Abstract
Altered tumor cell metabolism is now firmly established as a hallmark of human cancer. Downstream of oncogenic events, metabolism is re-wired to support cellular energetics and supply the building blocks for biomass. Rapid, uncontrolled proliferation results in tumor growth beyond the reach of existing vasculature and triggers cellular adaptations to overcome limiting nutrient and oxygen delivery. However, oncogenic activation and metabolic re-programming also elicit cell intrinsic stresses, independent of the tumor microenvironment. To ensure metabolic robustness and stress resistance, pro-growth signals downstream of oncogene activation or tumor suppressor loss simultaneously activate homeostatic processes. Here, we summarize recent literature describing the adaptive mechanisms co-opted by common oncogenes, including mTOR, MYC, and RAS. Recurrent themes in our review include: 1) coordination of oncogene-induced changes in protein and lipid metabolism to sustain endoplasmic reticulum homeostasis, 2) maintenance of mitochondrial functional capacity to support anabolic metabolism, 3) adaptations to sustain intracellular metabolite concentrations required for growth, and 4) prevention of oxidative stress. We also include a discussion of the hypoxia inducible factors (HIFs) and the AMP-dependent protein kinase (AMPK)—stress sensors that are co-opted to support tumor growth. Ultimately, an understanding of the adaptations required downstream of specific oncogenes could reveal targetable metabolic vulnerabilities.
Keywords: cancer metabolism, stress response, ER stress, autophagy, hypoxia inducible factors, AMPK
1. Introduction
Cellular transformation, driven by activation of oncogenes and loss of tumor suppressors, results in dysregulated cell proliferation. Activation of pro-growth pathways downstream of MYC, RAS, mTOR, and other oncogenes not only maintain cellular bioenergetics, but also rewire metabolism to generate the biomass needed for cell division; reviewed in [1, 2]. While proliferation of non-transformed cells is controlled by environmental cues such as growth factor signaling and nutrient availability, tumors often grow beyond the reach of perfusion by existing vasculature, generating a cell extrinsic source of stress that requires cancer cell adaptations for survival. These responses include homeostatic pathways to trigger angiogenesis, maintain energetics, overcome stress, and sustain viability. Thus, microenvironmental stresses can promote a transition from anabolism and growth to catabolism and homeostasis.
However, there may not be such a sharp demarcation between states of growth versus homeostasis in cancer cells. Instead, oncogene-induced anabolic processes generate cell intrinsic metabolic stresses that enhance their requirement for homeostatic (often catabolic) processes, even in the absence of microenvironmental perturbations. For example, increased protein synthesis consumes amino acids and increases load on the endoplasmic reticulum (ER). While heightened protein synthesis contributes to disease progression [3, 4], it also elicits vulnerabilities to proteotoxicity and ER stress. Additionally, metabolic changes associated with cellular transformation generate reactive oxygen species (ROS) [5]. While low levels of ROS have signaling functions that support tumor growth, excessive levels can damage mitochondria, thus impairing tumor cell anabolic metabolism, and lead to oxidative damage of DNA, proteins, and lipids.
To ensure metabolic robustness and stress resistance, pro-growth signals downstream of oncogene activation or tumor suppressor loss simultaneously activate homeostatic processes. Such processes maintain intracellular metabolite concentrations to support growth and viability, ensure mitochondrial quality to support anabolism and limit oxidative stress, and prevent ER stress. In this review, we summarize recent literature describing how oncogenes not only impart self-sufficiency of growth signals, but also stimulate adaptations to support such growth. Knowledge of these pathways can elucidate targetable tumor cell vulnerabilities. For a summary of the homeostatic processes discussed in this review, see Table 1.
Table 1.
Summary of adaptive processes co-opted by oncogenes to support growth.
| Pathway | Description | References |
|---|---|---|
| Autophagy | Facilitates lysosomal degradation of intracellular contents targeted to autophagasomes, including proteins, lipids, and organelles. Functions include maintenance of intracellular metabolite concentrations and clearance of damaged mitochondria or insoluble protein aggregates. | 12, 13 |
| Unfolded protein response (UPR) | A highly conserved response triggered by increased mis-folded protein load or disruption of ER membrane lipid composition. ER stress sensors including PERK, IRE-1α, and ATF6 initiate adaptations to restore ER homeostasis, but can trigger cell death if stress is irremediable. | 14–16 |
| NADPH-dependent redox control | NADPH provides reducing equivalents for biosynthesis and regeneration of reduced glutathione to control redox balance. Cellular sources of NADPH include oxidative pentose phosphate pathway, malic enzyme, and serine-dependent one carbon metabolism. | 35, 36, 38 |
| Hypoxia inducible factors (HIFs) | The cellular response to hypoxia is largely orchestrated by two oxygen-labile HIF-α subunits (HIF-1α and HIF-2α). Upon stabilization under low oxygen and heterodimerization with HIF-1β, the HIFs promote transcriptional adaptation to hypoxia. | 44–47 |
| AMP-activated protein kinase (AMPK) | A heterotrimeric Ser/Thr kinase complex that functions as an energy sensor. APMK is activated by increased AMP:ATP ratio and alters cellular metabolism to restore energy homeostasis. | 69–70 |
2. mTORC1
The mechanistic target of rapamycin (mTOR) is a Ser/Thr kinase that functions as a master regulator of cell growth, proliferation, and metabolism [6]. mTOR exists in two distinct complexes (mTORC1 and mTORC2), with different subunit composition and cellular functions. Our focus will be on mTORC1, which promotes protein synthesis, glycolysis, lipogenesis, and nucleotide biosynthesis [6–8]. mTORC1 functions as a nutrient and growth factor sensor in normal cells. Growth factor signaling and amino acid availability regulate mTORC1 via distinct mechanisms and both are required for mTORC1 activation; reviewed in [9]. Growth factors activate mTORC1 via the Rheb GTPase. In the absence of growth factors, Rheb is inactivated by a GTPase activating protein (GAP) complex consisting of TSC1 and TSC2 [10]. Many mTORC1 activities have been identified through examination of cells lacking the TSC1/2 tumor suppressor complex. Individuals with loss of function mutations in TSC1 or TSC2 develop a tumor syndrome consisting of malignancies in the kidney, skin, brain, and heart. However, mTORC1 hyperactivation is observed in a broad range of human malignancies and discoveries made in TSC-null cells are often generalizable to other cancers.
