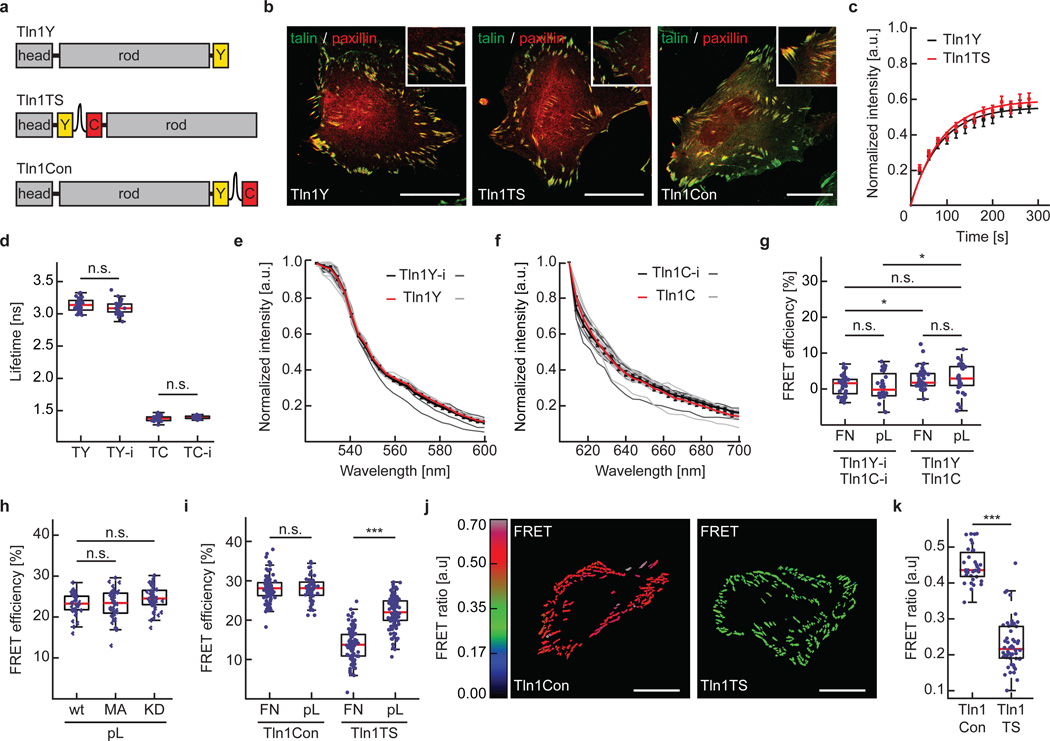
To examine talin forces in cells, we genetically inserted HP35-TS into the unstructured linker region connecting the head- and rod-domain of mouse talin-1 (Tln1TS)13. In parallel, we generated a C-terminally YPet-tagged control to test for talin function (Tln1Y), a C-terminally HP35-TS-tagged talin-1 to determine force-independent effects (Tln1Con; Fig. 2a) and intermolecular FRET controls in which the individual fluorophores were inserted into talin-1 (Tln1Y-i, Tln1C-i) or C-terminally attached (Tln1Y, Tln1C). Stable expression of these constructs in cells lacking talin-1 and talin-2 (Tln1−/−Tln2−/−) revealed proper subcellular localization as well as rescue of the severe cell adhesion and FAK activation defects of talin-deficient cells25 (Fig. 2b and Supplementary Fig. 2a, b). Furthermore, fluorescence recovery after photobleaching (FRAP) experiments demonstrated normal FA turnover rates of Tln1TS as compared to Tln1Y (Fig. 2c), together indicating that insertion of HP35-TS does not impair talin function. Next, we confirmed that the fluorescence lifetimes and the emission spectra of donor and acceptor fluorophores were unaffected by the insertion into talin-1 (Fig. 2d–f), and we quantified effects of intermolecular FRET (Fig. 2g), talin conformation (Fig. 2h), fluorescence intensity and temperature (Supplementary Fig. 2c, d) but found these confounding factors to be negligible in our experiments (see methods section for more detailed information). However, live cell FRET analysis by time correlated single-photon counting fluorescence lifetime microscopy (TCSPC-FLIM) or ratiometric imaging revealed an integrin-dependent reduction of energy transfer rates in Tln1TS cells indicating mechanical tension across talin-1 (Fig. 2i–k).
Figure 2.
Generation and evaluation of a talin-1 tension sensor. (a) Schematic illustrations of C-terminally YPet-tagged talin1 (Tln1Y), the talin1-HP35 tension sensor (Tln1TS) and the talin-1 zero force control (Tln1Con). (b) Representative images from 4 independent experiments showing Tln1−/−Tln2−/− cells expressing Tln1Y, Tln1TS or Tln1Con. Talin constructs are shown in green, paxillin in red; scale bars, 20 µm. (c) FRAP analyses demonstrating normal FA turnover rates of Tln1Y (black) and Tln1TS (red). Error bars indicate s.e.m; n=21 and 24 cells respectively for Tln1Y and Tln1TS, pooled from 5 independent experiments. (d) Live cell fluorescence lifetimes of internally and C-terminally tagged talin-1 constructs expressed in Tln1−/−Tln2−/− cells (n=29, 28, 30, 23 cells respectively from left to right, 3 independent experiments). (e) Live cell emission spectra of FA-localized Tln1Y-i (single measurements: dark gray lines, mean: black line) and Tln1Y (single measurements: light gray lines, mean: red line). Error bars indicate s.e.m; n=10 and 10 cells, 3 independent experiments. (f) Live cell emission spectra of FA-localized Tln1C-i (single measurements: dark gray lines, mean: black line) and Tln1C (single measurements: light gray lines, mean: red line). Error bars indicate s.e.m; n=10 (Tln1C-i) and 10 (Tln1C) cells, 3 independent experiments. (g) Intermolecular FRET analysis in Tln1−/−Tln2−/− cells co-expressing Tln1C-i/Tln1Y-i or Tln1C/Tln1Y on FN- or pL-coated glass coverslips (n=35, 28, 44, 36 cells respectively from left to right; 3 independent experiments). (h) No FRET efficiency differences in cells expressing Tln1TS (wt), Tln1TS-M319A (MA) and Tln1TS-K324D (KD) when seeded on pL-coated glass coverslips (n=39, 40, 42 cells respectively from left to right; 3 independent experiments) (i) Live-cell FLIM analysis demonstrating decreased FRET efficiency in Tln1TS cells when seeded on FN-coated surfaces indicating tension across talin-1 (n=35, 56, 102, 115 cells respectively from left to right; 5 independent experiments). (j) Representative ratiometric FA-FRET images of non-motile Tln1Con and Tln1TS cells confirming reduced FRET in Tln1TS cells; scale bars, 20 µm (3 independent experiments). (k) Mean ratiometric FA-FRET in Tln1Con and Tln1TS cells (n=32 and 47 cells respectively for Tln1Con and Tln1TS; 3 independent experiments). (d, g, h, i,:, Kolmogorov-Smirnov test, ***: p<0.001; **: p<0.01; *: p<0.05; not significant (n.s.): p > 0.05). Boxplots indicate the median (red line) as well as 25th and 75th percentiles; whiskers reach out to 2.7 standard deviations (σ). Statistic source data are available in Supplementary Table 1.
Talin-1 bears piconewton forces during cell adhesion
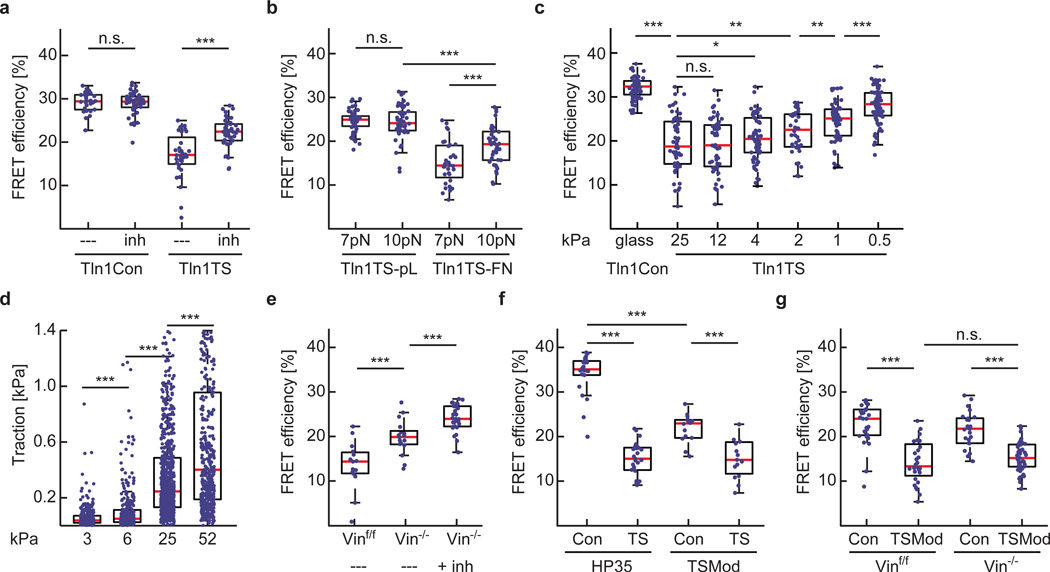
Next, we treated integrin-engaged Tln1TS and control cells with the Rock inhibitor Y-27632 to confirm that talin forces are actomyosin dependent. As expected, inhibitor treatment increased FRET efficiencies in Tln1TS cells but did not alter energy transfers in controls (Fig. 3a). Moreover, FRET efficiencies were specifically increased in integrin-engaged cells when the 9–11 pN-sensitive talin sensor (HP35st-TS) was used. Intriguingly, FRET was still lower than under control conditions indicating that a population of talin-1 molecules was subject to forces larger than 10 pN (Fig. 3b). To test whether talin establishes mechanical linkages also in softer environments, we analysed cells on matrices characterized by elastic moduli of 0.5–25 kPa. Tension across talin-1 was rather constant over a wide range of substrate rigidities and only gradually decreased on very soft substrates (Fig. 3c), while cells displayed the expected rigidity-dependent reduction in traction forces as described before11 (Fig. 3d). Thus, talin-1 mediates constitutive mechanical linkages, a significant subset of which experience forces of more than 7 pN and some even more than 10 pN.
