Abstract
Members of the receptor tyrosine kinases (RTK) regulate important cellular functions such as cell growth and migration, which are key steps in angiogenesis, organ morphogenesis, and in the unregulated states, cancer formation. One long-standing puzzle regarding RTKs centers on how the extracellular domain (ECD), which detects and binds to growth factors, is coupled with the intracellular domain (ICD) kinase activation. While extensive structural works on the soluble portions of RTKs have provided critical insights into RTK structures and functions, lack of a full-length receptor structure has hindered a comprehensive overview of RTK activation. In this study, we successfully purified and determined a 27-Å resolution structure of a full-length human Platelet-Derived-Growth-Factor Receptor Beta (PDGFRβ), in complex with its ligand PDGF-B. In the ligand-stimulated complex, two PDGFRβs assemble into a dimer via an extensive interface essentially running along the full-length of the receptor, suggesting that the membrane-proximal region, the transmembrane helix, and the kinase domain of PDGFRβ are involved in dimerization. Major structural differences are seen between the full-length and soluble ECD structures, rationalizing previous experimental data on how membrane-proximal domains modulate receptor ligand-binding affinity and dimerization efficiency. Also, in contrast to the two-fold symmetry of the ECD, the intracellular kinase domains adopt an asymmetric dimer arrangement, in agreement with prior observations for the closely related KIT receptor. In essence, the structure provides a first glimpse into how PDGFR ECD, upon ligand-stimulation, is coupled to its ICD kinase activation.
Keywords: receptor tyrosine kinase, membrane protein structure, electron microscopy, cancer, signal transduction
Graphical Abstract

Introduction
Platelet-Derived Growth Factors (PDGFs) are a family of four cystine-knot growth factors (PDGF-A, -B, -C and -D) that control the growth of connective tissue cells such as fibroblasts and smooth muscle cells 1; 2. By acting on these mesenchymal cells, PDGFs critically regulate embryonic development, especially the formation of vessels and organs (reviewed in 3). While PDGF-PDGFR signaling plays important roles during developmental stages, the expression of both PDGFs and PDGFRs are tightly controlled in adulthood. Enhanced PDGF-PDGFR signaling, except when happening briefly during wound repair, is generally considered abnormal, and is an important feature in a number of diseases involving proliferation, including many types of cancers, inflammation, pulmonary fibrosis and restenosis, and notably atherosclerosis 4.
The two receptors for PDGFs, PDGFRα and PDGFRβ, belong to the Class III receptor tyrosine kinases (RTKs), a clan of five members including PDGFRα and PDGFRβ, KIT, CSF1R (FMS) and FLT3 5. These Class III RTKs feature five immunoglobulin-like (Ig) domains in the extracellular segment (D1-D5), a single-pass transmembrane domain, and a split kinases domain that contains an insert of variable lengths 6; 7. Notably, PDGFRα and PDGFRβ bind to ligands of the cystine-knot fold rather than the helical-bundle fold as for KIT, CSF1R/FMS and FLT3 8; 9; 10; 11; 12; 13.
We have previously determined the crystal structure of PDGF-B in complex with the first three Ig domains of PDGFRβ, PDGFRβD1-D3. The structure shows how the D2 and D3 domains of the receptor form the primary ligand recognition sites. The overall assembly of the 2:2 PDGF-B/PDGFRβD1-D3 recognition complex resembles other ligand/Class III RTK complexes such as SCF/KIT and CSF1/CSF1R (FMS) 8; 9; 10; 12; 13. However, the overall domain arrangements required to relay ligand engagement at the extracellular side to the kinase activation in the intracellular side still remained unclear without further structural information from the full-length ligand-receptor complex. In this regard, this paper provides an initial structural characterization of a complex consisting of PDGF-B bound to its cognate receptor PDGFRβ in its entirety (PDGFRβFL). Combining with available crystal structures and the recently determined SCF/KIT EM structure 14, the current EM reconstruction provides insights into the general mechanism of Class III RTK activation.
Results and Discussion
FSEC screening for Class III RTKs
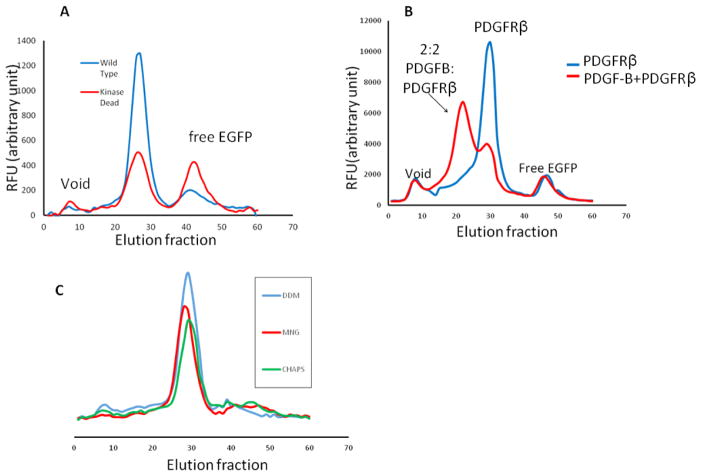
To ensure a native environment for folding and post-translational modifications, we employed the BacMam technology to express human RTKs in HEK293GnTI− cells. To effectively screen for expression constructs and purification conditions, we constructed a BacMam-based plasmid15, pFNsfEG, with a Gaussia luciferase secretion signal peptide, a FLAG tag, followed by a superfolder-EGFP (sfEGFP) for florescence-coupled size exclusion chromatography (FSEC) (Fig. S1A and S1B). The superfolder variant of EGFP was employed to withstand the oxidative extracellular environment that tends to denature traditional EGFP molecules 16. We systematically screened for PDGFRα, PDGFRβ, and CSF1R/FMS receptors for expression. Both the wild types (WT) and the kinase-dead (K634A) mutants were screened; we hypothesized that the kinase-dead mutants, due to lack of auto-phosphorylation and consequent kinase phosphorylation-induced receptor endocytosis/degradation, would give higher expression yields. Yet, among all constructs screened, the human PDGFRβ WT exhibits the highest expression level, and was therefore chosen for subsequent structural analysis. As expected, detergent-extracted receptor remains monomeric, and PDGF-B addition drives receptor dimerization (Fig. 1B). Also, detergent screen with FSEC shows that a wide variety of detergents were able to extract and solubilize human PDGFRβ with a monodisperse peak. Among the detergents screened, non-ionic detergent Lauryl Maltose Neopentyl Glycol (LMNG) yields the sharpest and the most symmetric gel-filtration peak, and therefore was chosen for downstream PDGFRβ solubilization and purification (Fig. 1C).
Figure 1. FSEC screening of PDGFRβ.
(A) WT versus kinase-dead mutant of PDGFRβ. (B) PDGF-B induced dimerization probed by FSEC. (C) Detergent screen of PDGFRβ.
Purification of PDGF-B/PDGFRβFL and negative-stain EM
LMNG detergent solubilized PDGF-B/PDGFRβFL complex was captured and eluted using anti-FLAG M2 immuno-affinity chromatography (Fig. S2A, S2B). The purified receptor retained capability for catalyzing auto-phosphorylation (Fig. S2C). Despite the size-exclusion step to get rid of high molecular weight aggregates, however, the sample still displayed low contrast and reformed aggregates under initial negative-stain EM analysis. We overcame the problem by using the GraFix17 method to slightly cross-link the complex; GraFix-treated sample exhibited significantly enhanced contrast and much improved structural integrity (Fig. S3A and S3B).