2.1. Sustaining mTORC1 driven protein synthesis
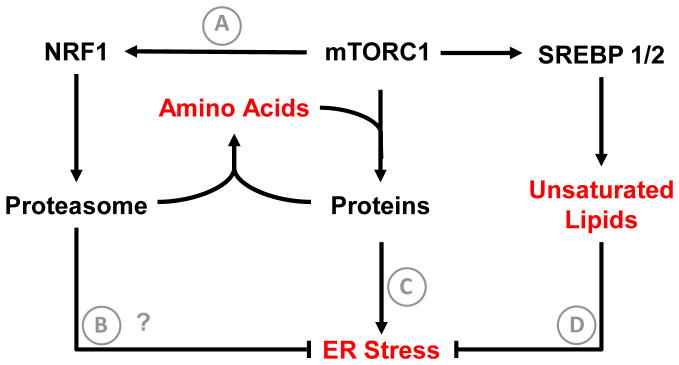
Perhaps one of the best-characterized functions of mTORC1 is increased protein synthesis through translation initiation and elongation (via phosphorylation of 4E-BP1 and S6K1). Protein synthesis consumes both energy and cellular amino acids. If heightened protein synthesis was not balanced with pathways to maintain cellular amino acid content, translation would not be sustainable and cells would become exhausted for amino acids that are also required to generate biomass. Recently, Zhang et al demonstrated that mTORC1 enhances proteasome activity to sustain intracellular amino acid levels [11] (Figure 1A). This occurs via activation of nuclear factor erythroid-derived 2-related factor (NRF1, encoded by NFE2L1) dependent upregulation of genes encoding proteasome components. The requirement for mTORC1 dependent proteasome activity is likely enhanced by the fact that mTORC1 suppresses autophagy, another primary mechanism for breakdown of intracellular proteins into constituent amino acids [12, 13]. While this study focused on maintenance of amino acid content, another function of the proteasome is suppression of proteotoxicity and ER stress (Figure 1B). Indeed, heightened protein synthesis in mTORC1-activated cells induces ER stress and triggers the unfolded protein response (UPR) [14–16] (Figure 1C), which includes proteasome-dependent clearance of misfolded proteins (ER associated protein degradation, ERAD) [16]. Thus, suppression of proteasome function in mTORC1 driven cancers could oppose tumor growth via exhaustion of intracellular amino acid concentrations and induction of ER stress. Indeed, the mechanism of action of the proteasome inhibitor Bortezomib in multiple myeloma derives, in part, from induction of cytotoxic ER stress [17].
Figure 1. mTORC1 driven adaptations to support tumor growth.
Activation of protein synthesis downstream of mTORC1 supports tumor progression, but also consumes amino acids at a rapid rate. To maintain amino acid concentrations required to sustain protein synthesis and anabolic metabolism, mTORC1 activates NRF1-dependepent proteasome biosynthesis (A). Another potential function of enhanced proteasome activity downstream of mTORC1 is maintenance of protein quality control (B), as proteasome dependent ER associated protein degradation (ERAD) is a part of the UPR. Increased protein synthesis also enhances ER stress (C). To expand the ER membrane and accommodate increased protein load, mTORC1 activates SREBP-dependent lipogenic gene expression. In particular, synthesis of unsaturated lipid by SCD1 is crucial to maintain ER lipid homeostasis and prevent ER stress (D). Functional outputs of tumor cell adaptations are shown in red. Question marks denote hypothesized mechanisms that have not been directly tested.
2.2. Coordinating protein and lipid metabolism to maintain ER homeostasis
The ER is a hub for both protein and lipid metabolism, and optimal ER function requires coordination of these processes (Figure 1D). In particular, increases in ER protein load require expansion of ER membrane via lipid synthesis, a process that is particularly important under ER stress [18]. In addition, altered ER lipid content can impair protein-folding capacity [19]. Multiple laboratories have identified the sterol regulatory element binding proteins (SREBP1/2) as mediators of fatty acid and sterol synthesis gene expression programs downstream of mTORC1 [20, 21]. Furthermore, Peterson et al demonstrated that the mechanism for this regulation involves mTORC1-dependent phosphorylation of lipin 1, which controls nuclear localization of mature SREBP [22]. mTORC1 activation not only enhances the quantity of fatty acids required for replication, but also the composition of the fatty acid pool that ultimately gives rise to phospholipid membranes within the cell. Proper desaturation of membrane lipids is absolutely required to sustain cell viability. The stearoyl CoA desaturase enzyme (SCD1) generates unsaturated lipids within the cell. This enzyme, which is an mTORC1/SREBP1 target gene, also requires oxygen for its enzymatic activity. Our lab demonstrated that under hypoxic conditions, where SCD1 is inhibited, mTORC1 activated tumor cells were uniquely sensitive to serum lipid deprivation and subsequent ER stress [23]. Notably cytotoxic ER stress was suppressed by providing cells with the unsaturated fatty acid oleic acid or by suppression of ER protein load using rapamycin or cyxloheximide. These findings were demonstrated in a variety of human cancer cells lines, revealing unsaturated lipid dependence as generalizable tumor cell vulnerability. Consistent with this finding, Griffiths et al showed that ablation of SREBP1 and 2, and thus SCD1 expression, elicited ER stress and cell death under conditions of serum lipid deprivation in various cancer cells [24].
While it may be attractive to target mTORC1 directly, there is growing evidence that it could be more effective to leave mTORC1 signaling intact and target resulting metabolic vulnerabilities [25]. For example, rapamycin analogs exhibit anti-tumor activity in various malignancies, but are often cytostatic and tumor regrowth commonly occurs upon therapy cessation [19]. The findings summarized in this section identify ER homeostasis as a prime target for mTORC1 driven malignancy, where heightened protein synthesis renders cells critically dependent on unsaturated lipid availability, and potentially proteasome function, for viability (Figure 1).
3. MYC
MYC activation has profound effects on tumor cell metabolism; reviewed in [26]. Here, we focus on three features of MYC activation: 1) heightened protein synthesis rate, 2) coordination of protein and lipid synthesis to support viability, and 3) maintenance of mitochondrial function to support growth.
