Abstract
Purpose
To evaluate the use of three EMR methods to quantify APT and NOE signals in human glioma.
Methods
Eleven patients with high-grade glioma were scanned at 3 T. aEMR2 (asymmetric magnetization-transfer or MT model to fit two-sided, wide-offset data), sEMR2 (symmetric MT model to fit two-sided, wide-offset data), and sEMR1 (symmetric MT model to fit one-sided, wide-offset data) were assessed. ZEMR and experimental data at 3.5 ppm and −3.5 ppm were subtracted to calculate the APT and NOE signals (APT# and NOE#), respectively.
Results
The aEMR2 and sEMR1 models provided quite similar APT# signals, while the sEMR2 provided somewhat lower APT# signals. The aEMR2 had an erroneous NOE# quantification. Calculated APT# signal intensities of glioma (~4%), much larger than the values reported previously, were significantly higher than those of edema and normal tissue. Compared to normal tissue, gadolinium-enhancing tumor cores were consistently hyperintense on the APT# maps and slightly hypointense on the NOE# maps.
Conclusion
The sEMR1 model is the best choice for accurately quantifying APT and NOE signals. The APT-weighted hyperintensity in the tumor was dominated by the APT effect, and the MT asymmetry at 3.5 ppm is a reliable and valid metric for APT imaging of gliomas at 3 T.
Keywords: MT, CEST, APT, NOE, brain glioma
INTRODUCTION
Chemical exchange saturation transfer (CEST) MRI is a novel molecular imaging technique by which to detect low-concentration, exogenous or endogenous targeted CEST agents (1–3), and probe the tissue microenvironment, such as the pH (4, 5). Amide proton transfer (APT) imaging (5), an important type of CEST MRI, is capable of detecting endogenous mobile proteins and peptides in tissue, such as those in the cytoplasm (6). Technically, the APT effect is usually quantified by a reduction in bulk water intensity, due to the chemical exchange of water protons with magnetically labeled backbone amide protons of endogenous mobile proteins and peptides at ~3.5 ppm downfield of the water resonance. Thus, specific protein information can be obtained indirectly though the bulk water signal usually used in water-based MR imaging. In recent years, there has been much progress in CEST applications, such as detecting and grading tumors (7–9), assessing tumor response to therapy (10–12), imaging the ischemic penumbra in acute stroke based on tissue acidosis (13–15), detecting neurotransmitters in the brain (16), imaging glucose uptake and metabolism in tumors (17, 18), detecting Parkinson’s disease (19), studying creatine kinetics (20), and characterizing pediatric brain development (21).
To demonstrate a specific CEST effect, a Z-spectrum is often generated by acquiring bulk water signal intensities as a function of radiofrequency (RF) saturation frequency offsets. In theory, the CEST enhancement displayed in the Z-spectrum depends on pool sizes, exchange rates, and relaxation times of exchangeable protons (22–24). CEST is often confounded by the effects of direct water saturation (DS) and the conventional semi-solid macromolecular magnetization transfer (MT) (25, 26). When the MT asymmetry analysis is used for quantification (5), the CEST signal is further confounded by the nuclear Overhauser enhancement (NOE) effects from non-exchangeable protons that resonate in the upfield region from water (27–32). The NOE signal in vivo may arise from the interaction between the water protons and mobile and relatively mobile proteins, peptides, metabolites, and lipids in tissue (33), by way of the intermolecular through-space dipolar coupling, the intramolecular NOE-relayed CEST process (28), and the intramolecular NOE-relayed spectral overlap with the water signal (34). The mobile biomolecules in tissue (T2 ~ 10 ms) detected either by CEST or NOE imaging have a relatively narrow RF saturation width (on the order of ppm), showing well-defined features in the Z-spectrum. Based on this, we hypothesize that the relatively less mobile biomolecules (T2 ~ 0.1‒1 ms) that have a relatively broad saturation width (tens of ppm) are responsible for the Z-spectrum asymmetry observed in CEST imaging (33). Note that the conventional MT imaging is sensitive to semi-solid macromolecules (T2 ~ 10 μs), with an even broad RF saturation width of hundreds of ppm (35, 36). Consequently, the Z-spectrum asymmetry analysis can remove the nearly symmetric DS and semi-solid MT effects (Supporting Fig. S1), whereas the asymmetric NOE signals from the mobile and relatively mobile proteins, peptides, metabolites, and lipids in tissue may remain in CEST imaging.
We recently introduced a new and straightforward fitting approach using extrapolated semi-solid MT reference (EMR) signals to better quantify the APT (APT#) and NOE (NOE#) signals (37). The method (called sEMR2) used the well-established, symmetric Henkelman’s two-pool MT model with a super-Lorentzian lineshape (35, 36) to fit the two-sided, wide-offset MT data. Further, it has been shown that four-pool CEST data fitting, using calculated semi-solid MT parameters as prior known information, could reduce the over-fitting errors for APT and NOE. In this current study, two more possible EMR approaches were assessed: aEMR2 (using a modified, asymmetric MT model (38) to fit the two-sided, wide-offset Z-spectrum data); and sEMR1 (using a symmetric MT model to fit the one-sided, wide-offset Z-spectrum data). The aEMR2 method assumes the chemical shift center mis-match between bulk water and semi-solid macromolecules (38). The sEMR1 method divides the asymmetric Z-spectrum observed into the conventional, symmetric, semi-solid Z-spectrum component (Supporting Fig. S1) and the upfield asymmetric NOE component of the aliphatic and olefinic protons of various relatively less mobile molecules. The purpose of this study was to assess the quantitative APT# and NOE# signals measured by three different EMR models and compare them with the commonly used MTRasym (3.5ppm) parameters in human gliomas at 3 Tesla.
METHODS
Theory
The reversible exchange process in a two-pool model can be depicted by the Bloch equations, modified with the coupling terms, which consist of a free bulk water proton pool (w) and a semi-solid macromolecular proton pool (m) (35, 36), as described in Supporting Theory. In the sEMR1 and sEMR2 models, the symmetric MT signal expression can be uniquely determined in terms of five combined model parameters (39, 40), R, T1m, T2m, and T1w/T2w, where T1w and T2w are the longitudinal and transverse relaxation times of the free water proton pool, respectively; T1m and T2m are the longitudinal and transverse relaxation times of the semi-solid macromolecular proton pool, respectively; and , is the fully-relaxed equilibrium magnetization value associated with the semi-solid macromolecular pool; and R is the rate constant describing the magnetization exchange between the two proton pools. After these five model parameters are obtained by fitting the observed wide-offset MT data, the EMR spectra (ZEMR) can be calculated with the corresponding RF irradiation amplitude, ω1, and frequency offset, Δm. For the aEMR2 model, the MT asymmetry can be described by assuming an average frequency difference between the semi-solid macromolecular protons and the free water protons, Δmw. The asymmetric MT signal expression can be determined in terms of six combined model parameters, R, T1m, T2m, , T1w/T2w and Δmw (38).
Patient Recruitment
The study was approved by the Johns Hopkins Institutional Review Board. Before involvement in this study, written, informed consent was obtained from all patients. Eleven patients (eight males, three females; median age, 50 years; age range, 30–65 years) with a confirmed high-grade glioma were recruited for this study.
