Abstract
Background & Aims
Little is known about the heritability of hepatic fibrosis, and the heritability of hepatic steatosis has not been systematically assessed in adults. We investigated the heritability of hepatic fibrosis and steatosis in a community-dwelling twin cohort.
Methods
We performed a cross-sectional analysis of a cohort of well-characterized twins residing in Southern California including 60 pairs of twins (42 monozygotic and 18 dizygotic; average age, 45.7±22.1 years; average body mass index, 26.4±5.7 kg/m2). We collected data on medical history, physical examinations, fasting laboratory test results, and liver health; all participants underwent an advanced magnetic resonance imaging (MRI) examination of the liver from January 2012 through January 2015. Hepatic steatosis was quantified non-invasively by MRI and determined based on the proton-density fat fraction (MRI-PDFF); liver fibrosis was measured based on stiffness measured by magnetic resonance elastography.
Results
Twenty-six of the 120 subjects (21.7%) had non-alcoholic fatty liver disease (defined as MRI-PDFF ≥ 5% after exclusion of other causes of hepatic steatosis). The presence of hepatic steatosis correlated between monozygotic twins (r2=0.70, P<.0001) but not between di-zygotic twins (r2=0.36, P=0.2). The level of liver fibrosis also correlated between monozygotic twins (r2=0.48, P<.002) but not between dizygotic twins (r2=.12, P=.7). In multivariable models adjusted for age, sex, and ethnicity, the heritability of hepatic steatosis (based on MRI-PDFF) was 0.52 (95% confidence interval, 0.31–0.73; P<1.1x10−11) and the heritability of hepatic fibrosis (based on liver stiffness) was 0.5 (95% confidence interval, 0.28–0.72; P<6.1 x 10−11).
Conclusions
A study of twins provides evidence that hepatic steatosis and hepatic fibrosis are heritable traits.
Keywords: genetic factors, fatty liver, NASH, NAFLD
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) is characterized by hepatic steatosis in individuals who consume little or no alcohol and who have no other identifiable causes of steatosis1. It is the most common cause of chronic liver disease in the United States1–4, affecting 80–100 million Americans, of whom about 18 million are thought to have nonalcoholic steatohepatitis (NASH), a more advanced form that may lead to progressive fibrosis, cirrhosis and hepatocellular carcinoma (HCC)5–9. Patients with hepatic fibrosis are at particularly high risk for developing cirrhosis and HCC, and require more intense monitoring and therapy6, 10, 11. Underpinning of genetic risk factors associated with hepatic steatosis and fibrosis in NAFLD is one of top research priorities in the field12, 13.
Hepatic steatosis is a key early event in the development of NAFLD whereas hepatic fibrosis is a later event that has prognostic significance in predicting long-term outcomes related to liver disease5, 10. Recent studies have suggested that there is a significant genetic association with presence of hepatic steatosis14–18. PNPLA-3 genotype has been linked to hepatic steatosis and also with features of NASH13, 19, 20. However, PNPLA-3 genotype explains 10–12% of the variance in the trait19. Therefore, 90% of variance in the trait remains to be elucidated. Although significant progress has been made in assessing genetic risk factors associated with hepatic steatosis there are limited human data in quantifying genetic risk factors associated with hepatic fibrosis in NAFLD. NAFLD is closely associated with metabolic traits21–24. However, heritability of NAFLD-associated hepatic steatosis in adults has not been systematically examined. Furthermore, there are no data whether hepatic fibrosis is a heritable trait. Liver biopsy will not be ethical in those without NAFLD, and assessment of twins with and without NAFLD and fibrosis would be needed to assess heritability of hepatic fibrosis. Therefore, accurate and precise non-invasive biomarkers were needed to document heritability of hepatic fibrosis.
Until recently, accurate and precise non-invasive quantification of hepatic fibrosis was not feasible, and therefore, heritability of hepatic fibrosis could not be examined. With the recent advances in magnetic resonance elastography (MRE), it has now become feasible to non-invasively assess hepatic fibrosis with increased accuracy and precision25–29.
Hence, utilizing a twin study design, we conducted a cross-sectional analysis of a prospective cohort study in community-dwelling monozygotic and dizygotic adult twins to examine the heritability of hepatic steatosis (as assessed by magnetic resonance imaging [MRI]) and hepatic fibrosis (as assessed by MRE).
METHODS
Setting and participants
This is a cross-sectional analysis of a prospective cohort study of twin-pairs residing in Southern California that was designed with the primary goal to study NAFLD. The cohort was derived from Newspaper advertisement and also access to twin-birth registry. Study participants were twin volunteers from urban Southern California (principally the San Diego area). All participants underwent a standardized clinical research visit including detailed medical history, past medical history, alcohol quantification using Skinner and Audit questionnaire, physical examination, and testing to rule out other causes of chronic liver diseases (see inclusion and exclusion criteria for further details), fasting laboratory tests (see biochemical and metabolic traits sub-section for further details), and then underwent an advanced MR examination of the liver between Jan 2012 and Jan 2015. Hepatic steatosis was quantified non-invasively by MRI-determined proton-density-fat-fraction (MRI-PDFF) and liver fibrosis by MRE-determined stiffness (MRE-stiffness) as previously published26, 30–33. Research visits and MRI procedures for each twin-pair were performed on the same day. Written informed consent was obtained from each participant, and the research protocol was approved by the UCSD institutional review board.
Inclusion and exclusion criteria
Inclusion criteria were as follows: Participants must be twins aged 18 years or older and willing and able to complete all research procedures and observations. Participants were fully informed and personally signed and dated the written Informed Consent and Health Insurance Portability and Accountability Act (HIPAA) provisions.
Exclusion criteria were as follows: Pregnancy or nursing at the time of study procedures; contraindications for MRI including severe claustrophobia, metal implants, or body circumference greater than the imaging chamber; use of steatogenic medications including amiodarone, methotrexate, glucocorticoids, L-asparaginase, and valproic acid for at least 3 months in the last 6 months; chronic diseases other than NAFLD that may be associated with hepatic steatosis including cystic fibrosis, human immunodeficiency virus, hepatitis C or B, Wilson’s disease, glycogen storage disease, lipodystrophy, celiac disease, or type 1 diabetes mellitus; significant alcohol consumption (defined as more than 10 g/day in females and more than 20 g/day in males, on average) for more than 3 consecutive months in the last 12 months or inability to reliably quantify alcohol consumption; prior bariatric surgery (eg, gastroplasty, roux-en-Y gastric bypass), low alpha-1-antitrypsin level and ZZ phenotype, dysbetalipoproteinemia, phenotypic hemochromatosis including presence of iron overload on MRI, polycystic liver disease, , clinical or laboratory evidence of systemic infectious disease, or clinical evidence of other causes of liver disease.
Definition of NAFLD
NAFLD was defined as presence of hepatic steatosis on MRI-PDFF ≥ 5% without any secondary causes of hepatic steatosis such as significant alcohol use or use of steatogenic medications or other causes of liver disease (please see exclusion criteria listed previously for further details); consistent with NAFLD practice guidelines1.
Assessment of twin-ship status
Detailed information regarding participants twin-ship status (mono- zygotic [MZ] or di-zygotic [DZ]) was obtained. Majority (34 twin-pairs) were diagnosed by a physician as either MZ or DZ by genetic testing. Participants were asked the following questions to further confirm twin-ship status by using the previously published and well-accepted questionnaire (Appendix III) developed by Boyd et al34.
