Abstract
Pyruvate kinase is an enzyme that catalyzes the conversion of phosphoenolpyruvate and ADP to pyruvate and ATP in glycolysis and plays a role in regulating cell metabolism. There are four mammalian pyruvate kinase isoforms with unique tissue expression patterns and regulatory properties. The M2 isoform of pyruvate kinase (PKM2) supports anabolic metabolism and is expressed both in cancer and normal tissue. The enzymatic activity of PKM2 is allosterically regulated by both intracellular signaling pathways and metabolites; PKM2 thus integrates signaling and metabolic inputs to modulate glucose metabolism according to the needs of the cell. Recent advances have increased our understanding of metabolic regulation by pyruvate kinase, raised new questions, and suggested the possibility of non-canonical PKM2 functions to regulate gene expression and cell cycle progression via protein-protein interactions and protein kinase activity. Here we review the structure, function, and regulation of pyruvate kinase and discuss how these properties enable regulation of PKM2 for cell proliferation and tumor growth.
Keywords: Cancer Metabolism, Pyruvate Kinase, PKM2
1. Introduction
Warburg, who first reported metabolic alterations in cancer tissue [1], argued that cancer was rooted in metabolic derangement due to loss of cellular respiratory capacity [1, 2]. Crabtree concurrently demonstrated a greater range of metabolic phenotypes across a broad array of tumor types, and noted that some tumors have high respiration rates [3]. These observations laid the foundation for later research on the proliferative metabolism of cancer cells, but a concrete connection between carcinogenic events and altered glucose metabolism was not possible until a role for oncogenes, protein kinases, and intracellular signaling pathways in driving cancer was defined decades later. In 1980, Eigenbrodt speculated that pyruvate kinase M2 (PKM2) was phosphorylated and inactivated by various newly-discovered protein kinases, including the src protein tyrosine kinase [4]. Eigenbrodt also suggested that inhibition of the pyruvate kinase step in glycolysis is necessary for channeling of metabolites into the pentose phosphate pathway to support nucleotide biosynthesis required by a rapidly dividing cell [5]. These two ideas – the connection of PKM2 to both cancer signaling and cancer metabolism – set the stage for current work on how PKM2 impacts cancer biology. Attention was increased because PKM2 is expressed in essentially all human cancers, and efforts have been made to use PKM2 as a cancer biomarker [6]. A direct connection between PKM2 and oncogenic signaling was made when it was shown that PKM2 interacts with peptides and proteins phosphorylated on tyrosine residues in the context of a src-like motif, and that these interactions facilitate tumor growth by inhibiting the enzyme to promote anabolic metabolism [7–10]. These observations provided a foundation for study of the metabolic needs of proliferating cells and informed efforts to understand how oncogenic signaling modulates metabolism to support the growth and survival of cancer cells.
2. Pyruvate kinase isoform nomenclature and expression patterns
There are four mammalian pyruvate kinase isoforms (PKM1, PKM2, PKR, PKL), and each isoform has had multiple designations over time in the literature [11]. Although a tissue may express more than one pyruvate kinase isoform, individual cells generally express only one isoform at appreciable levels; most adult tissues express PKM2, and expression of the other three isoforms is restricted to distinct tissues and cell types [12–14]. The PKM1 isoform is found in tissues with high catabolic demand, such as muscle, heart, and the brain. PKL is the major isoform in the liver and a minor isoform in the kidney, which also expresses PKM2. PKR is found exclusively in red blood cells. PKM2 is the embryonic isoform and is expressed in cancer. PKM2 is also expressed in normal proliferating cells, such as lymphocytes and the cells of the intestinal epithelium [13, 15], but lack of proliferation does not necessarily mean lack of PKM2 expression. For example, primary mouse embryonic fibroblasts retain PKM2 expression even upon replicative senescence, PKM2 is expressed in quiescent T cells, and PKM2 is found in many differentiated tissues, including white adipose tissue and the lung [13, 16, 17].
3. Pyruvate kinase genes, protein structure, and function
3.1 Alternative splicing of the PKM gene
The PKM gene is alternatively spliced to generate transcripts encoding either PKM1 or PKM2 (Figure 1). The human, rat, and mouse PKM genes each have 12 exons [18, 19]. The 9th and 10th exons are of identical length and are isoform-specific: exon 9 is specific to PKM1, exon 10 is specific to PKM2, and only one of the two exons is included in the properly-spliced final transcript. Therefore, splicing to produce PKM2 transcript requires repression of exon 9 and inclusion of exon 10 in the mature transcript (Figure 1). Three splicing factors – polypyrimidine tract binding protein (PTB), heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1), and heterogeneous nuclear ribonucleoprotein A2 (hnRNPA2) – act downstream of oncogenic signaling to repress inclusion of exon 9 [16, 20], while the serine/arginine-rich splicing factor 3 (SRSF3) binds within exon 10 to promote its inclusion in the transcript [21]. How mRNA splicing is controlled to generate a PKM1 transcript is much less well understood, in large part because PKM1 is normally only expressed at appreciable levels in differentiated tissues. Even the A172 glioblastoma cell line, which has been used to study PKM1 expression, expresses mostly PKM2 with only 5–15% of total PKM transcript being PKM1 [16, 21, 22]. Knockdown of the repressive factors (PTB, hnRNPA1, and hnRNPA2) in cell lines that normally express PKM2 facilitates an increase in PKM1 transcript production, but even these conditions result in <50% of PKM transcripts encoding PKM1 [16, 20]. Similarly, a mixture of PKM1 and PKM2 mRNAs is produced in SRSF3-knockdown cells [21]. A reduction in hnRNPA1 levels can also shift binding of this factor from exon 9 to exon 10 and contribute to exon 10 exclusion, thereby promoting PKM1 expression [23]. These studies argue that in the absence of factors that repress exon 9 inclusion, PKM1 may be the default splice product generated from pre-mRNA transcribed from the PKM gene. However, because studies of PKM mRNA splicing have focused primarily on cancer cells, it remains to be determined which regulatory elements determine exclusive PKM1 expression in cell types such as myotubes, skeletal muscle, or neurons that normally express only this isoform. Such regulatory elements would be expected to repress inclusion of exon 10 and/or actively promote inclusion of exon 9.
Figure 1. Generation of PKM1 and PKM2 by alternative splicing.
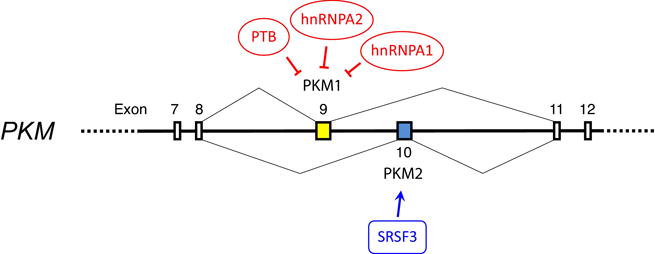
PKM1 and PKM2 are produced by alternative splicing from the PKM gene. PKM exon 9 is included in mature PKM1 mRNA, and exon 10 is included in mature PKM2 mRNA. Production of the M2 isoform of pyruvate kinase requires both repression of exon 9 and inclusion of exon 10. The splicing factors PTB, hnRNPA1, and hnRNPA2 repress inclusion of exon 9, while SRSF3 promotes inclusion of exon 10.
3.2 Protein structure of PKM1 and PKM2
Mammalian pyruvate kinase is a tetrameric protein of identical subunits, which are arranged in a dimer-of-dimers configuration (Figure 2). Each monomer contains one active site and is composed of three main domains – designated A, B, and C – plus a small N-terminal domain [24, 25]. The A domain is the largest domain and is a symmetric α8/β8 TIM barrel. The active site is located at one end of the barrel, in a cleft between the A and B domains. The B domain is mobile and closes on the active site upon binding of the Mg2+-ADP substrate complex [26]. The C domain is found on the opposite side of the A domain and contains the fructose-1,6-bisphosphate (FBP) binding pocket present in the PKM2, PKL, and PKR isoforms [27, 28]. FBP is a major allosteric activator of PKM2, PKL, and PKR and its function in regulating PK activity is discussed in section 3.4. The C domains form the dimer-dimer interface of the fully-associated tetramer, and the amino acids encoded by the alternatively spliced exon that distinguishes PKM1 from PKM2 are located in the C domain (Figure 3). Thus, differences in amino acids located at the dimer-dimer interface are responsible for differences in FBP binding and allosteric regulation of the PKM1 and PKM2 isoforms [28].