MRI Experiments
All patients were scanned on a Philips 3 T MRI scanner (Achieva 3.0 T; Philips Medical Systems, Best, The Netherlands) using a body coil for RF transmission and a 32-channel sensitivity-encoding coil for reception (Invivo, Gainsville, FL). Several standard MR images were acquired for reference, including T2-weighted (repetition time or TR = 2772 ms; echo time or TE = 80 ms; field of view or FOV = 212 × 190 mm2; slice thickness = 2.2 mm; slice spacing = 1 mm; matrix size = 512 × 512; 60 slices; and scan time = 3 min), fluid-attenuated inversion recovery (FLAIR: TR = 11 s; TE = 120 ms; inversion recovery time or TI = 2.8 s; FOV = 212 × 190 mm2; slice thickness = 2.2 mm; slice spacing = 1 mm; matrix size = 512 × 512; 60 slices; and scan time = 3 min), T1-weighted, and gadolinium-enhanced T1-weighted (three-dimensional, magnetization-prepared, rapid-gradient-echo sequence; TR = 3 s; TE = 3.7 ms; TI = 843 ms; FA = 8°; 150 slices; isotropic voxel = 1.1 mm3). T2-weighted and FLAIR imaging were acquired first to localize the tumor for APT imaging, and gadolinium (Gd)-enhanced T1-weighted imaging was the last sequence acquired.
CEST image data were obtained using a fat-suppressed, fast spin-echo pulse sequence using the following parameters: TR = 3 s; TE = 6.4 ms; FOV = 212 × 190 mm2; matrix size = 256 × 256; slice thickness = 4.4 mm; turbo-spin-echo factor = 45; and single slice acquisition. The RF saturation section included a series of four block RF saturation pulses (200 ms duration each and 2μT amplitude), each followed by a crusher gradient (10 ms duration and 10 mT/m strength). This interleaved approach was used to satisfy amplifier requirements regarding unblank time and duty cycle. The frequency sweep corresponded to a full Z-spectrum with 64 frequency offsets: off (S0 image), 0, ±0.5, …, and 14 ppm in intervals of 0.5 ppm. In order to obtain sufficient signal-to-noise ratios for the APT and NOE images, four acquisitions were placed at ±3.5 ppm. The water saturation shift-referencing (WASSR) method (26 offsets from 1.2 to −1.2 ppm at intervals of 0.125 ppm, one average, B1 = 0.5 μT) was used to determine B0 maps (41).
Data Processing and Fitting Procedure
All data processing was performed using Interactive Data Language (IDL, Version7; Exelis Visual Information Solutions, Inc., Boulder, CO) or MATLAB (The MathWorks, Inc., Natick, MA). Figure 1 shows the flow chart of the data processing procedures. First, the WASSR method, with Z1.2 ~ −1.2ppm, was used to correct for B0 field inhomogeneity effects (41). Z14 ~ −14ppm spectra were interpolated and aligned correspondingly on a pixel-by-pixel basis along the direction of the frequency offset axis. Then, the wide-offset data were fitted to Henkelman’s two-pool MT model with a super-Lorentzian lineshape for the aEMR2, sEMR2, and sEMR1 models. For the aEMR2 and sEMR2 models, data points of small frequency offsets between 7 and −7 ppm (Z′14 ~ −14ppm) were excluded prior to conventional MT modeling to avoid possible APT and mobile NOE contributions. For the sEMR1 model, data points of small frequency offsets between 7 and −14 ppm (Z14 ~ 7ppm) were excluded to avoid possible APT and NOE contributions, including NOEmobile and NOEless mobile effects.
Figure 1.
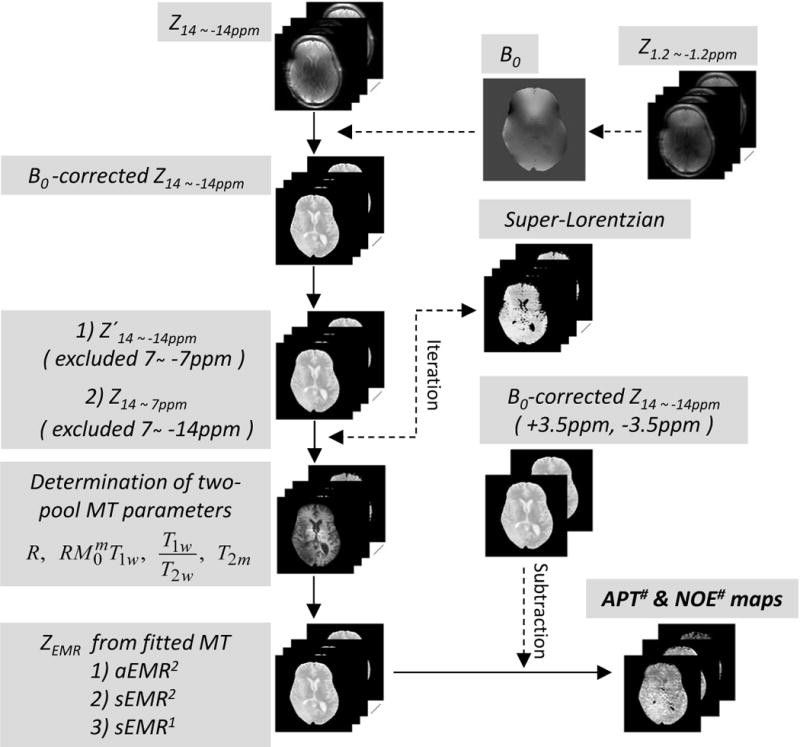
Flow chart of data processing procedures for APT# and NOE#. After B0 corrections with WASSR, the wide-offset dataset, Z′14 ~ −14ppm or Z14 ~ 7ppm, was fitted to the two-pool MT model with a super-Lorentzian lineshape, and R, T2m, , and T1w/T2w and Δmw (for aEMR2) were determined from the MT fitting routine. By subtracting experimental data (Z14 ~ −14ppm) from fitted EMR data, APT# and NOE# signals could be obtained.
Further, the independent semi-solid MT model parameters (R, T2m, , T1w/T2w, Δmw for aEMR2) were obtained by fitting all modified Z′14 ~ −14ppm and Z14 ~ 7ppm data to the two-pool semi-solid MT models (Eq. [S1] in Supporting Theory), based on the nonlinear least-squares fitting approach, which implemented the Levenberg-Marquardt algorithm. In this procedure, was conventionally normalized to 1 (35, 42). T1m was set as a constant value of 1.4 s (43) because it could not be determined well from fits. The super-Lorentzian function, characterized by one parameter, T2m, was evaluated by numerical integration. Fortunately, the singularity in the super-Lorentzian lineshape could be avoided during the MT fitting because the wide-frequency offsets were not defined at on-resonance. However, the super-Lorentzian value was extrapolated from 128 Hz to the asymptotic limit at the zero offset when drawing ZEMR curves. The quality of the estimated MT model parameters was evaluated by the root of the sum of the signal-normalized squared difference between the fitted and experimental data, and the χ2 goodness-of-fit metric. Finally, estimated baselines based on the three models were obtained, and APT# and NOE# were calculated by subtracting the corrected experimental data from the EMR data.
To quantitatively compare the APT# and NOE# signals in the glioma, regions of interest (ROIs) were analyzed. Three ROIs, enclosing the contralateral normal-appearing white matter (CNAWM), the glioma, and the edema, were carefully drawn on the conventional MR images. Data in graphs were presented as mean ±standard error. Statistical analysis was performed using one-way ANOVA, followed by Tukey’s post-hoc test. Statistical significance was considered at p < 0.05.
RESULTS
Figure 2 compares the average Z-spectra and MTRasym spectra of all ROIs from seven patients. The Z-spectra in the glioma and in the peritumoral edema overall showed higher signal intensities for all frequency offsets than those in the CNAWM. As reported previously (33), all MTRasym spectra were negative at the higher frequency (>5.5 ppm) due to the upfield NOEless mobile effects. The presence of the APT and other CEST effects became more pronounced between 1 ~ 4 ppm in the MTRasym spectra, and a higher MTRasym (3.5 ppm) signal was observed in the glioma, compared to the edema and the CNAWM.
