Abstract
In both Xiphophorus fishes and humans, males are reported to have a higher incidence of melanoma than females. To better understand sex specific differences in the molecular genetic response to UVB, we performed RNA-Seq experiments in skin of female and male Xiphophorus maculatus Jp 163 B following UVB doses of 8 or 16 kJ/m2 exposure. Male X. maculatus differentially express a significantly larger number of transcripts following exposure to 16 kJ/m2 UVB (1,293 genes) compared to 8 kJ/m2 UVB (324 genes). Female skin showed differential gene expression in a larger number of transcripts following 8 kJ/m2 UVB (765) than did males; however, both females and males showed similar numbers of differentially expressed genes at 16 kJ/m2 UVB (1,167 and1,293, respectively).
Although most modulated transcripts after UVB exposure represented the same dominant pathways in both females and males (e.g., DNA repair, circadian rhythm, and fatty acid biosynthesis), we identified genes in several pathways that exhibited opposite modulation in female vs. male skin (e.g., synaptic development, cell differentiation, wound healing, and glucose metabolism). The oppositely modulated genes appear related through uncoupling protein 3 (UCP3) that is involved with regulation of fatty acid oxidation and serves to balance glucose and lipid metabolism. Overall, these results identify gender specific differences in UVB induced genetic profiles in the skin of females and males and show female and male X. maculatus respond to UVB differently through pathways involved in reactive oxygen species, wound healing, and energy homeostasis.
Keywords: Ultraviolet light, UVB, gene expression, Xiphophorus, melanoma, RNA-Seq, transcriptome
Introduction
Although the effects of ultraviolet (UV) light induction of DNA damage have been studied and well documented, global shifts in genetic profiles that occur in skin upon UV exposure remain to be defined. Due to atmospheric filtering the primary UV radiation wavelengths reaching the earth’s surface include UVB (290- 320 nm), thought responsible for direct DNA damage in the outer dermal layers of animals, and UVA (320-400 nm) that was often considered weakly absorbed by DNA, but a source of free radicals that may produce oxidative DNA damage. Reports suggesting a role of various UV wavelengths in association with skin cancer are largely based on the genetics and enzymology of damage specific DNA repair mechanisms. However, it is clear that in addition to induction of DNA repair pathways, UV exposure induces a plethora of other genetic pathways involving functions such as cell cycle arrest (Gujuluva et al., 1994), apoptosis (Banerjee et al., 2005), immune response (Kripke, 1994), and fatty acid biosynthesis (Kim et al., 2010). As global genetic profiling techniques improve, primary concepts of the molecular genetic etiology of skin cancer are likely to be reformulated.
Both direct damage to DNA (i.e., primarily cyclobutane pyrimidine dimers and 6-4 pyrimidine pyrimidone photoproducts) and indirect DNA damage (due to photosensitizers and singlet oxygen reactions) have been associated with induced skin cancer in Xiphophorus interspecies hybrids (Fernandez et al., 2012; Meador et al., 2007; Mitchell et al., 2001; Setlow et al., 1993; Wood et al., 2006). It has been shown that melanoma development in Xiphophorus, like humans, has a male bias (Fernandez, 2010; Siciliano et al., 1971). However, differences in the global genetic response to UVB exposure in both females and males for this model has not been studied. Sex specific differences in both spontaneous tumorigenesis and in the response to various environmental stimuli have been documented in many fish models and in humans. For example, in medaka, (Oryzias latipes), spontaneous liver tumor incidence increased substantially with age and was shown higher in female fish compared to males (Masahito et al., 1989). Female and male guppies (Poecilia reticulata) and medaka were used to study sensitivity to three distinct toxins and it was determined that upon exposure to 2,2- bis(bromomethyl)-1,3-propanediol (BMP) males showed an increased incidence of hepatocellular adenomas and carcinomas compared to females for both fish types (Kissling et al., 2006). Human sex specific mortality rates for most types of cancer reveal men have a higher incidence of cancer and lower post diagnosis survival rates than females (Cook et al., 2011). This suggests gender specific assessments should be considered when using experimental tumor models to gain a better understanding of the underlying genetic mechanisms that might explain observed differences in tumor incidences.
In this study, we examine molecular genetic differences in the skin of female and male Xiphophorus maculatus Jp 163 B to UVB (311 nm) exposure. X. maculatus backcross hybrids were introduced as a melanoma model in the 1930’s and have since been used extensively to investigate the genetics underlying both spontaneous (Gordon, 1931) and induced melanoma (Nairn et al., 1996; Setlow and Woodhead, 1994; Setlow et al., 1989). It has been reported (Kazianis et al., 2001; Nairn et al., 1996; Walter and Kazianis, 2001) that when F1 interspecies hybrids produced from the cross (X. maculatus Jp 163 B × X. hellerii) are backcrossed to the X. hellerii parent, the backcross hybrid progeny are susceptible to UVB induced melanomagenesis where 18% of total progeny, or up to 40% of the pigmented progeny, develop tumors within 9 months.
Detailed assessment of the global effects of UV exposure on gene expression in the skin of female and male Xiphophorus fishes that have been utilized as an inducible experimental tumor model of melanoma has not been performed. Herein we present RNA-Seq results that identified shared and oppositely regulated responses by select sets of genes and pathways in the skin of female and male X. maculatus Jp 163 B. We show the overall skin response after UVB exposure in females and males is similar in magnitude at higher doses; however, females require a lower dose for onset of global genetic remodeling that is observed in both sexes at the higher dose. In addition, upon UV exposure male skin appears to shift toward energy metabolism via the AMPK pathway, while females significantly down regulate genes in this pathway.
Methods
2.1. Fish Used
All fish were supplied by the Xiphophorus Genetic Stock Center, Texas State University, San Marcos, TX 78666 (http://www.xiphophorus.txstate.edu). The Xiphophorus maculatus Jp 163 B (pedigree 104D) used in this study were mature 9-month-old female and male siblings derived from the same brood in the 104th inbred generation.
2.2. UV Exposure
UV exposure was performed as described previously (Walter et al., 2014; Yang et al., 2014). Briefly, fish were placed in 125 mL flasks containing 100 mL of filtered aquaria water and kept in the dark 12 hours prior to UV exposure. Fish were removed from the dark and transferred to UV transparent cuvettes (9 cm × 7.5 cm × 1.5 cm) containing 90 mL of filtered aquaria water. The exposure cuvettes were placed in a 77 cm (length) × 41 cm (height) × 36 cm (depth) wooden box. Air was circulated through the box with 15.5 cm high-speed fans mounted at the bottom and on each end of the box to maintain ambient temperature (approximately 24°C). The fish in the exposure cuvettes were suspended between two banks (total of 4 lights) unfiltered 24-inch narrow spectrum UVB lamps (Philips UVB TL 20W/01 RS SLV) emitting principally 311 nm light, that were mounted horizontally on each side of the exposure box. Fluence was determined to be 12.2 J m-2 s-1 on each side of the chamber using an IL-1400A Radiometer/Photometer coupled to a SEL 240/UVB detector containing a 280 nm Sharp Cutoff Filter (International Light, Newburyport, MA). Fish were exposed in duplicate (2 female and 2 male fish for each exposure) to 0 kJ/m2, 8 kJ/m2, or 16 kJ/m2 of UVB by varying the amount of time in the chamber. After exposure, the fish were returned to 125 mL flasks containing 100 mL of filtered aquaria water and returned to the dark for 6 hours to allow time for transcriptional remodeling before being euthanized and sacrificed for RNA isolation.
2.3. RNA Isolation and RNA-Seq
RNA was isolated from skin using a TRI Reagent (Sigma Inc., St Louis, MO, USA) extraction followed by the Qiagen RNeasy (Qiagen, Valencia, CA, USA) isolation protocol; each sample was independently maintained throughout isolation and sequencing. Briefly, the skin was flash frozen (ethanol-dry-ice bath) in 300 μL TRI Reagent immediately upon dissection. The skin was homogenized using a handheld tissue disruptor, and an additional 300 μL of TRI Reagent was added to the homogenate, followed by 120 μL of chloroform. The samples were vigorously shaken for 15 sec and then phases partitioned by centrifugation (12,000×g for 5 min at 4°C). The aqueous phase containing th e nucleic acids was transferred to a new microcentrifuge tube and extracted a second time (300 μL TRI Reagent and 60 μL chloroform). After extraction, the nucleic acids were precipitated with 500 μL 70% EtOH. The RNA was then purified using a Qiagen RNeasy mini RNA kit following the manufacturer’s protocol. Residual DNA was eliminated with on-column DNase treatment at 25°C f or 15 min. RNA quality was assessed by gel electrophoresis (1% TAE agarose gel in 1X TAE running buffer) and the concentration of RNA determined with a Qubit 2.0 fluorometer (Life Technologies, Grand Island, NY, USA).
