Abstract
We report the development of a general methodology to genotype HLA class I and class II loci. A Whole Genome Amplification (WGA) step was used as a sample sparing methodology. HLA typing data could be obtained with as few as 300 cells, underlining the usefulness of the methodology for studies for which limited cells are available. The next generation sequencing platform was validated using a panel of cell lines from the International Histocompatibility Working Group (IHWG) for HLA-A, -B, and -C. Concordance with the known, previously determined HLA types was 99%. We next developed a panel of primers to allow HLA typing of alpha and beta chains of the HLA DQ and DP loci and the beta chain of the DRB1 locus. For the beta chain genes, we employed a novel strategy using primers in the intron regions surrounding exon 2, and the introns surrounding exons 3 through 4 (DRB1) or 5 (DQB1 and DPB1). Concordance with previously determined HLA Class II types was also 99%. To increase throughput and decrease cost, we developed strategies combining multiple loci from each donor. Multiplexing of 96 samples per run resulted in increases in throughput of approximately 8-fold. The pipeline developed for this analysis (HLATyphon) is available for download at https://github.com/LJI-Bioinformatics/HLATyphon.
Keywords: HLA typing, Next generation sequencing, Sample Sparing, Pipeline
1 INTRODUCTION
The Human Leukocyte Antigen (HLA) complex on chromosome 6 is one of the most polymorphic regions in the human genome. The HLA genes play key function in immune activation and regulation, from infectious disease, transplantation, autoimmunity and cancer immunology [1]. Recent years have seen an explosive growth in the development of assays using Next Generation Sequencing (NGS). An obvious application for NGS is for HLA typing. However, surprisingly few groups or companies have published results from HLA typing using NGS [2, 3] that are validated on large reference panels.
The IMGT/HLA database currently defines nearly 9000 alleles for HLA Class I genes (HLA-A, -B, and – C), and nearly 3000 HLA Class II genes (HLA-DRB, -DQA/DQB, and DPA/DPB) [4]. With the explosion of whole genome sequencing projects, the number of novel HLA alleles is increasing at a phenomenal rate. Typically HLA has been typed in the clinic using sequence specific oligonucleotide (SSO) methods, sequence specific primer (SSP) methods, or Sanger sequencing [5]. These methods are reliable, but relatively costly and slow. For the research laboratory, HLA typing of research subjects is generally technically outside the expertise of the group, and most groups have outsourced their typing to commercial laboratories at considerable expense, thus limiting the number of subjects that can be typed. With the improvements and general availability of NGS, a HLA typing assay would allow for most laboratories to type subjects at a reasonable cost.
Recent improvements in NGS technologies have allowed for read lengths that are necessary for distinguishing between the two, often different, alleles expressed in an individual. Compared with the traditional Sanger method, NGS technologies produce many short reads, making the HLA genotyping process much cheaper and faster, as well as reducing ambiguous typing results of heterozygous samples in diploid genomes [6]. To date the hindrance for high-resolution HLA genotyping via NGS-based approach has been the allelic polymorphism of HLA genes. To overcome this issue a few purely computational means were developed: (1) De novo assembling the NGS short reads and mapping the resulting contigs against the HLA references, scoring each HLA allele on the basis of contigs depth of coverage, length and percent sequence identity [7, 8]; (2) Tree-based top down greedy algorithm by using hierarchical read weighting [9]; (3) Assuming the correct reference(s) should have more mapped reads than the incorrect one(s) and performing count reads maximization [10–13]. Although these methods could yield acceptable concordance rates, their schemes were using arbitrary and empirical criteria that might introduce bias in HLA genotype calling. Thus in this study we developed and validated an in-house HLA-typing pipeline on the basis of counting read depth rather than number of mapped reads with leveraging the NGS data of longer reads.
An important consideration in designing HLA-typing assays for practical clinical research studies is the limited sample availability. In cases where donors are affected by severe pathologies, or in longitudinal studies following donors after vaccination or in clinical studies, only a few million cells are available in total. Accordingly, sample sparing technologies are required, and as few PBMC as possible should be utilized for HLA typing determination, while still preserving accuracy, experimental convenience and contained costs. Whole Genome Amplification (WGA) is a technique developed around 20 years ago to allow for amplification of the subject’s genome in the case of limiting sample. Over the years, several types of WGA assays have been developed, generally either PCR based applications or isothermal amplification methods such as Multiple Displacement Amplification (MDA) [14]. Commercial products are available for both WGA methods, including REPLI-g (Qiagen) and PicoPlex WGA (Rubicon Genomics). In particular, MDA methods have been shown in many studies to be accurate and unbiased using limited samples [14, 15].
Accordingly, we sought here to validate HLA typing using limited PBMC amounts. Herein we report our results in developing a sample sparing HLA typing assay including an analysis pipeline (HLATyphon) using a MiSeq (Illumina).
2 MATERIAL AND METHODS
2.1 Sample preparation
DNA samples from cell lines with known HLA types were obtained from the International Histocompatibility Working Group (IHWG) using the SP Reference Panel, a combination of 51 DNA samples typed using the highest frequency sequence specific methods possible at the time of the 13th workshop (2007).