Figure 3.
Talin-1 mediates a constitutive mechanical linkage in FAs that is modulated by f-actin and vinculin association. (a) Treatment of cells with 10 µM Y-27632 (inh) induces an increase in average FRET efficiency specifically in Tln1TS cells (n=26, 42, 32 and 42 respectively from left to right; pooled from 3 independent experiments). (b) Comparing HP35-TS (7 pN) with HP35st-TS (10 pN)–based talin sensors in cells on FN- or pL-coated surfaces suggests that most talin-1 linkages experience force of more than 7 pN, some even more than 10 pN (n=52, 53, 32 and 40 cells respectively from left to right; 4 independent experiments). (c) FRET efficiencies in Tln1Con cells seeded on FN-coated glass coverslips (n=68) and in Tln1TS cells seeded on FN-coated 25 kPa (n=61), 12 kPa (n=58), 4 kPa (n=64), 2 kPa (n=40), 1 kPa (n=58) and 0.5 kPa (n=81) matrices; n represents the number of cells that were pooled from 5 independent experiments. (d) Rigidity-dependent traction force increase of Tln1Y cells seeded on FN-coated polyacrylamide gels with elastic moduli of 3.2 kPa (n=20), 6.3 kPa (n=16), 24.7 kPa (n=30) and 52 kPa (n=15); n represents the number of cells that were pooled from 3 independent experiments. Single data points represent traction forces from displacement of every hundredth bead. (e) Depletion of vinculin leads to an increase in FRET while treatment of Tln1TS-expressing Vin−/− cells with 10 µM Y-27632 further increases transfer rates indicating loss of vinculin leads to a reduction but not entire loss of talin tension (n=15, 17 and 29 cells respectively from left to right, pooled from 7 independent experiments). (f) The HP35-based sensor monitors talin-1 tension more efficiently than a biosensor using TSMod (n=22, 25, 14 and 15 cells respectively from left to right, 5 independent experiments). (g) TSMod does not properly resolve vinculin-dependent differences in talin-1 tension (n=29, 28, 25 and 41 cells respectively from left to right, 3 independent experiments). (a–c and e–g, Kolmogorov-Smirnov test; d, Wilcoxon–Mann–Whitney test. ***: p<0.001; **: p<0.01; *: p<0.05; not significant (n.s.): p > 0.05). Boxplots indicate the median (red line) as well as 25th and 75th percentiles; whiskers reach out to 2.7 standard deviations (σ). Statistic source data are available in Supplementary Table 1.
High talin tension depends on association with mechanically engaged vinculin
Vinculin is an adaptor protein thought to regulate force transmission in FAs12, 26, 27. As the talin-rod domain comprises eleven vinculin binding sites (VBS)13, we examined talin tension by transiently expressing Tln1TS in vinculin-expressing (Vinf/f) or vinculin-deficient (Vin−/−) cells. These experiments suggested that talin-1 tension was decreased in the absence of vinculin, whereas tension was restored by stable re-expression of full length vinculin (Vwt) but not by a vinculin truncation mutant (Vmut) unable to transduce mechanical forces12, 27, 28 (Supplementary Fig. 2e). To confirm these results, we generated Vinf/f and Vin−/− cells stably expressing Tln1TS and again observed higher FRET efficiencies in vinculin-deficient cells which could be further increased by Y-27632 treatment indicating that forces across talin-1 were reduced but not entirely lost in the absence of vinculin (Fig. 3e). To investigate this in more detail, we generated a talin tension sensor using an YPet/mCherry-version of our previously published 1–6 pN-sensitive probe12 (TSMod) and first analysed it in Tln1−/−Tln2−/− cells. Consistent with our Tln1TS measurements, this construct also indicated tension across talin-1, even though FRET efficiency differences were smaller due to TSMod’s rather narrow dynamic range (Fig. 3f). In contrast to the HP35-probes, however, TSMod-based sensors indicated very similar FRET values in Vinf/f and Vin−/− cells demonstrating that talin-1 is still subject to low forces between 1–6 pN in the absence of vinculin (Fig. 3g). Together, these results provide direct evidence that mechanical tension across talin-1 is determined by its association with f-actin and vinculin. While talin’s f-actin engagement is sufficient to establish mechanically resilient linkages that bear low pN forces, vinculin-binding appears to promote higher tension states. The results also underline the significance of the HP35-sensors which detect higher pN forces that cannot be resolved by TSMod.
The mechanical engagement of the talin-rod domain is dispensable for integrin activation but critical for vinculin recruitment to domains R1–R3
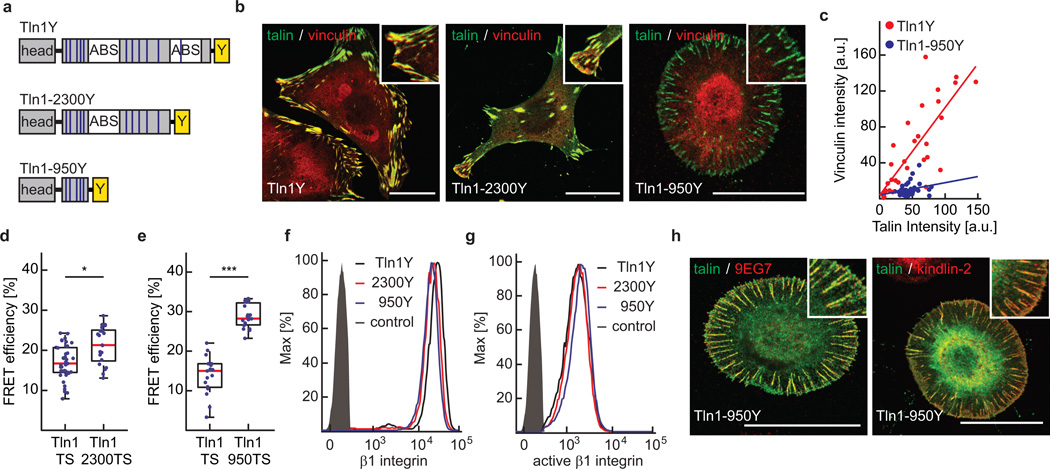
To elucidate whether vinculin recruitment to talin-1 depends on talin tension as proposed earlier15, 29, 30, we generated mutants in which the C-terminal actin-binding sites (ABS) and VBS of talin-1 were deleted to varying degrees and stably expressed them in Tln1−/−Tln2−/− cells (Fig. 4a). Removing talin’s dimerization domain as well as the entire C-terminal ABS (Tln1-2300) resulted in less efficient cell spreading and impaired formation of peripheral actin bundles but only slightly reduced forces across talin-1 and hardly influenced vinculin’s FA recruitment (Fig. 4b, d and Supplementary Fig. 3a). By contrast, deletion of the second ABS and six additional VBS (Tln1950) strongly impaired cell spreading and stress fibre formation. Tln1950 cells displayed normal surface levels of activated beta1 integrin and readily adhered to ECM substrates forming integrin-, talin-, and kindlin-2–positive adhesion sites (Fig. 4f–h). However, talin tension and vinculin recruitment were abolished even though Tln1950 still harboured five VBS in the talin-rod domains R1–R3 and vinculin was present at normal levels in the cytoplasm (Fig. 4b–e and Supplementary Fig. 3b, c). Together, the data indicate that vinculin association with talin-1’s N-terminal VBS requires preceding cytoskeletal engagement. This observation is consistent with a previously suspected positive feedback regulation of FA strengthening, where f-actin–dependent vinculin engagement promotes talin tension leading to further vinculin recruitment at R1–R3 and cell adhesion reinforcement15, 29–31.
Figure 4.
The talin-rod’s cytoskeletal engagement is essential for vinculin recruitment and talin tension but indispensable for integrin activation. (a) Schematic illustration of Tln1Y, Tln1-2300Y and Tln1950Y constructs; blue lines indicate VBS, white boxes ABS, yellow boxes the C-terminal YPet-tag. (b) Representative images from 3 independent experiments showing Tln1−/−Tln2−/− cells stably expressing Tln1Y, Tln1-2300Y and Tln1-950Y cells (green) stained with vinculin (red); scale bars, 20 µm. (c) Talin-1/vinculin FA co-localization analysis demonstrating the lack of vinculin recruitment to talin-positive adhesion sites in Tln1-950Y cells (n=32 (Tln1Y) and 33 (Tln1-950Y) FAs, pooled from 3 independent experiments). Pearson correlation coefficient (talin vs vinculin intensity): Tln1Y=0.8060, Tln1-950Y=0.2424. (d) Moderate reduction in talin-1 tension upon deletion of the dimerization domain and C-terminal ABS (Tln1-2300) (n=39 (Tln1TS) and 21 (Tln1-2300TS) cells, 4 independent experiments). (e) Loss of talin-1 tension in Tln1-950 cells (n=17 (Tln1TS) and 19 (Tln1-950TS), 4 independent experiments). (f, g) Representative FACS histograms of 4 independent experiments showing cells expressing Tln1Y (black), Tln1-2300Y (red) and Tln1950Y (blue) labelled for (f) beta1 integrin or (g) active beta1 integrin (9EG7); the negative control is shown in gray. Tln1950Y cells display normal integrin expression and activation. (h) Representative images from 3 independent experiments showing Tln1−/−Tln2−/− cells reconstituted with Tln1950Y (green) and labelled for active beta-1 integrin or kindlin-2 (red). The recruitment of kindlin-2 in Tln1950Y cells is consistent with normal integrin activation and cell adhesion; scale bars, 20 µm. (d, e, Kolmogorov-Smirnov test. ***: p<0.001; *: p<0.05). Boxplots indicate the median (red line) as well as 25th and 75th percentiles; whiskers reach out to 2.7 standard deviations (σ). Statistic source data are available in Supplementary Table 1.
