Abstract
Objectives
The role of promoter methylation in the development of mucoepidermoid carcinoma (MEC) has not been fully explored. In this study, we investigated the epigenetic landscape of MEC.
Methods
The Illumina HumanMethylation27 BeadChip array and differential methylation analysis were utilized to screen for epigenetic alterations in 14 primary MEC tumors and 14 matched normal samples. Bisulfite sequencing was used to validate these results, with subsequent quantitative Methylation-Specific PCR (qMSP) to validate chloride intracellular channel protein 3 (CLIC3) in a separate cohort. Furthermore, CLIC3 immunohistochemical (IHC) staining was performed in another separate cohort of MEC. Finally, clinical and pathological characteristics were statistically analyzed for correlation with methylation status of CLIC3 and CLIC3 IHC H-scores by Wilcoxon rank sum, Kruskall-Wallis, and X2 test tests.
Results
We obtained 6 significantly differentially methylated gene candidates demonstrating significant promoter hyper- or hypo- methylation from the array data. Using bisulfite sequencing, we found one gene, CLIC3, which showed differential methylation between MEC tumor and normal samples in a small validation cohort. qMSP analysis of the CLIC3 promoter in a separate validation set showed significantly lower methylation level in tumor than in normal. The level of CLIC3 methylation in MECs was not statistically correlated with clinical or pathological characteristics. However, IHC staining intensity and distribution of CLIC3 were significantly increased in MECs, compared with those of normal salivary gland tissues.
Conclusions
Hypomethylation of CLIC3 promoter and its overexpression are significant events in MEC. Its functional role and potential therapeutic utility in MEC are worthy of further exploration.
Keywords: Head and neck cancer, Mucoepidermoid carcinoma, DNA methylation, Array, Bisulfite sequencing, Methylation-Specific PCR, CLIC3, Immunohistochemistry, H-score
Introduction
Mucoepidermoid carcinoma (MEC) is the most common type of salivary gland malignancy in adults and children [1]. MECs arise more commonly in the parotid than the minor salivary glands, wherein the soft and hard palate are the main sites of involvement [2]. MEC is comprised of 3 main types of cells: squamous cells, mucus-secreting cells, and intermediate cells. MECs are classified into low, intermediate, and high grade based on histological and pathological characteristics. Low and intermediate grade MECs have more mucus-secreting and intermediate cells, while high grade MECs exhibit more squamous and intermediate cells [3]. Surgery remains the primary treatment modality for MECs, which generally carry a good prognosis. Surgery combined with post-operative radiotherapy is reserved for high grade MECs, but there are still generally poor results due to the local recurrence and metastasis. The use of chemotherapy in the postoperative setting remains controversial [4].
Several genetic alterations are associated with the carcinogenesis and metastasis of MEC [5]. The most well-known alteration is the t(11:19)(q21;p13.1), found in a minor salivary gland MEC and is thought to be an early and primary event in its tumorigenesis [6]. A novel fusion oncogene, CRTC1-MAML2, combines exon 1 of CRTC1 on chromosome 19p13 with exons 2-5 of the MAML2 gene on chromosome 11q21 and was confirmed in a child with lung MEC [7]. In one study, CRTC1-MAML2 gene fusions were found in all of the 15 (100%) low-grade MECs with few or no other genomic imbalances, whereas 3 of 13 (23%) high-grade MECs had the translocation with more genomic imbalances [8]. In another study on major and minor salivary gland MECs, MAML2 gene split with or without CRTC1/3-MAML2 fusion seemed to be associated with a favorable clinical outcome [9]. The CRTC1–MAML2 oncoprotein may upregulate the epidermal growth factor receptor (EGFR) ligand, amphiregulin (AREG), which activates the EGFR signaling pathway in an autocrine manner, ultimately stimulating MEC cell growth and survival [10].
Epigenetic changes can not only complement the possible mechanisms of carcinogenesis, but also become a potential method for the early detection, treatment, and prognostic assessment of the cancer patients. To date, only a few methylation studies have focused on MEC carcinogenesis. Protein levels of E-cadherin and secreted frizzled-related proteins (SFRPs) were down-regulated due to their promoter hypermethylation [11,12]. Promoter hypermethylation was also found in some tumor suppressor genes (TSGs), including p16INK4a, runt-related transcription factor 3 (RUNX3), O6-methylguanine-DNA methyltransferase (MGMT), retinoic acid receptor b2 (RARb2), RAS-associated domain family protein 1A (RASSF1), adenomatous polyposis coli (APC), and death-associated protein kinase (DAPK) [13-16]. However, there has not been a genome wide study looking at the profile of CpG methylation in the DNA promoter regions in MEC.