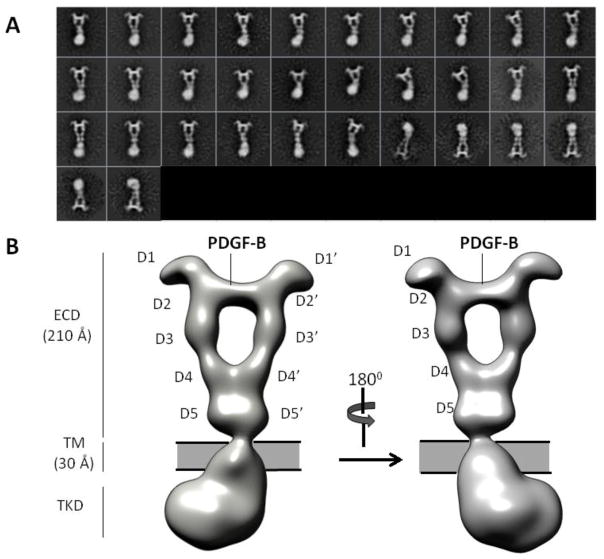
After 2D classifications of an initial small dataset (~2000 particles), we obtained initial models by the random-conical-tilt (RCT) approach (Fig. S4). Various RCT initial models were deemed similar as judged by visual inspection of reconstructed volumes and by analysis in the Xmipp package (Fig. S4, also Materials and Methods). The best initial model was further refined with the 3D classification/auto-refinement scheme in RELION to a resolution of 27-Å according to the “Gold-standard” Fourier shell correlation 0.143 (FSC 0.143) criterion (Fig. S6, Materials and Methods)18. This map reveals for the first time, the overall architecture of an intact ligand-bound PDGFRβ receptor dimer (Fig. 2B). The following discussion will highlight features unique to PDGFRβ, and at the same time provide a more refined view on the general structural features of Class III RTKs.
Figure 2. Overall architecture of PDGF-B/PDGFRβFL.
(A) Representative 2D class averages. (B) 3D Reconstruction.
Overall architecture of PDGF-B/PDGFRβFL complex
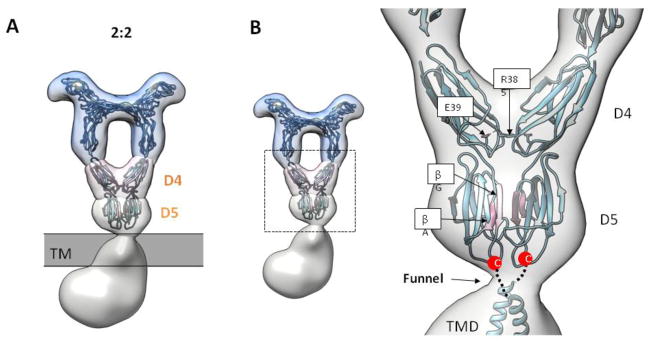
The architecture of the entire PDGF-B/PDGFRβFL complex comprises three layers: extracellular domain (ECD), transmembrane domain (TMD), and tyrosine kinase domain (TKD). The ECD layer exhibits a near 2-fold symmetry around an axis running along the vertical length of the entire ligand-receptor complex. Consistent with a model of ligand-mediated receptor dimerization, two receptor protomers are bridged by a divalent PDGF-B located at the center top portion of the complex. Characteristic of the ECD density is a large cavity bordered by PDGF-B at the top, two D3 domains at the sides, and two D4 domains at the bottom (Fig. 2A and B).
The ~21-nm long ECD can be further divided into three sub-layers: the head layer, the D4 layer, and the D5 layer. The head layer consists of receptor D1-D3 domains and is the main ligand-receptor interacting site. This layer displays the strongest signals in the 2D class averages and in our reconstructed EM map even under high map contour levels, and shows features consistent with the PDGF-B/PDGFRβD1-D3 crystal structure determined earlier (PDB ID: 3MJG)19 (Fig. 2A and B). The strong contour is reflective of the extensive ligand-receptor interface (~roughly 2,900-Å2) as shown in the crystal structure. Characteristically, the two D1 domains protrude from each side of the complex at the top of the head layer like two ears, and are the only parts of the whole complex that are not involved in either direct ligand-receptor recognition or receptor dimerization. Yet, the well-defined density for D1 suggests structural rigidity, consistent with the tight association between D1 and D2 (the D1-D2 module) seen in the crystal structure.
The D4 and D5 layers constitute the membrane-proximal part of the ECD, which has thus far not been structurally characterized for either PDGFRα or PDGFRβ. The D4 layer showcases two D4 domains engaged in receptor-receptor homotypic contact in a V-shaped conformation, confirming the conserved D4-D4 homotypic contact as predicted by earlier bioinformatics analysis and functional studies 20. Below the D4 layer, the ECD density tapers down to a close-knit density of the D5 layer. The shape of the D5 layer is most consistent with the two D5 domains also involved in close homotypic receptor interaction, a novel feature that has not been previously shown for the PDGFR receptor (more discussion in the section “Membrane-proximal D5 domain in full-length receptor dimer”).
The density below D5 comprises the TMD-TKD layers. Density segmentation analysis indicates that the enclosed volume of this region is too large for a single copy of TMD-TKD, and therefore most likely represents the volume of two interacting TMD-TKD regions from two receptor protomers. Taken together, except D1, all domains in PDGFRβ mediate dimerzation, either directly through receptor-receptor contacts (D4, D5, TMD, TKD), or indirectly through ligand binding (D2-D3). Importantly, receptor dimerization is driven entirely by PDGF-B binding, without which receptors are primarily monomeric as shown by FSEC (Fig. 1B). Formation of such an extensive dimer interface in a ligand-dependent manner indicates considerable coupling among disparate receptor domains in receptor activation. The following discussions will highlight examples of structural coupling as revealed by the EM structure.