Figure 2. Crystal structure of tetrameric PKM2 with bound ligands.
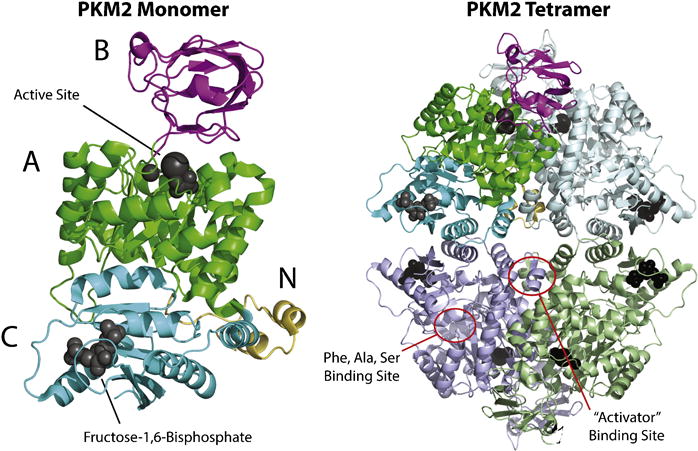
The ribbon structure of tetrameric PKM2 (PDB 3BJF [8]) is shown, with a single subunit colored to represent individual domains. The A, B, C, and N-terminal domains are depicted in green, magenta, cyan, and yellow, respectively. Bound ligands are shown as gray spheres. In this structure, the catalytic site is occupied by K+, Mg2+, and oxalate (a PEP mimetic), and FBP is bound at its allosteric pocket. The binding site for phenylalanine, alanine, and serine is empty; but this site of amino acid binding between the A and C domains is indicated. The binding site for small molecule activators is also indicated. The endogenous ligand binding in the “activator” site, if any, is unknown.
Figure 3. The isoform-specific portion of a PKM2 tetramer.
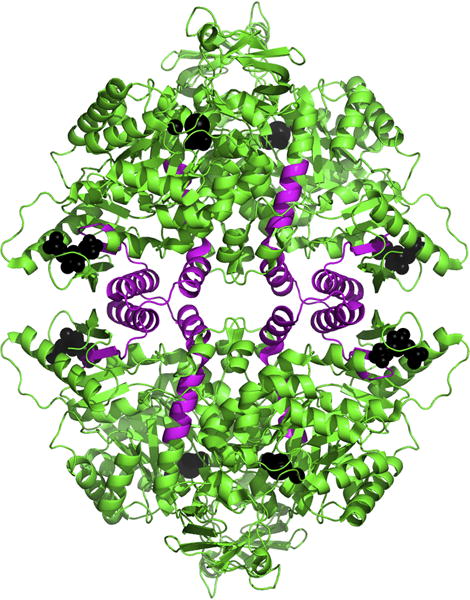
A ribbon structure of a PKM2 tetramer is shown in green. The parts of each subunit that are encoded by exon 10, and are therefore PKM2 isoform-specific, are highlighted in magenta. The PKM2-specific region of the protein forms the bulk of the interface between the dimer of dimers and also forms part of the FBP binding site. Ligands bound in the catalytic and FBP binding sites are shown as black spheres.
Pyruvate kinase is most active as a tetramer. Experiments involving progressive chaotropic denaturation of PKM1 – a constitutive tetramer – show that PKM1 dimers retain catalytic activity, but that monomers are inactive [29]. Unlike PKM1, PKM2 is not a constitutive tetramer; the PKM2 isoform is subject to reversible dissociation and inactivation when diluted in the absence of FBP [30]. When dissociated in the absence of FBP, the specific activity of PKM2 falls to only ~4% of that found in an FBP-activated tetramer [30]. Reversible activation of PKM2 by FBP allows dynamic regulation of its enzymatic activity, and this mode of regulation is conserved among most pyruvate kinases from bacteria and unicellular eukaryotes to metazoans.
3.3 Mechanism of catalysis
Pyruvate kinase catalyzes the direct transfer of phosphate from phosphoenolpyruvate (PEP) to ADP to produce ATP and pyruvate. This reaction is favorable due to the high energy of hydrolysis of PEP [31]. During catalysis, the active site is occupied by both substrates (PEP and ADP, which is complexed with Mg2+), one monovalent cation and one additional enzyme-bound divalent cation [32]. The latter two metal ions assist with substrate binding and coordination, and a catalytic lysine residue stabilizes the pentacoordinate transition state that exists as the phosphate is transferred directly from PEP to ADP. The conserved catalytic lysine is found at position 270 in mammals, 240 in S. cerevisiae, and 221 in Bacillus; substitution of the catalytic lysine with methionine in these proteins produces a properly-folded but catalytically-inactive version of the enzyme [33–35].
Transfer of the phosphate from PEP to ADP leaves the energetically less-stable enol form of pyruvate bound in the active site [36]. Tautomerization of enolpyruvate to the more stable keto form of pyruvate contributes to the favorable energetics of phosphate transfer from PEP to ADP. Tautomerization occurs when enolpyruvate accepts a proton from a water molecule that is held in position by conserved active site residues (T328 and S362 in humans) [27, 37, 38]. Following catalysis the products leave the active site, and neither substrate binding nor release of products is thought to be ordered [39].
In addition to using ADP as a substrate, PKM1 has been shown to accept other nucleotide substrates in vitro, including GDP, IDP, UDP, dADP, and CDP (in order of decreasing affinity) [40]. Compared to ADP, the enzyme displays a four-fold increase in Km for GDP [41], and a 15-fold increase in Km for dADP [40, 41]. Given the low affinity of PKM1 for substrates other than ADP and the relative abundance of ADP in the cytosol, the use of substrates other than ADP by PKM1 in cells is likely quite low. Less data for PKM2 is available, but PKM2 also exhibits decreased activity when presented with GDP and UDP [42]. Furthermore, the active site of PKM2 shares both primary and tertiary structure with the active site of PKM1 [43], making it likely that PKM1 and PKM2 exhibit similar nucleotide substrate specificities.
3.4 Kinetics and allosteric effectors
Mammalian pyruvate kinase isoforms exhibit similar kinetic parameters with respect to ADP, but they differ in their kinetics and allosteric regulation with respect to PEP [11, 13, 27, 43–58]. ADP is bound non-cooperatively and with high affinity by all pyruvate kinase isoforms, and this binding is generally insensitive to allosteric effectors. Most regulation of pyruvate kinase activity occurs via changes in PEP binding affinity. In the absence of allosteric activators the PKM2, PKL, and PKR tetramers have low affinity for PEP and exhibit positive cooperativity of PEP binding. The affinity of the enzyme for PEP at low PEP concentrations can be increased by binding of fructose-1,6-bisphosphate (FBP), an upstream glycolytic intermediate that is also thought to be the major allosteric activator of pyruvate kinase [56, 59]. Each PKM2, PKL, and PKR subunit contains one binding site that is specific for FBP and distinct from the active site (Figure 2); FBP binding acts at a distance to increase PEP binding affinity, promote tetramerization, and stabilize the enzyme tetramer in the active state. PKM1 does not bind FBP due to structural differences at the FBP binding pocket [27]; however, the PKM1 isoform naturally exists as a stable tetramer that has high constitutive activity rendering it insensitive to most of the allosteric effectors that act on PKM2, PKR, and PKL [43]. PKM1 and FBP-activated PKM2 have almost identical kinetic parameters [51], and both are locked into the same three-dimensional conformation [43] with the only structural differences occurring in areas of the protein that are encoded by isoform-specific exons: the FBP binding site and dimer-dimer interface (Figure 3).
Apart from FBP, PKM2 activity is regulated allosterically by numerous small molecules and metabolites [5, 6, 43]. While many metabolites have been shown to have some effect on PKM2 activity [43], most of these effects are modest and require high concentrations, calling into question their importance for the physiological regulation of PKM2. Some of the best-studied allosteric regulators are amino acids and other non-glycolytic metabolites, which together allow coordination of PKM2 activity with metabolic fluxes in other pathways.