Figure 2.
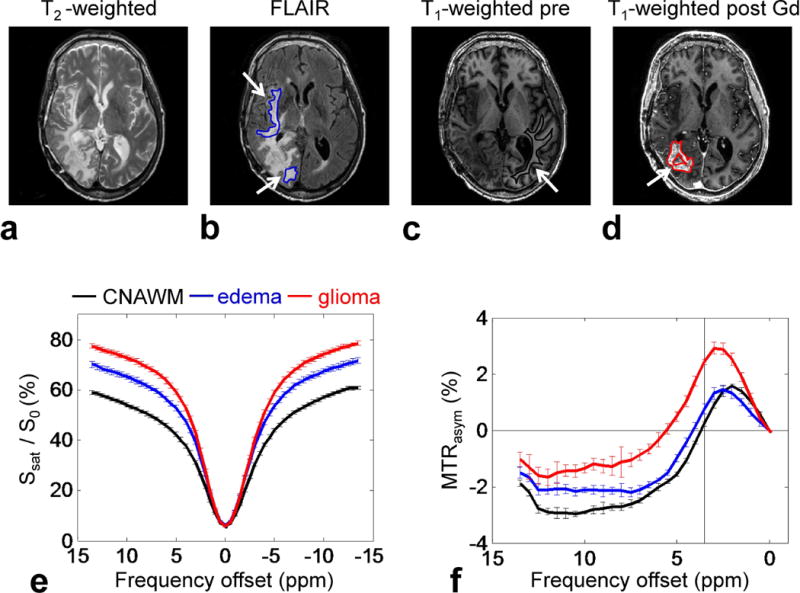
ROI analysis results from seven patients. a–d: Example of ROIs on conventional MR images for a patient with a glioblastoma (edema, blue; CNAWM, black; glioma, red). e–f: Average Z-spectra and MTRasym spectra for the CNAWM (black), edema (blue), and glioma (red).
Figure 3 shows the average two-pool MT-fitted results from the glioma (n = 11). Supporting Table S1 summarizes the fitted MT model parameters (R, T2m, , and T1w/T2w, Δmw for aEMR2). As reported previously (37), the use of the super-Lorentzian lineshape fits the lineshape of the curves quite well for wide-frequency offsets, yielding a close fit for Z′14 ~ −14ppm or Z14 ~ 7ppm. Based on Z′14 ~ −14ppm data (where frequency offsets between 7 and −7 ppm were excluded to remove CEST and most mobile NOE effects close to the water resonance), the aEMR2 model accurately predicted the behavior of the semi-solid MT system for wide-frequency offsets, while the sEMR2 model was associated with a deviation of one-to-two percentage points at both the positive and negative frequency offsets, primarily due to the asymmetric effect contributed by the upfield NOEless mobile signals. Based on Z14 ~ 7ppm data (where frequency offsets between 7 and −14 ppm were excluded to remove CEST, mobile NOE, and relatively less mobile NOE contributions), the sEMR1 model fitted the MT behavior very well. Notably, the ZEMR spectra with the sEMR1 model overall showed higher signal intensities for all negative frequency offsets, compared with the experimental Z14 ~ −14ppm in the ROI, providing further evidence regarding the presence of mobile NOE and relatively less mobile NOE contributions.
Figure 3.
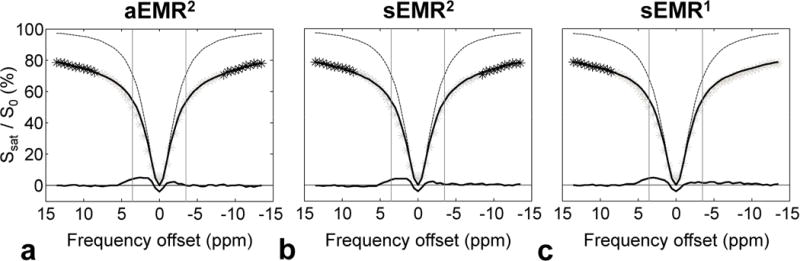
Average two-pool MT fitted results (solid lines) obtained with three EMR models, and experimental data (black asterisks) from the glioma (n = 11). a: aEMR2 fit using Z′14 ~ −14ppm data. b: sEMR2 fit using Z′14 ~ −14ppm data. c: sEMR1 fit using Z14 ~ 7ppm data. The Lorentzian lineshapes of bulk water protons (dashed lines) are shown for comparison. The downfield APT# and upfield NOE# features were obtained by subtracting the experimental Z-spectra (gray asterisks) from the corresponding EMR spectra.
The simulated conventional MT Z-spectra based on the aEMR2 model were asymmetric around the water resonance frequency, with lower signal intensities appearing on the negative frequency offset side, as shown in Fig. 4. According to the usual definition (Eq. [S5] in Supporting Theory), the aEMR2 asymmetry changed from 0 to a few negative percentage points at the offset of ~4 ppm (around −2.8% for the CNAWM, −2.4% for edema, −1.1% for glioma) and then remained relatively stable (around −2.5% for the CNAWM, −2% for edema, −1% for glioma) for the offsets far from water (>5 ppm). Therefore, the magnitude of the upfield NOEless mobile effects was dependent on the frequency offset and the tissue type. The maximal aEMR2 asymmetry was observed in the CNAWM, which had a chemical shift center about 1.55 ± 0.1 ppm upfield from the water. The chemical shift center was found to be 1.26 ± 0.3 ppm and 0.51 ± 0.4 ppm for the edema and the glioma, respectively.
Figure 4.
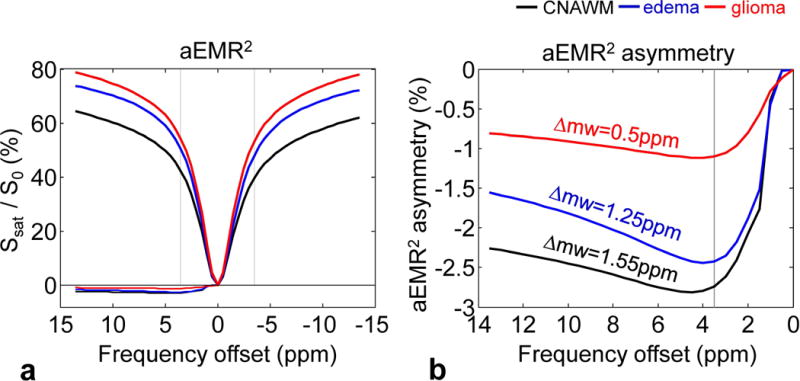
a: Average aEMR2 spectra and asymmetry spectra obtained from the CNAWM (black), the edema (blue), and the glioma (red) from 11 patients. The aEMR2 spectra are asymmetric with the lower signal intensities on the negative frequency offset. b: Detailed aEMR2 asymmetry spectra. The asymmetric effect is dependent on the frequency offset and different tissue type.
Figure 5 shows the experimentally measured downfield APT# and upfield NOE# signal features as a function of frequency offsets, which were obtained by subtracting the experimental data (gray asterisks in Fig. 3) from the corresponding ZEMR curves (solid lines in Fig. 3). The downfield CEST# signals may be attributable to amide protons (APT#, 3.5 ppm downfield from water), amine protons (2 ppm downfield from water), and other possible sources. The upfield NOE# signals may be attributable (according to the mobility) to mobile biomolecules (T2 ~ 10 ms) and less mobile biomolecules (T2 ~ 0.1–1 ms, namely, between semi-solid and mobile; for sEMR1 and sEMR2). Note that a large dip around water frequency was observed in all three EMR methods, which was caused by a non-zero saturation intensity at water frequency (Fig. 2e), due to the low signal-to-noise ratio and the remaining B0 inhomogeneity.