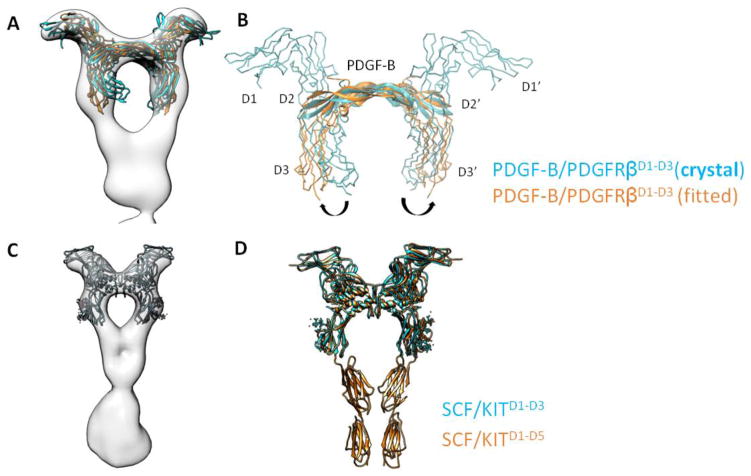
Rearrangement of ligand-engaging D2-D3 domains in full-length receptor
Notably, the orientation of the D3 in the full-length receptor differs from that of D3 in the earlier crystal structure of PDGF-B/PDGFRβD1-D3 containing only the first three Ig domains of PDGFRβ. In the crystal structure, the two D3 domains curve inward in an orientation that is incompatible with D4-D4 homotypic contacts shown in the EM reconstruction (Fig. 3A). We then performed flexible fitting aided by molecular dynamics (MDFF) to fit PDGF-B/PDGFRβD1-D3 crystal structure into the full-length EM map. In the simulation, the D3 domain undergoes a rotation around the D2-D3 hinge. Rotation of D3 is also accompanied by a minor rigid-body rotation of the D1-D2 module around the same D2-D3 hinge so that D2 and D3 can maintain a firm grip on PDGF-B (Fig. 3A and B). For PDGF-B, slight conformational changes are also observed in the receptor binding loops to accommodate the remodeled D2-D3 geometry. This finding highlights the potential flexibility of the D2-D3 hinge that becomes less flexible in the context of a full-length ligand-receptor dimer, which may explain our prior findings where we noticed a difference in PDGF-C binding affinities between PDGFRαD1-D3 versus a longer PDGFRαD1-D5 construct containing the entire receptor ECD19. Interestingly, we have previously found that Macrophage Colony-Stimulating Factor 1(M-CSF1) displays greater than 50 fold stronger affinity towards CSF1RD1-D5 than to CSF1RD1-D3, which may be explained by the similarly remodeled D2-D3 ligand-clamping site in the longer CSF1R D1-D5 construct8.
Figure 3. Conformation of D2-D3 in full-length receptor.
(A and B) Comparison between PDGFRβD1-D3 in crystal structure (cyan) versus PDGFRβD1-D3 after MDFF fitting in EM map (orange); the latter is representative of D2-D3 conformation in a full-length receptor. (C and D) Similar comparison for KIT receptor.
As a sharp comparison, the D1-D3 domains in SCF/KITD1-D3 adopt almost the same orientation as those in SCF/KITD1-D5, with a root-mean-square-deviation (r.m.s.d) of 0.9 Å for 256 residues compared (Fig. 3C and D). Accordingly, studies from Schlessinger’s group found no difference in measured affinities between the two KIT extracellular constructs towards SCF 21. What distinguishes KIT from PDGFR and CSF1R likely originates from the more extensive and less flexible D2-D3 inter-domain interface in KIT10. In this regard, free KIT D2-D3 already adopts a conformation primed for ligand binding. On the other hand, at least for PDGFR and CSF1R, although D2 and D3 are sufficient for ligand binding, the rest of the receptor, most likely the membrane-proximal D4-D5 domains, also affects ligand-binding affinity by helping fixate the conformation of the flexible ligand-binding D2-D3 domains.
Receptor homotypic contact reinforces receptor dimerization
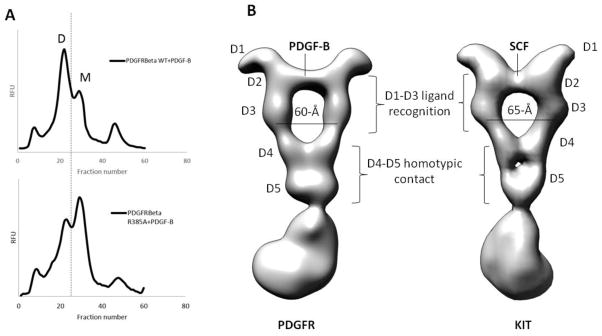
As shown in the EM reconstruction, PDGF receptor dimerization consists of two types of interactions: 1) dimeric PDGF ligand engaging the D2-D3 region and 2) homotypic receptor contacts involving the membrane-proximal D4-D5, TMD, and TKD regions. Because FSEC showed that ligand addition seems to be the primary driving force for receptor dimerization, we tested the additional role of homotypic contacts in PDGFR dimerization.
Based on sequence comparison with KIT, we designed full-length receptor mutants disrupting the conserved membrane-proximal D4-D4 homotypic interaction (R385A) identified in the SCF/KITD1-D5 crystal structure. After cell lysis, PDGF-B was added to membrane fractions to allow ligand-receptor complex formation in a native membrane environment before detergent extraction. As shown in FSEC, R385A mutant still leads to PDGF-induced dimerization. Yet, R385A mutation results in reduced receptor dimerization in comparison with the wild-type receptor (Fig. 4A), a finding that is independent of receptor expression level and amount of ligand added. The data demonstrate that ligand binding may represent the primary driving force for dimerization, which is further reinforced by membrane-proximal homotypic contacts. Although the molecular basis is not clear, but based on the discussion in the previous section suggesting how D2-D3 orientation is influenced by the presence or absence of D4-D5 region, the membrane-proximal contact might reinforce the optimal D2-D3 geometry most compatible in a 2:2 complex.
Figure 4. Influence of D4 on receptor dimerization, and common geometric requirement for Class III RTK dimerization.
(A) Influence of R385A that disrupts D4-D4 homotypic contact on receptor dimerization. D = ligand-induced dimer. M = receptor monomer. (B) PDGFR (left) versus KIT (right) full-length structure. Whereas D1-D3 are different for the two receptors to accommodate different ligands, the D4-D5 homotypic contact is conserved.
Common geometric requirement for Class III RTK activation
What distinguishes PDGFRs from other Class III RTK members (KIT, FMS/CSF1R, and FLT3) is that PDGFRs are activated by growth factors of the cystine-knot fold rather than the helical-bundle cytokine family. Reflective of the planar β-sheet conformation characteristic of the cystine-knot fold, the PDGF-B density appears to be flatter and more elongated in comparison with the more compact SCF density in the SCF/KITFL structure14 (Fig. 3A versus C, also Fig. 4B). Also, whereas PDGFR D1 is not involved in ligand contact, D1 in KIT receptor forms part of the ligand-interacting interface. Accordingly, there is a more prominent protruding D1 contour in the PDGF-B/PDGFRβFL reconstruction in comparison with the more retracted KIT D1 density. The differences highlight how Class III RTKs have evolved different D1-D3 modules for recognizing diverse ligands (Fig. 4B).
Despite differences at the ligand-engagement D1-D3 module, both KIT and PDGFRβ dimer ECD exhibit a signature “elbow” conformation due to a ~ 120-degree bent angle at the D3-D4 junction (Fig. 4B). From the high-resolution x-ray structure of SCF/KITD1-D5, the bent angle stems from specific interactions between D3 and D410. Significantly, this D3-D4 elbow leads to a separation of 60–65-Å between two D3 domains and anchors the downstream two D4 domains in a proper orientation that allows them to interact via a complementary interface (Fig. 4B). Similar D3-D4 junction elbow configuration also appears in the low-resolution EM reconstruction and SAXS models of M-CSF1/CSF1RD1-D5 and IL-34/CSF1RD1-D5 complexes 22, as well as in the x-ray model of FL/FLT3 complex11. Although we currently do not have a full-length structure of a ligand activated CSF1R, or FLT3, we expect that the characteristic D3-D4 conformation will hold for the full-length CSF1R and FLT3 as well.
