Abstract
The mechanisms by which n-3 polyunsaturated fatty acids (n-3 PUFA), abundant in fish oil, exert their anti-inflammatory effects have not been rigorously defined. We have previously demonstrated that n-3 PUFA decrease the amount of phosphatidylinositol-(4,5)-bisphosphate, [PI(4,5)P2], in CD4+ T cells, leading to suppressed actin remodeling upon activation. Since discrete pools of PI(4,5)P2 exist in the plasma membrane, we determined whether n-3 PUFA modulate spatial organization of PI(4,5)P2 relative to raft and non-raft domains. We used Förster resonance energy transfer (FRET) to demonstrate that lipid raft mesodomains in the plasma membrane of CD4+ T cells enriched in n-3 PUFA display increased co-clustering of Lck(N10) and LAT(ΔCP), markers of lipid rafts. CD4+ T cells enriched in n-3 PUFA also exhibited a depleted plasma membrane non-raft PI(4,5)P2 pool as detected by decreased co-clustering of Src(N15), a non-raft marker, and PH(PLC-δ), a PI(4,5)P2 reporter. Incubation with exogenous PI(4,5)P2 rescued the effects on the non-raft PI(4,5)P2 pool, and reversed the suppression of T cell proliferation in CD4+ T cells enriched with n-3 PUFA. Furthermore, CD4+ T cells isolated from mice fed a 4% docosahexaenoic acid (DHA)-enriched diet exhibited a decrease in the non-raft pool of PI(4,5)P2, and exogenous PI(4,5)P2 reversed the suppression of T cell proliferation. Finally, these effects were not due to changes to post-translational lipidation, since n-3 PUFA did not alter the palmitoylation status of signaling proteins. These data demonstrate that n-3 PUFA suppress T cell proliferation by altering plasma membrane topography and the spatial organization of PI(4,5)P2.
Keywords: Fluorescence resonance energy transfer, T cell, membrane, phosphatidylinositol, polyunsaturated fatty acids
Graphical Abstract
Introduction
The fluid-mosaic membrane concept first proposed in 1972 [1] has evolved from a composition of dynamic proteins dissolved in a viscous solution of lipids to a model where higher order structures involving protein-protein, protein-lipid, and lipid-lipid interactions, in conjunction with the membrane cytoskeleton network, coordinate the spatiotemporal organization of signaling events [1–4]. Lipid rafts are mesoscale domains enriched in sphingolipids, cholesterol, and saturated fatty acids that form in the plasma membrane as a result of favorable lipid-lipid interactions. Mounting evidence underscores the importance of the interaction between the lipids in the plasma membrane and protein function [5, 6]. From a biophysical perspective, changes to the plasma membrane properties such as bilayer thickness, membrane curvature, bilayer elasticity, and membrane stretching, can allosterically regulate membrane protein function [5, 6]. Recently, it has become evident that very modest changes to the plasma membrane can have biological effects. For example, in the fly photoreceptor system, activation of rhodopsin by light results in the hydrolysis of PI(4,5)P2 into diacylglycerol (DAG) [7]. Simplistic estimates suggest that the conversion of PI(4,5)P2 into DAG only reduces the membrane area by 0.8% and contracts the microvillus of the fly photoreceptor system by 0.2 nm [7]. However, this “small” change is sufficient to activate TRP channels in the fly photoreceptor system to modulate the photomechanical response, and lead to the micrometer contraction of the photoreceptor system [6, 7].
The protein and lipid composition of lipid rafts is highly heterogeneous and dynamic; lipid rafts are known to coalesce to form microscopic signaling platforms in response to several stimuli. This is most evident during CD4+ T cell activation, a process that involves major rearrangements of the T cell proteome and lipidome [8, 9]. Engagement of the TCR with an antigen peptide displayed by MHCII of an APC results in the formation of a higher ordered structure termed the immunological synapse (IS) [8–10]. Signaling at the IS is highly coordinated through the temporally ordered recruitment of signaling-propagating, and exclusion of signaling-terminating, proteins [11, 12]. Raft lipid constituents, including cholesterol, saturated phosphatidylcholine species, and sphingolipids, are also enriched at the IS [13–15]. Collectively, these findings underscore the importance of protein-protein, protein-lipid, and lipid-lipid interactions at mesoscale membrane domains for effective T cell activation [15].
Dietary fish oil, enriched in long chain n-3 polyunsaturated fatty acids (n-3 PUFA) such as eicosapentaenoic acid (EPA, 20:5 Δ5,8,11,14,17) and docosahexaenoic acid (DHA, 22:6 Δ4,7,10,13,16,19), has been shown to exert anti-inflammatory and immunosuppressive effects across a spectrum of human diseases [16–18]. Using two strategies for the enrichment of n-3 PUFA at the membrane, including dietary supplementation [19, 20] and the transgenic Fat-1 mouse model [21, 22], we have previously demonstrated that early signaling events critical to T cell activation are suppressed. In particular, plasma membrane enrichment of n-3 PUFA restricted phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2]-dependent actin remodeling, calcium signaling, and mitochondrial translocation to the IS, resulting in the suppression of T cell activation (lymphoproliferation) [14, 19, 23, 24]. Additional studies in Jurkat cells have shown that other chemicals such as 7-ketocholesterol that modulate the biophysical properties of the plasma membrane, can also suppress T cell activation [15, 25]. Together, these findings clearly demonstrate that by incorporating into the plasma membrane, n-3 PUFA can target a key lipid mediator, PI(4,5)P2, and affect cortical actin, two interplaying components regulating membrane-cytoskeletal interactions critical for lymphoproliferation.
From a biophysical perspective, DHA is highly disordered and has low affinity for saturated fatty acids and cholesterol, constituents of lipid rafts [26–29]. Previous findings indicate that the insertion of DHA-containing phospholipids into the plasma membrane enlarges lipid raft domains while concomitantly decreasing the amount of cholesterol and saturated fatty acids [30, 31]. We have previously demonstrated that n-3 PUFA increase the lipid order/rigidity of the plasma membrane in vivo [13, 14, 32], and affect downstream T cell responses to IL-2 [14, 21, 33]; therefore, we hypothesized that n-3 PUFA suppress T cell signaling and activation by modifying the membrane topography and spatial organization of PI(4,5)P2. Here we demonstrate that n-3 PUFA modify the biophysical properties of the plasma membrane and alter the spatial distribution of PI(4,5)P2, a critical phospholipid involved in the organization of membrane mesodomains and the membrane-associated cytoskeletal network. Of significance, n-3 PUFA-induced reorganization of membrane mesodomains results in suppression of clonal expansion upon activation of primary CD4+ T cells. These results highlight previously unappreciated effects of n-3 PUFA on membrane organization that, by modulating early TCR signaling, lead to suppression of T cell proliferation.
Materials and Methods
Plasmids
Enhanced cyan fluorescent protein (ECFP), and yellow fluorescent protein for energy transfer (YPet) were synthesized by Integrated DNA Technologies (Coralville, Iowa) into pIDTSMART and subcloned into pLenti vector using BamHI and NotI (New England Biolabs, Ipswich, MA). A NheI site was included immediately downstream of BamHI for insertion of additional fragments. cDNA containing the first 10 amino acids of Lck [Lck(N10)], the first 15 amino acids of Src [Src(N15)], the transmembrane domain of linker for activation of T cells [LAT(ΔCP)], and the PH domain of phospholipase-δ [PH(PLC-δ)], were synthesized and subcloned into pLenti using BamHI and NheI restriction sites, upstream and in-frame to ECFP or YPet. All plasmids were verified by sequencing (Eton Bioscience, San Diego, CA), and expressed as fusion probes in 293T cells followed by examination using microscopy and Western blotting. R40L mutant of PH(PLC-δ) was generated by using the Quikchange II Kit (Agilent, Santa Clara, CA), confirmed by sequencing, and expressed in 293T cells.