Figure 5.
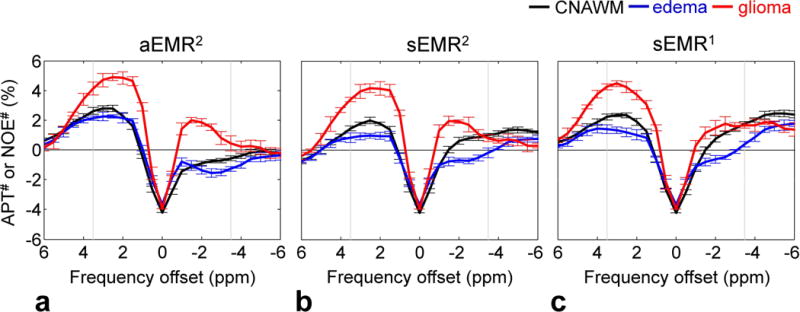
Downfield APT# and upfield NOE# signal features as a function of frequency offsets obtained with the aEMR2 (a), sEMR2 (b), and sEMR1 (c) models from the CNAWM (black), the edema (blue), and the glioma (red) from 11 patients. The results were obtained by subtracting the experimental Z-spectra (gray asterisks in Fig. 3) from the corresponding EMR spectra (solid lines in Fig. 3).
Figure 6 quantitatively compares the MTRasym (3.5ppm), APT#, NOE#, and δ (for aEMR2 only) in the CNAWM, peritumoral edema, and glioma across the three different EMR models used. Based on this, several important results can be observed. (i) The MTRasym (3.5ppm) signal of the glioma was significantly higher than those of the edema and the CNAWM (both p < 0.001), as expected. (ii) The APT# signals measured for all ROIs from the three EMR methods were overall larger than the corresponding NOE# signals, due to the use of the relatively larger RF power levels (2 μT), as reported previously (33). (iii) The downfield APT# signals of the glioma were significantly higher than those of the edema and the CNAWM for each of the three EMR models (all p < 0.05). The APT# signals seemed slightly lower in the edema than in the CNAWM, but the differences were statistically not significant. (iv) Interestingly, the APT# signals measured by the aEMR2 and sEMR1 were quite similar for all ROIs. However, the APT# signals measured by the sEMR2 were somewhat lower, particularly the values for the edema and the CNAWM, which were significantly lower than those by the aEMR2 (both p < 0.05). (v) The upfield NOE# signals were slightly lower in edema than in glioma and the CNAWM, and the differences were most statistically significant (p < 0.05). Note that the negative NOE# values in the CNAWM and the edema (with larger Δmw values), using the aEMR2 model, were observed due to the largely overestimated NOEless mobile that resulted from the oversimplified assumption of only one chemical shift center, Δmw.
Figure 6.
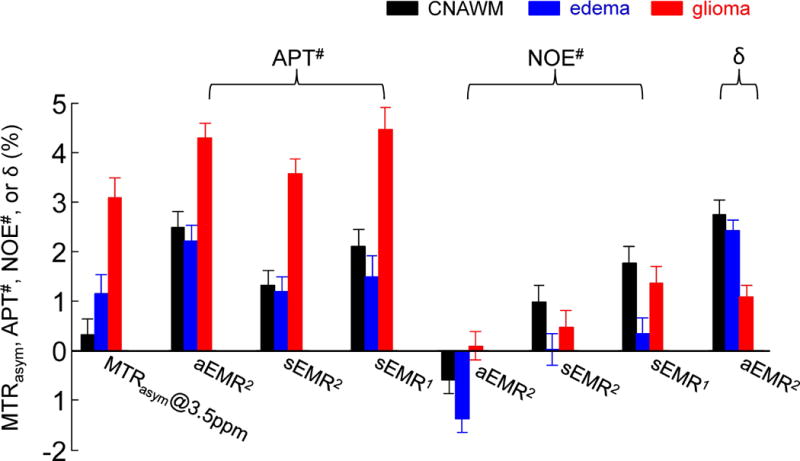
Average MTRasym (3.5ppm), APT#, NOE#, and δ (for aEMR2 only) signal intensities obtained with three EMR models from the CNAWM, edema, and glioma from 11 patients. APT# = ZEMR (+3.5ppm) − Z(+3.5ppm); NOE# = ZEMR (−3.5ppm) − Z(−3.5ppm); δ = ZEMR (3.5ppm) − ZEMR (−3.5ppm), for aEMR2 only. Error bars depict standard errors. Calculated APT# signal intensities of glioma (~4%) were much larger than the values reported previously (7).
Figure 7 further compares the image contrasts of MTRasym (3.5ppm), APT#, NOE#, and δ (for aEMR2 only) between the glioma (or the edema) and the CNAWM for the three EMR methods. (i) The downfield APT# image contrasts between the glioma and the CNAWM were consistently positive across all EMR models (p < 0.05). (ii) The upfield NOE# image contrast between the glioma and the CNAWM were negative for both sEMR2 and sEMR1. For the aEMR2 method, the δ image contrast between the glioma and the CNAWM was negative. The upfield NOE# image contrast between the glioma and the CNAWM was positive; however, the (NOE# + δ) image contrast between the glioma and the CNAWM also became negative. (iii) Interestingly, the downfield APT# and upfield NOE# image contrasts between the edema and the CNAWM were all negative across all EMR models, although some differences were not significant. (iv) Notably, the APT# image contrasts between the glioma and the CNAWM were consistently larger than the corresponding NOE# contrasts across all EMR models (p < 0.05). However, the APT# image contrast between the edema and the CNAWM were consistently smaller than corresponding NOE# contrast (all p < 0.05). (v) Finally, the MTRasym (3.5ppm) image contrasts between the glioma (or the edema) and the CNAWM were both positive (p < 0.01) (as described by Eq. [S6] in Supporting Theory).
Figure 7.
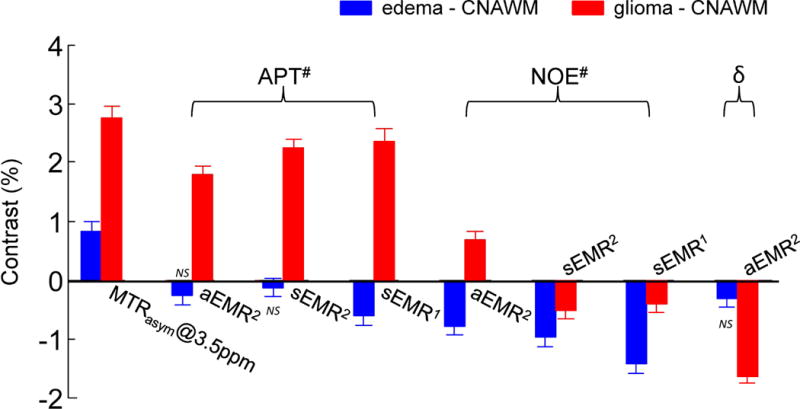
Average MTRasym (3.5ppm), APT#, NOE#, and δ (for aEMR2 only) image contrasts between the CNAWM and the glioma or the edema (values at the glioma/edema – values at the CNAWM). Error bars depict standard errors. All values are statistically significant (p < 0.05) unless otherwise indicated. NS indicates not statistically significant.