In essence, while the D1-D3 ligand-binding modules adopt various geometries to suit diverse ligands for different receptor families, a seemingly invariant D3-D4 conformation has been observed across Class III RTK members. This observation indicates that D3-D4 hinge might play an essential role in coupling the ligand-biding information at D3 to the D4 domain involved in homotypic interaction.
Rearrangement of membrane-proximal D5 domain in full-length receptor dimer
While the importance of D4-D4 homotypic contact is well established in Class III RTKs, the precise role of D5 domain remains to be clarified. In the SCF/KITD1-D5 X-ray crystal structure (PDB ID: 2E9W), both D5 domains do not have obvious interface and are more flexible than the D4 domains as evidenced by the weaker electron density and higher B-factor 10. Similarly, while low-resolution EM and SAXS structures of M-CSF1-bound CSF1RD1-D5 dimers demonstrate a conserved D4-D4 homotypic contact, a large 65-Å separation between the neighboring D5 domains is observed 23. The uncertainty regarding D5 conformations raises the question whether transmembrane and kinase domain dimerization is required for RTK activation.
In contrast to the aforementioned isolated receptor ECD structures, the reconstructed EM volume for the ligand-stimulated PDGFRβFL demonstrates a clear D5-D5 interaction (Fig. 5A). The compact and contiguous nature of the D5 layer density suggests that the D5-D5 interface is larger than the D4-D4 interface. Recent EM structure of SCF/KITFL also confirms our finding regarding the D5 homotypic contact14. Despite the low sequence similarity between KIT D5 and PDGFRβ D5, the same homotypic interaction found in both receptors is highly indicative of a universal activation mechanism for disparate Class III RTK members.
Figure 5. PDGFR D5 homotypic contact.
(A) Full-length PDGF-B/PDGFRβ structure stratified into different layers. The D4 and D5 layers highlight the homotypic contact established by MDFF simulation. (B) Closed-up view for the MDFF simulation, with the interface strands highlighted in pink.
To gain more structural insight, we generated a PDGFRβ D4-D5 dimer model using KIT D4-D5 (PDB ID: 2E9W) and fit it into our EM density with molecular dynamics/flexible fitting (MDFF). During the short 0.5-ns simulation process, the two D5 domains rotates around the D4-D5 linker and comes close together to form an interface mediated by strand βA and βG (Fig. 5B). Importantly, D5 domains of Class III RTKs are hotspots for cancer mutations. Several point mutations found in cancer patients can be mapped to strands βA that mediate the interface24. This suggests that cancer mutations in the D5 domains likely enforce receptor dimerization in a ligand-independent manner. The establishment of this novel D5-D5 interface therefore represents another level of conformation coupling in the activation of a full-length receptor.
Transmembrane domain dimerization in full-length receptor
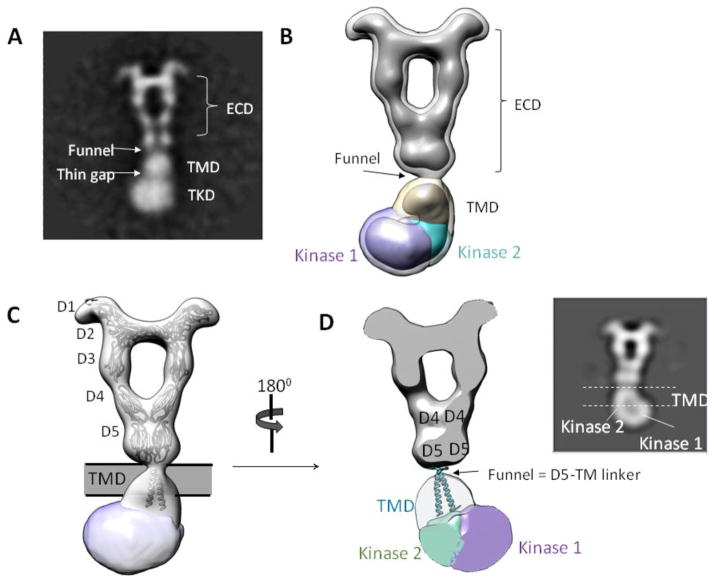
Although isolated RTK transmembrane helices oligomerize to different degrees in various reconstituted bicelle systems, TM dimerization in the context of a full-length receptor has not been demonstrated. Below the D5 layer of the current EM reconstruction, the density thins to a narrow “funnel”, suggesting that the two D5 C-termini are close to one another (Fig. 5B). In the flexible fitting model, the two D5 C-termini are within 16-Å, a distance that is close enough for downstream TM dimerization.
In the 2D class averages of PDGF-B/PDGFRβFL, the circular silhouette below D5 domains (the TMD layer) most likely represents the large LMNG detergent micelle surrounding two TM helices (Fig. 6A). In this manner, the “funnel” between the ECD and the TMD densities should represent the short linker that connects the ECD to the beginning of the transmembrane helix. Our 3D reconstruction provides evidence for our interpretation of the 2D analysis (Fig. 6B). Density analysis by SEGGER shows that the density below the ECD spans a vertical distance of ~30–35-Å consistent with the vertical length of a biological membrane bilayer (the TMD layer). Rigid-body docking of PDGFRβ TM dimer structure determined by NMR 25 nicely positions the structure into the density of the TMD layer (Fig. 6C), although the real TM conformation in the full-length receptor will likely vary from the NMR structure.
Figure 6. Conformation of TMD-TKD layer.
(A) 2D class average showing features of PDGF-B/PDGFRβ TMD-TKD. (B) EM reconstruction agrees with 2D class average. (C) Fitting of ECD and TMD atomic models. (D) Slice-through view of 3D map. The “funnel” likely represents the linker between ECD and the beginning of TM domain, and the TM helices likely cross each other near their N-terminal portions (near the funnel) and separate at the C-termini in order to connect to the two kinases located on each side of the TKD density. Inset: 2D view.
Even though the current resolution does not allow visualization of the exact conformation of the transmembrane helices, the restricted funnel shape of the ECD-TMD junction, as well as the gradual widening of the TMD density, implies that the two transmembrane helices should be held close together via interactions near their N-termini portions (Fig. 6C and 6D). The TM dimer crossing near the N-terminal ends has been an emerging feature observed in many published TM structures such as EGFR, EphA, and Growth Hormone Receptor (GHR) 26; 27; 28; 29. Potentially, this conformation allows separation of the C-terminal portions of the TM helices so that the downstream kinase domains have a sufficient range of motion to interact with one another during receptor activation.
Asymmetric dimer conformation of TKD
The bulky density below the TMD encloses a volume ~ 200,000 to 250,000-Å3, roughly corresponding to the volume of two kinase domains calculated from crystal structures of a single KIT kinase domain (one kinase domain ~ 80,000 to 100,000-Å3). Therefore, this bottom density most likely represents the volume of two interacting kinase domains from two receptor protomers (the TKD layer).