3.1. Sustaining MYC driven protein synthesis
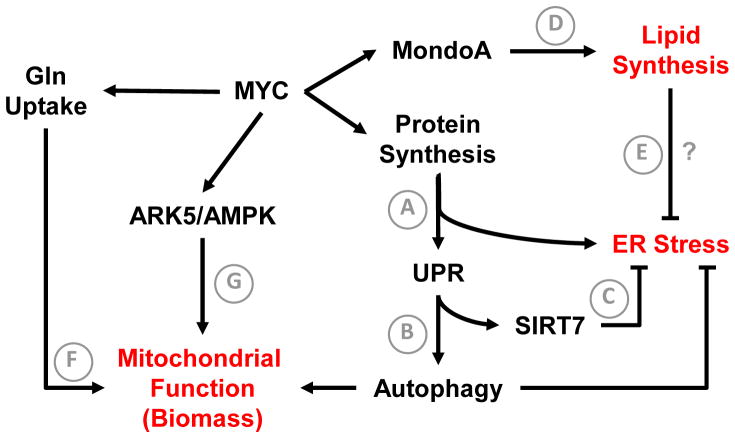
As a transcriptional regulator of the overall translational machinery, MYC enhances global protein synthesis when stimulated. This is required for transformation downstream of MYC, as haploinsufficiency of a ribosomal gene L24 reduced protein synthesis rates and inhibited tumor progression in the Eμ-Myc model of B-cell lymphoma [3]. However, increased translation rates also generate metabolic stresses that must be overcome to sustain tumor growth (Figure 2A). Recently, Hart et al demonstrated that MYC dependent protein synthesis triggers UPR-mediated cytoprotective autophagy to support cell viability [27]. Mechanistically, increased ER protein load triggered the PERK arm of the UPR, which was required for maintenance of autophagy (Figure 2B). Suppression of either PERK signaling or autophagy led to apoptosis in cell culture models and impaired xenograft tumor growth. Importantly, samples from patients with MYC driven lymphoma clearly exhibited evidence of an engaged MYC-PERK-autophagy axis. Autophagy mediates multiple cellular adaptations, including protein quality control, maintenance of intracellular metabolite concentrations, and mitochondrial quality control (via mitophagy) [12, 13]. Future studies elucidating the precise tumor-promoting mechanisms of PERK-dependent autophagy may reveal additional targetable vulnerabilities.
Figure 2. MYC driven adaptations to support tumor growth.
Activation of protein synthesis downstream of MYC supports tumor progression. An increase in ER protein load triggers the UPR (A), which limits ER stress by multiple mechanisms. Activation of PERK facilitates cytoprotective autophagy (B). IRE-1α signaling upregulates the NAD+ dependent histone deacetylase SIRT7 (C), which represses MYC-dependent transcription of ribosomal genes and limits protein synthesis rate to prevent ER stress. MYC activates lipogenic gene expression by upregulating MondoA-dependent lipid synthesis (D), which is required for viability in MYC-activated cells. A potential mechanism involves maintenance of ER lipid homeostasis to accommodate increased protein load and limit ER stress (E). MYC drives glutamine uptake and metabolism to support anabolic processes. (F). This process, termed glutamine-dependent anaplerosis, requires mitochondrial metabolism. MYC activates an ARK5-AMPK pathway to maintain mitochondrial function (G). ARK5 ablation is synthetically lethal to MYC activated cells. Functional outputs of tumor cell adaptations are shown in red. Question marks denote hypothesized mechanisms that have not been directly tested.
Shin et al reported additional cross talk between the UPR and MYC-driven translation [28]. These authors demonstrated that the NAD+ dependent histone deacetylase SIRT7 is induced by the IRE1α/XBP1 arm of the UPR and functions to dampen MYC dependent transactivation of ribosomal genes, limit ER protein load, and ameliorate ER stress (Figure 2C). SIRT7 enzymatic activity was required for this function. While this work focused on MYC in the context of fatty liver disease, it seems plausible that this signaling axis is engaged downstream of UPR activation in MYC transformed cells and required for ER stress prevention. These studies reveal that increased translation and ER stress may be therapeutic vulnerabilities in MYC driven malignancies. For example, PERK or autophagy inhibitors would be predicted to enhance cytotoxic ER stress, while suppression of SIRT7 deacetylase activity could result in unsustainable rates of translation and subsequent proteotoxicity.
3.2. Coordinating protein and lipid metabolism to maintain ER homeostasis
As discussed for mTORC1 (section 2.1), heightened protein synthesis renders tumor cells critically dependent on altered lipid metabolism to support ER homeostasis and viability. Recent work by Carroll et al sheds light on how MYC stimulates various homeostatic processes, including lipogenesis, to sustain cell viability [29] (Figure 2D). The authors demonstrate that MYC induces MondoA, a MYC superfamily member, which cooperates with MYC at a subset of loci, but also transactivates a variety of genes independently of MYC. Within the latter category are processes that limit metabolic stress downstream of MYC, including ER maintenance and lipid biosynthesis. Remarkably, MondoA ablation was selectively toxic in MYC activated cells. Additionally, expression of MondaA correlates with poor prognosis in multiple malignancies, including neuroblastoma, hepatocellular carcinoma, and colon carcinoma. The importance of MondoA dependent lipogenesis was underscored by the finding that provision of exogenous lipid in the form of the unsaturated fatty acid oleic acid was sufficient to rescue MondoA loss. While the authors did not address the mechanisms whereby lipid deprivation led to cell death in MondoA depleted cells, emerging data describing the importance of coordinating protein and lipid synthesis suggest that ER stress may be involved (Figure 2E). Lastly, because MondoA activity requires heterodimerization with MLX, targeted suppression of this pathway may be feasible.