Talin linkages are essential for extracellular rigidity sensing
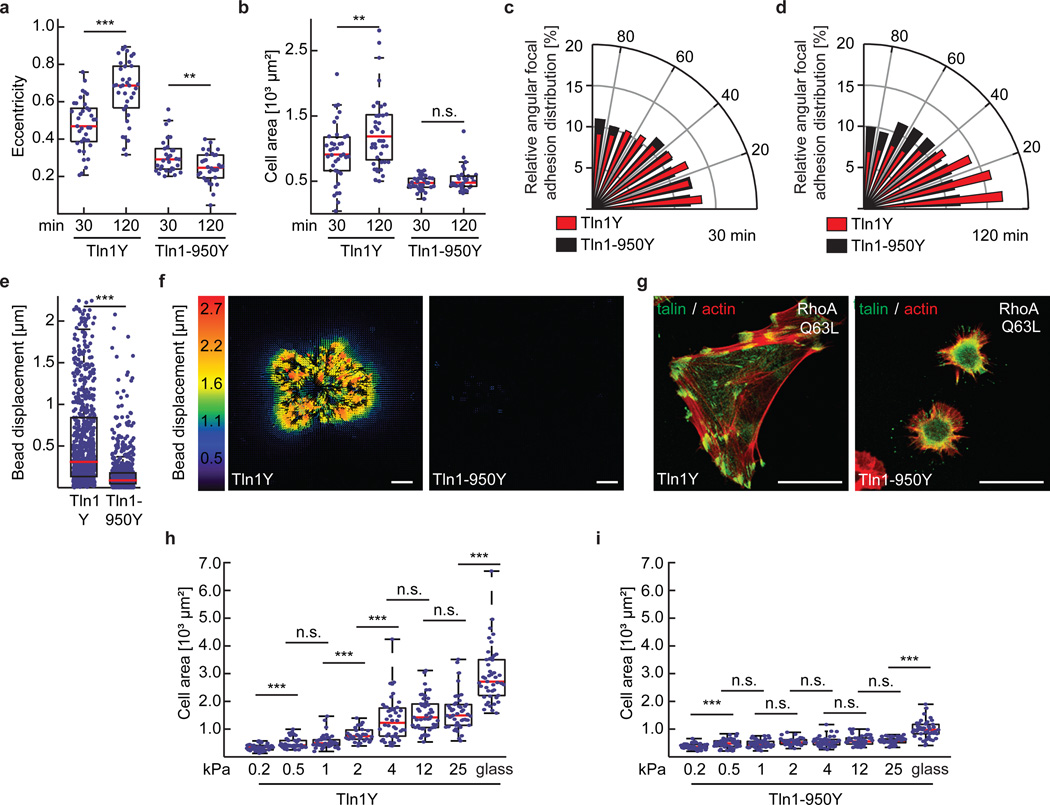
Throughout the experiments, we noticed that cell spreading and polarization, FA enlargement and the generation of traction forces – all processes that require mechanical stabilization of FAs – were strongly impaired in Tln1-950 cells (Fig. 5a–f). Moreover, Tln1950 cells failed to strengthen their FAs in response to increased intracellular contractile forces after expression of active RhoA (RhoA Q63L) (Fig. 5g). We therefore tested whether extracellular rigidity sensing, which appears to require FA strengthening9, is affected in cells lacking mechanically engaged talin-1. Indeed, Tln1950 cells seemed incapable of sensing and/or responding to different ECM rigidities while Tln1Y control cells reacted with the expected rigidity-dependent increase in cell spreading area (Fig. 5h, i). Thus, the mechanical engagement of the talin-rod domain with the actin cytoskeleton is indispensable for cell adhesion reinforcement and hence extracellular rigidity sensing.
Figure 5.
Cytoskeletal engagement of the talin-1 rod domain is indispensable for cell spreading, polarization, traction force generation and extracellular rigidity sensing. (a) Cellular eccentricity and (b) cell area of Tln1Y and Tln1-950Y cells after 30 min or 120 min of spreading on FN-coated glass coverslips; Tln1-950Y cells are unable to polarize and spread (in a and b: n=37, 36, 31 and 32 cells respectively from left to right, pooled from 3 independent experiments). (c, d) Relative angular FA distribution in Tln1Y cells (red) and Tln1950Y cells (black) after (c) 30 min and (d) 120 min of spreading on FN-coated glass coverslips indicating lack of polarization in Tln1950Y cells (in c and d: n=37 (Tln1Y 30 min), 36 (Tln1-950Y 30 min), 31 (Tln1Y 120 min) and 32 (Tln1-950Y 120 min) cells, 3 independent experiments). (e) Bead displacements observed under Tln1Y (n=21) and Tln1-950Y (n=22) cells cultured on 2 kPa polyacrylamide gels; data were pooled from 4 independent experiments. Tln1-950Y cells are characterized by significantly lower traction forces indicated by very small bead displacements. (f) Representative displacement images (4 independent experiments, corresponding quantification shown in e) of Tln1Y and Tln1-950Y cells indicating the virtual absence of traction forces in Tln1-950Y cells; scale bar, 10 µm. (g) Representative images from 3 independent experiments showing Tln1Y and Tln1-950Y cells (green) expressing active RhoAQ63L, stained for f-actin (red); Tln1-950Y cells fail to reinforce their FAs; scale bars, 20 µm. (h, i) Cell area of (h) Tln1Y and (i) Tln1-950Y cells after overnight culture on glass or FN-coated polyacrylamide gels with the indicated elastic moduli ranging from 0.2–25 kPa; Tln1Y (n=45, 45, 45, 32, 44, 47, 47 and 45 cells respectively from left to right, 3 independent experiments), Tln1-950Y (n=47, 49, 47, 32, 47, 46, 46 and 45 cells respectively from left to right, 3 independent experiments). Note that Tln1-950Y cells fail to distinguish rigidity differences. (a, b, h, i: two sided t-test; e, Wilcoxon–Mann–Whitney test. ***: p<0.001; **: p<0.01; *: p<0.05; not significant (n.s.): p > 0.05). Boxplots indicate the median (red line) as well as 25th and 75th percentiles; whiskers reach out to 2.7 standard deviations (σ). Statistic source data are available in Supplementary Table 1.
Integrin force transduction is talin isoform-specific
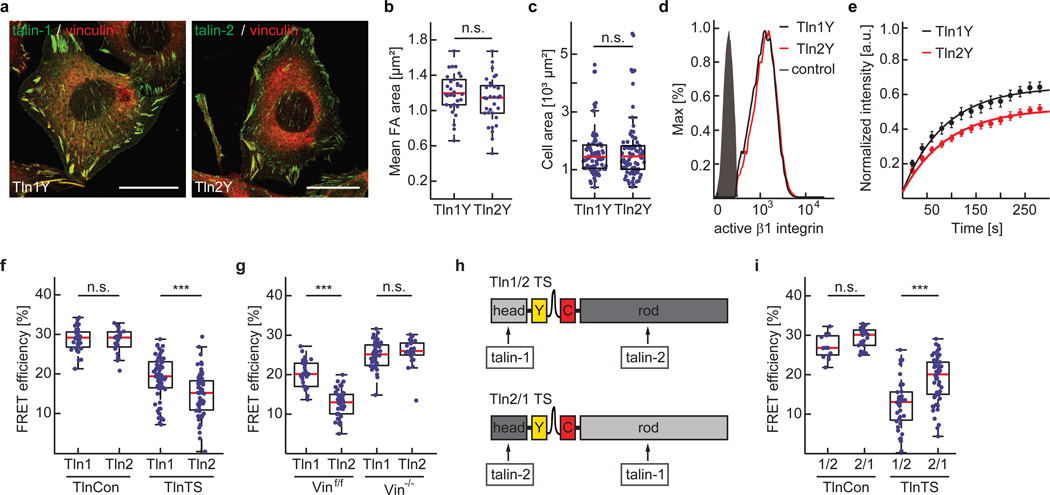
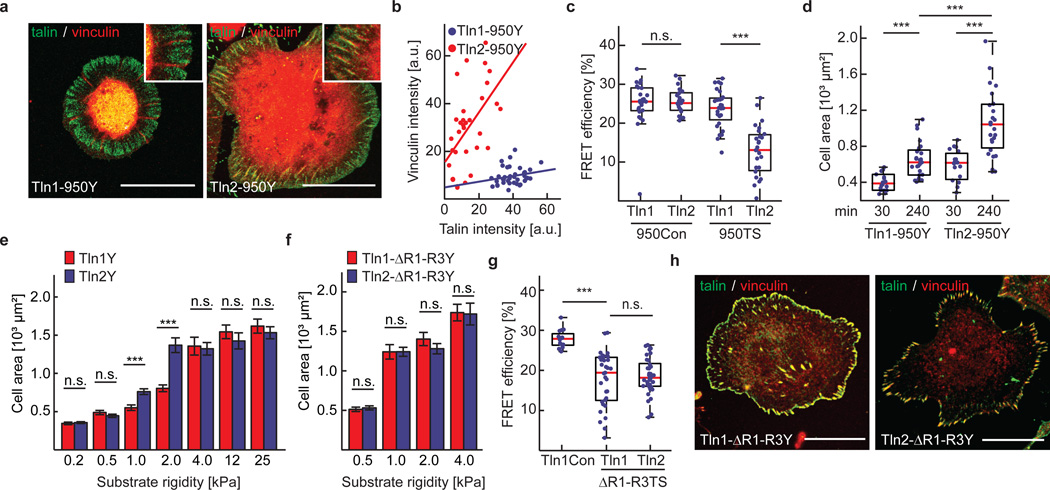
Mammals express two very similar talin isoforms, the ubiquitous talin-1 and the more restrictively expressed talin-2. It is unclear why certain tissues require the expression of a second talin isoform, as both proteins efficiently activate integrins and connect to the cytoskeleton13, 32. We therefore wanted to test whether mechanical forces may be differentially transduced by talin-1 and talin-2 and generated a whole set of human talin-1 and talin-2 expression constructs to stably reconstitute Tln1−/−Tln2−/− cells. While individual expression of the talin isoforms rescued the cell spreading, integrin activation and FAK phosphorylation phenotype of talin-deficient cells equally (Fig. 6a–d and Supplementary Fig. 4a–c), talin-2 was more immobilized in FAs (Fig. 6e and Supplementary Fig. 4d) and an increased number of talin-2 molecules were exposed to tension in integrin-engaged cells (Fig. 6f and Supplementary Fig. 4e–g). Elevated talin-2 tension levels were also observed in Vinf/f but not in Vin−/− cells emphasizing the important role of the talin-vinculin interaction for generating high talin forces (Fig. 6g). To confirm that differences in isoform-specific talin tension were mediated exclusively by the talin rod-domain, we engineered chimeric talin constructs, in which the C-terminal domains of talin-1 and talin-2 were exchanged (Fig. 6h). Stable expression of these constructs rescued the cell adhesion and cell spreading defect of talin-deficient cells, while FLIM experiments demonstrated that the tension increase was indeed entirely mediated by the rod-domain of talin-2 (Fig. 6i). Next, we generated a talin-2 truncation mutant lacking all C-terminal ABS but retaining the five VBS residing in R1–R3 (Tln2-950) analogous to the talin-1 construct described above. Remarkably, Tln2-950 FAs were vinculin-positive, exposed to mechanical tension and able to induce, albeit not completely rescue, cell spreading (Fig. 7a–d). Together, these results show that the two human talin isoforms bear mechanical forces differently; they also suggest that the f-actin-dependent vinculin recruitment to talin’s N-terminal rod-domain, specifically to domains R1–R3, is talin-1–specific.
