Abstract
Human tRNA3 Lys is the primer for reverse transcription of HIV; the 3′ end is complementary to the primer-binding site on HIV RNA. The complementarity ends at the 18th base, A58, which in tRNA3 Lys is modified to remove Watson–Crick pairing. Motivated to test the role of the modification in terminating the primer-binding sequence and thus limiting run-on transcription, we asked how the modification of RNA could be accomplished. tRNA m1A58 methyltransferase (m1A58 MTase) methylates N1 of A58, which is buried in the TψC-loop of tRNA, from cofactor S-adenosyl-L-methionine. This conserved tRNA modification is essential for stability of initiator tRNA in Saccharomyces cerevisiae. Reported here, three structures of human tRNA m1A58 MTase in complex with human tRNA3 Lys and the product S-adenosyl-L-homocysteine show a dimer of heterodimers in which each heterodimer comprises a catalytic chain, Trm61, and a homologous but noncatalytic chain, Trm6, repurposed as a tRNA-binding subunit that acts in trans; tRNAs bind across the dimer interface such that Trm6 from the opposing heterodimer brings A58 in to the active site of Trm61. T-loop and D-loop are splayed apart showing how A58, normally buried in tRNA, becomes accessible for modification. This result has broad impact on our understanding of the mechanisms of modifying internal sites in folded tRNA. The structures serve as templates for design of inhibitors that could be used to test tRNA m1A58 MTase's impact on retroviral priming and transcription.
Keywords: X-ray, RNA modification, S-adenosyl-L-methionine, 1-methyladenosine, HIV
Graphical abstract
Introduction
In all kingdoms of life, tRNA is extensively modified posttranscriptionally by an array of enzymes. Each enzyme is specific for one or a few tRNA sites. The modifications contribute to translational fidelity, recognition of aminoacyl-tRNA synthetases, tRNA stability and tRNA structure. Much research has focused on tRNA m1A58 methyltransferase (m1A58 MTase), which catalyzes the transfer of a methyl group from the cofactor S-adenosyl-L-methionine (SAM) to N1 of adenine 58 to give 1-methyladenosine (m1A). This modification is essential in Saccharomyces cerevisiae because it is required for the stability of initiator methionine tRNA [1].
m1A58 modification of tRNA3 Lys also has a role in replication of HIV in humans. Immunodeficiency viruses, such as HIV-1, coopt m1A58-modified tRNA3 Lys as a primer for the initiation of reverse transcription and rely on the modified base to terminate the base-paired region after 18 bases at the 3′ end [2]. Thus, m1A58 MTase may be a new human host enzyme that can be targeted for therapeutic intervention in HIV infection. However, m1A58 MTase has not yet been validated as a drug target. Contradictory reports as to whether A58 methylation is the sole determinant for termination of viral (+) strand strong-stop DNA synthesis and thus for reverse transcription exist. The mutation A58U inhibits replication of HIV in vivo [3]; on the other hand, a complementation system in which a HIV-1 proviral genome with altered primer-binding site uses Escherichia coli tRNA3 Lys added in trans to prime reverse transcription is insensitive to the A58U mutation in the E. coli tRNA3 Lys [4].
In humans, m1A58 MTase is composed of two subunits, a catalytic component, Trm61, and an RNA-binding component, Trm6 [5]. Bacterial and archaeal m1A58 MTases comprise only one subunit, called TrmI, and crystal structures of four such m1A58 MTases reveal that the enzymes assemble as homotetramers [6–9]. None of these structures include tRNA so that the mechanism and selectivity even for the bacterial enzymes is as yet undefined, although structure–function studies have pinpointed regions of tRNA that are essential for binding and catalysis [10].
In canonical L-shaped tRNA, A58 is buried in the core of tRNA where it forms a reverse Hoogsteen base pair with T54 [11]. In order for A58 to be modified, tRNA must adopt a tertiary structure in which A58 is accessible to the enzyme active site. Thus, predicting the binding mode of tRNA to m1A58 MTase is a daunting challenge that requires taking into account major conformational changes to the tRNA and perhaps also to the protein upon their association. Crystal structures of other enzymes that modify the core nucleosides of tRNA are mainly of the enzymes in apoform or in complex with minimal substrates that are tRNA fragments; these provide few clues to the mechanisms used to access the target nucleosides. Only one structure of a tRNA-core-modifying enzyme bound to a complete tRNA is available. The structure of the tRNA complex of archaeosine tRNA-guanine transglycosylase, which modifies position 15 in the D-loop of tRNA, shows that the tRNA has undergone a major reorganization of base pairing in order to detach the D-arm from the tRNA core [12].
Here we report three structures of human m1A58 MTase in complex with its cofactor and a cognate substrate, human tRNA3 Lys. The tRNAs are reconfigured into a novel conformation in which the D-arm and the T-arm are detached from each other and bind in separate pockets in the heterotetramer but in which the secondary structure of L-form tRNA is otherwise conserved. The structures reported here identify flexible points in tRNA and thus provide clues to the range of tRNA conformations recognized by other tRNA-modifying enzymes in order to access their targets.
Results
Evolution of diversity in the human versus bacterial and archaeal m1A58 MTases
The structure of human tRNA m1A58 MTase in complex with tRNA3 Lys and S-adenosyl-L-homocysteine (SAH) was determined at 2.2 Å resolution from a tetragonal crystal form with space group P43212. The first foothold into the structure of the RNA–protein complex was obtained after numerous trials by molecular replacement search using one of the two different dimers of protein subunits excised from the homotetrameric protein from the Thermus thermophilus apo-m1A58 MTase crystal structure (PDB ID: 2PWY) [6]. The pair of subunits that succeeded were those that shared the largest contact surface areas, consistent with their being the more stable basis for the tetrameric (quasi-tetrameric in human) structure (Supplementary Fig. 1a and b). The correct molecular replacement solution placed the Trm61/Trm6 dimer against a crystallographic 2-fold axis coincident with the 2-fold axis in the biological unit, a heterotetramer. The product complex of m1A58 MTase with SAH and m1A58–tRNA3 Lys was isomorphous with this human complex and was determined by molecular replacement using this structure.
The sequences of the catalytic human Trm61 and the binding Trm6 are 16% identical between them and the subunits have similar overall folds (Supplementary Fig. 1c and d), as predicted by fold-recognition sequence analysis [13]. However, the r.m.s.d. between the corresponding 218 Cα atoms of Trm6, and Trm61 is a large 2.6 Å, signaling the divergence of Trm6 toward an RNA-binding role, while the catalytic Trm61 remained more similar to the monomer within the bacterial homotetramer.
Initially, the structure of tRNA3 Lys was not visible in the replacement solutions; however, it was extracted through extensive cycles of rebuilding followed by difference map refinement [14], which progressively revealed the entire tRNA3 Lys and SAH at uniformly complete occupancy. The tRNA3 Lys conformation revealed the distortion necessary to expose N1 of A58 to the active site.
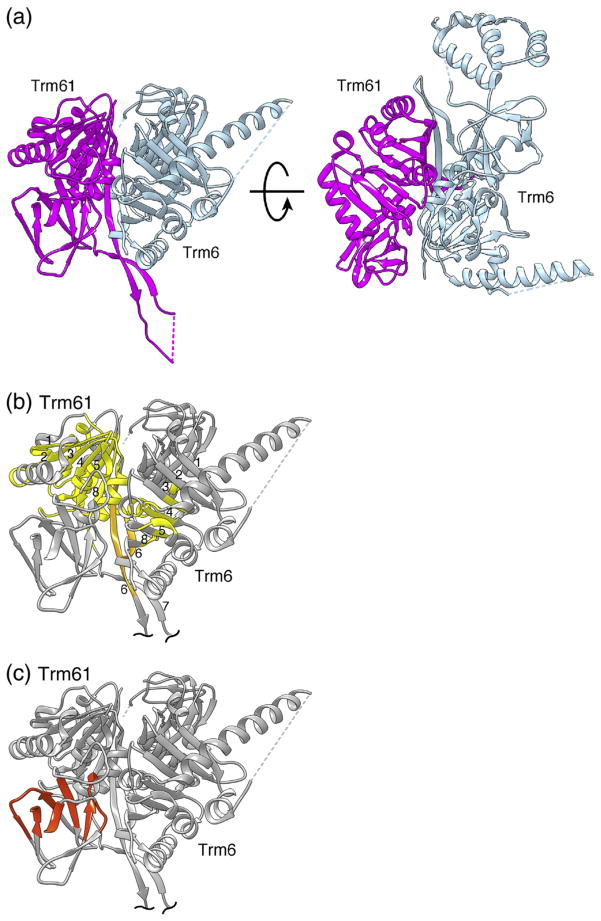
In contrast to the homotetrameric bacterial apoprotein, the human heterotetramer utilizes Trm61 as the catalytic subunit and Trm6 solely as a binding assist. However, both the Trm61 and Trm6 subunits consist of a small N-terminal β-sheet domain connected by a flexible linker to a C-terminal domain with a class I MTase fold (Supplementary Fig. 1). The MTase fold domain consists of a seven-stranded mixed β-sheet (β1–β6 and β8) sandwiched by helices (Fig. 1). As in the particular dimer within the T. thermophilus 222 symmetric homotetramer that shares the greatest contact (the formative dimer), the edges of the β-sheets of Trm6 and Trm61 (β6; Fig. 1) associate to generate a cross-subunit β-sheet, which is the conserved core of the formative dimer (Fig. 1). The interface area for this heterodimer is 2513 Å2, suggesting a strongly favorable association binding free energy of −25 kcal/mol [15].
