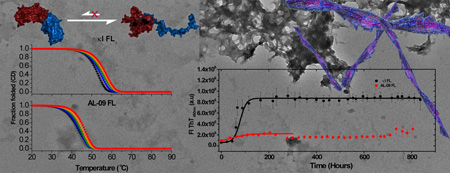
Abstract
Light chain (AL) amyloidosis is a fatal disease where monoclonal immunoglobulin light chains deposit as insoluble amyloid fibrils. For many years it has been considered that AL amyloid deposits are formed primarily by the variable domain, while its constant domain has been considered not to be amyloidogenic. However recent studies identify full length (FL) light chains as part of the amyloid deposits. In this report, we compare the stabilities and amyloidogenic properties of two light chains, an amyloid-associated protein AL-09 FL, and its germline protein κI O18/O8 FL (IGKV 1–33). We demonstrate that the thermal unfolding for both proteins is irreversible andand scan rate dependent, with similar stability parameters compared to their VL counterparts. In addition, the constant domain seems to modulate their amyloidogenic properties and affect the morphology of the amyloid fibrils. These results allow us to understand the role of the kappa constant domain in AL amyloidosis.
Keywords: Light chain amyloidosis, kinetic stability, amyloid fibril formation, irreversible thermal unfolding, protein aggregation
Graphical abstract
Introduction
Light chain (AL) amyloidosis is a fatal disease where monoclonal immunoglobulin light chains (LC) deposit in the human body as insoluble amyloid fibrils. The process of deposition causes tissue degeneration, organ failure, and death. An immunoglobulin LC consists of one variable (VL) and one constant domain (CL). The variable region presents somatic mutations and as a result, each AL amyloidosis patient has a unique monoclonal protein sequence secreted into circulation and later deposited in vital organs. Amyloidogenic light chain AL-09 is a patient derived protein that belongs to the κI O18/O8 (IGKV 1–33) germline sequence. In previous studies, the variable domain of AL-09 has been identified as an unstable protein that forms fibrils much faster than the germline variable domain [1, 2].
Our laboratory and others have demonstrated that in general, there is a direct correlation between immunoglobulin light chain VL thermodynamic stability and the protein’s propensity to form amyloid fibrils (for a review, see [3]), although we have recently reported that there is a thermal stability threshold for amyloid formation for some VL proteins [4] and a possible inverse correlation for some AL protein VLs and their mutants (Marin-Argany et al., under review). For many years, AL amyloidosis researchers have conducted their biochemical and biophysical studies with the variable domain. This decision was supported by early reports that stated that amyloid deposits from AL amyloidosis patients were formed primarily by the variable domain and small regions of the constant domain [5]. It has been considered that the variable domain somehow corresponds to the central region undergoing misfolding and aggregation in AL amyloidosis. However, in 2008, proteomic studies conducted with amyloid deposits from fat aspirates [6] found full length (FL) light chains as part of the amyloid deposits. This was later confirmed by a proteomics study using biopsy samples from affected tissues [7]. However, it is unclear what proportion of the amyloid deposit consists of FL light chains and what proportion is formed by fragments consisting mainly of the VL as those found in earlier reports. What is now clear after the Lavatelli and Vrana reports is that the structure and stability of FL light chains may play a role in the amyloid formation process in AL amyloidosis.
The participation of the CL in the pathophysiology of AL amyloidosis is not well understood, although it has been reported previously that the κI CL alone can form fibrils both in vivo [8] and in vitro under denaturing conditions [9]. The role of the CL in thermal stability has been characterized for a lambda-6a (IGLV 6–57) full length AL protein comparing its properties with its corresponding variable and constant domain proteins [10].
The purpose of the current study is to perform thermodynamic and fibril formation studies of FL amyloidogenic light chains, in order to compare them to the VL studies conducted previously [1]. This will allow us to determine the role that the CL plays in the protein stability and its ability to form amyloid fibrils. Our results show that the thermal unfolding of AL-09 FL and its germline protein κI O18/O8 FL is irreversible, precluding us from calculating thermodynamic parameters from equilibrium experiments, as has been reported before. Both FL proteins have characteristic scan rate dependencies during thermal denaturation. Both proteins are able to form amyloid fibrils with kinetics that are slower than their variable domain counterparts. Their pH dependencies for amyloid formation seem to be altered as well.
Materials and Methods
Protein preparation
The expression vectors for AL-09 FL and κI O18/O8 FL (herein called κI FL for simplicity) were expressed in Rosetta Gami cells as described previously [11]. Briefly, proteins were expressed as inclusion bodies that were solubilized using 8 M urea. Samples were dialyzed against 10 mM Tris-HCl and loaded onto a Superdex 75 column. SDS-PAGE gels were run to ensure the purity of the fractions.
Circular Dichroism Spectroscopy
CD spectroscopy was used to determine the global secondary structure after protein extraction/purification. Initial exploratory analysis was conducted using 10 µM AL-09 FL in 10 mM Tris-HCl pH 7.4 in 0.2 cm cuvettes for CD analysis. Far UV-CD spectra between 260 and 200 nm were acquired at 4°C, on a Jasco Spectropolarimeter 810 (Jasco, Inc., Easton, MD) prior to thermal denaturation. Temperature was regulated within ±0.002°C using a Peltier system.
A wavelength of 216 nm (indicative of β-sheet content) was used to follow protein unfolding from 4–90°C. Data was collected every 2°C for each of the five scan rates: 0.5, 1.0, 1.5, 2.0, and 2.5°C/min. Each scan rate was collected in triplicate. We observed distortions in the baseline of the unfolded state that we attributed to aggregation. For that reason, subsequent experiments were done with 2 µM AL-09 FL and κI FL in PBS buffer in a 1.0 cm cuvette under continuous stirring, for both Circular Dichroism and Fluorescence spectroscopy. For these sets of experiments, data was collected every 0.5°C, at the same scan rates listed above. Each scan rate was collected in triplicate. The Far UV-CD spectra and the thermal denaturation experiments were conducted as indicated above.
Fluorescence experiments
Intrinsic tryptophan fluorescence measurements were carried out using a PTI spectrofluorometer (Photon Technology International, Lawrenceville, NJ) using a quartz 1.0 cm micro-cuvette. Temperature was regulated within ± 0.2 °C using a Peltier system. An excitation wavelength of 295 nm and emission wavelength of 350 nm were used during thermal unfolding scans. Each data point was integrated for 20 seconds. Data was collected in triplicate for the same scan rates as for CD. Thermal unfolding data was normalized into a fraction folded percentage using linear folded and unfolded baselines [12].
Analysis and fitting of irreversible two-state transitions
Scan rate dependent thermal unfolding was previously used to calculate thermodynamic parameters [12]. Since the thermal unfolding of both FL proteins is irreversible and kinetically controlled, the apparent transition midpoint (Tmapp) depends on the applied scan rate and therefore is entirely determined by the kinetics of the formation of the final state. This circumstance makes a straightforward thermodynamic analysis, assuming equilibrium between states, impossible. Consequently, it is not possible to obtain meaningful thermodynamic parameters such as an equilibrium Tm or ΔH to describe the unfolding/refolding process.
However, by using a fully kinetic two-state model, explicitly considering the rate constant for irreversibility, stability parameters could be obtained with the appropriate data treatment, as reported by [12].