In this study, we assessed the 6 methylated genes identified by the Illumina (San Diego, CA) HumanMethylation27 BeadChip array in an attempt to identify possible biomarkers involved in MEC. Bisulfite sequencing in a small validation cohort of 8 MEC and 6 normal salivary gland frozen samples was used to identify differentially methylated genes. We further performed qMSP to examine the promoter methylation status of the selected highly altered genes in a separate, large validation cohort of 25 MEC and 17 normal salivary gland formalin-fixed paraffin embedded (FFPE) samples. We then analyzed the correlation between the promoter methylation status of CLIC3 and the clinical and pathological characteristics of 25 MEC patients. Furthermore, CLIC3 immunohistochemical staining (IHC) was performed in a second cohort of MEC and normal control FFPE samples. Finally, clinicopathologic characteristics were analyzed for correlation with CLIC3 IHC H-scores in a separate cohort.
Materials and Methods
Tissue samples
14 primary MEC samples and 14 normal salivary gland samples were obtained from collaborations with the University of Texas MD Anderson and the National Institutes of Dental and Craniofacial Research supported Salivary Gland Tumor Biorepository, under an IRB approved protocol. The samples were de-identified so that clinical data were not available. These samples were used for our whole genome study whereby the Illumina HumanMethylation27 BeadChip array was utilized.
A pilot cohort of 8 MEC and 6 normal salivary gland frozen samples, and a validation cohort of 25 MEC and 17 normal salivary gland FFPE samples were obtained from surgically treated patients at Department of Otolaryngology-Head and Neck Surgery (Johns Hopkins Medical Institution, Baltimore, MD, USA) with written informed consent approved by the Johns Hopkins Institutional Review Board. All of the cases received no pre-operative chemotherapy or radiotherapy. Histological grade was defined according to WHO classification [17]. Tumor staging was defined according to TNM classification [18]. A total of 58 MECs and 20 normal salivary gland tissues from China Medical University during January 2009 to July 2013 were obtained for IHC under an appropriate IRB approved protocol. All MEC cases were confirmed by a head neck pathologist. MEC patients received no systemic therapy before surgery. There were 35 males and 23 females. Patients' ages ranged from 29 to 78 years old.
Methylation Array
The Illumina HumanMethylation27 BeadChip array was performed at the Johns Hopkins Stanley Kimmel Comprehensive Cancer Center DNA sequencing core, per the manufacturer's instructions. The arrays were performed in a single batch. We applied empirical Bayes moderated t-statistics implemented in the R/Bioconductor package LIMMA [19] to compare DNA methylation beta values for probes in CpG Islands between the 7 fusion positive tumors or 7 fusion negative tumors to the set of normal samples. We then identified those genes with FDR adjusted p-values below 0.05 as gene targets and proceeded with further validation.
DNA extraction
Genomic DNA from FFPE samples were extracted as previously described [16,20]. The FFPE tissue blocks were de-paraffinized with xylene and digested with 50 μg/ml proteinase K (Boehringer, Mannheim, Germany) and 1% SDS at 48°C overnight. Total genomic DNA of FFPE samples and frozen samples were extracted with phenol/chloroform, precipitated with ethanol, and finally eluted in 60 μl low-salt Tris-EDTA buffer (EDTA 2.5 mM and Tris-HCl 10 mM, LoTE) and stored at -20°C until use.
Sodium Bisulfite Treatment
Sodium Bisulfite was used to convert unmethylated cytosines into uracils, leaving the methylated cytosines unchanged as previously described [21]. 2 μg of genomic DNA was treated with the EpiTect Bisulfite Kit (Qiagen, Valencia, California, USA) according to the manufacturer's instructions. Bisulfite conversion thermal cycler conditions were as follows: Denaturation, 95°C for 5 min, incubation, 60°C for 25 min, denaturation, 95°C for 5 min, incubation, 60°C for 85 min, denaturation, 95°C for 5 min, incubation, and 60°C for 175 min. The bisulfite-modified DNA was purified through a spin column and eluted in 60 μl Buffer EB and stored at -80°C until use.
Sodium Bisulfite Sequencing
Bisulfite sequencing primers were designed for CpG islands in the promoters or the first exon regions of the top 25 genes with MethPrimer online [22] without the consultation of the each probe sequence in the methylation array, so the CpGs amplified by BSP for each primer set were not necessarily the same CpGs in the each probe of the methylation array. The BSP primers designed for the top 25 genes are listed in the Table 1. Touch-down polymerase chain reactions (TD-PCRs) were performed as follows: an initial Taq activation, 95°C for 5 min; denaturation, 95°C for 30 s; annealing, 60°C for 1 min; extension, 72°C for 1 min. Touch-down annealing temperatures were changed from 60°C to 58°C, 56°C and 54°C, 2 cycles for each temperature, and finally to 52°C for the next 35 cycles.
Table 1.