Clinical research visit and laboratory tests
All participants underwent a uniform and standardized clinical research visit at the UCSD NAFLD Translational Research Unit. Participants underwent a detailed medical history, including history of liver disease and other co-morbid conditions, medication use, and alcohol consumption. The Alcohol Use Disorders Identification Test (AUDIT) questionnaire and Skinner Lifetime Drinking history were administered to record and quantify alcohol use. A physical exam including vital signs, height, weight and anthropometric measurements was performed by a trained investigator. Body mass index (BMI) was calculated by dividing body weight (in kilograms) by the square of the height (in meters). After completion of the above elements of the history and physical examination, participants had fasting laboratory work including complete blood count, screening etiologic tests (hepatitis B surface antigen, hepatitis C antibody, and iron panel including serum ferritin), clinical chemistry (creatinine, total protein, blood urea nitrogen (BUN), uric acid), hemoglobin A1c (HbA1c), hepatic panel (total bilirubin, direct bilirubin, aspartate aminotransferase (AST), alanine aminotransferase (ALT), alkaline phosphatase, gamma glutamyltransferase (GGT), albumin, prothrombin time (PT), and international normalized ratio (INR)), lipid profile and glucose-insulin levels.
Genotyping
Whole blood specimens collected during the research visit were utilized and DNA was extracted. Patatin-like phospholipase domain containing 3 genotyping was conducted and it’s association in explaining the variance in hepatic steatosis and hepatic fibrosis was examined. The genotyping was performed by Human Longevity Inc, San Diego, CA. Same-day clinical and MRI visits were performed with participants fasting. This allowed collection of fasting laboratory tests and reduced potential confounding factors on MRI results.
MRI protocol
MR imaging examinations were performed using a 3T research scanner (GE Signa EXCITE HDxt, GE Healthcare, Waukesha, WI) at the UCSD MR3T Research Laboratory. MRI was done in the supine position. Two MR techniques were performed: MRE to measure liver stiffness as a marker of fibrosis and an advanced MR fat quantification technique to measure the proton density fat fraction (MRI-PDFF) as marker of steatosis. The details of the MRI protocol are available in Appendix I as previously published35, 36.
Justification for not using liver biopsy for assessment of liver fat and fibrosis
Liver biopsy was not used for liver fat and fibrosis assessment because of following reasons: 1) It would be unethical to perform liver biopsy in normal participants who do not have a clinical indication of performing a liver biopsy1. Therefore, a non-invasive, and quantitative, method was needed to estimate liver fat and fibrosis37. 2) We have previously shown that MRI-PDFF is a more precise marker of liver fat than liver biopsy38. 3) Emerging data suggests that MRE-stiffness is the most accurate, currently available, non-invasive marker of liver fibrosis26, 29.
Rationale for using MRI-PDFF for hepatic steatosis quantification
Previously published studies from our group as well as others have shown that MRI-PDFF is an accurate, precise, and reproducible non- invasive biomarker for quantification of liver fat39. The variability of liver fat measurement by MRI- PDFF is <1% and it has robust correlation with MR spectroscopy (r2=0.99)31. It is superior to ultrasound and computed tomography for quantification of liver fat content40.
Rationale for using MRE-stiffness for hepatic fibrosis quantification
MRE-stiffness is better than available clinical prediction rules and ultrasound-based tests for quantification of hepatic fibrosis29, 41. Therefore, we utilized MRE to quantitatively assess hepatic fibrosis.
Primary outcome
There were two co-primary outcomes of interest including heritability of hepatic steatosis and heritability of hepatic fibrosis.
Statistical analysis
Data analyses were performed by a team led by an experienced statistical geneticist. In order to determine the relative influence of genetic and environmental factors on hepatic stetaosis, fibrosis and metabolic traits, we fit a univariate model to the data. We used a classical twin design of an AE model. The variance of each phenotype is decomposed into the proportion of total variance attributed to additive genetic (A) influences and unique environmental (E) influences42–44. Additive genetic influences can be estimated from twin data by contrasting the phenotypic correlation between MZ twins who generally share 100% of their genes, and DZ twins who on average share 50% of their segregating genes. Shared environment is assumed to induce a correlation of equal magnitude between both types of a twin pair. Unique environmental influences are assumed to be uncorrelated between the members of a twin pair. Measurement error is also included in the E term because it is also assumed to be uncorrelated between twins. A variance component model incorporating these assumptions can be used to estimate variance components capturing both the A and E terms42–44.
The proportion of a phenotype’s total variance attributable to additive genetic influences is considered the heritability (H) of the phenotype (H=A/(A+E)). The significance of genetic influences was tested by fixing the A parameter to zero, and then comparing the fit of the reduced model against the full model. Model comparisons were performed using the likelihood-ratio chi-square test (LRT), calculated as the difference in the -2 log likelihood (-2LL) of the reduced model from that of the full model. Significant LRT values indicate a significant change in model fit relative to the comparison model.
We tested the influence of PNPLA3 gene variant on MRI PDFF and MRE Stiffness phenotypes using the 'AE' twin models by treating PNPLA3 genotype as a covariate with additive genotype codings: CC=0.0, CG=0.5 and GG=1.0. We compared models with and without the PNPLA3 genotype included to obtain an estimate of the fraction of variation explained by the PNPLA3 genotype.
Sample-size estimation
Previous studies have suggested that the heritability of hepatic steatosis could range from 0.37 (derived from studies using ultrasound and serum ALT to assess hepatic steatosis) to almost 1.0 (derived from study using MRI to assess hepatic steatosis in obese Hispanic probands and their families 15, 18). We therefore assumed that the heritability of hepatic steatosis in our sample would be in the range of ~0.5. Using the classical ACE model, Visscher45 has shown that the number of twin pairs needed to detect an additive genetic (i.e., heritable) component in an ACE model of between 0.4 and 0.8 would require ~36–74 twin pairs depending on how many MZ twins were included in the sample at a power level of 0.95 and a type I error of 0.05. We were thus confident that our target of ~50 pairs (53 recruited and 48 used in the analysis) would be adequate to detect an appropriate heritability in this cohort.
RESULTS
Baseline characteristics
One hundred and forty two twins (71 pairs) underwent a detailed clinical research visit, physical exam, fasting biochemical assessment and advanced MRI and MRE examination, and 120 (42 MZ twin-pairs and 18 DZ twin-pairs) with paired clinical and MR data were included in the present study (derivation of cohort is described in Appendix 2). The average (± standard deviation [sd]) age and BMI was 45.7 (±22.1) years and 26.4 (± 5.7) Kg/m2, respectively. The detailed demographic, biochemical, and imaging profile of the entire cohort, stratified by presence (or absence) of NAFLD, is presented in Table 1. The prevalence of NAFLD in this twin cohort was 21.7% (26/120). The mean (± sd) of MRE-derived liver stiffness between the NAFLD versus (vs.) non-NAFLD group was 3.0 (± 1.23) vs. 2.1 (± .42) Kpa, and the median (interquartile range) of MRE-derived liver stiffness between the NAFLD versus non-NAFLD group was 2.5 (0.89) vs. 2.1 (0.64) Kpa, respectively.
Table 1.