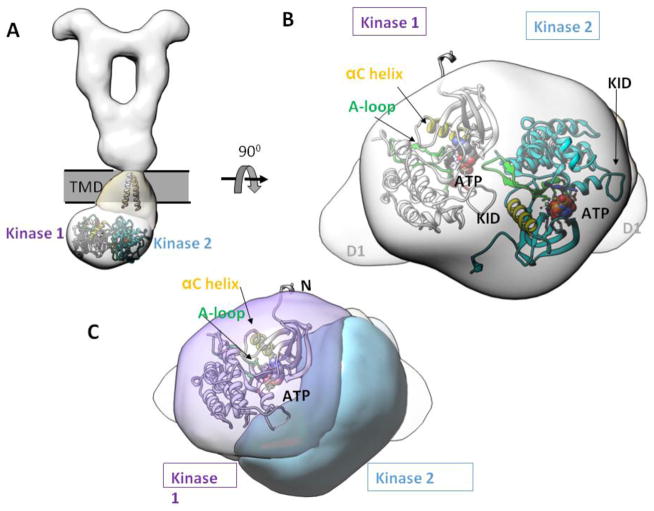
The reconstructed map shows that the TKD layer adopts a tilted conformation that does not follow the overall two-fold symmetry of the ECD, a feature that was also reported for SCF-stimulated full-length KIT14. Although the map is of limited resolution, density segmentation aided by rigid-body docking allows rough placement of two kinase domains. One kinase domain (“kinase 1” in Fig. 7) is displaced more laterally from the overall two-fold symmetry axis than the other kinase (“kinase 2” in Fig. 7). The only way to fit two kinase domain crystal structures into the density requires that the kinases arrange in an asymmetric fashion (Fig. 7A and B). Despite the low resolution that prevents detailed interpretations of the fitting, two findings are evident. Firstly, the kinase dimers are engaged in extensive dimer interface, which is indicative of some specific recognition event at the dimer interface (Fig. 7B and C). This notion is in contrast to the standard depiction where the kinase domains merely dangle off from the transmembrane helices and interact with one another only transiently. If so, the flexibility of the kinase domains would render a significantly weaker density in the TKD layer, due to averaging from all different conformations/orientations in 3D reconstruction. Secondly, rigid-body docking of multiple crystal structures of RTK kinase domains positions the catalytic site in close proximity to its partnering kinase (Fig. 7C). This suggests that the asymmetric kinase dimer arrangement may represent a natural consequence to the requirement for two receptors to phosphorylate one another. In this manner, one kinase plays the role as the “substrate” and other as the “enzyme”.
Figure 7. Asymmetric dimer of TKD.
(A) The atomic models of TM and TKD are fitted to show spatial relationships. (B) Bottom view of TKD asymmetric dimer. (C) Same view as in (B), but highlighting the catalytic site position of kinase 1, where the ATP binding pocket, the A-loop, and the αC helix are in close proximity to the neighboring kinase 2. KID = kinase insert domain. αC = control α-helix. A-loop = activation loop.
Discussion
RTKs have generally lagged behind other membrane proteins in terms structural determination due to difficulties in expression and the nearly impossible task of crystallization, which significantly hindered advances in the RTK field. In this study, we have determined the structure of a full-length PDGFRβ bound to its natural ligand PDGF-B. Along with the recently determined full-length KIT in complex with SCF, these structures demonstrate for the first time the architectures and domain arrangements of intact ligand-induced RTK dimers 14. Importantly, these structures underscore the possibility of and pave the way for obtaining higher resolution structures of RTKs in the future.
In the context of a full-length ligand-PDGFR complex, an extensive dimer interface is observed. Besides D1, all domains contribute to dimer interface, either indirectly through ligand engagement or directly via inter-receptor contacts. Importantly, despite the large total interface area for dimerization, receptors remain largely monomeric in the absence of ligand binding, as shown by FSEC (Fig. 1B). Thus, from ligand-binding to receptor activation, receptors likely go from a highly flexible, monomeric state to a rigid and energetically restrained state that is stabilized by multiple weak interactions that propagate from the ligand-biding domain to the kinase domain. This increase in receptor dimer stabilization likely involves morphing of D2-D3 for ligand binding, D3-D4 conformational coupling, D5/TM domain dimerization, and formation of asymmetric kinase domain dimer.
Several differences are seen between the full-length EM map and partial structures of soluble domains, and highlight the need to consider full-length RTK structures in the future. Firstly, in comparison with the crystal structure of PDGF-B/PDGFRβD1-D3, the D3s in the EM map (after flexible fitting) are farther apart and oriented differently to adopt an “elbow” conformation at the D3-D4 junction for downstream D4-D4 homotypic interface. Establishment of the D3-D4 elbow and the consequent ~ 60 to 65 Å separation between two D3 C-termini seem to be a common feature across Class III RTKs.
Secondly, the full-length EM map resolves the long-standing question on the conformation of D5 domains and oligomerization status of the transmembrane domain. The D5-D5 close association is clearly visible in the full-length PDGFR reconstruction. The presence of TM likely imposes a physical constraint on the conformational freedom in D5. Observed D5-D5 homotypic interface suggests that the effect of D5 cancer mutations in PDGFRs and KIT, as well as other members of Class III RTKs, is to drive D5-D5 interaction in a ligand-independent manner. Noticeably, receptor D5-D5 homotypic contact necessitates the ends of D5 to come close together, which would allow dimerization of the transmembrane helices. Interestingly, papillomavirus (BPV) E5 protein, which encodes a 44 amino-acid transmembrane helix, is known to facilitate PDGFRβ TM dimerization in a ligand-independent manner. In fact, E5 is so specific for PDGFRβ that it does not activate the related PDGFRα, nor other Class III RTKs. The specificity is still not well understood, but one of the E5-interacting residue on PDGFRβ, K531, is located right before the N-terminal start position of the TM domain; the specific interaction between E5 and PDGFRβ K531 likely drives the TM to adopt a dimer conformation similar to that seen in the current EM reconstruction 30.
Thirdly, while crystalized kinase domains of RTKs are largely monomeric, in a full-length receptor they form a close-knit dimer. It has been demonstrated with ample evidence that EGFR, FGFR, and KIT kinase domains adopt asymmetric kinase dimers during activation 14; 31; 32; 33; 34; here we show that PDGFR likely adopt asymmetric kinase dimer as well. Through crystallographic analysis of receptor ECDs, we have come to realize the surprising diversity of RTK extracellular assemblies in driving receptor dimerization. Likewise, although asymmetric kinase dimer arrangement may be a common activation mechanism, the detailed interface chemistries may differ among 50 families of RTKs. Further works on RTK kinase dimer arrangement at atomic detail will be required in order to translate into future drug design.
The current work also raises further questions for, in addition to providing a more refined view on, Class III RTK activation. For instance, the crystal structures of the FLT3 ECD in complex with its ligand, surprisingly, do not show homotypic interactions in D4 and D5; in particular, the D4 interface residues conserved in other Class III RTKs are missing in FLT3 11. Although it is possible that FLT3 uses a novel mechanism for dimerization and receptor activation, we suggest that, in the context of full-length FLT3, ligand-induced receptor dimerization might still enforce the D4-D4 and D5-D5 homotypic interactions for FLT3, albeit with a different interfaces than that used by KIT, PDGFRs, and CSF1R/FMS.
Also, class III RTKs are known for their pleiotropism in ligand recognition; whether different ligands of a common receptor induce the same receptor conformation is currently unknown. For instance, CSF1R (FMS) binds to two different cytokine ligands, M-CSF1 and IL-34, through different surface epitopes on D2 and D3 12. Low-resolution SAXS data on the entire ECD of CSF1R in complex with either M-CSF1 or IL-34 show a common assembly principle with divergent details22. In particular, while the same D4-D4 homotypic contact is observed, the orientations of the D1 and D5 domains are different. Notably, the two ligands have been found to induce different biological activities; whether such a difference is due to different ligand-receptor assembly is currently unknown. Future works on full-length CSF1R (FMS) with both ligands will be required to answer the question.