Bisulfite sequencing primer sequences for the significantly differentially methylated genes between either fusion-positive or fusion-negative mucoepidermoid carcinoma and normal controls. These genes in bold font are hypomethylated in their promoters.
| Gene | Forward Primer Sequence (5′ – 3′) | Reverse Primer Sequence (5′ – 3′) | Product Size (bp) | Tm (°C) |
|---|---|---|---|---|
| CLIC3 | TTTGTTTTTTGGTTTTTATTTATTA | AAAAACTCTTTAAATCTCCCTATAC | 281 | 53-54 |
| PREX1 | ATGTAGGGGTTTGGGGATTT | CCCTCCTCTAAACTTCAAATTTTATC | 173 | 59 |
| RANBP2 | TGTTAAGATATTTTTAGATTATAGGG | AATACCTACAAACCACTAAAACAAC | 195 | 52 |
| URI1 | AATGTGTTTATGAAATAGTTTTAGTA TATG | CCCTCTAAAAACTTACATTTTATACC | 258 | 54-55 |
| TMED2 | TTTTAGATTAGATTGGTTAATATGG | CCTCCTAAATAACTAAAATTACAAAC | 250 | 53-54 |
| SPINT2 | TGATTTAGGGATTAGTTTGGTATGG | CCACCCTTCCCTTAAACTTAATAAC | 237 | 59-60 |
Sequencing
After TD-PCRs, gel electrophoresis was used to confirm the efficiency of bisulfite sequencing products and primer specificity for each of the top 25 genes examined. If the efficiency and primer specificity were not good for some primer sets, the new primer sets were re-designed by MethPrimer and TD-PCRs were performed again. After the efficiency and specificity of each primer set were confirmed, the BSP product for each of these top 25 genes was gel purified and subsequently sent to GeneWiz (Frederick, MD) for sequencing. The sequence of BSP product of each gene in each sample was read with FinchTV software and then all sequences of BSP products for each gene in all samples were aligned by Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) with each gene's reference sequence from RefSeq to examine the methylation status of cytosines in the region checked.
Quantitative Methylation-specific PCR
We further assessed the methylation status of the gene CLIC3 in a separate, large validation cohort of 25 MECs and 17 normal salivary gland FFPE samples by SYBR-Green Methylation-Specific PCR as described before [23,24]. Primers were designed using the online qPCR assay design tool, PrimerQuest (http://www.idtdna.com/PrimerQuest/Home/Index). qMSP primer sequences of CLIC3 were forward 5′-TATCGGGTGGGCGGATT-3′ and reverse 5′-ACTACCCAACGCGCAAA-3′. β-actin was used for normalization. β-actin primer sequences were forward 5′-TGGTGATGGAGGTTTAGTAAG-3′ and reverse 5′ AACCAATAAAACCTACTCCTCCCTTAA-3′. qMSP reactions were performed in 384-well microtiter plates in the Applied Biosystems 7900HT Fast Real-Time PCR System and analyzed using SDS 2.3 (Applied Biosystems, Foster City, California). Each reaction volume was 10 μl, with components as previously described [16]. qMSP reactions were first performed for each primer set using the universally methylated, bisulfite converted human DNA (Zymo Research, Irvine, California) under these conditions (95 °C for 5 min, 45 cycles at 95°C for 15 s and 60°C for 45 s), followed by a melting curve to examine the specificity of each primer set. After the efficiency and specificity of each primer set was confirmed, qMSP reactions were done using FFPE samples (tumor and control) under the same reaction conditions above, followed by a melting curve. Standard curves were established for each plate with serial dilutions of universally methylated human DNA. Each DNA sample was run in triplicate at each condition (gene-specific primer pair or β-actin primer pair). For standard curves, each serially diluted, universally methylated DNA control sample was run in triplicate at each condition. In addition, water was used to replace universally methylated DNA control sample for no DNA template controls that were run in at least 9 wells at each condition. Data were obtained following 45 amplification cycles. All samples were within the range of sensitivity and reproducibility of the assay based on the amplification of the internal reference standard.
Correlation of Clinical and Pathological Characteristics with Methylation Status
Retrospective medical record abstraction was performed to determine the clinicopathologic variables of interest for the MEC patients included in the validation cohort. Descriptive variables were summarized with frequencies and proportions for categorical variables and median and interquartile range for continuous variables. Median methylation levels of 3 genes were compared between MEC tumors and normal salivary gland controls and then with the clinical and pathological characteristics of 25 MEC patients using the Wilcoxon rank sum nonparametric test for binary variables and the Kruskall-Wallis nonparametric test for categorical variables. Data analysis was performed using STATA 11.2 (College Station, TX, 2012).
Immunohistochemical Staining (IHC)
The IHC was done as previously reported [25]. The CLIC3 antibody (catalog number: sc-81872) was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). It was a mouse monoclonal IgG2a antibody that was raised against recombinant human CLIC3.