In order to determine and compare the stability of AL-09 FL and κI FL, analysis of thermal unfolding transitions was carried out using two different approaches:
-
Measurement of scan rates for the determination of Tm values at zero scan rate. This approach requires the determination of the transition midpoint of the thermal unfolding at different scan rates. The apparent transition midpoints, Tmapp, obtained from spectroscopic data at each scan rate were calculated by nonlinear least-squares fitting using the integrated van't Hoff equation as described previously [13]
The Tm values at zero scan rate (called Tmeq) were determined empirically from linear extrapolation of the thermal scan rate dependence of the apparent Tm to zero scan rate.
Fitting the data to an irreversible two-state model. For an irreversible two-state transition, unfolding is kinetically controlled by a first order rate constant that varies with temperature according to the Arrhenius equation. This procedure was published by Tischer et al. 2013 [12]. We will describe it below.
The activation energy of an unfolding reaction defined as N → U is ΔG‡ = −RTlnk‡; we expand k‡ as a function of temperature defining Δβ = (β − β*) = (1/RT − 1/RT*) using a Taylor series, where T* is the temperature at which ΔG‡ = 0.
| (1) |
Where we define , and The latter term corresponds to the enthalpy of activation and is the relationship that defines the heat capacity of activation. The inclusion of the second order parameter in this Taylor expansion is given only for the sake of completion.
Adapting the calculations from Tischer et al. 2013, we assume that the rate equation for the irreversible formation of U with respect to β is in terms of the population, PU.
| (2) |
where ∂PU/∂t = k(1 − PU); ∂t/∂T is the inverse of the thermal scan rate (υ) and ∂T/∂β = −RT2. Separation of variables followed by numerical integration results in the following expression for the unfolded state population, where (PN + PU = 1).
| (3) |
For spectroscopy we used the following function for the observed CD or fluorescence change.
| (4) |
Where ObsN(T) and ObsU(T) are linear baseline functions for the native and unfolded state observables.
This last equation was used to fit all thermal transitions using a Microsoft Excel script written by Tischer et al. that incorporates Boole’s method for the numerical integration [14]. Fitting results were minimized to the total standard deviation of the sum of squared residuals. The goodness of fit was calculated via the R2 coefficient of determination from the calculated total sum of squares SStot = Σij (yi − ȳ)2, regression of squares SSreg Σi (fi − ȳ)2 and the residual sum of squares SSerr = Σi (yi − fi)2, where yi and fi are individual observed and fit values at any given temperature and is the mean experimental observable for the entire transition.
| (5) |
The analysis results in the transition temperature T* at which the rate of unfolding is equal to unity and the enthalpy of the transition at T* that describes the temperature dependence of the rate constants for both heat-induced unfolding and refolding by cooling. Using this method it is possible to compare the T* and ΔH* values which are independent of the applied scan rate.
Amyloid fibril formation kinetics
Fibril formation kinetics experiments were conducted as reported previously [13]. Briefly, proteins were thawed at 4°C and filtered using 0.45 µm membranes. The proteins were then ultracentrifuged at 90,000 rpm (645019 ×g) for 3.3 h at 4°C in order to remove any preformed aggregates from soluble proteins. 10 µM and 20 µM ultracentrifuged protein solution was incubated at different pH values using 10 mM sodium Acetate, 10 mM Borate, 10 mM sodium Citrate (ABC) buffer in the presence of 0.15 M NaCl, 0.02% NaN3, and 10 µM Thioflavin T. All fibril formation assays were performed in triplicate using black 96-well polystyrene plates (Greiner, Monroe, NC). The plates were sealed and covered with a black polystyrene cover, sealed again with tape and incubated at 37°C under continuous orbital shaking (300 rpm). Fibril formation was monitored daily for 1 month (~750 h). The excitation wavelength used was 440 nm and the emission wavelength was 480 nm. The t50 value was obtained by fitting each independent kinetic trace to a sigmoidal function (defined as a Boltzmann function by the Origin software package http://www.originlab.com/www/helponline/Origin/en/UserGuide/Boltzmann.html).
| (6) |
Where A1 is the initial fluorescence value, A2 is the final fluorescence value, x0 is the center (t50 value), dx is defined as the time constant. A larger t50 value indicates a longer time required to form fibrils.
Electron Microscopy
A 3 µl fibril sample was placed on a 300 mesh copper formvar/carbon grid (Electron Microscopy Science, Hatfield, PA), and excess liquid was removed. The samples were negatively stained with 2% uranyl acetate, washed twice with H2O, and air-dried. Grids were analyzed on a Philips Tecnai T12 transmission electron microscope at 80 kV (FEI, Hillsboro, OR).
Results
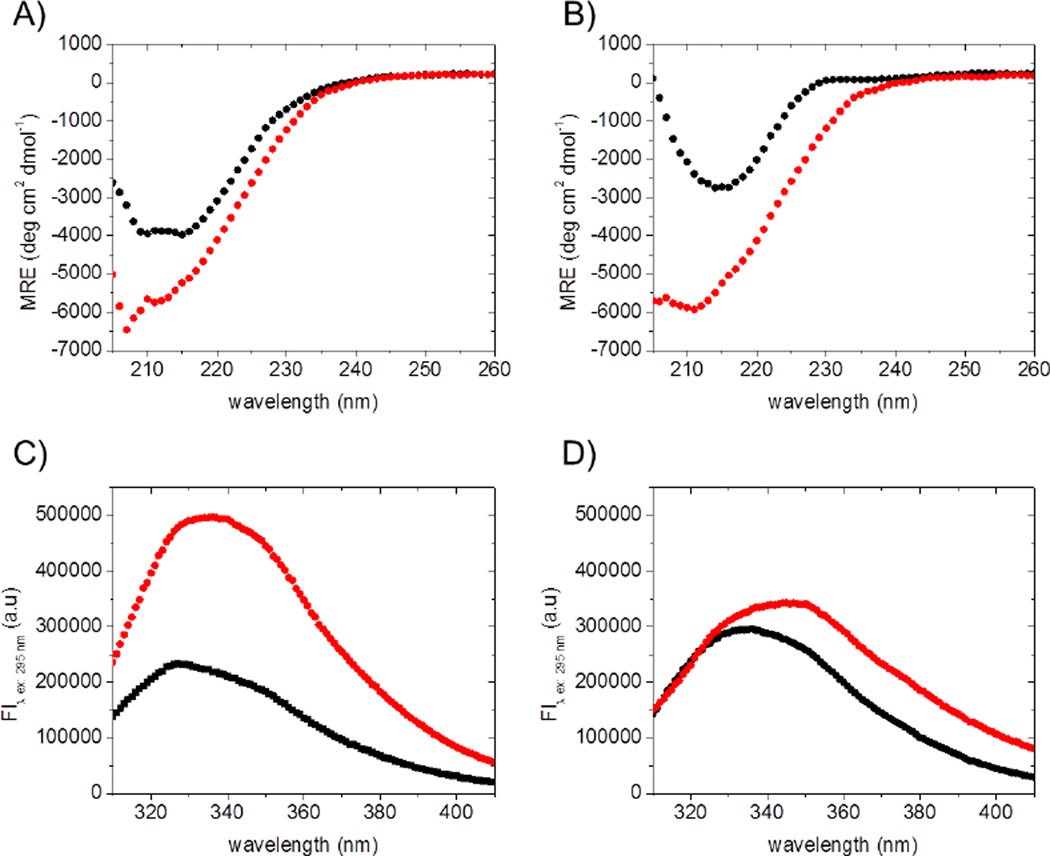
Far UV-CD spectra of AL-09 FL and κI FL were acquired to confirm that the proteins adopt the folded β-sheet conformation previously reported [11]. As reported before, the proteins were unable to refold after thermal denaturation. This was confirmed by the far UV-CD spectra (secondary structure) as well as by the intrinsic Tryptophan fluorescence spectra (tertiary structure), acquired before the thermal denaturation and after the protein was cooled down to 4°C from the unfolding experiment (Figure 1).