The allosteric inhibitor phenylalanine (Phe) reduces both PKM1 and PKM2 activity by decreasing affinity of the enzyme for PEP [60–62]. The Phe binding site is distinct from both the active site and the FBP binding site (Figure 2), and FBP can only partially activate Phe-bound PKM2 [63, 64]. Structural studies suggest that Phe binding stabilizes the tetramer in an inactive state, where the active site cleft is held in an open conformation [43]. This tetramer-stabilizing effect of Phe is demonstrated by greater resistance of Phe-bound PKM1 to chaotropic denaturation [65]. Phe binding thus does not oppose the tetramer-promoting activity of FBP, but rather stabilizes a pyruvate kinase tetramer in a less-active state.
The phenylalanine binding site can also be occupied by alanine [63], which is an allosteric inhibitor of PKM2 but not PKM1 [43]. In contrast to inhibition by phenylalanine, alanine inhibition appears to favor PKM2 dissociation to a dimeric form. Consistent with the ability of FBP to stabilize PKM2 tetramers, alanine inhibition can be fully overcome by FBP [66, 67]. Serine binds in the same pocket as alanine and phenylalanine, but serine acts as an allosteric activator of PKM2 [17, 68] with a reported AC50 of 1.3 mM [69]. Serine and alanine compete with phenylalanine for binding to PKM1 [63], but the effect of combinations of these allosteric effectors on PKM2 has not been explored.
The thyroid hormone triiodo-L-thyronine (T3) is an allosteric inhibitor of PKM2; it binds to the monomeric form of PKM2 and stabilizes the enzyme in its inactive, monomeric state [30, 43]. PKM2 inactivation by T3 is overcome in the presence of FBP, as FBP binding promotes tetramerization of the enzyme. T3 has a similar inhibitory effect on PKR [70], although the binding location of T3 on both isoforms is unknown.
PKM2 is activated by succinylaminoimidazolecarboxamide ribose-5′ phosphate (SAICAR), an intermediate metabolite in the de novo purine synthesis pathway [71]. Activation of PKM2 by SAICAR was first reported to occur during glucose starvation. The ability of SAICAR to activate pyruvate kinase is specific to PKM2, as SAICAR does not activate PKM1, PKL or PKR [71]. The reported EC50 for SAICAR activation of PKM2 is 300 μM; however, in vitro phosphorylation of PKM2 by ERK1/2 can increase affinity of PKM2 for SAICAR and lower the EC50 to 12 μM [72]. ERK-dependent phosphorylation occurs on PKM2 S37 [73], but both the location of the SAICAR binding site and how S37 phosphorylation affects PKM2 conformation are unknown. ERK-dependent SAICAR activation is reported to occur during conditions that favor cell proliferation, and it is posited to be important for sustained proliferative signaling involving PKM2 activity as a protein kinase [72]. This non-canonical protein kinase activity of PKM2 is discussed in section 4.3.
A number of synthetic small molecule allosteric activators of PKM2 have been identified through efforts to screen for modulators of enzyme activity [64, 74–77]. These activators all have EC50 values in the low nanomolar range, and structures of PKM2 crystalized with bound activator show that the activators bind at the subunit interface of the tetramer, with one binding site per dimer (Figure 2) [64, 76, 78]. This site is distinct from where other endogenous allosteric PKM2 modulators interact with the enzyme. Whether an endogenous ligand can occupy this same site is unknown. This binding modality likely differs from that of synthetic small molecule pyruvate kinase activators that target the yeast enzyme, as those activators are predicted to occupy the same site as FBP [79].
Oxalate can activate pyruvate kinase by yet another mechanism, via an interaction at the catalytic pocket. Oxalate is an analog of enolpyruvate and functions as a competitive inhibitor of pyruvate kinase [80]; however, at sub-saturating PEP concentrations, relatively low concentrations of oxalate can activate PKM2 by contributing to the positive cooperativity of PEP binding [27].
3.5 Regulation of PKM2 activity by intracellular signaling
PKM2 activity can be reduced by phosphotyrosine growth signaling in the cell [8, 81]. Interactions between PKM2 and phosphotyrosine-containing proteins and peptides cause PKM2 to release bound FBP. The phosphorylated protein inducing FBP release can be PKM2 itself [9], or any other protein with a phosphorylated tyrosine in a src-like motif that is a predicted target of many growth factor signaling proteins [8]. Because a tyrosine phosphorylated protein can interact with many PKM2 tetramers to release FBP, this allows relatively low stoichiometry of tyrosine phosphorylation to regulate PKM2 activity despite high PKM2 abundance. The interactions of PKM2 with negatively-charged phosphotyrosine depend on K433, a positively-charged amino acid located at the FBP binding site [8]. The side chain of K433 interacts directly with a phosphate group of bound FBP [27]. Interactions of PKM2 with phosphotyrosine promote FBP release in part by interfering with the PKM2-FBP interaction [8]. A K433E substitution is reported to disallow phosphotyrosine peptide binding and FBP release but not affect FBP binding [8]. A peptide containing phosphotyrosine was shown to bind to PKM2 by fluorescence polarization; this binding could be outcompeted by high levels of FBP, suggesting that FBP and phosphotyrosine peptides may bind competitively [82]. In vivo interactions between PKM2 and other proteins that are phosphorylated on tyrosine residues are likely transient, as a dominant phosphorylated binding partner has not been reported outside of specific contexts.
PKM2 is subject to direct phosphorylation on tyrosine residues, including Y105, allowing phosphorylated PKM2 to play a role in inactivating other PKM2 tetramers by stimulating FBP release [9]. Given that the apparent AC50 of FBP for PKM2 (7 μM) is approximately an order of magnitude lower than the reported FBP concentration in mammalian cells (80 μM) [43, 83, 84], sustained phosphotyrosine signaling would be needed to inactivate PKM2 by continually catalyzing FBP release. PKM2 would be expected to bind FBP and enter an active conformation once the growth signal has been withdrawn and phosphotyrosine levels fall. This regulation of PKM2 activity ensures that PKM2 activity is reduced only when growth signaling is active in the cell, effectively coupling the regulation of glucose metabolism to active tyrosine kinase-mediated signaling.
PKM2 is also subject to additional post-translational modifications that directly affect enzyme activity. Acetylation of PKM2 K305 occurs under nutrient-replete conditions and reduces pyruvate kinase activity by both lowering affinity for PEP and triggering degradation of the enzyme itself [85], and acetylation of PKM2 K433 disrupts FBP binding and reduces enzyme activity [86]. PKM2 activity is also reduced by oxidation of C358 by reactive oxygen species [87], and additional modifications such as prolyl-hydroxylation have also been reported [34]. These modifications appear to represent another way to affect PKM2 activity and coordinate the metabolic response to changing cell states.
3.7 Heterotetramerization of pyruvate kinase isoforms
In vitro experiments showed that PKM1 and PKL subunits are capable of forming functional heterotetramers [48] and PKM1 and PKM2 subunits also heterotetramerize [78]. Isolation and kinetic characterization of individual PKL-PKM1 heterotetramer species showed that increasing numbers of PKM1 subunits serve to progressively stabilize the tetramer in an active conformation, as supported by a progressive reduction in Km for PEP and reduction in cooperativity with respect to PEP binding [48, 50]. The distribution of heterotetramer species obtained from these experiments is consistent with random assortment of individual PKM1 and PKL monomers into all possible heterotetramer species, which indicates a lack of binding preference between the isozymes. The high degree of conservation among vertebrate pyruvate kinase isoforms is further highlighted by the ability of chicken PKM1 to form functional heterotetramers with bovine PKL [47]. This ability to heterotetramerize may explain why most individual cells restrict expression to a single isoform to retain the allosteric regulatory properties of that isoform. However, characterization of the tissue distribution of pyruvate kinase isoforms suggest that pyruvate kinase heterotetramers may exist in select situations in vivo. Co-expression of PKM2 and PKM1 occurs as PKM2 is replaced by PKM1 during development [16]; PKM1-PKM2 heterotetramers are evident in skeletal muscle and heart during this transition, which can last for several weeks in the rat [12]. Heterotetramers of PKL and PKM2 occur in rat and bovine fetal liver [12], and PKM1-PKM2 heterotetramers are also observed in bovine Purkinje fibers, great vessels, heart valves [12, 14]. Other adult tissues express more than one pyruvate kinase isoform, but heterotetramers are not observed in most cases, providing biochemical evidence that expression of each isoform is usually restricted to a particular cell type [12].