Scoring of CLIC3 IHC was performed as previously described [25]. Two head and neck pathologists from observed all sections in a double blinded fashion. The score depended on the intensity of cell staining and the percentage of stained cells. Staining intensity was measured as: no color as 0 (no staining), yellow as 1 (weak staining), brown as 2 (moderate staining), and dark brown as 3 (strong staining). The percentage of stained cells was: no stained cells as 0 (absent), 1% to 9% as 0.1 (focal), 10% to 49% as 0.5 (multiple), and ≥50% as 1.0 (diffuse). The product of the score of the intensity of cell staining multiplied by the score of percentage of stained cells was reported as an H-score in each field of view. Ten random view fields of high magnification were chosen in each slide for all biological samples, and three technical replicates for each sample were scored. The integrated H-score of all three slides for one sample that was ≥1.0 was deemed as positive expression, and <1.0 was deemed as negative expression. SPSS 17.0 software was used for statistical analysis. The CLIC3 H-scores detected by IHC were correlated with the clinicopathologic parameters of the MEC patients by X2 test [25].
Results
Results of the Illumina (San Diego, CA) HumanMethylation27 BeadChip Array
We computed differential methylation statistics for each probe of the 27K array in a CpG island to compare MECs to normal. Analyses were performed to compare the 7 fusion positive MECs to all 14 normal controls and to compare the 7 fusion negative MECs to all 14 normal controls. We obtained 6 genes that were significantly differentially methylated between the positive MECs and normal controls, which are listed in Table 1 along with the bisulfite sequencing primer sequences utilized. Of these six genes, CLIC3 had the lowest p-value (an adjusted p-value of 0.009). It was also the only gene that was significantly differentially expressed between the negative MECs and normal controls.
Methylation Status of the Top Gene, CLIC3, in the Test Set
Using bisulfite sequencing, we found in a pilot group of 8 primary MECs and 6 normal salivary gland samples where the CLIC3 gene showed differential methylation profiles between tumor and normal samples. Fourteen CpG sites were designed in the promoter of CLIC3, and methylation was noted in 2/8 (25.0%) tumors with 3 (2.7%) methylated CpG sites and 6/6 (100%) normal samples with 21 (25.0%) methylated CpG sites, suggesting hypomethylation in tumors.
Methylation Status of CLIC3 in the Validation Set
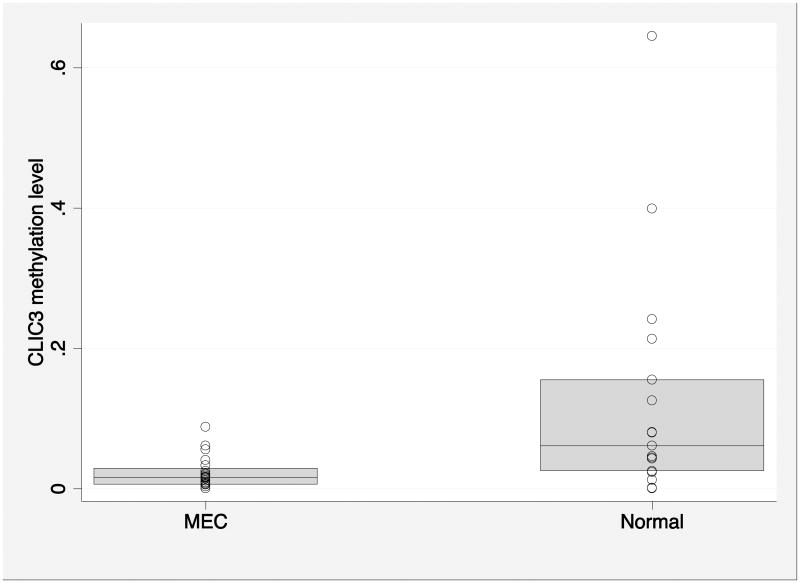
We further validated the methylation status of CLIC3 with the quantitative MSP in a larger cohort of 25 MEC patients and 17 normal salivary gland FFPE samples. One MEC sample was not evaluable for CLIC3 methylation level. Methylation of the CLIC3 promoter was significantly lower in tumors than normal tissues (median 0.0157 [IQR 0.0061-0.0291], compared with 0.0612 [IQR 0.0250-0.155], p=0.0019; Figure 1).
Figure 1.
Comparison of CLIC3 methylation level between MEC cases and normal salivary gland controls (p=0.0019). The shaded areas represent the interquartile.
MEC: mucoepidermoid carcinoma
Clinical and Pathological Data in the Validation Cohort for Methylation Status of CLIC3
The clinical and pathological parameters of the 24 MEC samples evaluable for CLIC3 methylation level are described in Table 2. The median age was 46 years and there were 10 males and 14 females. The majority of tumors arose from major salivary glands (n=17). There were no significant differences in the level of CLIC3 methylation by clinical or pathological characteristics (Table 2).