Figure 8 shows one example of the conventional MR images and quantitative APT# and NOE# maps for a patient with GBM. As reported before (7), the Gd-enhancing area (tumor core) on the post-Gd T1-weighted image was hyperintense on the MTRasym (3.5ppm) and APT# maps, but the glioma was seemingly hypointense on the NOE# maps. Therefore, the APT effect was the major contributor to the APT-weighted image contrast (based on MT asymmetry analysis) between the tumor and the normal brain tissue.
Figure 8.
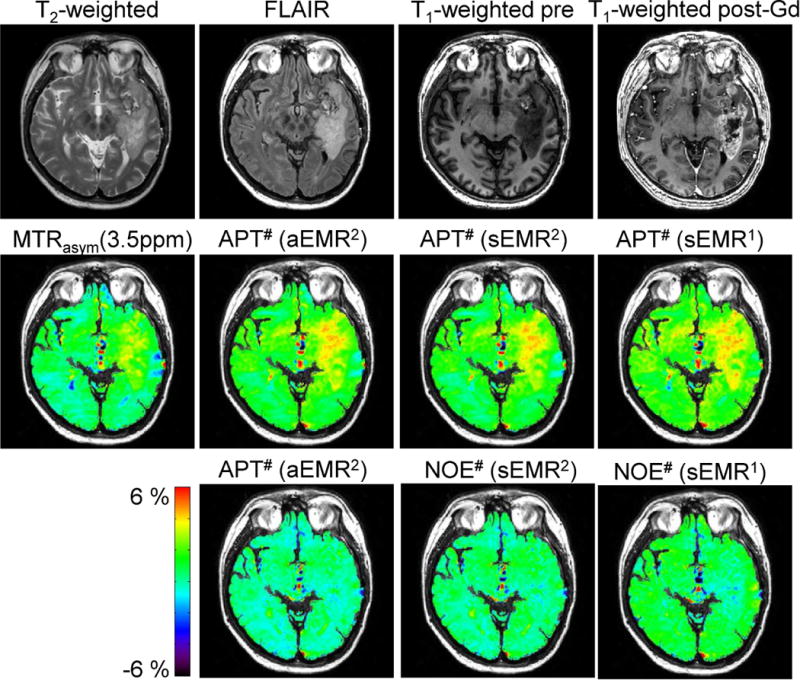
MTRasym (3.5ppm), APT#, and NOE# maps calculated with three EMR models and overlaid on a corresponding FLAIR image for a representative patient with a glioblastoma.
DISCUSSION
In this study, we investigated the mixed effects of conventional MT, APT, and NOE in human gliomas at 3 T and assessed three EMR approaches based on Henkelman’s two-pool MT model with a super-Lorentzian lineshape for the quantification of APT and NOE signals. The key point was to find the accurate EMR signals at 3.5 and −3.5 ppm as the reference signals by which to quantify APT and NOE. The quantitative results from the three EMR methods showed that the APT# signals were significantly higher in the glioma than in the edema and in the CNAWM. The APT# signals were overall larger than the NOE# signals at the RF power level of 2 μT, and were the major contributor to the APT-weighted image contrast in the glioma, compared to the CNAWM.
It has become clear recently that the observed Z-spectrum is asymmetric with the center of the Z-spectrum shifted slightly upfield from the water signal (38, 44, 45), resulting in a negative background signal for asymmetry analysis of Z-spectrum (5). The immobile semi-solid macromolecule protons associating with the conventional MT resonate over a very large spectral width and have very short T2 (~10 μs), thus corresponding to an extremely large saturation width of hundreds of ppm (based on the Bloch equations). Therefore, the semi-solid MT should appear to be nearly symmetric around the water resonance (Supporting Fig. S1), such that the sMER1 and sMER2 models can work. Further, the mobile molecules (T2 ~ 10 ms) lead to upfield well-defined NOE signals in the offset range of 0 to −5 ppm. Notably, the observed Z-spectrum asymmetry in tens of ppm may be attributable to some relatively less mobile (T2 ~ 0.1‒1 ms) proteins, peptides, lipids, and metabolites in tissue (33). The NOE# signals obtained by sEMR1 and sMER2 may include the NOEmobile and NOEless mobile contributions, while the NOE# signals obtained by aEMR2 may include the NOEmobile contribution only.
As shown in Figs. 6 and 7, all three EMR models resulted in quite similar APT# signals and contrasts. Roughly, for each ROI, the APT# signal intensities had a relationship: aMER2 ≈ sMER1 ≥ sMER2, because slightly lower ZEMR (3.5ppm) signals were obtained for sMER2 (Fig. 3b). However, these EMR models may show very different NOE# signals, as discussed above. Roughly, one had a relationship: aMER2 ≤ sMER2 ≤ sMER1. Using the aEMR2 method, the presence of a single resonance frequency difference, Δmw, is actually an oversimplified assumption. Although the model fitted the wide-offset data well (Fig. 3a), it gave inaccurate EMR signals (extra dips in Fig. 4b) around the water resonance, thus influencing the NOE# quantification. As shown in Fig. 5a, some negative NOE# values with aEMR2 were observed in the CNAWM and in edema (with larger Δmw values). These errors may be due to that fact that the single resonance frequency difference is used to define all macromolecular protons. Thus, the aEMR2 model may not be suitable for the quantification of the upfield NOE signals. As a result, the sEMR1 method (using the data points in the offset range between 14 and 7 ppm, and thus, excluding most APT and NOE contributions) would be the best choice for the accurate quantification of both the downfield APT and the upfield NOE signals.
It is known that the APT-weighted signal quantified by the MTR asymmetry analysis with the saturation signal at the other side of water (−3.5ppm) as a reference is an apparent APT signal that is contaminated with the NOE signal, as described early (46). The measured APT-weighted signal can be positive or negative, dependent on saturation power levels and durations, as well as on the B0 field strength (47). At 3 T, as shown in Fig. 6, the APT# signal in the tumor was larger than the NOE# signal at this relatively larger RF power applied (~ 2 μT), which is an optimal saturation power level for APT (48), rather than for NOE. The measured MTRasym(3.5ppm) signals in the glioma, the edema, and the CNAWM were all positive. In addition, the lower NOE# (or NOE# + δ for aEMR2) signal in the tumor relative to the normal brain tissue boosted the APT-weighted image contrast (Fig. 7). It is important to notice that our conclusion based on 3 T and specific experimental parameters cannot be generalized to other measurement conditions, particularly other field strengths (30, 49). In addition to the EMR method, several other alternative data processing or acquisition approaches are being developed for isolating the APT signal from various concurrent saturation effects (50–55).
Whether and how the T1 relaxation time of water (namely, T1w) in tissue affects the measured APT signal is currently an interesting research topic (37, 52, 53, 56, 57). As far as tumor tissue is concerned, it has long been known that higher water content and longer T1w relaxation time are usually simultaneously observed (58–60). Therefore, the effect of the increasing T1w on the measured APT signal may mostly or partially be compensated by the effect of the increasing water content, as pointed out previously (5). Based on this work (Fig. 6) and several previous reports at 3 T (46, 61), the measured APT# signals were significantly higher in the high-grade glioma than in the peritumoral edema (with comparable T1w) and in the CNAWM (with short T1w; see also Supporting Fig. S2). Thus, the influence of T1w on APT imaging in vivo is not as being linear as it appears in the equation (5). This may explain why the use of the newly introduced quantitative metric (AREX, simply multiplying by R1w) led to the unexpected result (AREXtumor ≈ AREXnormal) (52, 53), although the method can bring back the diluted effects of spillover and semi-solid MT on APT imaging. These authors further reported that their corrected APT measurement was consistent with the total protein measurement with the Bradford and bicinchoninic acid assay (BCA) methods (52). However, as pointed out in our recent studies (37, 57), it is important to keep in mind that APT imaging can in principle detect mobile proteins and peptides in tissue only, not the total protein (mobile and semi-solid) contents measured by the standard biochemical methods.