4. The role of PKM2 in cancer
4.1 Metabolic needs of proliferating cells
A proliferating cell must acquire and organize mass to replicate; this requires lipids for new membranes, carbohydrates for protein glycosylation, nucleotide precursors to support DNA replication and provide RNA for new ribosomes, and amino acids and other cellular building blocks [88, 89]. Generating and assembling these building blocks requires ATP; but biosynthesis also requires carbon and nitrogen precursors and the reducing equivalents necessary to process them [89, 90]. De novo lipid synthesis in particular requires significant expenditure of reducing power in the form of NADPH. Anabolic metabolism typically requires shunting of glucose carbon from glycolysis to biosynthetic pathways, most notably the pentose phosphate pathway (PPP). The PPP produces ribose-5-phosphate and phosphoribosyl pyrophosphate (PRPP), which is required for nucleotide biosynthesis. The oxidative PPP also generates reducing equivalents in the form of NADPH. Glucose carbon is shunted from glycolysis for serine and glycine biosynthesis, and dihydroxyacetone phosphate is used to make glycerol-3-phosphate that can be further processed for generation of lipids.
4.2 PKM2 favors anabolic metabolism
PKM2 controls the final step of glycolysis, and its regulation serves to integrate intracellular signaling inputs with the metabolic state of the cell. Down-regulation of PKM2 activity, and up-regulation of enzymes committing glucose to glycolysis have been thought to allow the accumulation of phosphorylated glycolytic intermediates, which are then available to spill into branching biosynthetic pathways [5]. Interestingly, cell culture experiments suggest a high ratio of pyruvate kinase activity to phosphofructokinase activity in neoplastic cells [91], suggesting that an expansion of glycolytic metabolites may not be due to a bottleneck at the pyruvate kinase step. More recent metabolite measurements have not detected significant changes in the pool sizes of glycolytic intermediates as a result of changes in pyruvate kinase activity [10, 78]; however, it is worth noting that changes in pool sizes do not necessarily correlate with obvious changes in metabolic fluxes [92–94]. Altering pyruvate kinase regulation in mouse embryonic fibroblasts through forced PKM1 expression arrests cell proliferation and results in discrete changes in metabolism that are limited to serine biosynthesis and nucleotide production. An inability of these cells to make deoxynucleotides appears to be most limiting following inappropriate PKM1 expression, as supplementation with thymine and other DNA bases can partially rescue cell proliferation [10]. These data suggest that the regulation of pyruvate kinase activity in proliferating cells may be particularly important to coordinate glucose metabolism with the synthesis of deoxynucleotides for DNA replication.
Regulation of PKM2 activity by tyrosine kinase signaling is important for metabolic changes that support proliferative metabolism and tumor growth in several contexts [7, 8]. Oxidation of PKM2 C358 reduces enzyme activity and promotes oxidative PPP flux, which helps to regulate redox status in the cell [87]. A reduction in PKM2 activity can also promote serine biosynthesis and support proliferation in conditions where serine is limiting [64, 95], and down-regulation of pyruvate kinase activity is important for nucleotide biosynthesis in non-transformed cells [10]. Experiments with small molecule activators of PKM2 showed that artificially increasing intracellular pyruvate kinase activity causes global metabolic changes and has an anti-proliferative effect [64, 76, 78]. The same effect is seen as a consequence of exogenous PKM1 expression, which also increases pyruvate kinase activity within the cell [78, 96]. Despite the utility of PKM2 in contributing to proliferative metabolism regulation, the PKM2 isoform is not required in a mouse breast cancer model [96]. Following PKM2 deletion, pyruvate kinase expression is extremely low in proliferating tumor cells suggesting that low pyruvate kinase activity is important to support cancer cell proliferation in tumors. In contrast, non-proliferating tumor cells were found to express PKM1, arguing that it is the ability to reactivate pyruvate kinase in non-proliferating tumor cells together with the need to inactivate the enzyme in proliferating cells that may be driving selection for PKM2 in cancer. These findings, together with the observation that many human breast cancers express little to no pyruvate kinase argue that PKM2 is not required for the proliferation of all cells. This argument is supported by data from other mouse models [97, 98]. Mutations of PKM2 consistent with decreased enzyme activity are also found in human cancers [96], raising the possibility that decreased PKM2 activity may provide a selective advantage for some tumor cells. Nevertheless, PKM2 deletion in mouse models of leukemia disrupts metabolism and prolongs disease latency [97], arguing that complete loss of PKM2 does not provide an advantage in all contexts. The ability of a proliferating cell to regulate pyruvate kinase activity in accordance with cell state appears to be differentially important for cell proliferation in different cancers.
4.3 Non-canonical pyruvate kinase functions of PKM2
Recent work has reported non-metabolic functions for PKM2; these activities serve to regulate gene expression or the cell cycle and are based on PKM2 protein-kinase function or protein-protein interactions involving PKM2.
A protein kinase activity of PKM2 has been reported, with PKM2 catalyzing transfer of a phosphate directly from PEP to serine, threonine or tyrosine residues on various protein substrates [72, 99, 100]. The protein kinase activity of PKM2 is competitively inhibited by ADP [72, 100], which suggests that the active site of PKM2 must accommodate binding of multiple phosphate acceptors, including ADP and target proteins, as well as the phosphate donor, PEP. Published phosphorylation targets include histone H3 T11 [100], STAT3 Y705 [99], BUB3 Y207 [101], MLC2 Y118 [102], and ERK1, with T202 being the likely residue [72]; additionally, a study claimed 149 proteins as potential substrates, with 91 of these being protein kinases themselves [72]. How the enzyme maintains selectivity among nucleotide-diphosphate substrates (see section 3.3) while accepting a wide variety of protein substrates at the same site is not clear, and the ability of PKM2 to directly phosphorylate protein using either PEP or ATP as a phosphate donor has been challenged by subsequent work using purified components [103].
The molecular mechanism underlying the protein kinase activity of PKM2 is unknown, as are the factors regulating the choice of substrate (i.e., ADP vs. protein). Although other PEP-dependent protein kinases have not been described in eukaryotes, PEP-dependent phosphorylation of proteins does occur in the bacterial phosphotransferase system, which transfers phosphate from PEP to hexose via a set of phospho-enzyme intermediates [104, 105]. Enzyme I of the phosphotransferase system transfers phosphate from PEP to another protein; however, it shares structural and mechanistic homology with pyruvate phosphate dikinase [106, 107], and is less informative in the case of PKM2. The possibility of contaminating ATP-dependent protein kinases and ADP, which is readily converted to ATP in the presence of PKM2 and PEP, is an additional factor that can confound the study of independent PKM2 protein kinase activity [103].
SAICAR, an intermediate in purine nucleotide synthesis, has been reported to activate both the pyruvate kinase and protein kinase activities of PKM2 [71, 72]. Activation of both activities by SAICAR implies that both activities depend on increased affinity of the enzyme for PEP, the phosphate donor. The low μM Km for PEP for the protein kinase activity of SAICAR-activated PKM2 has led to the conclusion that the active protein kinase state is the active, tetrameric R-state of PKM2 [72]. The site of SAICAR binding is unknown, but PKM2 mutants have shown that SAICAR activation is separable from FBP activation, which indicates that these effectors likely bind at different sites [71]. The Q393K substitution renders PKM2 insensitive to SAICAR [71]; this mutation changes a residue at the novel binding pocket that is bound by multiple classes of small molecule activators [64, 76, 78], suggesting that SAICAR may modulate the enzyme via an analogous mechanism to stabilize the enzyme tetramer. However, an ability of SAICAR to also promote PKM2 protein kinase activity contradicts other reports that argue PKM2 has protein kinase activity as a dimer [86, 99, 108]. A putative protein kinase activity of the PKM2 dimer is offered as an explanation for why PKM1 lacks protein kinase activity [99, 100, 102, 108], since the catalytic sites of PKM1 and PKM2 are identical in amino acid sequence and virtually identical in structure when in tetrameric form [43]. The PKM1 isoform and the active tetramer form of PKM2 also exhibit nearly identical kinetic parameters with respect to PEP and ADP [51]. Additional work will be required to reconcile these conflicting observations.