3.3. Maintaining mitochondrial function in MYC transformed cells
Despite the observation that MYC stimulates aerobic glycolysis (Warburg effect), mitochondrial function is vital in MYC transformed cells. First of all, MYC activation enhances dependence on exogenous glutamine to sustain concentrations of TCA cycle metabolites that are consumed for biosynthetic reactions (a process termed anaplerosis) (Figure 2F). MYC drives glutamine-dependent anaplerosis by upregulating genes that promote glutamine uptake (ASCT2 and SLC7A25) and conversion to glutamate (glutaminase, GLS) [30, 31]. Glutamine-dependent anaplerosis requires mitochondrial function, which MYC also supports through multiple mechanisms. First of all, MYC enhances mitochondrial functional capacity by activating peroxisome proliferator-activated receptor gamma coactivator 1α (PGC-1α) and transcription factor A, mitochondrial (TFAM), mediators of mitochondrial biogenesis and gene expression, respectively [32, 33]. MYC also sustains respiratory capacity of mitochondria. In a synthetic lethality screen for genes that are required for viability in MYC transformed cells, Liu et al identified AMPK-related kinase 5 (ARK5), which stimulates AMP-activated kinase (AMPK) to limit mTORC1 activity [34]. ARK5 function was required to sustain mitochondrial function, glutamine-dependent anaplerosis, and cell viability. These ARK5 functions depend on its ability to suppress mTORC1 (Figure 2G). However, the mechanism(s) by which ARK5/AMPK sustains mitochondrial function in MYC activated cells were unclear. One potential mechanism is AMPK-dependent clearance of damaged mitochondria via autophagy, which would be activated upon mTORC1 suppression. In this case, one would expect autophagy inhibition to phenocopy ARK5 ablation in MYC activated cells. Ultimately, direct inhibition of ARK5 or suppression of autophagy-mediated mitochondrial quality control may be tractable targets in MYC driven cancers.
4. RAS
Oncogenic activation of RAS commonly occurs via suppression of its GTPase activity and confers growth factor independent proliferation [35]. To support this commitment to rapid growth, RAS promotes stress resistance and metabolic robustness through a variety of pathways. Here, we summarize recent literature describing these adaptations.
4.1. Maintaining redox homeostasis in RAS transformed cells
A well-recognized feature of RAS activation is increased levels of mitochondria-derived reactive oxygen species (ROS) [5] (Figure 3A). While excessive oxidative stress can induce cell death, low levels of ROS have crucial signaling functions that support tumor growth, including modulation of MAPK and HIF pathways [36, 37]. Multiple mechanisms prevent excessive ROS accumulation in RAS transformed tumor cells.
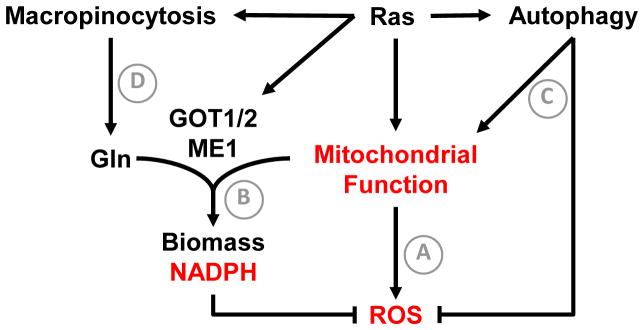
Figure 3. RAS driven adaptations to support tumor growth.
A well-recognized feature of RAS activated cells is increased levels of mitochondrial reactive oxygen species (A). While low levels of ROS promote tumor growth, RAS transformed cells must engage adaptations to prevent excessive oxidative damage. RAS drives glutamine-dependent NADPH generation via aspartate amino transferase (GOT1) and malic enzyme 1 (ME1). NADPH generated from this pathway is required to for redox balance in RAS transformed cells (B). RAS also stimulates high rates of basal autophagy (C), which are required to maintain mitochondrial function and limit oxidative stress. To sustain intracellular amino acid concentrations, such as glutamine, RAS transformed cells engage in macropinocytosis of extracellular protein (D). Note that both autophagy and macropinocytosis-dependent nutrient acquisition require lysosome function, revealing a potential vulnerability of RAS transformed cells. Functional outputs of tumor cell adaptations are shown in red.
NADPH is a source of reducing equivalents for biosynthetic reactions and maintenance of redox homeostasis [5]. Three sources of cellular NADPH have been described (Table 1) [5, 38]. Son et al demonstrated that RAS reprograms glutamine metabolism to support malic-enzyme 1 (ME1) dependent NADPH production in pancreatic cancer (Figure 3B). While most cells utilize glutamate dehydrogenase (GLUD1) to convert glutamine-derived glutamate into α-ketoglutarate, RAS transformed pancreatic cancer cells relied on a distinct pathway in which glutamate undergoes a transamination reaction with oxaloacetate (catalyzed by mitochondrial aspartate aminotransferase, GOT2) to generate aspartate and α-ketoglutarate. Aspartate is then exported to the cytosol where GOT1 generates oxaloacetate. Cytosolic oxaloacetate is metabolized to malate and then pyruvate. This last step, catalyzed by ME1, generates NADPH that is utilized to sustain levels of reduced glutathione for redox balance. RAS directly reprograms the metabolic fate of glutamine by downregulating GLUD1 and upregulating GOT1. This work follows reports that RAS preferentially enhances metabolism of glucose through the non-oxidative pentose phosphate pathway (PPP) to generate nucleotide precursors, rather than the NADPH generating oxidative PPP [39], likely explaining the enhanced requirement for glutamine-dependent NADPH generation. Importantly, RAS driven pancreatic cancer cells were sensitive to AOA (aspartate aminotransferase inhibitor) but not ECGG (glutamate dehydrogenase inhibitor). p53 loss commonly occurs alongside RAS activation in human malignancies. Notably, Jiang et al demonstrated that p53 suppressed ME1 and ME2 expression [40]. Collectively, these findings suggest a greatly enhanced requirement for malic enzyme activity in RAS/p53 mutated pancreatic cancers. Further studies are warranted to test the generalizability of this pathway within other RAS driven malignancies, such as lung cancer.
The critical role of redox balance in RAS transformed cells is further exemplified by their enhanced requirement for autophagy in lung and pancreatic cancers [41, 42] (Figure 3C). Multiple reports indicate that RAS activation induces a high level of basal autophagy, independent of starvation [42, 43]. A common endpoint of inhibiting autophagy in RAS transformed tumors is induction of oxidative stress, DNA damage, and accumulation of dysfunctional mitochondria [41, 42]. As the mitochondria serve both energetic and biosynthetic functions in tumor cells, maintenance of functional mitochondria is absolutely required for disease progression. Consistent with this idea, Guo et al indicate that chronic autophagy suppression diverts RAS driven tumors toward benign oncocytomas rather than carcinoma [41].