Alternatively, genomic DNA isolated from PBMC of study subjects by standard techniques (QIAmp; Qiagen, Valencia, CA) was used for HLA typing. All studies using human PBMCs were performed using anonymized samples following approved protocols from the relevant Research Ethics Committees, for which informed consent was obtained from all individual donors. These samples included four studies in San Diego, CA (University of CA and LJI). The cell lines used for the study include AMAI (IHW09010), AMALA (IHW09064), KAS011 (IHW09009), KOSE (IHW09056), KT17 (IHW09024), MGar (IHW09014), RML (IHW09016), RSH (IHW09021), DBB (IHW09052), M7 (IHW09215), and Priess (IHW09301).
WGA (REPLI-g; Qiagen, Valencia, CA) was used to allow typing of limiting amounts of samples. Input cell numbers ranged from 30 to 30,000 cells.
2.2 PCR amplification
Amplicons specific for HLA Class I genes were generated using primers described by Wang, et.al [11]. The primers, located in exons 1 and 7, generated a PCR product of approximately 2700 bp. For Class II, amplicons specific for exons 2 and 3 from the appropriate HLA Class II genes were generated by PCR using locus-specific primers designed to amplify most polymorphic HLA genes. The primers were designed using Primer3 (v.0.4.0) [16, 17], and compared to the consensus sequences present in the IMGT/HLA database [4]. For the beta chain genes (DRB1, DQB1, and DPB1), two primer sets are used to generate two amplicons spanning exon 2 and exons 3 through 4 (DRB1) or exons 3 though 5 (DQB1 and DPB1). For the alpha chain genes (DPA and DQA), a single set of primers was used to generate an amplicon spanning exons 2 through 4. A primer mix was prepared for each primer set. Each primer in the set was present in the mix at a concentration of 10 µM.
A 46 µl PCR master mix was prepared for each sample containing 10 µl of 5× Crimson LongAmp Taq Reaction Buffer (Crimson LongAmp Taq kit; New England BioLabs, Ipswich, MA), 1.5 µl of 10 mM dNTPs (New England Biolabs, Ispswich, MA), 2 µl (5 units) Crimson LongAmp Taq, 4 µl primer mix (final concentration 0.8 µM for each primer), and 28 µl nuclease-free water (Qiagen). Four µl of DNA (Qiagen purified or REPLI-G material) were added to 46 µl of the PCR master mix in a 96 well PCR tray. After mixing, the PCR was performed on a thermocycler (BioRad or Applied Biosystems). The thermal profile was 94°C for 2 m, followed by 40 cycles at 94°C for 30 s, 63°C for 30 s, and 68°C for 3 m. A finishing step of 68°C for 7 m completed the reaction. The PCR reaction products were analyzed for the correct size fragment on a 1% agarose gel electrophoresed at 125 volts for 30 minutes.
Amplicons of the correct size were then purified using Zymo DNA Clean-up Kit, according to the manufacturer’s instructions. The purified amplicons were quantitated by fluorescence using Qubit 2.0 (Life Technologies, Carlsbad, CA). The two amplicons for each beta chain gene were combined, and a single library was prepared for the set.
2.3 Sequencing Libraries
Sequencing libraries are prepared using Nextera XT reagents (Illumina, San Diego, CA). The Nextera XT kit utilizes an engineered transposome to simultaneously fragment, and tagment input DNA, adding unique adapter sequences and bar codes in the process. Briefly, 96 libraries were prepared in parallel, using 1 ng of input DNA amplicon(s) for each library. The two amplicons for each of the beta chains were combined, and one ng of the combined material was used. The protocol provided by the manufacturer was used for the tagmentation, PCR amplification and PCR clean-up steps. The libraries were purified using AMPure XP (Beckman Coulter, Brea, CA) with a ratio of 0.5:1 beads to DNA (volume:volume).
The libraries were individually normalized by fluorometric quantitation (Qubit), and pooled in equimolar amounts. The combined libraries were quantified and fragment size was determined using TapeStation (Agilent). Libraries with an average fragment size of at least 600 bp (range 300 to 1000 bp) and concentration of 2000 pmol/l were considered to pass QC. The pooled library was loaded at 5.4 pM on one MiSeq flowcell with 1% phiX spiked in (MiSeq Reagent Kit v3). Paired end sequencing was performed with 300 cycles in each direction.
2.4 Computational HLA Genotyping pipeline
For making HLA typing calls, a protocol similar to that of Wang et al [11] was employed, with several modifications described herein. All scripts were developed in Python 2.7 for execution on a Linux cluster through the Torque submission system. The MiSeq-generated reads were first trimmed to remove all bases with a low quality call (Q <=28) as well as those downstream. For a given locus, the trimmed reads were then mapped against all corresponding HLA cDNA references obtained from the IMGT/HLA database release 3.17.0 [4]. Bowtie2 [18] was used for mapping the reads, as it has internal support for ‘soft-clipping’ the reads at exon borders; a requirement for mapping reads derived from genomic DNA against cDNA references. Implausible reference sequences were then filtered out by applying the following criteria: read pairs must be mapped to the same exon, alignment length must be at least 50bp, reads cannot be clipped or contain gaps within an exon. The 5’ dynamic boundary of a given exon was defined as the first nucleotide with non-zero depth of coverage by performing a single 5’-to-3’ pass from the 5’ intron-exon boundary and the 3’ dynamic boundary was defined analogously. The total aligned bases (TAB) were counted on exons 2, 3, and 4 for loci A, B, C, and DRB1 and exons 2 and 3 for loci DPA1, DPB1, DQA1, and DQB1. Given the heterogenous nature of the final mapped read lengths, we found this metric to be more reliable than the total mapped reads. For each locus, the remaining plausible references were sorted in descending order of their TAB, and up to top 200 alleles were retained for further analysis. The union of the aligned reads for each pairwise combination from this pool was determined. Longer alignments were favored in cases where the same read mapped to both templates. The allele combination with the highest number of TAB would be the most probable genotype for the sample. In cases of ties, the pair with the highest joint allele frequency was selected [19] To test for the possibility of homozygosity, an empirically derived scaling factor of 1.05 was applied to the TAB of each individual plausible reference. If this quantity was greater than that of any of the pairwise combinations, the sample was determined to be homozygous. The HLATyphon software can be downloaded from: https://github.com/LJI-Bioinformatics/HLATyphon.