PKM2, but not PKM1, was also reported to activate gene expression by binding to and trans-activating the HIF1α transcription factor in cancer cells [34] and activated macrophages [109]. PKM2 has also been reported to interact with many proteins [6], including Oct4 [110] and DAPk [111]. In the context of EGFR-driven cancer, PKM2 localizes to the nucleus [73], where it is reported to play a role in cell cycle progression via β-catenin transactivation [112] and direct phosphorylation of histone H3 [100], mitotic progression and chromosome segregation via direct phosphorylation of the spindle checkpoint Bub3 [101], and cytokinesis via direct phosphorylation of MLC2 [102]. Phosphorylation of Bub3 by PKM2 is argued to be a late-evolving mitotic checkpoint, as this mechanism of cell cycle regulation is not found in S. cerevisiae [101]. Less attention has been paid to the role of PKM2 in normal tissues, and understanding the relative importance of both the metabolic and non-metabolic PKM2 functions in various tissues and contexts remains an active area of investigation.
5. PKM2: some outstanding questions
5.1 PKM2 regulation and lactate production: a case for channeling?
Two main fates of pyruvate in catabolic metabolism are reduction to lactate by lactate dehydrogenase (LDH) or oxidation to acetyl-CoA and CO2 by pyruvate dehydrogenase (PDH). Paradoxically, the level of pyruvate kinase activity in the cell correlates with pyruvate fate in several studies. Increasing pyruvate kinase activity with PKM2 activators or exogenous PKM1 expression favors greater oxygen consumption and reduced lactate production [7, 78]. Conversely, lactate production is increased when PKM2 activity is reduced as a result of intracellular signaling [81]. One explanation for these observations is substrate channeling. For instance, destabilization of the PKM2 tetramer may favor association with LDH and promote conversion of pyruvate to lactate. While cytosolic complexes of multiple glycolytic enzymes have been found [113, 114], conclusive evidence for channeling of pyruvate to LDH is lacking, and substrate channeling of glycolytic intermediates remains a controversial field with little definitive data [115]. A second hypothesis is that PKM2 activity affects lactate production by causing global changes in the metabolic network. Changes in pyruvate kinase activity affect other metabolic pathways, including the pentose phosphate pathway and serine biosynthesis [10, 64, 87], and changes in the cytosolic NAD+/NADH ratio can impact pyruvate to lactate conversion by LDH. Analysis of how PKM2 activity influences intracellular metabolic fluxes, and careful characterization of the NAD+/NADH ratio in cells may shed light on how pyruvate kinase activity influences the balance between lactate production and pyruvate oxidation.
5.2 Is a product of the PKM gene required?
Gene mapping efforts in mice revealed a spontaneous loss-of-function allele of pyruvate kinase (designated PK-3r) [116]. This mutation appeared to be a complete loss-of function because animals heterozygous for the PK-3r allele retained only 50–60% of normal pyruvate kinase activity in the PKM2-expressing kidney and PKM1-expressing heart [116, 117]. Mice homozygous for the PK-3r allele were never obtained from heterozygous crosses [116]. Lethality of PK-3r homozygous embryos occurred around the time of implantation [118], suggesting that a product of the Pkm gene – or another closely linked gene – is required during embryonic development; unfortunately, the molecular lesion associated with the PK-3r allele was not characterized beyond mapping, and the mice are no longer in existence. Regardless, the lack of compensatory upregulation of pyruvate kinase expression from the wild-type allele in heterozygous animals indicates a lack of intracellular feedback on expression from the Pkm gene in this setting. While pyruvate kinase acts as the ATP payoff step of glycolysis, there is some evidence for an alternative pathway to convert PEP to pyruvate that does not generate ATP but would be sufficient to allow continued glycolysis [119]. If the lethality of the PK-3r allele is due solely to loss of pyruvate kinase, an alternative pyruvate kinase activity does not appear capable of replacing PKM2 during embryogenesis. Nevertheless, tumor cells with undetectable levels of pyruvate kinase can be found in vivo [96]. Whether proliferation of these cells relies on an alternative to pyruvate kinase remains to be determined.
6. Conclusion
Of the four mammalian pyruvate kinase isoforms, PKM2 is most closely associated with proliferation and has been investigated for its role in cancer for almost 40 years. PKM2 is expressed in the developing embryo, in many adult tissues, and in cancer cells, but a full picture of how PKM2 benefits proliferating cells has yet to be painted. PKM2 is also expressed in many non-proliferating tissues, and its role in those contexts is largely unstudied. PKM2 enzymatic activity is heavily regulated by allosteric effectors and intracellular signaling, likely allowing metabolic flexibility and aiding cellular adaptation to changing conditions. Novel, non-canonical functions of PKM2 have also been reported; additional study will be required to fully understand the importance of PKM2 in the contexts of cancer and normal physiology.
Acknowledgments
This work was supported by NIH R01CA168653 and the Burroughs Wellcome Fund. WJI additionally acknowledges support from the Sara and Frank McKnight Fund for Biochemical Research. We thank Aaron M. Hosios, Talya L. Dayton, Katherine R. Mattaini, Lucas B. Sullivan, and Andrea J. Howell for helpful discussion and feedback on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Warburg OH, Posener K, Negelein E. Über den Stoffwechsel der Carcinomzelle. Biochem Z. 1924;152:309–44. [Google Scholar]
- 2.Warburg O. On the origin of cancer cells. Science. 1956;123:309–14. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 3.Crabtree HG. Observations on the carbohydrate metabolism of tumours. Biochem J. 1929;23:536–45. doi: 10.1042/bj0230536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eigenbrodt E, Glossmann H. Glycolysis – one of the keys to cancer? Trends Pharmacol Sci. 1980;1:240–5. [Google Scholar]
- 5.Eigenbrodt E, Reinacher M, Scheefers-Borchel U, Scheefers H, Friis R. Double role for pyruvate kinase type M2 in the expansion of phosphometabolite pools found in tumor cells. Crit Rev Oncog. 1992;3:91–115. [PubMed] [Google Scholar]
- 6.Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2011;43:969–80. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–3. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 8.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–6. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 9.Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lunt SY, Muralidhar V, Hosios AM, Israelsen WJ, Gui DY, Newhouse L, et al. Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Mol Cell. 2015;57:95–107. doi: 10.1016/j.molcel.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imamura K, Tanaka T. Pyruvate kinase isozymes from rat. Methods Enzymol. 1982;90(Pt E):150–65. doi: 10.1016/s0076-6879(82)90121-5. [DOI] [PubMed] [Google Scholar]
- 12.Cardenas JM, Dyson RD. Mammalian pyruvate kinase hybrid isozymes: tissue distribution and physiological significance. J Exp Zool. 1978;204:361–7. doi: 10.1002/jez.1402040307. [DOI] [PubMed] [Google Scholar]
- 13.Imamura K, Tanaka T. Multimolecular forms of pyruvate kinase from rat and other mammalian tissues. I. Electrophoretic studies. J Biochem. 1972;71:1043–51. doi: 10.1093/oxfordjournals.jbchem.a129852. [DOI] [PubMed] [Google Scholar]