Figure 1.
Far-UV CD spectra of (A) AL-09 FL and (B) κI FL and Tryptophan fluorescence emission spectra of (C) AL-09 FL and (D) κI FL in PBS (pH 7.4) at 4°C, before (black symbols) and after irreversible thermal unfolding (red symbols). In all cases, samples were prepared at 2.0 µM and measured using a 1 cm cuvette. Both heating and cooling were carried out at 0.5°C/min.
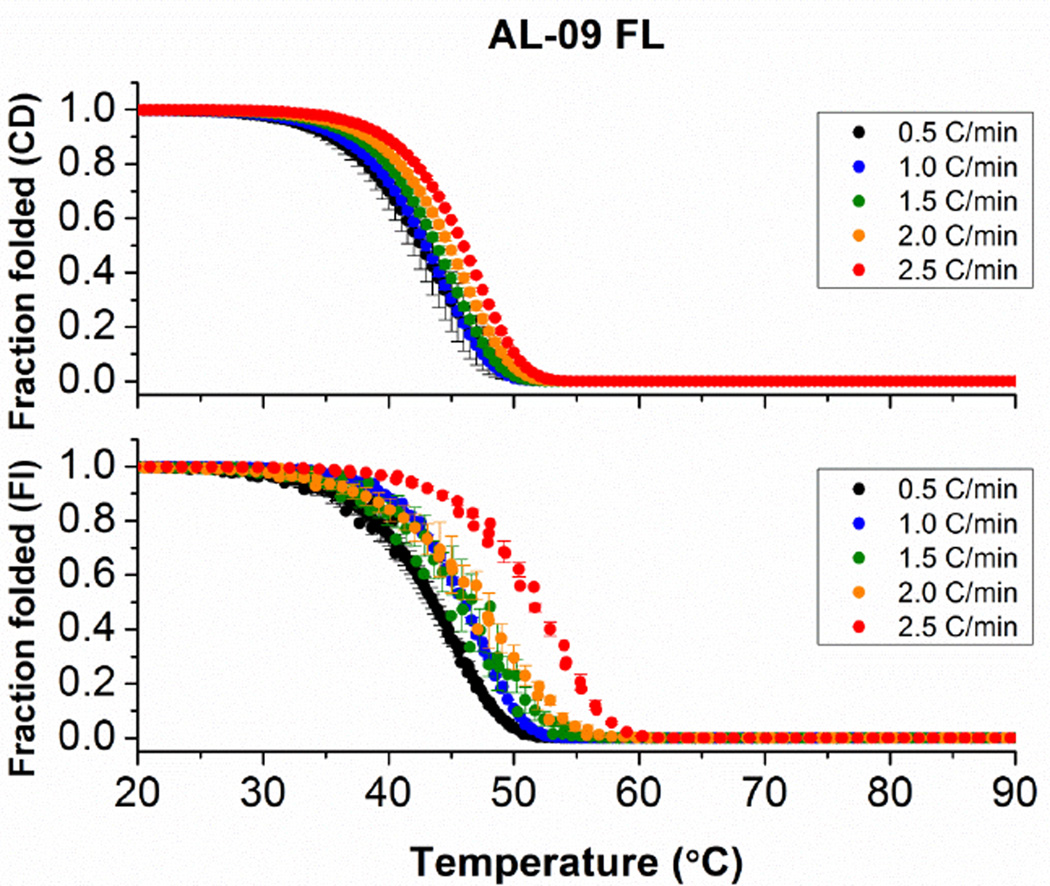
These results indicate that the thermal unfolding of both AL-09 FL and κI FL is irreversible and hence kinetically controlled. Therefore we have measured a scan rate dependency in order to observe the effect of decreasing scan rates on the apparent transition midpoint. Figure 2 demonstrates a strong dependency of the unfolding transition on the rate at which the temperature is changed, indicating that the unfolding of AL-09 FL is kinetically controlled.
Figure 2.
Thermal stability analysis of AL-09 FL following changes with (A) Circular Dichroism and (B) intrinsic fluorescence emission as a function of thermal scan rate. In all cases, melting curves were measured continuously from 4°C to 90°C every 0.5°C. Heating scan-rates employed are indicated in the figure. Samples were prepared at 2.0 µM and measured using a 1 cm cuvette and continuous stirring. Error bars correspond to the standard error obtained from 3 independent experiments.
As the scan rate increases, the apparent Tm also increases. Since the thermal unfolding of AL-09 FL and κI FL proteins is irreversible and kinetically controlled, data cannot be analyzed using an equilibrium two-state unfolding mechanism. The scan rate dependency is more significant when the thermal denaturation is followed by intrinsic Tryptophan fluorescence.
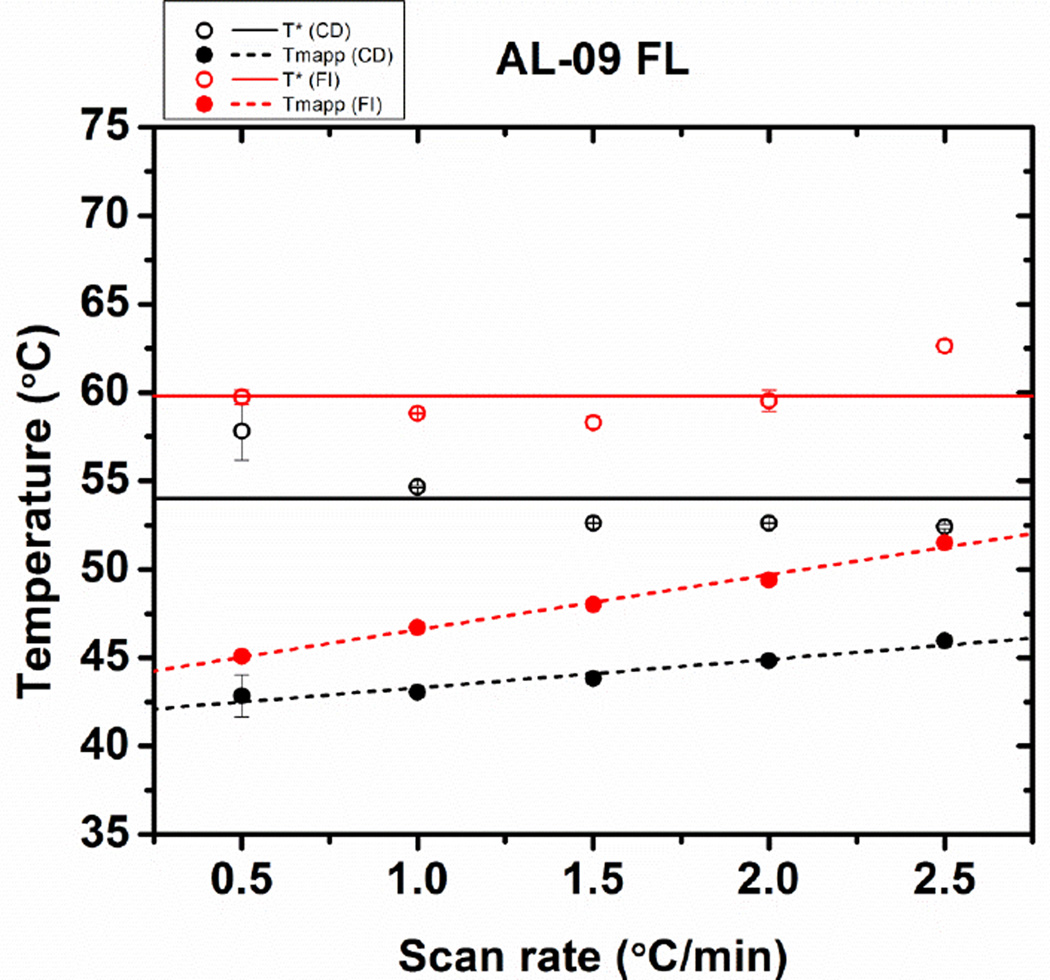
Instead, thermal transitions were analyzed in terms of the apparent Tm (Tmapp) and also by a two-state irreversible model described by Tischer et al [12]. The latter method allows us to calculate T* and ΔH* values which are independent of the scan rate as described in the materials and methods. The calculated parameters are shown in Figure 3, where we observe that the Tmapp increases as a function of scan rate for both CD and Fluorescence measurements. The Tmapp and T* values calculated at each scan rate are consistently lower for experiments conducted using CD.