Figure 6.
Talin-1 and talin-2 both rescue cell spreading and integrin activation but they transduce mechanical forces differently. (a) Representative images (3 independent experiments) of Tln1−/−Tln2−/− cells reconstituted with Tln1Y or Tln2Y (green) and labelled for vinculin (red). Cell lines are indistinguishable when cultured on plastic or glass coverslips; scale bars, 20 µm. (b) Mean FA area (n=30 (Tln1Y) and 34 (Tln2Y) cells; pooled from 4 independent experiments) and (c) mean cell area (n=77 (Tln1Y) and 77 (Tln2Y) cells; 4 independent experiments) determined from Tln1Y and Tln2Y cells seeded on FN-coated glass coverslips. (d) Representative FACS histogram of cells expressing Tln1Y (black) or Tln2Y (red) labelled for active beta1 integrin; the negative control is shown in gray. (4 independent experiments) (e) Normalized fluorescence recovery rates of Tln1Y (black, n=18 cells) and Tln2Y cells (red, n=17 cells) as determined by live cell FRAP experiments. Cells were pooled from 3 independent experiments; error bars indicate s.e.m. (f) FRET efficiencies in Tln1Con, Tln2Con, Tln1TS and Tln2TS cells (n=35, 25, 63 and 63 cells respectively from left to right, 3 independent experiments) indicating increased tension across talin-2. (g) Isoform-specific tension differences are abolished in vinculin-deficient cells (n=28, 41, 42 and 24 cells respectively from left to right; 3 independent experiments). (h) Schematic illustrations of chimeric talin-1–head/talin-2–rod (Tln1/2-TS) and talin-2–head/talin-1–rod (Tln2/1-TS) tension sensor constructs. (i) FRET analysis of chimeric talin constructs demonstrating that the isoform-specific tension increase is talin-rod–dependent (n=15, 24, 43 and 58 cells respectively from left to right; 7 independent experiments). (b, c two sited t-test; f, g, i, Kolmogorov-Smirnov test. ***: p<0.001; **: p<0.01; *: p<0.05; not significant (n.s.): p > 0.05). Boxplots indicate the median (red line) as well as 25th and 75th percentiles; whiskers reach out to 2.7 standard deviations (σ). Statistic source data are available in Supplementary Table 1.
Figure 7.
Talin-isoform–specific differences are mediated by domains R1–R3. (a) Representative images from 3 independent experiments showing that vinculin (red) colocalizes with Tln2-950Y but not with Tln1-950Y adhesion sites; scale bar, 20 µm. (b) Quantification of talin/vinculin co-localization in adhesion sites of Tln1-950Y (n=32 adhesions) and Tln2-950Y (n=38 adhesions) expressing cells; n values represent pooled adhesions from 3 independent experiments. Pearson correlation coefficient (talin vs vinculin intensity): Tln1-950Y=0.2214, Tln2-950Y=0.5323. (c) Deletion of C-terminal ABS abolishes tension across talin-1 but not across talin-2 (n=24, 21, 28 and 25 cells respectively from left to right; 5 independent experiments). (d) Cell area of Tln1-950Y and Tln2-950Y cells after 30 min and 240 min spreading on FN-coated glass coverslips (n=18, 27, 19 and 27 cells respectively from left to right; 3 independent experiments).(e) Cell area quantification of Tln1Y and Tln2Y cells seeded on FN-coated substrates of indicated stiffness; Tln2Y cells respond differently on 1 kPa and 2 kPa matrices (n=47, 49, 45, 46, 45, 48, 32, 30, 44, 47, 48, 48, 46 and 46 cells respectively from left to right; 3 independent experiments; data are means ± s.e.m.). (f) No differences in cell spreading of Tln1-ΔR1R3Y (n=50 (0.5 kPa), 50 (1 kPa), 53 (2 kPa) and 47 (4 kPa) cells) and Tln2-ΔR1R3Y cells (n= 52 (0.5 kPa), 47 (1 kPa), 47 (2 kPa) and 47 (4 kPa) cells). Cells were pooled from 3 independent experiments; data are means ± s.e.m. (g) FRET analysis of talin-deficient cells expressing Tln1-ΔR1R3TS and Tln2-ΔR1R3TS constructs seeded on FN-coated glass coverslips (n=16, 33 and 37 cells respectively from left to right; 3 independent experiments). (h) Representative images from 3 independent experiments showing Tln1-ΔR1R3Y and Tln2-ΔR1R3Y cells on FN-coated glass coverslips stained for vinculin; the talin signal is labelled in green, the vinculin signal is shown in red; scale bars, 20 µm. (c, g: Kolmogorov-Smirnov test; d–f: two sided t-test. ***: p<0.001; not significant (n.s.): p > 0.05). Boxplots indicate the median (red line) as well as 25th and 75th percentiles; whiskers reach out to 2.7 standard deviations (σ). Statistic source data are available in Supplementary Table 1.
Isoform–specific effects are mediated by the talin rod domains R1–R3
Since our data indicated that initial vinculin recruitment to talin-2 is independent of preceding force generation through f-actin association, we tested whether FA strengthening and cell spreading on low rigidity substrates, where actomyosin contractility is naturally reduced, is talin-isoform dependent. Indeed, talin-2 cells spread more efficiently than talin-1 cells on 1–2 kPa matrices but behaved like talin-1 expressing cells on very soft (0.2–0.5kPa) and more rigid (4–25 kPa) substrates (Fig. 7e). To test whether the observed isoform-specific effects are mediated by the talin rod domains R1–R3, we generated talin-1 and talin-2 expression constructs in which these domains were deleted (Tln1-ΔR1–R3). Indeed, Tln1−/−Tln2−/− cells expressing talin-1 or talin-2 ΔR1–R3 deletion constructs did not display differences in cell spreading on 1–2 kPa substrates and differences in talin tension were abolished (Fig. 7f, g). As vinculin was still efficiently recruited by the remaining VBS (Fig. 7h), we conclude that the observed talin isoform-specific effects are mediated by talin’s N-terminal rod domains R1–R3.
Discussion
Cells ability to efficiently sense their mechanical environment is critical for many developmental, homeostatic and pathological processes1–6. Yet the underlying molecular mechanisms have been difficult to elucidate because suitable methods to study force propagation across individual molecules in cells were missing. We therefore developed a biosensor that allows molecular force measurements at 1–6 pN and the technique has been widely used to determine forces across various proteins in living cells and even whole organisms12, 17, 18, 33–37. An obvious limitation of the method, however, has been its inability to resolve forces higher than 6 pN. In this work, we described the generation, single-molecule calibration and application of two biosensors that enable efficient measurements at 6–8 pN and 9–11 pN. The probes are characterized by sharp and very fast force-responses, they are reversible, benefit from an improved dynamic range and do not unravel until forces of about 35 pN are reached (Fig. 1).
Application of the biosensors revealed that the integrin activator talin establishes mechanical linkages during cell adhesion which are indeed subject to forces larger than 7 pN, while a small but significant fraction of talin molecules experiences forces of more than 10 pN. On the other hand, talin-tension dropped to relatively low levels in the absence of vinculin-binding and disappeared upon talin’s disengagement from the actin cytoskeleton (Fig. 2 and Fig. 3). Thus, talin bears a range of forces depending on the degree of its mechanical engagement and it should be worthwhile investigating how talin tension correlates with FA dynamics for instance during cell migration. Interestingly, the formation of force-bearing talin linkages occurred over a wide range of substrate rigidities and seemed inherently linked to the formation of enlarged FAs. Consistent with this observation, cells expressing a truncated talin mutant lacking all C-terminal f-actin-binding sites (Tln1-950) were unable to form large FAs; they failed to generate significant cellular traction forces and did not sense extracellular rigidity differences even though integrin activation was unaffected (Fig. 4 and Fig. 5). Thus, in addition to its crucial role as an integrin activator, talin is an indispensable mediator of integrin mechanosensing. This dual role distinguishes talin from kindlins, which are also critical for integrin activation but do not appear to mediate a direct connection to the actin cytoskeleton38.
