Abstract
For nearly a century developmental biologists have recognized that cells from embryos can differ in their potential to differentiate into distinct cell types. Recently, it has been recognized that embryonic stem cells derived from both mice and humans display two stable yet epigenetically distinct states of pluripotency, naïve and primed. We now show that nicotinamide-N-methyl transferase (NNMT) and metabolic state regulate pluripotency in hESCs. Specifically, in naïve hESCs NNMT and its enzymatic product 1-methylnicotinamide (1-MNA) are highly upregulated, and NNMT is required for low SAM levels and H3K27me3 repressive state. NNMT consumes SAM in naïve cells, making it unavailable for histone methylation that represses Wnt and activates HIF pathway in primed hESCs. These data support the hypothesis that the metabolome regulates the epigenetic landscape of the earliest steps in human development.
INTRODUCTION
Pluripotent stem cells are able to self-renew and have the capacity to regenerate all tissues in the body. These cells hold promise for understanding early human development as well as developing therapies in regenerative medicine. Recent findings have revealed that pluripotency does not represent a single defined state; diverse states of pluripotency, with differences in measurable characteristics relating to gene expression, epigenetics and cellular phenotype, provide an experimental system for studying potential key regulators that constrain or expand the developmental capacity of pluripotent cells1–4. Two stable pluripotent states have been derived in the mouse, and now in humans; preimplantation naïve and postimplantation primed ESC states5–12 . Since naïve, preimplantation human embryonic stem cells (hESCs) show higher developmental potential than postimplantation, primed hESCs8,12, it is critical to understand the key molecular differences between these pluripotent cell types.
Metabolic signatures are highly characteristic for a cell and may act as a leading cause for cell fate changes13–20. Recent data have shown that pluripotent stem cells have a unique metabolic pattern. The naïve to primed mouse ESC transition accompanies a dramatic metabolic switch from bivalent to highly glycolytic state20. However, primed state of inert mitochondria rapidly changes to highly respiring mitochondria during further differentiation. It is not yet understood how and why the pluripotent cells enter the highly glycolytic, metabolically cancer-like (Warburg effect) state and how a differentiating cell leaves this state.
In mouse embryonic stem cells (mESCs) threonine and S-adenosyl methionine (SAM) metabolism are coupled resulting in regulation of histone methylation marks21. Methionine and SAM are also required for the self renewal of hESCs, since depletion of SAM leads to reduced H3K4me3 marks and defects in maintenance of the hESC state22. SAM therefore is shown to be a key regulator for maintaining ESC undifferentiated state and regulating their differentiation. However, little is known about SAM levels or its regulation during the transition between naïve and primed human embryonic states. Recent derivation of naïve human ESCs allows a deeper analysis of the human naïve to primed transition6–12. These studies have already revealed that the epigenetic landscape changes from the naïve to primed state through increased H3K27me3 repressive methylation marks. However, the regulation of this process or the metabolomics of this transition have not been dissected.
We now show that the upregulation of H3K27me3 repressive epigenetic marks during naïve to primed hESC transition is controlled by the metabolic enzyme, NNMT. Knockdown of NNMT in naïve hESCs increased H3K27me3 repressive marks in developmental as well as key metabolic genes that regulate the metabolic switch in naïve to primed transition. CRISPR-Cas9 based NNMT KO naïve hESC lines show upregulation of SAM, H3K27me3 marks, HIF activation, Wnt repression and a general gene expression shift towards primed stage. These data show that NNMT consumes SAM in naïve cells, making it unavailable for histone methylation. Histone methylation further regulates the key signaling pathways important for the metabolic changes that are necessary for early human development.
RESULTS
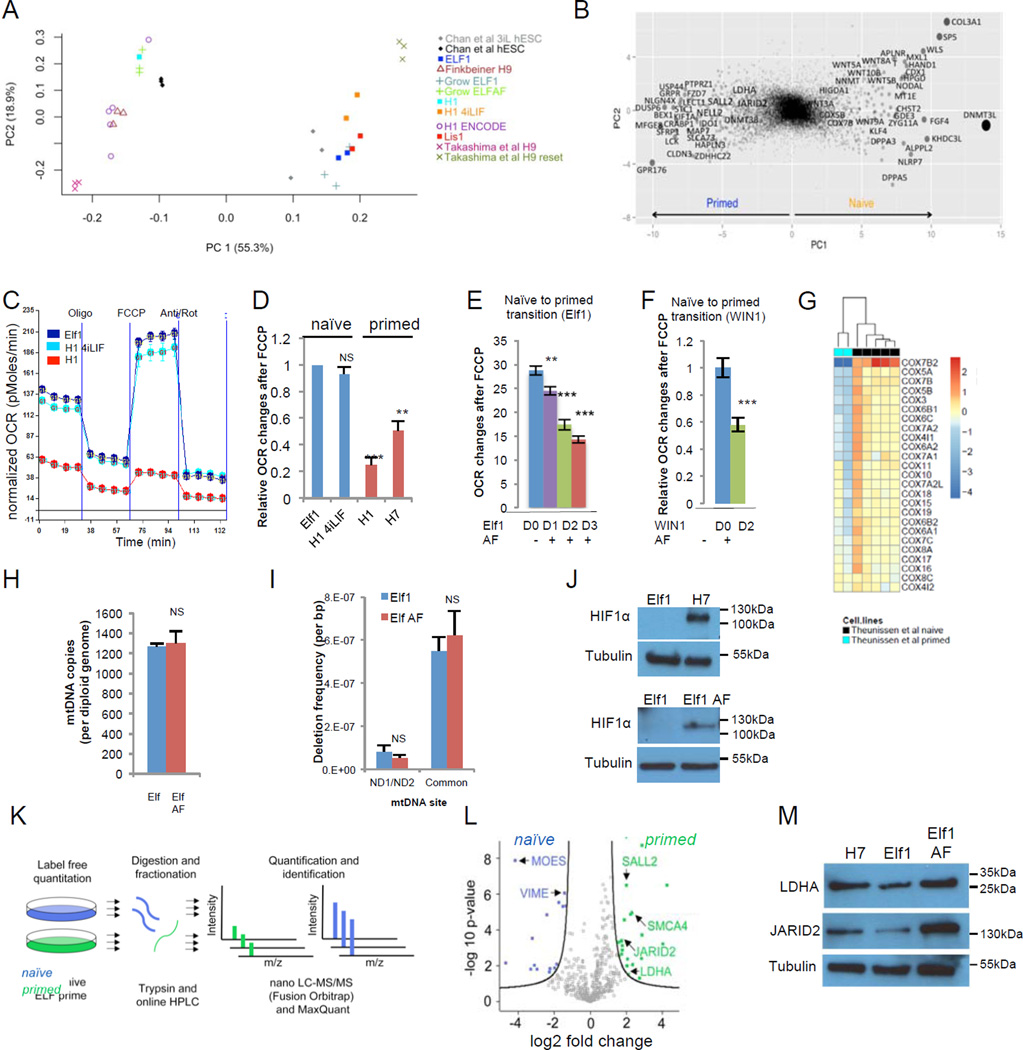
A dramatic metabolic switch occurs in mouse ESCs between pre-implantation (naïve) and post-implantation (primed) state20. Human naïve counterpart has been recently toggled or derived from embryos. Principal component analysis (PCA) of the expression signatures of these new cell types confirmed that all derived human naïve hESCs are in a significantly earlier stage than primed hESCs6,8–10,23(Fig.1A–B, Suppl.Fig.1A–C, Suppl.Table.1A). To assess the metabolic profiles of the human naïve and primed hESCs, we analyzed the cells’ oxygen consumption rates (OCR) using a SeaHorse extracellular flux analyzer. As seen previously in mouse ESCs20, we detected an increase in oxygen consumption rate after FCCP injection in the newly derived naïve hESCs (Elf112; WIN-110) while little increase was observed in primed hESCs (H1, H7) or cells transitioning to primed state (Elf1 AF, WIN1 AF) (Fig.1C–F, Suppl.Fig.1E–I). Likewise, cells “toggled” back to a more naïve state (H1 2iF12, H1 4iLIF) showed increased OCR in response to FCCP to a level similar to mESCs (Fig1A,C–D, Suppl.Fig1G,J–K). These results indicate that the primed hESCs have a lower mitochondrial respiration capacity than naïve hESCs.
Figure 1. Naïve and primed ESCs are metabolically different.
A: PCA of RNA-seq data from this study (Elf1, H1 4i–LIF, Lis1, H1) and other studies6,9,45–46,. ComBat was applied on the combined RNA-seq dataset. B: Genes contributing to principal components separating primed vs. naïve hESC. Size of dots are proportional to the square of PC1 value. Top contributing genes are darker. C: Metabolic profile of naïve (Elf1, H1-4iLIF) and primed (H1) hESCs. A trace of OCR changes is shown under a MitoStress protocol (s.e.m, n=6 biological replicates). D: Primed hESCs (H7 and H1) have reduced OCR changes in response to FCCP compared to naïve hESCs (Elf1 and H1-4iLIF), n=18 (H1, H1 4iLIF) or 24 (Elf1, H7) biological replicates; s.e.m.; p=0.122 for H14iLIFvs.Elf1, p=0.0001 for H1vs.Elf1, p=0.0014 for H7vs.Elf1; 2-tailed t-test. E-F: Transition of naïve hESCs Elf1 (E) and WIN1 (F) toward a more primed state by culture in ActivinA-FGF (AF) media reduced OCR changes in response to FCCP after 1 to 3 days (n=29 for Elf AF 1D, n=20 for Elf AF 2D, n=28 for Elf AF 3D, n=33 for Elf1, n=18 for WIN1 and WIN1 AF; s.e.m.; p=0.0013 for ElfAF1Dvs.Elf1, p<0.001 for ElfAF2Dvs.Elf1, ElfAF3Dvs. Elf1 and WIN1AFvs.WIN1; 2-tailed t-test). G: Heatmap log2 fold expression change of mitochondria complexes genes between primed and naïve stages10. H–I: Naïve hESCs (Elf1) and primed hESCs (Elf1 AF) have similar mitochondrial DNA copy number (H,n=3) and mitochondrial mutation frequencies (I,n=3). S.e.m.; p=0.7802 (H), p=0.37 and 0.6 (I); 2-tailed t-test. J: HIF1α protein is stabilized in primed hESCs (H7 and Elf1 AF). K: Proteomic workflow used to identify differentially regulated protein expression in primed vs. naive hESCs. L: Volcano plot of differentially expressed proteins in primed hESCs (right, green; Elf1 AF) vs naïve cells (left, blue, Elf1). Significant hits are shown (FDR<0.05). Proteins were quantified by nano-LC-MS/MS on a Fusion Orbitrap. M: JARID2 and LDHA proteins are upregulated in primed hESCs (Elf1 AF and H7) compared to naïve hESCs (Elf1), as revealed by Western blot analysis. Unprocessed original scans of blots are shown in Supplementary Suppl.Fig.9. For raw data, see Supplementary Table 4. n=number of biological replicates.
The higher mitochondrial capacity of naïve hESCs (Elf1) reflects neither more mature mitochondria12, nor an increase in mitochondrial DNA (mtDNA) copy number compared to primed hESCs (ElfAF, H7, H1) (Fig.1H, Fig.1L–M). Further, no obvious increase in mtDNA mutation frequency was detected in primed compared to naïve hESCs (Fig.1I, Suppl.Fig.1N–O), suggesting that reduction of oxidative respiration in primed hESCs is not caused by a deteriorating mitochondrial genome. However, consistent with the mouse data, RNA-seq data from our study and microarray or RNA-seq data from other studies3,6,8–10 showed that expression of most mitochondrial electron transport chain complex IV- Cytochrome c oxidase (COX) genes is significantly downregulated in the primed state compared to the naïve state (Fig.1G, Suppl.Table1B–D). Also, consistent with the mouse data20, HIF1α is stabilized in primed but not in naïve hESCs (Fig.1J), correlating with a significant change in expression of prolyl hydroxylase domain-containing protein 2, PHD2 (EGLN1), the primary regulator of HIF1α steady state levels24,25 (Suppl.Table.1B). Further support for HIF1α stabilization and activity at the primed state comes from our proteomic analysis revealing a significant increase in the protein expression of HIF targets, Ldha and Jarid2 at primed hESC state (Elf1 AF compared to Elf1; Fig.1K–L; Suppl.Table.1E; Suppl.Fig.1P, validation in Fig.1M).
Differential metabolites between naïve and primed embryonic stem cells
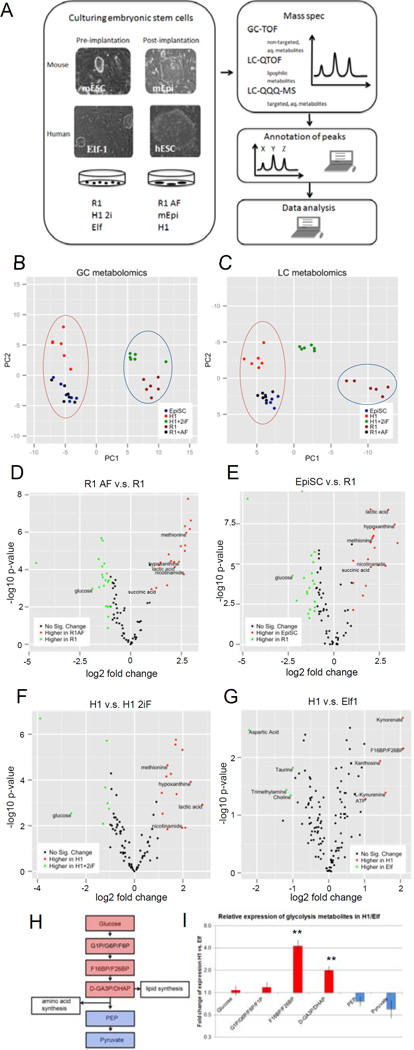
To search for critical metabolites that control the metabolic transitions between naïve and primed mouse and human ESCs, we performed metabolic profiling using GC-TOF, LC-QTOF and LC-QQQ mass spectrometry (MS) analysis (Fig.2A; Suppl.Table.1F–L).