2.5 SSO/SSP HLA typing
High resolution Luminex-based typing for HLA Class II was utilized according to the manufacturer’s instructions (Sequence-Specific Oligonucleotides (SSO) typing; One Lambda, Canoga Park, CA). Where indicated, PCR based methods were used to provide high resolution sub-typing. (Sequence- Specific Primer (SSP) typing; One Lambda, Canoga Park, CA).
3 RESULTS
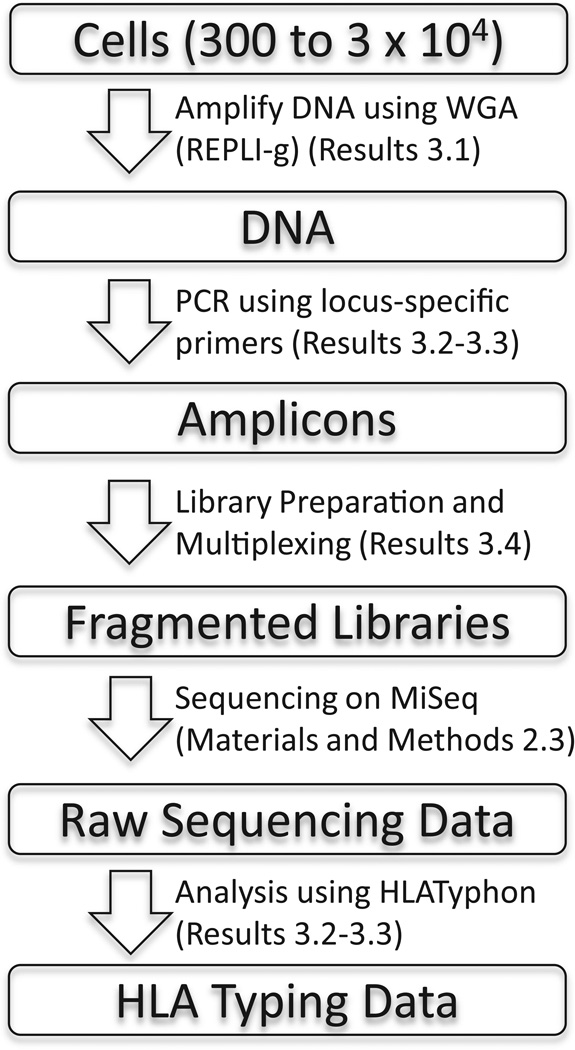
The workflow described in the following sections has been summarized in a schematic diagram (Figure 1).
Figure 1.
Schematic diagram describing the workflow described in the paper. The Results or Methods and Materials section where each step is described is indicated.
3.1 HLA typing of small number of PBMCs
Because samples for HLA typing are often limited, we focused on validating HLA typing methods using limited PBMC amounts. A Whole Genome Amplification (WGA) method using a commercial product developed by Qiagen, REPLI-g, was utilized for this purpose. This product has been used in similar applications [15, 20, 21], and for HLA typing [22–24]. We validated this assay in a comparative study using DNA prepared by standard methods (Qiagen) as well as REPLI-g, and titrated the number of input cells from 3×104 down to 30 cells. The samples using 3×104 and 300 cells were used to HLA type the donors using the SSO method.