Figure 3.
Comparison of the stability parameters of AL-09 FL obtained from fitting the thermal transitions in Figure 2. (A) Scan rate dependence of the T* values (open symbols and continuous regression lines) and apparent Tmapp values (solid symbols and discontinued lines), from far-UV CD data (black) or fluorescence emission (red). Error bars correspond to the standard error obtained from 3 independent experiments.
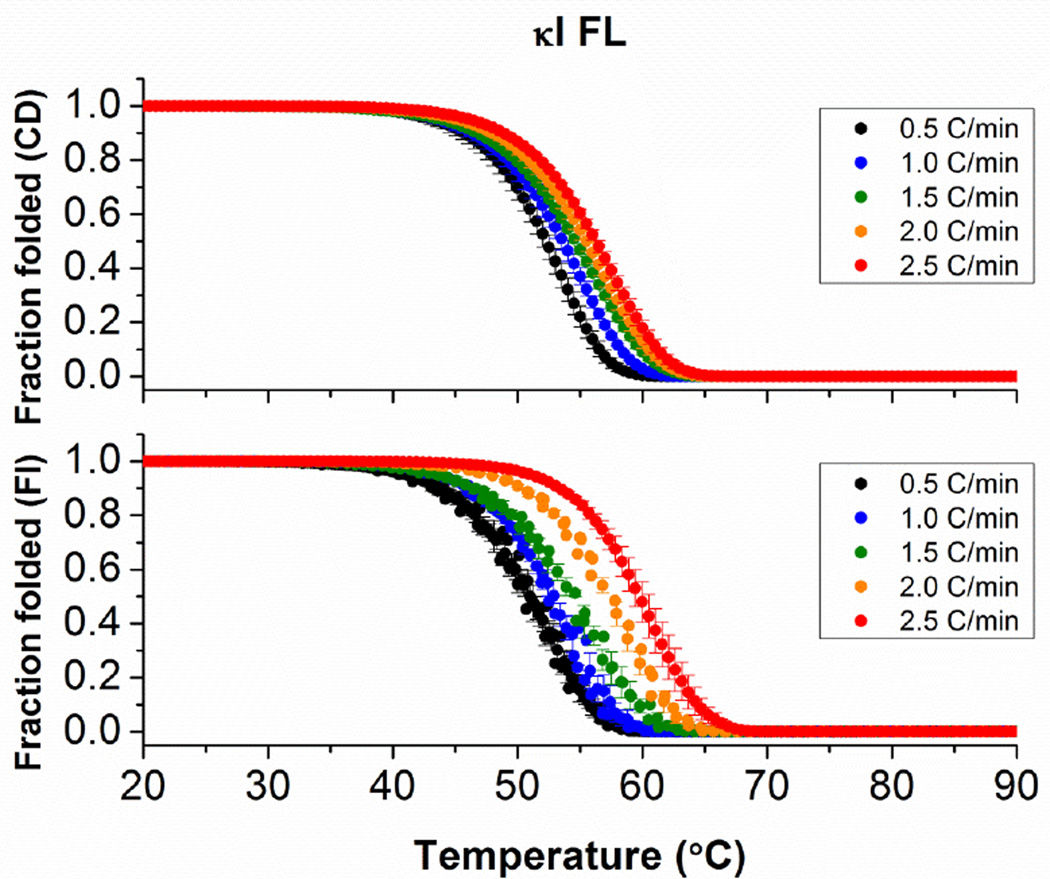
For κI FL, we observe a scan rate dependency on the protein unfolding transitions as well. As with AL-09 FL, the scan rate dependency is more significant when the thermal denaturation is followed by intrinsic Tryptophan fluorescence (Figure 4).
Figure 4.
Thermal stability analysis of κI FL following changes with (A) circular dichroism and (B) intrinsic fluorescence emission as a function of thermal scan rate. In all cases, melting curves were measured continuously from 4°C to 90°C every 0.5°C. Heating scan-rates employed are indicated in the figure. Samples were prepared at 2 µM and measured using a 1 cm cuvette and continuous stirring. Error bars correspond to the standard error obtained from 3 independent experiments.
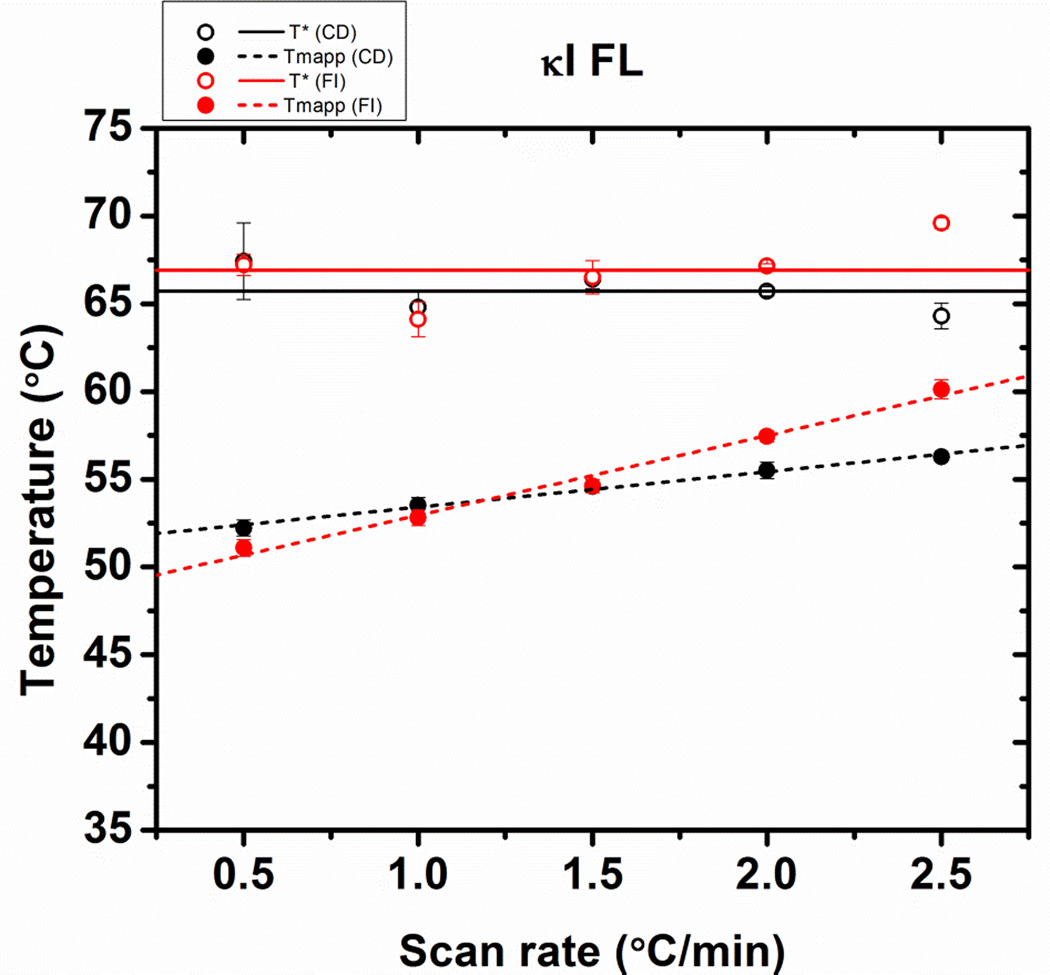
The Tmapp would remain constant at all scan rates only if the protein unfolding occurs in equilibrium and under reversible conditions. An extrapolation of the Tm to a scan rate of 0°C/min provides a value where reversible unfolding occurs under reversible conditions: 42.0 ± 0.1°C for AL-09 FL from both CD and fluorescence data. The same type of analysis has been conducted for κI FL (51.6 ± 0.8°C). We found that AL-09 FL is less thermodynamically stable than κI FL, which is consistent to the results obtained for their corresponding VL domains (AL-09 VL Tm= 41.1 ± 1.0°C; κI VL Tm= 54.7 ± 0.3°C [1, 13].