Prior to RNA sequencing, RNA quality scores were determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), and all samples had a RIN score above 8. The X. maculatus Jp 163 B skin RNA samples were sequenced (75 bp, paired-end [PE] reads) using the Illumina TruSeq Stranded mRNA Library Prep Kit on a HiSeq 2000 platform (Illumina, Inc., San Diego, CA, USA) at the M.D. Anderson Cancer Center Science Park Next-Generation Sequencing Facility (Smithville, TX).
The raw sequencing data were trimmed and filtered using the vendor supplied post sequence routines with a per position quality score cutoff of 20 with a custom Perl script, available upon request, (Garcia et al., 2012) that also removed low-scoring sections of each read while preserving the longest remaining fragments as previously described (Yang et al., 2014). The raw filtered reads were truncated based on similarity to library adaptor sequences using custom Perl scripts (Garcia et al., 2012) followed by merging of overlapping PE reads using FLASH (Magoč and Salzberg, 2011). FastQC was then used to visually assess the quality of the filtered reads to identify any potential deficiencies within the data for each sample (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
2.4. Computational and Pathway Analyses
GSNAP (Wu and Nacu, 2010) was used to map the filtered reads to the X. maculatus reference transcriptome (Ensembl v71) containing 20,219 transcripts. The percentage of reads mapped and unmapped as well as the nucleotide coverage was identified using SAMtools flagstat and depth respectively (Li et al., 2009). Differentially modulated genes were then determined using the R-Bioconductor (www.bioconductor.org) package DESeq (Anders and Huber, 2010) with a fold- change cutoff of ±2 (p-adj ≤ 0.05). DESeq was also used to identify differentially modulated transcripts between female basal (0kJ/m2) samples and male basal samples to rule out sex specific variation when comparing treated samples against the basal (0 exposure).
X. maculatus female and male responses to UVB were assessed using Venn diagrams generated with the Bioconductor R package VennDiagram (Hanbo Chen, 2011). Gene molecular function and/or biological processes of the gene classes identified in the Venn diagrams were analyzed using both the gene ontology (GO) enrichment in ConsensusPathDB (http://cpdb.molgen.mpg.de/CPDB) and DAVID (http://david.abcc.ncifcrf.gov).
In addition, pathway analysis was visualized using the Kegg pathway database (www.genome.jp/kegg/pathway.html) and heat maps were generated with Bioconductor using the R packages RColorBrewer version 1.0-5 (Neuwirth, 2011), gplots version 2.14.1 (Warnes, 2012) and Cluster 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/) to assess the overall shared genetic response between the female and male fish and identify potential gene ontology clusters. Heat maps were also used to qualify and cluster genes that did not segregate within Venn shared regions and/or appeared to exhibit opposite modulation in females and males in response to UVB exposure.
2.5. Quantitative real time PCR
Quantitative real time PCR (qRT-PCR) was performed as previously described (Yang et al., 2014; Walter et al., 2014). Briefly, transcripts (14 total) that exhibited differential modulation in response to UVB by RNA-Seq analyses were confirmed using qRT-PCR. ABI High Capacity cDNA Reverse Transcription kit (Applied Bioscience, Carlsbad, CA) was employed to generate cDNA from skin RNA. Briefly, cDNA synthesis was performed with 100 mM dNTPs and 10X RT random primers, RNase inhibitor, and Multiscribe Reverse Transcriptase mixed with 1.5 μg of RNA in a 20 μL reaction. Thermocycler conditions for cDNA synthesis were 25°C for 10 min, 37°C for 120 min, and 85°C for 5 min. Each cDNA reaction was diluted (480 μL of 0.1% DEPC water) to a final volume of 500 μL and quantified using a Qubit 2.0 fluorometer.
Each sample was analyzed with qRT-PCR on Applied Biosystems 7500 Fast Real Time PCR System (Applied Biosystems, Carlsbad, CA, USA). Primers used for qRT-PCR were designed using Geneious (Biomatters Ltd., Auckland, New Zealand) software version 6.1. Primer design included melting temperatures (Tm) between 60 and 62°C (50 mM Na, 3 mM K, and 0.8 mM dNTPs), lengths of 18–25 nt with 55–60% GC content, and not more than 1°C between Tms of each primer in a set. Target amplicons were limited to 100–150 bps in length, and were designed to amplify across at least one intron-exon junction. Primers specific for the Xiphophorus transcripts (Table 1) were tested for efficiency and only primers with efficiency values between 70 and 120% were utilized in downstream analyses. Each primer set was used to test the relative expression of each gene from cDNAs (15 ng) produced from X. maculatus skin in four qRT-PCR technical replicates. All plates were run using the SYBR Green protocol as previously described (Walter et al., 2014; Yang et al., 2014) wherein amplification for each target is normalized against the 18S rRNA transcript to determine ΔCT values. UV modulated fold changes are relative to 0-point control (ΔΔCT) values.
Table 1.
Gene targets and sequences for primers used in qRT-PCR data confirmation.
| Gene Name | Forward Sequence 5′-3′ | Reverse Sequence 5′-3′ |
|---|---|---|
| klhl38b | TTCATCTTCGTCCTGGGTGG | CGGCTTCCCATTGGTTGAAC |
| ucp3 | ATTGCTGACCTCATCACCTTTCC | GTTTTGATGGTTCCGAACACACC |
| inpp4aa | TGTGCGACTCCAAGCTGC | GGCGAGAGGGGGAAGAAAAC |
| ovgp1 | GGTGGGATTCGACAACAAGC | TCCAGAGACCAGACCAATGC |
| zmat3 | AAGATGGCAACGAGTACCGAC | TCGGCCTCGGGTTGTAATAC |
| tacr2 | CATGGTAGACTGGCCGGATG | GCAGGGGGAGCAAGTAGATC |
| itih4 | TTCCAGCTCCACTGCAACTG | GTCCTCTCCAGGGCAAAGTC |
| pip5kl1 | AGGACCTGAACTTTGAAGGCC | CCAGGAAGCTGGTGTCGATC |
| ptn | AACCACCTGCTGAGTTACGG | GATTGTTGTCCTGGGTTGGC |
| hmgala | GGTCCCCAACTCCAAAGAGG | CTGCGTTTTCCTCCTGCAG |
| wdr76 | TCGCTGCTGGAGACAAGTG | GGATGACACCTGGAGAACGC |
| hhipl1 | CTTAGCCACAGGAGTCCCAAG | CATTTTCCAGGTGGTGCTCG |
| cpd | CTCTCAGCCAGCTCTCCC | ACCAGCTCCTCGATGAAGG |
| 6-4 | TTGGGGCACTCAAAGACCTG | GAACACATCTTCTGGTTTCCCC |
Results
3.1 Next-Generation Sequencing Analysis
After filtering Illumina High-Seq raw sequencing data, the filtered reads were mapped to the X. maculatus reference transcriptome (Ensembl v71). The number of reads mapped was similar for both females and males and included ≈60% of the total raw reads after post sequence filtration (Table 2). Thus, differences observed between female and males are not due to discrepancies in the number of reads mapped to the X. maculatus reference transcriptome.
Table 2.