Table 2.
Correlation of CLIC3 methylation level with clinical and pathological patient characteristics.
| Characteristics | N (%) | Median CLIC3 methylation | p-value |
|---|---|---|---|
| Gender | 0.81 | ||
| Male | 10 (42) | 0.0146 | |
| Female | 14 (58) | 0.0164 | |
| Age | 0.286 | ||
| <40 | 8 (33) | 0.0189 | |
| 40-60 | 10 (42) | 0.0164 | |
| 60+ | 6 (25) | 0.0077 | |
| Smoking history | 0.11 | ||
| No | 20 (91) | 0.0157 | |
| Yes | 2 (9) | 0.0481 | |
| Site | 0.19 | ||
| Major salivary gland | 17 (71) | 0.0139 | |
| Minor salivary gland | 7 (29) | 0.0199 | |
| Overall stage | 0.27 | ||
| I | 11 (46) | 0.0094 | |
| II | 7 (29) | 0.0153 | |
| III-IV | 6 (25) | 0.0234 | |
| Margin status | 0.24 | ||
| Negative | 16 (73) | 0.0125 | |
| Positive | 6 (27) | 0.0198 | |
| Histologic grade | 0.90 | ||
| Low | 10 (42) | 0.0157 | |
| Intermediate | 8 (33) | 0.0131 | |
| High | 6 (25) | 0.0140 | |
| Perineural invasion | 0.46 | ||
| No | 12 (86) | 0.0125 | |
| Yes | 2 (14) | 0.0284 |
Detection of CLIC3 Protein Levels by IHC in another Validation Cohort
We collected a separate cohort of 58 MEC patients and 20 normal salivary gland FFPE samples, and CLIC3 IHC was performed on these samples. It was found that CLIC3 was significantly over-expressed in 50 out of 58 MEC samples, whereas no significant CLIC3 expression was found in 20 normal salivary gland samples, except for focal CLIC3 expression in ductal epithelial cells of some ducts in two cases (Figure 2 and Table 3, P<0.001). However, the over-expression of CLIC3 in these MEC samples was not found to correlate with any of the clinicopathologic parameters by X2 test (Table 4).
Figure 2. HE staining and CLIC3 immunohistochemical staining of normal salivary gland and mucoepidermoidal carcinoma tissue sections.
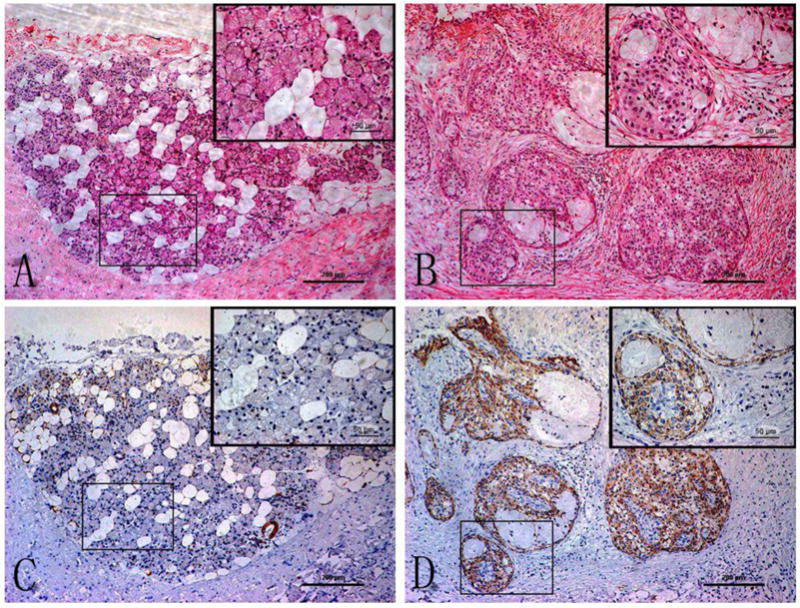
Panel A (normal) and B (MEC) are representative photomicrographs of H&E staining; Panel C (normal) and D (MEC) are representative photomicrographs of CLIC3 staining. Each panel was taken at low magnification (×100). The inset in each panel was taken from the boxed region of the same slide at high magnification (×400). The bar scale in each panel and each inset is 200 and 50 microns, respectively.