We observe that for κI FL, the apparent Tm and T* values obtained using CD and fluorescence overlap at the different scan rates (Figure 5), suggesting that for this protein, β-sheet content and intrinsic Tryptophan fluorescence change simultaneously as a function of temperature, while for AL-09 FL, it appeared that changes in tertiary and secondary structure content are uncoupled during the thermal unfolding transition.
Figure 5.
Comparison of the stability parameters of κI FL obtained from fitting the thermal transitions in Figure 2. (A) Scan rate dependence of the T* values (open symbols and continuous regression lines) and apparent Tm values (solid symbols and discontinued lines), from far-UV CD data (black) or fluorescence emission (red). Error bars correspond to the standard error obtained from 3 independent experiments.
It is important to note that LCs are formed by two independent globular domains whose structural and spectroscopic properties are almost the same. Each domain contains a single tryptophan, W35 in the VL and W148 in the CL, both located in the hydrophobic core. Structural models of κI LCs indicate that each Tryptophan is located very near to the intra-chain disulfide bond, resulting in a strong significantly quenching of the fluorescence signal in the native state [15]. Therefore, it can be used as a ‘reporter group’ for the conformational state of each domain. The presence of an intermediate is unlikely, or if present, it is at very low concentration that we do not observe a three-state unfolding transition as it has been shown in other two-domain unfolding reactions [16], it is worth to note that it is known that well-defined independent domains, can “overlap” thermodynamically [17].
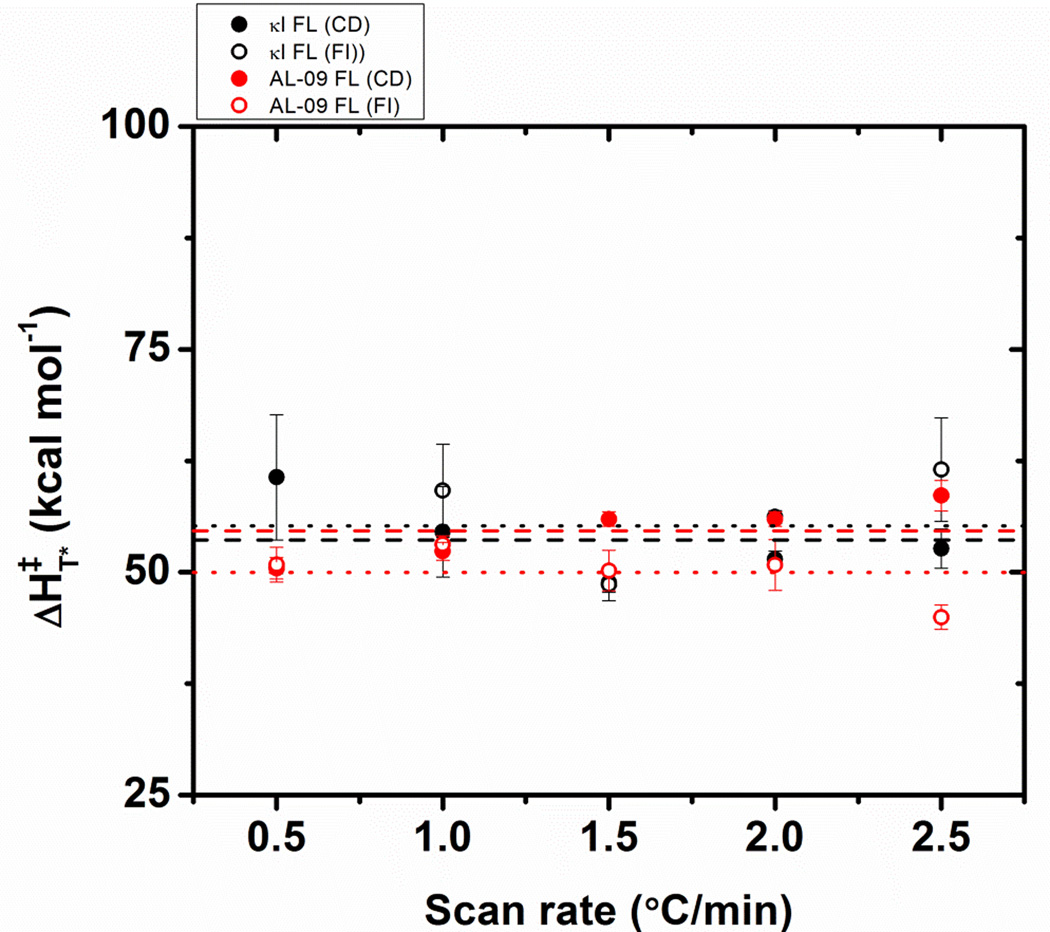
Figure 6 shows the ΔH‡T* as a function of scan rate for both AL-09 FL and κI FL. While the values for both proteins at the different scan rates oscillate around an average of 53.3 kcal/mol, κI FL presents higher ΔH‡T* values at slow scan rate values compared to AL-09 FL. This indicates that at slow scan rates, the rate of unfolding of AL-09 FL is faster than κI FL, as suggested by Tischer et al [12].
Figure 6.
Changes in the enthalpy reference independent of the applied scan rate ΔH‡, as a function of the scan rate. Lines are dashed for CD data and dotted for fluorescence data. Symbols are: κI FL (black); AL-09 FL (red). Data are the average of triplicates obtained from fitting each thermal unfolding transition using equation 4. Error bars correspond to the standard error obtained from 3 independent experiments.
We then tested the ability of the studied proteins to form amyloid fibrils at different solution conditions. We have reported previously that AL-09 VL is able to form amyloid fibrils in vitro with very fast rates and under a number of different conditions [1, 2]. We tested fibril formation for both proteins from pH 2–11; we are reporting results from three different pH conditions for simplicity (Figure 7).