Read depth and RNA-Seq statistics for UVB exposed female and male skin sample reads mapped to the X. maculatus Jp 163 B reference transcriptome of 20,219 transcripts.
| Sex | UV Dose (kJ/m2) | Raw Data
|
Processed Reads
|
Average Coverage (X) | |||
|---|---|---|---|---|---|---|---|
| Filtered Reads (M) | Read Length (B) | Reads Mapped (M) | Reads Unmapped (M) | % Mapped | |||
| Female | 0-A | 55.3 | 2.5 | 31.9 | 23.4 | 57.7 | 51 |
| 0-B | 58.9 | 2.6 | 34.6 | 24.3 | 58.7 | 54 | |
| 8-A | 66.2 | 3.0 | 39.2 | 27.0 | 59.2 | 60 | |
| 8-B | 49.1 | 2.2 | 29.0 | 20.1 | 58.9 | 47 | |
| 16-A | 46.8 | 2.1 | 28.3 | 18.4 | 60.6 | 44 | |
| 16-B | 56.1 | 2.5 | 34.0 | 22.1 | 60.6 | 53 | |
| Male | 0-A | 53.0 | 2.4 | 30.4 | 22.6 | 57.4 | 47 |
| 0-B | 41.3 | 1.8 | 24.1 | 17.1 | 58.5 | 39 | |
| 8-A | 47.7 | 2.1 | 27.7 | 20.0 | 58.0 | 45 | |
| 8-B | 49.4 | 2.2 | 29.2 | 20.2 | 59.1 | 46 | |
| 16-A | 46.6 | 2.1 | 27.6 | 18.9 | 59.4 | 43 | |
| 16-B | 52.6 | 2.1 | 31.4 | 21.1 | 59.8 | 49 | |
M = Million, B = Billion
Differences in unexposed X. maculatus female and male skin were determined by comparing unexposed (0 kJ/m2) female skin to unexposed male skin reads using DESeq. Seventy-one genes were identified as exhibiting differential expression (Table 3) in the unexposed fish based on gender (± 2-fold cut-off, p-adj < 0.05). These 71 genes did not readily cluster into distinct functional or GO groups but rather appeared to represent many disparate biological functions. None of the 71 genes (Table S1) are involved with previously reported male UVB responses (Yang, et al., 2014).
Table 3.
Number of modulated genes at ± 2-fold (p-adj < 0.05) in unexposed female and male X. maculatus samples (Table S1). Differential modulation is calculated as the ratio of female to male reads.
| Modulation Direction | 0 kJ/m2 |
|---|---|
| Up | 20 |
| Down | 51 |
| Total Modulation | 71 |
Overall in UVB exposed samples, females were observed to modulate significantly more genes at 8 kJ/m2 UVB than males (765 in female vs. 324 in males), however, the female and male responses following 16 kJ/m2 UVB were similar in the number of transcript differentially expressed (1,167 in females vs. 1,293 in males) (Table 4).
Table 4.
Number of modulated genes at ± 2-fold (p-adj < 0.05) in female and male X. maculatus when exposed to 8 or 16 kJ/m2 UVB.
| Sex | Modulation Direction | 8 kJ/m2 | 16 kJ/m2 |
|---|---|---|---|
| Female | Up | 450 | 696 |
| Down | 315 | 471 | |
|
|
|||
| Total Modulation | 765 | 1167 | |
| Male | Up | 176 | 688 |
| Down | 148 | 605 | |
|
|
|||
| Total Modulation | 324 | 1293 | |
3.2 Shared Genetic Response to UVB Exposure
The degree of genetic response and the levels of shared response in female and male X. maculatus skin after UVB exposure is presented in Figure 2. The total number of genes shared between females and males at both 8 kJ/m2 (141 genes or 29% up and 80 genes or 21% down) and 16 kJ/m2 (471 genes or 52% up and 231 genes or 27% down) UVB exposure are shown.
Figure 2.
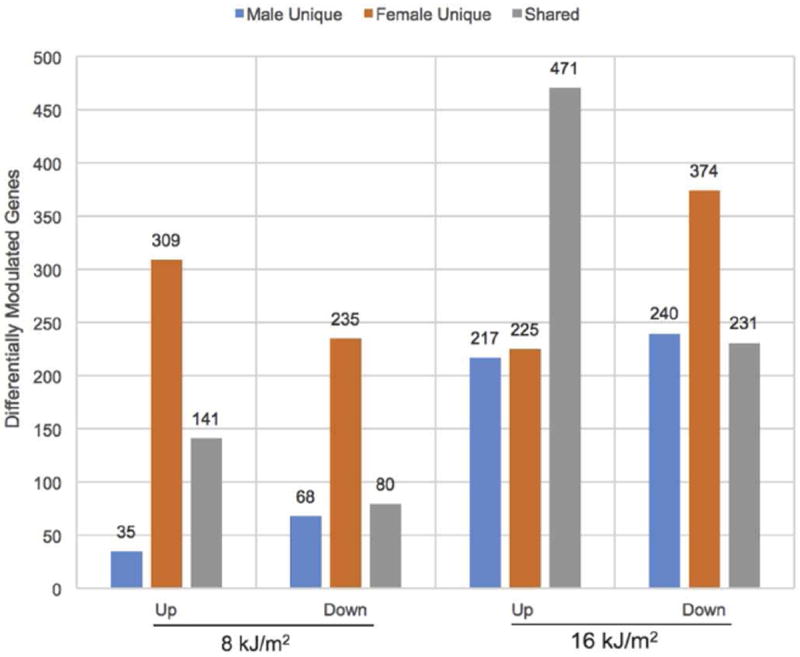
Bar graph showing ± 2-fold (p-adj ≤ 0.05) differentially expressed genes in X. maculatus female (orange) and male (blue) skin following 8 or 16 kJ/m2 UVB exposure. The grey shaded areas represent the numbers of shared genes responding to the indicated UVB exposure for female and male skin.
Although females at 8 kJ/m2 responded more robustly than males (450 up and 315 down modulated genes in females vs. 176 and 148 in males, respectively), the number of genes affected by UVB is similar for both genders at 16 kJ/m2 (696 genes up and 605 genes down in females vs. 688 up and 471 down modulated in males). In addition, the fraction of genes shared (30% up following 8 kJ/m2 compared to 52% following 16 kJ/m2) almost doubles in the up-modulated gene set (1.7 fold greater than 8 kJ/m2) following 16 kJ/m2 UVB exposure. However, the fraction of shared genes down modulated after UV exposure (20% in females and 27% in males) remains relatively constant.
The shared UVB responsive genes for female and male skin at each dose were compared to identify potential functional gene clusters. Figure 3 presents, 63 down and 134 up modulated genes (Table S2) that were shared in both UV doses and for both genders. The bracketed GO functional clusters in Figure 3 are identified.
Figure 3.
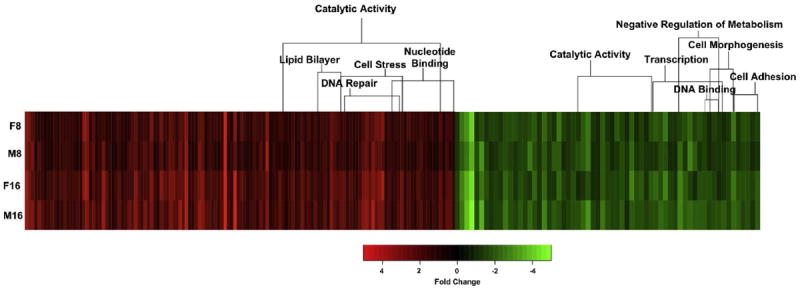
Heat map representing shared gene modulation in female and male X. maculatus skin at 8 (F8 and M8) and 16 kJ/m2 (F16 and M16). Shown are 134 up modulated (red) and 59 down-modulated (green) genes that exhibited a shared UVB response in the skin of both genders. Bracketed genes are functional clusters based on DAVID Gene Ontology analysis. Genes involved with cell stress, DNA repair, and nucleotide binding appear up modulated, while functional classes down modulated are cell morphogenesis, cell adhesion, and metabolism.
Although females modulated expression of more genes at 8 kJ/m2 UVB than males, the gender shared gene families at both 8 and 16 kJ/m2 UVB exposure are similar to those previously reported for X. maculatus males using the same dose regimen (Yang et al., 2014). ConsensusPathDB was used to identify potential genetic associations from the shared up-modulated genes and a± 2-fold cut-off was used for functional and pathway analysis. One hundred twenty-one shared genes were queried and associations were determined and linked based on functional and pathway analysis (Kamburov et al., 2009). The121 queried genes clustered into represented gene networks indicating that they were either related by protein interactions (orange lines), gene regulatory interactions (purple lines), or biochemical interactions (green lines) (Figure 4).
Figure 4.