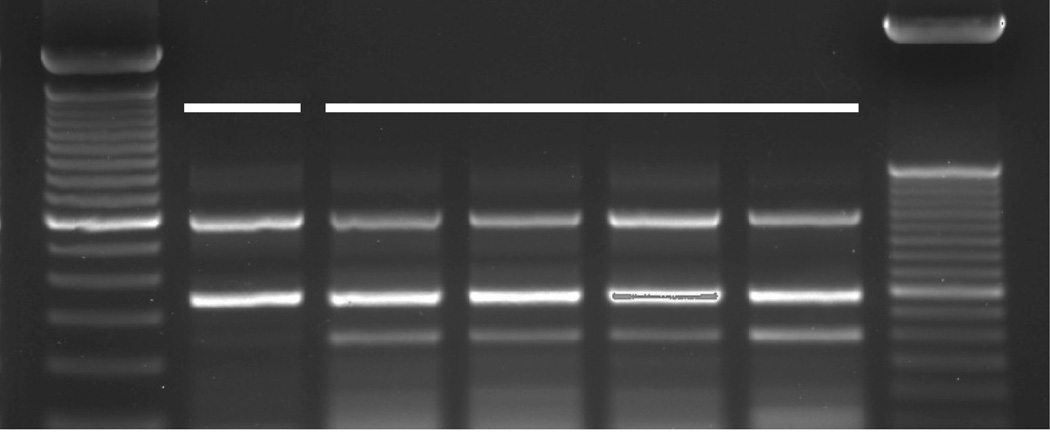
After DNA prep by standard methods or WGA, the first step was to generate a PCR product using locus specific primers. Figure 2 shows the gel of the PCR products obtained using HLA-A-specific primers. For the donor used in this study, the band pattern was identical in both size and intensity whether the DNA was prepared by standard methods, or using WGA by REPLI-g. The amplicons were subsequently used in a Luminex-based SSO assay to obtain the HLA-A type, and in both cases, the donor typed as HLA-A*01:01/A*43:01. Further validation using both SSO and SSP methods and additional donors showed identical results for HLA types across all loci (data not shown). The samples utilized in the multiplexing study described below were also prepared using WGA, and serve as a further validation of this method.
Figure 2.
DNA prepared by standard Qiagen methods and by REPLI-g WGA method give similar results when used in a PCR reaction with HLA-A specific primers. The number of cells prepared by each method is indicated below each lane. The expected sizes for the PCR products are approximately 600 and 350 base pairs when run out on a 2% agarose gel. The left ladder is 100 bp (major band at 600 bp), while the right ladder is 50 bp (major band at 350 bp; Qiagen Trackit DNA ladders). The band at 250 bp is a pseudogene often seen when using these primers, and does not interfere with HLA typing (One Lambda personal communication). The PCR products were subsequently used in a SSO HLA typing assay using Luminex (One Lambda) and by NGS sequencing, and the HLA type obtained was identical for both the standard prep using 5×106 cells and the REPLI-g prep using 300 cells. The donor typed as HLA-A*01:01/A*43:01.
3.2 Implementation of an HLA typing pipeline (HLATyphon)
As a first step toward the establishment of a general HLA typing method (HLATyphon), we began with a algorithm similar to that of Wang et al. [11] and further optimized it to deal with the comparatively short reads generated by the MiSeq and to remove intronic and intergenic reads. First, the Sequence Polymorphism (SP) Reference Panel was obtained from the International Histocompatibility Working Group [4]. The SP reference panel consists of samples from 51 donors typed using the highest frequency sequence specific methods possible at the time of the 13th Working Group (2007). From this panel, samples were chosen to serve for validation of Next Generation Sequencing (NGS) HLA typing methods. Using the primers defined in Wang et.al. [11], amplicons were generated for the HLA class I loci.
For HLA-A, -B, and –C, a total of 47 loci were chosen from the IHWG SP panel, ranging from 14–17 samples per locus (Table 1). These were selected as a standard against which to validate the HLATyphon results. Samples were also selected to maximize the diversity of HLA alleles types, and approximately one third (n=26) of the samples chosen were homozygous. The results of this initial validation are summarized in Table 1, while detailed results can be found in Supplemental Table 1. Concordance with the known, previously determined HLA types was 99% (93/94 calls) overall, similar to other groups’ published concordance rates [10, 11, 25]. The single incorrect call out of the 47 IHWG samples (94 possible calls) was the result of designating one homozygous sample as heterozygous. This suggests that further optimization of the scaling parameter for homozygosity, discussed in the methods, and/or an alternative strategy to further address homozygosity may further decrease the error rate.
Table 1.
Validation samples typed for HLA-A, -B, and –C using amplicons derived from previously published primers [11].
| Locus | Number of Samples | Correct Calls | Concordance Rate |
|---|---|---|---|
| A | 16 | 15.5 | 0.97 |
| B | 14 | 14 | 1.00 |
| C | 17 | 17 | 1.00 |
| Total | 47 | 46.5 | 0.99 |
3.3 Development of a panel of primers to allow HLA typing of HLA-DR, -DQ and -DP loci
To allow for complete typing of both the alpha and beta chains for both DQ and DP loci, as well as for the beta chain of the DRB1 locus, new sets of primers were developed. The primers used and the corresponding amplicon sizes are shown in Table 2. For DR, we focused on the DRB1 locus. Although DRB3, DRB4 and DRB5 loci are also important for antigen presentation, the development of primers for these loci is in progress.
Table 2.
HLA typing primers for HLA Class II loci.