Table 3. Expression of CLIC3 in MEC and normal salivary gland tissues.
| CLIC3 | ||||
|---|---|---|---|---|
|
|
||||
| Group | n | + | - | P-value * |
| MEC | 58 | 50 | 8 | <0.001 |
| Normal salivary gland | 20 | 0 | 20 | |
: X2 test
Table 4. Correlation between CLIC3 expression and clinicopathologic parameters in MEC.
| Parameters | n | CLIC3 | % positive | P value* | |
|---|---|---|---|---|---|
|
| |||||
| + | - | ||||
| Age | |||||
| <60 | 34 | 29 | 5 | 76.47% | 0.81 |
| ≥60 | 24 | 21 | 3 | 87.50% | |
| Sex | |||||
| Male | 35 | 29 | 6 | 82.85% | 0.361 |
| Female | 23 | 21 | 2 | 91.30% | |
| Site | |||||
| Major salivary gland | 42 | 38 | 4 | 90.47% | 0.127 |
| Minor salivary gland | 16 | 12 | 4 | 75.00% | |
| Tumor size | |||||
| <4 cm | 52 | 46 | 6 | 84.61% | 0.143 |
| ≥ 4 cm | 6 | 4 | 2 | 66.66% | |
| TNM stage | |||||
| I+II | 48 | 43 | 5 | 89.58% | 0.105 |
| III+IV | 10 | 7 | 3 | 70.00% | |
| Pathological grading | |||||
| High | 47 | 42 | 5 | 89.36% | 0.15 |
| Low | 11 | 8 | 3 | 72.72% | |
| Lymph node metastasis | |||||
| Yes | 8 | 7 | 1 | 87.50% | 0.909 |
| No | 50 | 43 | 7 | 86.00% | |
: X2 test
Discussion
In this study, 6 genes had significantly altered methylation between MEC tumors and normal controls and were validated. Of these 6 genes, only CLIC3 had significantly altered methylation in both the CRTC1-MAML2 fusion positive and negative tumor samples. The CLIC3 gene demonstrated hypermethylation in normal salivary glands and hypomethylation in MECs by bisulfite sequencing. In the FFPE validation cohort, only the methylation status of CLIC3 showed a significant difference, with hypomethylation in MECs and hypermethylation in normal tissues.
CLIC3 was identified as one of the chloride intracellular channel (CLIC) family members [26], which was not only highly expressed in placenta [27], but also overexpressed in lung and heart, with low expression in skeletal muscle, kidney, pancreas and brain. CLIC3 is located predominantly in the nucleus, and indirectly interacts with ERK7, whose function is to inhibit DNA synthesis, suggesting that CLIC3 may function to regulate cell growth [26]. Elevated levels of CLIC3 were associated with high-grade pancreatic ductal adenocarcinomas, locally invasive tumors and lymph node involvement, suggesting a role in invasion and lymph node metastasis. Reduced survival was highlighted in pancreatic cancers with high CLIC3 expression, and the worst outcomes occurred when both Rab25 and CLIC3 were upregulated. A high level of CLIC3 was an independent predictor of poor prognosis through multivariate Cox proportional-hazard regression analysis for those tumors [28].
CLIC3 was also identified as one of genes thought to indicate resistance to neratinib used in an ErbB2 positive breast tumor cell line SKBR-3 through a genome-wide RNAi screen [29]. An anti-angiogenic peptide, CLT1, induced tumor cell death in the presence of fibronectin in a mechanism that depends on up-regulation of tumor cell integrin α5β1 and CLIC3 in CLT1 responsive tumor cell lines. Knocking down CLIC3 significantly reduced CLT1-mediated cell death, and co-localization of CLIC3 with CLT1 in tumor cells suggested a mechanistic link between CLIC3 and CLT1 internalization [30]. In two estrogen receptor (ER)-negative breast cancer cell lines, CLIC3 controlled trafficking of pro-invasive matrix metalloproteinase MT1-MMP independently of Rab25, and high CLIC3 expression was associated with low survival rate of ER-negative early breast cancer patients [30]. Furthermore, a higher level of CLIC3 mRNA was identified in invasive breast carcinomas than normal breast tissue by interrogating the Oncomine database (http://www.oncomine.org), as reported in [31]. Overall, CLIC3 appears to be oncogenic in nature, but there have been no studies looking into its epigenetic regulation. In our small validation set, we found hypermethylation in the promoter region of CLIC3 in 6 normal samples, and hypomethylation in 8 primary MECs using bisulfite sequencing. In the further larger validation set, the quantitative methylation status of CLIC3 showed a statistically significant difference between MEC and normal samples, which suggests that CLIC3 has an oncogenic role. Because we were not able to extract good quality RNA samples from the matched FFPE samples that were used for quantitative MSP, and we did not have the frozen samples of these same cohorts used in qMSP assays, we were not able to do the RT-PCR to check CLIC3 mRNA levels in MEC.
Instead, we performed IHC staining of CLIC3 in 58 FFPE MEC and 20 normal salivary gland tissues. The results showed that 86.2% of 58 MEC samples were positively stained for CLIC3, whereas none of the 20 normal salivary glands were CLIC3 positive (P<0.001), suggesting that there was an association between methylation and protein expression. While this association does not definitively prove that promoter methylation is the sole regulator of gene expression in CLIC3, it does support the correlation between hypomethylation of CLIC3 in MEC samples detected by quantitative MSP (qMSP) and high expression of CLIC3 in MEC samples detected by IHC staining.