Intriguingly, integrin-dependent mechanosensing is talin isoform-specific (Fig. 6). While our data are consistent with the previously hypothesized tension-dependent vinculin recruitment to talin-1’s N-terminal rod domain15, 29–31, this mechanism does not seem to exist for talin-2. Instead, vinculin recruitment occurs even in the absence of C-terminal f-actin binding, which coincides with increased talin-2 FA immobilization and elevated average tension levels. As a consequence, talin-2 expressing cells spread more efficiently on 1–2 kPa surfaces than their talin-1 counterparts and it is interesting to note that talin-2 is expressed highest in brain tissue which is characterized by similar rigidities32, 39. In our experiments, the observed mechanical differences can be ascribed to talin’s R1–R3 (Fig. 7) which seemingly contrasts a previous study that attributed isoform-specific differences to distinct integrin–talin head-domain affinities40. It has to be noted, however, that the Tln1−/−Tln2−/− cells used here do not express integrin β1D, a muscle-specific integrin isoform for which especially high affinities to talin-2 have been reported40. Thus, a study using muscle-specific cell types or genetically-modified cell lines expressing distinct integrin receptor subtypes is needed.
Collectively, our experiments reveal that talins mediate a mechanical linkage which is essential to couple cell adhesion with integrin mechanosensing and is thus required for cells to detect tissue stiffness. It appears that differential expression of talin isoforms provides a means by which cells adjust to different ECM rigidities and the fact that integrins and at least one talin isoform are abundantly expressed in all cell types indicates that the observed mechanical linkages are relevant in many tissues. As tissue rigidity changes during development or with the onset of numerous disease states1–4, 6, it will be important to investigate the role of the individual talin-isoforms during these processes in more detail. The biosensors described in this study will allow the isoform-specific analysis of talin force transduction in many cell types and should be useful for such studies. Furthermore, they will be valuable to investigate effects of other talin interaction partners, such as RIAM or FAK, on molecular force propagation in FAs. Finally, application of the HP35 probes to other force-transducing proteins should allow the detailed analysis of many different mechanobiological processes that are subject to higher single pN forces in cells and whole organisms.
Methods
Antibodies and Reagents
The following antibodies were used: anti-paxillin [clone 349] (BD Transduction Laboratories, 610051; IF 1:400), anti-talin-1 [C45F1] (Cell Signaling, 4021; WB 1:1000), anti-talin-1/2 [8d4] (Sigma, T3287; WB 1:2000), anti-talin-2 [68E7] (Abcam, ab105458; WB 1:2000), anti-vinculin (hVIN-1) (Sigma, V9131; WB 1:4000 and IF 1:200), anti-tubulin [DM1A] (Sigma, T9026; WB 1:3000), anti-FAK (Millipore, 06–543; WB 1:2000 and IF 1:200), anti-pY397-FAK (Life Technologies, 44–624; WB 1:1000), anti-kindlin2 (Millipore, MAB2617; IF 1:400), anti-GFP (Abcam, ab290; WB 1:2000), anti-β1 integrin (Chemicon, MAB1997; FACS 1:400 and IF 1:200), anti-β1 integrin [9EG7] (PharMingen, 550531; FACS 1:200 and IF 1:200), anti-mouse IgG–horseradish peroxidase (HRP) conjugate (Bio-Rad, 170–6516; WB 1:15000), anti-rabbit IgG–HRP conjugate (Bio-Rad, 170–6515; WB 1:15000), anti-mouse IgG–Alexa Fluor-405 (Life Technologies, A31553; IF 1:400), anti-mouse IgG–Alexa Fluor-647 (Life Technologies, A31571; IF 1:400), anti-rat IgG–Alexa Fluor-647 (Life Technologies, A21247; IF 1:400). The following reagents were used: Alexa Fluor-568 phalloidin (Life Technologies, A12380; IF 1:400), Alexa Fluor-647 phalloidin (Life Technologies, A22287; IF 1:400), poly-L Lysine (Sigma, P4707), puromycin (Sigma, P8833), Y27632 (Sigma, Y0503), fibronectin (Calbiochem, 341631). Micro-patterned substrates (CYTOO), soft substrate dishes: Softview Easy Coat 0.2, 0.5, 1.0, 2.0, 4.0, 12 and 25 kPa dishes (Matrigen Life Technologies).
Generation of HP35-TS cDNA expression constructs
HP35-TS cDNA constructs were generated according to our published protocols17. In brief, restriction sites were added to YPet (amino acids (aa) 1–228) (5’Xho/3’BamHI) and mCherry (5’BamHI/3’NotI) cDNA by polymerase chain reaction (PCR) and PCR products were combined in a pBluescript SK(+) vector. The sequence encoding for the HP35 linker peptide (LSDEDFKAVFGMTRSAFANLPLWKQQNLKKEKGLF) was inserted between fluorophores using annealed oligonucleotides with 5’BglII/3’BamHI overhangs (forward primer: 5’aat tca gat ctC TCT CCG ATG AGG ACT TCA AAG CTG TGT TTG GCA TGA CCA GGA GCG CAT TTG CCA ACC TTC CTC TGT GGA AAC AGC AAC ACC TGA AGA AGG AAA AGG GAC TGT TCg3’; reverse primer: 5’-gat ccG AAC AGT CCC TTT TCC TTC TTC AGG TGT TGC TGT TTC CAC AGA GGA AGG TTG GCA AAT GCG CTC CTG GTC ATG CCA AAC ACA GCT TTG AAG TCC TCA TCG GAG AGa gat ctg-3’). The TS module containing the stable HP35st linker peptide (LSDEDFKAVFGMTRSAFANLPLWKQQALMKEKGLF) was created using the QuikChange II Site-Directed Mutagenesis kit (Agilent Technologies). For expression in cells, HP35(st)-TS cDNA was transferred into the pLPCX plasmid (Clontech) containing a modified multiple cloning site (pLPCXmod). To modify HP35(st)-TS for single-molecule calibration (smHP35-TS and smstHP35-TS), terminal cysteine residues to allow attachment of DNA oligonucleotides and a histidine-tag for protein purification were added by PCR; the cDNA was then transferred into pLPCXmod. The correct sequence of all constructs was confirmed by DNA sequencing (Eurofins Genomics).
Protein expression and purification
For protein expression, smHP35-TS or smHP35stab-TS were transiently transfected into HEK293 cells by CaPO4-precipitation as described before17. After 48 h, cells were detached, re-suspended in hypotonic lysis buffer, incubated for 20 min on ice and then homogenized with a Dounce homogenizer. Lysates were cleared by centrifugation and subjected to metal ion affinity chromatography (His-Trap, GE Healthcare), followed by ion-exchange chromatography (Sephadex, GE Healthcare). Purified samples were then concentrated to about 20 µM by membrane ultrafiltration (Vivaspin, GE Healthcare) and subjected to the protein-oligonucleotide binding reaction described below.
Assembling protein-DNA chimeras
To attach DNA handles to smHP35-TS or smHP35st-TS, cysteine-based chemistry was used according to previously published protocols41, 42. In brief, 34 base pair–long, lyophilized maleimide-modified single-stranded (ss) DNA oligonucleotides were dissolved in phosphate buffered saline (PBS, pH 6.7) and incubated overnight at 4 °C with the purified protein; the protein was kept in PBS supplemented with 0.2 mM Tris-2-carboxyethylphosphine (TCEP) to avoid oligomerization via disulfide bonds. Byproducts, namely unreacted oligonucleotides, unreacted protein as well as protein with only one attached DNA handle, were removed using metal ion affinity chromatography (His-Trap, GE Healthcare) in combination with size-exclusion chromatography (Superdex 200, GE Healthcare). Finally, double stranded (ds) DNA (λ-DNA, NEB) handles carrying a biotin- or a digoxigenin-modification at one end and a ss-overhang at the other were hybridized to complementary ssDNA oligonucleotides at either end of the protein resulting in a contour length of 185 nm for each handle.
Optical tweezers setup, sample preparation and measurement procedure
To calibrate the new HP35-TS biosensors, we performed single-molecule force spectroscopy measurements using a custom-built, dual-trap optical tweezers setup with back-focal plane detection as described previously43; for improved temporal resolution, quadrant photodiodes were used as position sensitive devices (QP154-Q-HVSD, Pacific Silicon Sensor). Data on the beads’ positions with respect to their trap centres as well as the distance between the two traps were sampled at 200 kHz and filtered at the Nyquist frequency. Each trap’s signal was corrected for cross-talk; trap stiffnesses calculated from corrected power spectra were determined to be about 0.37 pN/nm, while the error of the trap stiffness calibration is approximately 10 %. To prepare the sample, streptavidin-coated 1 µm-sized silica beads (Bangs Laboratories) were incubated with protein-DNA chimeras in PBS (pH 7.4). Next, functionalized anti-digoxigenin silica beads were added; glucose oxidase and catalase were used as an oxygen scavenger system as previously described41, 42. To obtain the dumbbell-like configuration schematically shown in Fig. 1b, one anti-digoxigenin and one streptavidin-functionalized bead were trapped, each in one of the two laser foci of the dual-trap. By moving the laser beam of one steerable laser focus, both beads were brought in close proximity until a single tether was successfully formed. Subsequently, repeated stretch and relax cycles were performed at constant velocities of 10–500 nm/s. Each cycle yielded typical force-extension traces where protein unfolding and refolding could be observed. Keeping the traps at constant distance allowed to record time traces of protein fluctuations at constant force bias; maximal forces of about 50 pN were reached. A step-by-step protocol describing the biosensor calibration can be found at Nature Protocol Exchange [reference required]
HP35-TS and HP35st-TS force–extension relation
To generate force-extension calibration curves (Fig. 1i, dashed lines), we converted our force-distance measurements and complete fits to the data into corresponding force-extension curves (Supplementary Note). The gain in extension caused by the dsDNA handles was subtracted using the parameters supplied by the extensible worm-like chain fit (eWLC-fit; see Supplementary Note) so that the remaining force-extension relation is characteristic for HP35(st)-TS and the thermal motion of the system. By averaging multiple smoothed force-extension traces of one single molecule, sub-nanometer resolution was obtained (Fig. 1d, squares).