Generation of Lentivirus
Lentivirus was generated as previously described with minor modifications [34, 35]. Individual pLenti contructs were co-transfected with pLP-1, pLP-2, and pVSV-G (Life Technologies, Grand Island, NY) into HEK293T/17 (ATCC CRL-11268) using calcium phosphate precipitation. Medium containing the virus was harvested three times within 72 hrs of transfection. Lentivirus was then concentrated using a concentrator (Clontech, Mountain View, CA), resuspended in RPMI media (Irvine Scientific, Santa Ana, CA) containing 5% fetal bovine serum (Irvine Scientific), 1% glutamax (Life Technologies), and 1% penicillin-streptomycin (Life Technologies), and stored at −80 °C.
Animal Husbandry and CD4+ T Cell Isolation
Animal protocol for this study (AUP #2011-176) was approved by the Institutional Animal Care and Use Committee at Texas A&M University following the U.S. Public Health Service guidelines. Fat-1 transgenic mice on a C57BL/6 background, generously provided by Dr. Jing X. Kang (Department of Medicine, Harvard University, [22]), were bred, genotyped, and phenotyped as previously described [14, 23, 24]. Wild type and Fat-1 litter-mate controls were fed a 10% safflower diet (Research Diets, New Brunswick, NJ) ad libitum in a 12:12 light:dark cycle. In diet experiments, 4 to 6 weeks old C57BL/6 mice were fed either an n-6 PUFA enriched 5% corn oil diet, a 0.95% DHA triglyceride-, or a 4% DHA triglyceride-enriched diet as previously described [13, 23]. Briefly, diets differed only in their oil composition, either (control) 5% corn oil by weight or a mixture of 57% pure DHA triglyceride (Martek, Columbia, MD) and corn oil (0.95:3:05, or 4:1 w/w). Additional diet components, expressed as g/100 g, were 20 casein, 42 sucrose, 22 corn starch, 6 cellulose, 3.5 AIN-76 mineral mix, 1 AIN-76 vitamin mix, 0.3 DL-methionine, 0.2 choline chloride, and 5 dietary oil (Table 1). Mice were fed ad libitum for 6 to 8 weeks using a 12:12 light:dark cycle [19, 20, 36] and diet was changed daily to prevent the formation of oxidative byproducts.
Table 1.
Composition
| g/100 g | 5% Corn Oil | 0.95% DHA Triglyceride | 4% DHA Triglyceride |
|---|---|---|---|
| Casein | 20 | 20 | 20 |
| Sucrose | 42 | 42 | 42 |
| Corn Starch | 22 | 22 | 22 |
| Cellulose | 6 | 6 | 6 |
| AIN-76 Mineral Mix | 3.5 | 3.5 | 3.5 |
| AIN-76 Vitamin Mix | 1 | 1 | 1 |
| DL-Methionine | 0.3 | 0.3 | 0.3 |
| Choline Chloride | 0.2 | 0.2 | 0.2 |
| Corn Oil | 5 | 4.05 | 1 |
| Safflower Oil | |||
| DHA Triglyceride | 0 | 0.95 | 4 |
CD4+ T cells were purified by removing the spleen aseptically and labeling with magnetic CD4 (L3T4) microbeads according to manufacturer’s protocol (Miltenyi Biotec, Auburn, CA). CD4+ T cells were cultured in complete RPMI media containing 20 μM 2-mercaptoethanol (Sigma Aldrich, St Louis, MO), until the start of the downstream assay (within hrs). For longer experiments (within 48 hrs), 20 ng/mL of rIL-2 (eBioscience, San Diego, CA) was added to the complete RPMI media. For rescue experiments using exogenous PI(4,5)P2, cells were treated after lentiviral transduction with PI(4,5)P2 dissolved in 1X PBS supplemented with 0.0025% of protein stabilizer (Echelon Biosciences, Salt Lake City, UT) for 1 hr, at 37 °C, 5% CO2, collected, and washed prior to initiation of FRET experiments.
Lentiviral Transduction
CD4+ T cells were resuspended to a final concentration of 7.5 × 106 cells/mL, seeded into a 24 well plate and incubated with lentivirus for 48 hrs before imaging in the presence of 2 μg/mL polybrene (Sigma Aldrich). Typically ECFP-containing lentivirus was added 8 hrs prior to YPet lentivirus to optimize the level of the fluorophores for imaging. After 48 hrs of incubation, CD4+ T cells were collected, washed, resuspended in 300 μL of Leibovitz’s media, and seeded onto poly-L-lysine coated 35 mm glass bottom dishes (MatTek, Ashland, MA) for imaging.
Microscopy and Image Analysis
FRET experiments were conducted using the Zeiss LSM 510 META laser scanning confocal microscope with an excitation at 458 nm for ECFP, and 514 nm for YPet using an argon laser. Emissions were detected between 480 – 520 nm for ECFP, and between 535 – 590 nm for YPet. Samples were imaged with a 63X oil objective with a numerical aperture of 1.2, and recorded in 8-bit mode. Images were processed using the accompanying software and values were exported and analyzed in Microsoft Excel.
FRET was analyzed by acceptor photobleaching to detect the increase in ECFP signal. A region of interest (ROI) consisting of 4 μm × 3 μm was drawn around the plasma membrane, and photobleaching was conducted by illuminating the ROI for 16 sec with the laser. For each cell, four images were recorded: i) YFPPRE, YPet fluorescence before photobleaching; ii) CFPPre, ECFP fluorescence before photobleaching; iii) YFPPost, YPet fluorescence after photobleaching; iv) CFPPost, ECFP fluorescence after photobleaching. Corrections to the fluorescence intensities were also made by conducting acceptor photobleaching on cells expressing only the ECFP and YPet probes to determine the bleaching of ECFP during photobleaching (CFPBleach), and the bleed-through of YPet into the CFP channel (CFPYFP). Corrections to the CFP intensities before and after photobleaching were calculated as follows:
| Eq. 1 |
| Eq. 2 |
To determine the FRET efficiency (E%), the corrected CFP values were used in the following equation:
| Eq. 3 |
To determine fluorescence recovery after photobleaching (FRAP), the following equation was used to analyze cells expressing only the YPet probe where F(t) is fluorescence intensity at time (t), F(∞) is the fluorescence intensity after recovery, F(0) is the initial fluorescence intensity, and k is the rate of fluorescence intensity recovery after photobleaching:
| Eq. 4 |
At least 4 animals per genotype/treatment, and at least 10 cells per animal were analyzed for each FRET combination.
CFSE Assay
After CD4+ T cell isolation, cells were labeled with CFSE as previously described [14, 21]. In brief, 5 μM of CFSE (Life Technologies) in DMSO was added into 1 × 106 cells/mL in 1X PBS (Life Technologies) containing 0.1% BSA (Roche Diagnostics, Indianapolis, IN), incubated at 37 °C for 30 min, rapidly chilled by adding an excess of ice-cold complete RPMI, and incubated for 5 min on ice. Cells were collected, washed with 1X PBS, and resuspended to a final cell concentration of 2 x 106 cells/mL. 2×105 cells were cultured in 96 well plates either unstimulated or stimulated with plated anti-CD3 (Clone 145-2C11, 0.2 μg/mL, eBiosciences) and soluble anti-CD28 (Clone 37.51, 1 μg/mL, eBiosciences) for 72 hrs, at 37 °C. On the day of analysis, cells were collected, washed with 1X PBS, and resuspended in staining buffer (eBioscience) containing 1 μg/mL of propidium iodide (PI, Miltenyi Biotec). Samples were processed using a BD Accuri C6 flow cytometer (BD Bioscience, San Joe, CA). The Proliferation Index, representing the sum of cells in all generations divided by the theoretical parental population at the start of the experiment, was calculated using ModFitLT 3.2 [37].
Membrane Phospholipid Analysis
Phospholipid fatty acid composition was analyzed as previously described [21]. Briefly, total lipids were extracted from CD4+ T cells using Folch (chloroform:methanol, 2:1 v/v), and separated by thin layer chromatography (TLC) on silica gel 60G plates using the developing solvent of choloroform/methanol/acetic acid/water (90:8:1:0.8, v/v). The plate was then sprayed with 0.1% 8-anilino-naphthalene-sulfonic acid and visualized under ultraviolet light. Total phospholipids were scraped from the plates, spiked with heptadecanoic acid (17:0), and transesterified in the presence of 6% methanol-HCl. Afterwards, fatty acid methyl esters were extracted using hexane and 0.1 M potassium chloride and analyzed by gas chromatography.