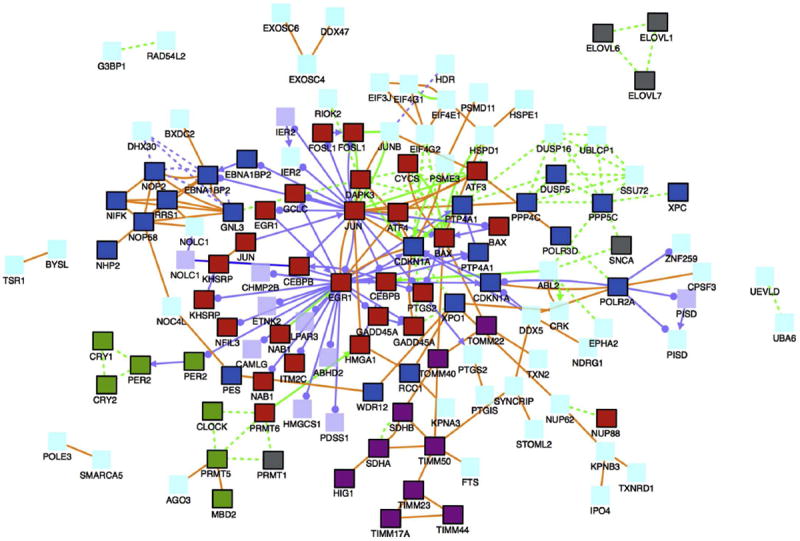
Gene and protein associations between shared female and male up-modulated genes (± 2-fold) in X. maculatus skin after 16 kJ/m2 UVB exposure. The gene network is based on the ConsensusPathDB (CPDB; Max Planck Institute for Molecular Genetics) gene analysis software. In total, 121 genes were queried and associations were found as follows: protein interactions (orange lines), gene regulatory interaction (purple lines), and biochemical interaction (green lines). Red boxes are genes involved in apoptosis, green boxes are genes involved with circadian rhythm, grey boxes are genes involved with fatty acid biosynthesis, dark purple boxes are mitochondria associated genes, and dark blue boxes are genes involved with an induced response to toxins, extracellular stimulus, or UV exposure including DNA repair. Other queried genes are represented by light purple boxes and proteins by light blue boxes.
The female and male up-modulated shared genetic response at 16 kJ/m2 UVB is dominated by 4 pathways: apoptosis (red boxes), cell cycle repression due to DNA damage (dark blue boxes), circadian rhythm (green boxes), and fatty acid/lipid biosynthesis (grey boxes, Figure 4). Many expected light responsive genes and pathways, such as circadian rhythm, DNA repair and apoptosis, were observed following 16 kJ/m2 UVB.
3.3 Opposing Genetic Responses to UVB Exposure
Figure 2 shows the number of genes at 8 and 16 kJ/m2 UVB that are not shared between females (544, 599) and males (103, 457). To identify these sex specific genes, the lists of oppositely modulated (p-adj < 0.05 and ± 2-fold) genes at both 8 and 16 kJ/m2 UVB were cross-compared. For example, the ± 2-fold up modulated genes unique to males at 8 kJ/m2 were compared to all of the down modulated genes in females including those that were excluded from prior analysis due to the statistical cut-off (p-adj < 0.05). Only those genes that were oppositely modulated in females and males at both doses were included for functional analysis (Table S3). Genes that showed opposing trends in modulation in females and males were clustered using the DAVID GO and Kegg employed to identify potential genetic pathways, as shown in Figure 5.
Figure 5.
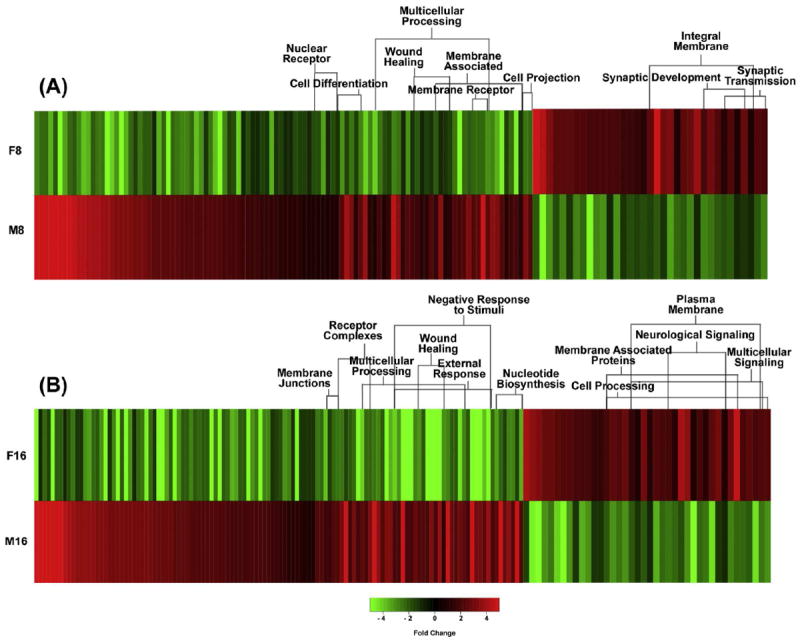
Heat map representing genes that exhibit opposing modulation in female and male X. maculatus skin following UVB exposure. Panel A shows the opposing modulated genes (141) after 8 kJ/m2 (F8 and M8) UVB exposure while panel B shows opposing modulated genes (160) after 16 kJ/m2 (F16 and M16) UVB exposure.
Following 8 kJ/m2, 141 genes showed opposing modulation in female andmale fish skin, while 160 genes were oppositely modulated following 16 kJ/m2 UVB. Of the gene clusters identified, the primary unshared genetic responses were involved in wound healing, neurological synapse, cell differentiation, and multicellular processing (Figure 5). At 8 kJ/m2, females showed an up modulation of synaptic development, and synaptic transmission, and a down modulation of wound healing, cell differentiation, and cell projection. At 16 kJ/m2, females showed an up modulation of membrane associated proteins, multicellular signaling, and neurological signaling, and a down modulation of wound healing, multicellular processing, external response, and nucleotide biosynthesis. Males had the opposite genetic response to females at both 8 and 16 kJ/m2 with an up modulation of wound healing, cell differentiation, multicellular processing and external response. In addition, the males had a down modulation of synaptic development and synaptic transmission at 8 kJ/m2, and a down modulation of neurological signaling, multicellular signaling, and cell processing at 16 kJ/m2.
3.4 Confirmation of RNA-Seq Results
To validate the RNA-Seq data, qRT-PCR was performed on 14 selected transcripts for both males and females, and 13 of those confirmed the RNA-Seq trends for females and 12 confirmed for males. Transcripts were selected to validate the different observed data trends and GO categories observed; shared UV responsive genes (4 transcripts), oppositely modulated at one dose 8 or 16 kJ/m2 (2 transcripts), and oppositely modulated at both UVB doses (8 transcripts). Statistical validations were performed using a t-test (p-value < 0.05) for exposed versus unexposed; fold-changes reported as ≤ ± 1.6 fold were determined to be statistically insignificant compared to the unexposed samples (Figure 6).
Figure 6.
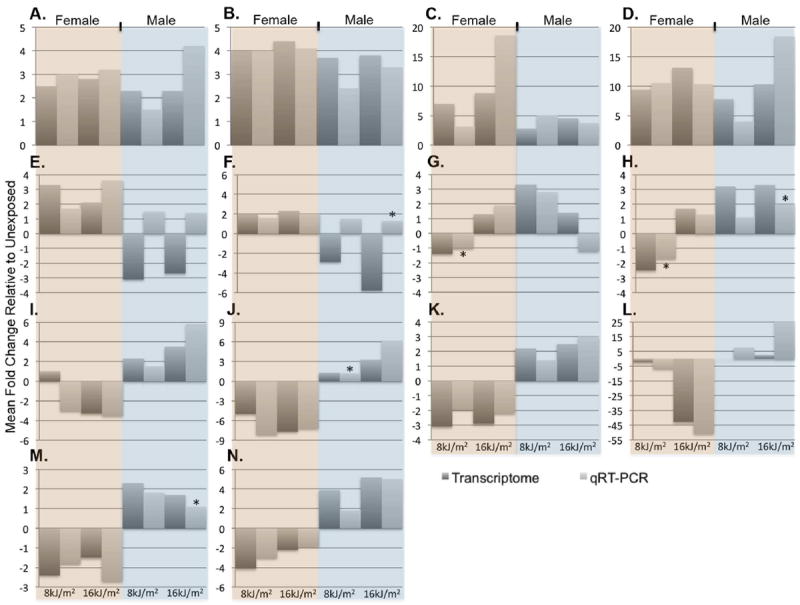
Comparison of RNA-Seq and qRT-PCR data. RNA-Seq (dark gray bars), qRT-PCR light gray bars) run on 14 selected transcripts for female (orange) and male (blue) X. maculatus skin samples. Each bar represents 3 biological replicates averaged and reported as fold change differences relative to 3 unexposed biological replicates. Of the transcripts tested, 13 of 14 gene targets confirmed the RNA-Seq data in females and were determined to be statistically significant p-value < 0.05 (* denotes those that were not statistically significant); 12 of 14 transcripts were confirmed in males. Genes tested, A- cpd, B- 6-4 PHR, C- hmgala, D- wdr76, E- ch211-170j16, F- ch211-136a13, G- tacr2, H- pip5kl1, I- klh38b, J- ucp3, K- zmat3, L- zgc-112265, M- inpp4aa and N- ovgp1.