Evaluating the mechanical stability of genetically-encoded fluorophores
Fluorophore stability is critical to any FRET-based force sensor as unfolding of donor or acceptor fluorophore would prohibit quantitative measurements. We therefore set up experiments to specifically examine HP35-TS fluorophore stability under force. As expected, fluorophore unfolding occurred only if at least 35 pN were reached/exceeded during the pulling measurements (Fig. 1e). To examine fluorophore stability under constant force, we exposed HP35-TS to 24 pN for more than 5 min but also did not observe any fluorophore unfolding (Fig. 1f). We also note that the observed average contour length of 363 nm was actually less than the expected theoretical 370 nm (Supplementary Note) indicating that both fluorophores are properly folded in HP35-TS. Altogether, these experiments suggest that the employed fluorophores in HP35-TS are insensitive to the mechanical forces the biosensors are supposed to measure.
Generation of talin expression constructs
Talin-1 expression constructs are based on murine talin-1 cDNA (accession number: X56123). To generate C-terminal fusion constructs (Tln1Y, Tln1C and Tln1Con), restriction sites (5’EcoRI/3’NotI) along with 5’ Kozak-sequence (ACC ATG) were added and the 3’ stop codon was removed by PCR. In parallel, restriction sites (5’NotI/3’ClaI) and stop codon were added to YPet, mCherry or HP35-TS cDNA by PCR and these fragments were combined with talin cDNA using NotI/ClaI restriction sites. For transient or stable expression in cells, constructs were cloned into pLPCXmod. To insert individual fluorophores (Tln1Y-i, Tln1C-i) or tension sensors into talin-1, we generated a short linker encoding for 5’SalI/3’NotI restriction sites after the codon corresponding to aa 447 in murine talin-1 cDNA by overlap extension PCR; cDNAs of the individual fluorophores or the HP35-TS were inserted using 5’XhoI/3’NotI restriction sites. Point mutations (M319A, K324D) were introduced into talin-1 cDNA using the QuikChange II Site-Directed Mutagenesis kit (Agilent Technologies); deletion mutants (Tln1-2300 and Tln1950) were generated by PCR amplification of the respective cDNA fragments. As we only had access to human talin-2 cDNA, we generated expression constructs based on human talin-1 (BC042923) and human talin-2 (NM015059) to allow a direct comparison of both isoforms. Based on aa homology, HP35-TS(st) was inserted into talin-2 after the codon corresponding to aa 450. In the chimeric talin-1/2 constructs, the talin-1 head-domain (aa 1–447) was fused to the talin-2 rod-domain (aa 451–2542); in the talin-2/1 construct, the talin-2 head-domain (aa 1–450) was fused to the talin-1 rod-domain (aa 448–2541). To delete domains R1–R3, sequences corresponding to aa 482–911 for talin-1 and aa 485–914 for talin-2 were removed.
Generation of vinculin and RhoA cDNA expression constructs
To generate vinculin expression constructs, human full-length vinculin cDNA was isolated from an ImaGene cDNA library (BG284191) and 5’ApaI/3’XbaI restriction sites as well as Kozak-sequence were added by PCR; also the vinculin truncation mutant (encoding for aa 1–883) was amplified by PCR. A TagBFP-fluorophore was added C-terminally to each construct using 5’XhoI/3’NotI (vinculin truncation mutant) or 5’XbaI/3’NotI (full length vinculin) restriction sites, and the final cDNAs were transferred into a pLPCXmod by 5’ApaI/3’NotI. The active RhoA expression construct was based on a previously described EGFP-tagged RhoQ63L cDNA44. To allow fluorescence analysis in the presence of YPet-tagged talin-1 constructs, the N-terminal EGFP was exchanged for TagBFP using 5’HindIII/3’EcoRI restriction sites.
Generation of talin- and vinculin-deficient cell lines and stable protein re-expression
To generate cells in which both talin-1 and talin-2 are genetically inactivated, talin-2 knockout mice (Tln2−/−)45 were intercrossed with mice in which the talin-1 gene is flanked with loxP sites (Tln1f/f)46. Mice with the Tln1f/fTln2−/− genotype were used to isolate fibroblastoid cells from kidneys of a three week-old mouse and cells were subsequently immortalized with the SV40 large T antigen. Talin1 was abrogated by adenoviral transduction of Cre recombinase and clonal cell lines were isolated. To generate vinculin-deficient cells, SV40 large T immortalized fibroblast, in which the vinculin gene is flanked with loxP sites (Vinf/f), were transduced with Cre recombinase and clonal cell lines were isolated (Vin−/−). Vinculin cDNA constructs were transiently transduced using Lipofectamine 2000 (Invitrogen) and talin double knockout cells (Tln1−/−Tln2−/−) were stably transduced by the phoenix cell transfection system as described earlier17. After infection, cells were puromycin-selected and stable protein expression was confirmed by western blotting using standard protocols. When necessary, cells were sorted by fluorescence-activated cell sorting (FACS) using a FACSAria cell sorter™ IIu (BD Biosciences) to isolate cells with comparable expression levels. Cell lines were freshly generated for this work and thus are not listed in the database of commonly misidentified cell lines maintained by ICLAC and NCBI Biosample; the cell lines have not been authenticated.
Cell culture conditions and immunostaining protocol
Cell lines were cultured in high glucose DMEM-GlutaMAX™ medium (Life Technologies) supplemented with 10 % fetal bovine serum (FBS, Life Technologies) and 1 % penicillin/streptomycin (P/S, Life Technologies) (growth medium). For live cell imaging, DMEM without phenol red containing 4.5 mg ml− glucose, 25 mM HEPES, 2 mM glutamine (Life Technologies) was used and supplemented with 10 % FBS and 1 % P/S (imaging medium). For cell staining, cells were seeded on FN-coated (10 µg/ml) glass slides (Menzel, #1.5) and allowed to spread overnight, if not indicated otherwise. Cells were fixed in 4 % paraformaldehyde (PFA) for 10 min at room temperature and immunostainings were performed as described before17. Samples were mounted in Prolong Gold (Life Technologies) and stored at 4 °C. Images were acquired using a LSM780 confocal scanning microscope equipped with a 100× oil objective (Plan-APOCHROMAT, NA=1.46). For image acquisition of cells on soft substrates, a Leica TCS SP5 X confocal microscope equipped with a 40× long-distance water objective (APO 40×/1.10 W CORR C S2) was used.
Fluorescence-activated cell sorting (FACS)
Integrin surface expression was determined via fluorescence-activated cell sorting (FACS) using a BD FACS Canto™ II instrument (BD Biosciences). Cell stainings with anti-integrin antibodies were performed in 5 % BSA solution containing 1 mM MgCl2, 1 mM CaCl2 and 0.02 % NaN3. To control for unspecific antibody labelling, cells lacking β1, β2, β3 and β7 integrin subunits were used47. Surface expression of integrins was quantified by geometric mean fluorescence intensity using FlowJo software (Tree Star).
Fluorescence recovery after photobleaching (FRAP) experiment and analysis
To allow FRAP analysis of cells with comparable FA sizes and states, cells were seeded on FN-coated micro-patterned substrates (CYTOO); under these conditions, cells adapt highly similar morphologies and develop very regular FAs of comparable size, shape and intensity. For each experiment, Tln1Y, Tln2Y and Tln1TS cells were seeded on Y-shaped micro-patterned substrates for at least 4 h and were analysed at 37 °C and 5 % CO2 on a Leica SP8 confocal laser scanning microscope equipped with a 63× water objective (HCX PL APO, NA=1.2). Cells of comparable intensity were excited at 514 nm with a laser power of 5 % to record two pre-bleach images within an interval of 10 s. Selected FAs were then photo-bleached using a laser power of 100 % for 1 s and post-bleach images were acquired every 20 s for 260–280 s; the fluorescence intensity was recorded between 530–570 nm. Next, fluorescence intensity data were imported into ImageJ and analysed using the ImageJ plugin ‘FRAP profiler’. Only data with comparable initial mean intensities and sufficient initial photo-bleaching were processed in MATLAB (Mathworks). To determine fluorescence recovery, we assumed a reaction-dominated model as described before48, 49 and fitted the data according to equation 1 (eq. 1). Only data sets with a fitting quality of R2=0.98 were considered for further analysis.
| (eq. 1) |
where t is time, koff the rate constant and A the mobile fraction. The recovery half-time τ1/2 was calculated according to eq. 2.
| (eq. 2) |
Time-correlated single photon counting fluorescence lifetime microscopy (TCSPC-FLIM)
TCSPC-FLIM experiments were performed using a confocal microscope (Leica TCS SP5 X) equipped with a pulsed white light laser (WLL, 80 MHz repetition rate, NKT Photonics), a FLIM X16 TCSPC detector (LaVision Biotech) and a 63× water objective (HCX PL APO CS, NA=1.2); a bandpass filter 545/30 (AHF Analysentechnik) was used to block photons emitted by the acceptor fluorophore. Images were acquired with a scanning velocity of 400 Hz, a spatial resolution of 512×512 pixels and resulting image field coverage of 123.02×123.02 µm2. The detection covered a time window of 12.24 ns after the excitation pulse with a temporal resolution of 0.08 ns. For each experimental condition 15–20 cells were recorded and each experiment was repeated at least 3–5 times. Data analysis was conducted by a custom-written MATLAB program calculating the FRET efficiency E according to eq. 3, where τD is the mean donor lifetime and τDA is the lifetime of the donor in presence of an acceptor fluorophore. For more detailed information see our previously published protocols17.
| (eq. 3) |
Measurement of fluorophore emission spectra in FAs of living cells
To examine whether the photo-physical properties of donor or acceptor fluorophore are affected by their insertion into talin, Tln1−/−Tln2−/− cells were reconstituted with constructs in which the individual fluorophores had been inserted into talin-1 (Tln1Y-i, Tln1C-i) or were C-terminally attached (Tln1Y, Tln1C). Cells were seeded on FN–coated coverslips and the emission spectra of integrated or C-terminally tagged fluorophores from FAs of living cells were measured. The emission spectrum of YPet (excitation: 508 nm; 525–605 nm; 10 nm detection band width) and mCherry (excitation 587 nm; 610–710 nm; 20 nm detection band width) were recorded using a confocal microscope equipped with an acousto-optical beam splitter (Leica TCS SP5 X) and a pulsed white light laser (WLL, 80 MHz repetition rate, NKT Photonics). Only FA-specific signal was processed in the subsequent data analysis.