Based on a long continuous-wave (CW) RF saturation (typically used in animal MRI scanners), the MT process can approach a steady state. However, if this is not the case, the longitudinal magnetization of the free bulk water protons would not reach a steady state. In Supporting Table S2, we compared MT model parameters fitted under the steady-state and the non-steady-state conditions. Our results show that only a small difference exists in these MT parameters observed from two saturation conditions. Therefore, the simple steady-state expression (Eq. [S1] in Supporting Theory) can always be used to fit the MT model parameters for the EMR signals.
CONCLUSIONS
Three possible EMR approaches (aEMR2, sEMR2, and sEMR1) were assessed to quantitatively measure pure APT and NOE signals in normal tissue, edema, and glioma. Results showed that all EMR models can be used to accurately calculate the APT and other CEST signals downfield from the water resonance, but the sEMR1 approach (using a symmetric MT model to fit the one-sided, wide-offset Z-spectrum data) could more accurately quantify the upfield NOE signals. The tumor cores identified by Gd-enhanced T1-weighted MR images were consistently hyperintense on the APT# maps and slightly hypointense on the NOE# maps. Thus, the strong APT effect in the tumor would be the major contributor to the APT-weighted image contrast (based on MT asymmetry analysis) between the tumor and normal brain tissue. These quantitative results have clearly demonstrated the reliability and validity of the MT asymmetry at 3.5 ppm [namely, MTRasym (3.5ppm)] as a metric for APT imaging of malignant gliomas at 3 Tesla.
Supplementary Material
Supporting Table S1 Fitted two-pool MT model parameters for the CNAWM (C), the edema (E), and the glioma (G) (mean ± standard deviation).
Supporting Table S2 Fitted two-pool MT model parameters under the steady state and non-steady state condition for the CNAWM (mean ± standard deviation).
Supporting Fig. S1. CEST experimental experiments on a phantom with the egg white solution and semi-solid agar, and a healthy human subject. Unlike the in vivo case, the pure semi-solid MT (such as agar) was almost symmetric around the water signal (with −0.0003% asymmetry at 100 ppm, −0.2% asymmetry from 60 to 40 ppm).
Supporting Fig. S2. Comparison of the APT# signals in normal-appearing gray matter (NAGM), normal-appearing white matter (NAWM), peritumoral edema (hyperintensity in FLAIR), and Gd-enhanced tumor area. The fact that the APT signal in the Gd-enhanced tumor region was significantly higher than in the normal tissue and in the edema region showed that water T1 is not a dominating contributor to APT signals.
Acknowledgments
The authors thank Ms. Mary McAllister for editorial assistance. This work was supported in part by grants from the National Institutes of Health (R01EB009731, R01CA166171, R01NS083435, R21EB015555, R01EB015032, and P41EB015909).
Footnotes
Additional Supporting Information may be found in the online version of this article.
Supporting Theory
Conventional MT Models;
APT-Weighted Imaging Signal and Contrast;
MT Model under a Non-Steady-State (NS) Condition
References
- 1.Ward KM, Aletras AH, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST) J Magn Reson. 2000;143:79–87. doi: 10.1006/jmre.1999.1956. [DOI] [PubMed] [Google Scholar]
- 2.Zhang S, Winter P, Wu K, Sherry AD. A novel europium(III)-based MRI contrast agent. J Am Chem Soc. 2001;123:1517–1578. doi: 10.1021/ja005820q. [DOI] [PubMed] [Google Scholar]
- 3.Goffeney N, Bulte JWM, Duyn J, Bryant LH, van Zijl PCM. Sensitive NMR detection of cationic-polymer-based gene delivery systems using saturation transfer via proton exchange. J Am Chem Soc. 2001;123:8628–8629. doi: 10.1021/ja0158455. [DOI] [PubMed] [Google Scholar]
- 4.Aime S, Barge A, Delli Castelli D, Fedeli F, Mortillaro A, Nielsen FU, Terreno E. Paramagnetic Lanthanide(III) complexes as pH-sensitive chemical exchange saturation transfer (CEST) contrast agents for MRI applications. Magn Reson Med. 2002;47:639–648. doi: 10.1002/mrm.10106. [DOI] [PubMed] [Google Scholar]
- 5.Zhou J, Payen J, Wilson DA, Traystman RJ, van Zijl PCM. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nature Med. 2003;9:1085–1090. doi: 10.1038/nm907. [DOI] [PubMed] [Google Scholar]
- 6.Zhou JY, Yan K, Zhu H. A simple model for understanding the origin of the amide proton transfer MRI signal in tissue. Appl Magn Reson. 2012;42:393–402. doi: 10.1007/s00723-011-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou J, Zhu H, Lim M, Blair L, Quinones-Hinojosa A, Messina AA, Eberhart CG, Pomper MG, Laterra J, Barker PB, van Zijl PCM, Blakeley JO. Three-dimensional amide proton transfer MR imaging of gliomas: Initial experience and comparison with gadolinium enhancement. J Magn Reson Imaging. 2013;38:1119–1128. doi: 10.1002/jmri.24067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dula AN, Arlinghaus LR, Dortch RD, Dewey BE, Whisenant JG, Ayers GD, Yankeelov TE, Smith SA. Amide proton transfer imaging of the breast at 3 T: Establishing reproducibility and possible feasibility assessing chemotherapy response. Magn Reson Med. 2013;70:216–224. doi: 10.1002/mrm.24450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Togao O, Yoshiura T, Keupp J, Hiwatashi A, Yamashita K, Kikuchi K, Suzuki Y, Suzuki SO, Iwaki T, Hata N, Mizoguchi M, Yoshimoto K, Sagiyama K, Takahashi M, Honda H. Amide proton transfer imaging of adult diffuse gliomas: correlation with histopathological grades. Neuro Oncol. 2014;16:441–448. doi: 10.1093/neuonc/not158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J, Tryggestad E, Wen Z, Lal B, Zhou T, Grossman R, Wang S, Yan K, Fu D-X, Ford E, Tyler B, Blakeley J, Laterra J, van Zijl PCM. Differentiation between glioma and radiation necrosis using molecular magnetic resonance imaging of endogenous proteins and peptides. Nature Med. 2011;17:130–134. doi: 10.1038/nm.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong X, Liu L, Wang M, Ding K, Fan Y, Ma B, Lal B, Tyler B, Mangraviti A, Wang S, Wong J, Laterra J, Zhou J. Quantitative multiparametric MRI assessment of glioma response to radiotherapy in a rat model. Neuro Oncol. 2014;16:856–867. doi: 10.1093/neuonc/not245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sagiyama K, Mashimo T, Togao O, Vemireddy V, Hatanpaa KJ, Maher EA, Mickey BE, Pan E, Sherry AD, Bachoo RM, Takahashi M. In vivo chemical exchange saturation transfer imaging allows early detection of a therapeutic response in glioblastoma. Proc Natl Acad Sci (USA) 2014;111:4542–4547. doi: 10.1073/pnas.1323855111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun PZ, Zhou J, Sun W, Huang J, van Zijl PCM. Detection of the ischemic penumbra using pH-weighted MRI. J Cereb Blood Flow Metab. 2007;27:1129–1136. doi: 10.1038/sj.jcbfm.9600424. [DOI] [PubMed] [Google Scholar]