Discussion
While Xiphophorus UVB induced melanoma and gene expression have been performed (Kazianis et al., 2001; Nairn et al., 1996; Yang et al., 2014), sex specific responses in the skin of this model species has yet to be determined. Hormonal differences have been observed and linked to a sex specific bias in female vs. male tumor incidence for Xiphophorus backcross hybrids exposed to UVB (Mitchell et al., 2014; Patton et al., 2010; Siciliano et al., 1971). This gender difference in UVB induced melanoma incidence in the Xiphophorus model, mirrors human incidence thus suggesting a molecular genetic disposition and response to UVB that has yet to be defined.
Shared Genetic Response to UVB Exposure
Following UVB exposure, the overall male and female shared genetic response observed at 16 kJ/m2 is consistent with previous data reported in mammals and fish including modulation of known UVB responsive genes such as cdkn1a, xpc, egr1, gadd45a, and circadian rhythm genes, clock and per2 (de Feraudy et al., 2010; Matsumoto et al., 2015; Meador et al., 2007; Sherr and Roberts, 1999; Yang et al., 2014). Both females and males show a greater genetic response at 16 kJ/m2 than at 8 kJ/m2. This trend was previously observed in male X. maculatus skin following UVB exposure at doses of 8, 16 and 32 kJ/m2 (Yang et al., 2014) where the number of modulated genes doubled from 8 kJ/m2 to the 16 kJ/m2 UVB dose. The experiments using male fish presented in Yang et al. (2014) were independent having been performed over one year prior to the data presented herein; yet 93% of the male fold-changes for up-modulated transcripts, and all (100%) of the down modulated transcripts, are in agreement at the 8 kJ/m2 dose. In addition, 89% of the up-modulated transcripts and 100% of down-modulated transcripts observed in the experiments reported herein were also similarly observed for the 16 kJ/m2 dose in the previous report (Yang et al., 2014).
While the majority of the genes in females and males were shared for both UVB doses tested, females exhibited a more robust initial response at 8 kJ/m2 than did males (Figure 2). Excessive UV exposure can lead to acute erythema (sunburn), and it has been documented in burn situations that circulating sex hormones, estrogen and testosterone, can impact the immune response (Bird et al., 2008). It has also been reported that during the reproductive years, females have a more robust response to injury than males, due to circulating adrenal glucocorticoids, growth hormones, and estrogen (Grossman, 1989). Furthermore, it is known the female immune response to injury in mammals is more robust than for males (Bouman et al., 2005), and that UV exposure can lead to an inflammatory response (D’Orazio et al., 2013; Yang et al., 2014). Collectively these data, and the robust female response at 8 kJ/m2, support the idea that Xiphophorus female skin may serve as a more sensitive biological dosimeter for UVB exposure than males.
Although females have a more robust response than males at 8 kJ/m2, the males show a quantitatively and qualitatively similar genetic response following 16 kJ/m2 UVB. One shared pathway induced in both females and males at 16 kJ/m2 are genes involved in fatty acid biosynthesis (elovl, prmt1, and snca genes) (Figure 4). Fatty acids are essential structural components of the cell membrane and also involved in various cellular processes including signaling (Graber et al., 1994). It has been reported that an increase in free fatty acids may play a key role in cell cycle repression and apoptosis (Artwohl et al., 2004; Kim et al., 2010). Our data also indicate the ribosome biogenesis genes nop58, nop2, rrs1, nop2, nhp2 and ebna1bp2 are up modulated in both females and males. This may be due to DNA damage induced cell cycle arrest since it has been shown that ribosome biogenesis may lead to cell cycle arrest through interaction with p53 (Takagi et al., 2005).
Based upon pathway analysis of up modulated transcripts (e.g., phosphoinositide 3-kinase [PI3K] receptors grasp and lpar3 at 16 kJ/m2) both females and males utilize the PI3K signaling pathway for cell cycle regulation. UV exposure has been reported to increase oxidative stress (Svobodová et al., 2011) and uptake glucose through the PI3K pathway (Higaki et al., 2008), ultimately playing a key role in the G2/M cell cycle transition (Liang and Slingerland, 2003).
Overall, given the observed up modulation of genes known to be involved with apoptosis (Bhinge et al., 2007; Carey et al., 2013; Chen et al., 2010; Duan et al., 2013; Fedele et al., 2001; Fleischer and Rebollo, 2004; Franklin et al., 2002; Hartman et al., 2004; Jin et al., 2011; Pawlowski and Kraft, 2000; Shi et al., 2015; Thyss et al., 2005), and concurrent up modulation of genes involved with fatty acid biosynthesis, ribosome biogenesis, and the cellular stress response, one shared primary response for both females and males is to arrest the cell cycle. This is consistent with the concept of UVB damaged cells to either repair DNA damage caused by UVB exposure or induce apoptosis in cells containing extensive DNA damage at the higher dose (e.g. at 16 kJ/m2).
Unique Genetic Response to UVB Exposure
Differences in gene families modulated between the sexes following UVB exposure to skin cluster into several interesting pathways (Figure 5). For example, female X. maculatus down modulate genes involved in the mitochondrial uncoupling protein 3 (ucp3) pathway whereas males up modulate ucp3 in response to UVB exposure. Uncoupling the electron transport chain results in the reduction of ROS by bypassing CoQH to reduce the production of O-·2 inside the mitochondria (Korshunov et al., 1997; Skulachev, 1998, 1996). In an attempt to reduce oxidative damage after UVB exposure, males may be increasing free fatty acid biosynthesis and subsequently using these products to uncouple electron transport and thereby up-regulate ucp3 to bypass CoQH to reduce ROS, whereas females do not.
In addition, our data suggest that male X. maculatus skin up modulates wound healing genes in response to 8 and 16 kJ/m2 UVB (Figure 5). For example, serpinc1, serpind1, proca, plg, cfi, f7, and cpb2 are all involved with the coagulation pathway, and all are oppositely modulated in females (down) and males (up). UVB exposure leads to erythema and inflammation that could produce a wound healing response (Cadet et al., 2015). There has been speculation that wound healing may be involved in cancer (Dvorak, 1986; Martins-Green et al., 1994) and a male sex bias for skin cancer is well established (Thomas-Ahner et al., 2007). An interesting avenue for future research might be to test the idea that UVB exposure of male X. maculatus may produce a higher incidence of skin cancer than females due, in part, to the higher expression of wound healing pathways.