FRET control experiment I – effects of intermolecular FRET
To test for effects of intermolecular FRET (i.e. energy transfer between adjacent molecules), Tln1−/−Tln2−/− cells were co-transfected with constructs in which the individual fluorophores were inserted into talin-1 or C-terminally attached. These cells were then seeded on FN- or pL-coated glass slides and FRET efficiencies were determined using TCSPC-FLIM. We did not observe differences when pL and FN conditions were compared indicating that effects of intermolecular FRET in these experiments are negligible. However, a slight increase of intermolecular FRET in C-terminally-tagged talin controls was detectable, presumably due to talin dimerization which is mediated at talin’s C-terminus (Fig. 2g); this may explain the slightly increased FRET efficiency values determined in Tln1Con cells as compared to Tln1TS cells on pL (e.g. Fig. 2i).
FRET control experiment II – effects of talin’s inter-domain association
To examine whether Tln1TS FRET is affected by the inter-domain association between the talin head- and the talin rod-domain50, we inserted point mutations (K324D and M319A) into Tln1TS that were previously described to abolish the intramolecular interaction50, 51. Since talin is expected to predominantly exist in an auto-inhibited conformation in the cytoplasm52, we hypothesized that the high FRET efficiencies observed in cells on pL should be significantly reduced by these activating point mutations if FRET of the biosensor was sensitive to conformational changes. However, FRET efficiencies of Tln1TS-M319A, Tln1TS-K324D and Tln1TS were indistinguishable when cells were seeded on pL (Fig. 2h). This indicates that effects of the inter-domain association on Tln1TS FRET are negligible.
FRET control experiment III – evaluating effects of fluorescence intensity and temperature
To examine whether the observed FRET effects are fluorescence intensity–dependent, FRET efficiencies were plotted over mean fluorescence intensities and the Pearson’s correlation coefficient (PCC) was determined (Supplementary Fig. 2c); however, no correlation was observed. As HP35 unfolding shows a small but significant temperature dependency (see Supplementary Note and Supplementary Fig. 1i) we also tested how moderate changes in temperature affect FRET measurements. Thus, FRET ratio measurements of Tln1Con or Tln1TS cells were performed at 30 °C (the temperature at which the single-molecule calibration was performed) and 37 °C (the temperature used during FRET ratio measurements; Supplementary Fig. 2d).
Isolating FA-specific signals for FLIM and ratiometric FRET analysis
To isolate the FA-specific signals from FLIM data sets, images were imported into MATLAB and regions of interest (ROIs) were manually set to exclude cytoplasmic background and signals from adjacent cells. A three level multi-Otsu thresholding algorithm was applied and the highest intensities were defined as FA signal. After conversion into a binary image, these FA masks were used to calculate the mean FA FRET efficiency per cell. To isolate FA signals for morphological analysis and ratiometric FRET measurements, fluorescence images were imported to MATLAB followed by manual ROI selection to analyse individual cells. Cytosolic background was subtracted from the donor image by convolving the image (Gaussian structure element; width: 25, height: 2) and applying a top-hat filtering step (disk SE; radius: 7 pixel) as described before53. Obtained individual FA masks were then used to calculate FA mean intensity of donor and acceptor signal. For mean acceptor values 1.7 times larger than manually determined average background signal, individual FA ratiometric FRET values were calculated by mean intensity acceptor/donor division. Step-by-step protocols describing the live cell FLIM and FRET experiments can be found at Nature Protocol Exchange [reference required].
Morphological FA analysis and FA co-localization analysis
For cell size and polarization analysis, cells were allowed to spread on FN-coated glass coverslips or on FN-coated Softview Easy Coat dishes (Matrigen Life Technologies). At the indicated time points, cells were fixed and stained. Images of phalloidin-stained cells were then used to determine the cell area and the cell’s major principal axis. Using Otsu thresholding as a cut-off criterion, binarized cellular shape masks were created and mask orientation was calculated to define the reference major principal axis. Subsequently, individual FA masks were analysed with respect to their eccentricity and area. The FA orientation with respect to cell major principal axis was used to evaluate cell polarization. To quantify subcellular co-localization of vinculin with talin, cells were fixed and immunostained for vinculin as described above. Intensity line plots across at least 25 FA were defined in ImageJ and evaluated in MATLAB. To find the local maximum of individual FAs, a Gaussian fit was applied in the talin channel. Intensity values for both channels at the fitted maximum ±2 pixels were averaged to minimize local errors.
Traction force microscopy
Traction force microscopy was performed using a SP8 confocal laser scanning microscope equipped with a 63× glycerol objective (HC PL APO, NA=1.3). Fluorescent beads of 0.2 µm diameter (FluoSpheres [625/645], Life Technologies) were incorporated into polyacrylamide gels with defined elastic properties that were produced according to established protocols54. Young's moduli of the substrates were calculated from Acrylamide/Bis-Acrylamide concentrations as previously described55 and obtained moduli were checked by measuring the indentation profile of a coloured bead that was placed on the gel surface as described in56; the analysis of traction force microscopy data was performed as described before57.
Sources of noise and data interpretation
Live cell FRET experiments are inherently noisy because the energy transfer rate between donor and acceptor fluorophore does not only depend on the fluorophore separation distance but also on other factors18, 58 including temperature, pH or ion-concentration. Furthermore, targeting a biosensor to a specific subcellular location can further complicate the experiment as effects through intermolecular FRET or molecular crowding may become more prominent. In addition, the biosensor expression level and the used cell type, in particular potentially high expression levels of the endogenous protein, need to be taken into account. Finally, chromatic aberrations, laser fluctuations, the Poisson statistics of photon arrival at the detector or other technical limitations contribute to the noise in FRET measurements. It is also important to note that in contrast to the single-molecule calibration experiments described in Fig. 1, the live-cell FRET experiments presented in Figs. 2–4 and Fig. 6–7 are based on bulk measurements, in which the signal from hundreds of FAs containing many molecules is averaged to calculate a mean FRET efficiency per cell. As a consequence, only an average force per molecule can be calculated.
Statistical analyses
Error bars represent the standard error of te mean (s.e.m.) if not indicated otherwise; to confirm that data are normally distributed, the Lilliefors test was used. Statistical significance is given by a p-value calculated from a two-sample Kolmogorov-Smirnov test using the default significance level of α = 0.05. Additional testing was performed with a two-sided t-test as indicated. The following nomenclature was used in all figures:***: p<0.001; **: p<0.01; *: p<0.05; not significant (n.s.): p > 0.05. Boxplots were generated using the MATLAB function boxplot()or the origin Lab software indicating the median as well as 25th and 75th percentiles; whiskers reach out to 2.7 standard deviations (σ). Statistic source data are available in Supplementary Table 1.
Computational codes
Mechanical fits were performed using previously published custom-written code24, 42; analysis software code runs on: IGOR Pro 6.31, 64-bit. Software for FA-FRAP and TCSPC-FLIM analyses was generated specifically for the project and can be used in MATLAB. The data analysis algorithm for ratiometric FRET analyses is based on a previously published Focal Adhesion Tracking algorithm53. All software is available upon request.
Supplementary Material
Acknowledgments
C. G. is supported by the German Research Council (DFG, GR3399/21 and GR3399/5–1), the Collaborative Research Centre SFB863 (B9) and a Paul Gerson Unna Research Group of the Max Planck Förderstiftung. A.M. is supported by the Nanosystems Initiative Munich (NIM); M.R. is supported by the German Research Council through the Collaborative Research Centre SFB863 (A2). R.Z. is supported by R01-DK083187, R01-DK075594, R01-DK069221 and VA Merit Award 1I01BX002196. B.S. acknowledges funding from the DAAD. The authors thank R. Fässler for continuous support and helpful discussions; F. Schnorrer for critically reading the manuscript; A. Lambacher and the MPIB core facility for technical support; G. Zoldak for helpful discussion, A. Sechi for sharing data analysis software and D. Critchley for providing talin-2 cDNA.
Footnotes
Author contributions. C.G. and K.A. initiated the project, generated cell lines, performed cellular and biochemical experiments and analysed data; P.R. generated cell lines, performed cellular experiments, wrote data analysis software and analysed data. A.M. and M.R. performed the single-molecule calibration and theoretical modelling. A.C.-G. created talin expression constructs, C. K. generated vinculin expression constructs and cell lines, C.K. and Ch.K wrote data analysis software. K.A., C.K. and B.S. performed traction force microscopy experiments and analyses. R.Z. provided genetically modified talin-cells. C.G. wrote the manuscript with the input from all authors.
Competing financial interest. The authors declare no competing financial interest.