In essence, we have shown in this work how mammalian cells can be used to screen and purify recombinant human RTKs, and how disparate domains of a physiologically relevant RTK are coupled upon ligand stimulation. This work also shows how EM as an emerging tool to deliver structural insight into a class of membrane protein that has historically defied crystallographic analysis. We expect that, through further improvement in full-length RTK purification/reconstitution, higher resolution structures will be attainable in the near future to answer in more definitive terms the puzzling features of RTK activation.
Materials and Methods
Fluorescence-coupled size-exclusion chromatography (FSEC)
A FSEC vector based on BacMam technology was constructed in a manner similar to that reported recently 15. We engineered a new version of the vector with a novel variant of EGFP—Superfolder EGFP (sfEGFP) that has been reported to have a more robust folding in harsh environment than traditional EGFP 16. The new vector, harboring a secretion signal followed by a FLAG tag and sfEGFP, is from now on referred to as pFNsfEG (Fig. S1). FSEC experiments were performed as described earlier15. In brief, pFNsfEG plasmids with inserted targets of interest were transfected with FuGENE HD in suspension HEK293 GnTI- cells in a 6-well plate according to the instructions from the manufacturer (Promega). After 4–6 hours of transfection, the media was exchanged into serum-free media supplemented with 10mM sodium butyrate, and incubated for 24–28 hours in 37°C before harvesting. The cells were lysed by dounce homogenization in the lysis buffer (20mM Tris, 1mM EDTA, Roche EDTA-free protease inhibitor, pH 7.6), and the membrane fractions were pelleted by centrifuging at 60,000 rpm using a Beckmann table-top ultracentrifuge for one hour. To extract membrane proteins, the membrane pellets were resuspended in solubilization buffer (10mM Hepes, 150mM NaCl, and 1% detergent of choice in the presence of protease inhibitors) and stirred in 4°C overnight. The following day, insoluble materials were pelleted by ultracentrifugation at 60,000 rpm for one hour. The clarified sample (~500μL) volume was loaded into a Superose 6 size-exclusion column pre-equilibrated with FSEC buffer (10mM Hepes, 150mM NaCl, 0.05% DDM), and fractions of 250μL were collected. Fluorescence signal was recorded in a 96-well flat-bottom black plate (Corning) with excitation 485/emission 510.
To assess ligand-induced receptor dimerization, 200–500 μL of purified ligand in HBS was used to resuspend the membrane pellet, and detergent at 1% final concentration was added to the sample and incubated overnight in 4°C. Other procedures were the same as FSEC for unstimulated receptors.
BacMam virus generation
For protein expression, pFNsfEG vector was modified to remove the N-terminal sfEGFP, but leave the secretion signal, the FLAG tag, and the MCS intact (referred to as pF-BM vector) (Fig S1). BacMam virus was generated as described19.
PDGF-B expression
The amplified BacMam viruses for human PDGF-B were used to transduce suspension HEK293 GnTI− cells at a density of 1.5 × 106 cells/ml. After 5–6 hours, media were exchanged to serum-free media supplemented with 10mM sodium butyrate to boost protein expression. The conditioned media was harvested after 72 hours and buffer exchanged into HBS by a tangential concentrator.
PDGFRβ expression and PDGF-B/PDGFRβ purification
The amplified BacMam viruses were used to transduce suspension HEK293 GnTI− cells at a density of 1.5 × 106 cells/ml. After 5–6 hours, media were exchanged to serum-free media supplemented with 10mM sodium butyrate to boost protein expression. Cells were pelleted 24–28 hours later and washed with PBS + 1mM EDTA, and lysed by dounce homogenizer in lysis buffer (20mM Tris, 1mM EDTA, Roche EDTA-free, protease inhibitor cocktail, pH 7.6). To prevent free-cysteines in the cytoplasmic domain from cross-linking but leave the extracellular disulfide bonds intact, 2mg/mL iodoacetamide was added to the lysis buffer. After dounce-homogenization, cell debris and organelles were removed by centrifuge at 3,000 rpm for 10 minutes. The douncing and low-speed centrifugation step was repeated to maximize cell lysis.
The supernatant from the low-speed centrifugation step was then ultracentrifuged at 40,000 rpm for 1 hour in a Ti 45 fixed-angle rotor (Beckmann). To form PDGF-B/PDGFRβ complex, the pellet was re-suspended in ~150-mL PDGF-B conditioned media (without serum) and incubated overnight to ensure receptor dimerization by PDGF-B. Next day, 1 % LMNG was added to the suspension, and stirred for 1–1.5 hour in room temperature for an hour in the presence of protease inhibitor (Roche). The insoluble materials were discarded by ultracentrifugation at 40,000 rpm for 1 hr, and the supernatant containing detergent-solubilized ligand-receptor complex was further purified by anti-FLAG M2 affinity column (Sigma). After washing with running buffer (10mM Hepes, 150mM NaCl, 0.05% sodium azide, 0.01% LMNG), the complex was eluted by addition of 100-μg/mL FLAG peptide (Sigma). The proteins were further purified with size exclusion columns (Superose 6, GE Life Sciences) pre-equilibrated with running buffer.
Gradient Ultracentrifugation and Fixation (GraFix)
GraFix protocol was carried out as described previously 17. In brief, purified ligand-receptor complex was concentrated to ~200 μL with total protein less than 200 pmol to prevent cross-linking between particles. The glycerol gradient of 10–25% was chosen based on the molecular weight of our complex (~300kD). To set up the gradient, a top layer solution and a bottom layer solution were prepared. The top layer contains HBS+10% glycerol+0.01% LMNG, and the bottom layer consists of HBS+25% glycerol + 0.1% gluteraldehyde + 0.01% LMNG. After filtering the solutions, 2mL of top solution was injected gently first in an ultracentrifuge tube, followed by 2mL of bottom solution. Caution was taken to avoid introducing bubbles. To form a linear gradient, the tube was sealed with parafilms and gently placed horizontally in room temperature for 2 hours. Afterwards, the tube was put in a stand-up position and cooled in 4°C for one additional hour. Prior to sample loading, a 200 μL cushion layer containing HBS + 5% glycerol + 0.01% LMNG was gently pipetted on top of the top layer solution, followed by 200 μL of samples. Ultracentrifugation is performed with SW 55 Ti (Beckmann) at 40,000 rpm for 16 hours. After ultracentrifugation, 200 μL fraction was collected and analyzed on a 4–20% gradient SDS denaturing gel. Fractions containing the ~300kDa complex moved about two-thirds of the way down the gradient (corresponding to ~ 20% glycerol and 0.0067% glutaraldehyde) and were pooled together for EM analysis (Fig. S3A). 80mM glycine was added to the pooled fractions to stop further cross-linking.