- 14.Tee YK, Harston GW, Blockley N, Okell TW, Levman J, Sheerin F, Cellerini M, Jezzard P, Kennedy J, Payne SJ, Chappell MA. Comparing different analysis methods for quantifying the MRI amide proton transfer (APT) effect in hyperacute stroke patients. NMR Biomed. 2014;27:1019–1029. doi: 10.1002/nbm.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tietze A, Blicher J, Mikkelsen IK, Ostergaard L, Strother MK, Smith SA, Donahue MJ. Assessment of ischemic penumbra in patients with hyperacute stroke using amide proton transfer (APT) chemical exchange saturation transfer (CEST) MRI. NMR Biomed. 2014;27:163–174. doi: 10.1002/nbm.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai KJ, Haris M, Singh A, Kogan F, Greenberg JH, Hariharan H, Detre JA, Reddy R. Magnetic resonance imaging of glutamate. Nature Med. 2012;18:302–306. doi: 10.1038/nm.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan KWY, McMahon MT, Kato Y, Liu GS, Bulte JWM, Bhujwalla ZM, Artemov D, van Zijl PCM. Natural D-glucose as a biodegradable MRI contrast agent for detecting cancer. Magn Reson Med. 2012;68:1764–1773. doi: 10.1002/mrm.24520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker-Samuel S, Ramasawmy R, Torrealdea F, Rega M, Rajkumar V, Johnson SP, Richardson S, Gonçalves M, Parkes HG, Årstad E, Thomas D, Pedley RB, Lythgoe MF, Golay X. In vivo imaging of glucose uptake and metabolism in tumors. Nature Med. 2013;19:1067–1072. doi: 10.1038/nm.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Peng S, Wang R, Chen H, Su W, Zhao X, Zhou J, Chen M. Chemical exchange saturation transfer MR imaging of Parkinson’s disease at 3 Tesla. Eur Radiol. 2014;24:2631–2639. doi: 10.1007/s00330-014-3241-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haris M, Singh A, Cai K, Kogan F, McGarvey J, Debrosse C, Zsido GA, Witschey WR, Koomalsingh K, Pilla JJ, Chirinos JA, Ferrari VA, Gorman JH, Hariharan H, Gorman RC, Reddy R. A technique for in vivo mapping of myocardial creatine kinase metabolism. Nature Med. 2014;20:209–214. doi: 10.1038/nm.3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H, Zhao X, Zhou J, Peng Y. Amide proton transfer (APT) MR imaging of the brain in Children at 3T; Proc 99th Ann Meeting RSNA; Chicago, IL. 2013; pp. SSQ17–07. [Google Scholar]
- 22.Zhou J, Wilson DA, Sun PZ, Klaus JA, van Zijl PCM. Quantitative description of proton exchange processes between water and endogenous and exogenous agents for WEX, CEST, and APT experiments. Magn Reson Med. 2004;51:945–952. doi: 10.1002/mrm.20048. [DOI] [PubMed] [Google Scholar]
- 23.Zhou J, van Zijl PC. Chemical exchange saturation transfer imaging and spectroscopy. Progr NMR Spectr. 2006;48:109–136. [Google Scholar]
- 24.Vinogradov E, Sherry AD, Lenkinski RE. CEST: From basic principles to applications, challenges and opportunities. J Magn Reson. 2013;229:155–172. doi: 10.1016/j.jmr.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun PZ, Zhou J, Huang J, van Zijl P. Simplified quantitative description of amide proton transfer (APT) imaging during acute ischemia. Magn Reson Med. 2007;57:405–410. doi: 10.1002/mrm.21151. [DOI] [PubMed] [Google Scholar]
- 26.Zaiss M, Schmitt B, Bachert P. Quantitative separation of CEST effect from magnetization transfer and spillover effects by Lorentzian-line-fit analysis of z-spectra. J Magn Reson. 2011;211:149–155. doi: 10.1016/j.jmr.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 27.Ling W, Regatte RR, Navon G, Jerschow A. Assessment of glycosaminoglycan concentration in vivo by chemical exchange-dependent saturation transfer (gagCEST) Proc Natl Acad Sci (USA) 2008;105:2266–2270. doi: 10.1073/pnas.0707666105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones CK, Huang A, Xu J, Edden RA, Schar M, Hua J, Oskolkov N, Zaca D, Zhou J, McMahon MT, Pillai JJ, van Zijl PC. Nuclear Overhauser enhancement (NOE) imaging in the human brain at 7T. Neuroimage. 2013;77:114–124. doi: 10.1016/j.neuroimage.2013.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin T, Wang P, Zong X, Kim S-G. MR imaging of the amide-proton transfer effect and the pH-insensitive nuclear overhauser effect at 9.4 T. Magn Reson Med. 2013;69:760–770. doi: 10.1002/mrm.24315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu D, Zhou J, Xue R, Zuo Z, An J, Wang DJJ. Quantitative characterization of nuclear Overhauser enhancement and amide proton transfer effects in the human brain at 7 Tesla. Magn Reson Med. 2013;70:1070–1081. doi: 10.1002/mrm.24560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mougin O, Clemence M, Peters A, Pitiot A, Gowland P. High-resolution imaging of magnetisation transfer and nuclear Overhauser effect in the human visual cortex at 7 T. NMR Biomed. 2013;26:1508–1517. doi: 10.1002/nbm.2984. [DOI] [PubMed] [Google Scholar]
- 32.Zu Z, Xu J, Li H, Chekmenev EY, Quarles CC, Does MD, Gore JC, Gochberg DF. Imaging amide proton transfer and nuclear overhauser enhancement using chemical exchange rotation transfer (CERT) Magn Reson Med. 2014;72:471–476. doi: 10.1002/mrm.24953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J, Hong X, Zhao X, Gao JH, Yuan J. APT-weighted and NOE-weighted image contrasts in glioma with different RF saturation powers based on magnetization transfer ratio asymmetry analyses. Magn Reson Med. 2013;70:320–327. doi: 10.1002/mrm.24784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu J, Zhou J, Cai C, Cai S, Chen Z. Observation of true and pseudo NOE signals using CEST-MRI and CEST-MRS sequences with and without lipid suppression. Magn Reson Med. 2014 doi: 10.1002/mrm.25277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henkelman RM, Huang X, Xiang Q-S, Stanisz GJ, Swanson SD, Bronskill MJ. Quantitative interpretation of magnetization transfer. Magn Reson Med. 1993;29:759–766. doi: 10.1002/mrm.1910290607. [DOI] [PubMed] [Google Scholar]
- 36.Stanisz GJ, Kecojevic A, Bronskill MJ, Henkelman RM. Characterizing white matter with magnetization transfer and T(2) Magn Reson Med. 1999;42:1128–1136. doi: 10.1002/(sici)1522-2594(199912)42:6<1128::aid-mrm18>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 37.Heo H-Y, Zhang Y, Lee D-H, Hong X, Zhou J. Quantitative assessment of amide proton transfer (APT) and nuclear Overhauser enhancement (NOE) imaging with extrapolated semi-solid magnetization transfer reference (EMR) signals: Application to a rat glioma model at 4.7 T. Magn Reson Med. 2015 doi: 10.1002/mrm.25581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hua J, Jones CK, Blakeley J, Smith SA, van Zijl PCM, Zhou J. Quantitative description of the asymmetry in magnetization transfer effects around the water resonance in the human brain. Magn Reson Med. 2007;58:786–793. doi: 10.1002/mrm.21387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morrison C, Henkelman RM. A model for magnetization transfer in tissues. Magn Reson Med. 1995;33:475–482. doi: 10.1002/mrm.1910330404. [DOI] [PubMed] [Google Scholar]