Fig. 1.
Structure of the human m1A58 MTase dimer. (a) Ribbon drawing of the tight Trm61/Trm6 heterodimer of human m1A58 MTase shown in two views. The views are related by an approximately 90° counterclockwise rotation around the horizontal axis. In the view on the left, the insert in the N-terminal domain of Trm6 is behind the clipping plane, thus not visible. The catalytic subunit, Trm61, is magenta and the noncatalytic subunit, Trm6, is gray-blue. (b) Structurally highly conserved core, with respect to T. thermophilus m1A58 MTase, spanning the C-terminal domains of the human m1A58 MTase heterodimer. The core is colored gold except for the β6 strands that form the dimer interface, which are highlighted in light orange. β-Strands in the cross-subunit β-sheet of the C-terminal domains are numbered as in TrmI [6]. (c) Conserved structural core of the N-terminal RNA-binding domain of Trm61, colored orange-red.
The interface between heterodimers that forms the human heterotetramer is also conserved with respect to that of the T. thermophilus homotetramer but is more extensive because it includes an insert in Trm61. In both Trm6 and Trm61, β6 extends beyond the central β-sheet and forms an antiparallel β-ribbon with β7 (Fig. 1). In the heterotetramer, this β-strand forms an extended antiparallel β-sheet connecting all four subunits, which is stabilized by conserved salt bridges between Trm61 Glu232 and Trm6′ Arg431 (in our nomenclature, a prime indicates a residue or subunit from the opposite heterodimer) (Fig. 2a). The interface between opposing subunits Trm61 and Trm6′ (and Trm61′ and Trm6) is 1484 Å2 and the estimated binding free energy for the Trm61/Trm6′ dimer is −13.5 kcal/mol [15]. This interface is almost twice the area seen in the corresponding T. thermophilus m1A58 MTase interface, again reflecting divergence to a more differentiated and stable assembly.
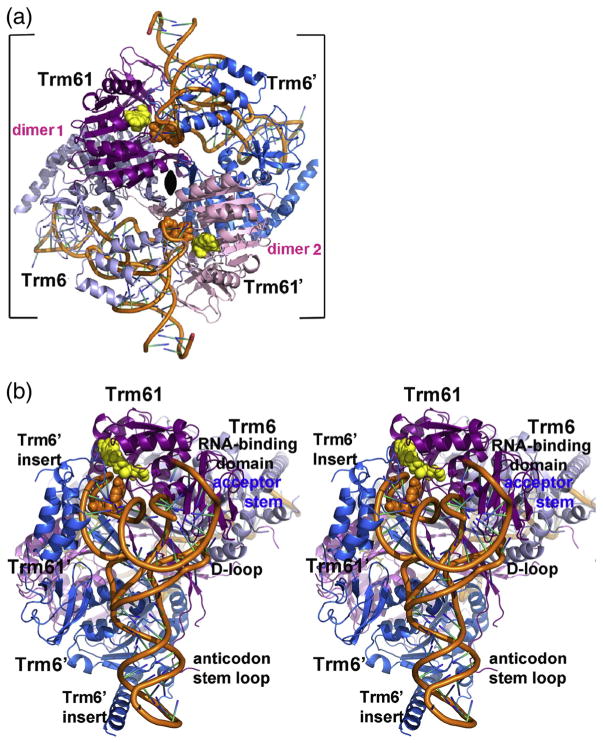
Fig. 2.
Quaternary structure of human m1A58 MTase. (a) Ribbon drawing of tRNA3 Lys-bound human m1A58 MTase heterotetramer viewed looking down the crystallographic 2-fold that coincides with the molecular 2-fold of the heterotetramer in the tetragonal crystal form. The 2-fold axis is indicated by the black oval. The tight dimers are colored magenta and gray-blue (“dimer 1”) and pink and blue (“dimer 2”). The tRNA-binding sites span the tight heterodimers: Trm61 and Trm6′ contribute 1522 Å2 and 1673 Å2, respectively, to the tRNA interface. The target nucleotide and the cofactor product SAH are shown in orange and yellow space-filling spheres, respectively. (b) Stereo plot of the heterotetramer viewed looking into the tRNA-binding site. The color scheme is the same as in (a). The view is related to the view in (a) by a 45° clockwise rotation about the crystallographic 2-fold and an approximately 90° rotation about the vertical axis in the plane of the paper.
A second crystal form of the tRNA m1A58 MTase–tRNA3 Lys-SAH complex was obtained under identical conditions, in a P212121 space group, which diffracted to 3.6 Å resolution and was determined by molecular replacement from the tetragonal form. In this crystal form, the heterodimers were not constrained to be identical by crystallographic symmetry. The modest resolution of the P212121 crystal structure (Table 1) precluded accurately defining the conformations of side chains and nucleic acid bases; however, the backbone trace of the protein and tRNA could readily be discerned and accurately placed, revealing clear conformational differences between the two heterodimers. The differences were confined to inserts in the Trm6 subunits and to the TψC-loops and D-loops of the tRNAs; the structural cores and the subunit interfaces of the P212121 form were the same as in the tetragonal crystal form.
Table 1.
Data collection and refinement statistics (molecular replacement).
| m1A58 MTase–tRNA-SAH | m1A58 MTase–tRNA-SAM | m1A58 MTase–tRNA-SAH | |
|---|---|---|---|
| Data collection | |||
| Space group | P43212 | P43212 | P212121 |
| Cell dimensions | |||
| a, b, c (Å) | 137.2, 137.2, 177.0 | 136.8, 136.8, 177.2 | 128.2, 137.4, 157.6 |
| Resolution (Å) | 97.0–2.0 (2.05–2.0)a | 54.1–2.1 (2.15–2.10) | 59.4–3.6 (3.69–3.6) |
| I/σI | 18.6 (1.0) | 16.7 (0.9) | 6.4 (1.0) |
| Completeness (%) | 99.9 (99.9) | 99.9 (100) | 98.5 (98.1) |
| Redundancy | 8.1 (7.6) | 8.1 (8.2) | 4.3 (4.3) |
| CC1/2 (%)b | 100 (38.6) | 99.9 (35.5) | 99.3 (30.4) |
| Refinement | |||
| Resolution (Å) | 74.4–2.2 | 54.1–2.3 | 59.4–4.05 |
| I/σIc | 24.3 (2.5) | 21.5 (2.0) | 8.6 (2.0) |
| Rmeasd (%) | 6.1 (86.4) | 7.9 (115.0) | 19.4 (98.7) |
| CC1/2 (%) | 100 (82.6) | 99.9 (72.1) | 99.6 (59.7) |
| No. of reflections | 85,931 | 74,959 | 23,182 |
| Rwork/Rfree | 18.8/21.4 | 19.2/22.6 | 21.0/24.8 |
| No. of atoms | |||
| Protein | 5088 | 5181 | 9637 |
| Ligand/ion | 1713 | 1763 | 3254 |
| Water | 416 | 309 | 0 |
| B-factors | |||
| Protein, Trm61 | 46 | 50 | 110 |
| Protein, Trm6 | 63 | 68 | 119 |
| tRNA | 64 | 68 | 137 |
| Water | 52 | 53 | — |
| r.m.s.d. Values | |||
| Bond lengths (Å) | 0.005 | 0.005 | 0.003 |
| Bond angles (°) | 0.88 | 0.79 | 0.65 |
Values in parentheses are for highest-resolution shell.