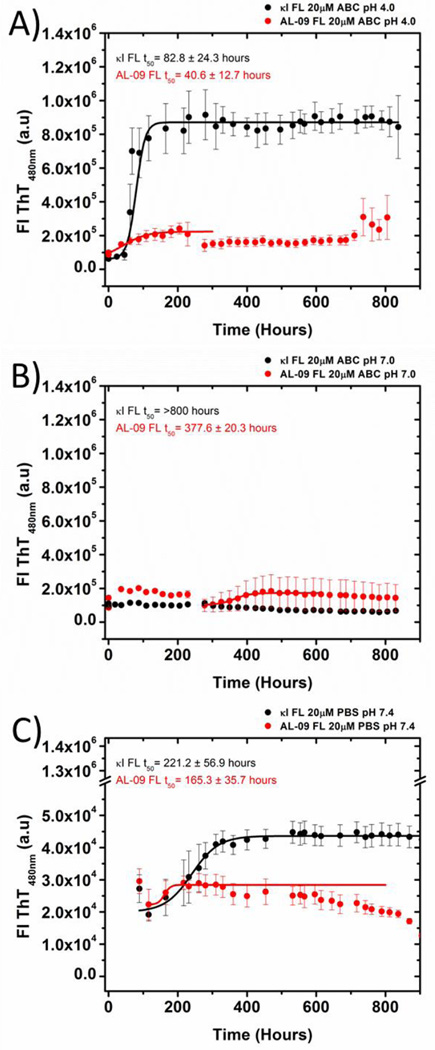
Figure 7.
Kinetics of fibril formation using (A) 10 mM (Acetate/Borate/Citrate) ABC buffer, pH 4.0,(B) 10 mM ABCbuffer, pH 7.0, (C) PBS buffer, pH 7.4. All traces are averages of three independent experiments. Solid lines represent fitting as reported before [13]. Data were collected for 800 hours. The last data points were not considered in the fitting due to the decrease in the ThT fluorescence observed attributed to inner filter effects. Error bars correspond to the standard error of the average of 3 independent experiments.
At pH 4.0, both AL-09 FL and κI FL present very fast kinetics, equivalent to the fast kinetics observed for AL-09 VL at pH 2.0; with t50 values of 40.6±12.7 hours for AL-09 FL and 82.8 ± 24.3 hours for κI FL. We observe that AL-09 FL presents lower ThT fluorescence enhancement compared to κI FL at the end of the reaction. At pH 7.0, using the buffer system of 10 mM Acetate/10 mM Borate/10 mM Citrate, we observe that κI FL does not present any discernable ThT fluorescence enhancement and that AL-09 presents delayed kinetics (t50=377.6 ± 20.3 hours) compared to the reaction at pH 4.0. We conducted amyloid fibril formation at pH 7.4 using PBS. We decided to use two buffer systems comparing pH 7.0 and pH 7.4 because our laboratory is avoiding use of Tris-HCl buffer above room temperature due to the pH changes that can occur with Tris as a function of temperature [18].
The reactions in PBS for both proteins showed ThT fluorescence enhancement for both proteins, although the t50 values indicate that the kinetics are slower than the reactions at pH 4.0 for both proteins; with values of 165.3 ± 35.7 hours for AL-09 FL and 221.2 ± 56.9 hours for κI FL. We observe again lower ThT fluorescence enhancement for AL-09 FL compared to κI FL.
In general, the reactions for AL-09 and κI FL proteins presented minimal ThT fluorescence enhancement compared to the fluorescence enhancement observed with their VL counterparts. There are two explanations for this result. One possibility is that the amyloid formation reactions with FL proteins do not have the same amount of amyloid formed at the end of the reactions. The other explanation is that the structure of the amyloid formed by FL proteins does not allow as many ThT binding sites as the structure for VL amyloid or that the affinity of each binding site is different [19]. In addition, it is possible that ThT molecules bound to the different fibrils have different fluorescence quantum yields. Thus, samples were analyzed by electron microscopy to determine the presence amyloid fibrils or any other kind of aggregate at the end of the fibril formation reaction.
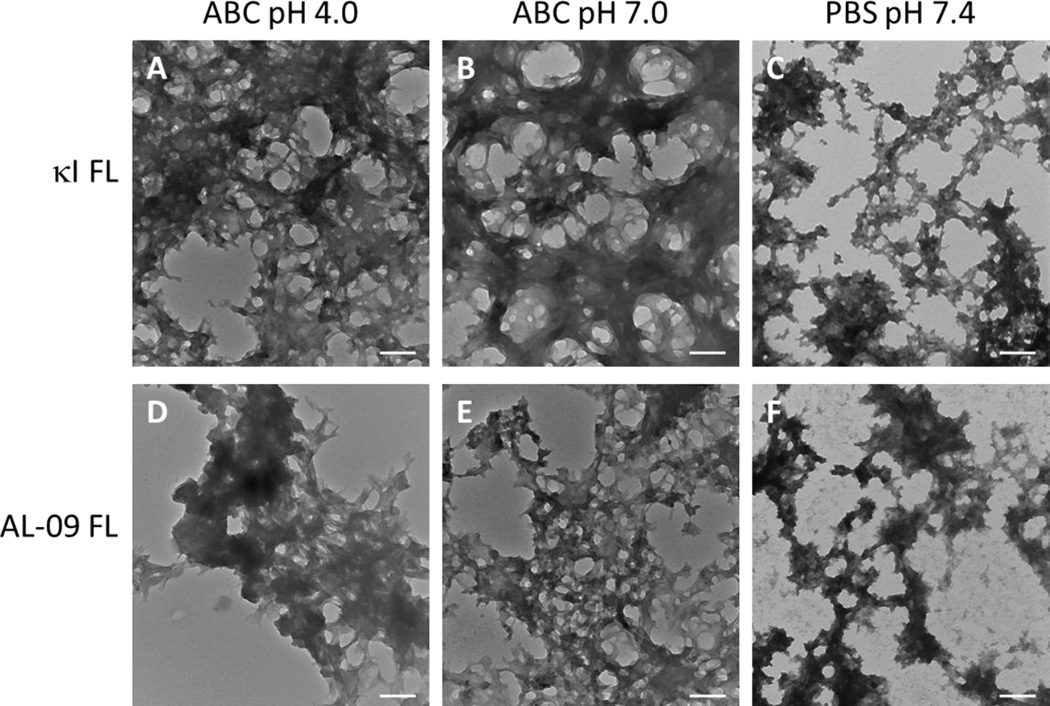
Transmission electron microscopy images (TEM) show that both FL proteins are capable of forming dense bundles of fibrils, similar to those found with their VL proteins (Figure 8). Fibrils formed in ABC buffer at pH 4.0 are better defined and can present more elongation compared to fibrils obtained at pH 7.0 and pH 7.4. While κI FL samples present more individual fibrillar species, AL-09 FL fibrils coalesce into highly dense zones, hampering the identification of individual fibrils.
Figure 8.
TEM images of fibrils at the endpoint of the reaction. Comparison of the morphology of amyloid fibrils, at the end of the fibril reaction. κI FL in (A) Acetate/Borate/Citrate buffer pH 4.0, (B) ABC buffer pH 7.0, (C) PBS pH 7.4 and AL-09 FL in (D) ABC pH 4.0, (E) Acetate/Borate/Citrate buffer pH 7.0, and (F) PBS pH 7.4. Scale bar represent 200 nm.
Fibrils formed in Acetate/Borate/Citrate buffer at pH 7.0 (Figure8B and 8E) showed the presence of fibrils that exhibit a tendency to recruit material at its periphery, creating a “fuzzy” area between the fibrils, where the uranyl acetate accumulate, probably due to the presence of non-fibrillar amorphous aggregates. Fibrils formed in PBS (Figure8C and 8F) revealed a different morphology; although they retain some amount of fibrillarlike structure, we observe the presence of amorphous aggregates tightly attached to the fibrillary bundles.