Figure 2. Metabolomic analysis of naïve and primed ESCs.
A: Scheme of mass spectrometry experiments performed for metabolites on mouse and human naïve (pre-implantation) and primed (post-implantation) ESCs. B-C: naïve and primed stem cells can be clearly separated based on their metabolic profiles. (B) PCA plot of water-soluble untargeted GC-MS metabolomics data. The first principal component (PC), which separates the primed cell types (left) from the naïve cell types (right) explained 50.5% of total variance. (C) PCA plot of untargeted LC metabolomics data. 3 clusters are along the first PC: primed cells (left), primed cells toggled back to naïve cells (middle) and naïve cells (right). The first PC explained 68.2% of total variance. D–G: volcano plots of differentially abundant metabolites between primed and naïve cells in mESCs (D, E) detected by GC-TOF, and hESCs (F: GC-TOF, G: LC-QQQ-MS). x-axis is log2 fold change of abundance, y-axis is negative log10 of p-value. Metabolites of biological interest for further analysis are labeled. H: Visualization of the glycolysis pathway and connections to lipid and amino acid synthesis. I: Fold change of glycolysis metabolites (n=3, s.e.m.; Glucose (p=0.6630), G1P-G6P-F6P-F1P (p=0.3713), F16BP-F26BP (p=0.0070), D-GA3P-DHAP (p=0.0058), PEP (p=0.1925), Pyruvate (p=0.1416); 2-tailed t-test) after log2 transformation and mean centering in H1 vs. Elf1 detected by targeted LC-QQQ-MS. For raw data, see Supplementary Tables 1 and 4. n=number of biological replicates.
PCA of the metabolite data reveals a difference in metabolite profiles between naïve and primed cells, regardless of species (Fig.2B–F, Suppl.Fig.2). Multiple naïve and primed cell lines from human and mouse separated clearly by naïve vs. primed state based on the PCA plot of GC metabolomics data (Fig.2B, D–F; Suppl.Fig.2A–C). Stearic acid and cholesterol are the metabolites that contribute the most to the separation within the first PC, indicating that when ESCs transition from naïve to primed state, a major switch occurs in the lipid metabolism. A similar trend of naïve and primed ESC separation is observed in PCA plots of the LC metabolomics data (Fig.2C, Suppl.Fig2D). However, H1 2iF, which is a primed cell line “toggled” towards the naïve state, and Elf1, the naïve hESC line, clustered midway between mouse naïve cell line (R1) and primed cell lines (R1AF, EpiSC, Elf1AF, H7 and H1) in LC analysis, suggesting that H1 2iF and Elf1 have not reached the same naïve state as observed in mouse, with respect to lipid signature (Fig.2C; Fig.2D).
In addition, targeted analysis of metabolites was performed using LC-QQQ-MS with naïve Elf1 and primed H1 hESCs (Fig.2G, Suppl.Table.1H–I) and using GC-TOF with naive Elf1 and primed ElfAF (Suppl.Fig.2B, Suppl.Table.1J). Metabolites upregulated in the primed state include fructose(1,6/2,6)-bisphosphate (F16BP or F26BP), lactate, methionine, nicotinamide and kynurenine (Fig.2F–G; Suppl.Fig.2B). Upregulation of F16BP is in concord with highly active glycolysis, however, phosphoenolpyruvate (PEP), a downstream metabolite of F16BP, does not increase in primed hESCs (Fig.2H–I). Intermediates prior to PEP can be conserved for biosynthetic purposes: 3-phosphoglycerate (3PG) can be diverted to serine and glycine synthesis, which can supply one-carbon units to multiple methylation reactions; dihydroxyacetone phosphate (DHAP) can be converted to glycerol, which serves as the backbone of glycerolipids. Therefore we tested potential changes in lipid and fatty acid metabolism and amino acid pathways (Fig.2H).
Differential Fatty Acid metabolism in Naïve and Primed ESCs
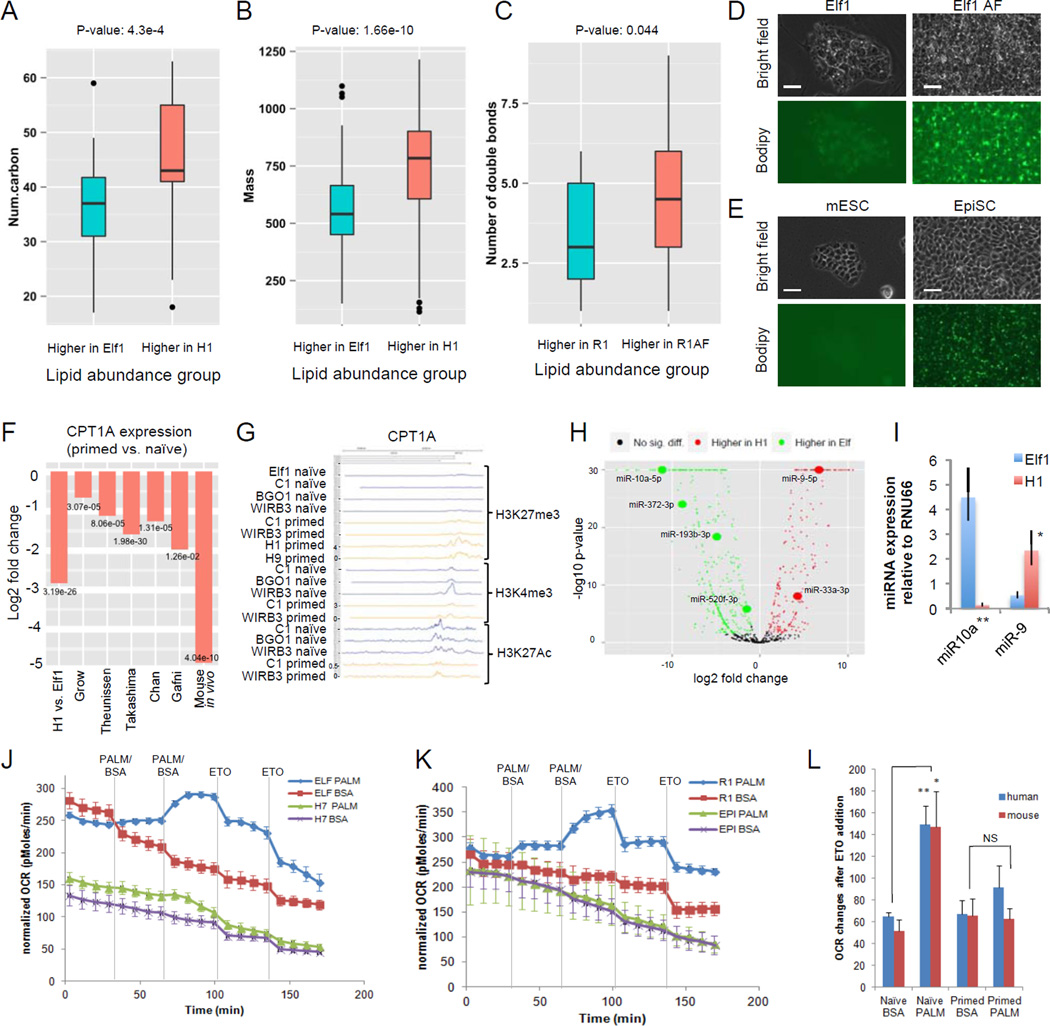
Further lipid analysis was performed using a LC-QTOF instrument on naïve Elf1 and primed H1 cells (Suppl.Table.1K) as well as non-targeted LC-QTOF analysis on Elf1, Elf AF, R1 and mEpi ESCs (Suppl.Table.1L). For 119 lipidomic features with identified molecular formulae and structures, lipids more abundant in H1 have higher numbers of carbons (Wilcoxon rank sum test p-value 4.30e-4, Fig.3A). For 320 features with just identified mass, lipids more abundant in H1 are heavier (p-value=1.66e-10, Fig.3B). Non-targeted LC-QTOF also showed that lipids more abundant in mEpi than in R1 have a significantly higher number of carbons (p-value=0.012, Suppl. Fig.3G). When sorting based on level of unsaturation lipids more abundant in primed R1AF have a higher number of double bonds than lipids more abundant in naïve R1 (p-value=0.044, Fig.3C), which can also be observed in primed Elf1 AF compared to naïve Elf1 (p-value=7.35e-5, Suppl.Fig.3F).
Figure 3. Primed ESCs accumulate lipids while naïve ESCs use fatty acids as a source of energy.
A–B: More abundant lipids in primed hESCs (H1) have more carbon atoms (A) and larger mass (B) than more abundant lipids in naïve hESCs (Elf1). C: More abundant lipids in primed mESCs (R1AF) are more unsaturated than more abundant lipids in naïve mESCs (R1). n=6, p-values Wilcoxon ranksum test. Boxes represent median, 25th and 75th quantiles. Whiskers extend 1.5 IQR above 75th quantile and below 25th quantile. Dots represent values beyond whiskers. D-E: BODIPY 493/503staining shows an increase of lipid droplet accumulation in primed human (Elf1, D) and mouse (EpiSCs, E) ESCs compared to naïve human (Elf1 AF, D) and mouse (R1, E) ESCs. Scale bar, 50µm. F: CPT1A is downregulated in human and mouse primed ESCs compared to naïve ESCs in our study and others. n= from left to right (primed, naïve): 2,1; 3,3; 2,5; 3,3; 3,3; 3,9; 3,2. Negative binomial test p-values are shown. G: ChIP-seq analysis of CPT1A gene shows more repressive H3K27me3 marks and less active H3K4me3 and H3K27ac marks in primed hESCs (C1, WIBR38, H1, H945) than naïve hESCs (Elf112; naïve C1, naïve BGO1, naïve WIBR38). H: volcano plot, microRNA expression in naïve hESCs (Elf1) and primed hESCs (H1, ENCODE, suppl. table 1M). I: qPCR expression of hsa-miR-9 and hsa-miR-10a (predicted to target CPT1A and FASN respectively). hsa-miR-10a is 34-fold higher, and hsa-miR-9 is 4-fold lower in Elf1 vs.H1 (n=3, s.e.m; miR-10a: p=0.004, miR-9: p=0.022; 2-tailed t-test). J-L: Seahorse palmitate assay shows that naïve human and mouse ESCs use fatty acids as a source of energy. A trace of OCR changes after palmitate or BSA vehicle addition, followed by ETO in human ESCs (naïve Elf1 and primed H7, J) and mouse ESCs (naïve R1 and primed EpiSCs, K). n=4 for Elf1BSA, R1BSA, EpiBSA, n=5 for Elf1PALM, R1PALM, EpiPALM, n=6 for H7BSA, H7PALM; s.e.m. Changes after ETO injections were quantified in L (Elf1BSA (n=12), R1BSA (n=12), EpiBSA (n=12), Elf1PALM (n=15), R1PALM (n=15), EpiPALM (n=15), H7BSA (n=18), H7PALM (n=18), s.e.m.; naïve hESCs: p=0,0096, primed hESCS: p=0.354, naïve mESCs: p=0.03, primed mESCs: p=0.88, 2-tailed t-test). For raw data, see Supplementary Table 4. n=number of biological replicates.
In concordance with the significant increase in long carbon chain lipids observed in primed mouse and human ESCs, we also detected a significant increase in accumulation of lipid droplets in the primed state, as observed by Oil Red O and BODIPY staining (Fig.3D–E, Suppl.Fig.3A–B). These data indicate an increased synthesis and-or decreased beta-oxidation in primed cells. Interestingly, several of the enzymes involved in fatty acid transport into the mitochondria and fatty acid beta-oxidation are significantly downregulated in primed human ESCs, as well as in mouse in vivo post-implantation state (Suppl.Fig3H–I). Carnitine acyltransferase 1 (CPT1) transfers long chain acyl groups to carnitine, facilitating the initial step in acyl transfer to the mitochondrial matrix. Interestingly, the rate limiting fatty acid transporter CPT1A is downregulated in both mouse in vivo post-implantation and human primed ESC state compared to all analyzed naïve states (Fig.3F). The decrease in CPT1A expression in the primed state could be due to increased H3K27me3 and decreased H3K4me3 and H3K27ac marks observed in CPT1A promoter in primed hESCs6,8,12 (Fig3G; Suppl.Fig3C). Several of the miRNAs predicted to target CPT1A and other enzymes involved in beta-oxidation are up-regulated in primed hESCs (e.g., miR-9, miR-33a-3p, Fig.3H; Suppl.Table.1M). Moreover, microRNAs predicted by Targetscan and miRTarBase to target enzymes involved in fatty acid synthesis were downregulated in primed cells (e.g., miR-10a and miR-193, Fig.3H). Concomitantly, key fatty acid synthesis genes were up in primed H1 hESCs compared to naïve Elf1 state (SLC25A1, ACLY, ACACA, FASN, and SREBP-1c; Suppl.Fig3D). We further validated some of the miRNAs by qPCR analysis and showed that miR-9, predicted to target CPT1A, was upregulated, while miR-10a, predicted to target SREBP-1c (a regulator of fatty acid and cholesterol synthesis), was downregulated in the human primed state (Fig.3I).
To test the level of fatty acid beta-oxidation in naïve and primed human and mouse ESCs, we performed a palmitate-oxidation assay in the Seahorse metabolic flux analyzer26. Importantly, both mouse and human naïve ESCs were capable of utilizing palmitate as an energy source, while primed mouse or human ESCs were not (Fig.3J–L; Suppl.Fig.3E). This result suggests that primed human and mouse ESCs are not capable of significant beta-oxidation and, in combination with increased fatty acid synthesis, may explain the accumulation of lipids observed in this state (Fig.3D–E, Suppl.Fig.3A–B).