- 14.Strandholm JJ, Dyson RD, Cardenas JM. Bovine pyruvate kinase isozymes and hybrid isozymes. Electrophoretic studies and tissue distribution. Arch Biochem Biophys. 1976;173:125–31. doi: 10.1016/0003-9861(76)90242-3. [DOI] [PubMed] [Google Scholar]
- 15.Netzker R, Greiner E, Eigenbrodt E, Noguchi T, Tanaka T, Brand K. Cell cycle-associated expression of M2-type isozyme of pyruvate kinase in proliferating rat thymocytes. J Biol Chem. 1992;267:6421–4. [PubMed] [Google Scholar]
- 16.Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci U S A. 2010;107:1894–9. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eigenbrodt E, Leib S, Kramer W, Friis RR, Schoner W. Structural and kinetic differences between the M2 type pyruvate kinases from lung and various tumors. Biomed Biochim Acta. 1983;42:S278–82. [PubMed] [Google Scholar]
- 18.Noguchi T, Inoue H, Tanaka T. The M1- and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem. 1986;261:13807–12. [PubMed] [Google Scholar]
- 19.Takenaka M, Noguchi T, Sadahiro S, Hirai H, Yamada K, Matsuda T, et al. Isolation and characterization of the human pyruvate kinase M gene. Eur J Biochem. 1991;198:101–6. doi: 10.1111/j.1432-1033.1991.tb15991.x. [DOI] [PubMed] [Google Scholar]
- 20.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–8. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z, Chatterjee D, Jeon HY, Akerman M, Vander Heiden MG, Cantley LC, et al. Exon-centric regulation of pyruvate kinase M alternative splicing via mutually exclusive exons. J Mol Cell Biol. 2012;4:79–87. doi: 10.1093/jmcb/mjr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z, Jeon HY, Rigo F, Bennett CF, Krainer AR. Manipulation of PK-M mutually exclusive alternative splicing by antisense oligonucleotides. Open Biol. 2012;2:120133. doi: 10.1098/rsob.120133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen M, David CJ, Manley JL. Concentration-dependent control of pyruvate kinase M mutually exclusive splicing by hnRNP proteins. Nat Struct Mol Biol. 2012;19:346–54. doi: 10.1038/nsmb.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stammers DK, Muirhead H. Three-dimensional structure of cat muscle pyruvate kinase at 3-1 A resolution. J Mol Biol. 1977;112:309–16. doi: 10.1016/s0022-2836(77)80146-0. [DOI] [PubMed] [Google Scholar]
- 25.Muirhead H, Clayden DA, Barford D, Lorimer CG, Fothergill-Gilmore LA, Schiltz E, et al. The structure of cat muscle pyruvate kinase. EMBO J. 1986;5:475–81. doi: 10.1002/j.1460-2075.1986.tb04236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen TM, Benning MM, Rayment I, Reed GH. Structure of the bis(Mg2+)-ATP-oxalate complex of the rabbit muscle pyruvate kinase at 2.1 A resolution: ATP binding over a barrel. Biochemistry. 1998;37:6247–55. doi: 10.1021/bi980243s. [DOI] [PubMed] [Google Scholar]
- 27.Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44:9417–29. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- 28.Jurica MS, Mesecar A, Heath PJ, Shi W, Nowak T, Stoddard BL. The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate. Structure. 1998;6:195–210. doi: 10.1016/s0969-2126(98)00021-5. [DOI] [PubMed] [Google Scholar]
- 29.Cottam GL, Hollenberg PF, Coon MJ. Subunit structure of rabbit muscle pyruvate kinase. J Biol Chem. 1969;244:1481–6. [PubMed] [Google Scholar]
- 30.Ashizawa K, McPhie P, Lin KH, Cheng SY. An in vitro novel mechanism of regulating the activity of pyruvate kinase M2 by thyroid hormone and fructose 1, 6-bisphosphate. Biochemistry. 1991;30:7105–11. doi: 10.1021/bi00243a010. [DOI] [PubMed] [Google Scholar]
- 31.Mazurek S, Grimm H, Boschek CB, Vaupel P, Eigenbrodt E. Pyruvate kinase type M2: a crossroad in the tumor metabolome. Br J Nutr. 2002;87(Suppl 1):S23–9. [PubMed] [Google Scholar]
- 32.Gupta RK, Oesterling RM. Dual divalent cation requirement for activation of pyruvate kinase; essential roles of both enzyme- and nucleotide-bound metal ions. Biochemistry. 1976;15:2881–7. doi: 10.1021/bi00658a028. [DOI] [PubMed] [Google Scholar]
- 33.Bollenbach TJ, Mesecar AD, Nowak T. Role of lysine 240 in the mechanism of yeast pyruvate kinase catalysis. Biochemistry. 1999;38:9137–45. doi: 10.1021/bi990690n. [DOI] [PubMed] [Google Scholar]
- 34.Luo W, Hu H, Chang R, Zhong J, Knabel M, O’Meally R, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–44. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakai H. Mutagenesis of the active site lysine 221 of the pyruvate kinase from Bacillus stearothermophilus. J Biochem. 2005;137:141–5. doi: 10.1093/jb/mvi027. [DOI] [PubMed] [Google Scholar]
- 36.Seeholzer SH, Jaworowski A, Rose IA. Enolpyruvate: chemical determination as a pyruvate kinase intermediate. Biochemistry. 1991;30:727–32. doi: 10.1021/bi00217a022. [DOI] [PubMed] [Google Scholar]
- 37.Susan-Resiga D, Nowak T. Proton donor in yeast pyruvate kinase: chemical and kinetic properties of the active site Thr 298 to Cys mutant. Biochemistry. 2004;43:15230–45. doi: 10.1021/bi049864d. [DOI] [PubMed] [Google Scholar]
- 38.Susan-Resiga D, Nowak T. The proton transfer step catalyzed by yeast pyruvate kinase. J Biol Chem. 2003;278:12660–71. doi: 10.1074/jbc.M300257200. [DOI] [PubMed] [Google Scholar]
- 39.Ainsworth S, MacFarlane N. A kinetic study of rabbit muscle pyruvate kinase. Biochem J. 1973;131:223–36. doi: 10.1042/bj1310223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plowman KM, Krall AR. A kinetic study of nucleotide interactions with pyruvate kinase. Biochemistry. 1965;4:2809–14. doi: 10.1021/bi00888a035. [DOI] [PubMed] [Google Scholar]
- 41.Hohnadel DC, Cooper C. The effect of structural alterations on the reactivity of the nucleotide substrate of rabbit muscle pyruvate kinase. FEBS Lett. 1973;30:18–20. doi: 10.1016/0014-5793(73)80609-x. [DOI] [PubMed] [Google Scholar]
- 42.Mazurek S, Grimm H, Wilker S, Leib S, Eigenbrodt E. Metabolic characteristics of different malignant cancer cell lines. Anticancer Res. 1998;18:3275–82. [PubMed] [Google Scholar]
- 43.Morgan HP, O’Reilly FJ, Wear MA, O’Neill JR, Fothergill-Gilmore LA, Hupp T, et al. M2 pyruvate kinase provides a mechanism for nutrient sensing and regulation of cell proliferation. Proc Natl Acad Sci U S A. 2013;110:5881–6. doi: 10.1073/pnas.1217157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akhtar K, Gupta V, Koul A, Alam N, Bhat R, Bamezai RN. Differential behavior of missense mutations in the intersubunit contact domain of the human pyruvate kinase M2 isozyme. J Biol Chem. 2009;284:11971–81. doi: 10.1074/jbc.M808761200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berglund L, Humble E. Kinetic properties of pig pyruvate kinases type A from kidney and type M from muscle. Arch Biochem Biophys. 1979;195:347–61. doi: 10.1016/0003-9861(79)90360-6. [DOI] [PubMed] [Google Scholar]
- 46.Bücher T, Pfleiderer G. Pyruvate kinase from muscle: Pyruvate phosphokinase, pyruvic phosphoferase, phosphopyruvate transphosphorylase, phosphate—transferring enzyme II, etc. Phosphoenolpyruvate+ ADP⇌ Pyruvate+ ATP. Methods Enzymol. 1955;1:435–40. [Google Scholar]
- 47.Cardenas JM, Blachly EG, Ceccotti PL, Dyson RD. Properties of chicken skeletal muscle pyruvate kinase and a proposal for its evolutionary relationship to the other avian and mammalian isozymes. Biochemistry. 1975;14:2247–52. doi: 10.1021/bi00681a032. [DOI] [PubMed] [Google Scholar]
- 48.Cardenas JM, Dyson RD. Bovine pyruvate kinases. II. Purification of the liver isozyme and its hybridization with skeletal muscle pyruvate kinase. J Biol Chem. 1973;248:6938–44. [PubMed] [Google Scholar]
- 49.Cardenas JM, Dyson RD, Strandholm JJ. Bovine pyruvate kinases. I. Purification and characterization of the skeletal muscle isozyme. J Biol Chem. 1973;248:6931–7. [PubMed] [Google Scholar]