The AMP-activated protein kinase [AMPK] pathway is intimately involved in mediation of glucose uptake, insulin secretion, and fatty acid oxidation (Winder and Hardie, 1999). A known target for AMPK is carnitine palmitoyltransferase (CPT). When acetyl-CoA carboxylase is phosphorylated, it is converted into malonyl-CoA, which inhibits CPT. This inhibition prevents CPT from transporting fatty acids into the mitochondria for oxidation (Bezaire et al., 2005; Piiper et al., 2002). In our data, CPT is down regulated 2.6 fold in females, but not in males, perhaps suggesting females are not oxidizing fatty acids as well as males in the mitochondria. Furthermore, Senese et al. (2010) report that a decrease in ucp3 modulation results in a decrease in AMPK signaling (Senese et al., 2011), andfemales exhibit substantial (7.7 fold) down regulation of ucp3 at 16kJ/m2. We also observe up modulation of genes involved with insulin pathways in males. For example, glut4 and gck are in pathways down modulated in females but up modulated in males, suggesting males may be increasing glucose uptake for energy production relative to females. As an example, gck is down in females about 20 fold, but up modulated in males 7 fold. It has been suggested that an increase in UCP3, due to an increase in intracellular free fatty acids, up regulates the AMPK pathway in order to switch from glucose metabolism to free fatty acid metabolism in order to reduce reactive oxygen species (ROS) (Criscuolo et al., 2006). We postulate that males may increase glucose absorption by increasing insulin production in response to UVB. This may result in an increase in fatty acid metabolism through ucp3 uncoupling and the AMPK pathway in an attempt to reduce cell damage by UVB induced ROS.
Although both females and males up modulate free fatty acid synthesis, only the males up modulate ucp3 and the AMPK pathway to help dissipate ROS. In addition, females seem to have a more robust response to lower doses of UVB (e.g. 8kJ/m2) than males. While it is unclear what pathways are being utilized by females to reduce UVB induced damage, it appears they are not modulating pathways, induced by males, to shut down cellular respiration and deplete carbon energy sources. This may be because females repair damage at lower doses and do not reach a build up in ROS incurred in males. This is another possible reason why males are more prone to UVB induced damage and subsequent melanoma incidence than females and may be investigated in future experiments.
Supplementary Material
Figure 1.

Representative female (top) and male (bottom) X. maculatus Jp 163 B employed in UV exposures.
Figure 7.
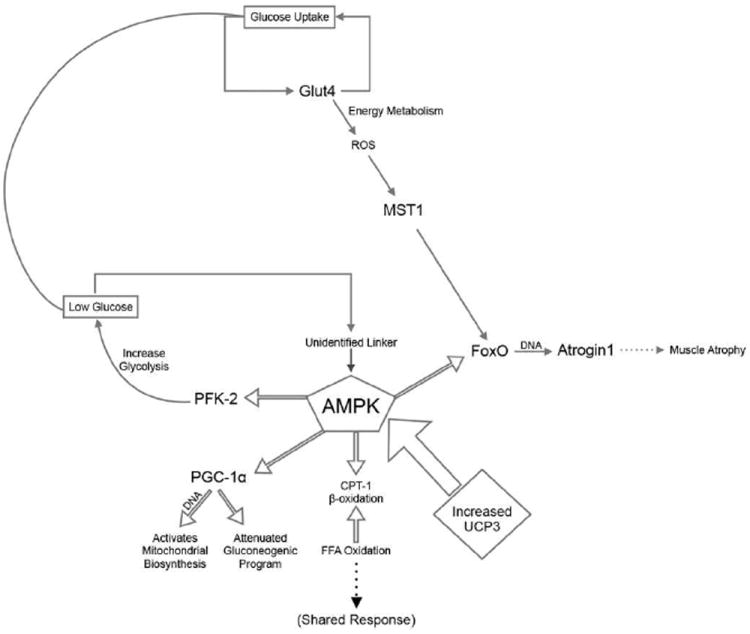
Proposed opposing modulation of the AMPK pathway for energy homeostasis by utilizing the AMPK pathway. All genes listed in the pathway were identified as being oppositely modulated in X. maculatus females and male skin following 16 kJ/m2 UVB irradiation. The modulated genes were not only located linearly within the ucp3 to AMPK pathway, but in addition other genes were found to be oppositely modulated within pathways known to be linked to AMPK lending support to the data. Within these oppositely modulated genes the pfk-2 gene helps regulate the uptake and replenishment of glucose. Through glut4, ROS trigger mst1 and feed into the FoxO pathway ultimately leading to muscle atrophy.
Acknowledgments
The authors would like to thank the staff of the Xiphophorus Genetic Stock Center, Texas State University, for maintaining the pedigreed fish lines and caring for the animals used in this study. We wish to acknowledge the assistance of Drs. Tzinzuni Garcia, Yingjia Shen, and Kuan Yang for developing some of the bioinformatics data analysis pipelines utilized in this project. Funding for these studies was provided in part by the NIH, ORIP grants R24-OD-011120 (RBW), R24-OD-011199 (RBW), CPRIT Core Facility Support grant RP120348 (JS), and Texas State University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artwohl M, Roden M, Waldhäusl W, Freudenthaler A, Baumgartner-Parzer SM. Free fatty acids trigger apoptosis and inhibit cell cycle progression in human vascular endothelial cells. FASEB J. 2004;18:146–148. doi: 10.1096/fj.03-0301fje. [DOI] [PubMed] [Google Scholar]
- Banerjee G, Gupta N, Kapoor A, Raman G. UV induced bystander signaling leading to apoptosis. Cancer Lett. 2005;223:275–284. doi: 10.1016/j.canlet.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Bezaire V, Spriet LL, Campbell S, Sabet N, Gerrits M, Bonen A, Harper M-E. Constitutive UCP3 overexpression at physiological levels increases mouse skeletal muscle capacity for fatty acid transport and oxidation. FASEB J. 2005;19:977–979. doi: 10.1096/fj.04-2765fje. [DOI] [PubMed] [Google Scholar]
- Bhinge AA, Kim J, Euskirchen GM, Snyder M, Iyer VR. Mapping the chromosomal targets of STAT1 by Sequence Tag Analysis of Genomic Enrichment (STAGE) Genome Res. 2007;17:910–6. doi: 10.1101/gr.5574907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird MD, Karavitis J, Kovacs EJ. Sex differences and estrogen modulation of the cellular immune response after injury. Cell Immunol. 2008;252:57–67. doi: 10.1016/j.cellimm.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouman A, Heineman MJ, Faas MM. Sex hormones and the immune response in humans. Hum Reprod Update. 2005;11:411–423. doi: 10.1093/humupd/dmi008. [DOI] [PubMed] [Google Scholar]
- Brand MD. Uncoupling to survive? The role of mitochondrial inefficiency in ageing. Experimental Gerontology. 2000:811–820. doi: 10.1016/s0531-5565(00)00135-2. [DOI] [PubMed] [Google Scholar]
- Cadet J, Douki T, Ravanat J-L. Oxidatively Generated Damage to Cellular DNA by UVB and UVA Radiation. Photochem Photobiol. 2015;91:140–155. doi: 10.1111/php.12368. [DOI] [PubMed] [Google Scholar]
- Carey KT, Tan KH, Ng J, Liddicoat DR, Godfrey DI, Cole TJ. Nfil3 is a glucocorticoid-regulated gene required for glucocorticoid-induced apoptosis in male murine T cells. Endocrinology. 2013;154:1540–52. doi: 10.1210/en.2012-1820. [DOI] [PubMed] [Google Scholar]
- Chen L, Wang S, Zhou Y, Wu X, Entin I, Epstein J, Yaccoby S, Xiong W, Barlogie B, Shaughnessy JD, Zhan F. Identification of early growth response protein 1 (EGR-1) as a novel target for JUN-induced apoptosis in multiple myeloma. Blood. 2010;115:61–70. doi: 10.1182/blood-2009-03-210526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook MB, McGlynn KA, Devesa SS, Freedman ND, Anderson WF. Sex disparities in cancer mortality and survival. Cancer Epidemiol Biomarkers Prev. 2011;20:1629–1637. doi: 10.1158/1055-9965.EPI-11-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo F, Mozo J, Hurtaud C, Nübel T, Bouillaud F. UCP2, UCP3, avUCP, what do they do when proton transport is not stimulated? Possible relevance to pyruvate and glutamine metabolism. Biochim Biophys Acta. 2006;1757:1284–1291. doi: 10.1016/j.bbabio.2006.06.002. [DOI] [PubMed] [Google Scholar]