Detection of Protein Palmitoylation
15-azidopentadecanoic acid (Life Technologies, Grand Island, NY) dissolved in DMSO (50 mM) was added to each cell culture containing 20 × 106 CD4+ T cells to a final concentration of 50 μM, and incubated for 24 h. Cells were then collected, washed, and lysed in homogenization buffer composed of 50 mM Tris-HCl, pH = 8, 1% SDS (w/v), 100 μM of sodium orthovanadate, 1X sigma protease cocktail, and 250 units of benzonase nuclease. Protein lysates from two cultures were pooled and concentrated using centrifugal concentrator (3 kDa cut-off, Millipore, Billerica, MA). Protein concentration was subsequently determined using the bicinchoninic acid assay (Thermo Scientific, Rockford, IL) and protein concentration was adjusted to 4 μg/μL for Click chemistry. Click chemistry was performed using the Click-iT kit (Life Technologies) according to the manufacturer’s protocol. Briefly, 200 μg of cellular protein was combined with 100 μL of Click-iT reaction buffer containing PEG4 carboxamide-propargyl biotin (Life Technologies), 10 μL of ddH2O, vortexed for 5 sec, before addition of CuSO4 and component C. The reaction was vortexed for 5 sec, and incubated for 3 min at 23 °C. Finally, component D was added to the reaction and incubated for at 23 °C for 30 min, protected from light. After the reaction, protein was precipitated according to the manufacturer’s protocol, and resuspended in a homogenization buffer containing 50 mM Tris-HCl, pH = 7.5, 15 mM EGTA, pH = 7.5, 100 mM NaCl, 0.1% Triton X-100, 500 μM of sodium orthovanadate, 1 Sigma protease cocktail, and 10 mM β-mercaptoethanol. The protein concentration was determined by the Commassie Plus Protein assay (Pierce, Rockford, IL). Samples were then loaded onto 4–20% Tris-glycine polyacrylamide precast gel (Invitrogen) and separated for 2.5 h at 4 ºC. Proteins were then transferred onto polyvinylidene fluoride membranes at 400 mA for 90 min. Membranes were blocked at room temperature for 1 h in 4% nonfat dry milk. Primary antibody against biotin (mouse anti-biotin, Jackson ImmunoResearch, West Grove, PA) was incubated overnight at 4 ºC. Membranes were subsequently washed and incubated with secondary horseradish peroxidase conjugated rabbit anti-mouse IgG (KPL) and developed using Pierce SuperSignal West Femto maximum sensitivity substrate. Membranes were then scanned using Fluor-S Max MultiImager system (BioRad, Hercules, CA) and analyzed using QuantityOne (BioRad).
For immunoprecipitation, 100 μg of protein was combined with rabbit polyclonal IgG anti-Lck (Millipore) for 90 min at 4 °C prior to addition of Protein G Dynabeads (Life Technologies) for overnight incubation at 4 °C. The following day, the protein-antibody complex was collected using DynaMagnet, and boiled in the presence of 2X pyronin before immunoblotting as described above.
Statistical Analysis
All experiments were conducted with at least four animals, and data were expressed as mean ± sem unless otherwise noted. Using GraphPad Prism (La Jolla, CA), data were analyzed by two-tailed Student’s t-test between genotypes with a single treatment, one-way ANOVA between multiple groups, or by two-way ANOVA between genotypes and treatments. In cases where P < 0.05 in ANOVA, a Tukey post-hoc test was conducted. P < 0.05 was considered to be statistically significant.
Results
Optimization of FRET Experiments
We have previously shown that n-3 PUFA perturb the membrane lipid bilayer by increasing the liquid order of plasma membrane in CD4+ T cells isolated from mice fed a diet enriched in n-3 PUFA, e.g., DHA [13]. To further probe the effects of n-3 PUFA on plasma membrane mesodomains, we performed a series of FRET experiments utilizing the raft markers Lck(N10) and LAT(ΔCP), in which the 10 N-terminal amino acids of Lck and the transmembrane domain LAT are fused to fluorescent reporters. For comparative purposes, we also investigated the properties of the non-raft marker Src(N15), which contains the 15 N-terminal amino acids of c-Src. These peptides are well-described probes used to target lipid raft and non-raft regions of the plasma membrane, and have been shown to localize predominantly to the plasma membrane in Jurkat and primary T cells (Fig S1A; [38–41]).
Mammals are unable to convert n-6 PUFA into n-3 PUFA, thus we utilized primary CD4+ T cells isolated from Fat-1 transgenic mice, a model in which the plasma membrane is enriched with n-3 PUFA upon feeding a diet high in n-6 PUFA [21, 22]. We utilized a lentivirus system to transduce the reporter plasmids in order to avoid activating the CD4+ T cells with antibodies or chemicals [38]. Using this system, the presence of n-3 PUFA did not affect lentiviral transduction efficiency (Fig S1B).
We initially assessed FRET efficiency by performing a series of control experiments (Fig S2). For this purpose, FRET standards, C5Vand C32V were expressed in 293T cells (Fig S2A; [42]). These constructs were generated by positioning Cerulean (C) relative to Venus (V) with either a 5- or 32-amino acid linker; thus C5V should have higher FRET efficiency relative to C32V [42]. We then determined the FRET efficiencies of the Lck(N10)/Lck(N10) FRET pair in both 293T cells and primary CD4+ T cells to evaluate the change associated with photobleaching the whole cell versus a region of interest (ROI). FRET by acceptor photobleaching has been used for both the C5V and C32V standards [43], and for measuring membrane markers in Jurkat T cells [38, 41]. No statistical differences between the two approaches were observed (Figs S2B&C). Collectively, these data indicate that the diffusion of YPet outside the ROI is slow enough to accurately measure FRET efficiency immediately after photobleaching. Furthermore, the lack of any difference between whole cell versus plasma membrane targeted regions of interest suggests that the probes are localized predominantly to the plasma membrane (Fig 1A) and not in endosomes. For example, non-interacting probes localized to internal membranes would decrease the overall FRET detected in whole cells when compared to a ROI at the plasma membrane [44]. We subsequently optimized the iterations of photobleaching in an ROI in order to ensure maximum photobleaching without prolonged laser exposure, thereby minimizing effects on cell viability (Fig S2D).