| Locus | Primer Name | Primer Sequence | Amplicon Size (bp) |
|---|---|---|---|
| DRB1 Exon 2 | DRB1 I1 F1 | GCCATCGCTTTCACTGCTCT | 1074 |
| DRB1 I1 F2 | GCCATCACTTTCACTGCTCT | ||
| DRB1 I2 R1 | ACCCACCTCCCTTGTCACCT | ||
| DRB1 I2 R2 | ACCCCCCTCCCACGTCACCT | ||
| DRB1 Exon 3 | DRB1 I2 F1 | TCAAGGTCAGAGCCTGGGTTT | 1398 |
| DRB1 I2 F2 | TCAAGGCCAGAGCCTGGGTTT | ||
| DRB1 I2 F3 | TCAAGGCCAGAGCCTAGGTTT | ||
| DRB1 I4 R1 | TCTCTGCAGGCCACAAGCTA | ||
| DRB1 I4 R2 | TCTCTGTAGGCCACAAGCTA | ||
| DQA | DQA I1 F1 | TGCCAGGCACTCAGGAAATA | 2489 |
| DQA R1 | CACTTCCCAATTCCCCTACAAC | ||
| DQB Exon 2 | DQB E2 F3 | AATCAGCCCGACTGCCTCTT | 1018 |
| DQB E2 F4 | AATCAACCCGACTGCCTCTT | ||
| DQB E2 R8 | TGGGGCAGCCCTAACTCC | ||
| DQB E2 R9 | TGGAACAGCCCTAACTCC | ||
| DQB E2 R10 | TGGAGCAGCCCTAACTCC | ||
| DQB Exon 3 | DQB E3 F1 | CTTTCCACTCTGGTTCCAAGGAG | 1762 |
| DQB E3 F2 | CTTTCCACTCTGGTCCAAGGAG | ||
| DQB E3 F3 | CTTTCCATTCTGGTTCCAAGGAG | ||
| DQB E5 R1 | GCACAAAGTGGGCATCATCC | ||
| DPA | DPA I1 F1 | TGGTGTTGCTCCTTCTTCTTCC | 1287 |
| DPA I1 F2 | TCCCCATATGTCCTTCCTTTGA | ||
| DPA R1 | CACAGAGCACAGTCTCCGTTGT | ||
| DPB Exon 2 | DPB F1 | GGTGGGAAGATTTGGGAAGAAT | 692 |
| DPB I2 R1 | TGCCATCTCCACCTCCATCT | ||
| DPB Exon 3 | DPB I2 F1 | CGCCACTGCATTCCAGACTT | 1849 |
| DPB R3 | TGCTAACGAAACACAGCAAATG | ||
| DPB R4 | TGCTAACAAAACACAGCAAATG |
For the beta chain genes, our strategy was to design primers in the intron regions surrounding exon 2, and the introns surrounding exons 3 through 4 (DRB1) or exons 3 through 5 (DQB1 and DPB1). This strategy allowed us to avoid sequencing the very large intron 2, and thus increased the number of reads for each gene. The primers were targeted to the least polymorphic regions of the introns, as based on the reference sequences in the IMGT HLA database, and primers were generated to cover all currently described variants. To date, we have been able to generate amplicons for approximately 800 donor samples using these primer sets.
The results of validation of HLA typing of the 58 samples using amplicons derived from primers designed specifically for this study are summarized in Table 3 and detailed results are shown in Supplemental Table 1. We found that when these primers were used to generate and sequence amplicons from the reference panel, the concordance rate was at 99% (115/116). A single sample was miscalled for a DRB1 allele, and as previously described for the Class I genes, the issue involved homozygosity. In this case, the donor was originally typed as a heterozygote, but HLATyphon called the donor a homozygote.
Table 3.
Validation samples typed using amplicons derived from primers designed specifically for this study.
| Locus | Number of Samples | Correct Calls | Concordance Rate | |
|---|---|---|---|---|
| DPA1 | 9 | 9 | 1.00 | |
| DPB1 | 12 | 12 | 1.00 | |
| DQA1 | 8 | 8 | 1.00 | |
| DQB1 | 12 | 12 | 1.00 | |
| DRB1 | 17 | 16.5 | 0.97 | |
| Total | 58 | 57.5 | 0.99 | |
3.4 Increased efficiency by multiplexing
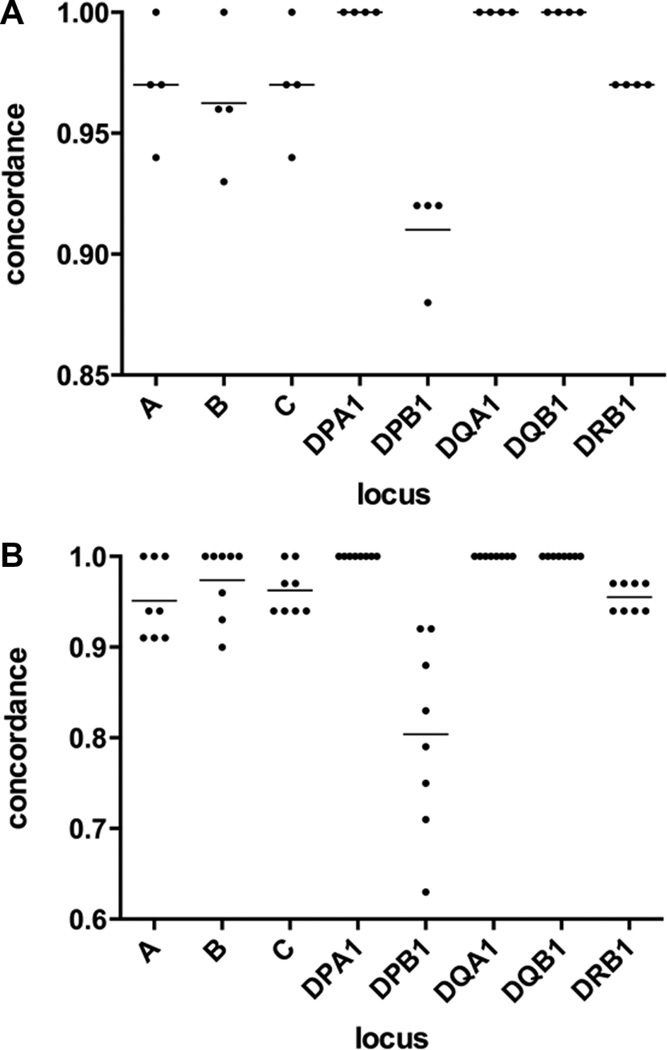
Our initial sequencing strategy used multiplexing of 96 individual loci samples in a single run using unique barcodes. To increase throughput, and thereby decrease cost, we examined alternative strategies; either including additional barcodes or combining multiple loci from each donor. To test these approaches, we utilized the data from the validation study described above. Read pairs were randomly distributed into four equal portions, and reanalysed with HLATyphon. The calls generated using a quarter of the reads were remarkably similar to the initial data (Figure 3A and Supplemental Tables 2 & 3, indicating that typing using 384 barcodes should be possible. The only locus with a marked decrease in concordance was DPB1, which was due to the relatively low number of reads mapping to exon 3 of this locus in the full dataset.