CLIC3 methylation was not significantly associated with clinical or pathological characteristics in this small study. Additional research is indicated to elucidate any clinical significance of CLIC3 methylation levels in MEC and whether there is increased expression detected.
In summary, CLIC3 is a promising epigenetic biomarker in MEC. The presence of CLIC3 hypomethylation does appear to be significant in MEC as well as its overexpression on IHC. Therefore, its functional role in MEC is worthy of further exploration, and we hope to pursue this in future studies.
Highlights.
The HumanMethylation27 BeadChip array was used in a cohort of mucoepidermoid carcinoma.
The increased protein expression of CLIC3 via IHC was noted in MEC versus normal salivary gland tissues.
CLIC3 was found to have significant hypomethylation in these MEC tumors.
Acknowledgments
This work was supported in part by NIH NIDCR U54- RDCRC, OD-08-001
Footnotes
Conflict of Interest: All authors declare no conflict of interest that could inappropriately influence this work
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McHugh JB, Visscher DW, Barnes EL. Update on selected salivary gland neoplasms. Arch Pathol Lab Med. 2009;133(11):1763–74. doi: 10.5858/133.11.1763. [DOI] [PubMed] [Google Scholar]
- 2.Devaraju R, Gantala R, Aitha H, Gotoor SG. Mucoepidermoid carcinoma. BMJ Case Rep. 2014:bcr-2013–202776. doi: 10.1136/bcr-2013-202776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seifert G, Brocheriou C, Cardesa A, Eveson JW. WHO International Histological Classification of Tumours. Tentative Histological Classification of Salivary Gland Tumours. Pathol Res Pract. 1990;186(5):555–81. doi: 10.1016/S0344-0338(11)80220-7. [DOI] [PubMed] [Google Scholar]
- 4.McHugh CH, Roberts DB, El-Naggar AK, Hanna EY, Garden AS, Kies MS, et al. Prognostic factors in mucoepidermoid carcinoma of the salivary glands. Cancer. 2012;118(16):3928–36. doi: 10.1002/cncr.26697. [DOI] [PubMed] [Google Scholar]
- 5.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87(2):159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 6.El-Naggar AK, Lovell M, Killary AM, Clayman GL, Batsakis JG. A mucoepidermoid carcinoma of minor salivary gland with t(11;19)(q21;p131) as the only karyotypic abnormality. Cancer Genet Cytogenet. 1996;87(1):29–33. doi: 10.1016/0165-4608(95)00266-9. [DOI] [PubMed] [Google Scholar]
- 7.Serra A, Schackert HK, Mohr B, Weise A, Liehr T, Fitze G. t(11;19)(q21;p12∼p1311) and MECT1-MAML2 fusion transcript expression as a prognostic marker in infantile lung mucoepidermoid carcinoma. J Pediatr Surg. 2007;42(7):E23–29. doi: 10.1016/j.jpedsurg.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 8.Jee KJ, Persson M, Heikinheimo K, Passador-Santos F, Aro K, Knuutila S, et al. Genomic profiles and CRTC1-MAML2 fusion distinguish different subtypes of mucoepidermoid carcinoma. Mod Pathol. 2013;26(2):213–22. doi: 10.1038/modpathol.2012.154. [DOI] [PubMed] [Google Scholar]
- 9.Noda H, Okumura Y, Nakayama T, Miyabe S, Fujiyoshi Y, Hattori H, et al. Clinicopathological significance of MAML2 gene split in mucoepidermoid carcinoma. Cancer Sci. 2013;104(1):85–92. doi: 10.1111/cas.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z, Chen J, Gu Y, Hu C, Li JL, Lin S, et al. Aberrantly activated AREG-EGFR signaling is required for the growth and survival of CRTC1-MAML2 fusion-positive mucoepidermoid carcinoma cells. Oncogene. 2014;33(29):3869–77. doi: 10.1038/onc.2013.348. [DOI] [PubMed] [Google Scholar]
- 11.Shieh YS, Shiah SG, Jeng HH, Lee HS, Wu CW, Chang LC. DNA methyltransferase 1 expression and promoter methylation of E-cadherin in mucoepidermoid carcinoma. Cancer. 2005;104(5):1013–21. doi: 10.1002/cncr.21278. [DOI] [PubMed] [Google Scholar]
- 12.Lee CH, Hung YJ, Lin CY, Hung PH, Hung HW, Shieh YS. Loss of SFRP1 expression is associated with aberrant beta-catenin distribution and tumor progression in mucoepidermoid carcinoma of salivary glands. Ann Surg Oncol. 2010;17(8):2237–46. doi: 10.1245/s10434-010-0961-z. [DOI] [PubMed] [Google Scholar]