4.2. Feeding the RAS-driven cancer
The enhanced growth rate driven by RAS necessitates increased uptake of nutrients to sustain cell proliferation and viability. RAS is known to promote angiogenesis to maintain tumor perfusion [35]. Recent literature suggests that RAS transformed cells also engage in scavenging of both proteins and lipids from the extracellular space to support growth. Commisso et al demonstrate that macropinocytosis-dependent engulfment of extracellular protein (ie. Albumin) can yield intracellular amino acids, including glutamine [44] (Figure 3D). In addition, Kamphorst et al demonstrate that RAS transformed cells take up high levels of serum unsaturated lysophospholipids (phospholipids with one acyl chain) [45]. An understanding of the mechanisms driving lipid scavenging and the function of this phenotype in RAS driven cancers could guide the development of additional therapeutic opportunities. Potential tumor-promoting functions for lipid uptake would include maintenance of ER homeostasis and a decreased reliance on de novo lipid synthesis, which is a heavy NADPH consuming process. The latter would leave more NADPH available for redox balance. Furthermore, macropinocytosis and autophagy both require lysosome function to yield intracellular metabolites, suggesting another vulnerability of RAS driven cancers.
5. HIFs and AMPK: stress sensors that are co-opted for tumor growth
5.1. Hypoxia inducible factors
Cellular responses to low oxygen are primarily orchestrated by the hypoxia inducible factors (HIFs), which include two oxygen labile subunits (HIF-1α and HIF-2α) that independently heterodimerize with ARNT/HIF-1β to mediate adaptive transcriptional changes [46–48]. HIFs mediate broad metabolic changes, including induction of aerobic glycolysis, autophagy, and altered lipid metabolism [46, 49]. Interestingly, multiple oncogenes elicit hypoxia-independent stabilization of HIF-α subunits and co-opt their downstream metabolic adaptations. For example, mTORC1 activation enhances expression of HIF-1α [20, 50], while mTORC2 activation promotes HIF-2α expression [50]. Furthermore, elevated ROS levels are sufficient to inhibit the prolyl-hydroxylase (PHD) enzymes required for HIF degradation. Normoxic stabilization of HIF-1α by elevated ROS, downstream of either mTORC1, MYC, or RAS, promoted tumor growth in various malignancies, including lung cancer, glioma, and lymphoma [51–54].
5.1.1 Hypoxia inducible factors in clear cell renal cell carcinoma
The roles of HIF-1α and HIF-2α in tumor progression are complex and likely context dependent. Nonetheless, there are many examples in which metabolic changes downstream of the HIF pathway can support tumor growth. HIF-1α induces aerobic glycolysis and glutamine dependent lipid synthesis [55–57], while HIF-2α has a prominent role in rewiring lipid metabolism in hepatocytes and clear cell renal cell carcinoma (ccRCC) [58–60]. HIF functions have been extensively studied in the context of ccRCC, where loss of the pVHL E3 ubiquitin ligase results in normoxic stabilization of HIF-1α and HIF-2α in approximately 90% of cases. Functional distinctions between HIF-1α and HIF-2α are evident in ccRCC, where genetic and functional data implicate HIF-1α as a tumor suppressor [61–63], while HIF-2α is absolutely required for disease [64]. Well-characterized tumor promoting functions of HIF-2α include cyclin D1 expression [65], mTORC1 activation [66], and secretion of growth factors that promote proliferation (TGFα) and angiogenesis (VEGFA) [49, 67].
Recent work from our laboratory revealed a surprising functional connection between two hallmarks of ccRCC—HIF-2α activation and enhanced lipid storage [60]. The lipid droplet is functionally and physically associated with the ER, as both protein and lipid species are exchanged between these two organelles [68, 69]. We found that HIF-2α promoted lipid storage by upregulating the lipid droplet coat protein PLIN2. Mechanistically, PLIN2 dependent lipid storage suppressed cytotoxic ER stress responses that otherwise result from elevated protein synthetic activity in ccRCC. A majority of human ccRCC samples exhibit mTORC1 activation [70]. Consistent with our discussion of coordinating protein and lipid metabolism in mTORC1 activated cells (section 2.2), suppression of ER protein load via mTORC1 inhibitors or cycloheximide ameliorated ER stress and cell death in PLIN2 depleted cells. Furthermore, our results revealed ER stress as a targetable vulnerability created by inhibition of HIF-2α-dependent lipid storage, including enhanced sensitivity to the proteasome inhibitor Bortezomib. As HIF-2α specific inhibitors are currently being tested for ccRCC therapy (NCT02293980), our work provides a proof of principle for combining these agents along with ER stress inducers. Future investigations into the specific ER lipid alterations that occur upon PLIN2 depletion may also reveal additional mechanisms for targeting ER homeostasis in ccRCC.
5.2. AMP activated protein kinase
The AMP activated protein kinase (AMPK) is a sensor of cellular energy status; reviewed in [71, 72]. Similar to the HIFs, AMPK has context specific roles in cancer. Tumor suppressive attributes of AMPK include the ability to inhibit mTORC1 and activate p53 [71]. However, many studies provide examples of tumor promoting functions for AMPK, owing to its ability to overcome metabolic stresses typical of advanced tumors. We summarize recent examples here.
5.2.1. Translational control by AMPK in cancer
Translational elongation is often constitutively activated downstream of oncogenic signaling, including mTORC1 and RAS [73]. This process requires activity of elongation factor 2 (EF2), which mediates the translocation stage of translational elongation [73]. Protein synthesis is a costly process with respect to energy and biomass. As a result, in normal cells, EF2 activity is suppressed by inactivating phosphorylation via the EF2 kinase (EF2K) when growth factors or nutrients are limiting, a process involving activating phosphorylation by AMPK [74, 75]. On the other hand, oncogene activation suppresses EF2K activity by various mechanisms, including 1) inhibitory phosphorylation by S6K1 (downstream of mTORC1) or RSK (downstream of RAS) and 2) loss of AMPK-dependent activation of EF2K. Faller et al recently revealed a tumor promoting function of mTORC1-mediated translation elongation in a mouse model of colon cancer [4]. Thus, as in the case of MYC-dependent transformation (section 3.1), heightened translation is a tumor-promoting process downstream of mTORC1 activation.