Pearson correlation coefficient between average intensities of randomly chosen subsets of measurements for unique reflections.
Data statistics for reflections used in final refinements.
Multiplicity-independent R-factor Rmeas = n1/2(n − 1)[ΣΣ|Ii(hkl) − AvgI(hkl)|/ΣΣIi(hkl)] where summations are over all n measurements
of all unique reflections.
2-Fold related tRNAs bind across the tetramer interface
Two molecules of tRNA bind at sites that span the tetramer interface generated by the crystallographic 2-fold: each tRNA binds almost exclusively to a single Trm61/Trm6′ pair (Fig. 2a). The anticodon stem–loop, comprising one branch of the L-shaped tRNA structure, is approximately parallel with the central β-sheet of the Trm6′ C-terminal domain, packing against the loops at the ends of the β-strands but forming few hydrogen bonds with protein residues (Fig. 2b and Supplementary Fig. 3). The amino-acid-accepting branch of the tRNA lies across the catalytic subunit Trm61. At one end of this branch, directly above the anticodon stem, the target base, A58, in the TψC-loop binds to the active-site pocket of Trm61. A helical insert in the N-terminal domain of the Trm6′ subunit packs against the TψC stem–loop (Fig. 2b). At the far end of the amino-acid-accepting branch, the acceptor stem binds to the N-terminal domain of Trm61 (Fig. 2b). This tRNA binding mode indicates that the heterotetramer is the only active species and shows how both subunits are required for orientating tRNA for catalysis at Trm61 [5,16].
The tRNA-binding site explains the results of mutations of eukaryotic m1A58 MTases. In the S. cerevisiae ortholog of human m1A58 MTase, mutations of several conserved residues in the β-barrel core of the tetramer interface impair tRNA binding and catalysis without disrupting the oligomerization of the enzyme [16]. Our structures show that these mutated residues are clustered near residues that are part of the D-loop and TψC-loop interfaces. For example, in Trm61, mutated Tyr238(261) (numbers in parentheses correspond to the S. cerevisiae sequence) is adjacent to Asn239 at the interface with the D-loop. In Trm6, Tyr433(422) and Pro442(431) are adjacent to Gln434 and His441, respectively, both of which donate hydrogen bonds to the phosphate group of the target base, A58. Therefore, the mutations may decrease tRNA binding affinity by changing the tRNA interface structure. These results are evidence that the crystal structure corresponds to a productive m1A58 MTase–substrate–cofactor complex.
Trm6 is less conserved with respect to TrmI than Trm61 and lacks a cofactor-binding pocket
Trm61 is the catalytic subunit of human tRNA m1A58 MTase and most of its three-dimensional structure is highly conserved with respect to TrmI. Difference-distance analysis identifies a conserved structural core of 136 residues in the C-terminal MTase domains of Trm61 and TrmI whose α-carbons can be aligned against each other with an r.m.s.d. of ±1 Å (Fig. 1) [17,18]. A 24-amino-acid insert between β6 and β7 in Trm61 packs against Trm6′ in the tetramer, extending the tetramer interface but not interacting with other parts of Trm61.
Relative to apo-TrmI, the small N-terminal domain of Trm61 has undergone a rigid-body shift to bind to the tRNA (Fig. 2b and Supplementary Fig. 1a). The N-terminal domain is composed of six antiparallel ß-strands and a short a-helix.
The cofactor-binding site is highly conserved in Trm61, and even in the initial map generated with the homodimeric molecular replacement solution, the Trm61 subunit could be distinguished from Trm6 by density for SAH in the cofactor site. SAM-binding MTases are characterized by several conserved sequence motifs, four of which (motifs I, II, III and IV) line the SAM-binding site [19,20] (Supplementary Fig. 1d). In Trm61, SAH makes equivalent hydrogen bonds with residues from motifs I, II and IV to those in TrmI [6] (Supplementary Fig. 2).
The Trm6 structure has diverged from that of TrmI, with several variations in orientations of the helices that pack against the central β-sheet and altered conformations of some of the protein segments that bind to tRNA (Supplementary Fig. 1a). Trm6 lacks the conserved sequence motifs that line the SAM--binding pocket in SAM-dependent MTases (Supplementary Fig. 1d). In particular, a conserved Asp in motif IV (Asp181 in Trm61), which has been implicated in catalysis [6], is an Ala in Trm6. The cofactor-binding pocket is occluded by inserted residue Arg377, which hydrogen bonds to the tRNA backbone. Thus, Trm6 cannot have methylase activity and only has a tRNA-binding role.
Trm6 has an 83-residue insert in its N-terminal domain and a 98-residue insert in the middle of the C-terminal domain, both of which contribute to tRNA binding. The first insert forms a well-ordered three-helix bundle (Fig. 2b and Supplementary Fig. 1b) that interfaces with the TψC-loop. The second insert is mostly disordered but the C-terminal end of the insert forms a long helix that abuts the anticodon stem. Hence, this structure provides a compelling case of the repurposing of a subunit through evolution, from a catalytic subunit into an orienting element that optimizes substrate selectivity.
tRNA structure is refolded upon binding to a large electropositive surface
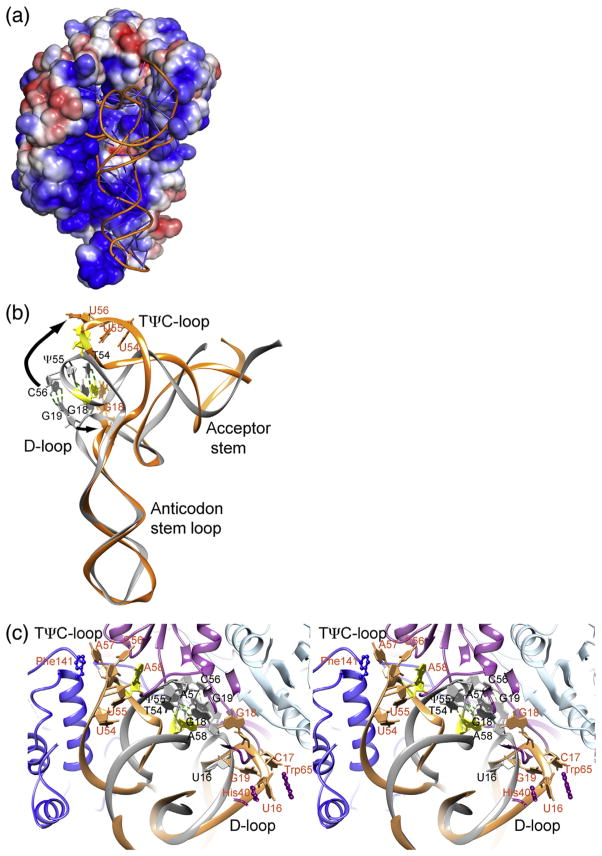
Binding to m1A58 MTase refolds tRNA such that A58 in the TψC-loop moves ~ 20 Å from its buried position in free tRNA into the active site. In canonical L-shaped free tRNA, G18 and G19 in the D-loop are base paired with ψ55 and C56 in the TψC-loop [11,21]. The tight association of the D-loop and the TψC-loop, stabilized by these base pairs, renders bases in these loops inaccessible to modification [11,21]. Binding to approximately 3200 Å2 of electropositive surface on Trm61/ Trm6′ (Fig. 3a) distorts tRNA by hinge bending to an “angled” L-shaped tertiary structure. The TψC and D stem–loops splay apart, away from the protein at the top of the anticodon stem, to make A58 accessible to the active site on Trm61 (Figs. 2b and 3b and c).