- 13.Guo XL, Sun SZ, Wang WX, Wei FC, Yu HB, Ma BL. Alterations of p16INK4a tumour suppressor gene in mucoepidermoid carcinoma of the salivary glands. Int J Oral Maxillofac Surg. 2007;36(4):350–3. doi: 10.1016/j.ijom.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Sasahira T, Kurihara M, Yamamoto K, Bhawal UK, Kirita T, Kuniyasu H. Downregulation of runt-related transcription factor 3 associated with poor prognosis of adenoid cystic and mucoepidermoid carcinomas of the salivary gland. Cancer Sci. 2011;102(2):492–7. doi: 10.1111/j.1349-7006.2010.01787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams MD, Chakravarti N, Kies MS, Maruya S, Myers JN, Haviland JC, et al. Implications of methylation patterns of cancer genes in salivary gland tumors. Clin Cancer Res. 2006;12(24):7353–8. doi: 10.1158/1078-0432.CCR-06-1272. [DOI] [PubMed] [Google Scholar]
- 16.Durr ML, Mydlarz WK, Shao C, Zahurak ML, Chuang AY, Hoque MO, et al. Quantitative methylation profiles for multiple tumor suppressor gene promoters in salivary gland tumors. PloS One. 2010;5(5):e10828. doi: 10.1371/journal.pone.0010828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnes L, Eveson J, Reichart P, Sidransky D. WHO Classification of Tumors: Pathology and Genetics of the Head and Neck Tumors. Lyon: IARC Press; 2005. [Google Scholar]
- 18.Sobin LH, Gospodarowicz M, Wittekind C. Classification of Malignant Tumours. 7th. New York: Wiley-Blackwell; 2010. [Google Scholar]
- 19.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43 doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shao C, Tan M, Bishop JA, Liu J, Bai W, Gaykalova DA, et al. Suprabasin is hypomethylated and associated with metastasis in salivary adenoid cystic carcinoma. PloS One. 2012;7(11):e48582. doi: 10.1371/journal.pone.0048582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93(18):9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18(11):1427–31. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 23.Tan M, Shao C, Bishop JA, Feng Z, Trock BJ, Westra WH, et al. Aquaporin-1 promoter hypermethylation is associated with improved prognosis in salivary gland adenoid cystic carcinoma. Otolaryngol Head Neck Surg. 2014;150(5):801–7. doi: 10.1177/0194599814521569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shao C, Sun W, Tan M, Glazer CA, Bhan S, Zhong X, et al. Integrated, genome-wide screening for hypomethylated oncogenes in salivary gland adenoid cystic carcinoma. Clin Cancer Res. 2011;17(13):4320–30. doi: 10.1158/1078-0432.CCR-10-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Zhou XQ, Li JY, Cheng JF, Zeng XN, Li X, et al. Prognostic significance of ERCC1 expression in postoperative patients with gastric cancer. Chin J Cancer Res. 2014;26(3):323–30. doi: 10.3978/j.issn.1000-9604.2014.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian Z, Okuhara D, Abe MK, Rosner MR. Molecular cloning and characterization of a mitogen-activated protein kinase-associated intracellular chloride channel. J Biol Chem. 1999;274(3):1621–7. doi: 10.1074/jbc.274.3.1621. [DOI] [PubMed] [Google Scholar]
- 27.Money TT, King RG, Wong MH, Stevenson JL, Kalionis B, Erwich JJ, et al. Expression and cellular localisation of chloride intracellular channel 3 in human placenta and fetal membranes. Placenta. 2007;28(5-6):429–36. doi: 10.1016/j.placenta.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Dozynkiewicz MA, Jamieson NB, Macpherson I, Grindlay J, van den Berghe PV, von Thun A, et al. Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Dev Cell. 2012;22(1):131–45. doi: 10.1016/j.devcel.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seyhan AA, Varadarajan U, Choe S, Liu W, Ryan TE. A genome-wide RNAi screen identifies novel targets of neratinib resistance leading to identification of potential drug resistant genetic markers. Mol Biosyst. 2012;8(5):1553–70. doi: 10.1039/c2mb05512k. [DOI] [PubMed] [Google Scholar]
- 30.Knowles LM, Zewe J, Malik G, Parwani AV, Gingrich JR, Pilch J. CLT1 targets bladder cancer through integrin α5β1 and CLIC3. Mol Cancer Res. 2013;11(2):194–203. doi: 10.1158/1541-7786.MCR-12-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macpherson IR, Rainero E, Mitchell LE, van den Berghe PV, Speirs C, Dozynkiewicz MA, et al. CLIC3 controls recycling of late endosomal MT1-MMP and dictates invasion and metastasis in breast cancer. J Cell Sci. 2014;127(Pt 18):3893–901. doi: 10.1242/jcs.135947. [DOI] [PubMed] [Google Scholar]