Baseline characteristics stratified by NAFLD status in the twin cohort
| Overall | Twins with NAFLD (MRI-PDFF ≥ 5%) | Twins without NAFLD (MRI-PDFF < 5%) | p-value | |
|---|---|---|---|---|
| N | 120 | 26 | 94 | |
| Demographics | ||||
| Age (years) | 45.7 (22.1) | 54.9 (17.3) | 43.2 (22.7) | .0163 |
| Sex (% male) | 33 (27.5%) | 11 (42.3%) | 22 (23.4%) | .0561 |
| Race | .0858 | |||
| White | 94 (78.3%) | 18 (69.2%) | 76 (80.9%) | |
| Hispanic | 18 (15.0%) | 5 (19.2%) | 13 (13.8%) | |
| Asian | 6 (5.0%) | 1 (3.9%) | 5 (5.3%) | |
| Hawaiian/Pac Islander | 2 (1.7%) | 2 (7.7%) | 0 (0%) | |
| Physical | ||||
| Height (cm) | 165.5 (8.3) | 167.0 (10.7) | 165.1 (7.5) | .4187 |
| Weight (kg) | 72.5 (18.0) | 88.8 (20.5) | 68.0 (14.3) | <.0001 |
| Body mass index (kg/m2) | 26.4 (5.7) | 31.5 (4.8) | 24.9 (5.1) | <.0001 |
| Systolic blood pressure (mm Hg) | 125.8 (19.7) | 135.5 (16.7) | 123.1 (19.6) | .0038 |
| Diastolic blood pressure (mm Hg) | 78.9 (12.4) | 82.9 (13.0) | 77.8 (12.1) | .0603 |
| Waist circumference (cm) | 89.5 (12.8) | 100.0 (10.6) | 86.5 (11.8) | <.0001 |
| Hip circumference (cm) | 100.2 (11.4) | 107.5 (11.6) | 98.1 (10.5) | .0001 |
| Laboratory data | ||||
| Glucose (mg/dL) | 89.7 (19.1) | 101.6 (34.3) | 86.4 (9.7) | .0023 |
| Insulin (U/L) | 8.4(5.5) | 12.6 (7.1) | 7.2(4.4) | .0006 |
| Hemoglobin A1c | 5.8 (0.5) | 6.1(0.7) | 5.7(0.3) | .0017 |
| HOMA-IR | 1.9 (1.4) | 3.1 (1.9) | 1.6 (1.1) | .0002 |
| AST (U/L) | 23.4 (9.9) | 27.0 (16.6) | 22.4 (6.7) | .1459 |
| ALT (U/L) | 23.1 (19.8) | 35.0 (32.8) | 19.8 (10.4) | .0012 |
| Alkaline Phosphatase (U/L) | 68.8 (19.4) | 69.7 (15.1) | 68.5 (21.0) | .5998 |
| Total bilirubin (mg/dL) | 0.5(0.3) | 0.5(0.4) | 0.5(0.2) | .7444 |
| Direct bilirubin (mg/dL) | 0.1(0.1) | 0.1(0.1) | 0.1(0.1) | .1112 |
| Albumin (g/dL) | 4.5 (0.3) | 4.5(0.2) | 4.5(0.4) | .3529 |
| GGT (U/L) | 23.8 (20.4) | 33.1 (29.6) | 21.2 (16.2) | .0058 |
| Total cholesterol (mg/dL) | 194.3 (40.9) | 200.6 (36.1) | 192.5 (42.2) | .2190 |
| HDL-cholesterol (mg/dL) | 65.3 (21.8) | 51.0 (15.9) | 69.3 (21.6) | <.0001 |
| LDL-cholesterol (mg/dL) | 110.3 (34.8) | 119.8 (30.7) | 107.5 (35.6) | .0308 |
| Trigylcerides (mg/dL) | 94.1 (55.1) | 150.7 (71.1) | 78.0 (36.4) | <.0001 |
| White blood cell count (x103/uL) | 5.8 (1.3) | 6.4 (1.5) | 5.6 (1.2) | .0126 |
| Hemoglobin (g/dL) | 13.7 (1.2) | 14.0 (1.4) | 13.6 (1.2) | .1287 |
| Hematocrit (%) | 40.7 (3.3) | 41.5 (3.8) | 40.5 (3.2) | .1020 |
| Platelet count (x103/uL) | 251.7 (51.2) | 253.0 (57.7) | 251.4 (49.6) | .9123 |
| INR | 1.1 (0.3) | 1.1 (0.4) | 1.1 (0.3) | .4965 |
| Ferritin (ng/mL) | 101.9 (93.9) | 134.4 (144.4) | 92.6 (72.0) | .1702 |
| Imaging data | ||||
| MRI-PDFF (%) | 4.156 (4.25) | 10.740 (5.08) | 2.334 (0.85) | <.0001 |
| MRE (Kpa) | 2.339 (0.77) | 3.004 (1.23) | 2.148 (0.42) | <.0001 |
| PNPLA3 phenotype (N=87) | 0.8360 | |||
| CC | 39 (44.8%) | 9 (45.0%) | 30 (47.8%) | |
| CG | 38 (43.7%) | 8 (40.0%) | 30 (44.8%) | |
| GG | 10 (11.5%) | 3 (15.0%) | 7 (10.4%) | |
Footnote: Mean value provided with standard deviation in parenthesis, unless otherwise noted as N(%). Differences between participants with and without NAFLD were evaluated with t tests or Wilcoxon Mann–Whitney for continuous variables and χ2 or Fishers exact tests for categorical variables.
Abbreviations for table: NAFLD, non-alcoholic fatty liver disease; HOMA-IR, homeostatic model of insulin resistance; AST, aspartate aminotransferase; ALT, alanine aminotransferase; GGT, gamma- glutamyl transpeptidase; LDL, low-density lipoprotein; HDL, high-density lipoprotein; INR, international normalized ratio; MRI, magnetic resonance imaging; PDFF, proton-density-fat-fraction.
Significant p-values <0.05
Twin-ship correlation by hepatic steatosis
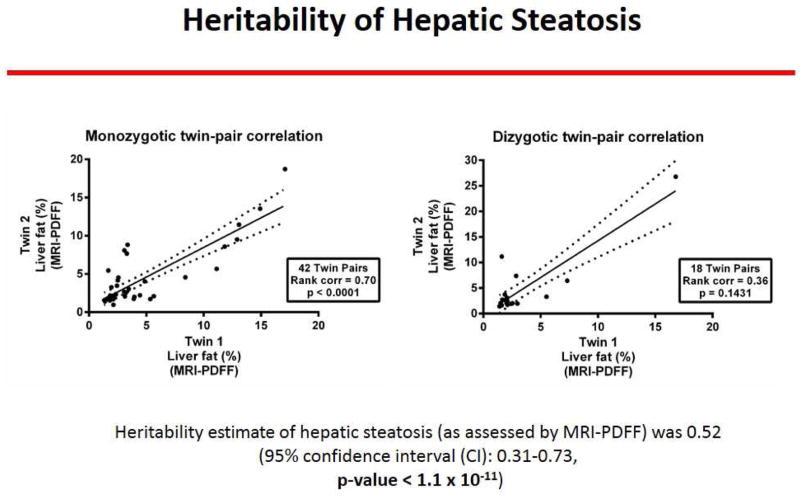
The MZ twin-pairs showed a robust correlation in hepatic steatosis as quantified by MRI-PDFF (r2 of 0.70, p- value <0.0001) but not the di-zygotic twin-pairs (r2 of 0.36, p-value .2) as shown in figure 1.
Figure 1. Twin-ship correlation by hepatic steatosis assessed by MRI.
The mono-zygotic twin-pairs showed a robust correlation in hepatic steatosis as quantified by MRI-PDFF (r2 of 0.70, p-value <0.0001) but not the di-zygotic twin-pairs (r2 of 0.36, p-value .2); demonstrating that hepatic steatosis is a heritable trait.