- D’Orazio J, Jarrett S, Amaro-Ortiz A, Scott T. UV Radiation and the Skin. Int J Mol Sci. 2013;14:12222–48. doi: 10.3390/ijms140612222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Feraudy S, Ridd K, Richards LM, Kwok P-Y, Revet I, Oh D, Feeney L, Cleaver JE. The DNA damage-binding protein XPC is a frequent target for inactivation in squamous cell carcinomas. Am J Pathol. 2010;177:555–562. doi: 10.2353/ajpath.2010.090925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X-H, Chang J-R, Zhang J, Zhang B-H, Li Y-L, Teng X, Zhu Y, Du J, Tang C-S, Qi Y-F. Activating transcription factor 4 is involved in endoplasmic reticulum stress-mediated apoptosis contributing to vascular calcification. Apoptosis. 2013;18:1132–44. doi: 10.1007/s10495-013-0861-3. [DOI] [PubMed] [Google Scholar]
- Dvorak HF. Tumors: wounds that do not heal: similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315:1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- Fedele M, Pierantoni GM, Berlingieri MT, Battista S, Baldassarre G, Munshi N, Dentice M, Thanos D, Santoro M, Viglietto G, Fusco A. Overexpression of proteins HMGA1 induces cell cycle deregulation and apoptosis in normal rat thyroid cells. Cancer Res. 2001;61:4583–90. [PubMed] [Google Scholar]
- Fernandez AA. A cancer-causing gene is positively correlated with male aggression in Xiphophorus cortezi. J Evol Biol. 2010;23:386–396. doi: 10.1111/j.1420-9101.2009.01914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AA, Paniker L, Garcia R, Mitchell DL. Recent advances in sunlight-induced carcinogenesis using the Xiphophorus melanoma model. Comp Biochem Physiol C Toxicol Pharmacol. 2012;155:64–70. doi: 10.1016/j.cbpc.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer A, Rebollo A. Induction of p53-independent apoptosis by the BH3-only protein ITM2Bs. FEBS Lett. 2004;557:283–287. doi: 10.1016/s0014-5793(03)01519-9. [DOI] [PubMed] [Google Scholar]
- Franklin CC, Krejsa CM, Pierce RH, White CC, Fausto N, Kavanagh TJ. Caspase-3-Dependent Cleavage of the Glutamate-L-Cysteine Ligase Catalytic Subunit during Apoptotic Cell Death. Am J Pathol. 2002;160:1887–94. doi: 10.1016/S0002-9440(10)61135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia TI, Shen Y, Crawford D, Oleksiak MF, Whitehead A, Walter RB. RNA-Seq reveals complex genetic response to deepwater horizon oil release in Fundulus grandis. BMC Genomics. 2012;13:474. doi: 10.1186/1471-2164-13-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon M. Hereditary basis of melanosis in hybrid fishes. Am J Cancer. 1931;15:1495–1523. [Google Scholar]
- Graber R, Sumida C, Nunez EA. Fatty acids and cell signal transduction. J Lipid Mediat Cell Signal. 1994;9:91–116. [PubMed] [Google Scholar]
- Grossman C. Possible underlying mechanisms of sexual dimorphism in the immune response, fact and hypothesis. J Steroid Biochem. 1989;34:241–251. doi: 10.1016/0022-4731(89)90088-5. [DOI] [PubMed] [Google Scholar]
- Gujuluva CN, Baek JH, Shin KH, Cherrick HM, Park NH. Effect of UV-irradiation on cell cycle, viability and the expression of p53, gadd153 and gadd45 genes in normal and HPV-immortalized human oral keratinocytes. Oncogene. 1994;9:1819–1827. [PubMed] [Google Scholar]
- Hanbo Chen, B PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011;12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman MG, Lu D, Kim M-L, Kociba GJ, Shukri T, Buteau J, Wang X, Frankel WL, Guttridge D, Prentki M, Grey ST, Ron D, Hai T. Role for activating transcription factor 3 in stress-induced beta-cell apoptosis. Mol Cell Biol. 2004;24:5721–32. doi: 10.1128/MCB.24.13.5721-5732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck DE, Vetrano AM, Mariano TM, Laskin JD. UVB light stimulates production of reactive oxygen species: Unexpected role for catalase. J Biol Chem. 2003;278:22432–22436. doi: 10.1074/jbc.C300048200. [DOI] [PubMed] [Google Scholar]
- Higaki Y, Mikami T, Fujii N, Hirshman MF, Koyama K, Seino T, Tanaka K, Goodyear LJ. Oxidative stress stimulates skeletal muscle glucose uptake through a phosphatidylinositol 3-kinase-dependent pathway. Am J Physiol Endocrinol Metab. 2008;294:E889–E897. doi: 10.1152/ajpendo.00150.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanowicz LR, Stafford JL, Patiño R, Bengten E, Miller NW, Blazer VS. Channel catfish (Ictalurus punctatus) leukocytes express estrogen receptor isoforms ERα and ERβ2 and are functionally modulated by estrogens. Fish Shellfish Immunol. 2014;40:109–19. doi: 10.1016/j.fsi.2014.06.021. [DOI] [PubMed] [Google Scholar]
- Jin Y, Wang C, Liu X, Mu W, Chen Z, Yu D, Wang A, Dai Y, Zhou X. Molecular characterization of the microRNA-138-Fos-like antigen 1 (FOSL1) regulatory module in squamous cell carcinoma. J Biol Chem. 2011;286:40104–9. doi: 10.1074/jbc.C111.296707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamburov A, Wierling C, Lehrach H, Herwig R. ConsensusPathDB--a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37:D623–8. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazianis S, Gimenez-Conti I, Setlow RB, Woodhead AD, Harshbarger JC, Trono D, Ledesma M, Nairn RS, Walter RB. MNU induction of neoplasia in a platyfish model. Lab Invest. 2001;81:1191–1198. doi: 10.1038/labinvest.3780333. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Kim M-K, Jin X-J, Oh J-H, Kim JE, Chung JH. Skin Aging and Photoaging Alter Fatty Acids Composition, Including 11,14,17- eicosatrienoic Acid, in the Epidermis of Human Skin. J Korean Med Sci. 2010;25:980–983. doi: 10.3346/jkms.2010.25.6.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissling GE, Bernheim NJ, Hawkins WE, Wolfe MJ, Jokinen MP, Smith CS, Herbert RA, Boorman GA. The utility of the guppy (Poecilia reticulata) and medaka (Oryzias latipes) in evaluation of chemicals for carcinogenicity. Toxicol Sci. 2006;92:143–156. doi: 10.1093/toxsci/kfj181. [DOI] [PubMed] [Google Scholar]
- Korshunov SS, Skulachev VP, Starkov AA. High protonic potential actuates a mechanism of production of reactive oxygen species in mitochondria. FEBS Lett. 1997;416:15–18. doi: 10.1016/s0014-5793(97)01159-9. [DOI] [PubMed] [Google Scholar]
- Kovacs D, Raffa S, Flori E, Aspite N, Briganti S, Cardinali G, Torrisi MR, Picardo M. Keratinocyte growth factor down-regulates intracellular ROS production induced by UVB. J Dermatol Sci. 2009;54:106–113. doi: 10.1016/j.jdermsci.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Kripke ML. Ultraviolet radiation and immunology: something new under the sun--presidential address. Cancer Res. 1994;54:6102–6105. [PubMed] [Google Scholar]
- Krötz F, Sohn H-Y, Pohl U. Reactive oxygen species: players in the platelet game. Arterioscler Thromb Vasc Biol. 2004;24:1988–96. doi: 10.1161/01.ATV.0000145574.90840.7d. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. J Gerontol. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Slingerland J. Multiple Roles of the PI3K/PKB (Akt) Pathway in Cell Cycle Progression. Cell Cycle. 2003;2:336–342. [PubMed] [Google Scholar]
- Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. J Gerontol. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-Green M, Boudreau N, Bissell MJ. Inflammation is responsible for the development of wound-induced tumors in chickens infected with Rous sarcoma virus. Cancer Res. 1994;54:4334–4341. [PubMed] [Google Scholar]