Figure 3.
Sampling of validation data. Concordance rates using limited data sets for each locus are shown. Individual data points are plotted with a line representing the mean for each locus. A. Read pairs were randomly distributed into 4 sets and run through the HLA-typing pipeline. B. Read pairs were randomly distributed into 8 sets, followed by pooling reads for all loci and running through the HLA-typing pipeline.
To evaluate the possibility of multiplexing further by pooling amplicons from each donor, a similar approach was used. In this case, read pairs were distributed into 8 equal-sized sets and reads from each locus were pooled into sets of virtual individuals. In this case, each pool of reads were run through HLATyphon without specifying the locus and the concordance rates were calculated (Figure 3B and Supplemental Tables 4 & 5). Again, the HLA typing determination was remarkably similar to that of the full set of reads with individual amplicons. The reason the concordance rate is lower, as compared to the multiplexing by barcode approach, is twofold: 1) this approach uses half of the reads of the aforementioned approach and 2) it is possible for the same reads to map to references of more than one locus, as the HLATyphon software does not attempt to align to unique regions only..
Next, the accuracy of the increased multiplexing was tested experimentally. Using an input of approximately 5 × 104 cells from each of 11 IHWG subjects, DNA was prepared using WGA as described above. Amplicons were then generated for HLA A, B, C, DRB1, DQA/B and DPA/B loci from this DNA. These amplicons were combined into a single sample for each donor and the library was prepared from the combined samples. These samples were then sequenced in a run along with an additional 85 samples so that the number of reads for each sample was at the level of a 96 sample multiplexed run. The data are shown in Table 4. The concordance rate for the typing of these samples was 94% (162/172 loci). At the locus level (two digits), the concordance rate increased to 97%.
Table 4.
Concordance of sample data generated from combining amplicons from all loci for individual donors.
| Concordance | |||
|---|---|---|---|
| Locus | 4 digit level |
2 digit level |
|
| A | 0.95 | 0.95 | |
| B | 0.91 | 0.95 | |
| C | 0.91 | 1.00 | |
| DPA1 | 1.00 | 1.00 | |
| DPB1 | 0.86 | 0.86 | |
| DQA1 | 0.95 | 1.00 | |
| DQB1 | 1.00 | 1.00 | |
| DRB1 | 0.95 | 1.00 | |
| Total | 0.94 | 0.97 | |
4 DISCUSSION
We report herein the development of a HLA genotyping assay using NGS. HLA typing is of great significance for basic and translational research. Knowledge of the HLA type of individuals whose immune responses is assessed is crucial to enable the use of HLA binding predictions, as a means to identify epitopes and detect responses [26]. HLA typing data is also necessary for determining the fine specificity (also known as HLA restriction) of responding T cells [27]. Knowledge of HLA restriction is in turn key to manufacture of HLA tetramers [28–30], one of the key approaches to enumeration and phenotypic characterization of antigen specific responses. HLA typing data is also useful to assess association between specific HLA types and susceptibility or resistance to infectious disease [31, 32]. There is also strong evidence of HLA linkage to disease and autoimmunity [33]. In addition, HLA genotypes are used to match donor and recipients during transplantation to reduce the risk of transplant rejection [34].
Critical for the development of this assay was the design and validation of specific primers that allowed for the typing of HLA class II loci. Although other groups have published NGS HLA typing protocols, primer sequences have rarely been disclosed. Our strategy was to target areas of low variability in the introns surrounding exon 2 and exons 3 through 5, and then prepare a cocktail of primers for each locus. The resulting primer sets have been utilized in HLA genotyping of more than 800 subjects to date. A similar primer set for the DRB3, DRB4 and DRB5 loci are currently in development.
Other groups have described the generation of chimeric PCR products particularly from the DRB loci[3, 35, 36]. These chimeric products may be formed at very low levels when degenerate primers designed to amplify DRB1 as well as DRB3, DRB4, and DRB5 and other DRB pseudo genes are used [3]. We believe this issue is minimal when using our primer sets because our primers were designed to specifically amplify DRB1, and not DRB3, DRB4, or DRB5 products. In addition, we did not see any indications of this phenomenon in our validation samples, where the correspondence to previous data was 99%.