Negative-stain electron-microscopy
The grid was prepared as described 35. Briefly, 400-mesh copper grids (EMS) were coated with a thin layer (~2.2 nm) of continuous carbon film (home-made). After drying overnight, a carbon-coated grid was glow-discharged, and 5-μL sample directly from GraFix (~ 20 μg/mL) was then loaded on to the grid, which was then dipped briefly in 5 drops of distilled water before staining with two drops of 0.75% (w/v) uranyl formate (UF). Images were acquired using a Gatan 1400 TEM operated at 120 kV, with nominal magnification at 30k using a 4k × 4k CCD camera, corresponding to 3.71-Å/pixel. Defocus value ranges between 1 and 2 μm.
Image processing – reconstruction of initial model
We employed the random-conical tilt (RCT) method for initial model generation. For RCT, tilt-pair micrographs were recorded at 0° and 55°. 2,830 particle pairs were manually selected in Xmipp 36. The untitled particles were classified initially into 16 classes, with about 100–200 particles in each class. Classes with fuzzy averages and no identifiable features were thrown out, and the remaining good particles were subject to another round of 2D classification. After multiple rounds of 2D classification and data cleaning, a final dataset consists of 1,859 particles in 9 classes. The homogeneity of each class was analyzed by the principle component analysis (PCA) and was shown to be rather pure.
Five best classes (class 1–4, 9), based on the quality of their class averages and homogeneity, were chosen for initial model generation in Xmipp. To improve accuracy in 3D reconstruction, only particles in the “stable cores” for each class were chosen. The 5 reconstructed initial models were visually inspected and compared in Chimera. Because classes 1, 2 and 9 displayed similar volumes, the particles in the corresponding classes were combined. Similarities between the volumes from classes 1, 2 and 9 were further confirmed by comparing a similarity score between back-projections of the models with different class averages (Screen classes in Xmipp). The best initial volume (class 2) was chosen and refined via projection-matching against 771 particles (from particles of the combined classes). The improved initial model was low-pass filtered at 25 Å and masked for subsequent 3D classification and refinement procedures.
Image processing – 3D reconstruction
A separate data set was collected, in which 10,477 particles were manually picked and subject to 5 cycles of 2D classification in Xmipp (Fig. S5). To obtain a better handle on noise and quality of reconstruction, we pursued RELION 3D classification/refinement workflow. The initial RCT (after projection matching against the smaller dataset used for RCT) was masked and low-pass filtered to 100-Å for input into RELION. Auto-refine against 8,570 particles was performed to generate a “consensus model” for subsequent 3D classification with minimal initial model bias. From the consensus and low-passed structural template, 3D classification and refinement schemes are outlined in the Supplemenal Information (Fig. S6).
Density map analysis
Rigid body docking, as well as map visualization and analysis, was done in UCSF Chimera 37. Map segmentation analysis was performed using the SEGGER module also implemented in Chimera. Based on density threshold, the tyrosine kinase domain (TKD) was automatically segmented into two volumes, each of which corresponds to the volume of a single kinase domain. For rigid-body docking of kinase domain crystal structures, the two segmented kinase domain densities were saved as a single MRC volume stack file. One copy of the Insulin Receptor kinase domain (PDB ID: 1IR3) was first manually docked into the density, followed by placement of a second copy. Because the normal “Fit in Map” command produces significant clashes between the two copies, we opted for “sequential docking” implemented in Chimera, which produces a much better fit with noapparent clashes between the two copies of molecules.
Molecular Dynamics/Flexible Fitting (MDFF) simulation
Molecular modeling of the entire ECD was done by MDFF. The crystal structure of PDGF-B/PDGFRβD1-D3 (PDB ID: 3MJG) was used as the initial model, and PDGFRβ D4D5 is modeled from KIT D4D5 (PDB ID: 2E9W). The structure parameter files are produced in VMD 38 via the plug-in “Automatic PSF Builder.” Rigid-body docking of both PDGF-B/PDGFRβD1-D3 and PDGFRβ D4D5 homology model was performed in the Situs package39. Symmetry is imposed on PDGF-B/PDGFRβD1-D3 and D4D5 homology model to not only prevent over-fitting, but also preserve the R385/E390A salt-bridge during the simulation process. The EM map was converted to the force field potential using the MDFF commands implemented in VMD. The molecular dynamics simulation was performed in NAMD2.140 using the CHARMM force-field, and was run on the Northwestern computing cluster. All simulations were performed in 0.5 ns time-scale.
Supplementary Material
Highlights.
Full-length transmembrane RTK structures are currently lacking
Purification and visualization of PDGF-stimulated full-length PDGFRβ by electron-microscopy
Domain organizations that differ from crystal structures of soluble domains
Asymmetric kinase domain dimer arrangement in full-length receptor
Demonstration of extracellular coupling with intracellular kinase activation
Acknowledgments
We thank Jonathan Remis from Northwestern Cryo-EM facility for training and technical support. We also thank Sarah Rice and Kasia Soczek for their help on GraFix. Po-Han Chen is supported by a National Cancer Institute F30 pre-doctoral fellowship and a scholarship from the Malkin Scholars Program of Northwestern University Cancer Center. Xiaolin He is supported by National Institute of Health Grant R01 GM098259-04.
Abbreviations
- EM
electron-microscopy
- FSEC
fluorescence-coupled size-exclusion chromatography
- GraFix
Gradient and Fixation
- Ig
immunoglobulin
- M-CSF
macrophage colony stimulating factor
- MDFF
molecular dynamics/flexible fitting
- PDGF
Platelet-derived growth factor
- PDGFR
PDGF receptor
- RCT
random conical tilt
- RTK
receptor tyrosine kinase
- SCF
stem cell factor
- sfEGFP
superfolder enhanced green fluorescence protein
Footnotes
- SCF/KITFL (EMDB ID: 2648).