Fig. 3.
tRNA structure in the P43212 crystal form of the human m1A58 MTase–tRNA3 Lys-SAH complex. (a) Surface representation of the tetramer colored by electrostatic potential (−3 kT/e to 3 kT/e). The view is the same as in Fig. 2b. (b) Cartoon of the tRNA3 Lys structure in the m1A58 MTase complex (orange) superimposed on tRNAPhe (PDB ID: 1EHZ) (gray), which has the canonical L-shaped tRNA structure. The view of tRNA is related to the view in (a) by an approximately 90° counterclockwise rotation around the vertical axis in the plane of the paper. In the canonical structure, G18 and G19 from the D-loop are interdigitated with A57 and A58 from the TψC-loop and base paired with U55 and C56, respectively, while in the bound tRNA structure, the D-loop and the TψC-loop are splayed apart, as shown by the arrows. G18, G19 and nucleotides 54–58 are plotted as sticks with filled base and sugar rings. A58, the target, is colored yellow. Labels for the apo-tRNAPhe are black and those for the enzyme-tRNA3 Lys tRNA complex are dark orange. (c) Stereo cartoon plot of the m1A58 MTase complex superimposed with tRNAPhe (in gray), viewed looking into the tRNA-binding site as in (a). The colors for the complex are lighter shades of those used in Fig. 2. The A58 base is yellow. Nucleotides in the D-loop and in the TψC-loop are drawn as sticks with filled base and sugar rings. Side chains of Trm61 His40 and Trp65, which anchor the D-loop by stacking with U16, and Trm6 Phe141, which stacks against A57, are shown in a ball-and-stick form. Labels for the apo-tRNAPhe are black and those for the enzyme-tRNA3 Lys tRNA complex are dark orange.
The remarkable opening of tRNA is accomplished by only a few localized changes in the tRNA backbone, particularly in the tertiary interactions between stem–loop structures. Reorientation of U47 and C48 in the variable region alters the angle between the acceptor stem and the anticodon stem–loop. G15 flips out of the tRNA core, breaking the base pair between G15 and C48, and rearrangement of the rest of the D-loop (nucleotides 16–20) breaks the base pairing between G18 and G19 with U55 and C56. Nucleotides 55–60 in the TψC-loop also change their conformation to orient A58, the target, in the active site of Trm61. Base pairing of nucleotides outside of the D-loop and the TψC-loop is completely conserved with respect to canonical L-shaped tRNA. The electropositive surface that binds tRNA is highly complementary in shape to the distorted L-form of tRNA, thus maximizing the binding interaction energy that drives the necessary tertiary structure rearrangement in tRNAs. The conformational change to tRNA is likely to result strictly from binding to the enzyme, not from crystal packing, since it is seen in both crystal forms of the m1A58 MTase–tRNA3 Lys-SAH complex. The major packing interface in the P43212 crystal form comprises 750 Å2 on the anticodon stem–loop, which does not change conformation upon binding to the enzyme. There are no crystal contacts to the D-loop or the TψC-loop.
Nucleotides 17–19 in the D-loop are the most disordered region in the bound tRNA. Nucleotides 17–19 could not be fitted to density with a single conformation in the P43212 crystal form. The P212121 crystal form has an entire heterotetramer in the asymmetric unit; thus, the structures of the two bound tRNA molecules are not constrained by crystallographic symmetry and are treated independently during refinement. In the P212121 structure, nucleotides 17–19 are in different conformations in the two tRNAs. There are no hydrogen bonds between the protein and the G18 or G19 base to replace the disrupted hydrogen bonds with the TψC-loop. However, U16 is very well ordered by stacking interactions with His40 and Trp65 from the N-terminal domain of Trm61, thus helping to fix the D-loop in its binding pocket on the tetramer (Fig. 3c).
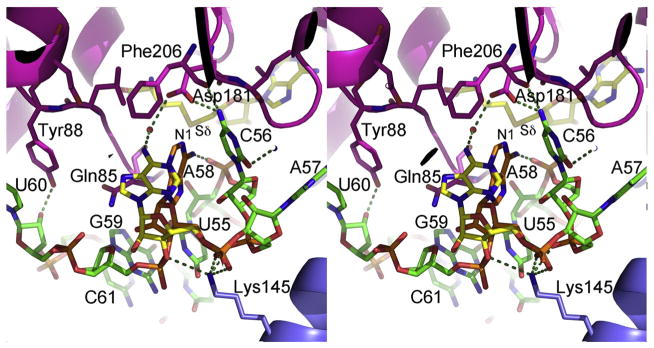
In the TψC-loop, the C56 base, which base pairs with G19 in canonical L-shaped tRNA, is inserted into a pocket in the active-site cavity where it makes ideal hydrogen bonds with Asp181 Oδ1, Gln212 Nε2 and the backbone carbonyl of Phe206 (Fig. 4). C56 also donates a water-mediated hydrogen bond to Ser207 Oγ. No other tRNA base in the complex is as extensively hydrogen bonded to the protein. The hydrogen bond interactions help compensate for the disrupted hydrogen bonds between the D-loop and the TψC-loop and help position the TψC-loop in the active-site cavity.
Fig. 4.
Active-site structure. Stereo plot of the active site of human m1A58 MTase. The protein is shown in ribbons with selected side chains in the active-site cavity plotted in stick form. The color scheme for the protein is the same as in Fig. 2. The tRNA is shown in sticks with carbons in green, oxygen atoms in red, nitrogen atoms in blue and phosphorous atoms in orange, except for the target, A58. The major and minor conformations of A58 are shown with carbon atoms colored orange and yellow, respectively. SAH is plotted in sticks with yellow carbons. The red sphere is a water molecule. Hydrogen bonds are shown with broken lines.
Binding to the enzyme leaves the most variable length subdomains of tRNA, namely the D-loop and the variable loop between the anticodon stem–loop and the TψC stem–loop, exposed to solvent; thus, inserts in these regions can be accommodated in the complex. This binding mode and the paucity of hydrogen bond interactions between the protein and the tRNA bases explain why the enzyme accepts most human tRNAs as substrates. Of the 20 human tRNA sequences in the transfer RNA database, 18 have the m1A58 modification [22]. Microarray assays for m1A58 modification in several human cell lines showed that 9 out of 40 nuclear-encoded tRNAs were consistently hypomodified [23]; however, the hypomodified tRNAs had no obvious common features that would make them poorer substrates for m1A58 MTase.
Structure of the active site
In addition to the three hydrogen bonds with the C56 base, the protein makes 22 hydrogen bonds with the sugar-phosphate backbone of the TψC-loop, more than the total number of hydrogen bonds to the remainder of the tRNA (Supplementary Fig. 3). These hydrogen bonds stabilize a new conformation of the TψC-loop with A58 positioned at the active site of the enzyme. The new loop conformation is also stabilized by an intra-RNA hydrogen bond between N6 of A58 and a phosphate oxygen of C56. A similar stabilizing hydrogen bond to N6 is used by the 30S ribosome MTase NpmA to orient its target base, A1408, for catalysis. In the case of NpmA [24], however, rearrangement of the RNA upon binding is limited to flipping of A1408 into the active site, and the hydrogen bond to A1408 is between N6 and a backbone carbonyl of the protein. For NpmA, the hydrogen bond is important mainly for proper alignment of A1408 N1 against the SAM, while in m1A58 MTase, the hydrogen bond helps stabilizes a major reorganization of the TψC-loop.
There are no hydrogen bonds between the protein and the A58 base, which has two dominant conformations (Fig. 4). In the major conformation, the base sits directly below the methionine moiety of SAH with the N6 amino group donating a hydrogen bond to a C56 phosphate oxygen atom (Fig. 4). In this conformation, N1, the target for methylation, is 5.1 Å from Sδ of the product SAH and 3.4 Å from the projected position of the Sδ-methyl group. The mutual orientations of A58 and SAH may resemble a product complex after methyl transfer: N6 is closer to the projected position of the methyl of a substrate SAM (2.4 Å) than N1, and the N1 lone electron pair is no longer inline with the Sδ–CH3 bond. Difference electron density around G59 and U60 in the final structure suggests that these nucleotides may move between multiple conformations though they could not justifiably be refined as such.
The target base has moved away from the cofactor in this complex of m1A58 MTase with the substrate tRNA and the product of methyl transfer SAH. The only hydrogen-bonding side chains close to the target-binding site are Asp181 and Gln85. Of these, Asp181 is best positioned to interact with the A58 base: in the crystal structure, Asp181 Oδ1 is 3.3 Å from A58 N1. Mutation of the corresponding Asp to Ala in T. thermophilus TrmI causes a 300-fold reduction in kcat [6]. Asp181 may facilitate methyl transfer by increasing the nucleophilicity of A58 N1 and also contributes to cofactor binding [6] and hydrogen bonding to C56.