Differential amino acid metabolism in naïve and primed ESCs
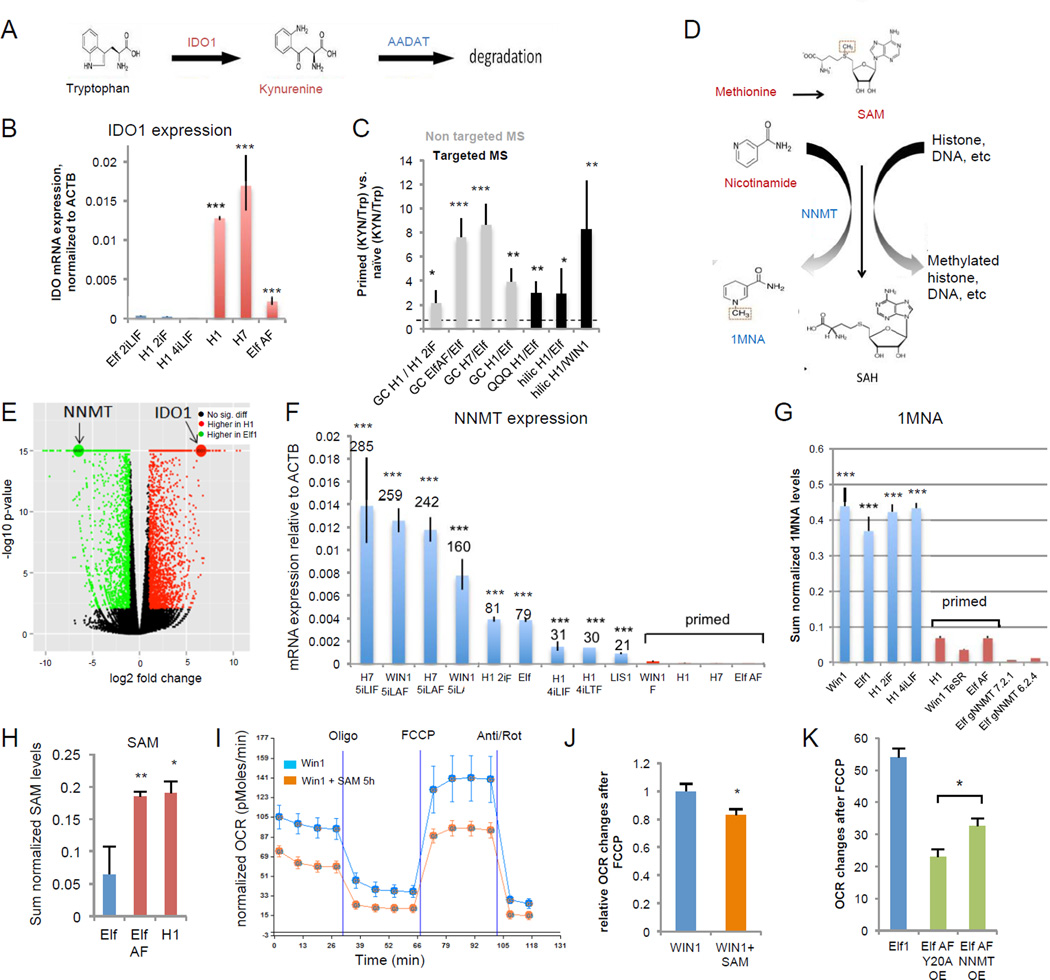
In addition to glycolysis and fatty acid metabolism, primed cells show changes in amino acid metabolism pathways. In primed vs. naïve hESCs we observed a large enrichment of the tryptophan degradation product kynurenine, which can act as a ligand for the nuclear receptor AHR27 (Fig.4A,C). Interestingly, tryptophan is shown to be critical for primed hESCs growth22. Accordingly, RNAseq and qPCR data show a large increase of the tryptophan metabolizing enzyme IDO1 in primed hESCs compared to naïve hESC and in vivo 8 cell human embryo23 (Fig.1B, Fig.4BE,; Suppl.Fig.4A). IDO1 levels quickly drop during differentiation, indicating a specific function for IDO1 in primed state28 (Suppl.Fig.4B–C, Suppl.Table.1N, 12,70).
Figure 4. Amino acids methionine and tryptophan are differentially regulated in naïve and primed hESCs.
A: Model of Tryptophan-Kynurenine pathway. B: IDO1 is highly expressed in primed hESCs as compared to naïve hESCs (qPCR, n=3 for H14iLIF, ElfAF, n=4 for H12iF, H1, H7, n=5 for Elf 2iLIF; s.e.m.; ***p<0.001; 2-tailed t-test). C: The kynurenine vs. tryptophan ratio is higher in primed than naïve hESCs, as detected by targeted (n=6) and non-targeted (HILIC: n=4, QQQ n=3) mass spectrometry. s.e.m.; *p<0.05, **p<0.01, ***p<0.001; 1-tailed t-test,). D: Model of SAM pathway and NNMT. Metabolites in red are up-regulated in primed hESCs. Metabolites and enzymes in blue are up-regulated in naïve hESCs. E: Volcano plot of RNA-seq data from naïve hESCs (Elf1) and primed hESCs (H1). Genes with greater than 2-fold change and FDR<0.05 are colored. NNMT and IDO1 are among the most differentially expressed genes. F: NNMT is highly up-regulated in naïve hESCs compared to primed hESCs (qPCR). Numbers indicate fold changes of naïve hESCs compared to H1 and H7 primed hESCs. (n=3 for WIN1, H75iLAF, Elf1, H14iLIF, H14iLTF, LIS1, WIN1F, Elf1AF, n=4 for WIN15iLA, H12iF, H1, H7, n=5 for H75iLIF; s.e.m.; ***p<0.001; 2-tailed t-test) G: Naïve hESCs (n=4 each) have higher amounts of the 1-MNA, than primed hESCs (n=4 for H1, n=6 for WIN1TeSR, ElfAF) (s.e.m. ***p<0.001, 2-tailed t-test). 1-MNA was not detected in Elf1 CRISPR-Cas9 KO mutant of NNMT (gNNMT 7.2.1, n=6; gNNMT 6.2.4, n=6). H: SAM levels are higher in primed hESCs (H1 n=4, Elf AF n=6) than in naïve hESCs (Elf1 n=4) (s.e.m.; p=0.0089 for ElfAF vs. Elf1, p=0.0376 for H1 vs. Elf1; 2-tailed t-test). I-J: SAM induces a “primed-like” metabolic profile in naïve hESCs. Addition of SAM (500µM) for 5h in media without methionine reduces OCR changes in response to FCCP in naïve hESCs (WIN1). A Seahorse trace is shown in I (n=6; s.e.m). OCR changes after FCCP are quantified in J (n=23; s.e.m.; p=0.017). K: Overexpression of NNMT delays the metabolic transition from naïve to primed (n=4; s.e.m.; p=0.028, 2-tailed t-test). For raw data and exact p values, see Supplementary Table 4. n=number of biological replicates.
Interestingly, a strong upregulation of 1-methyl-nicotinamide (1-MNA), a product of Nicotinamide N-methyltransferase (NNMT) activity was observed in all naïve compared to primed hESC (Fig.4G). Methionine and nicotinamide downregulation along with 1-MNA upregulation in the naïve state correlates with upregulation of NNMT, shown previously to create a metabolic methyl sink, thereby promoting epigenetic remodeling in cancer28 (Fig.4D–G, Suppl.Fig.4G). Primed hESCs show an increase in SAM levels compared to the naïve state (Fig.4H; Suppl.Fig4H; Suppl.Table.1O). The increase in SAM correlates with the sharp decrease in NNMT enzyme levels observed in primed hESCs6,8,11 (Fig.4H,E–F), suggesting that SAM levels may be reduced in the naïve state by high NNMT activity. Further, significant expression changes of NNMT among various tissues reveal that NNMT is dynamically regulated during development and suggest that NNMT might act as regulator of SAM levels also in a developmental context, not only in cancer29 (Suppl.Fig.4D–F, Suppl.Table.1N). Accordingly, we show that high levels of SAM induce a primed-like metabolic profile in naïve hESCs while overexpression of NNMT, but not the mutated form of NNMT delays the naïve to primed hESC metabolic switch (Fig.4H–K, Suppl.Fig.4I).
NNMT regulates repressive histone modifications
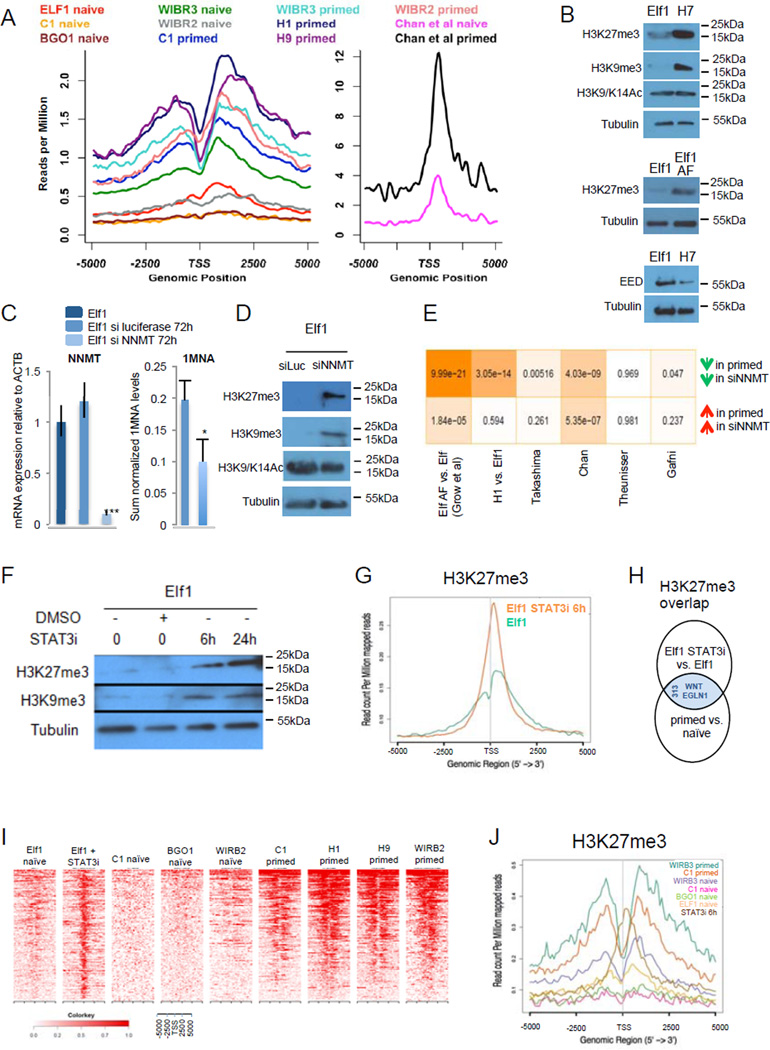
Reduction of NNMT levels during the naïve to primed transition correlates with a significant increase in SAM levels and in H3K27me3 histone methylation marks in 648 developmentally regulated genes6,8,10,12 (Fig.5A). Moreover, Western blot analysis revealed an overall increase of H3K27me3 and H3K9me3 marks in primed hESCs compared to naïve hESCs, while the H3K9 and H3K14 acetylation marks remained unchanged (Fig.5B, Suppl.Fig.5H). ChIP-seq analysis of other marks (H3K4me1, H3K4me3 and H3K27ac) did not show significant change between primed and naïve hESCs6,8 (Suppl.Fig.5A–G). RNA-seq analysis of histone methyltransferases and histone demethylases involved in H3K27 and H3K9 methyl marks did not show changes in expression levels that could explain the significant increase in repressive methylation marks observed at the primed state (Suppl.Fig.5I–J). Furthermore, we showed by Western analysis that the protein levels of the polycomb repressive complex 2 (PRC2) regulator, EED, are not increased in primed state compared to naïve state (Fig.5B).
Figure 5. High NNMT expression in naïve hESCs regulates histone methylation status.
A: H3K27me3 reads mapped 5kb around transcription start sites (TSS) of 648 developmental genes were plotted for Ware et al., Gafni et al., Theunissen et al., Bernstein et al. (left panel) and Chan et al (right panel) ChIP-seq data sets. B: Western blot analyses show higher H3K27me3 and H3K9me3 in primed hESCs (H7, Elf1 AF) than naïve hESCs (Elf1). C: qPCR analysis shows a knock-down regulation of NNMT using siRNA (50 nM, 72h) in naïve hESCs (Elf1), inducing a decrease of 1-MNA levels (qPCR n=3; s.e.m., p=0.001, 2-tailed t-test; HILIC n=4, s.e.m., p=0.039 1-tailed t-test) D: Western blot analysis of histone marks in Elf1 cells treated with siRNA against NNMT or siRNA against luciferase as a control. E: Hypergeometric test p-values for the overlap between genes expressed higher (lower) in siNNMT compared to siLUC and genes expressed higher (lower) in primed lines compared to naïve lines from multiple studies. Color shade is proportional to negative log10 of p-values. siLUC transcriptomic signature has significant overlap with the ELFAF vs. Elf1 data set. F: Western blot analysis of histone modifications after treatment of Elf1 cells with 100 µM of STAT3 inhibitor. G: 6h treatment with STAT3 inhibitor (100 µM) in Elf1 cells increases H3K27me3 marks, as shown by ChipSeq analysis on all genes. H: WNT ligands and EGLN1 are among the 313 overlapping genes with increased H3K27me3 mark in primed vs. naïve hESCs (8,10,64), and Elf1 cells treated for 6h with 100uM STAT3 inhibitor vs. Elf1 cells. I: Windowed chromatin heatmaps of H3K27me3 profile +/− 5kb of promoters of the 313 overlapping genes with increased H3K27me3. J: H3K27me3 reads from ChIP-seq data mapped 5kb around TSS were plotted for naïve hESCs (C1, WIBR3, BGO18, and Elf112.), primed hESCs (C1, WIBR38, H112) and naïve hESCs Elf1 treated for 6h with 100µM of STAT3 inhibitor. Unprocessed original scans of blots are shown in Supplementary Fig.9. For raw data, see Supplementary Table 4. n= number of biological replicates.