In this study we developed and validated an in-house HLA-typing pipeline (HLATyphon) on the basis of counting read depth rather than number of mapped reads with leveraging the NGS data of longer reads. Recent improvements in NGS technologies have allowed for read lengths that are necessary for distinguishing between the two, often different, alleles expressed in an individual. Compared with the traditional Sanger method, NGS technologies produce many short reads. This makes the HLA genotyping process less expensive and faster, as well as reducing ambiguous typing results of heterozygous samples in diploid genomes.
Several computational approaches to HLA typing have been reported: (1) De novo assembling the NGS short reads and mapping the resulting contigs against the HLA references, scoring each HLA allele on the basis of contigs depth of coverage, length and percent sequence identity [7, 8]; (2) Tree-based top down greedy algorithm using hierarchical read weighting [9]; (3) Assuming the correct reference(s) should have more mapped reads than the incorrect one(s) and performing count reads maximization [10–13]. Each of these approaches has merit. The main contribution of our paper was to test performance of our approach on a large reference panel and to disclose our novel primer sequences, so that our current study will serve as a common reference point.
HLATyphon achieved more than 99% concordance rate on IHWG samples, similar to another recently published MiSeq-based HLA-typing algorithm [25]. Additional improvements might include: factoring in the linkage information (especially for DRB1), controlling for differential amplicon quantities in class II, and improvements in the identification of DPB1 alleles, which are currently not completely understood.
Sample size is also a critical factor, as for many samples, a very limited amount of sample is available and what is available is needed for research purposes. To address this issue, we have applied whole genome amplification to these samples. The robustness of this procedure allows to reliably testing as few as 300 PBMC, easily obtained by scraping vials of frozen PBMC, thus underlying the value of this approach for studies where the availability of sample available for HLA typing is limiting. We now routinely test for HLA types using approximately 10 µl of cells obtained by scraping the top of a frozen vial of cells, which we estimate may contain approximately 5×104 cells. This typing configuration has been used to successfully type over 1500 samples using both traditional SSO/SSP methods and NGS methods.
In conclusion, we report here a NGS method for HLA typing. The method described discloses novel primers and computational approaches, and also addressed several issues, in particular reliability, cost and sample availability, as critical factors in allowing large scale HLA typing. We anticipate that description of our methodology will facilitate implementation of similar procedures in a broad base of laboratories involved in HLA typing determinations.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Abigail Conroy for technical assistance. This work was supported by National Institutes of Health Contract numbers HHSN272201400045C, N01-AI-900042C, AI-900044C, AI-900048C, and AI-100275 (to A.S.).
ABBREVIATIONS
- WGA
Whole genome amplification
- IHWG
International Histocompatibility Working Group
- TAB
total aligned bases
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Trowsdale J, Knight JC. Major histocompatibility complex genomics and human disease. Annual review of genomics and human genetics. 2013;14:301. doi: 10.1146/annurev-genom-091212-153455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Santis D, Dinauer D, Duke J, Erlich HA, Holcomb CL, Lind C, et al. 16(th) IHIW : review of HLA typing by NGS. International journal of immunogenetics. 2013;40:72. doi: 10.1111/iji.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erlich H. HLA DNA typing: past, present, and future. Tissue antigens. 2012;80:1. doi: 10.1111/j.1399-0039.2012.01881.x. [DOI] [PubMed] [Google Scholar]
- 4.Robinson J, Halliwell JA, McWilliam H, Lopez R, Parham P, Marsh SG. The IMGT/HLA database. Nucleic acids research. 2013;41:D1222. doi: 10.1093/nar/gks949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunn PP. Human leucocyte antigen typing: techniques and technology, a critical appraisal. International journal of immunogenetics. 2011;38:463. doi: 10.1111/j.1744-313X.2011.01040.x. [DOI] [PubMed] [Google Scholar]
- 6.Grumbt B, Eck SH, Hinrichsen T, Hirv K. Diagnostic applications of next generation sequencing in immunogenetics and molecular oncology. Transfusion medicine and hemotherapy : offizielles Organ der Deutschen Gesellschaft fur Transfusionsmedizin und Immunhamatologie. 2013;40:196. doi: 10.1159/000351267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warren RL, Choe G, Freeman DJ, Castellarin M, Munro S, Moore R, et al. Derivation of HLA types from shotgun sequence datasets. Genome medicine. 2012;4:95. doi: 10.1186/gm396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu C, Yang X, Duffy B, Mohanakumar T, Mitra RD, Zody MC, et al. ATHLATES: accurate typing of human leukocyte antigen through exome sequencing. Nucleic acids research. 2013;41:e142. doi: 10.1093/nar/gkt481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim HJ, Pourmand N. HLA haplotyping from RNA-seq data using hierarchical read weighting. Plos One. 2013;8:e67885. doi: 10.1371/journal.pone.0067885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erlich RL, Jia X, Anderson S, Banks E, Gao X, Carrington M, et al. Next-generation sequencing for HLA typing of class I loci. BMC genomics. 