- 50.Hubbard DR, Cardenas JM. Kinetic properties of pyruvate kinase hybrids formed with native type L and inactivated type M subunits. J Biol Chem. 1975;250:4931–6. [PubMed] [Google Scholar]
- 51.Ibsen KH, Chiu RH, Park HR, Sanders DA, Roy S, Garratt KN, et al. Purification and properties of mouse pyruvate kinases K and M and of a modified K subunit. Biochemistry. 1981;20:1497–506. doi: 10.1021/bi00509a014. [DOI] [PubMed] [Google Scholar]
- 52.Ikeda Y, Tanaka T, Noguchi T. Conversion of Non-allosteric Pyruvate Kinase Isozyme into an Allosteric Enzyme by a Single Amino Acid Substitution. J Biol Chem. 1997;272:20495–501. doi: 10.1074/jbc.272.33.20495. [DOI] [PubMed] [Google Scholar]
- 53.Imamura K, Taniuchi K, Tanaka T. Multimolecular forms of pyruvate kinase. II. Purification of M2-type pyruvate kinase from Yoshida ascites hepatoma 130 cells and comparative studies on the enzymological and immunological properties of the three types of pyruvate kinases, L, M1, and M2. J Biochem. 1972;72:1001–15. doi: 10.1093/oxfordjournals.jbchem.a129962. [DOI] [PubMed] [Google Scholar]
- 54.Strandholm JJ, Cardenas JM, Dyson RD. Pyruvate kinase isozymes in adult and fetal tissues of chicken. Biochemistry. 1975;14:2242–6. doi: 10.1021/bi00681a031. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka T, Harano Y, Sue F, Morimura H. Crystallization, characterization and metabolic regulation of two types of pyruvate kinase isolated from rat tissues. J Biochem. 1967;62:71–91. doi: 10.1093/oxfordjournals.jbchem.a128639. [DOI] [PubMed] [Google Scholar]
- 56.Taylor CB, Bailey E. Activation of liver pyruvate kinase by fructose 1,6-diphosphate. Biochem J. 1967;102:32C–3C. doi: 10.1042/bj1020032c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cardenas JM. Pyruvate kinase from bovine muscle and liver. Methods Enzymol. 1982;90(Pt E):140–9. doi: 10.1016/s0076-6879(82)90120-3. [DOI] [PubMed] [Google Scholar]
- 58.Kahn A, Marie J. Pyruvate kinases from human erythrocytes and liver. Methods Enzymol. 1982;90(Pt E):131–40. doi: 10.1016/s0076-6879(82)90119-7. [DOI] [PubMed] [Google Scholar]
- 59.Koler RD, Vanbellinghen P. The mechanism of precursor modulation of human pyruvate kinase I by fructose diphosphate. Adv Enzyme Regul. 1968;6:127–42. doi: 10.1016/0065-2571(68)90010-1. [DOI] [PubMed] [Google Scholar]
- 60.Carminatti H, Jimenez de Asua L, Leiderman B, Rozengurt E. Allosteric properties of skeletal muscle pyruvate kinase. J Biol Chem. 1971;246:7284–8. [PubMed] [Google Scholar]
- 61.Vijayvargiya R, Schwark WS, Singhal RL. Pyruvate kinase: modulation by L-phenylalanine and L-alanine. Can J Biochem. 1969;47:895–8. doi: 10.1139/o69-140. [DOI] [PubMed] [Google Scholar]
- 62.Weber G. Inhibition of human brain pyruvate kinase and hexokinase by phenylalanine and phenylpyruvate: possible relevance to phenylketonuric brain damage. Proc Natl Acad Sci U S A. 1969;63:1365–9. doi: 10.1073/pnas.63.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams R, Holyoak T, McDonald G, Gui C, Fenton AW. Differentiating a ligand’s chemical requirements for allosteric interactions from those for protein binding. Phenylalanine inhibition of pyruvate kinase Biochemistry. 2006;45:5421–9. doi: 10.1021/bi0524262. [DOI] [PubMed] [Google Scholar]
- 64.Kung C, Hixon J, Choe S, Marks K, Gross S, Murphy E, et al. Small molecule activation of PKM2 in cancer cells induces serine auxotrophy. Chem Biol. 2012;19:1187–98. doi: 10.1016/j.chembiol.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Consler TG, Lee JC. Domain interaction in rabbit muscle pyruvate kinase. I. Effects of ligands on protein denaturation induced by guanidine hydrochloride. J Biol Chem. 1988;263:2787–93. [PubMed] [Google Scholar]
- 66.Schulz J, Sparmann G, Hofmann E. Alanine-mediated reversible inactivation of tumour pyruvate kinase caused by a tetramer-dimer transition. FEBS Lett. 1975;50:346–50. doi: 10.1016/0014-5793(75)80524-2. [DOI] [PubMed] [Google Scholar]
- 67.Sparmann G, Schulz J, Hofmann E. Effects of L-alanine and fructose (1,6-diphosphate) on pyruvate kinase from ehrlich ascites tumour cells. FEBS Lett. 1973;36:305–8. doi: 10.1016/0014-5793(73)80397-7. [DOI] [PubMed] [Google Scholar]
- 68.Ibsen KH, Marles SW. Inhibition of chicken pyruvate kinases by amino acids. Biochemistry. 1976;15:1073–9. doi: 10.1021/bi00650a018. [DOI] [PubMed] [Google Scholar]
- 69.Chaneton B, Hillmann P, Zheng L, Martin AC, Maddocks OD, Chokkathukalam A, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–62. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fanjul AN, Farias RN. Cold-sensitive cytosolic 3,5,3′-triiodo-L-thyronine-binding protein and pyruvate kinase from human erythrocytes share similar regulatory properties of hormone binding by glycolytic intermediates. J Biol Chem. 1993;268:175–9. [PubMed] [Google Scholar]
- 71.Keller KE, Tan IS, Lee YS. SAICAR stimulates pyruvate kinase isoform M2 and promotes cancer cell survival in glucose-limited conditions. Science. 2012;338:1069–72. doi: 10.1126/science.1224409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Keller KE, Doctor ZM, Dwyer ZW, Lee YS. SAICAR induces protein kinase activity of PKM2 that is necessary for sustained proliferative signaling of cancer cells. Mol Cell. 2014;53:700–9. doi: 10.1016/j.molcel.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012;14:1295–304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boxer MB, Jiang JK, Vander Heiden MG, Shen M, Skoumbourdis AP, Southall N, et al. Evaluation of substituted N,N′-diarylsulfonamides as activators of the tumor cell specific M2 isoform of pyruvate kinase. J Med Chem. 2010;53:1048–55. doi: 10.1021/jm901577g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jiang JK, Boxer MB, Vander Heiden MG, Shen M, Skoumbourdis AP, Southall N, et al. Evaluation of thieno[3,2-b]pyrrole[3,2-d]pyridazinones as activators of the tumor cell specific M2 isoform of pyruvate kinase. Bioorg Med Chem Lett. 2010;20:3387–93. doi: 10.1016/j.bmcl.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parnell KM, Foulks JM, Nix RN, Clifford A, Bullough J, Luo B, et al. Pharmacologic activation of PKM2 slows lung tumor xenograft growth. Mol Cancer Ther. 2013;12:1453–60. doi: 10.1158/1535-7163.MCT-13-0026. [DOI] [PubMed] [Google Scholar]
- 77.Walsh MJ, Brimacombe KR, Veith H, Bougie JM, Daniel T, Leister W, et al. 2-Oxo-N-aryl-1,2,3,4-tetrahydroquinoline-6-sulfonamides as activators of the tumor cell specific M2 isoform of pyruvate kinase. Bioorg Med Chem Lett. 2011;21:6322–7. doi: 10.1016/j.bmcl.2011.08.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8:839–47. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bond CJ, Jurica MS, Mesecar A, Stoddard BL. Determinants of allosteric activation of yeast pyruvate kinase and identification of novel effectors using computational screening. Biochemistry. 2000;39:15333–43. doi: 10.1021/bi001443i. [DOI] [PubMed] [Google Scholar]
- 80.Reed GH, Morgan SD. Kinetic and magnetic resonance studies of the interaction of oxalate with pyruvate kinase. Biochemistry. 1974;13:3537–41. doi: 10.1021/bi00714a020. [DOI] [PubMed] [Google Scholar]
- 81.Varghese B, Swaminathan G, Plotnikov A, Tzimas C, Yang N, Rui H, et al. Prolactin inhibits activity of pyruvate kinase M2 to stimulate cell proliferation. Mol Endocrinol. 2010;24:2356–65. doi: 10.1210/me.2010-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Allali-Hassani A, Wasney GA, Chau I, Hong BS, Senisterra G, Loppnau P, et al. A survey of proteins encoded by non-synonymous single nucleotide polymorphisms reveals a significant fraction with altered stability and activity. Biochem J. 2009;424:15–26. doi: 10.1042/BJ20090723. [DOI] [PubMed] [Google Scholar]