Overexpression of NNMT in primed cells reduced H3K27me3 marks (Suppl.Fig.4I). The direct reduction of NNMT levels in naïve hESCs significantly reduced the enzymatic product, 1-MNA, reduced naïve hESC enriched microRNAs and increased H3K27me3 and H3K9me3 marks, as analyzed by Western blots, while H3K9 and H3K14 acetylation marks did not change (Fig.5C–D,Suppl.Fig.5K–L). RNA-seq analysis of NNMT knockdown identified a significant overlap between genes changed in NNMT KD and naïve to primed transition (Fig.5E). In particular, the most significant overlap is observed between genes up-regulated by NNMT knockdown and genes expressed higher in Elf AF vs. Elf1. Therefore, based on transcriptome signatures, NNMT knockdown samples do not only show increased repressive H3K27me3 marks but are also moving towards the primed state.
To analyze the early NNMT responsive genes for H3K27me3 marks, we altered the NNMT regulator, LIF-STAT pathway. In naïve hESCs LIF activates STAT330. Activated STAT is previously shown to bind NNMT promoter and activate its transcription31. H1 toggled to more naïve state using 2iF12 without external LIF addition also has high level of NNMT (Fig.4F) and activated LIF-STAT pathway, suggesting that LIF pathway is endogenously activated in the naïve hESC state (Suppl.Fig.5M). We show that treating naïve hESCs with a STAT3 inhibitor affects NNMT expression and the repressive histone methylation pattern. qPCR analysis showed a reduction of NNMT expression on Elf1 cells as early as 6h after STAT3 inhibitor addition (Suppl.Fig.5N).Importantly, reduction of NNMT in naïve hESCs by STAT3 inhibitor also increased H3K27me3 and H3K9me3 marks, as shown by Western blot analysis (Fig.5F). We characterized H3K27me3 in naïve hESCs by ChIP-seq analysis and observed a significant increase in H3K27me3 marks at promoters after 6h STAT3 inhibitor treatment (Fig.5G; Suppl.Table.1P–Q). Interestingly, over 25% (313 genes) of genes with primed-enriched H3K27me3 marks already showed increased H3K27me3 marks after 6 hour STAT3 inhibitor treatment (Fig.5H; Suppl.Table1R). Windowed heatmap and average profile of ChIP-seq signal revealed a dramatic increase in H3K27me3 marks close to the transcription start site of these 313 genes in the naïve to primed transition as well as after 6h STAT3 inhibitor treatment5,9 (Fig.5I–J). Among these genes are components of the WNT pathway and regulators of HIF (Fig.5H, Suppl.Table1R).
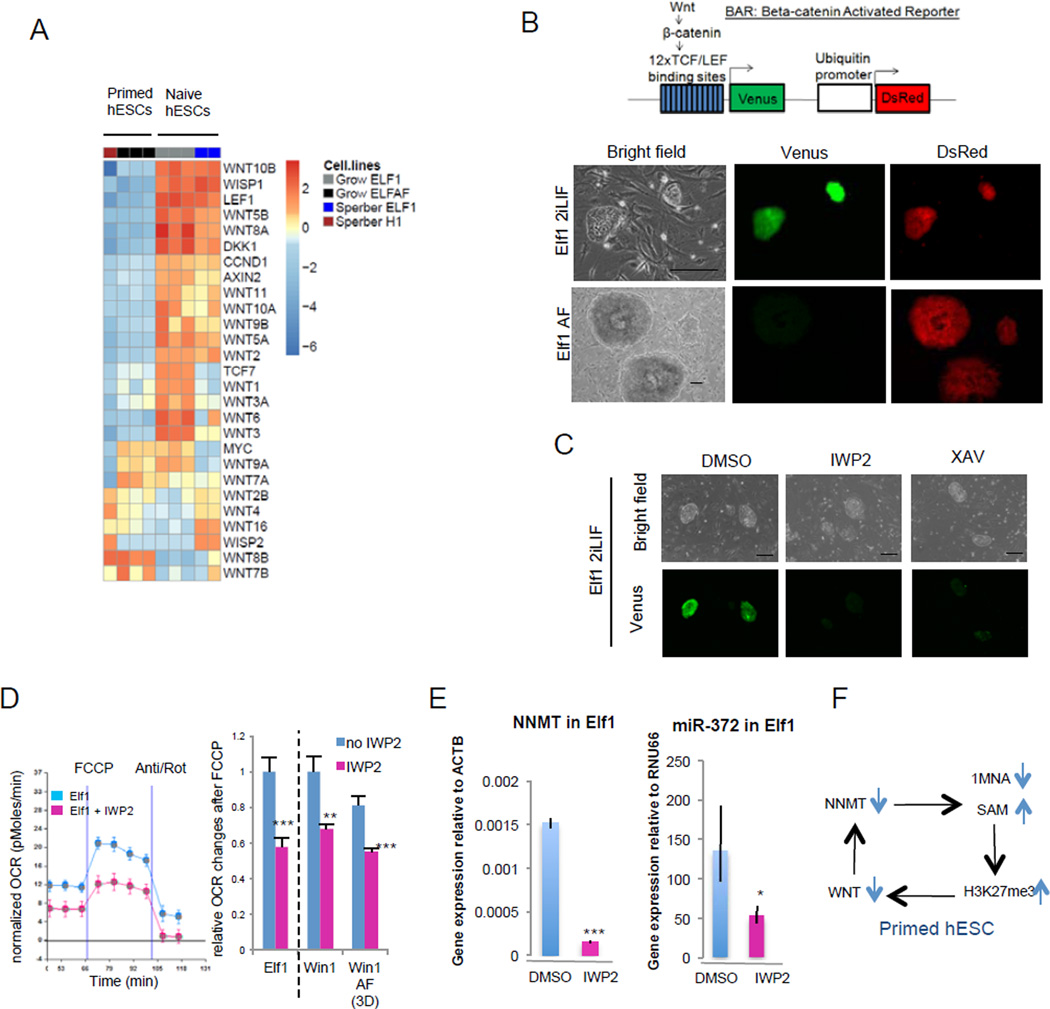
The majority of Wnt ligands and target genes are downregulated in primed compared to naïve hESCs, suggesting that the Wnt pathway might be inactivated during the naïve to primed transition (Fig.6A, Suppl.Fig.6A–D; Fig.1B, 1L, Vime32). Previous studies have revealed that in human and mouse primed ESCs the Wnt pathway is not active and forced activation of the pathway leads to differentiation33,34. We now show using a Wnt-pathway activity reporter33 that while the reporter is not activated in primed hESCs, strong activation is observed in naïve hESCs (Fig.6B, Suppl.Fig.6F). We furthermore show that the Wnt-activity in naïve hESCs is dependent on β-catenin since siRNA(β-cat) or XAV939 treatment dramatically downregulated the reporter activity (Fig.6C, Suppl.Fig.6E). In addition, the Wnt ligand is produced by the naïve hESCs since IWP2, an inhibitor that represses Wnt palmitylation also represses the reporter activity in naïve cells (Fig.6C). Inhibition of Wnt in naïve hESCs reduces expression of the naïve hESC enriched markers, NNMT and microRNA miR-37212 and accelerates the transition toward the primed metabolic state (Fig.6D–F, Suppl.Fig.6G). These data reveal that the robust Wnt activity in naïve hESCs is among the earliest responders to the repressive H3K27me3 marks during naïve to primed hESC transition.
Figure 6. WNT pathway is active in naïve hESCs.
A: Heatmap of gene expression of WNT ligands and WNT targets in primed hESCs (H1, Elf1 AF) and naïve hESCs (Elf1). B: Wnt is activated in naïve hESCs. Endogenous Wnt signaling in naïve (Elf1) and primed (Elf1 AF) BAR-reporter cells. Scale bars represent 200µm. C: Wnt inhibitor IWP2 (2µM) and Wnt antagonist XAV939 (5µM) inhibit the reporter activity in naïve Elf1 cells after 72h. Scale bars represent 200µm. D: Wnt inhibition by IWP2 (2µM, 48h) decreases OCR changes after FCCP in naïve hESCs (Elf1, WIN1) and in naïve hESCs transitioning to primed (WIN1 AF). A trace of OCR changes is presented in Elf1 (n=8 for Elf1, n=6 for Elf1+IWP2; s.e.m.). OCR changes after FCCP were quantified (n=8 for Elf1, n=6 for Elf1+IWP2, WIN1, WIN1+IWP2, WIN1AF, n=7 for WIN1AF+IWP2; s.e.m.; p=0.0009 for Elf1+IWP2 vs. Elf1, p=0.0084 for WIN1+IWP2 vs. WIN1, p=0.0006 for WIN1AF+IWP2 vs. WIN1AF; 2-tailed t-test). E: Wnt inhibition by IWP2 (2µM, 72h) downregulates NNMT and miR-372 expression in naïve hESCs (Elf1) as shown by qPCR analysis. (n=3; s.e.m.; p=0.04 for miR-372, p=6.44E-06 for NNMT; 1-tailed t-test). F: Model of self-reinforcing loop between WNT and NNMT in primed hESCs. For raw data, see Supplementary Table 4. n= number of biological replicates.
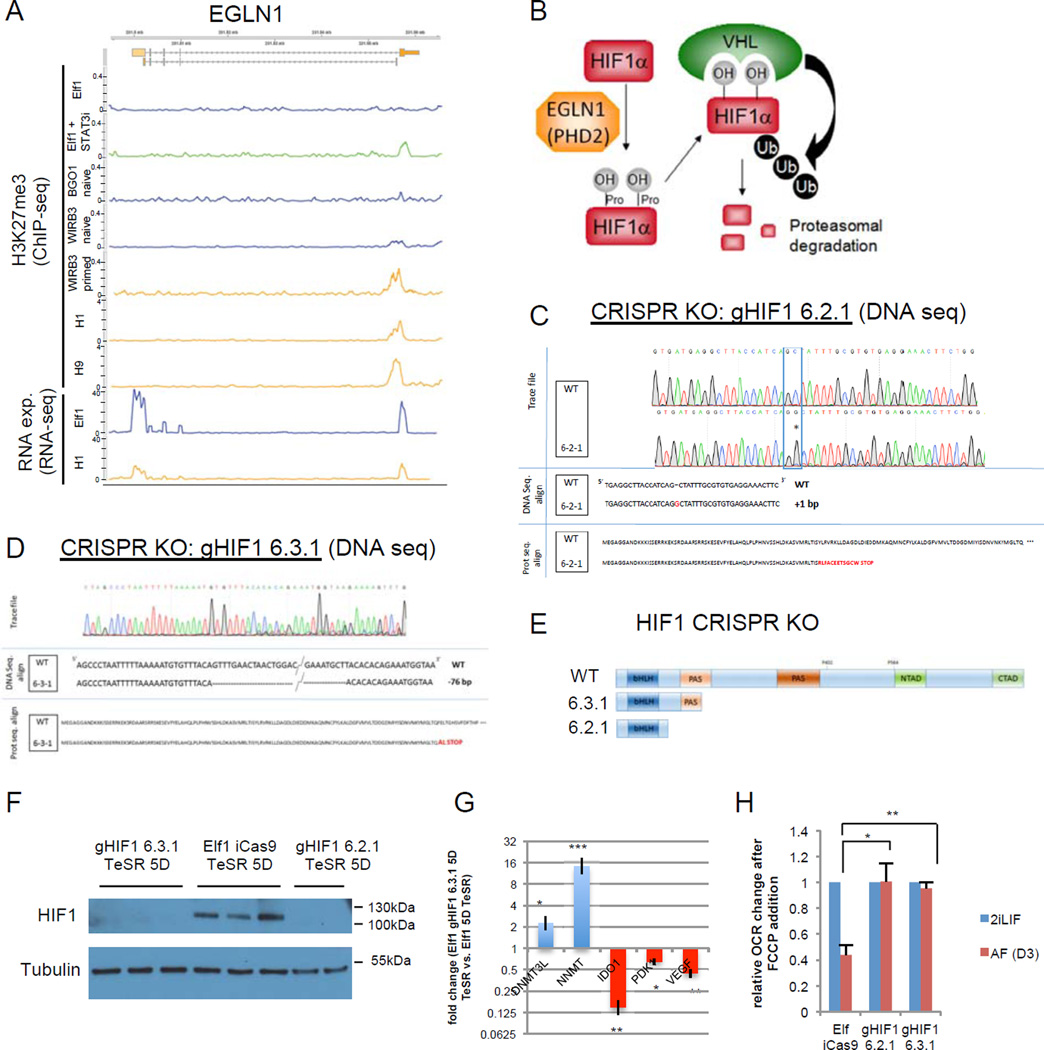
A significant increase of H3K27me3 marks was observed in the metabolic gene, prolyl hydroxylase 2 (EGLN1), ECHS1, HIGD1 and miR-193 (Fig.5H, Fig.7A) promoters in the primed state as well as in STAT3 inhibitor treated Elf1 cells, vs. naïve state (Suppl.Table.1R). The increased repressive H3K27me3 mark in EGLN1 promoter correlated with the observed reduced gene expression in the primed state (Fig.7A). Since EGLN1 induces HIF1α VHL and ubiquitination-dependent degradation, its repression in the primed state (Fig.7A) could cause HIF1α stabilization in primed hESC stage (Fig.7B; Fig.1J). Furthermore, ectopic HIF1α stabilization accelerates primed hESC markers (Suppl.Fig.7A–C). To test whether HIF1α KO affects naïve to primed transition in hESCs, we generated CRISPR-Cas9 based HIF1α KO lines and analyzed their capacity to develop from pre- to post-implantation stage in human development (Fig.7C–F, Suppl.Fig.7D). Importantly, the gene expression analysis revealed that naïve markers were upregulated and primed markers downregulated, suggesting that HIF1α KO naïve hESCs are not capable of transitioning to primed stage (Fig.7G). Furthermore, Seahorse flux analysis showed that HIF1α KO hESCs do not display OCR reduction, suggestion that they are defective in metabolic switch normally observed in naïve to primed hESC transition (Fig.7H, Suppl.Fig.7E). These data show that HIF1α is required for naïve to primed hESC transition.