Twin-ship correlation by hepatic fibrosis
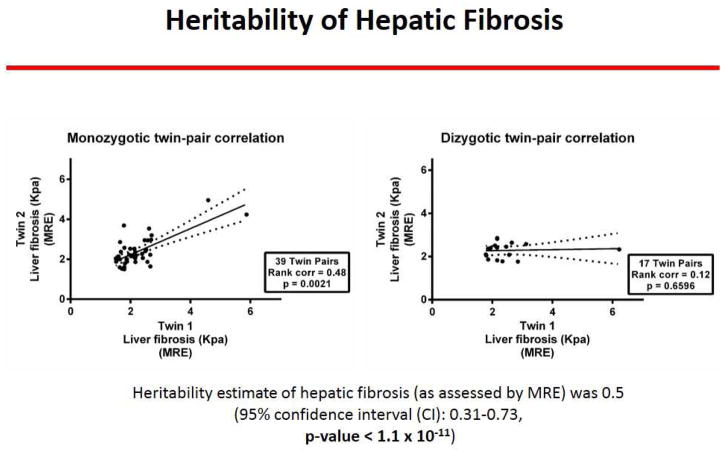
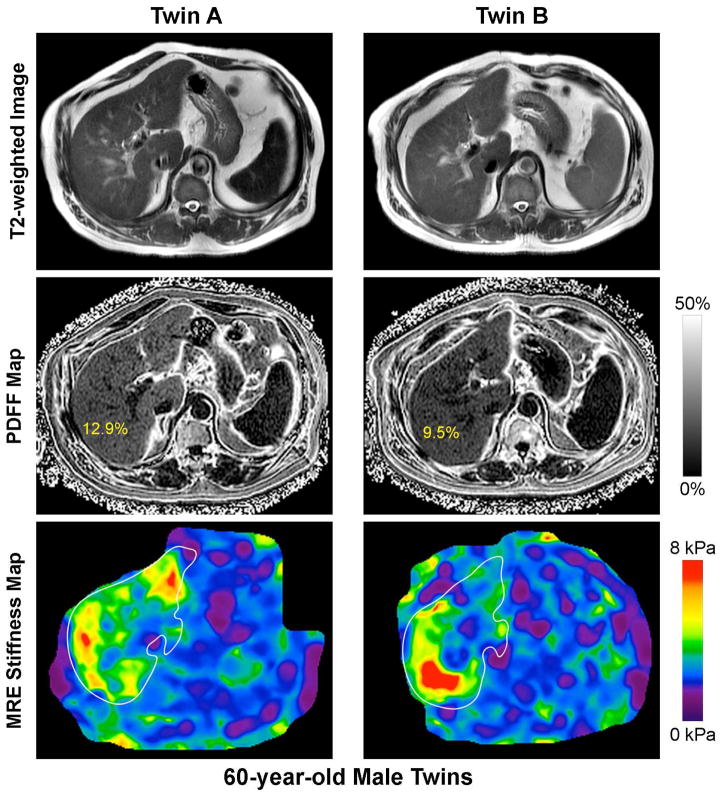
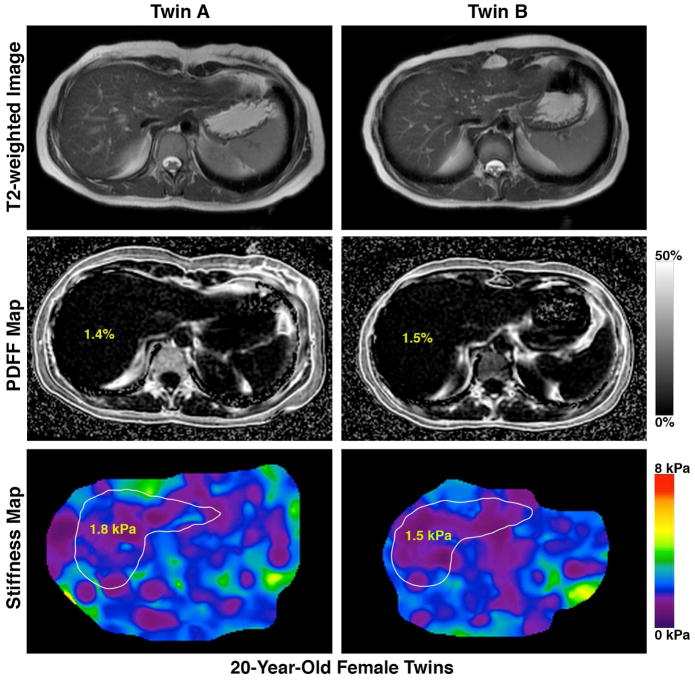
Similar to the twin-ship correlation for hepatic steatosis, the MZ twin-pairs showed a robust correlation in liver fibrosis as quantified by MRE-stiffness (r2 of 0.48, p-value <0.002) but not the di-zygotic twin-pairs (r2 of .12, p-value .7), as shown in figure 2. We show an example each of a twin-pair that is concordant for presence of NAFLD and advanced fibrosis (figure 3A), a twin-pair that is concordant for the absence of NAFLD (figure 3B), and a twin-pair that is discordant for NAFLD (figure 3C).
Figure 2. Twin-ship correlation by hepatic fibrosis assessed by MRE.
The mono-zygotic twin-pairs showed a robust correlation in liver fibrosis as quantified by MRE-stiffness (r2 of 0.48, p-value <0.002) but not the di-zygotic twin-pairs (r2 of .12, p-value .7); demonstrating that hepatic fibrosis is a heritable trait.
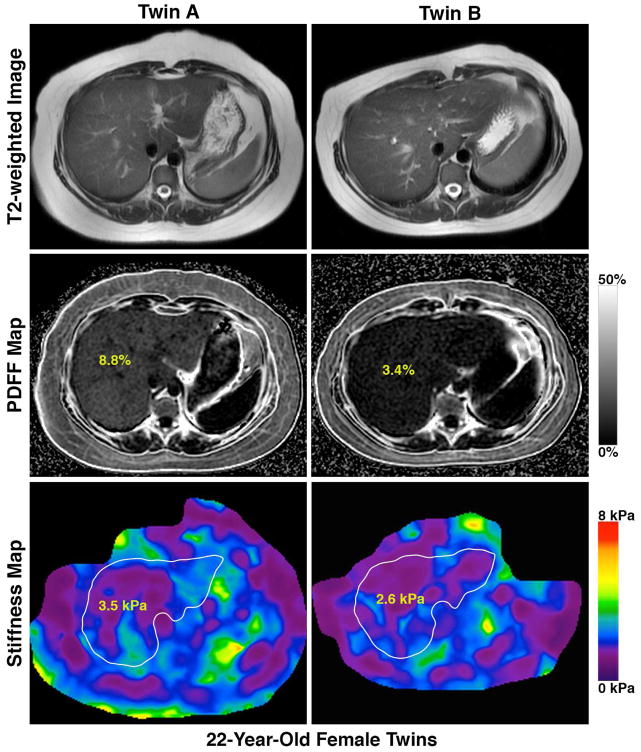
Figure 3. Novel MRI-PDFF map and MRE-map demonstrating the detailed phenotyping of the twins based upon the presence (or absence) of hepatic steatosis.
Footnote: A twin-pair that is concordant for presence of NAFLD and advanced fibrosis (figure 3A), a twin-pair that is concordant for the absence of NAFLD (figure 3B), and a twin-pair that is discordant for NAFLD (figure 3C)
Heritability of hepatic steatosis and hepatic fibrosis
The heritability estimates of metabolic traits and hepatic steatosis and hepatic fibrosis are provided in Table 2. The heritability (95% confidence interval) of hepatic steatosis was 0.87 (95% CI: 0.80–0.93), which was statistically and clinically significant with a p-value < 2.2 x 10−11. In multivariable-adjusted models after adjustment for age, sex and ethnicity, the results remained statistically significant with an h2 of 0.52 (95% CI: 0.31–0.73), which was statistically and clinically significant with a p-value of 1.1 x 10−11. The heritability (95% confidence interval) of hepatic fibrosis was 0.67 (95% CI: 0.52–0.83), which was statistically significant with a p-value <2.2 x 10−16. In multivariable-adjusted models after adjustment for age, sex and ethnicity, the results remained statistically significant with a h2 of 0.50 (95% CI: 0.28–0.72) with a p-value of 6.1 x 10−11.