The product complex shows an initial step in product release
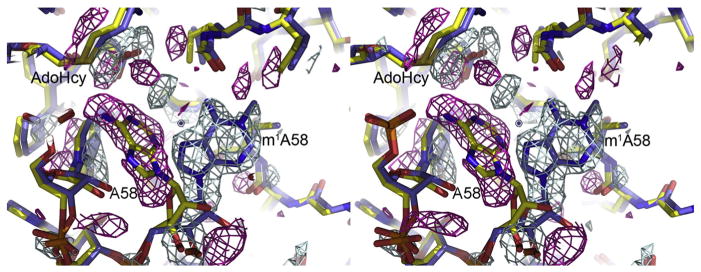
When m1A58 MTase is cocrystallized with tRNA3Lys and SAM, methyl transfer occurs and a product complex is obtained. Crystals of the product complex are isomorphous with crystals of the m1A58 MTase–tRNA3 Lys-SAH complex. In the product complex, SAH is bound in the cofactor-binding site and the methylated adenine base has rotated out of the active site (Fig. 5). A minor conformation of the methylated A58, equivalent to the minor conformation of A58 in the m1A58 MTase–tRNA3 Lys-SAH complex, is also present in the crystal structure. Except for the methylated, rotated target base, the structures of the product complex and the substrate-SAH complex are the same. This result suggests that rotation of the methylated A58 base out of the active site is an initial step in product release.
Fig. 5.
Product complex structure. Close-up view of the active site of the product complex of m1A58 MTase (sticks coloredcoded by atom type: oxygen, red; nitrogen, dark blue; carbon, blue) overlaid with the substrate complex (sticks colorcoded oxygen, red; nitrogen, dark blue; carbon, yellow) and superimposed on an (F1 − F2)αcalc difference map, which sensitively shows the differences between the two structures. The amplitudes for the difference map are the differences between the product complex (F1) and substrate complex (F2) structure factor amplitudes and the phases are from the substrate complex. Positive density, shown in blue contours, delineates the methylated A58 base. Negative density is shown with magenta contours. Each structure also had a common minor conformation of A58, shown in Fig. 4, but not plotted here. Density for the minor conformer cancels out in the (F1 − F2)αcalc ma.
Hydrogen bonds between the Trm6 insert and the TψC-loop contribute to orientation of the target base
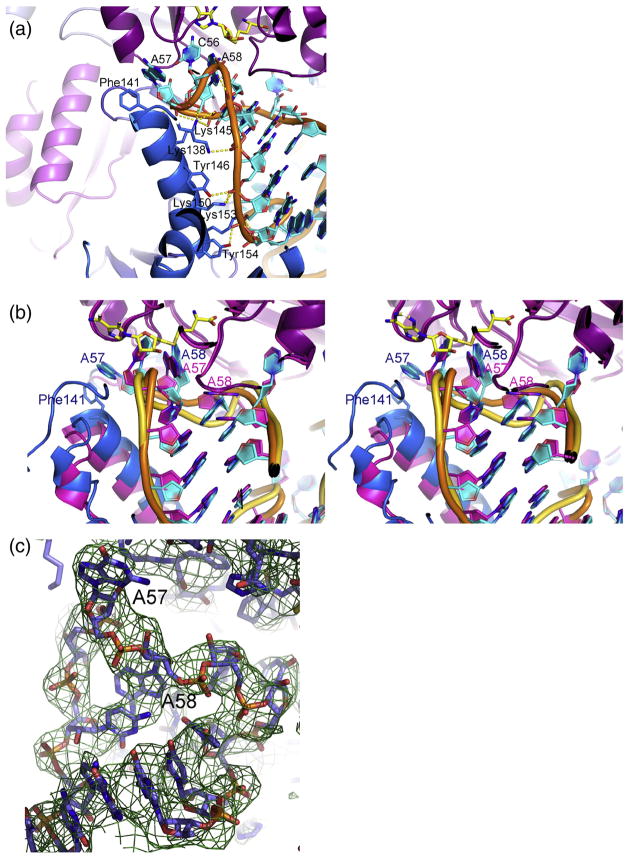
Electrostatic interactions with the positive surface on m1A58 MTase likely drive the distortions in the tRNA structure that induce the TψC-loop to detach from the D-loop and bind near the active site of the enzyme. Subsequently, hydrogen bonding between the TψC-loop backbone and the N-terminal domain of Trm6 (Figs. 4 and 6a) fine-tunes the TψC-loop conformation to orient the target base for catalysis, as observed in the P212121 crystal form of the m1A58 MTase–tRNA3 Lys-SAH complex.
Fig. 6.
Interactions of an insert in Trm6 with the TψC-loop. (a) Cartoon plot of the TψC stem–loop in the m1A58 MTase–tRNA3 Lys-SAH complex from the P43212 crystal structure, highlighting the hydrogen bonds between an insert in Trm6 (blue) and the backbone of the TψC stem–loop. Hydrogen-bonding side chains from the insert are shown as sticks and hydrogen bonds are shown as broken lines. SAH is shown as sticks with yellow carbons. (b) Superposition of the two crystallographically independent active sites of the P212121 crystal form of the m1A58 MTase–tRNA3 Lys-SAH complex, shown in cartoon rendition. In one active site, the Trm6 insert is optimally positioned to make the hydrogen bonds to the TψC-loop shown in (a) and A58 is in a productive orientation (blue insert, cyan RNA bases, orange RNA backbone). In the other active site, the Trm6 insert is rotated away from the TψC-loop and A57, A58 and G59 are in a nonproductive conformation with A57 instead of A58 in the active site (magenta insert, magenta bases and gold RNA backbone). Labels for the nonproductive active site are magenta and those for the productive active site are dark blue. (c) Nonproductive conformation of the TψC-loop from the P212121 crystal structure overlaid with a (2Fo −Fc)αcalc composite omit map.
Unlike the P43212 crystal form, the P212121 crystal form contains a heterotetramer in the asymmetric unit, with two crystallographically independent active sites. The P212121 heterotetramer is asymmetric primarily due to a shift in the N-terminal domain: while one Trm6 subunit has the same structure as in the heterodimer of the P43212 crystal form, in the second, the N-terminal domain is rotated a few degrees away from the tRNA and Trm6 residues 78–132, which contain all of the N-terminal domain insert except the third helix, are poorly ordered. Consequently, residues 140–154, the third helix in the Trm6 insert of the N-terminal domain, are too far apart from the tRNA to form hydrogen bonds. Hence, the energetics of this attachment may be relatively weak. The tRNA base A57 displaces A58 in the active site, suggesting that this structure may be along a trajectory toward product release. The splayed-apart tRNA conformation and the locations of the D-loop and the TψC-loop in their respective binding pockets are conserved. Thus, the asymmetric tetramer allows dissection of the role of the Trm6 N-terminal domain in orienting the target A58 base into the active site for catalysis.
Prominent density peaks for the electron-dense phosphates in the tRNA3 Lys accurately map out the tRNA fold, revealing two snapshots of the tRNA as it binds to the enzyme (Supplementary Fig. 4). One heterodimer is essentially the same as that in the P 43212 complex , where the TψC - loop conformation positions A58 in the active site in a nearly optimum orientation for catalysis. In the other heterodimer, the Trm6 N-terminal domain is rotated away from tRNA, and the target base, A58, and flanking nucleotides A57 and G59 assume a conformation with A57 rather than A58 in the active site (Fig. 6b and c). However, although A57 is located at the active site, it does not appear to be properly positioned for catalysis and is not likely targeted for methylation. In the first, more catalytically competent heterodimer complex, accurately defined at 2.2 Å in the P43212 crystal form, the TψC-loop A57 is rotated out of the loop where it has perpendicular aromatic stacking interactions with Phe141, while A58 binds next to C56 in the active-site cavity. This arrangement is stabilized by hydrogen bonds from Lys145 Nζ to the phosphate oxygen atoms of both A57 and A58 and a salt bridge between Lys145 Nζ and the phosphate oxygen of G59 (Fig. 4). Both Phe141 and Lys145 are too far from the tRNA to provide these stabilizing interactions when the N-terminal domain is rotated away from the tRNA. Thus, the N-terminal domain insert in Trm6 may contribute to target specificity.