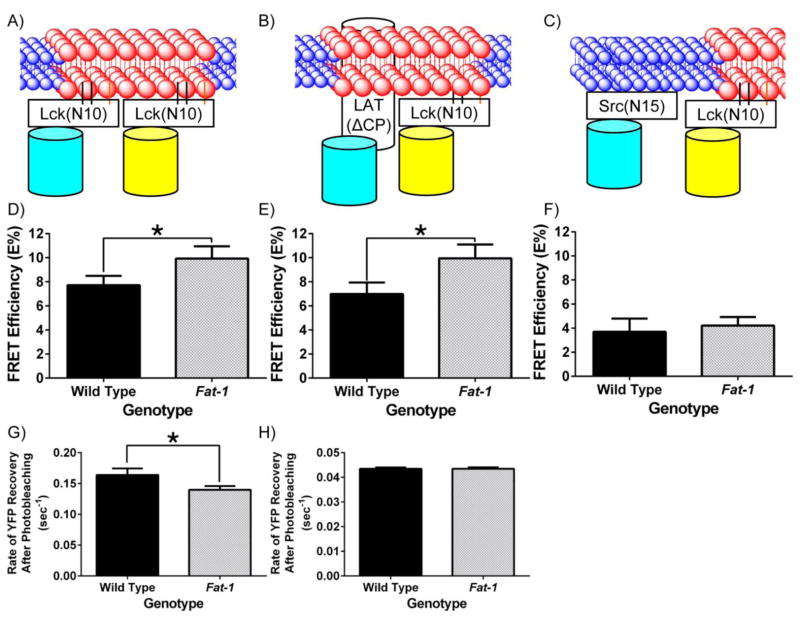
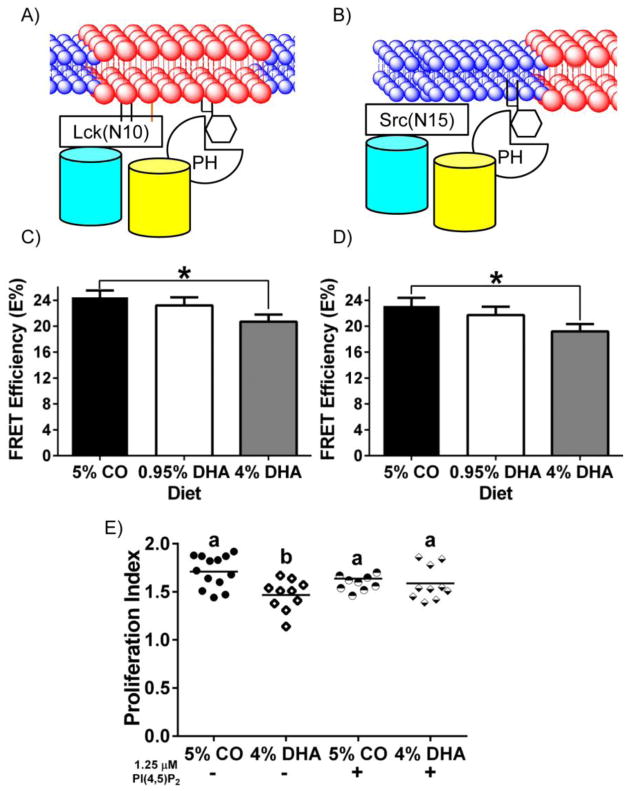
Fig 1. n-3 PUFA perturb the biophysical properties of the plasma membrane detected by increased interaction of fluorescent lipid raft markers.
(A, B, and C) Schematic diagrams of the interaction between neighboring fluorescent probes targeted to the raft or the non-raft membrane fraction of the plasma membrane. Lck(N10) and Src(N15) consist of the first 10 and 15 N-terminal amino acids of Lck and Src, respectively. LAT(ΔCP) consists of the first 36 amino acids, representing the extracellular, transmembrane, and the membrane anchoring portions of LAT. Splenic CD4+ T cells were incubated with lentivirus containing (D) Lck(N10)/Lck(N10), (E) Lck(N10)/LAT(ΔCP), or (F) Lck(N10)/Src(N15) before FRET by acceptor photobleaching (n = 4 mice per genotype). FRET efficiency (E%) was determined by comparing the CFP intensity before and after acceptor photobleaching, and corrected as described in the Materials and Methods (*P < 0.05 between genotypes, two-tailed Student’s t-test). The rate of recovery after photobleaching was also determined for CD4+ T cells expressing (G) Lck(N10)-YPet and (H) LAT(ΔCP)-YPet (*P < 0.05 between genotypes, two-tailed Student’s t-test).
Using Raft and Non-Raft Markers to Probe the Effects of n-3 PUFA on CD4+ T Cell Plasma Membrane Mesodomains
We hypothesized that the presence of n-3 PUFA in the plasma membrane would increase the FRET efficiency between the lipid raft markers since increased liquid order would favor coalescence of lipid raft mesodomains. Therefore, we measured the FRET efficiency between the various raft and non-raft markers (Figs 1A–C). Consistent with our observations that n-3 PUFA increase the liquid order of the plasma membrane, the incorporation of n-3 PUFA into the plasma membrane increased the FRET efficiencies of the lipid raft markers Lck(N10)/Lck(N10), and Lck(N10)/LAT(ΔCP) (Figs 1D&E), compared to the control noninteracting markers Lck(N10)/Src(N15) (Fig 1F). Since the diffusion of membrane proteins can be influenced by whether they are localized in lipid raft mesodomains [45], we also determined the rate of fluorescence recovery after photobleaching (FRAP) of the raft markers Lck(N10) and LAT(ΔCP). FRAP of Lck(N10) in CD4+ T cells enriched with n-3 PUFA exhibited a 20% decrease relative to the FRAP of Lck(N10) expressed in wild type CD4+ T cells (Fig 1G). We did not observe a difference in the FRAP of LAT(ΔCP) between genotypes, perhaps due to the slow rate of recovery exhibited by LAT(ΔCP) (Fig 1H). Collectively, the results from the FRET and FRAP experiments indicate that n-3 PUFA alter the dynamics of the lipid raft mesodomains in the plasma membrane.
Incorporation of n-3 PUFA into the CD4+ T Cell Plasma Membrane Decreases the Non-Raft Pool of PI(4,5)P2
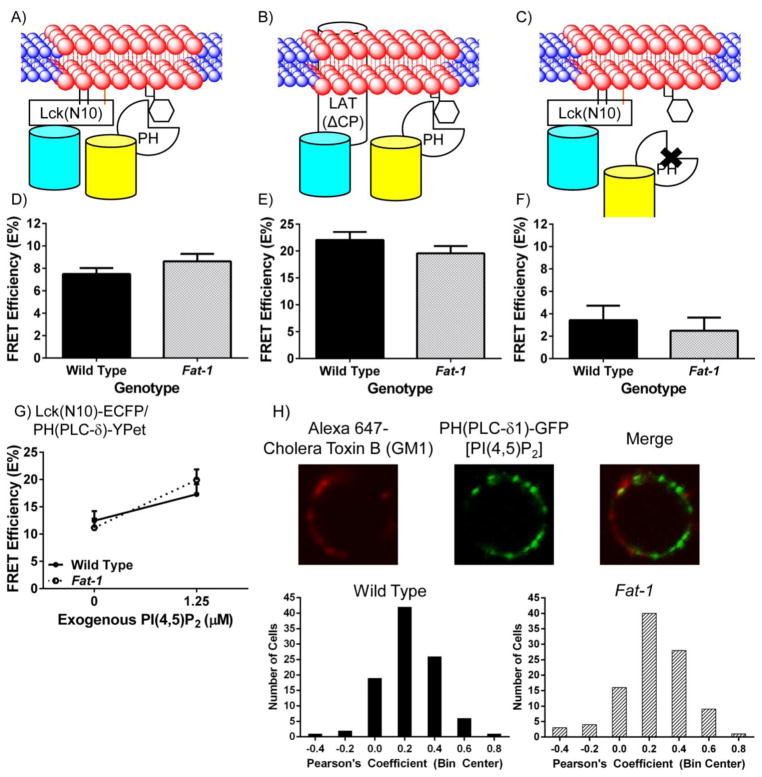
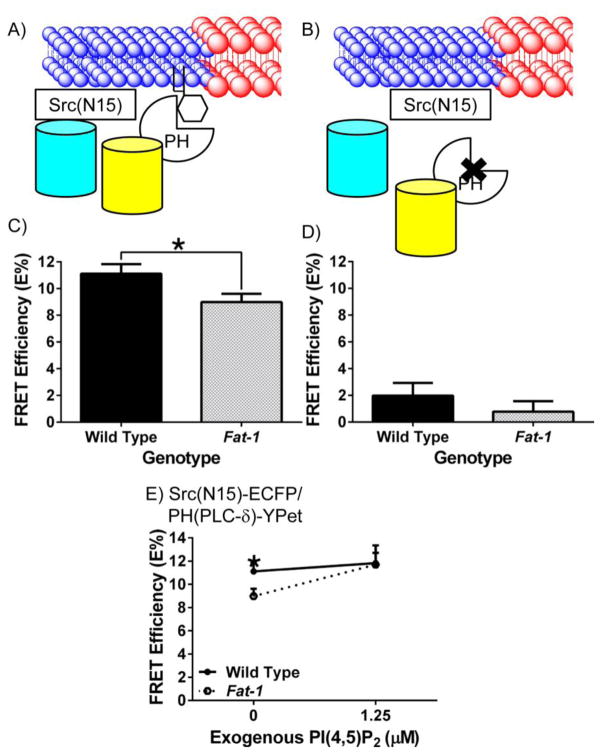
Since we have shown that the lipid raft mesodomains in the plasma membrane are perturbed by the presence of n-3 PUFA, and PI(4,5)P2 couples the meso-scale raft domains with membrane cytoskeleton compartments [2–4], we hypothesized that incorporation of n-3 PUFA into the plasma membrane would alter the spatial organization of PI(4,5)P2 relative to raft and non-raft mesodomains. Utilizing the raft markers [Lck(N10) and LAT(ΔCP)] and the non-raft marker [Src(N15)] in combination with the PI(4,5)P2 reporter PH(PLC-δ), which has been validated previously to target PI(4,5)P2 [35, 46], we tested our hypothesis in a series of FRET experiments (Figs 2&3). Although n-3 PUFA increased the FRET efficiencies between lipid raft markers (Figs 1D&E), the FRET efficiency between PH(PLC-δ) and either Lck(N10) or LAT(ΔCP) was not affected in CD4+ T cells enriched with n-3 PUFA (Figs 2D&E). This was corroborated by measuring the co-localization between GM1, a well-characterized lipid raft marker [47], and PH(PLC-δ), which showed no difference in the presence of n-3 PUFA (Fig 2H). The FRET efficiency between PH(PLC-δ) and the non-raft marker Src(N15), however, was decreased by 20% in CD4+ T cells enriched with n-3 PUFA (Figs 3A&C). To verify the specificity of our PH(PLC-δ) probe, we performed FRET experiments using the PHR40L(PLC-δ) mutant to abolish binding of the PH domain to PI(4,5)P2 [35, 46] in conjunction with the raft marker Lck(N10) or the non-raft marker Src(N15). As expected, binding of the R40L mutated PH domain to PI(4,5)P2 exhibited markedly decreased FRET efficiencies in both wild type and Fat-1 CD4+ T cells (Figs 2F&3D). These results suggest that the incorporation of n-3 PUFA into the plasma membrane not only influences the biophysical properties of the phospholipid bilayer, but also alters the spatial organization of PI(4,5)P2 by depleting the non-raft pool of PI(4,5)P2.