References
- 1.Heisenberg CP, Bellaiche Y. Forces in tissue morphogenesis and patterning. Cell. 2013;153:948–962. doi: 10.1016/j.cell.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 2.Wozniak MA, Chen CS. Mechanotransduction in development: a growing role for contractility. Nat. Rev. Mol. Cell Biol. 2009;10:34–43. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franze K, Janmey PA, Guck J. Mechanics in neuronal development and repair. Annu. Rev. Biomed. Eng. 2013;15:227–251. doi: 10.1146/annurev-bioeng-071811-150045. [DOI] [PubMed] [Google Scholar]
- 4.Ingber DE. Mechanobiology and diseases of mechanotransduction. Ann. Med. 2003;35:564–577. doi: 10.1080/07853890310016333. [DOI] [PubMed] [Google Scholar]
- 5.Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 7.Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell. 2012;151:1513–1527. doi: 10.1016/j.cell.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoen I, Pruitt BL, Vogel V. The Yin-Yang of Rigidity Sensing: How Forces and Mechanical Properties Regulate the Cellular Response to Materials. Annu Rev Mater Res. 2013;43:589–618. [Google Scholar]
- 9.Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman BD, Grashoff C, Schwartz MA. Dynamic molecular processes mediate cellular mechanotransduction. Nature. 2011;475:316–323. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elosegui-Artola A, et al. Rigidity sensing and adaptation through regulation of integrin types. Nature materials. 2014;13:631–637. doi: 10.1038/nmat3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grashoff C, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Critchley DR. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu. Rev. Biophys. 2009;38:235–254. doi: 10.1146/annurev.biophys.050708.133744. [DOI] [PubMed] [Google Scholar]
- 14.Kanchanawong P, et al. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.del Rio A, et al. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J, et al. Talin determines the nanoscale architecture of focal adhesions. Proceedings of the National Academy of Sciences of the United States of America. 2015 doi: 10.1073/pnas.1512025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Austen K, Kluger C, Freikamp A, Chrostek-Grashoff A, Grashoff C. Generation and analysis of biosensors to measure mechanical forces within cells. Methods Mol. Biol. 2013;1066:169–184. doi: 10.1007/978-1-62703-604-7_15. [DOI] [PubMed] [Google Scholar]
- 18.Cost AL, Ringer P, Chrostek-Grashoff A, Grashoff C. How to Measure Molecular Forces in Cells: A Guide to Evaluating Genetically-Encoded FRET-Based Tension Sensors. Cell Mol Bioeng. 2015;8:96–105. doi: 10.1007/s12195-014-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo J, Sachs F, Meng F. Fluorescence-based force/tension sensors: a novel tool to visualize mechanical forces in structural proteins in live cells. Antioxid. Redox Signal. 2014;20:986–999. doi: 10.1089/ars.2013.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Ha T. Defining single molecular forces required to activate integrin and notch signaling. Science. 2013;340:991–994. doi: 10.1126/science.1231041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blakely BL, et al. A DNA-based molecular probe for optically reporting cellular traction forces. Nat. Methods. 2014;11:1229–1232. doi: 10.1038/nmeth.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan Y, Kollman PA. Pathways to a protein folding intermediate observed in a 1-microsecond simulation in aqueous solution. Science. 1998;282:740–744. doi: 10.1126/science.282.5389.740. [DOI] [PubMed] [Google Scholar]
- 24.Zoldak G, Stigler J, Pelz B, Li H, Rief M. Ultrafast folding kinetics and cooperativity of villin headpiece in single-molecule force spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:18156–18161. doi: 10.1073/pnas.1311495110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, et al. Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nat. Cell Biol. 2008;10:1062–1068. doi: 10.1038/ncb1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carisey A, et al. Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr. Biol. 2013;23:271–281. doi: 10.1016/j.cub.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dumbauld DW, et al. How vinculin regulates force transmission. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9788–9793. doi: 10.1073/pnas.1216209110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humphries JD, et al. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol. 2007;179:1043–1057. doi: 10.1083/jcb.200703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hytonen VP, Vogel V. How force might activate talin's vinculin binding sites: SMD reveals a structural mechanism. PLoS Comp. Biol. 2008;4:e24. doi: 10.1371/journal.pcbi.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao M, et al. Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci. Rep. 2014;4:4610. doi: 10.1038/srep04610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciobanasu C, Faivre B, Le Clainche C. Actomyosin-dependent formation of the mechanosensitive talin-vinculin complex reinforces actin anchoring. Nat. Commun. 2014;5:3095. doi: 10.1038/ncomms4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Praekelt U, et al. New isoform-specific monoclonal antibodies reveal different sub-cellular localisations for talin1 and talin2. Eur. J. Cell Biol. 2012;91:180–191. doi: 10.1016/j.ejcb.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borghi N, et al. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krieg M, Dunn AR, Goodman MB. Mechanical control of the sense of touch by beta-spectrin. Nat. Cell Biol. 2014 doi: 10.1038/ncb2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paszek MJ, et al. The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature. 2014;511:319–325. doi: 10.1038/nature13535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai D, et al. Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell. 2014;157:1146–1159. doi: 10.1016/j.cell.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conway DE, et al. Fluid shear stress on endothelial cells modulates mechanical tension across VE-cadherin and PECAM-1. Curr. Biol. 2013;23:1024–1030. doi: 10.1016/j.cub.2013.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moser M, Legate KR, Zent R, Fassler R. The tail of integrins, talin, and kindlins. Science. 2009;324:895–899. doi: 10.1126/science.1163865. [DOI] [PubMed] [Google Scholar]
- 39.Miller K, Chinzei K, Orssengo G, Bednarz P. Mechanical properties of brain tissue in-vivo: experiment and computer simulation. J. Biomech. 2000;33:1369–1376. doi: 10.1016/s0021-9290(00)00120-2. [DOI] [PubMed] [Google Scholar]
- 40.Anthis NJ, Wegener KL, Critchley DR, Campbell ID. Structural diversity in integrin/talin interactions. Structure. 2010;18:1654–1666. doi: 10.1016/j.str.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 41.Cecconi C, Shank EA, Dahlquist FW, Marqusee S, Bustamante C. Protein-DNA chimeras for single molecule mechanical folding studies with the optical tweezers. Eur. Biophys. J. 2008;37:729–738. doi: 10.1007/s00249-007-0247-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stigler J, Ziegler F, Gieseke A, Gebhardt JC, Rief M. The complex folding network of single calmodulin molecules. Science. 2011;334:512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 43.von Hansen Y, Mehlich A, Pelz B, Rief M, Netz RR. Auto- and cross-power spectral analysis of dual trap optical tweezer experiments using Bayesian inference. Rev. Sci. Instrum. 2012;83:095116. doi: 10.1063/1.4753917. [DOI] [PubMed] [Google Scholar]
- 44.Subauste MC, et al. Rho family proteins modulate rapid apoptosis induced by cytotoxic T lymphocytes and Fas. J. Biol. Chem. 2000;275:9725–9733. doi: 10.1074/jbc.275.13.9725. [DOI] [PubMed] [Google Scholar]
- 45.Conti FJ, Monkley SJ, Wood MR, Critchley DR, Muller U. Talin 1 and 2 are required for myoblast fusion, sarcomere assembly and the maintenance of myotendinous junctions. Development. 2009;136:3597–3606. doi: 10.1242/dev.035857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monkley SJ, et al. Disruption of the talin gene arrests mouse development at the gastrulation stage. Dev. Dyn. 2000;219:560–574. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1079>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 47.Schiller HB, et al. beta1- and alphav-class integrins cooperate to regulate myosin II during rigidity sensing of fibronectin-based microenvironments. Nat. Cell Biol. 2013;15:625–636. doi: 10.1038/ncb2747. [DOI] [PubMed] [Google Scholar]
- 48.Sprague BL, McNally JG. FRAP analysis of binding: proper and fitting. Trends Cell Biol. 2005;15:84–91. doi: 10.1016/j.tcb.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 49.Carisey A, Stroud M, Tsang R, Ballestrem C. Fluorescence recovery after photobleaching. Methods Mol. Biol. 2011;769:387–402. doi: 10.1007/978-1-61779-207-6_26. [DOI] [PubMed] [Google Scholar]
- 50.Goult BT, et al. The structure of an interdomain complex that regulates talin activity. J. Biol. Chem. 2009;284:15097–15106. doi: 10.1074/jbc.M900078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goksoy E, et al. Structural basis for the autoinhibition of talin in regulating integrin activation. Mol. Cell. 2008;31:124–133. doi: 10.1016/j.molcel.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goult BT, et al. Structural studies on full-length talin1 reveal a compact auto-inhibited dimer: implications for talin activation. J. Struct. Biol. 2013;184:21–32. doi: 10.1016/j.jsb.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wurflinger T, Gamper I, Aach T, Sechi AS. Automated segmentation and tracking for large-scale analysis of focal adhesion dynamics. J. Microsc. 2011;241:37–53. doi: 10.1111/j.1365-2818.2010.03404.x. [DOI] [PubMed] [Google Scholar]
- 54.Yeung T, et al. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 55.Boudou T, Ohayon J, Picart C, Pettigrew RI, Tracqui P. Nonlinear elastic properties of polyacrylamide gels: implications for quantification of cellular forces. Biorheology. 2009;46:191–205. doi: 10.3233/BIR-2009-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lembong J, Sabass B, Sun B, Rogers ME, Stone HA. Mechanics regulates ATP-stimulated collective calcium response in fibroblast cells. J R Soc Interface. 2015;12:20150140. doi: 10.1098/rsif.2015.0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sabass B, Gardel ML, Waterman CM, Schwarz US. High resolution traction force microscopy based on experimental and computational advances. Biophys. J. 2008;94:207–220. doi: 10.1529/biophysj.107.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jares-Erijman EA, Jovin TM. FRET imaging. Nat. Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.