However, Leprivier et al demonstrated that constitutive activation of translation elongation is a liability under conditions of nutrient deprivation [76]. While acute oncogene activation suppressed the ability of AMPK to activate EF2K, advanced tumors often reactivate the capacity for AMPK to stimulate EF2K-dependent phosphorylation of EF2. This suppression of translation elongation promotes survival in the setting of nutrient deprivation. Advanced human medulloblastoma exhibit greater EF2 phosphorylation compared to normal tissue. Additionally, heightened EF2K expression correlates with poor prognosis in multiple human malignancies, including medulloblastoma and glioma. This suggests that targeting EF2K could be a strategy to enhance metabolic stress in advanced human malignancies. However, like AMPK itself, the role of the EF2K pathway in cancer is context dependent and therapeutic tractability must be determined empirically.
5.2.2. Recruitment of AMPK by oncogene activation or tumor suppressor loss
Similar to the HIF proteins, known oncogenic/tumor suppressive pathways can mediate their metabolic changes though AMPK signaling. Tennakoon et al reported that AMPK was activated in human prostate cancer [77]. Functional studies revealed that androgen signaling promoted AMPK activation and subsequent metabolic adaptations, including mitochondrial biogenesis and enhanced oxidation of glucose and fatty acids. These metabolic alterations were mediated by AMPK-dependent activation of PGC-1α, a master regulator of mitochondrial biogenesis. Importantly, they showed that androgen-induced proliferation required this AMPK/PGC-1α axis.
Similarly, Yan et al recently reported activation of the AMPK/PGC-1α pathway in patients with the hereditary tumor syndrome Birt-Hogg-Dube (BHD) [78]. BHD patients carry germ line mutations in Folliculin (FLCN) and are predisposed to renal cell carcinoma, skin tumors, and lung cysts. FLCN was previously demonstrated to interact with AMPK. In this study, the authors showed that loss of FLCN function activated AMPK-dependent metabolic changes, including mitochondrial biogenesis and enhanced glucose oxidation. The growth advantage derived from metabolic reprogramming due to FLCN loss required activation of HIF-1α downstream of AMPK/PGC-1α driven mitochondrial biogenesis and ROS formation. Furthermore, human BHD tumors exhibit increased mitochondrial content and activation of HIF-1α activity. Interestingly, enhanced HIF-1α activity was not associated with elevated protein levels. Thus, ROS-dependent suppression of PHD function is not sufficient to explain HIF activation. The asparagine hydroxylase factor inhibiting HIF (FIH) is another mechanism for regulation of HIF activity, which functions though suppression of HIF transactivation capacity on chromatin. Interestingly, Masson et al previously reported that FIH is more potently suppressed by reactive oxygen species than the PHD enzymes [79], suggesting a potential explanation for these findings.
Highlighting the complexity and context dependent nature of AMPK function in cancer, Faubert et al demonstrated that AMPK deletion enhanced HIF-1α dependent aerobic glycolysis (Warburg effect) and tumor progression in the context of Myc-driven lymphoma [80]. Thus, while AMPK activity could restrain the heightened anabolic metabolism that is necessary for initial tumor development, it could become crucial in more advanced tumors that experience metabolic stress. Mutations in AMPK subunits are rare in human cancer, suggesting that it may have some tumor promoting functions. Ultimately, evidence of AMPK activation in human cancers could provide insight into the metabolic dependencies of such tumors, including increased mitochondrial respiration and adaptive suppression of translation elongation.
6. Concluding remarks
Altered tumor cell metabolism is now firmly established as a hallmark of human cancer. We have highlighted examples of the vulnerabilities created by heightened anabolic metabolism and summarized recent literature that describes oncogene-induced adaptive processes co-opted to overcome these metabolic challenges. Knowledge of these adaptive functions has the potential to guide additional therapies, and future studies are warranted to examine if a therapeutic window for targeting these processes is available.
Recurrent themes in our review of the literature include maintenance of ER homeostasis and mitochondrial function as crucial adaptations for supporting tumor growth. Experience with proteasome inhibitors in multiple myeloma, where enhanced immunoglobulin synthesis renders tumor cells more sensitive to ER stress-inducing agents, suggests that ER homeostasis can be targeted therapeutically. Additionally, inhibitors of the PERK arm of the UPR have been developed [81]. While on target toxicity within the endocrine pancreas has been noted with these inhibitors [81, 82], the finding that MYC activation enhances the requirement for PERK-mediated adaptations suggests that a therapeutic window may be possible. Multiple studies have revealed the importance of maintaining ER lipid homeostasis to prevent cytotoxic ER stress that would otherwise occur downstream of oncogene-induced protein synthesis. Thus, disruption of the balance between protein and lipid synthesis (or uptake) may be another targetable tumor cell vulnerability.
The antitumor effect of inhibiting autophagy has been demonstrated in many preclinical models and numerous clinical trials in human malignancies are under-way [12]. From the work reviewed here, RAS and MYC driven malignancies would be predicted to be sensitive to such therapies. In the case of RAS driven pancreatic cancer cells, the mitochondria are vital for glutamine-dependent NADPH generation and redox balance. Perhaps compromised mitochondrial function in autophagy-inhibited cells would enhance the anti-tumor effect of inhibiting glutamine uptake and metabolism. While a role for autophagy in MYC transformed cells has been demonstrated, a greater understanding of the mechanisms by which autophagy supports tumor growth would yield similar opportunities to target tumor cell metabolic vulnerabilities.