Fig 2. n-3 PUFA do not affect the interaction of fluorescent raft and PI(4,5)P2 markers detected using FRET in Fat-1 CD4+ T cells.
(A and B) Schematic diagrams of the interaction between neighboring fluorescent probes targeted to the raft membrane fraction of the plasma membrane, and PH(PLC-δ), a PI(4,5)P2 reporter. (C) A control PHR40L(PLC-δ) was also expressed to test the specificity of the PH(PLC-δ) reporter. CD4+ T cells were incubated with lentivirus containing (D) Lck(N10)/PH(PLC-δ) (n = 8 per genotype), (E) LAT(ΔCP)/PH(PLC-δ) (n = 6 per genotype), or (F) Lck(N10)/PHR40L(PLC-δ) (n = 4 per genotype) before FRET by acceptor photobleaching. FRET efficiency (E%) was determined as described in Fig 1 (*P < 0.05 between genotypes, two-tailed Student’s t-test). (G) CD4+ T cells were isolated and transduced with Lck(N10)-ECFP and PH(PLC-δ)-YPet for 48 hrs prior to incubation with 1.25 μM PI(4,5)P2 or PBS (0 μM) for one hr. Cells were analyzed as described in Fig 1. (H) Colocalization between the lipid raft marker GM1 and PI(4,5)P2 is not altered in the presence of n-3 PUFA. Splenic CD4+ T cells were positively selected and incubated with lentivirus containing pLenti-PH(PLC-δ)-GFP for 48 hrs (n = 4 mice per genotype). Cells were collected, washed, labeled with Alexa 647 Cholera Toxin B (final concentration 10 μg/mL) and crosslinked with anti-Cholera Toxin B [Vybrant Lipid Raft Labeling Kit (Life Technologies)] according to the manufacturer’s protocol. Cells were imaged using a Zeiss 510 LSM confocal microscope. The plasma membrane region of the cell was drawn and Pearson’s coefficient was determined using NIS-Elements (Nikon).
Fig 3. n-3 PUFA decrease the interaction of fluorescent non-raft and PI(4,5)P2 markers detected using FRET in Fat-1 CD4+ T cells.
(A and B) Schematic diagrams of the interaction between neighboring fluorescent probes targeted to the non-raft membrane fraction of the plasma membrane, and PH(PLC-δ), a PI(4,5)P2 reporter, or the control PHR40L(PLC-δ). CD4+ T cells were incubated with lentivirus containing (C) Src(N15)/PH(PLC-δ) (n = 7 per genotype), or (D) Src(N15)/PHR40L(PLC-δ) (n = 4 per genotype) before FRET by acceptor photobleaching. FRET efficiency (E%) was determined as described in Fig 1 (*P < 0.05 between genotypes). (E) CD4+ T cells were isolated and transduced with Src(N15)-ECFP and PH(PLC-δ)-YPet for 48 hrs prior to incubation with 1.25 μM PI(4,5)P2 or PBS (0 μM) for one hr. Cells were analyzed as described in Fig 1. A two-tailed t-test was used to compare FRET efficiencies within specific concentrations (*P < 0.05 at specific concentrations of PI(4,5)P2).
We have previously demonstrated that the incorporation of n-3 PUFA into the plasma membrane of CD4+ T cells decreased the level of PI(4,5)P2 by 50%, resulting in suppressed actin remodeling upon anti-CD3/anti-CD28 stimulation [23]. The suppression of PI(4,5)P2-dependent actin remodeling was reversed by incubation with exogenous PI(4,5)P2. However, recent studies have suggested that different pools of PI(4,5)P2 are responsible for differential activation of different signaling pathways, such as calcium signaling and actin remodeling [48–50]. Therefore, we hypothesized that co-incubation with exogenous PI(4,5)P2 and CD4+ T cells enriched in n-3 PUFA would rescue the non-raft pool of PI(4,5)P2. FRET efficiencies using the lipid raft marker Lck(N10) and PI(4,5)P2 marker PH(PLC-δ) before and after incubation with exogenous PI(4,5)P2 did not differ between wild type and CD4+ T cells enriched with n-3 PUFA (Fig 2G). In contrast, using the non-raft marker Src(N15) and PH(PLC-δ), the FRET efficiency of CD4+ T cells enriched with n-3 PUFA increased to the level of wild type CD4+ T cells (Fig 3E). These data indicate that exogenous PI(4,5)P2 increases the non-raft pool of PI(4,5)P2 in CD4+ T cells enriched with n-3 PUFA to a level comparable to wild type cells.
Exogenous PI(4,5)P2 Reverses the Suppression of Proliferation in CD4+ T Cells Enriched with n-3 PUFA
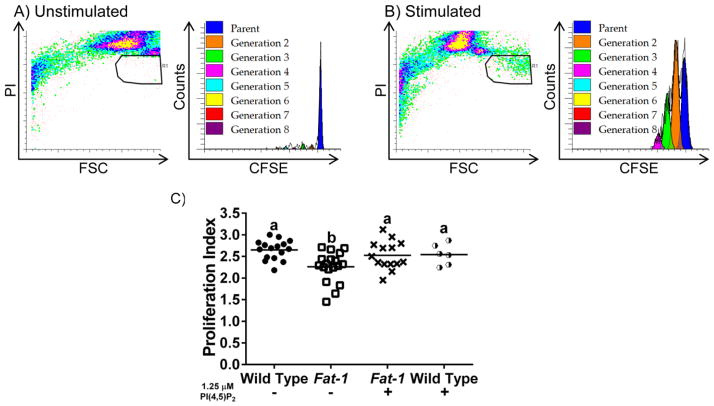
We have previously shown that CD4+ T cells enriched with n-3 PUFA exhibit suppressed proliferation following anti-CD3/anti-CD28 stimulation [14, 19]. Since exogenous PI(4,5)P2 restores the non-raft pool of PI(4,5)P2 (Fig 3E), and can rescue PI(4,5)P2-dependent actin remodeling [23], we hypothesized that exogenous PI(4,5)P2 would also reverse the suppressed proliferative phenotype in CD4+ T cells enriched with n-3 PUFA. Using CFSE as a marker of T cell proliferation, and PI fluorescence as a marker of cell viability, cell division was determined based on the number of distinct CFSE fluorescence intensity peaks (Fig 4). The proliferation index of each population was determined using the generational data, providing an indicator of the proliferative capacity of the population [37]. CD4+ T cells enriched with n-3 PUFA exhibited a suppressed (~20% lower) T cell proliferative capacity compared to wild type (Fig 4C). Importantly, the suppressed proliferation recovered to wild type levels upon pre-treatment with exogenous PI(4,5)P2. These findings demonstrate that i) incorporation of n-3 PUFA into the plasma membrane of CD4+ T cells not only alters the spatial organization of PI(4,5)P2 by decreasing the non-raft pool of PI(4,5)P2, but also suppresses T cell proliferation; and ii) replenishment of the non-raft pool of PI(4,5)P2 (Fig 3E) and rescue of PI(4,5)P2-dependent actin remodeling in CD4+ T cells enriched with n-3 PUFA [23], restore full lymphoproliferative capacity.