- Masahito P, Aoki K, Egami N, Ishikawa T, Sugano H. Life-span studies on spontaneous tumor development in the medaka (Oryzias latipes) Jpn J Cancer Res. 1989;80:1058–1065. doi: 10.1111/j.1349-7006.1989.tb02259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Fischer ES, Yasuda T, Dohmae N, Iwai S, Mori T, Nishi R, Yoshino K-I, Sakai W, Hanaoka F, Thomä NH, Sugasawa K. Functional regulation of the DNA damage-recognition factor DDB2 by ubiquitination and interaction with xeroderma pigmentosum group C protein. Nucleic Acids Res. 2015;43:1700–1713. doi: 10.1093/nar/gkv038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meador JA, Walter RB, Mitchell DL. Induction, Distribution and Repair of UV Photodamage in the Platyfish, Xiphophorus signum. Photochem Photobiol. 2007;72:260–266. doi: 10.1562/0031-8655(2000)072<0260:idarou>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Mitchell DL, Fernandez AA, Garcia R, Paniker L, Lin K, Hanninen A, Zigelsky K, May M, Nuttall M, Lo H-H, Person MD, Earley R. Acute exposure to ultraviolet-B radiation modulates sex steroid hormones and receptor expression in the skin and may contribute to the sex bias of melanoma in a fish model. Pigment Cell Melanoma Res. 2014;27:408–417. doi: 10.1111/pcmr.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DL, Meador JA, Byrom M, Walter RB. Resolution of UV-Induced DNA Damage in Xiphophorus Fishes. Mar Biotechnol. 2001;3:S61–S71. doi: 10.1007/s101260000000. [DOI] [PubMed] [Google Scholar]
- Nairn RS, Morizot DC, Kazianis S, Woodhead AD, Setlow RB. Nonmammalian models for sunlight carcinogenesis: genetic analysis of melanoma formation in Xiphophorus hybrid fish. Photochem Photobiol. 1996;64:440–448. doi: 10.1111/j.1751-1097.1996.tb03089.x. [DOI] [PubMed] [Google Scholar]
- Neuwirth E. RColorBrewer: ColorBrewer palettes. R package version 1.0- 5 2011 [Google Scholar]
- Patton EE, Mitchell DL, Nairn RS. Genetic and environmental melanoma models in fish. Pigment Cell Melanoma Res. 2010;23:314–337. doi: 10.1111/j.1755-148X.2010.00693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski J, Kraft AS. Bax-induced apoptotic cell death. Proc Natl Acad Sci. 2000;97:529–531. doi: 10.1073/pnas.97.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piiper A, Dikic I, Lutz MP, Leser J, Kronenberger B, Elez R, Cramer H, Müller-Esterl W, Zeuzem S. Cyclic AMP induces transactivation of the receptors for epidermal growth factor and nerve growth factor, thereby modulating activation of MAP kinase, Akt, and neurite outgrowth in PC12 cells. J Biol Chem. 2002;277:43623–30. doi: 10.1074/jbc.M203926200. [DOI] [PubMed] [Google Scholar]
- Senese R, Valli V, Moreno M, Lombardi A, Busiello RA, Cioffi F, Silvestri E, Goglia F, Lanni A, de Lange P. Uncoupling protein 3 expression levels influence insulin sensitivity, fatty acid oxidation, and related signaling pathways. Pflugers Arch. 2011;461:153–64. doi: 10.1007/s00424-010-0892-3. [DOI] [PubMed] [Google Scholar]
- Setlow RB, Grist E, Thompson K, Woodhead AD. Wavelengths effective in induction of malignant melanoma. Proc Natl Acad Sci U S A. 1993;90:6666–6670. doi: 10.1073/pnas.90.14.6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow RB, Woodhead AD. Temporal changes in the incidence of malignant melanoma: Explanation from action spectra. Mutat Res Fundam Mol Mech Mutagen. 1994;307:365–374. doi: 10.1016/0027-5107(94)90310-7. [DOI] [PubMed] [Google Scholar]
- Setlow RB, Woodhead AD, Grist E. Animal model for ultraviolet radiation-induced melanoma: platyfish-swordtail hybrid. Proc Natl Acad Sci U S A. 1989;86:8922–8926. doi: 10.1073/pnas.86.22.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Shi S, Zhao J, Yang L, Nie X, Han J, Ma X, Wan C, Jiang J. KHSRP participates in manganese-induced neurotoxicity in rat striatum and PC12 cells. J Mol Neurosci. 2015;55:454–65. doi: 10.1007/s12031-014-0367-7. [DOI] [PubMed] [Google Scholar]
- Siciliano MJ, Perlmutter A, Clark E. Effect of sex on the development of melanoma in hybrid fish of the Genus Xiphophorus. Cancer Res. 1971;31:725–729. [PubMed] [Google Scholar]
- Skulachev VP. Role of uncoupled and non-coupled oxidations in maintenance of safely low levels of oxygen and its one-electron reductants. Q Rev Biophys. 1996;29:169–202. doi: 10.1017/s0033583500005795. [DOI] [PubMed] [Google Scholar]
- Skulachev VP. Uncoupling: new approaches to an old problem of bioenergetics. Biochim Biophys Acta - Bioenerg. 1998;1363:100–124. doi: 10.1016/s0005-2728(97)00091-1. [DOI] [PubMed] [Google Scholar]
- Sun X, Wray C, Tian X, Hasselgren P-O, Lu J. Expression of uncoupling protein 3 is upregulated in skeletal muscle during sepsis. Am J Physiol Endocrinol Metab. 2003;285:E512–20. doi: 10.1152/ajpendo.00446.2002. [DOI] [PubMed] [Google Scholar]
- Svobodová AR, Galandáková A, Sianská J, Doležal D, Ulrichová J, Vostálová J. Acute exposure to solar simulated ultraviolet radiation affects oxidative stress-related biomarkers in skin, liver and blood of hairless mice. Biol Pharm Bull. 2011;34:471–479. doi: 10.1248/bpb.34.471. [DOI] [PubMed] [Google Scholar]
- Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Thomas-Ahner JM, Wulff BC, Tober KL, Kusewitt DF, Riggenbach JA, Oberyszyn TM. Gender differences in UVB-induced skin carcinogenesis, inflammation, and DNA damage. Cancer Res. 2007;67:3468–3474. doi: 10.1158/0008-5472.CAN-06-3798. [DOI] [PubMed] [Google Scholar]
- Thyss R, Virolle V, Imbert V, Peyron J-F, Aberdam D, Virolle T. NF-kappaB/Egr-1/Gadd45 are sequentially activated upon UVB irradiation to mediate epidermal cell death. EMBO J. 2005;24:128–37. doi: 10.1038/sj.emboj.7600501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle A, Oliver J, Roca P. Role of uncoupling proteins in cancer. Cancers (Basel) 2010;2:567–91. doi: 10.3390/cancers2020567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter DJ, Boswell M, Boswell W, Walter RB. Characterization and differential expression of CPD and 6-4 DNA photolyases in Xiphophorus species and interspecies hybrids. Comp Biochem Physiol Part C Toxicol Pharmacol. 2014;163:77–85. doi: 10.1016/j.cbpc.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter RB, Kazianis S. Xiphophorus interspecies hybrids as genetic models of induced neoplasia. ILAR J. 2001;42:299–321. doi: 10.1093/ilar.42.4.299. [DOI] [PubMed] [Google Scholar]
- Warnes GR. gplots: Various R programming tools for plotting data. J Phycol 2012 [Google Scholar]
- Winder WW, Hardie DG. AMP-activated protein kinase, a metabolic master switch: possible roles in type 2 diabetes. Am J Physiol -- Leg Content. 1999;277:E1–10. doi: 10.1152/ajpendo.1999.277.1.E1. [DOI] [PubMed] [Google Scholar]
- Wood SR, Berwick M, Ley RD, Walter RB, Setlow RB, Timmins GS. UV causation of melanoma in Xiphophorus is dominated by melanin photosensitized oxidant production. Proc Natl Acad Sci U S A. 2006;103:4111–4115. doi: 10.1073/pnas.0511248103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. J Gerontol. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Boswell M, Walter DJ, Downs KP, Gaston-Pravia K, Garcia T, Shen Y, Mitchell DL, Walter RB. UVB-induced gene expression in the skin of Xiphophorus maculatus Jp 163 B. Comp Biochem Physiol - C Toxicol Pharmacol. 2014;163:86–94. doi: 10.1016/j.cbpc.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeretssian G, Doiron K, Shao W, Leavitt BR, Hayden MR, Nicholson DW, Saleh M. Gender differences in expression of the human caspase-12 long variant determines susceptibility to Listeria monocytogenes infection. Proc Natl Acad Sci U S A. 2009;106:9016–9020. doi: 10.1073/pnas.0813362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