Acknowledgments
This work was supported by NIH grant 2-P01-CA104838 to M.C.S, and NIH fellowship 5-F30-CA177106 to B.Q.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–5. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faller WJ, Jackson TJ, Knight JR, Ridgway RA, Jamieson T, Karim SA, et al. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature. 2015;517:497–500. doi: 10.1038/nature13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabharwal SS, Schumacker PT. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles’ heel? Nat Rev Cancer. 2014;14:709–21. doi: 10.1038/nrc3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–8. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, et al. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339:1320–3. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- 9.Chantranupong L, Wolfson RL, Sabatini DM. Nutrient-Sensing Mechanisms across Evolution. Cell. 2015;161:67–83. doi: 10.1016/j.cell.2015.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:13571–6. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Nicholatos J, Dreier JR, Ricoult SJ, Widenmaier SB, Hotamisligil GS, et al. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 2014;513:440–3. doi: 10.1038/nature13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White E. The role for autophagy in cancer. The Journal of clinical investigation. 2015;125:42–6. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kenific CM, Debnath J. Cellular and metabolic functions for autophagy in cancer cells. Trends in cell biology. 2015;25:37–45. doi: 10.1016/j.tcb.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozcan U, Ozcan L, Yilmaz E, Duvel K, Sahin M, Manning BD, et al. Loss of the tuberous sclerosis complex tumor suppressors triggers the unfolded protein response to regulate insulin signaling and apoptosis. Molecular cell. 2008;29:541–51. doi: 10.1016/j.molcel.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke HJ, Chambers JE, Liniker E, Marciniak SJ. Endoplasmic reticulum stress in malignancy. Cancer cell. 2014;25:563–73. doi: 10.1016/j.ccr.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 16.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–6. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 17.Obeng EA, Carlson LM, Gutman DM, Harrington WJ, Jr, Lee KP, Boise LH. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107:4907–16. doi: 10.1182/blood-2005-08-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–6. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu S, Yang L, Li P, Hofmann O, Dicker L, Hide W, et al. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature. 2011;473:528–31. doi: 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Molecular cell. 2010;39:171–83. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell metabolism. 2008;8:224–36. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–20. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young RM, Ackerman D, Quinn ZL, Mancuso A, Gruber M, Liu L, et al. Dysregulated mTORC1 renders cells critically dependent on desaturated lipids for survival under tumor-like stress. Genes & development. 2013;27:1115–31. doi: 10.1101/gad.198630.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffiths B, Lewis CA, Bensaad K, Ros S, Zhang Q, Ferber EC, et al. Sterol regulatory element binding protein-dependent regulation of lipid synthesis supports cell survival and tumor growth. Cancer & metabolism. 2013;1:3. doi: 10.1186/2049-3002-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medvetz D, Priolo C, Henske EP. Therapeutic targeting of cellular metabolism in cells with hyperactive mTORC1: a paradigm shift. Molecular cancer research : MCR. 2015;13:3–8. doi: 10.1158/1541-7786.MCR-14-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hart LS, Cunningham JT, Datta T, Dey S, Tameire F, Lehman SL, et al. ER stress-mediated autophagy promotes Myc-dependent transformation and tumor growth. The Journal of clinical investigation. 2012;122:4621–34. doi: 10.1172/JCI62973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin J, He M, Liu Y, Paredes S, Villanova L, Brown K, et al. SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell reports. 2013;5:654–65. doi: 10.1016/j.celrep.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carroll PA, Diolaiti D, McFerrin L, Gu H, Djukovic D, Du J, et al. Deregulated Myc requires MondoA/Mlx for metabolic reprogramming and tumorigenesis. Cancer cell. 2015;27:271–85. doi: 10.1016/j.ccell.2014.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–5. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009;15:6479–83. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, et al. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Molecular and cellular biology. 2005;25:6225–34. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes & development. 2004;18:357–68. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 34.Liu L, Ulbrich J, Muller J, Wustefeld T, Aeberhard L, Kress TR, et al. Deregulated MYC expression induces dependence upon AMPK-related kinase 5. Nature. 2012;483:608–12. doi: 10.1038/nature10927. [DOI] [PubMed] [Google Scholar]
- 35.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–74. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8788–93. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finkel T. Signal transduction by reactive oxygen species. The Journal of cell biology. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–70. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang P, Du W, Mancuso A, Wellen KE, Yang X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature. 2013;493:689–93. doi: 10.1038/nature11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes & development. 2013;27:1447–61. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes & development. 2011;25:717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes & development. 2011;25:460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–7. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamphorst JJ, Cross JR, Fan J, de Stanchina E, Mathew R, White EP, et al. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8882–7. doi: 10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Molecular cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Molecular cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 49.Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2012;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toschi A, Lee E, Gadir N, Ohh M, Foster DA. Differential dependence of hypoxia-inducible factors 1 alpha and 2 alpha on mTORC1 and mTORC2. The Journal of biological chemistry. 2008;283:34495–9. doi: 10.1074/jbc.C800170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blum R, Jacob-Hirsch J, Amariglio N, Rechavi G, Kloog Y. Ras inhibition in glioblastoma down-regulates hypoxia-inducible factor-1alpha, causing glycolysis shutdown and cell death. Cancer research. 2005;65:999–1006. [PubMed] [Google Scholar]
- 52.Chen C, Pore N, Behrooz A, Ismail-Beigi F, Maity A. Regulation of glut1 mRNA by hypoxia-inducible factor-1. Interaction between H-ras and hypoxia. The Journal of biological chemistry. 2001;276:9519–25. doi: 10.1074/jbc.M010144200. [DOI] [PubMed] [Google Scholar]