Fig 4. Exogenous PI(4,5)P2 rescues the n-3 PUFA-induced suppression of T cell proliferation.
Representative scatterplots and CFSE profiles of (A) unstimulated, and (B) anti-CD3/anti-CD28 stimulated CD4+ T cell cultures. Splenic CD4+ T cells were stained with CFSE, then cultured (n = 4 mice per genotype) either in unstimulated or stimulated conditions for 72 hrs in triplicate. Cells were collected and gated by propidium iodide staining to exclude dead cells. (C) Cell proliferation was analyzed using the Proliferation Wizard in ModFitLT3.2 to determine the proliferation index. Different letters represent statistically significant differences between the groups after one-way ANOVA followed by Tukey post-hoc test (P < 0.05).
CD4+ T cells Isolated from Mice fed a 4% DHA Triglyceride-Enriched Diet Exhibit Decreased Raft PI(4,5)P2, non-Raft PI(4,5)P2, and Lymphoproliferation which can be Rescued by Exogenous PI(4,5)P2
Since the predominant species of PI(4,5)P2 is 1-stearoyl-2-arachidonoyl phosphatidylinositol [51], and we have previously demonstrated that CD4+ T cells isolated from mice fed a 4% DHA triglyceride diet had approximately 25% lower levels of PI(4,5)P2 [23], we characterized the fatty acid profiles of CD4+ T cell phospholipid from C56BL/6 mice fed either a 5% corn oil (CO)-enriched diet (control), a 0.95% DHA-triglyceride-enriched diet, or a 4% DHA-triglyceride-enriched diet and isolated splenic CD4+ T cells (Fig 5). In particular, total phospholipids isolated from the 4% DHA group exhibited a dramatic 95% decrease in the amount of arachidonic acid (20:4, n-6), and a concomitant increase in DHA (22:6, n-3) compared to the control CO and 0.95% DHA groups.
Fig 5. Dietary DHA reduces membrane a rachidonic acid levels.
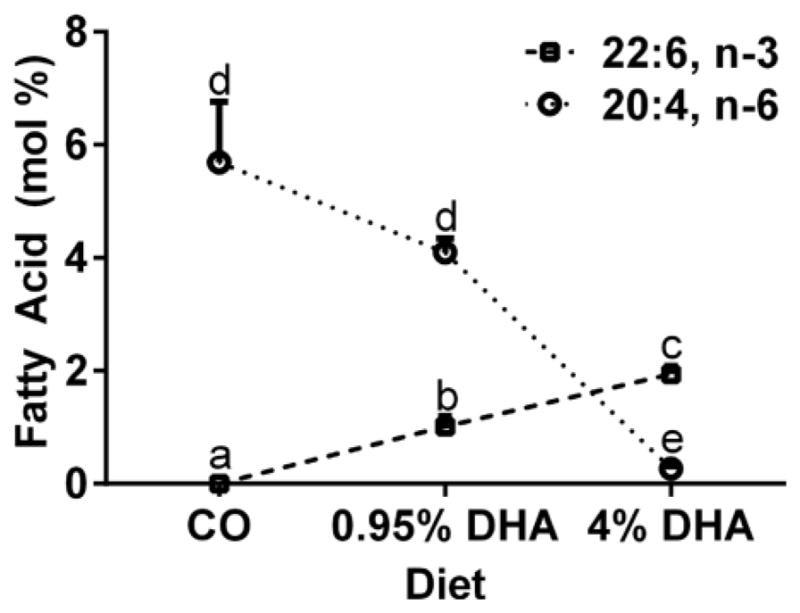
Levels of DHA (22:6, n-3) and arachidonic acid (20:4, n-6) in CD4+ T cell phospholipids isolated from mice fed 5% corn oil (CO), 0.95% DHA, or 4% DHA-enriched diet are shown. Fatty acids from CD4+ T cell total phospholipids were extracted, isolated, and characterized as previously described [19, 21]. Data represent mean ± sem (n = 4 per diet), and different letters represent statistically significant differences between the diet groups per fatty acid (P < 0.05). Note that other n-3 PUFA such as EPA (20:5, n-3) and DPA (22:5, n-3) were not detected in CD4+ T cell total phospholipids.
In order to determine whether dietary n-3 PUFA, can recapitulate the Fat-1 CD4+ T cell phenotype, mice were fed the aforementioned control CO-enriched diet, 0.95% DHA-triglyceride, or 4% DHA-triglyceride enriched diet. Using the raft and non-raft probes of Lck(N10) and Src(N15), respectively, in conjunction with the PI(4,5)P2 probe PH(PLC-δ), we detected decreased FRET efficiencies between CD4+ T cells isolated from mice fed the control diet and the 4% DHA-triglyceride-enriched diet using the combination of Lck(N10)/PH(PLC-δ) and Src(N15)/PH(PLC-δ), suggesting that both the lipid raft and non-raft pools of PI(4,5)P2 are perturbed (Figs 6A–D). The decrease in arachidonic acid in the phospholipid fraction (Fig. 5) was correlated with the decrease in PI(4,5)P2 in both the raft and non-raft fractions (Figs 6C&D). In addition, decreased PI(4,5)P2 was also associated with suppressed lymphoproliferation (Fig 6E), similar to Fat-1 CD4+ T cells. Importantly, incubation of CD4+ T cells with exogenous PI(4,5)P2 reversed the DHA-mediated suppression of lymphoproliferation caused by decreased PI(4,5)P2, demonstrating that decreased status of PI(4,5)P2 directly suppresses lymphoproliferation (Fig 6E). Collectively, these results demonstrate that enrichment of the CD4+ T cell plasma membrane with n-3 PUFA decreases the level of phospholipid arachidonic acid, perturbs both raft and non-raft pool of PI(4,5)P2 and suppresses lymphoproliferation that can be rescued by exogenous PI(4,5)P2.
Fig 6. Dietary DHA decreases the interaction of fluorescent raft and non-raft probes with PI(4,5)P2 marker detected using FRET, resulting in suppressed lymphoproliferation that can be rescued by exogenous PI(4,5)P2 in CD4+ T cells.
(A and B) Schematic diagrams of the interaction between neighboring fluorescent probes targeted to the raft or the non-raft membrane fraction of the plasma membrane, and PH(PLC-δ), a PI(4,5)P2 reporter. CD4+ T cells were incubated with lentivirus containing (C) Lck(N10)/PH(PLC-δ) (n = 4 per diet), or (D) Src(N15)/PH(PLC-δ) (n = 4 per diet) before FRET by acceptor photobleaching. FRET efficiency (E%) was determined as described in Fig 1 (*P < 0.05 between genotypes). (E) Splenic CD4+ T cells were stained with CFSE, then cultured (n = 6 mice per diet) either in unstimulated or stimulated conditions for 72 hrs in triplicate. Cells were collected and gated by propidium iodide staining to exclude dead cells. Proliferation Index was calculated using ModFitLT 3.2 [37]. Different letters represent statistically significant differences between the groups after one-way ANOVA followed by Tukey post-hoc test (P < 0.05).