These results suggest a binding mechanism for tRNA that involves a dynamic trajectory. We hypothesize that, first, the tRNA docks to the C-terminal domains of Trm61 and Trm6, which separates the TψC-loop and the D-loop and directs the TψC-loop to the active site. Subsequently, the Trm6 N-terminal domain insert closes over the TψC-loop and stabilizes a productive conformation of the loop through hydrogen bonds to the tRNA backbone. This conformation is characterized by A58 being productively aligned with the cofactor in the active site. Product release would reverse these steps. It is common for RNA modification enzymes to have small RNA-binding domains that interface with the RNA substrate and, which in some cases, contribute to substrate specificity [25]. These RNA-binding modules are flexibly linked to the catalytic domains and, in the absence of substrate, adopt a variety of positions with respect to the rest of the protein. Thus, shifting of the domains to bind to RNA is generally thought to be part of the substrate-binding process.
In the P212121 crystal form of the m1A58 MTase complex, the shifted position of the disordered Trm6 insert may allow a picture of the tRNA in the process of binding or release from the enzyme. A symmetry-related molecule interfaces with the Trm6 N-terminal domain in the nonproductive heterodimer, stabilizing it in an orientation rotated approximately 9° from its position in the productive heterodimer, a conformational difference that propagates to the Trm6 insert and prevents its close interaction with the TψC-loop. Crystal-packing interactions may have led to the shifted position.
Discussion
m1A58 MTase uses induced fit to access its target base
Structure–function experiments to determine the importance of tRNA architecture for the activity of tRNA-modifying enzymes in Xenopus laevis are consistent with the refolded tRNA that we see in our structures. These experiments show that the canonical L-shaped structure of tRNA is not compatible with catalysis in m1A58 MTase: tRNA mutations that disrupt the core structure actually increase m1A58 MTase activity [26]. Indeed, a tRNA fragment containing only the T-arm and acceptor stem is as good a substrate in vivo as the whole tRNA [26]. Consistent with this result, the great majority of the protein–tRNA hydrogen bonds in our structures involve the 3′ half of the tRNA, nucleotides 43–73. However, the protein–tRNA interface extends over all subdomains of the tRNA. We propose that m1A58 MTase utilizes this extensive tRNA-binding site to distort the tRNA structure in order to make the TψC-loop accessible for modification. Binding energy to distorted tRNA (estimated in PISA [15] to be 45 kcal/mol) would compensate for disruption of D-loop–TψC-loop interactions. Archaeosine tRNA-guanine transglycosylase, which modifies G15 in the D-loop of tRNA, does not distort L-shaped tRNA upon binding. Rather, it binds to the λ-form, in which base pairing is rearranged to expose G15 for modification [12]. In contrast to these two enzymes that modify sites in the tRNA core, the SAM-dependent tRNA MTase TrmD methylates N1 of G37 after a simple base flip of G37 into the active site [27]. G37 is part of the anticodon loop, which is both solvent accessible and flexible.
Distortion of tRNA upon binding is not unprecedented: aminoacyl-tRNA assumes a highly distorted structure when simultaneously binding to the decoding site of the ribosome and to EF-Tu [28]. A crystal structure of unmodified apo-tRNAPhe has a slightly different conformation than tRNAPhe with posttranscriptional modifications, differing by the angle between the anticodon stem–loop and the acceptor arm of the tRNA [29]. This result suggests that tRNA is inherently flexible, which may contribute to regioselectivity of some tRNA-modifying enzymes.
Heterotetrameric m1A58 MTases diverged from a primordial homotetrameric enzyme
Mutational experiments indicate that bacterial and archaeal m1A58 MTases probably bind to tRNA in a similar manner as does human m1A58 MTase. A primary difference is that the bacterial and archaeal species can bind tRNA against any one of four active sites. These homotetrameric enzymes are evolutionary forbears of human m1A58 MTase. They have similar electropositive surfaces at their putative alternate tRNA-binding sites, and they bind up to two tRNA molecules simultaneously per tetramer [6,30]. As in human m1A58 MTase, they are active against tRNA fragments comprising the 3′ half of the molecule [10,31], and the TψC-loop, acceptor stem and variable loop between the anticodon stem and the T-arm were identified as the major determinants of substrate specificity [10]. The C56 base, which is triply hydrogen bonded to the protein in our structures, is critical for activity in T. thermophilus m1A58 MTase [10]. Finally, when T. thermophilus m1A58 MTase acts on whole tRNAs, both Km and Vmax are lower than when it acts on tRNA fragment substrates, consistent with the proposal that tRNA binding to a large surface compensates for the energy required to detach the TψC-loop from the tRNA core [10].
A major difference between these enzymes and the human ortholog is that they are composed of a single subunit, TrmI. As such, each tRNA-binding site is symmetric and can bind tRNA in equivalent 2-fold related orientations. A second difference is that they have no structure equivalent to the Trm6 N-terminal-domain insert that binds to the T-arm, yet most target A58 exclusively. However, the archaeal Pyrococcus abyssi TrmI methylates both A57 and A58 [32]. These results indicate that the insert may contribute to but is not required for target site specificity.
Is m1A58 MTase a half-of-the-sites enzyme?
In the tetragonal crystal form of m1A58 MTase–tRNA 3 Lys -SAH, the heterodimers in the dimer-of-dimers complex are related by crystallographic symmetry, thus necessarily identical. In the orthorhombic structure of m1A58 MTase–tRNA3 -Lys-SAH, which contains a complete heterotetramer in the asymmetric unit, the heterodimers are not equivalent and the TψC-loops in the two tRNA are diverged to different extents from a catalytically competent conformation: in one heterodimer, A58 is close to optimally aligned with the extrapolated position of the cofactor methyl, while in the other heterodimer, A57 instead of A58 is positioned in the active site. The Trm6 N-terminal domain in the less optimal heterodimer has not moved into contact with the T-arm, and an insert in the N-terminal domain, which interacts with the T-arm through numerous hydrogen bonds in the P43212 complex, is largely disordered. The asymmetric heterotetramer may reflect the solution structure, opening the possibility that human m1A58 MTase is a half-of-the-sites enzyme that methylates A58 in only one active site at a time. Optimized binding for catalysis at one heterodimer may transmit structural changes to the second heterodimer that preclude productive binding there in exchange for higher affinity or specificity at the productive site. Subsequent to methylation at the productive site, optimization of binding at the opposite site may aid in product release.
Conclusion
We report the first structures of a m1A58 MTase bound to a substrate tRNA. Our structures of the human m1A58 MTase in complex with its cognate tRNA3 Lys substrate show how the enzyme makes A58, which is buried in the canonical structure of tRNA, accessible for modification by breaking ternary interactions between the D-loop and the TψC-loop. It identifies the regions throughout the protein that are important for binding and prying open the tRNA and particularly regions at the interface with the TψC-loop that determine specificity for A58 alone. Eukaryotic m1A58 MTases are heterotetramers composed of two distantly diverged but structurally homologous subunits. One subunit of a primordial homodimer diverged to Trm6, which specializes in orientation of the substrate for presentation to the conserved active site of the second subunit, Tmr61.
Materials and Methods
Expression and purification of m1A58 MTase
The expression plasmid, pET17b-6HisTrm6-Trm61, was isolated from a strain kindly provided from Charles S. McHenry and was modified to contain a 3C protease cleavage sequence C-terminal to the 6His tag. m1A58 MTase was expressed in E. coli BL21(DE)3 cells for 3 h at 37 °C after induction with 1 mM isopropyl-β-D-thiogalactoside at an OD600 of 0.6. Cells were lysed using an EmulsiFlex-C3 homogenizer (Avestin) in buffer containing 50 mM Hepes (pH 7.5), 500 mM NaCl, 30 mM imidazole and 5 mM β-mercaptoethanol. The lysate was cleared by centrifugation for 25 min at 32,000g, and the supernatant was filtered through a 0.45-μm filter and was loaded on a 1-ml HisTrap Ni-NTA column (GE Healthcare). After washing with 50 ml of 50 mM Hepes (pH 7.5), 500 mM NaCl, 5 mM β-mercaptoethanol and 30 mM imidazole, we eluted protein with a linear imidazole gradient (30–400 mM) in the same buffer.