- 53.Gao P, Zhang H, Dinavahi R, Li F, Xiang Y, Raman V, et al. HIF-dependent antitumorigenic effect of antioxidants in vivo. Cancer cell. 2007;12:230–8. doi: 10.1016/j.ccr.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Faubert B, Vincent EE, Griss T, Samborska B, Izreig S, Svensson RU, et al. Loss of the tumor suppressor LKB1 promotes metabolic reprogramming of cancer cells via HIF-1alpha. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2554–9. doi: 10.1073/pnas.1312570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun RC, Denko NC. Hypoxic regulation of glutamine metabolism through HIF1 and SIAH2 supports lipid synthesis that is necessary for tumor growth. Cell metabolism. 2014;19:285–92. doi: 10.1016/j.cmet.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–4. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gameiro PA, Yang J, Metelo AM, Perez-Carro R, Baker R, Wang Z, et al. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell metabolism. 2013;17:372–85. doi: 10.1016/j.cmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rankin EB, Rha J, Selak MA, Unger TL, Keith B, Liu Q, et al. Hypoxia-inducible factor 2 regulates hepatic lipid metabolism. Molecular and cellular biology. 2009;29:4527–38. doi: 10.1128/MCB.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walter KM, Schonenberger MJ, Trotzmuller M, Horn M, Elsasser HP, Moser AB, et al. Hif-2alpha promotes degradation of Mammalian peroxisomes by selective autophagy. Cell metabolism. 2014;20:882–97. doi: 10.1016/j.cmet.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 60.Qiu B, Ackerman D, Sanchez DJ, Li B, Ochocki JD, Grazioli A, et al. HIF-2alpha dependent lipid storage promotes endoplasmic reticulum homeostasis in clear cell renal cell carcinoma. Cancer discovery. 2015 doi: 10.1158/2159-8290.CD-14-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen C, Beroukhim R, Schumacher SE, Zhou J, Chang M, Signoretti S, et al. Genetic and functional studies implicate HIF1alpha as a 14q kidney cancer suppressor gene. Cancer discovery. 2011;1:222–35. doi: 10.1158/2159-8290.CD-11-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raval RR, Lau KW, Tran MG, Sowter HM, Mandriota SJ, Li JL, et al. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Molecular and cellular biology. 2005;25:5675–86. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maranchie JK, Vasselli JR, Riss J, Bonifacino JS, Linehan WM, Klausner RD. The contribution of VHL substrate binding and HIF1-alpha to the phenotype of VHL loss in renal cell carcinoma. Cancer cell. 2002;1:247–55. doi: 10.1016/s1535-6108(02)00044-2. [DOI] [PubMed] [Google Scholar]
- 64.Kondo K, Kim WY, Lechpammer M, Kaelin WG., Jr Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 2003;1:E83. doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schodel J, Bardella C, Sciesielski LK, Brown JM, Pugh CW, Buckle V, et al. Common genetic variants at the 11q13. 3 renal cancer susceptibility locus influence binding of HIF to an enhancer of cyclin D1 expression. Nature genetics. 2012;44:420–5. S1–2. doi: 10.1038/ng.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Elorza A, Soro-Arnaiz I, Melendez-Rodriguez F, Rodriguez-Vaello V, Marsboom G, de Carcer G, et al. HIF2alpha acts as an mTORC1 activator through the amino acid carrier SLC7A5. Molecular cell. 2012;48:681–91. doi: 10.1016/j.molcel.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 67.Gunaratnam L, Morley M, Franovic A, de Paulsen N, Mekhail K, Parolin DA, et al. Hypoxia inducible factor activates the transforming growth factor-alpha/epidermal growth factor receptor growth stimulatory pathway in VHL(−/−) renal cell carcinoma cells. The Journal of biological chemistry. 2003;278:44966–74. doi: 10.1074/jbc.M305502200. [DOI] [PubMed] [Google Scholar]
- 68.Walther TC, Farese RV., Jr Lipid droplets and cellular lipid metabolism. Annual review of biochemistry. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, et al. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Developmental cell. 2013;24:384–99. doi: 10.1016/j.devcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haddad AQ, Kapur P, Singla N, Raman JD, Then MT, Nuhn P, et al. Validation of mammalian target of rapamycin biomarker panel in patients with clear cell renal cell carcinoma. Cancer. 2014 doi: 10.1002/cncr.28976. [DOI] [PubMed] [Google Scholar]
- 71.Faubert B, Vincent EE, Poffenberger MC, Jones RG. The AMP-activated protein kinase (AMPK) and cancer: many faces of a metabolic regulator. Cancer letters. 2015;356:165–70. doi: 10.1016/j.canlet.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 72.Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nature reviews Molecular cell biology. 2012;13:251–62. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruggero D. Translational control in cancer etiology. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Browne GJ, Finn SG, Proud CG. Stimulation of the AMP-activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. The Journal of biological chemistry. 2004;279:12220–31. doi: 10.1074/jbc.M309773200. [DOI] [PubMed] [Google Scholar]
- 75.Horman S, Browne G, Krause U, Patel J, Vertommen D, Bertrand L, et al. Activation of AMP-activated protein kinase leads to the phosphorylation of elongation factor 2 and an inhibition of protein synthesis. Curr Biol. 2002;12:1419–23. doi: 10.1016/s0960-9822(02)01077-1. [DOI] [PubMed] [Google Scholar]
- 76.Leprivier G, Remke M, Rotblat B, Dubuc A, Mateo AR, Kool M, et al. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell. 2013;153:1064–79. doi: 10.1016/j.cell.2013.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tennakoon JB, Shi Y, Han JJ, Tsouko E, White MA, Burns AR, et al. Androgens regulate prostate cancer cell growth via an AMPK-PGC-1alpha-mediated metabolic switch. Oncogene. 2014;33:5251–61. doi: 10.1038/onc.2013.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yan M, Gingras MC, Dunlop EA, Nouet Y, Dupuy F, Jalali Z, et al. The tumor suppressor folliculin regulates AMPK-dependent metabolic transformation. The Journal of clinical investigation. 2014;124:2640–50. doi: 10.1172/JCI71749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Masson N, Singleton RS, Sekirnik R, Trudgian DC, Ambrose LJ, Miranda MX, et al. The FIH hydroxylase is a cellular peroxide sensor that modulates HIF transcriptional activity. EMBO Rep. 2012;13:251–7. doi: 10.1038/embor.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Faubert B, Boily G, Izreig S, Griss T, Samborska B, Dong Z, et al. AMPK is a negative regulator of the Warburg effect and suppresses tumor growth in vivo. Cell metabolism. 2013;17:113–24. doi: 10.1016/j.cmet.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Atkins C, Liu Q, Minthorn E, Zhang SY, Figueroa DJ, Moss K, et al. Characterization of a novel PERK kinase inhibitor with antitumor and antiangiogenic activity. Cancer research. 2013;73:1993–2002. doi: 10.1158/0008-5472.CAN-12-3109. [DOI] [PubMed] [Google Scholar]
- 82.Gao Y, Sartori DJ, Li C, Yu QC, Kushner JA, Simon MC, et al. PERK is required in the adult pancreas and is essential for maintenance of glucose homeostasis. Molecular and cellular biology. 2012;32:5129–39. doi: 10.1128/MCB.01009-12. [DOI] [PMC free article] [PubMed] [Google Scholar]