Palmitoylation is not affected by n-3 PUFA in unstimulated primary CD4+ T cells
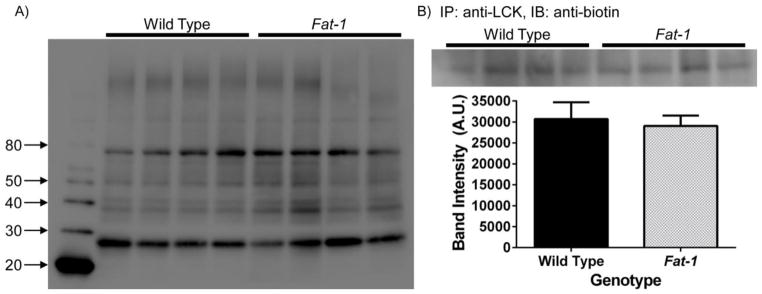
An alternative explanation for the observed results using Lck(N10) and LAT(ΔCP) probes in our FRET experiments could be that the presence of n-3 PUFA alter the palmitoylation of these probes. Indeed, it has been shown that EPA can decrease the palmitoylation of the Src family kinase Fyn in COS-1 cells [52]. To investigate the effect of n-3 PUFA on protein palmitoylation, CD4+ T cells were isolated from wild type and Fat-1 mice and subsequently incubated the azido-palmitic acid, followed by Click chemistry, before analysis by immunoblotting. Contrary to previous results suggesting that n-3 PUFA decrease the palmitoylation of signaling proteins [52], global palmitoylation status was not affected in Fat-1 CD4+ T cells (Fig 7A). To examine whether palmitoylation was affected in specific proteins, we performed an immunoprecipitation of LCK in order to isolate and detect its palmitoylation status. Consistent with the global palmitoylation signature, n-3 PUFA did not affect the palmitoylation status of LCK in CD4+ T cells (Fig 7B). These results suggest that the effects of n-3 PUFA on plasma membrane organization are independent of protein palmitoylation.
Fig 7. Palmitoylation status of membrane proteins is not altered in Fat-1 CD4+ T cells.
(A) 1.5 × 107 splenic CD4+ T cells were positively selected and seeded into a 35 mm culture dish with 4 mL of RPMI media supplemented with 2% FBS and rIL-2 overnight. Cells were treated with 50 μM of azido-palmitic acid for 24 h and 2 wells were pooled per sample (n = 2 mice per sample, n = 4 samples per genotype). Subsequently, 100 μg of protein lysate was used for Click chemistry and palmitoylation was detected by immunoblotting. Numbers on the left side represent molecular weight markers. (B) Palmitoylation of LCK is not affected in Fat-1 CD4+ T cells. Cells were selected and processed as in (A), and samples were subsequently immunoprecipitated (IP) using polyclonal rabbit IgG anti-LCK. Palmitoylated LCK was then detected by immunoblotting (IB) using an anti-biotin antibody and subsequently quantified.
Discussion
Given their widespread consumption and clinical use, it is important to elucidate the mechanisms by which n-3 PUFA exert their anti-inflammatory properties. Previous studies have examined the extrinsic effects of n-3 PUFA downstream of TCR engagement with MHCII antigen presentation [14, 19, 23, 24]. However, a number of outstanding issues concerning effects on membrane hierarchical organization and linkage to cell function remain to be addressed. In the present study, using both genetic and dietary approaches, we have demonstrated for the first time that membrane incorporation of n-3 PUFA perturbs the spatial organization of a critical second messenger, PI(4,5)P2 in CD4+ T cells, resulting in suppressed lymphoproliferation. Importantly, exogenous incubation of PI(4,5)P2 with CD4+ T cells enriched in n-3 PUFA rescued both the spatial perturbation of PI(4,5)P2 and suppressive effects on lymphoproliferation. These novel findings provide insight into the molecular mechanisms by which n-3 PUFA alter host inflammation, and contribute to the rational application of dietary lipids in the complementary and alternative medicine armamentarium.
Previous works have investigated the effects of n-3 PUFA on the localization and acylation of LCK and LAT in Jurkat T cells [52–55]. These earlier studies using the immortalized Jurkat T cells and ex vivo incubation of n-3 PUFA provided the impetus to study the effects of n-3 PUFA in primary mouse CD4+ T cells, where the plasma membrane is enriched with n-3 PUFA by either a genetic (Fat-1) or dietary (4% DHA-triglyceride) model [52–55]. Gu et al. has previously shown that incubation of mouse Pten-null prostate cells with DHA resulted in increased DHA at the sn-2 position of PI(4,5)P2, changing the localization of PI(3,4,5)P3 and downstream AKT signaling [56]. We have previously investigated the fatty acid species of PI(4,5)P2 using mass spectrometry, and we did not detect enrichment of DHA at the sn-2 position of PI(4,5)P2 in primary mouse CD4+ T cells [23]. Thus, our data suggest that the non-raft pool of PI(4,5)P2 is decreased in CD4+ T cells enriched with n-3 PUFA, and this is not due to a shift in localization.
Since lipid raft mesodomains in the plasma membrane play a critical role in initiating T cell activation [15, 25], and n-3 PUFA are incorporated into the plasma membrane, we hypothesized that n-3 PUFA suppress T cell activation by altering i) the biophysical properties of lipid raft mesodomains; and ii) the spatial organization of PI(4,5)P2 relative to lipid raft and non-raft mesodomains. By using lipid raft and non-raft markers in a series of FRET experiments, we demonstrate for the first time in primary CD4+ T cells that n-3 PUFA alter the topology of lipid raft mesodomains. The increase in FRET efficiency between the lipid raft markers observed in CD4+ T cells enriched in n-3 PUFA (Figs 1D&E) can be interpreted either as an increase in the size or the stability of the lipid raft mesodomains. In both cases, the enhancement of lipid raft mesodomains would be expected to decrease the average diffusion of the lipid raft marker, since it has been shown that an association with lipid raft mesodomains retards the diffusion rate of these markers [57, 58]. This is consistent with the FRAP data reported in Fig 1G. From an immunological perspective, this perturbation in plasma membrane biophysics is associated with a suppression of T cell proliferation observed in Fig 4C [14, 21]. Interestingly, other forms of perturbation to plasma membrane topology can change the threshold for T cell activation. For example, incubation of Jurkat T cells with 7-ketocholesterol decreased the membrane order and resulted in a suppression of downstream T cell signaling [25].
Since the organization of lipid raft mesodomains is closely linked with the actin cytoskeleton, and the actin cytoskeleton is regulated by PI(4,5)P2 [59], we hypothesized that the incorporation of n-3 PUFA into the plasma membrane of CD4+ T cells would alter the spatial organization of PI(4,5)P2. Indeed, we have previously shown that n-3 PUFA reduced the amount of total PI(4,5)P2 by 50% in CD4+ T cells and this reduction was correlated with a suppression of PI(4,5)P2-dependent actin remodeling necessary for proper T cell activation [23]. Surprisingly, the increase in FRET efficiency between lipid raft markers in the presence of n-3 PUFA did not correlate with a change in the lipid raft pool of PI(4,5)P2 (Figs 1&2); rather, the non-raft pool of PI(4,5)P2 was decreased (Fig 3C). In the context of PI(4,5)P2-dependent actin remodeling, this was not entirely unexpected, as this remodeling has been shown in other cell types to be regulated by a specific PIP5K isoform (PIP5Kγ) which is capable of synthesizing the non-raft pool of PI(4,5)P2 [48–50]. Furthermore, perturbations in the raft and non-raft pools of PI(4,5)P2 using a targeted phosphatase in Jurkat T cells produced different actin-dependent phenotypes [60]. These studies demonstrate the compartmentalization of PI(4,5)P2 in the plasma membrane (raft and non-raft), and the importance of non-raft PI(4,5)P2 in regulating actin remodeling. It is also noteworthy that incubation with exogenous PI(4,5)P2 increased PI(4,5)P2 mass back to wild type levels (Fig 3E), which is consistent with the rescue of PI(4,5)P2-dependent actin remodeling upon stimulation [23].