Fractions containing m1A58 MTase were pooled and dialyzed against 50 mM Hepes (pH 7.5), 150 mM NaCl and 1 mM tris(2-carboxyethyl)phosphine for 16 h at 4 °C in the presence of 3C protease. The protein was concentrated and loaded on a Superdex 200 (10/30) column equilibrated in 50 mM Hepes (pH 7.5) and 150 mM NaCl. Peak fractions of m1A58 MTase were pooled, concentrated to 13 mg/ml, flash frozen in liquid nitrogen and stored at −80 °C.
In vitro transcription of tRNA3 Lys
tRNA3 Lys was in vitro transcribed from a PCR-amplified template using the T7 promoter and polymerase system, as well as HDV ribozyme as described in Walker et al. [33]. An extra base at the 5′ end was set to G to increase the yield of the in vitro transcription reaction. After in vitro transcription, RNA was extracted by phenol-chloroform, ethanol precipitated using sodium acetate and applied to denaturing urea polyacrylamide gel electrophoresis. RNA was visualized by UV shadowing and the band corresponding to tRNA3 Lys was excised from the gel. RNA was gel extracted over night at room temperature in 50 mM Tris–HCl (pH 7.5), 300 mM NaCl and 10 mM ethylenedi-aminetetraacetic acid, and the solution was passed through a 0.22-μm filter. After ethanol precipitation using sodium acetate, RNA was resuspended in water to a concentration of 7.6 mg/ml. Purified tRNA3 Lys was stored at −20 °C.
Crystallization and data collection
Two crystal forms of the m1A58 MTase–tRNA3 Lys-cofactor complexes were grown at 16 °C by the hanging-drop vapor diffusion method against 0.1 M Na acetate (pH 4.8–5.0), 2% (w/v) polyethylene glycol 4000 and 15% (v/v) methyl-2,4-pentanediol. A 1-μl protein solution [4.8 mg/ml protein (27.7 μM tetramer), 66.4 μM tRNA3 Lys, 2 mM SAM or SAH, 1 mM MgCl2 and 50 mM Hepes (pH 7.5)] was mixed with a 1-μl-well solution and suspended over a 1000-μl reservoir solution. Crystals appeared after 4 days and reached a maximum size of 100–300 μm in about a week. Before data collection, tetragonal crystals were briefly soaked in cryoprotectant consisting of reservoir solution with 25% (v/v) methyl-2,4-pentanediol; data for the orthorhombic crystal form were collected without additional cryoprotection. Data were collected at the Lawrence Berkeley National Laboratory Advanced Light Source Beamline 8.3.1 at −170 °C using λ = 1.11587 Å. Data were processed using XDS [34]. One crystal was used for each data set.
High-resolution cutoffs for the data sets were determined based on CC1/2 statistics [35]. The CC1/2 cutoff values for the highest-resolution bins of the tetragonal substrate complex, the product complex and the orthorhombic complex were 39%, 35% and 30%, respectively; thus, these resolution shells contained a significant number of observed reflections, which contributed to signal in the density maps, and we used all the processed data for structure determination. For the final refinements, we truncated the data at resolutions where the average intensity was greater than or equal to 2σ.
Structure solution and refinement
The structure of the P43212 crystal form of m1A58 MTase–tRNA3 Lys-SAH (2.0 Å) was solved by molecular replacement using a dimer of T. thermophilus m1A58 MTase [6] (PDB ID: 2PWY) as a search model, using Phaser [36] from the PHENIX [37] suite of programs. The molecular replacement solution was very incomplete, lacking both tRNA and the protein inserts, and substantial regions of Trm61 had to be rebuilt in different conformations. Density fitting with Coot [38] was alternated with refinement of atomic positions, individual B-factors and occupancies against a maximum likeli-hood target in phenix.refine [37]. The RCrane [39] extension in Coot was used to aid refinement of the phosphate backbone conformation of the tRNA. Hydrogen atoms were included in riding positions during refinement. Refinement was guided by evaluation of the structures in MolProbity [40]. Residues 254–260 in Trm61 and residues 1–17, 81–87, 272–339 and 464–497 from Trm6 are missing in the final structure because there was no density for them. The structure refined at 2.2 Å has R/Rfree = 18.8/21.5%, 97% of residues in the most favored regions of the Ramachandran plot, no Ramachandran outliers, eight poor rotamers and a clash score of 7.2 overlaps/1000 atoms.
The structure of the P212121 crystal form of m1A58 MTase–tRNA3 Lys-SAH was determined by molecular re placement using the P43212 structure as search model. The structure of the product complex was solved by isomorphous replacement using the structure of the P43212 crystal form of m1A58 MTase–tRNA3 Lys-SAH. Both structures were refined by the same methods as the tetragonal m1A58 MTase–tRNA3 Lys-SAH structure. The product complex structure is missing residues 254–259 from Trm61 and residues 1–17, 81–87, 272–339 and 464–497 from Trm6. The structure refined at 2.3 Å has R/ Rfree = 19.2/22.6%, and there are 96% of residues in the most favored regions of the Ramachandran plot, no Ramachandran outliers, five poor rotamers and a clash score of 7.5 overlaps/1000 atoms. The P212121 complex structure is missing residues 254–260 from both Trm61 subunits; residues 1–17, 79–87, 272–339 and 464–497 from one Trm6 subunit; and residues 1–17, 78–132, 272–339 and 464–497 from the other Trm6 subunit. The structure refined at 4.05 Å has R/Rfree = 21.0/24.8%, and there are 93% of residues in the most favored regions of the Ramachandran plot, four Ramachandran outliers, 18 poor rotamers and a clash score of 1.3 overlaps/1000 atoms.
Structure analysis
Conserved domains between structures were identified by difference-distance methods using RAPIDO [18]. The PISA server [15] was used to analyze the structural and chemical properties of protein–tRNA and protein–protein interfaces and to estimate dissociation energies of complexes. Sequences of TrmI, Trm6 and Trm61 were aligned using CLUSTALW and edited by hand based on comparison of the structures. Figures 2, 3a, 4, 5 and 6 and Supplementary Figs. 1b and 2 were prepared using PyMOL (The PyMOL Molecular Graphics System, Version 1.4.1, Schrodinger, LLC); Supplementary Fig. 1d was prepared with ESPript [41]. The electrostatic surface in Fig. 3a was calculated in PyMOL using APBS [42]. Figures 1 and 3b and c and Supplementary Fig. 1a and c were made with the UCSF Chimera package [43,44].
Supplementary Material
Highlights.
How tRNA m1A58 MTase binds tRNA was unknown.
A structure of the human enzyme heterotetramer bound to two tRNAs was determined.
One subunit is catalytic, and the other one has diverged to have only a tRNA-binding role.
tRNA is distorted upon binding, allowing the target to bind to the active site.
Results show how a tRNA-modifying enzyme accesses a base buried in the tRNA core.
Acknowledgments
We thank Charles McHenry for providing the pET17b-6HisTrm6-Trm61 expression strain plasmid. We thank John Pak, Hemant Kumar and James Holton for assistance with data collection and Bryan Schmidt for critical reading of the manuscript. This work was supported by National Institutes of Health (GM51232 and GM0082250 to R.M.S.) and Deutsche Forschungsgemeinschaft (CZ 205/1-1 to N.C.). Chimera is developed by the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco (supported by National Institute of General Medical Sciences grant P41-GM103311). We are grateful to the University of California Office of the President, Multicampus Research Programs and Initiatives grant MR-15-338599 and the Program for Breakthrough Biomedical Research, which is partially funded by the Sandler Foundation for support of Advanced Light Source Beamline 8.3.1.
Glossary
- m1A58 MTase
tRNA m1A58 methyltransferase
- m1A
1-methyladenosine
- SAM
S-adenosyl-L-methionine
- SAH
S-adenosyl-L-homocysteine
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2015.10.005.
Footnotes
Accession codes
Coordinates and structure factors for the P43212 substrate/SAH complex, the P212121 substrate/SAH complex and the product complex have been deposited in the Protein Data Bank with PDB IDs: 5CCB, 5CD1 and 5CCX, respectively.