Figure 7. HIF1α is required for naïve to primed hESC transition.
A: screen shot of RNA expression and H3K27me3 marks of EGLN1 (PHD2) in naïve hESCs [Elf112, WIRB3 naïve and BGO1 naïve8)], primed hESCs [WIRB3 primed8, H1 and H964 and Elf1 treated with STAT3 inhbitor (100 µM) for 6h. B: HIFα is hydroxylated on prolyl residues by EGLN1 (PHD2), leading to VHL-mediated proteolysis. C-D: Sequencing trace files, DNA sequences and protein sequences of HIF1α CRISPR-Cas9 knock-out (KO) mutant clones (gHIF1 6.2.1, C; gHIF1 6.3.1, D). E: schematic representation of wild type HIF1α protein and proteins resulting from CRISPR-Cas9 knock-out (KO) mutants gHIF1 6.2.1 and gHIF1 6.3.1. bHLH= basic helix-loop-helix domain, PAS= Per-Arnt-Sim domain, NTAD= N-terminus transcriptional activation domain, CTAD= C-terminus transcriptional activation domain. F: HIF1α is not expressed in CRISPR-Cas9 KO mutants. Western blot analysis of HIF1α expression in cells pushed toward the primed stage by culture in TeSR1 for 5 days in wild type Elf1 cells (iCas9 Elf1), and two CRISPR-Cas9 KO mutants of HIF1α (gHIF1 6.2.1 and gHIF1 6.3.1). G: qPCR analysis of hESCs transitioning to primed reveals that naïve markers (DNMT3L and NNMT) are still expressed higher in Elf1 HIF1α CRISPR-Cas9 KO cells compared to wild type Elf1, while primed marker IDO1 and HIF target genes (PDK1 and VEGFA) are downregulated (n=3; s.e.m.; p=0.024 for DNMT3L, p=0.0005 for NNMT, p=0.001 for IDO1, p=0.12 for PDK1, p=0.004 for VEGFA; 2-tailed t-test). H: KO of HIF1α prevents the metabolic switch occurring during the transition of hESCs from naïve to primed state as shown by measuring OCR after FCCP addition using SeaHorse. n=3 for gHIF1 6.3.1 2iLIF and AF and n=4 for Elf iCas9 and gHIF1 6.2.1 2iLIF and AF; s.e.m.; p=0.0117 for gHIF1 6.2.1 vs. Elf iCas9, p=0.0032 for gHIF1 6.3.1 vs. Elf iCas9; 2-tailed t-test. Unprocessed original scans of blots are shown in Supplementary Fig.9. For raw data, see Supplementary Table 4. n= number of biological replicates.
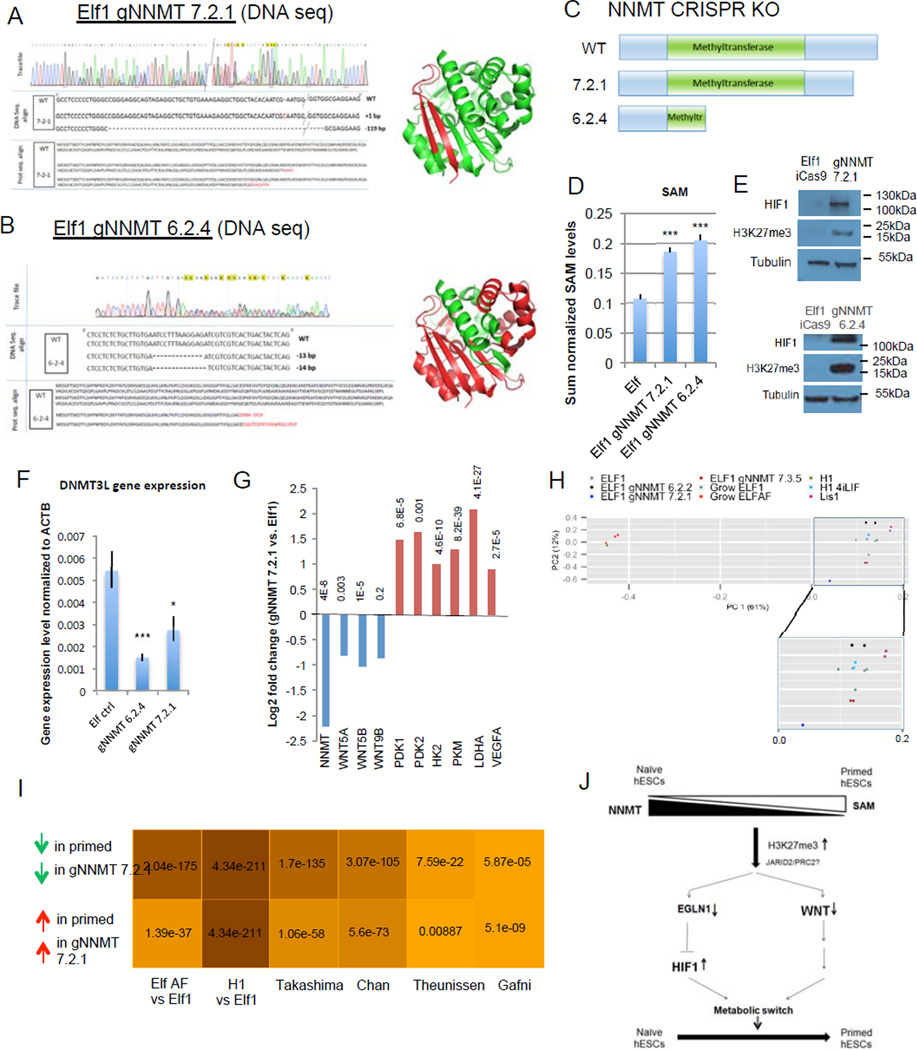
To validate the NNMT based regulation of Wnt and HIF pathways in naïve to primed transition (Fig.5H) we generated CRISPR-Cas9 based NNMT KO mutant lines that lack NNMT enzymatic activity as shown by lack of 1-MNA production and increase in SAM levels (Fig.8A–D; Suppl.Fig.8A–D8G,; Fig.4G). As observed with NNMT KD experiments (Fig.5D), NNMT KO lines showed significant increase in H3K27me3 marks (Fig.8E). This epigenetic modification was accompanied with dramatic stabilization of HIF1α protein and downregulation of naïve hESC marker DNMT3L (Fig.8E–F). Furthermore, genome wide RNA sequencing results revealed that NNMT KO line 7-2-1 shows upregulation of HIF pathway genes, downregulation of Wnt pathway (Fig.8G) and a genome wide gene expression signature indicative of a general shift towards primed stage, even in the presence of primed stage inhibitors (2iLIF) (Fig.8H–I, Suppl.Fig.8E–F8H,). Downregulation was also observed among a significant fraction of the 313 genes with increased repressive H3K27me3 marks in primed stage and after STAT3 inhibitor treatment (hypergeometric test p-value <0.0036). In summary, these data show that NNMT affects naïve to primed hESC transition through epigenetic alterations that repress Wnt pathway and activate HIF pathway.
Figure 8. NNMT affects naïve to primed hESC transition by repressing Wnt pathway and activating HIF pathway.
A–B: Sequencing trace files, DNA sequences, protein sequences and 3D protein structures predicted from sequence (Pymol) of various NNMT CRISPR-Cas9 KO mutant clones (gNNMT 7.2.1, A; gNNMT 6.2.4, B). Green color represents the truncated NNMT protein in the CRISPR-Cas9 mutant. C: Schematic representation of wild type NNMT protein and proteins resulting from the CRISPR-Cas9 KO mutants gNNMT 7.2.1 and gNNMT 6.2.4. D: Elf1 NNMT CRISPR-Cas9 KO cells have higher amounts of SAM than wild type Elf1 cells (n=6; s.e.m.; p=1.23E-05 for gNNMT7.2.1, p=5.47E-06 for gNNMT6.2.4; 2-tailed t-test). E: Western blot analysis reveals higher HIF1α expression and H3K27me3 marks in Elf1 CRISPR-Cas9 KO mutants gNNMT 7.2.1 and gNNMT 6.2.4 compared to control Elf1 (iCas9) cells. F: qPCR analysis of the naïve marker DNMT3L in wild type Elf1 cells (n=6) and Elf1 CRISPR-Cas9 KO mutants gNNMT 7.2.1 (n=5) and gNNMT 6.2.4 (n=3). s.e.m.; p=0.0009 for gNNMT 6.2.4 vs. Elf1, p=0.027 for gNNMT 7.2.1 vs. Elf1; 2-tailed t-test. G: log2 fold expression change of NNMT, WNT ligands and HIF target genes in Elf1 CRISPR-Cas9 KO gNNMT 7.2.1 compared to wild type Elf1 cells (RNAseq). H: PCA plot of CRISPR NNMT knockout line and different naïve and primed lines sequenced in this study. gNNMT 6.2.2 and gNNM 7.3.5 are heterozogous controls. PC1 (x-axis) explains majority of the variation in the data (61%), and the gNNMT 7.2.1 knockdown line moved along x-axis substiantially away from other naïve lines and toward the primed state. I: Hypergeometric test p-values for the overlap between genes expressed higher (lower) in gNNMT 7.2.1 compared to Elf1 and genes expressed higher (lower) in primed lines compared to naïve lines from multiple studies. Color shade is proportional to negative log10 of p-values. gNNMT 7.2.1 transcriptomic signature has significant overlap with all published primed transcriptomic datasets, supporting its transition toward a primed stage. J: Model of the intricate relationship between metabolism and epigenetic in hESCs. Unprocessed original scans of blots are shown in Supplementary Fig.9. For raw data, see Supplementary Table 4. n= number of biological replicates.
DISCUSSION
We show that human naïve and primed ESCs display distinct metabolic profiles and the switching between these metabolic states is regulated by NNMT, which controls the amount of SAM available for PRC2 dependent H3K27me3 histone methylation. Repressive histone methylation then controls the primed hESC specific metabolism through the Wnt and HIF pathways (Fig.8J). The naïve to primed hESC transition shows a reduction in Wnt signaling, electron transport chain activity, and fatty acid beta-oxidation and increase in mechanisms involved in lipid biosynthesis and HIF1α stabilization. In naïve hESCs NNMT and its enzymatic product 1-MNA are highly upregulated, while the substrates, nicotinamide and SAM are downregulated, correlating with reduced H3K27me3 marks. Inhibition of the NNMT regulator, STAT3, in naïve hESCs increases H3K27me3 repressive marks in developmental and metabolic genes, including Wnt signaling and the HIF1 repressor, prolyl hydroxylase EGLN1. Further validations using Wnt pathway inhibitors reveal Wnt activity critical for naïve state. HIF1α KO mutant line generated by CRISPR-Cas9 system is incapable of the naïve to primed metabolic and fate switch, showing that HIF1α is required for naïve to primed hESC transition. NNMT KO naïve hESC lines show increased H3K27me3 marks, HIF1α stabilization and Wnt ligand reduction, all indications of transition towards primed stage, even in the presence of naïve state stabilizers (MEK and GSK3 inhibitors and LIF). These data show that NNMT consumes SAM in naïve cells, making it unavailable for histone methylation that represses Wnt pathway and electron transport chain activity and activates HIF pathway and lipid synthesis, facilitating the metabolic switch in the naïve to primed hESC transition (Fig.8J). Therefore, differential metabolites between pluripotent states control epigenetic dynamics and signaling.
Primed ESCs are dependent on glycolysis14,17,35–38. We now show that while early glycolysis metabolites are upregulated, the downstream metabolites are downregulated in primed state hESCs, suggesting that metabolites are being channeled off to increase the amount of glycerol backbone available for biosynthesis of lipids in primed cells, or for the one-carbon cycle for methylation reactions by SAM. SAM can also be regulated by NNMT, whose enzymatic product, 1-MNA is dramatically increased in naïve vs primed hESC. Since NNMT is considered to create a methyl-sink28, reduction of NNMT in primed hESC can make SAM available as a substrate for DNA and histone methylation. We show that a difference in SAM levels between naïve and primed hESCs correlates with dramatic changes in H3K27me3 marks6,8,10–12 and reveal NNMT as a key regulator of these changes. while previous studies have shown SAM dependent regulation of histone methylation in stable primed hESCs, the effect was mainly observed in H3K4me3, not in H3K27me3 marks22. It is plausible that H3K27me3 marks, once established are less dynamic in primed hESCs than H3K4me3 marks39.
While H3K27me3 marks are reduced in naïve compared to primed hESCs, the enzymes required for this methylation (EZH2 and EED) are not downregulated. We now show that high NNMT activity in naïve hESCs sequesters the methylation substrate, SAM, thereby repressing H3K27me3 marks. Furthermore the PRC2 recruiting protein Jarid2 is upregulated in primed hESCs compared to naïve, which may give further specificity to PRC2 action in naïve to primed hESC transition40,41(Fig.1B, 1L–M, 8J).