- 83.Srivastava DK, Bernhard SA. Metabolite transfer via enzyme-enzyme complexes. Science. 1986;234:1081–6. doi: 10.1126/science.3775377. [DOI] [PubMed] [Google Scholar]
- 84.Srivastava DK, Bernhard SA. Enzyme-enzyme interactions and the regulation of metabolic reaction pathways. Curr Top Cell Regul. 1986;28:1–68. doi: 10.1016/b978-0-12-152828-7.50003-2. [DOI] [PubMed] [Google Scholar]
- 85.Lv L, Li D, Zhao D, Lin R, Chu Y, Zhang H, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42:719–30. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lv L, Xu YP, Zhao D, Li FL, Wang W, Sasaki N, et al. Mitogenic and oncogenic stimulation of K433 acetylation promotes PKM2 protein kinase activity and nuclear localization. Mol Cell. 2013;52:340–52. doi: 10.1016/j.molcel.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–83. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–64. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 89.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 90.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Board M, Humm S, Newsholme EA. Maximum activities of key enzymes of glycolysis, glutaminolysis, pentose phosphate pathway and tricarboxylic acid cycle in normal, neoplastic and suppressed cells. Biochem J. 1990;265:503–9. doi: 10.1042/bj2650503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Heinrich R, Rapoport TA. A linear steady-state treatment of enzymatic chains. General properties, control and effector strength. Eur J Biochem. 1974;42:89–95. doi: 10.1111/j.1432-1033.1974.tb03318.x. [DOI] [PubMed] [Google Scholar]
- 93.Heinrich R, Rapoport TA. A linear steady-state treatment of enzymatic chains. Critique of the crossover theorem and a general procedure to identify interaction sites with an effector. Eur J Biochem. 1974;42:97–105. doi: 10.1111/j.1432-1033.1974.tb03319.x. [DOI] [PubMed] [Google Scholar]
- 94.Kacser H, Burns JA. The control of flux. Symp Soc Exp Biol. 1973;27:65–104. [PubMed] [Google Scholar]
- 95.Ye J, Mancuso A, Tong X, Ward PS, Fan J, Rabinowitz JD, et al. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc Natl Acad Sci U S A. 2012;109:6904–9. doi: 10.1073/pnas.1204176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155:397–409. doi: 10.1016/j.cell.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang YH, Israelsen WJ, Lee D, Yu VW, Jeanson NT, Clish CB, et al. Cell-state-specific metabolic dependency in hematopoiesis and leukemogenesis. Cell. 2014;158:1309–23. doi: 10.1016/j.cell.2014.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cortes-Cros M, Hemmerlin C, Ferretti S, Zhang J, Gounarides JS, Yin H, et al. M2 isoform of pyruvate kinase is dispensable for tumor maintenance and growth. Proc Natl Acad Sci U S A. 2013;110:489–94. doi: 10.1073/pnas.1212780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012;150:685–96. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jiang Y, Li X, Yang W, Hawke DH, Zheng Y, Xia Y, et al. PKM2 regulates chromosome segregation and mitosis progression of tumor cells. Mol Cell. 2014;53:75–87. doi: 10.1016/j.molcel.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jiang Y, Wang Y, Wang T, Hawke DH, Zheng Y, Li X, et al. PKM2 phosphorylates MLC2 and regulates cytokinesis of tumour cells. Nat Commun. 2014;5:5566. doi: 10.1038/ncomms6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hosios AM, Fiske BP, Gui DY, Vander Heiden MG. Lack of evidence for PKM2 protein kinase activity. Mol Cell. 2015 doi: 10.1016/j.molcel.2015.07.013. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kundig W, Ghosh S, Roseman S. Phosphate bound to histidine in a protein as an intermediate in a novel phospho-transferase system. Proc Natl Acad Sci U S A. 1964;52:1067–74. doi: 10.1073/pnas.52.4.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Postma PW, Lengeler JW. Phosphoenolpyruvate:carbohydrate phosphotransferase system of bacteria. Microbiol Rev. 1985;49:232–69. doi: 10.1128/mr.49.3.232-269.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu LF, Saier MH., Jr On the evolutionary origins of the bacterial phosphoenolpyruvate:sugar phosphotransferase system. Mol Microbiol. 1990;4:1219–22. doi: 10.1111/j.1365-2958.1990.tb00698.x. [DOI] [PubMed] [Google Scholar]
- 107.Liao DI, Silverton E, Seok YJ, Lee BR, Peterkofsky A, Davies DR. The first step in sugar transport: crystal structure of the amino terminal domain of enzyme I of the E. coli PEP: sugar phosphotransferase system and a model of the phosphotransfer complex with HPr. Structure. 1996;4:861–72. doi: 10.1016/s0969-2126(96)00092-5. [DOI] [PubMed] [Google Scholar]
- 108.Gao X, Wang H, Yang JJ, Chen J, Jie J, Li L, et al. Reciprocal regulation of protein kinase and pyruvate kinase activities of pyruvate kinase M2 by growth signals. J Biol Chem. 2013;288:15971–9. doi: 10.1074/jbc.M112.448753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Palsson-McDermott EM, Curtis AM, Goel G, Lauterbach MA, Sheedy FJ, Gleeson LE, et al. Pyruvate Kinase M2 Regulates Hif-1alpha Activity and IL-1beta Induction and Is a Critical Determinant of the Warburg Effect in LPS-Activated Macrophages. Cell Metab. 2015;21:65–80. doi: 10.1016/j.cmet.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee J, Kim HK, Han YM, Kim J. Pyruvate kinase isozyme type M2 (PKM2) interacts and cooperates with Oct-4 in regulating transcription. Int J Biochem Cell Biol. 2008;40:1043–54. doi: 10.1016/j.biocel.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 111.Mor I, Carlessi R, Ast T, Feinstein E, Kimchi A. Death-associated protein kinase increases glycolytic rate through binding and activation of pyruvate kinase. Oncogene. 2012;31:683–93. doi: 10.1038/onc.2011.264. [DOI] [PubMed] [Google Scholar]
- 112.Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, et al. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–22. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Simon M, Spahr PF, Dainous F. The proteins associated with the soluble form of p36, the main target of the src oncogene product in chicken fibroblasts, are glycolytic enzymes. Biochem Cell Biol. 1989;67:740–8. doi: 10.1139/o89-111. [DOI] [PubMed] [Google Scholar]
- 114.Mazurek S, Hugo F, Failing K, Eigenbrodt E. Studies on associations of glycolytic and glutaminolytic enzymes in MCF-7 cells: role of P36. J Cell Physiol. 1996;167:238–50. doi: 10.1002/(SICI)1097-4652(199605)167:2<238::AID-JCP7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 115.Mendes P, Kell DB, Welch GR. Metabolic channeling in organized enzyme systems: experiments and models. Advances in Molecular and Cell Biology. 1995;11:1–19. [Google Scholar]
- 116.Johnson FM, Chasalow F, Anderson G, Macdougal P, Hendren RW, Lewis SE. A variation in mouse kidney pyruvate kinase activity determined by a mutant gene on chromosome 9. Genet Res. 1981;37:123–31. doi: 10.1017/s0016672300020103. [DOI] [PubMed] [Google Scholar]
- 117.Peters J, Andrews SJ. The Pk-3 gene determines both the heart, M1, and the kidney, M2, pyruvate kinase isozymes in the mouse; and a simple electrophoretic method for separating phosphoglucomutase-3. Biochem Genet. 1984;22:1047–63. doi: 10.1007/BF00499631. [DOI] [PubMed] [Google Scholar]
- 118.Lewis SE, Johnson FM. Dominant and recessive effects of electrophoretically detected specific locus mutations. In: de Serres FJ, Sheridan W, editors. Utilization of Mammalian Specific Locus Studies in Hazard Evaluation and Estimation of Genetic Risk. New York: Plenum Press; 1983. pp. 267–78. [Google Scholar]
- 119.Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–9. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]