Table 2.
Heritability estimates in the twins: Unadjusted, age-sex adjusted and age-sex-ethnicity adjusted models
| Proportions of phenotypic variance explained | |||
|---|---|---|---|
|
| |||
| Un-adjusted | Age-sex adjusted | Age-sex-ethnicity adjusted | |
| h2 | h2 | h2 | |
| Height (cm) | 0.93 | 0.90 | 0.89 |
| Weight (kg) | 0.92 | 0.51 | 0.91 |
| BMI (kg/m2) | 0.85 | 0.84 | 0.67 |
| Waist (cm) | 0.77 | 0.74 | 0.74 |
| Hip (cm) | 0.81 | 0.59 | 0.50 |
| Systolic BP (mmHg) | 0.55 | 0.52 | 0.52 |
| Diastolic BP (mmHg) | 0.44 | 0.42 | 0.41 |
| Glucose (mg/dL) | 0.41 | 0.31 | 0.29 |
| Albumin (g/dL) | 0.49 | 0.49 | 0.45 |
| Total Bilirubin (mg/dL) | 0.73 | 0.62 | 0.71 |
| AST (U/L) | 0.68 | 0.55 | 0.54 |
| ALT (U/L) | 0.75 | 0.72 | 0.50 |
| ALP (U/L) | 0.71 | 0.50 | 0.50 |
| Cholesterol (mg/dL) | 0.51 | 0.42 | 0.35 |
| HDL (mg/dL) | 0.81 | 0.75 | 0.73 |
| LDL (mg/dL) | 0.42 | 0.40 | 0.37 |
| Triglycerides (mg/dL) | 0.68 | 0.51 | 0.65 |
| Ferritin (ng/dL) | 0.18 | 0.49 | 0.50 |
| GGT (U/L) | 0.58 | 0.49 | 0.50 |
| Hemoglobin A1c | 0.48 | 0.37 | 0.26 |
| WBC count (x103/μL) | 0.64 | 0.50 | 0.63 |
| Hemoglobin (g/dL) | 0.66 | 0.50 | 0.52 |
| Hematocrit (%) | 0.64 | 0.50 | 0.46 |
| Platelet count (x103/μL) | 0.65 | 0.63 | 0.50 |
| INR | 0.55 | 0.50 | 0.49 |
| HOMA-IR | 0.63 | 0.46 | 0.42 |
| Insulin (U/L) | 0.71 | 0.61 | 0.55 |
| MRI-PDFF (%) | 0.87 | 0.71 | 0.52 |
| MRE Stiffness (kPa) | 0.67 | 0.51 | 0.50 |
Abbreviations for table: h2, heritability estimate; BP, blood pressure; HOMA-IR, homeostatic model of insulin resistance; AST, aspartate aminotransferase; ALT, alanine aminotransferase; ALP, alkaline phosphatase; GGT, gamma-glutamyl transpeptidase; LDL, low-density lipoprotein; HDL, high-density lipoprotein; WBC, white blood cell; INR, international normalized ratio; MRI, magnetic resonance imaging; PDFF, proton-density-fat-fraction; MRE, magnetic resonance elastography
Finally, heritability of hepatic steatosis and hepatic fibrosis models were compared with and without the PNPLA3 genotype included to obtain an estimate of the fraction of variation explained by the PNPLA3 genotype (Table 3). Models that included PNPLA3 genotype suggested that the genotype was not a statistically significant predictor of MRI PDFF or MRE Stiffness. As a result, the percentage of variation explained by the PNPLA3 genotype was effectively zero. This does not represent a true estimate but rather suggests that the study was underpowered to detect an effect of genotype on the trait.
Table 3.
Heritability of hepatic steatosis and hepatic fibrosis un-adjusted and multivariable-adjusted models and by PNPLA-3 genotype status.
| Hepatic steatosis (MRI-PDFF ) | |||
|---|---|---|---|
| Heritability | 95% confidence interval | p-value | |
| Unadjusted | 0.87 | 0.80–0.93 | <2.2 x 10−16 |
| Sex + age | 0.71 | 0.57–0.85 | <2.2 x 10−16 |
| Sex + age + race | 0.52 | 0.31–0.73 | 1.1 x 10−11 |
| SNP % Var | SNP p-value | ||
| PNPLA3 genotype (SNP) | 2.0% | 0.81 | |
| Sex + age + SNP | 21.0% | 0.78 | |
| Sex + age + race + SNP | 0.03% | 0.98 | |
| Hepatic fibrosis (MRE Stiffness) | |||
|---|---|---|---|
| Heritability | 95% confidence interval | p-value | |
| Unadjusted | 0.67 | 0.52–0.83 | <2.2 x 10−16 |
| Sex + age | 0.51 | 0.30–0.73 | 1.8 x 10−11 |
| Sex + age + race | 0.50 | 0.28–0.72 | 6.1 x 10−11 |
| SNP % Var | SNP p-value | ||
| PNPLA3 genotype (SNP) | 1.0% | 0.43 | |
| Sex + age + SNP | 22.0% | 0.63 | |
| Sex + age + race + SNP | 0.0% | 0.84 | |
We adjusted the models with PNPLA-3 genotype status to examine the percentage of the each trait is explained by the PNPLA-3 genotype and whether it is a significant association. Addition of PNPLA-3 genotype did not reveal significant association. Hence the percentage contribution of PNPLA-3 genotype on heritability of liver fat and liver fibrosis could be documented. The study was underpowered (n=89) to detect the genotype on trait effect.
Abbreviations: MRI-PDFF, magnetic resonance imaging- proton density fat fraction; MRE, magnetic resonance elastography; PNPLA-3, patatin-like phospholipase domain containing 3
DISCUSSION
Main findings
Utilizing a well-phenotyped, prospectively assessed, cohort of community-dwelling twins, this study provides evidence that both hepatic steatosis and hepatic fibrosis are heritable traits. Previous studies have provided some evidence on heritability of hepatic steatosis; the demonstration of heritability of hepatic fibrosis is a novel finding and has not been previously documented. These data have widespread implications for developing targeted approaches for hepatic fibrosis as genetic as well as epigenetic therapeutic targets may be exploited in the treatment of NASH related fibrosis.
In context with previously published literature
Previous studies have suggested that heritability of hepatic steatosis ranges from 0 (no heritability) to 1 (100% heritable). Utilizing data from 331 twins derived from a population-based cohort of 4929 individuals, Makkoken et al. demonstrated that approximately 60% of the variation in serum ALT is genetically determined14. They verified the association between serum ALT and hepatic steatosis by cross-validating the serum ALT with hepatic steatosis assessment by MR spectroscopy in 66 individuals14. Tarnoki et al. evaluated the heritability of NAFLD in 208 Hungarian twins but found that it was not heritable45. However, hepatic steatosis assessment was performed using ultrasonography, which lacks sensitivity and accuracy and especially fails to detect liver fat when it is between 5%–20%. Using MRI-PDFF to assess hepatic steatosis, Schwimmer et al. found that the heritability of hepatic steatosis approached 100%15. However, this study was conducted in overweight children and their family members who were predominantly of Hispanic ethnicity. While the authors concluded that NAFLD was highly heritable trait, their study population may have caused the heritability to be overestimated. Wagenknecht et al. examined NAFLD heritability in 794 Hispanic American and 347 African American adults, concluding that NAFLD was modestly heritable17; the use of computerized tomography in this study limits hepatic steatosis quantification 47, and the generalizability of the study is limited to that of Hispanic and African Americans. Finally, Brouwers et al. investigated the heritability of fatty liver—as measured with ultrasonography and serum ALT—in those with familial combined hyperlipidemia18, revealing a 20–36% heritability of NAFLD in this genetic background. Despite these seminal observations on the heritability of NAFLD, the heritability of NAFLD remained uncertain due to the aforementioned study limitations, therefore, we conducted this study in adult, community-dwelling twins using MRI to accurately assess and quantify hepatic steatosis.