In this study we show a direct impact of SAM levels and NNMT function on histone marks in naïve hESC,, revealing that changes in the metabolic profile of hESCs shape the epigenetic landscape during development. While previous studies have revealed the importance of Wnt and HIF pathways in naïve or primed pluripotent stem cells34,38,42–44, we now show that these pathways are regulated by metabolite levels. We propose that the availability of SAM triggers the cascade by activating PRC2 and thereby increasing repressive H3K27me3 epigenetic marks in the promoters of key regulators of naïve to primed transition, HIF repressor and Wnt ligands (Fig.8J). These studies pave the way for further understanding and utilization of metabolite specific molecular mechanisms for cell fate changes in general.
MATERIALS AND METHODS
Culture of primed and naïve embryonic stem cells
Primed human ESCs [H1 (WA-01, WiCell) and H7 (WA-07, WiCell)] and naïve hESCs [Elf-1(NIHhESC-12–0156, University of Washington), WIN1(NIHhESC-14–0299, MIT)] and LIS18 (Weizmann Institute of Science) were cultured as previously described10,12. Briefly, the cells were cultured on a feeder layer of irradiated primary mouse embryonic fibroblasts (MEF) in hESC media (Supplemental table 3). One passage prior to the experiments, the cells were transferred to growth factor reduced Matrigel (Becton Dickinson, Moutainview, CA) in MEF conditioned media (CM). Reverse toggling of H1 and H7 cells was performed using supplemented media (Supplemental table 3). H1 cells were pushed toward a more naïve state by culture in 4iLIF (modified from Gafni et al: 1µM GSK3 inhibitor (CHIR99021), 1µM of MEK inhibitor (PD0325901), 5µM JNK inhibitor (SP600125), 2µM p38 inhibitor (BIRB796), 10ng/mL human LIF, 5ng/mL IGF and 10ng/mL bFGF for 3 passages (Suppl.Fig.1D). In addition, human naïve cells (Elf1 and WIN1) were pushed toward a more primed state by culturing them in either TeSR1 (STEMCELL technologies), or bFGF (10ng/mL) with or without Activin A (10ng/mL) for 3 passages (Elf1 AF, WIN1 AF, WIN1 F, WIN1 TeSR). Mouse ESCs (R1, EpiSC, R1 AF) were cultured as described in Supplemental table 3. All cells were grown at 37 degres, 5%CO2 and 5% O2. Cells have been karyotyped by Diagnostic Cytogenetics,inc. and were tested for mycoplasma detection using MycoAlert detection kit (Lonza).
OCR and ECAR measurement using Seahorse Cellular Flux assays
Naïve and primed ESCs were seeded onto 96-well Seahorse plates pre-coated with Matrigel at 25×104 or 40×104 cells/well. Culture media were exchanged for base media (unbuffered DMEM (Sigma D5030) supplemented with sodium pyruvate (Gibco, 1mM) and with 25mM glucose (for MitoStress assay), 25mM glucose and 50µM carnitine (for palmitate assay), or 2mM glutamine (for glucose stress assay) 1 h prior to the assay. Substrates and selective inhibitors were injected during the measurements to achieve final concentrations of glucose (2.5mM), 4-(trifluoromethoxy)phenylhydrazone (FCCP, 300nM-500nM), oligomycin (2.5 µM), antimycin (2.5µM), rotenone (2.5µM), palmitate (50µM in BSA), BSA and ETO (50µM). The OCR and ECAR values were normalized to the number of cells present in each well, quantified by the Hoechst staining (HO33342; Sigma-Aldrich). Changes in OCR and ECAR in response to substrates and inhibitors addition were defined as the maximal change after the chemical injection compared to the last OCR value before the injection.
Mitochondrial DNA mutation frequency and copy number analysis
The DNA of Elf1 and H7 cells was isolated using DNAzol (Invitrogen). TaqMan primers were used to quantify mitochondrial and genomic DNA (Suppl.Table.2). The ratio of mtDNA to genomic DNA was determined using a standard curve for each primer. Each reaction contained 2 ng of DNA extract, 1x TaqMan Universal PCR Master Mix No AmpErase UNG, 500nM of each primer, and 200nM of the TaqMan probe. Using 7300 real time PCR system (Applied Biosystems), the reactions were amplified by incubation at 50°C for 2 minutes, 95°C for 10 minutes, and then 40 cycles of 15 seconds at 95°C followed by 1 minute at 60°C where the intensity of fluorescence was measured. Naïve hESCs (Elf1) and primed hESCs (H1 and Elf1 AF) cells were grown in triplicate for mutation analysis. Elf1 were analyzed between passage 19 and 23, Elf1AF were analyzed at passage 25 and H1 at passage 65All lines were grown on Matrigel for the last passage prior to analysis.
DNA was isolated from hESCs with the DNeasy Blood and Tissue Kit (QIAGEN). Rare mutation-bearing molecules were selectively enriched through endonucleolytic destruction of wild-type target sites by sequential additions of TaqI (New England Biolabs). Complete cleavage of wild-type TaqI sites was verified by PCR followed by restriction digest and gel electrophoresis. Reaction droplets were prepared as described previously51, with primers and probe sets specific to each target region (Supplementary Table 2). Fragments for point mutation detection and mtDNA copy number measurement were amplified as follows: 95°C for 10 min, followed by 40 cycles of 94°C for 30 s, and 60°C for 1 min. For digital deletion detection, thermal cycling was as follows: 95°C for 10 min, followed by 50 cycles of 94°C for 30 s, and 63.5°C for 2 min. The thermally cycled droplets were analyzed by flow cytometry in a QX100™ Droplet Digital™ Reader (Bio-Rad) for fluorescence analysis and quantification of mutation frequencies and mtDNA copy number, as described elsewhere51
Proteomics
Naïve hESCs (Elf1 2iLIF) and primed hESCs (Elf1 AF) were washed in 1xPBS and flash frozen. Cell pellets were lysed in 1M urea, 50mM ammonium bicarbonate, pH 7.8, and heated to 50°C for 20min. Normalized quantities of protein were reduced, alkylated, and digested overnight with trypsin. The resulting peptides were desalted on Waters Sep-Pak C18 cartridges. Peptides were measured by nano-LC-MS/MS on a Thermo Scientific Fusion Orbitrap. Peptides were separated by reverse phase chromatography in a 180min gradient (1–45% acetonitrile). The Fusion was operated in data-dependent mode with the following settings: 60000 resolution, 400–1600 m/z full scan, Top Speed 3 seconds, and an 1.8 m/z isolation window. Identification and label free quantification of peptides was done with MaxQuant 1.552 using a 1% false discovery rate (FDR) against the human Swiss-Prot/TrEMB database downloaded from Uniprot on October 11th, 2013. We analyzed two biological and three technical replicates per condition. Proteins that were significantly regulated between conditions were identified using a permutation-based t-test (S1, FDR 5%) in Perseus 1.4.1.3.
Non-targeted GC-TOF and LC-QTOF analysis for metabolites in mouse and human ESCs
For the first set of experiments (Fig.2B–F) 5–20 million cells per replicate (grown on “ghost”: irradiated MEFs lysed using a detergent solution 0.5% Triton and 0.034% (v/v) NH4OH (Sigma-Aldrich) to retain their extracellular matrix) were scraped in PBS, pelleted in 6 replicates per condition, frozen at −80 °C. For the first set of experiments cells were thawed on ice and mixed with 2mL of ice-cold degassed acetonitrile then vortexed for 20 sec and sonicated for 5 min.1mL material was taken and centrifuged for 5min at 14,000 rcf. Supernatant was divided into a 500µL (GC-TOF) and a 250µL (LC-QTOF) aliquot. Lyophilized aliquots were resuspended in 500µL of 1:1 acetonitrile:H2O and centrifuged. Supernatants were lyophilized until analysis. For the second set of experiments cells were thawed on ice and extracted as previously reported53. The upper layer was used for LC/MS analysis and bottom layer used for GC/MS analysis, both layers were evaporated to dryness.
Mass spectrometry-based untargeted metabolomics
Primary metabolites were derivatized in 40ug/mL methoxyamine/pyridine and subsequently trimethylsilylated. GC-TOF MS analysis was performed as previously described54,55 using a Leco Pegasus IV time of flight MS coupled to an Agilent 6890 GC equipped with a 30m long 0.25mm id Rtx5Sil-MS column and a Gerstel MPS2 automatic liner exchange system. Data were processed by ChromaTOF and BinBase filtering for metabolite identifications54.
LC-QTOF MS was used for analysis of complex lipids. For the first set of experiments, lyophilized material was redissolved in 100µL initial LC gradient solvent and analyzed within 24 hrs. HILIC and Reversed Phase LC-QTOF analysis and data processing was performed as previously described55 using an Agilent 1200 series HPLC equipped with either Agilent Zorbax Eclipse Plus C18 2.1×150mm column for Reversed Phase or a Waters 1.7µm Acquity BEH HILIC 2.1×150mm column. LC eluents were analyzed with an Agilent 6530 accurate mass Q-TOF mass spectrometer. For the second set of experiments the lipid extracted phase was analyzed as previously described53. Method blanks and human pooled plasma samples were used as QC controls. MZmine 2.10 was used to process the raw data and metabolites were reported when present in 50% of the samples in each group. Annotations were made based on in house accurate mass and retention time library created using LipidBlast, described previously54,56.
Multivariate Analysis of Primed vs Naïve Metabolomes
Metabolite measurements of known and unknown GC metabolites and known lipid metabolites (from ESI(+) and ESI(−) modes) were submitted using R to DeviumWeb (v 0.3.2)53 and normalized using unit norm normalization. O-PLS-DA, a multivariate classification model, was used to identify differences between primed and naïve human and mouse cells. Robust model performance statistics were generated by 100 rounds of Monte Carlo cross validation using training and testing compared to the permuted model (random chance).
LC-QTOF for Elf1 and H1 hESCs
For lipid extraction Elf1 and H1 cells were grown on matrigel for one passage. Cells were washed with PBS and 37 °C deionized water followed by the addition of 0.5ml of a −75 °C solution of internal standards54 and incubation on dry ice for 15 min. Cells were scraped into eppendorf tubes and 1ml of chloroform was added, followed by 15 min incubation on dry ice and spun for 5 min at 4 °C at 18000 rcf, after which the lower phase was collected and stored at −80 °C.
LC-QTOF-MS experiments were performed using an Agilent 1200 SL LC system coupled online with an Agilent 6520 Q-TOF mass spectrometer. Each sample (4 µL for positive ESI ionization, 8 µL for negative ESI ionization) was injected onto an Agilent Zorbax 300 SB-C8 column, which was heated to 50 °C.
Targeted LC-QQQ-MS analysis for water soluble metabolites
Targeted LC-QQQ-MS analysis was performed to detect a different set of water soluble metabolites to complement the other non-targeted mass spec experiments. Elf1 and H1 cells were grown on matrigel for one passage, washed with PBS and ice cold deionized water followed by the addition of a −75 °C 0.75 mL 9:1 methanol:chloroform solution. The plates were incubated on dry ice for 15 min before scraping into eppendorf tubes, which were spun at 18000 rcf for 5 min at 4 °C. All soluble extract was transferred into a new eppendorf tube and vacuum dried. Samples were stored in −80°C.
Chromatography conditions: dried samples were reconstituted in 200 µL 5 mM ammonium acetate in 40% water/60% acetonitrile + 0.2% acetic acid, and filtered through 0.45 µm PVDF filters (Phenomenex, Torrance, CA) prior to LC-MS analysis. LC-MS/MS was performed using an Agilent 1260 LC AB-Sciex 5500 QQQ MS. Both chromatographic separations were performed in HILIC mode on two SeQuant ZIC-cHILIC columns. The mobile phase was composed of Solvents A (5 mM ammonium acetate in 90%H2O/ 10% acetonitrile + 0.2% acetic acid) and B (5 mM ammonium acetate in 90%acetonitrile/ 10% H2O + 0.2% acetic acid). The chromatographic separation, MS ionization and data acquisition was performed using an AB Sciex QTrap 5500 mass spectrometer equipped with electrospray ionization (ESI) source. The instrument was controlled by Analyst 1.5 software. Targeted data acquisition was performed in multiple-reaction-monitoring (MRM) mode. The extracted MRM peaks were integrated using MultiQuant 2.1 software
Targeted HILIC-QTOF mass spectrometry metabolite quantifications of methionine metabolites
Cells were grown on matrigel for one passage, scraped and washed with PBS at room temperature, pelleted and flash frozen in liquid nitrogen. Samples were extracted by adding 1mL cold 3:1 cold methanol:water to the cell pellet, vortexed, placed at −20°C for 30 minutes, and centrifuged for 10 minutes at 14000 rcf. The supernatant was transferred then centrifuged again, then supernatant was evaporated to dryness. Samples were re-suspended in 80:20 acetonitrile:water containing Val-Try-Val. Standard curve dilutions for quantifications were prepared using mixture of 1-methylnicotinamide HCl (1-MNA), S-methyl-5’-thioadenosine (MTA), S-adenosyl methionine (SAM), S-adenosyl homocysteine (SAH), Methionine, Kynurenine and Tryptophan (Sigma).