Interestingly, while κI FL presents no discernable ThT fluorescence enhancement at pH 7.0, some amyloid fibrils were observed by TEM, although not as prominent and abundant as with the AL-09 FL sample.
Taken together, these results indicate that the presence of the constant domain have a significant effect on the stability of full length light chains, as well as formation and to a lower extent, the appearance of the amyloid fibrils.
Discussion
In this study we have characterized the thermodynamic and amyloid formation properties of the full length properties of two immunoglobulin light chains. Our results show that the both AL-09 and κI FL present similar thermodynamic stability parameters compared to their VL counterparts. In the case of κI FL, the thermodynamic parameters calculated in this study suggest that the VL domain is slightly more stable than the FL protein. Our results show that for AL-09 and κI FL, the presence of the CL domain did not have any stabilizing effect. Our results differ from what was found with a λ6 (IGLV 6–57) full length protein [10]. Klimtchuk and co-workers characterized the thermal unfolding properties of the VL, CL, and the FL versions of protein AL-01-095 (comparing their results to the urine-derived Bence Jones protein (BJP)). Their results show that the CL domain (LC3*04 or IGLC3) is more stable than any of the other proteins, suggesting that the CL domain in this λ6 (IGLV 6–57) protein is destabilized by the VL domain. Sequence alignment of the kappa constant domain (IGKC) and the constant domain from AL-01-095 (IGLC3) found that the proteins are 41% identical. The level of sequence identity among the 5 lambda constant domains (IGLC) is quite high (93–99%), varying among them by a maximum of 7 amino acids. Our sequence comparisons between IGKC and IGLC indicate that the differences in stability behavior observed between Klimtchuk and co-workers and our current study could be easily explained by the differences in the sequence of the constant domains included in each study. In addition, the high sequence identity among IGLC sequences suggests that their properties are probably very similar.
Pioneering studies have been done using LCs from urine-derived BJP, many of them from multiple myeloma patients, however only a few works have been done using AL light chain proteins. It is important to point to the fact that that every LC is unique and AL amyloidosis is a very heterogeneous condition; in fact, “every patient could be considered to have their own disease since symptoms and outcome vary enormously” [20]. This could explain the differences found between our study and Klimtchuk’s study.
The FL proteins characterized in this study do not present reversible folding transitions, in contrast with their VL counterparts [1]. This has been observed previously with urine derived light chain proteins (BJPs) [10, 21, 22] and the recombinant version of protein AL-01-095. We performed a scan rate dependency study on AL-09 FL and κI FL to determine their thermodynamic parameters. We found that AL-09 FL and κI FL present similar thermodynamic stability compared to AL-09 VL (based on their Tm values). If we compare ΔH values, we find that AL-09 FL and κI FL have similar ΔH‡ values between each other at each scan rate and that these values are lower compared to the ΔHvan’t Hoff reported for AL-09 VL and κI VL. While these two sets of ΔH values are not truly comparable, it offers a view of the enthalpic contribution to the folding reaction of these two sets of proteins.
The thermal unfolding transitions followed by far-UV CD and intrinsic Tryptophan fluorescence presented a different degree of scan-rate dependence. This difference indicates that the unfolding -of AL-09 FL and to a lesser extent, κI FL- is a complex, uncoupled process that cannot be solely accounted for by the independent unfolding of the two protein domains.
Fibril formation for FL proteins occurred for both proteins with some similarities and differences from what was observed with their VL counterparts. One common feature we observed with VL and FL proteins is that low pH solution conditions (in this study, pH 4.0) promote fibril formation. This seems to be the case for both proteins. One notorious difference is that the presence of the CL makes κI FL to have a larger ThT fluorescence enhancement over AL-09 FL.
At pH 7.0 and pH 7.4, we observed some notorious stochastic behavior in the kinetics of fibril formation. Using the Acetate/Borate/Citrate buffer at pH 7.0, we were able to observe ThT fluorescence enhancement with AL-09 FL but not κI FL. In contrast, we observe ThT fluorescence enhancement at pH 7.4 in PBS buffer for both proteins.
We suggest that the Acetate/Borate/Citrate buffer presents properties that may cause different interactions at pH 7.0 in the case of κI FL, were not optimal for amyloid formation. These interactions may not be happening with phosphate buffer. Phosphate is a common anion used in the different experimental studies done with the Hofmeister series [23] and it is usually ranked as one of the anions that increase surface tension. Acetate is usually included in the Hofmeister series [24], while citrate is not commonly included and its position varies depending on the study [25, 26]; borate has been only found in very few studies [27]. Given the position of phosphate in the Hofmeister series, we propose that acetate and phosphate have similar effects on the protein electrostatic interactions, while citrate may have a neutral or deleterious effect with respect to phosphate and acetate. AL-09 FL is a highly amyloidogenic protein not affected by solution conditions in general, so the only protein that showed some effect between Acetate/Borate/Citrate and PBS was κI FL.
As aggregate populations observed during amyloid formation are heterogeneous and changing as a function of time, it is widely accepted that amyloid fibrils are formed following a nucleation-polymerization model; described by two phases: the so-called nucleation/lag phase, where “nucleus” or oligomers prone to aggregate are formed and accumulated; and the polymerization/elongation phase, where the “nucleus”, acting as a seed, elongates from small oligomers to fibrils by the recruiting and addition of monomers [3, 28, 29]. In vivo, it has been found that biological surfaces play a role in the amyloid formation process and involve a variety of accessory molecules such as lipids from cell lipidic-bilayer membranes [28] and glycosaminoglycans [30], to name a few. In addition, a body of data emerging from in vitro studies depict amyloid fibril growth as a hierarchical process with several “more-or-less” defined steps. However, it is posible that the reaction may progress into “dead end” routes, resulting in off-pathway aggregates such as amorphous aggregates or protofibrillar species [28, 29].
In this study, the mixture of clustered amyloid fibrils together with amorphous aggregation observed in the amyloid formation reactions suggests that either amorphous aggregation is a prerequisite for amyloid formation and the FL reactions are not as efficient as the VL reactions or there are possibly some off-pathway aggregation reactions occurring for FL.
One possibility for the difference in ThT fluorescence enhancement observed between AL-09 FL and κI FL is that AL-09 FL is forming fibrils very rapidly and saturating the ThT fluorescence signal through an inner filter effect. We based this possibility on the TEM images we acquired for both proteins in which we see no discernable differences in the fibril density between the two proteins. It is possible that the constant domain constitute the “fuzziness” between fibrils identified by TEM, as a result of recruiting material from the periphery during the fibril elongation. The presence of this material could preclude the ThT binding to amyloid fibrils and therefore affect the quantum yield of the ThT bonded to the fibrils.
In summary, the stability of the full length light chains, AL-09 FL and κI FL tested in this study are comparable to that reported for their variable domains, however the thermal unfolding process is strongly affected -becoming irreversible- by the presence of the constant domain which indicates that the (un)folding is kinetically controlled. In addition, the constant domain appears to play an important role in the modulation of the amyloidogenic properties of AL LCs.
Therefore, we conclude that the low thermodynamic stability is not sufficient for amyloid formation and other factors -like the kinetic stability or structural features- modulate the amyloid fibril formation process.
Highlights.
Thermal unfolding of light chains is irreversible and kinetically controlled
Both light chains present a scan rate dependence on the unfolding transition
The kappa constant domain presents similar stability to the variable domains.