None of the prior studies examined the heritability of hepatic fibrosis. Furthermore, this study confirmed that hepatic steatosis is closely linked to metabolic traits. Our study found that hepatic fibrosis had robust correlation in MZ-twins but not in DZ-twins. It fits well with previous research that genetic risk factors and both epigenetic and genetic factors may be linked to disease progression in NAFLD33, 46, 47. We recently showed that serum microRNA (miR) profiling can explain discordancy between MZ-twins with and without hepatic steatosis. In addition, we showed that miR may themselves be heritable33. It also confirms the prior observation that higher insulin resistance and diabetes is associated with advanced NAFLD9, 48–50.
Strengths and limitations
The strengths of this study are several: 1. Well-characterized cohort with detailed and comprehensive quantification of hepatic steatosis by MRI-PDFF and detailed fat mapping of the liver. 2. Comprehensive quantification of hepatic fibrosis MRE-stiffness. Among all non-invasive methods to assess liver fibrosis, MRE has the highest accuracy for the diagnosis of advanced fibrosis and cirrhosis 28. Liver biopsy assessment is unethical in normal controls without NAFLD and assessment of heritability in twins can only be accomplished in studies that include both affected and unaffected twins. Therefore, current study design and study aims could only be accomplished using non-invasive tests that are accurate and robust to detect and diagnose both hepatic steatosis and hepatic fibrosis. The utilization of advanced MRI and MRE and their application was both required as well as adds to the novelty of the approach. 3. Twin-study design, which allowed us to examine the heritability of hepatic steatosis and fibrosis and their association with metabolic traits. 4. Presence of other causes of hepatic steatosis such as excess alcohol use and medications and viral hepatitis were excluded. 5. Documentation of heritability of hepatic fibrosis for the first time in a well-characterized cohort.
Limitations of the study include that liver biopsy could not be used to document features of NAFLD that currently cannot be assessed by a non-invasive test including inflammation, ballooning, fat droplet size, steatosis zonality, and presence of NASH. However, we and others have shown that MRI-PDFF is an accurate, repeatable and reproducible biomarker for diagnosis and quantification of liver fat, and may even be better than a liver biopsy assessment for assessing quantity of liver fat. Similarly, recent studies from our group as well as others have shown that MRE is an accurate, repeatable and reproducible biomarker for diagnosis and quantification of hepatic fibrosis. It is now considered the best non-invasive modality to quantify and detect fibrosis in NAFLD. Liver biopsy examination would be unethical in this study as majority of individuals would not have an indication for a liver biopsy. Therefore, the study took leverage from the innovative application of MRI-PDFF and MRE to tease out the heritability of hepatic steatosis and fibrosis non-invasively. With the advent of these advanced MR techniques, it now became feasible to examine and document the heritability of hepatic fibrosis which was hitherto unknown. PNPLA3 genotype was not a statistically significant predictor of MRI PDFF or MRE Stiffness. We acknowledge, however, that we may have been underpowered to detect an effect of genotype on the trait. Larger studies are needed to explore the effect of genes in explaining the heritability of hepatic steatosis and hepatic fibrosis.
CONCLUSIONS
We conclude that both hepatic steatosis and hepatic fibrosis in NAFLD are heritable. Both hepatic steatosis and hepatic fibrosis are highly correlated in MZ-twins but not in DZ-twins. These data have widespread implications for developing targeted approaches for hepatic fibrosis, as it is plausible that common mechanisms underlying hepatic fibrosis are genetically or even epigenetically mediated and targeting those my help treat hepatic fibrosis in NAFLD. Further studies are needed to detect specific genes and genetic pathways that may be responsible for genetic susceptibility of hepatic fibrosis, and transition from non-fibrotic NAFLD to fibrotic NAFLD.
Acknowledgments
The manuscript is dedicated in memory of the late Dr. Daniel O’Connor who inspired RL to initiate a twin cohort to study NAFLD.
Funding support: The study was conducted at the Clinical and Translational Research Institute, University of California at San Diego. RL is supported in part by the American Gastroenterological Association (AGA) Foundation – Sucampo – ASP Designated Research Award in Geriatric Gastroenterology and by a T. Franklin Williams Scholarship Award; Funding provided by: Atlantic Philanthropies, Inc, the John A. Hartford Foundation, the Association of Specialty Professors, and the American Gastroenterological Association and grant K23-DK090303. The authors would like to acknowledge Human Longevity, Inc. for performing the genomic sequencing.
Abbreviations and key words
- NAFLD
Nonalcoholic fatty liver disease
- NASH
Nonalcoholic Steatohepatitis
- NASH- CRN
Nonalcoholic Steatohepatitis Clinical Research Network
- MRI
Magnetic Resonance Imaging
- PDFF
proton density fat fraction
Appendix I MRI Protocol
MRE was performed as previously described 1–4 using commercially available software and hardware (Resoundant Inc., Rochester, MN). Briefly, an acoustic passive driver is secured with an elastic band over the body wall anterior to the liver and connected by a flexible plastic tube to an acoustic active driver outside the MRI room. Continuous vibrations at 60 Hz are generated by the active driver and delivered by the tube to the passive driver, which then transmits the vibrations into the body, thereby producing shear waves in the liver. A 2D gradient-recalled-echo MRE pulse sequence is performed while the vibrations are transmitted, and four non-contiguous axial slices (10-mm thick, 10-mm interslice gap) are acquired in a 16-second breathhold through the widest transverse dimension of the liver. Acquisition parameters include repetition time 50 ms, echo time 20.2 ms, flip angle 30°, matrix 256x64, field of view 48 x 48 cm, one signal average, receiver bandwidth ± 33 kHz (confirm), parallel imaging acceleration factor 2. By utilizing oscillating motion-sensitizing gradients that encode tissue motion into the phase of the MR signal, this sequence generates images (called wave images) that depict the shear waves within the liver. The sequence is repeated a total of four times, adjusting the phase relationship (phase offset) between the vibrations and the oscillating motion-sensitizing gradients, thereby producing, at each slice location, wave images at four evenly spaced time points over the wave cycle. Total acquisition time (four 16-second breathholds with short recovery in between) is about two minutes.
The wave images at each slice location then are processed automatically on the scanner computer using specialized software (called an inversion algorithm) to generate quantitative cross-sectional maps (called elastograms) depicting the stiffness of tissue. Four elastograms are generated, one at each of the four slice locations. These maps display stiffness with a color scale in units of kilopascals (kPa).
The elastograms were transferred offline for analysi5, 6. A trained image analyst (six months experience with MRE) in the MR3T research laboratory manually drew regions of interest (ROI) on the elastograms using a custom software package. ROIs were drawn at each of the four slice locations in portions of the liver in which the corresponding wave images showed clearly observable wave propagation, avoiding liver edges, large blood vessels, and artifacts. The mean liver stiffness was calculated by averaging the per-pixel stiffness values across the ROIs at the four slice locations, and the results were outputted automatically to an electronic spreadsheet.