Hydrophilic interaction chromatography (HILIC) analysis of standard curve and samples was performed using an Agilent 1290 Infinity Ultrahigh Pressure Liquid Chromatography stack equipped with an auto-sampler (4°C) using 5 µL injections into an Acquity UPLC BEH Amide column (Waters Corporation). Mobile phases were prepared with 10mM ammonium formate and 0.125% formic acid in either 100% LCMS grade water for mobile phase A or 95:5 acetonitrile:water for mobile phase B. Metabolites were detected and quantified by an Agilent 6530 accurate mass quadrupole time-of-flight (QTOF) mass spectrometer with a jet stream ESI source in positive ion mode. Mass calibration was maintained by constant reference ion infusion, with MS data collected at 4 spectra/s. Data files were analyzed using Agilent Mass Hunter TOF Quantitative Analysis software. Peak filtering was performed manually to eliminate peaks with a signal to noise ratio of less than 3. Retention times and major adducts for each compound are as follows: 1-MNA (m/z 137.0715) 6.345 min, MTA (m/z 297.0896) 2.583 min M+H, tryptophan (m/z 204.0899) 6.904 min M+H, kynurenine (m/z 208.0848) 6.971 min. M+H & M+Na, methionine (m/z 149.0511) 7.493 min M+H & M+2Na+H, SAH (m/z 384.1216) 8.810 min M+H, SAM (m/z 399.1451) 9.768 min. Metabolites at undetectable levels and metabolites whose levels saturated the system were given the lowest and highest detectable values respectively in subsequent analysis
Metabolite levels were sum-normalized for each sample using the methionine metabolite values (methionine, nicotinamide, MTA, 1-MNA, SAM and SAH). P-values were calculated using a 1-tailed t-test.
Transcriptomic data analysis
RNA-seq data processing was performed according to Takashima et al9. Raw RNA-seq reads from this study and 3 other studies (Chan et al6) and Takashima et al9, Yan et al23) were aligned to hg19/GRCh37 with STAR aligner57. Transcript quantification was performed with htseq-count from HTSeq package58 using GENCODE v15. Differential expression analysis was performed with DESeq after filtering out genes whose total read count across samples are below the 40th quantile of all genes.
Size factors used to normalize by library size were computed using the DESeq package59. Reads were further normalized by gene length.
Affymetrix Human Gene Array 1.0 ST arrays from Gafni et al8 were processed with oligo package60 and normalized using Robust Multi-array Average61. Multiple probes mapping into the same gene were summarized into a single expression value by taking the max. Affymetrix PrimeView arrays from Theunissen et al10 were processed with Affy package62 and normalized with RMA. Microarray differential expression analysis was performed using the limma package. RNA-seq and microarray data were combined as previously described9, expression levels were converted to log2 fold change relative to the mean of human embryo-derived PSC samples within each study. One-to-one orthologous genes between mouse-human were mapped as previously described9. PCA plot of all samples from all studies were generated using the princomp function from R stats package.
An alternative PCA analysis was performed where ComBat tool63 was applied to correct for batch effects and naïve samples were not normalized to primed samples. PCA was applied on batch effects-corrected, gene-wise mean-centered expression values.
Global metabolomic data analysis
Alll global metabolomic data was mean-centered within each sample prcomp function in R is used for Principle Component Analysis of metabolomics data. Differentially abundant metabolites were defined as metabolites with 2 fold change in abundance and Benjamini-Hochberg adjusted false discovery rate <0.2.
For the lipidomics data, features missing in more than half of all samples (4 or more out of 6) were removed from further analysis. Missing values were replaced with minimum detected values within each sample before mean-centering.
ChIP-seq data analysis
ChIP-seq data of H3K27me3 H3K4me3, H3K9me3 and H3K27ac modifications from Chan et al6, Gafni et al8, Theunissen et al10 and Bernstein et al64 was downloaded from Array Express, GEO, and ENCODE project website. Reads were aligned to hg19 using Bowtie version 1.0.0. allowing 1 mismatch (−N 1). ngsplot was used to generate plots of reads around 5 KB of transcription start sites of a priori defined developmental genes. Reads of replicate samples for the same cell type were merged for ngsplot. Reads with mapping quality above 20 were used by ngsplot. Differentially marked genomic regions were identified with diffReps version 1.55.465 and annotated to the closest genes. Genes associated with at least one significant genomic region (FDR less than 0.05 and fold change >1) were classified as differentially marked. When a gene is annotated with multiple significant genomic regions, the most significant one is assigned to that gene.
Lipid droplet visualization using Oil Red O and bodipy staining
Naïve and primed ESCs were fixed with 4% PFA at room temperature for 10min, washed twice with PBS and stained with Oil Red O dye (Sigma) for 10min at rt. Alternatively, lipid droplets were stained using BODIPY 493/503 (Molecular Probes) for 15min on a rocking platform at rt. Pictures were taken using a fluorescent microscope (Leica). Lipid droplet analysis at in vivo post-implantation stage has proven to be difficult.
Protein extraction and Western blot analysis
Cellular extracts were prepared using a lysis buffer containing 20 mM Tris HCl (pH 7.5), 150 mM NaCl, 15% glycerol, 1% Triton, 25mM β-glycerolphosphate, 50mM NaF, 10mM NaPyrophosphate, orthovanadate, PMSF (all chemicals are from Sigma-Aldrich, St. Louis, MO), Protease inhibitor cocktail (Roche) and 2% SDS. 25 U of Benzonase® Nuclease (EMD Chemicals, Gibbstown, NJ) and 20mM of DTT (Sigma) were added to the lysis buffer right before use. 15 µg of protein (determied by Bradford) was loaded, separated by 4–20% SDS-PAGE, and transferred to polyvinylidene difluoride membranes, blocked with 5% nonfat dry milk for 60 minutes at rt, and incubated overnight at 4°C with primary antibody. After incubation for one hour with horseradish peroxidase-conjugated secondary antibodies, they were visualized by enhanced chemiluminescence (Millipore Corp, Billerica, MA). Antibodies used in this study are: H3K27me3 (1/1000, Abcam, ab6002), H3K9me3 (1/1000, Abcam, ab8898), H3K9/14Ac (1/1000, Cell Signaling, 9677s), EED (1/1000, gift from Dr. Bomsztyk66), HIF1α (1/2000, BD Biosciences, 610958), LDHA (1/1000, Cell Signaling, 3582), JARID2 (1/1000, Cell Signaling, 13594), pSTAT3 (1/1000, Abcam, Ab76315) and γ-tubulin (1/10000, Promega, G712A).
RNA extraction and qPCR analysis
RNA was extracted using trizol and analyzed by SYBRgreen qPCR with the 7300 real time PCR system (Applied Biosystems)67 and TaqMan qPCR (Applied Biosystems). Primers used are listed in Suppl.Table.2.
qPCR of miRNAs was conducted using TaqMan miRNA assays (Applied Biosystems). Raw Ct values for miRNAs were normalized to RNU66. Linear expression values for all qPCR experiments were calculated using the 2(−ΔCt) method. P-values were calculated using a student's t-test.
ChIP-seq experiment
Naïve hESCs Elf1 2iLIF grown on matrigel were treated with 100µM of STAT3 inhibitor (Selleckchem) for 6h or 24h and analyzed for methylation marks by Western blot and ChIP Seq. For ChIP-seq analysis, cells were crosslinked and chromatin processed as previously described67 with minor modifications. Briefly, cells were harvested with accutase and crosslinked in suspension with 1% formaldehyde solution for 10min at room temperature. Reaction was quenched with glycine and crosslinked cells were rinsed with ice-cold PBS. Nuclei were isolated and chromatin sonicated using a Covaris E210 to approximately 200–500bp size range. ChIP-seq was conducted as previously described67 with minor modifications. Briefly, magnetic Dynabeads were incubated overnight rotating at 4C with antibody against H3K27me3 (Active Motif, cat # 39155). Sonicated chromatin from approximately 200 thousand cells was added to the bead-bound-antibodies and incubated at 4C rotating overnight. Beads were washed and bound chromatin was eluted from beads and reverse crosslinked overnight. Purified DNA was prepared for next-generation sequencing via end repair, A-tailing, ligation of custom Y-adapters and PCR amplification to generate final DNA library following gel size selection.
Generation of BAR-Elf reporter cell line
Elf1 cells grown in naïve media (2iL or 2iLIF) or primed media (AF) were infected with BAR reporter lentivirus33,68 and seeded onto matrigel-coated plates in MEF-CM with 10µM Y-27632, and 1µM Thiazovivin (ROCK inhibitors, Tocris). Transduced Elf cells were cultured for a week on matrigel, then passaged onto MEF plates for further selection and expansion. Single Elf1 naïve reporter cells were harvested via TrypLE Express and FACS sorted for the population with both Venus and DsRed positive signals. DsRed positive colonies of Elf1 primed reporter cells were transferred onto MEF plates, and the same positive selection was repeated 1–2 more rounds. Negative colonies were removed as a negative selection.
Manipulation of Wnt pathway
Wnt secretion and signaling were inhibited in naïve hESCs (Elf1, WIN1) by treatment with IWP2 (2µM, Torcis) or XAV939 (5µM, Sigma). Wnt pathway was activated in primed Elf1 AF reporter cells using a GSK3 inhibitor, CHIR99021 (72h, 10µM, AxonMedChem). Both IWP2 and CHIR99021 were reconstituted in DMSO.
Production of Conditioned Medium (LCM and Wnt3A–CM )
L and L-Wnt3A cells (ATCC) were cultured in 15cm plate in 10% FBS/DMEM media until ∼90% confluent. Medium was collected every 48hrs for three batches. Biological activity of secreted Wnt3A in the individual batches of the conditional medium was confirmed in 293T–BAR reporter cells33, then batches were pooled and filtered. Primed (Elf1 AF) reporter cells were grown on matrigel with 50% LCM or 50% Wnt3A–CM for 3 days prior taking bright field and fluorescent pictures (Leica microscope).
RNA interference experiments
Naïve Elf1 2iLIF cells were transfected on matrigel in MEF-CM supplemented with ROCK inhibitors (Torcis) using Lipofectamine RNAiMAX (Life Technologies). siRNA targeting NNMT (Hs-NNMT-8) was purchased from Qiagen as Flexitube siRNA premix, and siRNA targeting luciferase was used as control. siRNAs against NNMT and luciferase were used at 50nM final concentration. Protein and RNA were extracted 72h after transfection. siRNA targeting beta-catenin (Invitrogen, CTNNB1, Silencer Select ID s437) and Silencer Select Negative Control 1 (Invitrogen) were transfected in naïve Elf1 2iLIF cells at 10nM final concentration following a reverse transfection protocol. Bright field and fluorescence images were taken after 3 days. Efficacy was confirmed by qPCR analysis.
Overexpression of NNMT
Naïve hESCs were transfected with NNMT overexpression construct or inactive NNMT mutant overexpression construct (Y20A)28. Cells were plated the following day into matrigel coated Seahorse plates with primed hESC media (conditioned media + AF). Mitostress protocol in Seahorse flux analyzer was performed 2 days later. Alternatively, primed hESCs Elf1 AF were transfected with NNMT overexpression construct28 and proteins were extracted 3 days later for detection of H3K27me3 marks by Western blot analysis.
Treatment of naïve hESCs with the metabolite SAM
WIN1 cells were seeded in Seahorse plates 2 days prior change of media with media without L-methionine (Sigma-Aldrich 0422 supplemented with 0.584 gm/L L-glutamine (Invitrogen)) and addition of SAM (500µM), 5hours later Seahorse mitostress protocol was performed.
HIF1α overexpression in naïve hESCs
Naïve hESCs (Elf1) were infected with a non-degradable form of HIF1α over-expressing construct (Addgene plasmid 19005, Yan et al.) or a pBABE empty vector construct in presence of 4 ng/ml Polybrene (Invitrogen). 24 days later RNA and proteins were harvested or media was changed into naïve hESC media with LIF but without GSK3 and MEK inhibitors (2i). Pictures were taken 3 days after infection and primed-like morphology of colonies quantified.
CRISPR-Cas9 based NNMT and HIF1 KO lines
Six gRNAs were designed for each of the genes NNMT and HIF1α using the GECKO library and Zhang website (mit.edu) and annealed and ligated into the LentiCRISPRv2 (Addgene.com), which was previously digested with BsmB1 and dephosphorylated. The validated constructs were transfected using GeneJuice to naïve hESCs iCas9 Elf169 treated with doxycycline (2 µg/ml) for 1 or 2 days before and during transfection. On Day 3 the cells were trypsinized and replated on MEF-coated plates. On Day 6 single colonies were picked and amplified as described in Suppl.Fig8. Genomic DNA was harvested using DNAzol reagent (Invitrogen). NNMT KO 6-2–4 mutant was generated by transfecting in vitro transcribed gRNA to naïve hESC Elf1 iCas9 cells69. T7-gRNA in vitro transcription (IVT) template (120bp) was generated using T7 promoter-gRNA forward primer with a reverse primer against the scaffold. T7-gRNA PCR products were used as templates for IVT (MAXIscript T7 kit, Applied Biosystems). iCas9 Elf1 were treated with doxycycline (2µg/ml) for 1 or 2 days before and during transfection. For transfection, cells were dissociated with Trypsin, replated onto MEF-coated plates, and transfected in suspension with gRNAs using Lipofectamine RNAiMAX (Life Technologies). gRNA was added at a 10 nM final concentration. A second transfection was performed after 24 hours. Two days after the last gRNA transfection, iCas9-Elf1 cells were dissociated into single cells and replated onto MEF coated plates. Single colonies were randomly selected and amplified. Genomic DNA was harvested using DNAzol. Genomic regions flanking the CRISPR target sites were PCR amplified, purified and send to Genewiz, Inc for sequencing. Alternatively, Samtools mpileup and BCF tools were used to identify variants based on aligned RNA-seq BAM files.
Teratoma formation
Naïve hESCs H1 4iLIF were cultured on Matrigel-coated plates. 2 × 106 cells were resuspended in Matrigel supplemented with a cocktail of prosurvival factors70 and injected into the femoral muscle of female 43–49 day-old SCID-Beige mice (Charles River, Wilmington, MA). Palpable tumor masses developed in approximately 5 weeks. The tumor bearing mice were sacrificed, tumor tissue was fixed in 10% formalin for 24h and stored in 70% ethanol until paraffin imbedding. Five µm sections of the tumor were stained with hematoxylin and eosin (Suppl.Fig.1D) Experiment was performed in compliance with ethical regulations, IACUC protocol # 4152-01.