- 40.Sled JG, Pike GB. Quantitative imaging of magnetization transfer exchange and relaxation properties in vivo using MRI. Magn Reson Med. 2001;46:923–931. doi: 10.1002/mrm.1278. [DOI] [PubMed] [Google Scholar]
- 41.Kim M, Gillen J, Landman BA, Zhou J, van Zijl PC. Water saturation shift referencing (WASSR) for chemical exchange saturation transfer (CEST) experiments. Magn Reson Med. 2009;61:1441–1450. doi: 10.1002/mrm.21873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramani A, Dalton C, Miller DH, Tofts PS, Barker GJ. Precise estimate of fundamental in-vivo MT parameters in human brain in clinically feasible times. Magn Reson Imaging. 2002;20:721–731. doi: 10.1016/s0730-725x(02)00598-2. [DOI] [PubMed] [Google Scholar]
- 43.Ethofer T, Mader I, Seeger U, Helms G, Erb M, Grodd W, Ludolph A, Klose U. Comparison of longitudinal metabolite relaxation times in different regions of the human brain at 1.5 and 3 Tesla. Magn Reson Med. 2003;50:1296–1301. doi: 10.1002/mrm.10640. [DOI] [PubMed] [Google Scholar]
- 44.Pekar J, Jezzard P, Roberts DA, Leigh JS, Frank JA, McLaughlin AC. Perfusion imaging with compensation for asymmetric magnetization transfer effects. Magn Reson Med. 1996;35:70–79. doi: 10.1002/mrm.1910350110. [DOI] [PubMed] [Google Scholar]
- 45.Swanson SD, Pang Y. MT is symmetric but shifted with respect to water; Proc 11th Annual Meeting ISMRM; Toronto. 2003; p. 660. [Google Scholar]
- 46.Zhou J, Blakeley JO, Hua J, Kim M, Laterra J, Pomper MG, van Zijl PCM. Practical data acquisition method for human brain tumor amide proton transfer (APT) imaging. Magn Reson Med. 2008;60:842–849. doi: 10.1002/mrm.21712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou J, Hong X, Zhao X, Gao J-H, Yuan J. APT-weighted and NOE-weighted image contrasts in glioma with different RF saturation powers based on magnetization transfer ratio asymmetry analyses. Magn Reson Med. 2013;70:320–327. doi: 10.1002/mrm.24784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao X, Wen Z, Huang F, Lu S, Wang X, Hu S, Zu D, Zhou J. Saturation power dependence of amide proton transfer image contrasts in human brain tumors and strokes at 3 T. Magn Reson Med. 2011;66:1033–1041. doi: 10.1002/mrm.22891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin T, Wang P, Zong XP, Kim SG. Magnetic resonance imaging of the Amine-Proton EXchange (APEX) dependent contrast. NeuroImage. 2012;59:1218–1227. doi: 10.1016/j.neuroimage.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desmond KL, Moosvi F, Stanisz GJ. Mapping of amide, amine, and aliphatic peaks in the CEST spectra of murine xenografts at 7 T. Magn Reson Med. 2014;71:1841–1853. doi: 10.1002/mrm.24822. [DOI] [PubMed] [Google Scholar]
- 51.Cai K, Singh A, Poptani H, Li W, Yang S, Lu Y, Hariharan H, Zhou XJ, Reddy R. CEST signal at 2ppm (CEST@2ppm) from Z-spectral fitting correlates with creatine distribution in brain tumor. NMR Biomed. 2015;28:1–8. doi: 10.1002/nbm.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu J, Zaiss M, Zu Z, Li H, Xie J, Gochberg DF, Bachert P, Gore JC. On the origins of chemical exchange saturation transfer (CEST) contrast in tumors at 9. 4 T NMR Biomed. 2014;27:406–416. doi: 10.1002/nbm.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaiss M, Windschuh J, Paech D, Meissner J, Burth S, Schmitt B, Kickingereder P, Wiestler B, Wick W, Bendszus M, Schlemmer H, Ladd ME, Bachert P, Radbruch A. Relaxation-compensated CEST-MRI of the human brain at 7 T: Unbiased insight into NOE and amide signal changes in human glioblastoma. Neuroimage. 2015 doi: 10.1016/j.neuroimage.2015.02.040. http://dx.doi.org/10.1016/j.neuroimage.2015.1002.1040. [DOI] [PubMed]
- 54.Lee JS, Regatte RR, Jerschow A. Isolating chemical exchange saturation transfer contrast from magnetization transfer asymmetry under two-frequency rf irradiation. J Magn Reson. 2012;215:56–63. doi: 10.1016/j.jmr.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee JS, Xia D, Ge Y, Jerschow A, Regatte RR. Concurrent saturation transfer contrast in in vivo brain by a uniform magnetization transfer MRI. Neuroimage. 2014;95:22–28. doi: 10.1016/j.neuroimage.2014.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zong XP, Wang P, Kim SG, Jin T. Sensitivity and source of amine-proton exchange and amide-proton transfer magnetic resonance imaging in cerebral ischemia. Magn Reson Med. 2014;71:118–132. doi: 10.1002/mrm.24639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yan K, Fu Z, Yang C, Zhang K, Jiang S, Lee DH, Heo HY, Zhang Y, Cole RN, Van Eyk JE, Zhou J. Assessing amide proton transfer (APT) MRI contrast origins in 9L gliosarcoma in the rat brain using proteomic analysis. Mol Imaging Biol. 2015 doi: 10.1007/s11307-11015-10828-11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Damadian R. Tumor detection by nuclear magnetic resonance. Science. 1971;171:1151–1153. doi: 10.1126/science.171.3976.1151. [DOI] [PubMed] [Google Scholar]
- 59.Saryan LA, Hollis DP, Economou JS, Eggleston JC. Nuclear magnetic resonance studies of cancer. IV. Correlation of water content with tissue relaxation times. J Natl Cancer Inst. 1974;52:599–602. doi: 10.1093/jnci/52.2.599. [DOI] [PubMed] [Google Scholar]
- 60.Neeb H, Zilles K, Shah NJ. A new method for fast quantitative mapping of absolute water content in vivo. Neuroimage. 2006;31:1156–1168. doi: 10.1016/j.neuroimage.2005.12.063. [DOI] [PubMed] [Google Scholar]
- 61.Wen Z, Hu S, Huang F, Wang X, Guo L, Quan X, Wang S, Zhou J. MR imaging of high-grade brain tumors using endogenous protein and peptide-based contrast. NeuroImage. 2010;51:616–622. doi: 10.1016/j.neuroimage.2010.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Table S1 Fitted two-pool MT model parameters for the CNAWM (C), the edema (E), and the glioma (G) (mean ± standard deviation).
Supporting Table S2 Fitted two-pool MT model parameters under the steady state and non-steady state condition for the CNAWM (mean ± standard deviation).
Supporting Fig. S1. CEST experimental experiments on a phantom with the egg white solution and semi-solid agar, and a healthy human subject. Unlike the in vivo case, the pure semi-solid MT (such as agar) was almost symmetric around the water signal (with −0.0003% asymmetry at 100 ppm, −0.2% asymmetry from 60 to 40 ppm).
Supporting Fig. S2. Comparison of the APT# signals in normal-appearing gray matter (NAGM), normal-appearing white matter (NAWM), peritumoral edema (hyperintensity in FLAIR), and Gd-enhanced tumor area. The fact that the APT signal in the Gd-enhanced tumor region was significantly higher than in the normal tissue and in the edema region showed that water T1 is not a dominating contributor to APT signals.