2011;12:42. doi: 10.1186/1471-2164-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C, Krishnakumar S, Wilhelmy J, Babrzadeh F, Stepanyan L, Su LF, et al. High-throughput, high-fidelity HLA genotyping with deep sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8676. doi: 10.1073/pnas.1206614109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Major E, Rigo K, Hague T, Berces A, Juhos S. HLA typing from 1000 genomes whole genome and whole exome illumina data. Plos One. 2013;8:e78410. doi: 10.1371/journal.pone.0078410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, Kohlbacher O. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Bourcy CF, De Vlaminck I, Kanbar JN, Wang J, Gawad C, Quake SR. A quantitative comparison of single-cell whole genome amplification methods. Plos One. 2014;9:e105585. doi: 10.1371/journal.pone.0105585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han T, Chang CW, Kwekel JC, Chen Y, Ge Y, Martinez-Murillo F, et al. Characterization of whole genome amplified (WGA) DNA for use in genotyping assay development. BMC genomics. 2012;13:217. doi: 10.1186/1471-2164-13-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 17.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3--new capabilities and interfaces. Nucleic acids research. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Meth. 2012;9:357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez-Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic acids research. 2011;39:D913. doi: 10.1093/nar/gkq1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rykalina VN, Shadrin AA, Amstislavskiy VS, Rogaev EI, Lehrach H, Borodina TA. Exome Sequencing from Nanogram Amounts of Starting DNA: Comparing Three Approaches. Plos One. 2014;9:e101154. doi: 10.1371/journal.pone.0101154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treff NR, Su J, Tao X, Northrop LE, Scott RT., Jr Single-cell whole-genome amplification technique impacts the accuracy of SNP microarray-based genotyping and copy number analyses. Molecular human reproduction. 2011;17:335. doi: 10.1093/molehr/gaq103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Creary LE, Girdlestone J, Zamora J, Brown J, Navarrete CV. Molecular typing of HLA genes using whole genome amplified DNA. Transfusion. 2009;49:57. doi: 10.1111/j.1537-2995.2008.01943.x. [DOI] [PubMed] [Google Scholar]
- 23.Khan F, Liacini A, Arora E, Wang S, Assad M, Doulla J, et al. Assessment of fidelity and utility of the whole-genome amplification for the clinical tests offered in a histocompatibility and immunogenetics laboratory. Tissue antigens. 2012;79:372. doi: 10.1111/j.1399-0039.2012.01857.x. [DOI] [PubMed] [Google Scholar]
- 24.Ndlovu BG, Danaviah S, Moodley E, Ghebremichael M, Bland R, Viljoen J, et al. Use of dried blood spots for the determination of genetic variation of interleukin-10, killer immunoglobulin-like receptor and HLA class I genes. Tissue antigens. 2012;79:114. doi: 10.1111/j.1399-0039.2011.01807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lange V, Bohme I, Hofmann J, Lang K, Sauter J, Schone B, et al. Cost-efficient high-throughput HLA typing by MiSeq amplicon sequencing. BMC Genomics. 2014;15:63. doi: 10.1186/1471-2164-15-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sette A, Rappuoli R. Reverse vaccinology: developing vaccines in the era of genomics. Immunity. 2010;33:530. doi: 10.1016/j.immuni.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKinney DM, Southwood S, Hinz D, Oseroff C, Arlehamn CS, Schulten V, et al. A strategy to determine HLA class II restriction broadly covering the DR, DP, and DQ allelic variants most commonly expressed in the general population. Immunogenetics. 2013;65:357. doi: 10.1007/s00251-013-0684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nepom GT. MHC class II tetramers. Journal of immunology. 2012;188:2477. doi: 10.4049/jimmunol.1102398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newell EW, Sigal N, Nair N, Kidd BA, Greenberg HB, Davis MM. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nature biotechnology. 2013;31:623. doi: 10.1038/nbt.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newell EW, Klein LO, Yu W, Davis MM. Simultaneous detection of many T-cell specificities using combinatorial tetramer staining. Nature methods. 2009;6:497. doi: 10.1038/nmeth.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Apps R, Qi Y, Carlson JM, Chen H, Gao X, Thomas R, et al. Influence of HLA-C expression level on HIV control. Science. 2013;340:87. doi: 10.1126/science.1232685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiskopf D, Angelo MA, de Azeredo EL, Sidney J, Greenbaum JA, Fernando AN, et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2046. doi: 10.1073/pnas.1305227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai S, Santamaria P. MHC Class II Polymorphisms, Autoreactive T-Cells, and Autoimmunity. Frontiers in immunology. 2013;4:321. doi: 10.3389/fimmu.2013.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marks C. Immunobiological determinants in organ transplantation. Annals of the Royal College of Surgeons of England. 1983;65:139. [PMC free article] [PubMed] [Google Scholar]
- 35.Holcomb CL, Rastrou M, Williams TC, Goodridge D, Lazaro AM, Tilanus M, et al. Nextgeneration sequencing can reveal in vitro-generated PCR crossover products: some artifactual sequences correspond to HLA alleles in the IMGT/HLA database. Tissue antigens. 2014;83:32. doi: 10.1111/tan.12269. [DOI] [PubMed] [Google Scholar]
- 36.Danzer M, Niklas N, Stabentheiner S, Hofer K, Proll J, Stuckler C, et al. Rapid, scalable and highly automated HLA genotyping using next-generation sequencing: a transition from research to diagnostics. BMC genomics. 2013;14:221. doi: 10.1186/1471-2164-14-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.