Embryonic body (EB) formation
H1 4iLIF naïve-toggled cells were trypsinized and transferred into low attachment plates in differentiation medium (DMEM, high Glucose, 20% FBS, 0.1 mM NEAA, 2 mM L-Glutamine, 1 mM Sodium Pyruvate, 1% Pen/Strep, 0.1 mM β-mercaptoethanol). The medium was changed every other day and whole genome RNA seq was performed after 2 weeks (Suppl,Table1S).
Accession numbers
RNA-seq and ChIP-Seq data sets generated for this study are available from the NCBI GEO database under accession number GSE60955. Gene expression and ChIP-Seq data from Grow et al45 (GSE63570), Gafni et al8 (GSE52824), Chan et al6 (E-MTAB-2031 and E-MTAB-2041), Theunissen et al10 (GSE59435), and Takashima et al9 (GSE60945) were also analyzed. mRNA-seq, microRNA-seq and ChIP-Seq of H1 and ChIP-Seq of H9 hESC cell lines from the ENCODE project were downloaded from the ENCODE project website hosted at UCSC genome brower (https://genome.ucsc.edu/ENCODE/). RNA-seq of multiple lineages derived from H1 hESC were downloaded from release 9 of the Roadmap Epigenome project (http://www.genboree.org/EdaccData/Current-Release/).
Reproducibility of experiments
Number of independent experiments for each figure panel is described in the corresponding figure legend and raw data is available in the “Statistics source data” supplemental table 4. For HIF1α, LDHA and JARID2 western blots in Elf1, Elf1 AF and H7, there were 3 independent experiments (Fig.1J, 1M). For BODIPY 493/503 staining and Oil Red O of lipid droplets in primed and naïve human (Fig.3D, Suppl.Fig.3A–B) and mouse (Figure 3E) ESCs, there were 4 independent experiments. For western blots of histone marks and EED in Elf1, Elf1AF and H7 (Fig.5B) there were 3 to 8 independent experiments. For Western blots of histone marks in Elf1 and Elf1 treated with siRNA against NNMT or luciferase control (Fig.5D) and Elf1 treated with STAT3 inhibitor or DMSO control (Fig.5F) there were 3 independent experiments. For Wnt sensor analysis in naïve and primed cells, there were 3 independent experiments (Fig.6B–C, Suppl.Fig.6E–F) For western blot analysis of HIF1α expression in cells pushed toward the primed stage (Fig.7F), there were 3 independent experiments. For western blot of HIF1α expression and H3K27me3 marks in Elf1 CRISPR-Cas9 KO mutants and control cells (Fig8E), there were 4 independent experiments. For teratoma generated from H1 4iLIF (Suppl.Fig.1D), there were 3 independent experiments. For western blot of H3K27me3 in Elf1 AF cells overexpressing NNMT (Suppl.Fig.4I), there were 3 independent experiments. For western blot of H3K27me3 mark in naïve WIN1 and WIN1 TeSR, there were 3 independent experiments (Suppl.Fig.5H). For western blot of HIF1α and H3K27me3 in WIN1 cells after transfection with siRNA against NNMT or luciferase (Suppl.Fig.5L) there were 3 independent experiments. For western blot of phosphorylated STAT3 in H1 and H1 2iF (Suppl.Fig.5M), there were 2 independent experiments. For morphology of Elf1 transfected with EV or HIF1α OE, there were 3 independent experiments (Suppl.Fig.7B).
There is no estimate of variation in each group of data and the variance is similar between the groups. No statistical method was used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment. RNA samples with 260nm/280nm <1.80 were discarded.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Ruohola-Baker laboratory for helpful discussions throughout this work. We thank Danijel Djukovic and John K. Meissen for help with mass spectrometry, Daniel Jones for help with RNA-seq analysis, Dr. Wenyu Zhou, Angel Nelson, Sandra Shannon, Sonia Sidhu, Christopher Cavanaugh, Yuejuan Zhang, William Heath, Kendl Sankary, Emily Engelhart and Karan Toor for technical help, Kam Au for performing karyotype analysis, Dr. Anup Madan for performing RNA-seq, Dr. Piper Treuting for teratoma analysis, Dr. Karol Bomsztyk for providing EED antibody, Dr. Benjamin Cravatt for providing NNMT overexpression constructs and Dr. Rudolf Jaenisch and Dr. Jacob Hanna for providing WIN1 cells and LIS1 cells, respectively. This work is supported in part by the University of Washington's Proteomics Resource (UWPR95794). RTM is an Investigator, and AR and ZX are Associates, of the HHMI. This work was supported by fellowship from the American Heart Association to JM, AG-NS-0577-09 from the Ellison Medical Foundation for JB, Schultz Fellowship for HS, grants from the National Institutes of Health 1U24DK097154 for OF, RO1ES019319 for JB, R01GM097372, R01GM97372-03S1 and R01GM083867 for HRB, 1P01GM081619 for CTB, RM, CW, AM and HRB, and the NHLBI Progenitor Cell Biology Consortium (U01HL099997; UO1HL099993) for HRB.
REFERENCES
- 1.Buecker C, et al. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Factor DC, et al. Epigenomic Comparison Reveals Activation of "Seed" Enhancers during Transition from Naive to Primed Pluripotency. Cell Stem Cell. 2014;14:854–863. doi: 10.1016/j.stem.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tesar PJ, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 4.Wu J, et al. An alternative pluripotent state confers interspecies chimaeric competency. Nature. 2015;521:316–321. doi: 10.1038/nature14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brons IG, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 6.Chan YS, et al. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell. 2013;13:663–675. doi: 10.1016/j.stem.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 7.Duggal G, et al. Alternative Routes to Induce Naive Pluripotency in Human Embryonic Stem Cells. Stem Cells. 2015;33:2686–2698. doi: 10.1002/stem.2071. [DOI] [PubMed] [Google Scholar]
- 8.Gafni O, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 9.Takashima Y, et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Theunissen TW, et al. Systematic Identification of Culture Conditions for Induction and Maintenance of Naive Human Pluripotency. Cell Stem Cell. 2014 doi: 10.1016/j.stem.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valamehr B, et al. Platform for Induction and Maintenance of Transgene-free hiPSCs Resembling Ground State Pluripotent Stem Cells. Stem Cell Reports. 2014;2:366–381. doi: 10.1016/j.stemcr.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ware CB, et al. Derivation of naive human embryonic stem cells. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1319738111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bracha AL, Ramanathan A, Huang S, Ingber DE, Schreiber SL. Carbon metabolism-mediated myogenic differentiation. Nat Chem Biol. 2010;6:202–204. doi: 10.1038/nchembio.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folmes CD, et al. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greer SN, Metcalf JL, Wang Y, Ohh M. The updated biology of hypoxia-inducible factor. EMBO J. 2012;31:2448–2460. doi: 10.1038/emboj.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathieu J, et al. Hypoxia-inducible factors have distinct and stage-specific roles during reprogramming of human cells to pluripotency. Cell Stem Cell. 2014;14:592–605. doi: 10.1016/j.stem.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panopoulos AD, et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168–177. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rafalski VA, Mancini E, Brunet A. Energy metabolism and energy-sensing pathways in mammalian embryonic and adult stem cell fate. J Cell Sci. 2012;125:5597–5608. doi: 10.1242/jcs.114827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yanes O, et al. Metabolic oxidation regulates embryonic stem cell differentiation. Nat Chem Biol. 2010;6:411–417. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou W, et al. HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shyh-Chang N, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shiraki N, et al. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 2014;19:780–794. doi: 10.1016/j.cmet.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 23.Yan L, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20:1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 24.Berra E, et al. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simonson TS, et al. Genetic evidence for high-altitude adaptation in Tibet. Science. 2010;329:72–75. doi: 10.1126/science.1189406. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen-Tran DH, et al. Molecular mechanism of sphingosine-1-phosphate action in Duchenne muscular dystrophy. Dis Model Mech. 2014;7:41–54. doi: 10.1242/dmm.013631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Opitz CA, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478:197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- 28.Ulanovskaya OA, Zuhl AM, Cravatt BF. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat Chem Biol. 2013;9:300–306. doi: 10.1038/nchembio.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kraus D, et al. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature. 2014;508:258–262. doi: 10.1038/nature13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graf U, Casanova EA, Cinelli P. The Role of the Leukemia Inhibitory Factor (LIF) - Pathway in Derivation and Maintenance of Murine Pluripotent Stem Cells. Genes (Basel) 2011;2:280–297. doi: 10.3390/genes2010280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomida M, Ohtake H, Yokota T, Kobayashi Y, Kurosumi M. Stat3 up-regulates expression of nicotinamide N-methyltransferase in human cancer cells. J Cancer Res Clin Oncol. 2008;134:551–559. doi: 10.1007/s00432-007-0318-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilles C, et al. Transactivation of vimentin by beta-catenin in human breast cancer cells. Cancer Res. 2003;63:2658–2664. [PubMed] [Google Scholar]
- 33.Davidson KC, et al. Wnt/beta-catenin signaling promotes differentiation, not self-renewal, of human embryonic stem cells and is repressed by Oct4. Proc Natl Acad Sci U S A. 2012;109:4485–4490. doi: 10.1073/pnas.1118777109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.ten Berge D, et al. Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat Cell Biol. 2011;13:1070–1075. doi: 10.1038/ncb2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prigione A, Adjaye J. Modulation of mitochondrial biogenesis and bioenergetic metabolism upon in vitro and in vivo differentiation of human ES and iPS cells. Int J Dev Biol. 2010;54:1729–1741. doi: 10.1387/ijdb.103198ap. [DOI] [PubMed] [Google Scholar]
- 36.Varum S, et al. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One. 2011;6:e20914. doi: 10.1371/journal.pone.0020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J. 2011;30:4860–4873. doi: 10.1038/emboj.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou W, et al. Assessment of hypoxia inducible factor levels in cancer cell lines upon hypoxic induction using a novel reporter construct. PLoS One. 2011;6:e27460. doi: 10.1371/journal.pone.0027460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trojer P, Reinberg D. Histone lysine demethylases and their impact on epigenetics. Cell. 2006;125:213–217. doi: 10.1016/j.cell.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 40.Escobar TM, et al. miR-155 activates cytokine gene expression in Th17 cells by regulating the DNA-binding protein Jarid2 to relieve polycomb-mediated repression. Immunity. 2014;40:865–879. doi: 10.1016/j.immuni.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landeira D, Fisher AG. Inactive yet indispensable: the tale of Jarid2. Trends Cell Biol. 2011;21:74–80. doi: 10.1016/j.tcb.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blauwkamp TA, Nigam S, Ardehali R, Weissman IL, Nusse R. Endogenous Wnt signalling in human embryonic stem cells generates an equilibrium of distinct lineage-specified progenitors. Nat Commun. 2012;3:1070. doi: 10.1038/ncomms2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clevers H, Loh KM, Nusse R. Stem cell signaling An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346:1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 44.Mazumdar J, et al. O2 regulates stem cells through Wnt/beta-catenin signalling. Nat Cell Biol. 2010;12:1007–1013. doi: 10.1038/ncb2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grow EJ, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.ENCODE Project Consotrium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chadwick LH. The NIH Roadmap Epigenomics Program data resource. Epigenomics. 2012;4:317–324. doi: 10.2217/epi.12.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolfgang MJ, et al. The brain-specific carnitine palmitoyltransferase-1c regulates energy homeostasis. Proc Natl Acad Sci U S A. 2006;103:7282–7287. doi: 10.1073/pnas.0602205103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stadler B, et al. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev. 2010;19:935–950. doi: 10.1089/scd.2009.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu Y, et al. A rat RNA-Seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat Commun. 2014;5:3230. doi: 10.1038/ncomms4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor SD, et al. Targeted enrichment and high-resolution digital profiling of mitochondrial DNA deletions in human brain. Aging Cell. 2014;13:29–38. doi: 10.1111/acel.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 53.Liesenfeld DB, et al. Metabolomics and transcriptomics identify pathway differences between visceral and subcutaneous adipose tissue in colorectal cancer patients: the ColoCare study. Am J Clin Nutr. 2015;102:433–443. doi: 10.3945/ajcn.114.103804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kind T, et al. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem. 2009;81:10038–10048. doi: 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meissen JK, et al. Induced pluripotent stem cells show metabolomic differences to embryonic stem cells in polyunsaturated phosphatidylcholines and primary metabolism. PLoS One. 2012;7:e46770. doi: 10.1371/journal.pone.0046770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kind T, et al. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat Methods. 2013;10:755–758. doi: 10.1038/nmeth.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 62.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 63.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 64.Bernstein BE, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen L, et al. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One. 2013;8:e65598. doi: 10.1371/journal.pone.0065598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Denisenko ON, Bomsztyk K. The product of the murine homolog of the Drosophila extra sex combs gene displays transcriptional repressor activity. Mol Cell Biol. 1997;17:4707–4717. doi: 10.1128/mcb.17.8.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sperber H, et al. miRNA sensitivity to Drosha levels correlates with pre-miRNA secondary structure. RNA. 2014;20:621–631. doi: 10.1261/rna.043943.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Biechele TL, Adams AM, Moon RT. Transcription-based reporters of Wnt/beta-catenin signaling. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.prot5223. [DOI] [PubMed] [Google Scholar]
- 69.Gonzalez F, et al. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–226. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mathieu J, et al. Hypoxia induces re-entry of committed cells into pluripotency. Stem Cells. 2013;31:1737–1748. doi: 10.1002/stem.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.