References
- 1.Yin M, Talwalkar JA, Glaser KJ, et al. Assessment of hepatic fibrosis with magnetic resonance elastography. Clin Gastroenterol Hepatol. 2007;5:1207–1213. e2. doi: 10.1016/j.cgh.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J, Talwalkar JA, Yin M, et al. Early detection of nonalcoholic steatohepatitis in patients with nonalcoholic fatty liver disease by using MR elastography. Radiology. 2011;259:749–56. doi: 10.1148/radiol.11101942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim D, Kim WR, Talwalkar JA, et al. Advanced Fibrosis in Nonalcoholic Fatty Liver Disease: Noninvasive Assessment with MR Elastography. Radiology. 2013 doi: 10.1148/radiol.13121193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venkatesh SK, Yin M, Ehman RL. Magnetic resonance elastography of liver: technique, analysis, and clinical applications. J Magn Reson Imaging. 2013;37:544–55. doi: 10.1002/jmri.23731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noureddin M, Lam J, Peterson MR, et al. Utility of magnetic resonance imaging versus histology for quantifying changes in liver fat in nonalcoholic fatty liver disease trials. Hepatology. 2013;58:1930–40. doi: 10.1002/hep.26455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Permutt Z, Le TA, Peterson MR, et al. Correlation between liver histology and novel magnetic resonance imaging in adult patients with non-alcoholic fatty liver disease - MRI accurately quantifies hepatic steatosis in NAFLD. Aliment Pharmacol Ther. 2012;36:22–9. doi: 10.1111/j.1365-2036.2012.05121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
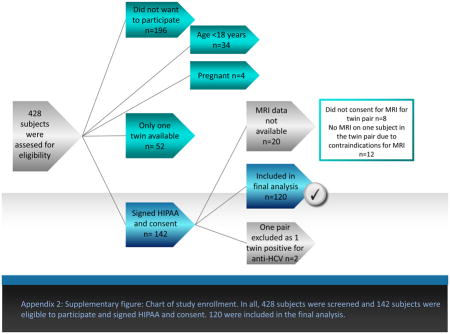
Appendix 2: Supplementary figure: Chart of study enrollment. In all, 428 subjects were screened and 142 subjects were eligible to participate and signed HIPAA and consent. 120 were included in the final analysis
Appendix III Questions used to determine zygosity
-
Were you and your twin “as alike as two peas in a pod”?
As alike as two peas in a pod
Usual sibling similarity Quite different
-
Were you and your twin mixed up as children?
Yes, very often
Now and then Never
-
In that case, by whom were you mixed up?
Parents
Teachers
Others Nobody
Footnotes
Potential competing interests: none
Role of study sponsor: The study sponsor(s) had no role in the study design, collection, analysis, interpretation of the data, and/or drafting of the manuscript.
All authors report that no conflicts of interest exist.
Author contributions:
Rohit Loomba- study concept and design, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript, obtained funding, study supervision, approved final submission
Nicholas Schork, Chi-Hua Chen- Analysis and interpretation of data, statistical analysis, critical revision of the manuscript, approved final submission
David A. Brenner, Ana Bhatt, Brandon Ang, Phirum Nguyen, Carolyn Hernandez, Lisa Richards, Joanie Salotti, Steven Lin, Ekihiro Seki, Karen E Nelson - critical revision of the manuscript, approved final submission
Claude Sirlin – MR imaging, MRI analyses, critical revision of the manuscript, approved final submission
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chalasani N, Younossi Z, Lavine JE, et al. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American Gastroenterological Association, American Association for the Study of Liver Diseases, and American College of Gastroenterology. Gastroenterology. 2012;142:1592–609. doi: 10.1053/j.gastro.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Ratziu V, Bellentani S, Cortez-Pinto H, et al. A position statement on NAFLD/NASH based on the EASL 2009 special conference. J Hepatol. 2010;53:372–84. doi: 10.1016/j.jhep.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Loomba R, Sanyal AJ. The global NAFLD epidemic. Nat Rev Gastroenterol Hepatol. 2013;10:686–90. doi: 10.1038/nrgastro.2013.171. [DOI] [PubMed] [Google Scholar]
- 4.Williamson RM, Price JF, Glancy S, et al. Prevalence of and risk factors for hepatic steatosis and nonalcoholic Fatty liver disease in people with type 2 diabetes: the Edinburgh Type 2 Diabetes Study. Diabetes Care. 2011;34:1139–44. doi: 10.2337/dc10-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh S, Allen AM, Wang Z, et al. Fibrosis progression in nonalcoholic fatty liver vs nonalcoholic steatohepatitis: a systematic review and meta-analysis of paired-biopsy studies. Clin Gastroenterol Hepatol. 2015;13:643–54. e1–9. doi: 10.1016/j.cgh.2014.04.014. quiz e39–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ekstedt M, Hagstrom H, Nasr P, et al. Fibrosis stage is the strongest predictor for disease-specific mortality in NAFLD after up to 33 years of follow-up. Hepatology. 2015;61:1547–54. doi: 10.1002/hep.27368. [DOI] [PubMed] [Google Scholar]
- 7.Adams LA, Lymp JF, St Sauver J, et al. The natural history of nonalcoholic fatty liver disease: a population-based cohort study. Gastroenterology. 2005;129:113–21. doi: 10.1053/j.gastro.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 8.Angulo P, Bugianesi E, Bjornsson ES, et al. Simple noninvasive systems predict long-term outcomes of patients with nonalcoholic fatty liver disease. Gastroenterology. 2013;145:782–9. e4. doi: 10.1053/j.gastro.2013.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhala N, Angulo P, van der Poorten D, et al. The natural history of nonalcoholic fatty liver disease with advanced fibrosis or cirrhosis: an international collaborative study. Hepatology. 2011;54:1208–16. doi: 10.1002/hep.24491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angulo P, Kleiner DE, Dam-Larsen S, et al. Liver Fibrosis, but No Other Histologic Features, Associates With Long-Term Outcomes of Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology. 2015 doi: 10.1053/j.gastro.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loomba R, Chalasani N. The Hierarchical Model of NAFLD: Prognostic Significance of Histologic Features in NASH. Gastroenterology. 2015 doi: 10.1053/j.gastro.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 12.Friedman SL. Liver fibrosis in 2012: Convergent pathways that cause hepatic fibrosis in NASH. Nat Rev Gastroenterol Hepatol. 2013;10:71–2. doi: 10.1038/nrgastro.2012.256. [DOI] [PubMed] [Google Scholar]
- 13.Speliotes EK, Butler JL, Palmer CD, et al. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology. 2010;52:904–12. doi: 10.1002/hep.23768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makkonen J, Pietilainen KH, Rissanen A, et al. Genetic factors contribute to variation in serum alanine aminotransferase activity independent of obesity and alcohol: a study in monozygotic and dizygotic twins. J Hepatol. 2009;50:1035–42. doi: 10.1016/j.jhep.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 15.Schwimmer JB, Celedon MA, Lavine JE, et al. Heritability of nonalcoholic fatty liver disease. Gastroenterology. 2009;136:1585–92. doi: 10.1053/j.gastro.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roh YS, Loomba R, Seki E. The TM6SF2 variants, novel genetic predictors for nonalcoholic steatohepatitis. Gastroenterology. 2015;148:252–4. doi: 10.1053/j.gastro.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagenknecht LE, Scherzinger AL, Stamm ER, et al. Correlates and heritability of nonalcoholic fatty liver disease in a minority cohort. Obesity (Silver Spring) 2009;17:1240–6. doi: 10.1038/oby.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brouwers MC, Cantor RM, Kono N, et al. Heritability and genetic loci of fatty liver in familial combined hyperlipidemia. J Lipid Res. 2006;47:2799–807. doi: 10.1194/jlr.M600312-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–5. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rotman Y, Koh C, Zmuda JM, et al. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology. 2010;52:894–903. doi: 10.1002/hep.23759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loomba R, Abraham M, Unalp A, et al. Association between diabetes, family history of diabetes, and risk of nonalcoholic steatohepatitis and fibrosis. Hepatology. 2012;56:943–51. doi: 10.1002/hep.25772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loomba R, Rao F, Zhang L, et al. Genetic covariance between gamma-glutamyl transpeptidase and fatty liver risk factors: role of beta2-adrenergic receptor genetic variation in twins. Gastroenterology. 2010;139:836–45. 845 e1. doi: 10.1053/j.gastro.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neuschwander-Tetri BA, Clark JM, Bass NM, et al. Clinical, laboratory and histological associations in adults with nonalcoholic fatty liver disease. Hepatology. 2010;52:913–24. doi: 10.1002/hep.23784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noureddin M, Yates KP, Vaughn IA, et al. Clinical and histological determinants of nonalcoholic steatohepatitis and advanced fibrosis in elderly patients. Hepatology. 2013 doi: 10.1002/hep.26465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Talwalkar JA, Yin M, et al. Early detection of nonalcoholic steatohepatitis in patients with nonalcoholic fatty liver disease by using MR elastography. Radiology. 2011;259:749–56. doi: 10.1148/radiol.11101942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loomba R, Wolfson T, Ang B, et al. Magnetic resonance elastography predicts advanced fibrosis in patients with nonalcoholic fatty liver disease: A prospective study. Hepatology. 2014 doi: 10.1002/hep.27362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asrani SK, Talwalkar JA, Kamath PS, et al. Role of Magnetic Resonance Elastography in compensated and decompensated liver disease. J Hepatol. 2013 doi: 10.1016/j.jhep.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim D, Kim WR, Talwalkar JA, et al. Advanced fibrosis in nonalcoholic fatty liver disease: noninvasive assessment with MR elastography. Radiology. 2013;268:411–9. doi: 10.1148/radiol.13121193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui J, Ang B, Haufe W, et al. Comparative diagnostic accuracy of magnetic resonance elastography vs. eight clinical prediction rules for non-invasive diagnosis of advanced fibrosis in biopsy-proven non-alcoholic fatty liver disease: a prospective study. Aliment Pharmacol Ther. 2015;41:1271–80. doi: 10.1111/apt.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin SC, Heba E, Wolfson T, et al. Noninvasive Diagnosis of Nonalcoholic Fatty Liver Disease and Quantification of Liver Fat Using a New Quantitative Ultrasound Technique. Clin Gastroenterol Hepatol. 2014 doi: 10.1016/j.cgh.2014.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le TA, Chen J, Changchien C, et al. Effect of colesevelam on liver fat quantified by magnetic resonance in nonalcoholic steatohepatitis: a randomized controlled trial. Hepatology. 2012;56:922–32. doi: 10.1002/hep.25731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loomba R, Sirlin CB, Ang B, et al. Ezetimibe for the treatment of nonalcoholic steatohepatitis: assessment by novel magnetic resonance imaging and magnetic resonance elastography in a randomized trial (MOZART trial) Hepatology. 2015;61:1239–50. doi: 10.1002/hep.27647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zarrinpar A, Gupta S, Maurya MR, et al. Serum microRNAs explain discordance of non-alcoholic fatty liver disease in monozygotic and dizygotic twins: a prospective study. Gut. 2015 doi: 10.1136/gutjnl-2015-309456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyd NF, Dite GS, Stone J, et al. Heritability of mammographic density, a risk factor for breast cancer. N Engl J Med. 2002;347:886–94. doi: 10.1056/NEJMoa013390. [DOI] [PubMed] [Google Scholar]
- 35.Permutt Z, Le TA, Peterson MR, et al. Correlation between liver histology and novel magnetic resonance imaging in adult patients with non-alcoholic fatty liver disease - MRI accurately quantifies hepatic steatosis in NAFLD. Aliment Pharmacol Ther. 2012;36:22–9. doi: 10.1111/j.1365-2036.2012.05121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel NS, Doycheva I, Peterson MR, et al. Effect of Weight Loss on Magnetic Resonance Imaging Estimation of Liver Fat and Volume in Patients With Nonalcoholic Steatohepatitis. Clin Gastroenterol Hepatol. 2014 doi: 10.1016/j.cgh.2014.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castera L, Vilgrain V, Angulo P. Noninvasive evaluation of NAFLD. Nat Rev Gastroenterol Hepatol. 2013;10:666–75. doi: 10.1038/nrgastro.2013.175. [DOI] [PubMed] [Google Scholar]
- 38.Noureddin M, Lam J, Peterson MR, et al. Utility of magnetic resonance imaging versus histology for quantifying changes in liver fat in nonalcoholic fatty liver disease trials. Hepatology. 2013;58:1930–40. doi: 10.1002/hep.26455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reeder SB. Emerging quantitative magnetic resonance imaging biomarkers of hepatic steatosis. Hepatology. 2013;58:1877–80. doi: 10.1002/hep.26543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reeder SB, Cruite I, Hamilton G, et al. Quantitative assessment of liver fat with magnetic resonance imaging and spectroscopy. J Magn Reson Imaging. 2011;34:729–49. doi: 10.1002/jmri.22580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rinella ME, Loomba R, Caldwell SH, et al. Controversies in the Diagnosis and Management of NAFLD and NASH. Gastroenterol Hepatol (N Y) 2014;10:219–27. [PMC free article] [PubMed] [Google Scholar]
- 42.Martin NG, Eaves LJ, Kearsey MJ, et al. The power of the classical twin study. Heredity (Edinb) 1978;40:97–116. doi: 10.1038/hdy.1978.10. [DOI] [PubMed] [Google Scholar]
- 43.Tambs K, Moum T, Eaves LJ, et al. Genetic and environmental contributions to the variance of body height in a sample of first and second degree relatives. Am J Phys Anthropol. 1992;88:285–94. doi: 10.1002/ajpa.1330880303. [DOI] [PubMed] [Google Scholar]
- 44.Kendler KS, Heath AC, Neale MC, et al. A population-based twin study of alcoholism in women. JAMA. 1992;268:1877–82. [PubMed] [Google Scholar]
- 45.Tarnoki AD, Tarnoki DL, Bata P, et al. Heritability of non-alcoholic fatty liver disease and association with abnormal vascular parameters: a twin study. Liver Int. 2012;32:1287–93. doi: 10.1111/j.1478-3231.2012.02823.x. [DOI] [PubMed] [Google Scholar]
- 46.Weber S, Gressner OA, Hall R, et al. Genetic determinants in hepatic fibrosis: from experimental models to fibrogenic gene signatures in humans. Clin Liver Dis. 2008;12:747–57. vii. doi: 10.1016/j.cld.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 47.Day CP. Genetic studies to identify hepatic fibrosis genes and SNPs in human populations. Methods Mol Med. 2005;117:315–31. doi: 10.1385/1-59259-940-0:315. [DOI] [PubMed] [Google Scholar]
- 48.Bazick J, Donithan M, Neuschwander-Tetri BA, et al. Clinical Model for NASH and Advanced Fibrosis in Adult Patients With Diabetes and NAFLD: Guidelines for Referral in NAFLD. Diabetes Care. 2015;38:1347–55. doi: 10.2337/dc14-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrison SA, Oliver D, Arnold HL, et al. Development and validation of a simple NAFLD clinical scoring system for identifying patients without advanced disease. Gut. 2008;57:1441–7. doi: 10.1136/gut.2007.146019. [DOI] [PubMed] [Google Scholar]
- 50.Arulanandan A, Ang B, Bettencourt R, et al. Association Between Quantity of Liver Fat and Cardiovascular Risk in Patients with Nonalcoholic Fatty Liver Disease Independent of Nonalcoholic Steatohepatitis. Clin Gastroenterol Hepatol. 2015 doi: 10.1016/j.cgh.2015.01.027. [DOI] [PubMed] [Google Scholar]