References
- 1.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marquet R, Isel C, Ehresmann C, Ehresmann B. tRNAs as primer of reverse transcriptases. Biochimie. 1995;77:113–124. doi: 10.1016/0300-9084(96)88114-4. [DOI] [PubMed] [Google Scholar]
- 3.Renda MJ, Bradel-Tretheway B, Planelles V, Bambara RA, Dewhurst S. Inhibition of HIV type 1 replication using lentiviral-mediated delivery of mutant tRNA(Ly-s3)A58U. AIDS Res Hum Retrovir. 2004;20:1324–1334. doi: 10.1089/aid.2004.20.1324. [DOI] [PubMed] [Google Scholar]
- 4.Yu W, McCulley A, Morrow CD. Mutations in the TPsiC loop of E. coli tRNALys,3 have varied effects on in trans complementation of HIV-1 replication. Virol J. 2007;4:5. doi: 10.1186/1743-422X-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozanick S, Krecic A, Andersland J, Anderson JT. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA. 2005;11:1281–1290. doi: 10.1261/rna.5040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barraud P, Golinelli-Pimpaneau B, Atmanene C, Sanglier S, Van Dorsselaer A, Droogmans L, Dardel F, Tisne C. Crystal structure of Thermus thermophilus tRNA m1A58 methyltransferase and biophysical characterization of its interaction with tRNA. J Mol Biol. 2008;377:535–550. doi: 10.1016/j.jmb.2008.01.041. [DOI] [PubMed] [Google Scholar]
- 7.Gupta A, Kumar PH, Dineshkumar TK, Varshney U, Subramanya HS. Crystal structure of Rv2118c: An AdoMet-dependent methyltransferase from Mycobacterium tuberculosis H37Rv. J Mol Biol. 2001;312:381–391. doi: 10.1006/jmbi.2001.4935. [DOI] [PubMed] [Google Scholar]
- 8.Guelorget A, Roovers M, Guerineau V, Barbey C, Li X, Golinelli-Pimpaneau B. Insights into the hyperthermostability and unusual region-specificity of archaeal Pyrococcus abyssi tRNA m1A57/58 methyltransferase. Nucleic Acids Res. 2010;38:6206–6218. doi: 10.1093/nar/gkq381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuratani M, Yanagisawa T, Ishii R, Matsuno M, Si SY, Katsura K, Ushikoshi-Nakayama R, Shibata R, Shirouzu M, Bessho Y, Yokoyama S. Crystal structure of tRNA m(1)A58 methyltransferase TrmI from Aquifex aeolicus in complex with S-adenosyl-L-methionine. J Struct Funct Genom. 2014;15:173–180. doi: 10.1007/s10969-014-9183-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takuma H, Ushio N, Minoji M, Kazayama A, Shigi N, Hirata A, Tomikawa C, Ochi A, Hori H. Substrate tRNA recognition mechanism of eubacterial tRNA (m1A58) methyltransferase (TrmI) J Biol Chem. 2015;290:5912–5925. doi: 10.1074/jbc.M114.606038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, Klug A. Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature. 1974;250:546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- 12.Ishitani R, Nureki O, Nameki N, Okada N, Nishimura S, Yokoyama S. Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell. 2003;113:383–394. doi: 10.1016/s0092-8674(03)00280-0. [DOI] [PubMed] [Google Scholar]
- 13.Bujnicki JM. In silico analysis of the tRNA:m1A58 methyltransferase family: Homology-based fold prediction and identification of new members from Eubacteria and Archaea. FEBS Lett. 2001;507:123–127. doi: 10.1016/s0014-5793(01)02962-3. [DOI] [PubMed] [Google Scholar]
- 14.Chambers JL, Stroud RM. Difference Fourier refinement of the structure of DIP-trypsin at 1.5 Å with a minicomputer technique. Acta Crystallogr, Sect B: Struct Sci Cryst Eng Mater. 1977;33:1824–1837. [Google Scholar]
- 15.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 16.Ozanick SG, Bujnicki JM, Sem DS, Anderson JT. Conserved amino acids in each subunit of the heteroligomeric tRNA m1A58 MTase from Saccharomyces cerevisiae contribute to tRNA binding. Nucleic Acids Res. 2007;35:6808–6819. doi: 10.1093/nar/gkm574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mosca R, Brannetti B, Schneider TR. Alignment of protein structures in the presence of domain motions. BMC Bioinf. 2008;9:352. doi: 10.1186/1471-2105-9-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosca R, Schneider TR. RAPIDO: A Web server for the alignment of protein structures in the presence of conformational changes. Nucleic Acids Res. 2008;36:W42–W46. doi: 10.1093/nar/gkn197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malone T, Blumenthal RM, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J Mol Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 20.Schluckebier G, O'Gara M, Saenger W, Cheng X. Universal catalytic domain structure of AdoMet-dependent methyltransferases. J Mol Biol. 1995;247:16–20. doi: 10.1006/jmbi.1994.0117. [DOI] [PubMed] [Google Scholar]
- 21.Kim SH, Quigley GJ, Suddath FL, McPherson A, Sneden D, Kim JJ, Weinzierl J, Rich A. Three-dimensional structure of yeast phenylalanine transfer RNA: Folding of the polynucleotide chain. Science. 1973;179:285–288. doi: 10.1126/science.179.4070.285. [DOI] [PubMed] [Google Scholar]
- 22.Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37:D159–D162. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saikia M, Fu Y, Pavon-Eternod M, He C, Pan T. Genome-wide analysis of N1-methyl-adenosine modification in human tRNAs. RNA. 2010;16:1317–1327. doi: 10.1261/rna.2057810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunkle JA, Vinal K, Desai PM, Zelinskaya N, Savic M, West DM, Conn GL, Dunham CM. Molecular recognition and modification of the 30S ribosome by the aminoglycoside-resistance methyltransferase NpmA. Proc Natl Acad Sci U S A. 2014;111:6275–6280. doi: 10.1073/pnas.1402789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hur S, Stroud RM, Finer-Moore J. Substrate recognition by RNA 5-methyluridine methyltransferases and pseudouridine synthases: A structural perspective. J Biol Chem. 2006;281:38969–38973. doi: 10.1074/jbc.R600034200. [DOI] [PubMed] [Google Scholar]
- 26.Grosjean H, Edqvist J, Straby KB, Giege R. Enzymatic formation of modified nucleosides in tRNA: Dependence on tRNA architecture. J Mol Biol. 1996;255:67–85. doi: 10.1006/jmbi.1996.0007. [DOI] [PubMed] [Google Scholar]
- 27.Ito T, Masuda I, Yoshida K, Goto-Ito S, Sekine S, Suh SW, Hou YM, Yokoyama S. Structural basis for methyl-donor-dependent and sequence-specific binding to tRNA substrates by knotted methyltransferase TrmD. Proc Natl Acad Sci U S A. 2015;112:E4197–E4205. doi: 10.1073/pnas.1422981112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FV, Weir JR, Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Byrne RT, Konevega AL, Rodnina MV, Antson AA. The crystal structure of unmodified tRNAPhe from Escherichia coli. Nucleic Acids Res. 2010;38:4154–4162. doi: 10.1093/nar/gkq133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guelorget A, Barraud P, Tisne C, Golinelli-Pimpaneau B. Structural comparison of tRNA m(1)A58 methyltransferases revealed different molecular strategies to maintain their oligomeric architecture under extreme conditions. BMC Struct Biol. 2011;11:48. doi: 10.1186/1472-6807-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamazaki N, Hori H, Ozawa K, Nakanishi S, Ueda T, Kumagai I, Watanabe K, Nishikawa K. Substrate specificity of tRNA (adenine-1-)-methyltransferase from Thermus thermophilus HB27. Biosci Biotechnol Biochem. 1994;58:1128–1133. doi: 10.1271/bbb.58.1128. [DOI] [PubMed] [Google Scholar]
- 32.Hamdane D, Guelorget A, Guerineau V, Golinelli-Pimpaneau B. Dynamics of RNA modification by a multi-site-specific tRNA methyltransferase. Nucleic Acids Res. 2014;42:11697–11706. doi: 10.1093/nar/gku820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker SC, Avis JM, Conn GL. General plasmids for producing RNA in vitro transcripts with homogeneous ends. Nucleic Acids Res. 2003;31:e82. doi: 10.1093/nar/gng082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kabsch W. Xds. Acta Crystallogr, Sect D: Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: A comprehensive Python-based system for macromolecular structure solution, Acta Crystallogr. Sect D: Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr, Sect D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Keating KS, Pyle AM. RCrane: Semi-automated RNA model building. Acta Crystallogr, Sect D: Biol Crystallogr. 2012;68:985–995. doi: 10.1107/S0907444912018549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr, Sect D: Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 42.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 44.Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.