Presence of the constant domain modulates light chains amyloidogenicity
Acknowledgements
This work was supported by National Institutes of Health R01 GM 071514 (MRA). We are also thankful for the financial support offered by Dr. Morie Gertz and the Seidler Professorship, the Mayo Foundation, and the generosity of amyloidosis patients and their families.
Abbreviations, symbols and terms
- LC
Light chain
- VL
variable Light chain domain
- CL
Constant domain
- FL
Full length light chain
- BJP
Bence-Jones proteins
- ThT
Thioflavin T
- T*
temperature reference independent of the applied scan rate
- ΔH‡
enthalpy reference independent of the applied scan rate
- Tm
melting temperature
- Tmapp
apparent melting temperature
- CD
Circular dichroism
- FI
fluorescence intensity
- t50
the time it takes to complete 50% of the fibril formation reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Baden EM, et al. Altered dimer interface decreases stability in an amyloidogenic protein. The Journal of biological chemistry. 2008;283(23):15853–15860. doi: 10.1074/jbc.M705347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin DJ, Ramirez-Alvarado M. Comparison of amyloid fibril formation by two closely related immunoglobulin light chain variable domains. Amyloid : the international journal of experimental and clinical investigation : the official journal of the International Society of Amyloidosis. 2010;17(3–4):129–136. doi: 10.3109/13506129.2010.530081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blancas-Mejia LM, Ramirez-Alvarado M. Systemic amyloidoses. Annual review of biochemistry. 2013;82:745–774. doi: 10.1146/annurev-biochem-072611-130030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poshusta TL, et al. Thermal stability threshold for amyloid formation in light chain amyloidosis. International journal of molecular sciences. 2013;14(11):22604–22617. doi: 10.3390/ijms141122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glenner GG, et al. Physical and chemical properties of amyloid fibers. II. Isolation of a unique protein constituting the major component from human splenic amyloid fibril concentrates. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 1969;17(12):769–780. doi: 10.1177/17.12.769. [DOI] [PubMed] [Google Scholar]
- 6.Lavatelli F, et al. Amyloidogenic and associated proteins in systemic amyloidosis proteome of adipose tissue. Molecular & cellular proteomics : MCP. 2008;7(8):1570–1583. doi: 10.1074/mcp.M700545-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vrana JA, et al. Classification of amyloidosis by laser microdissection and mass spectrometry-based proteomic analysis in clinical biopsy specimens. Blood. 2009;114(24):4957–4959. doi: 10.1182/blood-2009-07-230722. [DOI] [PubMed] [Google Scholar]
- 8.Solomon A, et al. Light chain-associated amyloid deposits comprised of a novel kappa constant domain. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(16):9547–9551. doi: 10.1073/pnas.95.16.9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamoto K, et al. The amyloid fibrils of the constant domain of immunoglobulin light chain. FEBS Letters. 2010;584(15):3348–3353. doi: 10.1016/j.febslet.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 10.Klimtchuk ES, et al. The critical role of the constant region in thermal stability and aggregation of amyloidogenic immunoglobulin light chain. Biochemistry. 2010;49(45):9848–9857. doi: 10.1021/bi101351c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levinson RT, et al. Role of mutations in the cellular internalization of amyloidogenic light chains into cardiomyocytes. Scientific reports. 2013;3:1278. doi: 10.1038/srep01278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tischer A, Cruz MA, Auton M. The linker between the D3 and A1 domains of vWF suppresses A1-GPIbalpha catch bonds by site-specific binding to the A1 domain. Protein science : a publication of the Protein Society. 2013;22(8):1049–1059. doi: 10.1002/pro.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blancas-Mejia LM, et al. Kinetic control in protein folding for light chain amyloidosis and the differential effects of somatic mutations. Journal of molecular biology. 2014;426(2):347–361. doi: 10.1016/j.jmb.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atkins PW. Physical Chemistry. New York: Freeman and Company; 1994. The rates of chemical Reactions; pp. 861–897. [Google Scholar]
- 15.Wall J, et al. Thermodynamic instability of human lambda 6 light chains: correlation with fibrillogenicity. Biochemistry. 1999;38(42):14101–14108. doi: 10.1021/bi991131j. [DOI] [PubMed] [Google Scholar]
- 16.Kong F, King J. Contributions of aromatic pairs to the folding and stability of long-lived human gammaD-crystallin. Protein science : a publication of the Protein Society. 2011;20(3):513–528. doi: 10.1002/pro.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batey S, Nickson AA, Clarke J. Studying the folding of multidomain proteins. HFSP journal. 2008;2(6):365–377. doi: 10.2976/1.2991513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Good NE, et al. Hydrogen ion buffers for biological research. Biochemistry. 1966;5(2):467–477. doi: 10.1021/bi00866a011. [DOI] [PubMed] [Google Scholar]
- 19.Lindberg DJ, et al. Steady-state and time-resolved Thioflavin-T fluorescence can report on morphological differences in amyloid fibrils formed by Abeta(1–40) and Abeta(1–42) Biochemical and biophysical research communications. 2015;458(2):418–423. doi: 10.1016/j.bbrc.2015.01.132. [DOI] [PubMed] [Google Scholar]
- 20.Enqvist S, Sletten K, Westermark P. Fibril protein fragmentation pattern in systemic AL-amyloidosis. The Journal of pathology. 2009;219(4):473–480. doi: 10.1002/path.2607. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y, et al. Thermodynamic modulation of light chain amyloid fibril formation. The Journal of biological chemistry. 2000;275(3):1570–1574. doi: 10.1074/jbc.275.3.1570. [DOI] [PubMed] [Google Scholar]
- 22.Sikkink LA, Ramirez-Alvarado M. Biochemical and aggregation analysis of Bence Jones proteins from different light chain diseases. Amyloid : the international journal of experimental and clinical investigation : the official journal of the International Society of Amyloidosis. 2008;15(1):29–39. doi: 10.1080/13506120701815324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cacace MG, Landau EM, Ramsden JJ. The Hofmeister series: salt and solvent effects on interfacial phenomena. Quarterly reviews of biophysics. 1997;30(3):241–277. doi: 10.1017/s0033583597003363. [DOI] [PubMed] [Google Scholar]
- 24.Collins KD. Ion hydration: Implications for cellular function, polyelectrolytes, and protein crystallization. Biophysical chemistry. 2006;119(3):271–281. doi: 10.1016/j.bpc.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Cremer PS. Interactions between macromolecules and ions: The Hofmeister series. Current opinion in chemical biology. 2006;10(6):658–663. doi: 10.1016/j.cbpa.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 26.Zhao H, et al. Hofmeister series of ionic liquids: kosmotropic effect of ionic liquids on the enzymatic hydrolysis of enantiomeric phenylalanine methyl ester. Tetrahedron: Asymmetry. 2006;17:377–383. [Google Scholar]
- 27.Erichsen Jones JR. A study of the relative toxicity of anions with Polycelis nigra as test animal. J. Exp. Biol. 1941:170–181. [Google Scholar]
- 28.Cecchi C, Stefani M. The amyloid-cell membrane system. The interplay between the biophysical features of oligomers/fibrils and cell membrane defines amyloid toxicity. Biophysical chemistry. 2013;182:30–43. doi: 10.1016/j.bpc.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nature reviews. Molecular cell biology. 2014;15(6):384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 30.Blancas-Mejia LM, et al. Differential effects on light chain amyloid formation depend on mutations and type of glycosaminoglycans. The Journal of biological chemistry. 2015;290(8):4953–4965. doi: 10.1074/jbc.M114.615401. [DOI] [PMC free article] [PubMed] [Google Scholar]