- PDGF-B/PDGFRβD1-D3 (PDB ID: 3MJG)
- M-CSF1/BARF1 (PDB ID: 4FA8)
- SCF/KITD1-D5 (PDB ID: 2E9W)
- Insulin Receptor kinase domain (PDB ID: 1IR3)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kohler N, Lipton A. Platelets as a source of fibroblast growth-promoting activity. Exp Cell Res. 1974;87:297–301. doi: 10.1016/0014-4827(74)90484-4. [DOI] [PubMed] [Google Scholar]
- 2.Ross R, Glomset J, Kariya B, Harker L. A platelet-dependent serum factor that stimulates the proliferation of arterial smooth muscle cells in vitro. Proc Natl Acad Sci U S A. 1974;71:1207–10. doi: 10.1073/pnas.71.4.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22:1276–312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ostman A. PDGF receptors-mediators of autocrine tumor growth and regulators of tumor vasculature and stroma. Cytokine Growth Factor Rev. 2004;15:275–86. doi: 10.1016/j.cytogfr.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–34. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coussens L, Van Beveren C, Smith D, Chen E, Mitchell RL, Isacke CM, Verma IM, Ullrich A. Structural alteration of viral homologue of receptor proto-oncogene fms at carboxyl terminus. Nature. 1986;320:277–80. doi: 10.1038/320277a0. [DOI] [PubMed] [Google Scholar]
- 7.Rosnet O, Marchetto S, deLapeyriere O, Birnbaum D. Murine Flt3, a gene encoding a novel tyrosine kinase receptor of the PDGFR/CSF1R family. Oncogene. 1991;6:1641–50. [PubMed] [Google Scholar]
- 8.Chen X, Liu H, Focia PJ, Shim AH, He X. Structure of macrophage colony stimulating factor bound to FMS: diverse signaling assemblies of class III receptor tyrosine kinases. Proc Natl Acad Sci U S A. 2008;105:18267–72. doi: 10.1073/pnas.0807762105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, Chen X, Focia PJ, He X. Structural basis for stem cell factor-KIT signaling and activation of class III receptor tyrosine kinases. EMBO J. 2007;26:891–901. doi: 10.1038/sj.emboj.7601545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuzawa S, Opatowsky Y, Zhang Z, Mandiyan V, Lax I, Schlessinger J. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130:323–34. doi: 10.1016/j.cell.2007.05.055. [DOI] [PubMed] [Google Scholar]
- 11.Verstraete K, Vandriessche G, Januar M, Elegheert J, Shkumatov AV, Desfosses A, Van Craenenbroeck K, Svergun DI, Gutsche I, Vergauwen B, Savvides SN. Structural insights into the extracellular assembly of the hematopoietic Flt3 signaling complex. Blood. 2011;118:60–8. doi: 10.1182/blood-2011-01-329532. [DOI] [PubMed] [Google Scholar]
- 12.Ma X, Lin WY, Chen Y, Stawicki S, Mukhyala K, Wu Y, Martin F, Bazan JF, Starovasnik MA. Structural basis for the dual recognition of helical cytokines IL-34 and CSF-1 by CSF-1R. Structure. 2012;20:676–87. doi: 10.1016/j.str.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 13.Liu H, Leo C, Chen X, Wong BR, Williams LT, Lin H, He X. The mechanism of shared but distinct CSF-1R signaling by the non-homologous cytokines IL-34 and CSF-1. Biochim Biophys Acta. 2012;1824:938–45. doi: 10.1016/j.bbapap.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Opatowsky Y, Lax I, Tome F, Bleichert F, Unger VM, Schlessinger J. Structure, domain organization, and different conformational states of stem cell factor-induced intact KIT dimers. Proc Natl Acad Sci U S A. 2014;111:1772–7. doi: 10.1073/pnas.1323254111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goehring A, Lee CH, Wang KH, Michel JC, Claxton DP, Baconguis I, Althoff T, Fischer S, Garcia KC, Gouaux E. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc. 2014;9:2574–85. doi: 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 17.Stark H. GraFix: stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods Enzymol. 2010;481:109–26. doi: 10.1016/S0076-6879(10)81005-5. [DOI] [PubMed] [Google Scholar]
- 18.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–30. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shim AH, Liu H, Focia PJ, Chen X, Lin PC, He X. Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex. Proc Natl Acad Sci U S A. 2010;107:11307–12. doi: 10.1073/pnas.1000806107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y, Yuzawa S, Schlessinger J. Contacts between membrane proximal regions of the PDGF receptor ectodomain are required for receptor activation but not for receptor dimerization. Proc Natl Acad Sci U S A. 2008;105:7681–6. doi: 10.1073/pnas.0802896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemmon MA, Pinchasi D, Zhou M, Lax I, Schlessinger J. Kit receptor dimerization is driven by bivalent binding of stem cell factor. J Biol Chem. 1997;272:6311–7. doi: 10.1074/jbc.272.10.6311. [DOI] [PubMed] [Google Scholar]
- 22.Felix J, Elegheert J, Gutsche I, Shkumatov AV, Wen Y, Bracke N, Pannecoucke E, Vandenberghe I, Devreese B, Svergun DI, Pauwels E, Vergauwen B, Savvides SN. Human IL-34 and CSF-1 establish structurally similar extracellular assemblies with their common hematopoietic receptor. Structure. 2013;21:528–39. doi: 10.1016/j.str.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 23.Elegheert J, Desfosses A, Shkumatov AV, Wu X, Bracke N, Verstraete K, Van Craenenbroeck K, Brooks BR, Svergun DI, Vergauwen B, Gutsche I, Savvides SN. Extracellular complexes of the hematopoietic human and mouse CSF-1 receptor are driven by common assembly principles. Structure. 2011;19:1762–72. doi: 10.1016/j.str.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verstraete K, Savvides SN. Extracellular assembly and activation principles of oncogenic class III receptor tyrosine kinases. Nat Rev Cancer. 2012;12:753–66. doi: 10.1038/nrc3371. [DOI] [PubMed] [Google Scholar]
- 25.Muhle-Goll C, Hoffmann S, Afonin S, Grage SL, Polyansky AA, Windisch D, Zeitler M, Burck J, Ulrich AS. Hydrophobic matching controls the tilt and stability of the dimeric platelet-derived growth factor receptor (PDGFR) beta transmembrane segment. J Biol Chem. 2012;287:26178–86. doi: 10.1074/jbc.M111.325555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arkhipov A, Shan Y, Das R, Endres NF, Eastwood MP, Wemmer DE, Kuriyan J, Shaw DE. Architecture and membrane interactions of the EGF receptor. Cell. 2013;152:557–69. doi: 10.1016/j.cell.2012.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Endres NF, Das R, Smith AW, Arkhipov A, Kovacs E, Huang Y, Pelton JG, Shan Y, Shaw DE, Wemmer DE, Groves JT, Kuriyan J. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell. 2013;152:543–56. doi: 10.1016/j.cell.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chavent M, Chetwynd AP, Stansfeld PJ, Sansom MS. Dimerization of the EphA1 receptor tyrosine kinase transmembrane domain: Insights into the mechanism of receptor activation. Biochemistry. 2014;53:6641–52. doi: 10.1021/bi500800x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brooks AJ, Dai W, O’Mara ML, Abankwa D, Chhabra Y, Pelekanos RA, Gardon O, Tunny KA, Blucher KM, Morton CJ, Parker MW, Sierecki E, Gambin Y, Gomez GA, Alexandrov K, Wilson IA, Doxastakis M, Mark AE, Waters MJ. Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science. 2014;344:1249783. doi: 10.1126/science.1249783. [DOI] [PubMed] [Google Scholar]
- 30.DiMaio D, Petti LM. The E5 proteins. Virology. 2013;445:99–114. doi: 10.1016/j.virol.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bae JH, Boggon TJ, Tome F, Mandiyan V, Lax I, Schlessinger J. Asymmetric receptor contact is required for tyrosine autophosphorylation of fibroblast growth factor receptor in living cells. Proc Natl Acad Sci U S A. 2010;107:2866–71. doi: 10.1073/pnas.0914157107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bae JH, Schlessinger J. Asymmetric tyrosine kinase arrangements in activation or autophosphorylation of receptor tyrosine kinases. Mol Cells. 2010;29:443–8. doi: 10.1007/s10059-010-0080-5. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–49. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 34.Chen H, Xu CF, Ma J, Eliseenkova AV, Li W, Pollock PM, Pitteloud N, Miller WT, Neubert TA, Mohammadi M. A crystallographic snapshot of tyrosine trans-phosphorylation in action. Proc Natl Acad Sci U S A. 2008;105:19660–5. doi: 10.1073/pnas.0807752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheres SH, Nunez-Ramirez R, Sorzano CO, Carazo JM, Marabini R. Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc. 2008;3:977–90. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 38.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–8. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 39.Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys Rev. 2010;2:21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.