In our diet model, the 4% DHA-triglyceride contributed approximately 5% total energy (kcals) to the diet. We demonstrated that phospholipid levels of arachidonic acid were reduced by 95% (Fig 5), which was associated with a decrease in both the raft and non-raft fractions of PI(4,5)P2 (Figs 6C&D) and a reduction in lymphoproliferation (Fig 6E). This observation is consistent with previous reports indicating that membrane phosphoinositide arachidonic acid levels are antagonized by n-3 PUFA across multiple cell types and tissues [19, 61]. Interestingly, the suppression of lymphoproliferation was reversed by incubation with exogenous PI(4,5)P2, demonstrating that PI(4,5)P2 is a direct target of DHA at the plasma membrane capable of modulating T cell proliferation. With respect to dose, in a typical Greenland Inuit diet, n-3 PUFA constitutes approximately 2.7 – 6.3% of daily energy [62–64]. Thus, a 4% DHA diet is physiologically relevant based on daily energy intake. It is interesting to note that the phospholipid level of arachidonic acid in Fat-1 CD4+ T cells was only reduced by 25% [21], and this reduction was correlated with a decrease in only the non-raft fraction of PI(4,5)P2, suggesting a dose-dependent differential regulation of the raft and non-raft pool of PI(4,5)P2. Further studies are required to address the dose-related effects of diet on different phospholipid pools.
Exogenous incubation of PI(4,5)P2, which enriches the plasma membrane pool of PI(4,5)P2 (Figs 2G&3E), has been employed to rescue PI(4,5)P2-dependent processes, such as insulin response in adipocytes [65, 66] and actin remodeling upon CD4+ T cell activation [23]. Since we have previously demonstrated that incorporation of n-3 PUFA into the T cell plasma membrane suppresses lymphoproliferation upon stimulation [14, 21], we hypothesized that the rescue of the non-raft pool of PI(4,5)P2 would also reverse the suppression of T cell proliferation, linking the biochemistry and cellular biology of the T cell membrane with a highly relevant immunological end point. Using the CFSE assay to measure T cell proliferation, we demonstrate that rescue of the non-raft pool of PI(4,5)P2 in CD4+ T cells enriched with n-3 PUFA also restores T cell proliferation back to wild type levels.
Previous experiments examining the perturbations of membranes by phytochemicals suggest that small changes to the cell membrane caused by amphiphilic phytochemicals can manifest itself in large changes to protein function [67–69]. As an example, the phytochemicals capsaicin, curcumin, epigallocatechin gallate (EGCG), genistein, and resveratrol only increased the average area per lipid, or decreased bilayer thickness, by at most 3%. Yet, these compounds were able to promiscuously modify channel proteins from five different classes [67]. These data clearly demonstrate that modest changes to the plasma membrane can have significant biological effects. Similarly, DHA has been found to decrease the elastic coefficient of the plasma membrane by only 10%, yet this modest decrease is sufficient to alter protein structure, as detected by gramicidin channel function [68, 69]. Consistent with the observation that small perturbations to the plasma membrane can have significant biological effects, we observed modest changes in FRET signals (3 – 5%) at the plasma membrane enriched with n-3 PUFA, which was associated with the suppression of T cell lymphoproliferation.
Since n-3 PUFA in general are considered anti-inflammatory in part by suppressing CD4+ T cell activation and differentiation into the pro-inflammatory Th1 and Th17 CD4+ T cell effector subsets, one could surmise that n-3 PUFA might negatively influence host immune response against infectious diseases. Indeed, we have demonstrated using a guinea pig model of mycobacterial infection that animals fed a diet enriched with n-3 PUFA were more susceptible to a low-dose pulmonary challenge of virulent M. tuberculosis [70]. This phenotype was also recapitulated in the Fat-1 mouse model [33]. Furthermore, exposing SMAD3-/- mice fed a high dose of fish oil (2.25% - 6%) to Helicobacter hepaticus worsened colonic inflammation, and lead to increased post-infection mortality [71]. These observations highlight the context-dependent, anti-inflammatory effects of n-3 PUFA.
In conclusion, this study highlights how n-3 PUFA, found in dietary fish oil, directly affect the lipid-lipid interactions fundamental to the formation of lipid raft mesodomains in the plasma membrane, thereby perturbing downstream signals required for T cell proliferation. The evolution of the lipid raft model to include nanoclustering of proteins that are highly regulated by both lipids and cortical actin cytoskeleton demonstrates the intricacies of organizing signaling components at the cell surface. Our novel findings, that n-3 PUFA perturb the spatial organization of PI(4,5)P2 at the cell surface, leading to suppressed actin remodeling and lymphoproliferation, further support the concept that n-3 PUFA can be used rationally for the treatment of chronic inflammatory diseases in which T cells are inappropriately activated.
Supplementary Material
Highlights.
The mechanisms of n-3 PUFA action at the plasma membrane are addressed.
n-3 PUFA increased the co-clustering of lipid raft markers in CD4+ T cells.
n-3 PUFA decreased the non-raft pool of PI(4,5)P2 is in CD4+ T cells.
n-3 PUFA suppressed lymphoproliferation, which was rescued by exogenous PI(4,5)P2.
Protein palmitoylation was unaffected by n-3 PUFA in CD4+ T cells.
Acknowledgments
The authors would like to thank Dr. Jing X. Kang for the Fat-1 breeder mice, and Evelyn Callaway for animal husbandry. We also thank DSM-Martek Biosciences for providing the DHA enriched algal oil. T.Y.H. was supported by pre-doctoral fellowships from the National Science and Engineering Research Council (NSERC) of Canada and Texas A&M University System Health Science Center Microbial Pathogenesis Training Grant. We also acknowledge support from U.S. Department of Agriculture CSREES (Cooperative State Research, Education, and Extension Service) Special Grant, “Designing Foods for Health” 2008-34402-17121] and NIH grants CA129444 and P30ES023512. Confocal and multiphoton microscopy was performed in the Texas A&M University College of Veterinary Medicine & Biomedical Sciences Image Analysis Laboratory, supported by NIH-NCRR (1S10RR22532-01).
Abbreviations
- CO
corn oil
- CFSE
carboxyfluorescein succinimidyl ester
- DAG
diacylglycerol
- DHA
docosahexaenoic acid
- ECFP
enhanced cyan fluorescent protein
- EGCG
epigallocatechin gallate
- EPA
eicosapentaenoic acid
- FRAP
fluorescence recovery after photobleaching
- FRET
Förster resonance energy transfer
- IS
immunological synapse
- n-3 PUFA
n-3 polyunsaturated fatty acids
- PI
propidium iodide
- PI(4,5)P2
phosphatidylinositol-(4,5)-bisphosphate
- ROI
region of interest
- TLC
thin layer chromatography
- YPet
yellow fluorescent protein for energy transfer
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 2.Kusumi A, Fujiwara TK, Chadda R, Xie M, Tsunoyama TA, Kalay Z, Kasai RS, Suzuki KG. Dynamic organizing principles of the plasma membrane that regulate signal transduction: commemorating the fortieth anniversary of Singer and Nicolson's fluid-mosaic model. Annu Rev Cell Dev Biol. 2012;28:215–250. doi: 10.1146/annurev-cellbio-100809-151736. [DOI] [PubMed] [Google Scholar]
- 3.Nicolson GL. The Fluid-Mosaic Model of Membrane Structure: Still relevant to understanding the structure, function and dynamics of biological membranes after more than 40years. Biochim Biophys Acta. 2014;1838:1451–1466. doi: 10.1016/j.bbamem.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 4.Simons K, Gerl MJ. Revitalizing membrane rafts: new tools and insights. Nat Rev Mol Cell Biol. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 5.Andersen OS, Koeppe RE., 2nd Bilayer thickness and membrane protein function: an energetic perspective. Annu Rev Biophys Biomol Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 6.Anishkin A, Loukin SH, Teng J, Kung C. Feeling the hidden mechanical forces in lipid bilayer is an original sense. Proc Natl Acad Sci U S A. 2014;111:7898–7905. doi: 10.1073/pnas.1313364111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardie RC, Franze K. Photomechanical responses in Drosophila photoreceptors. Science. 2012;338:260–263. doi: 10.1126/science.1222376. [DOI] [PubMed] [Google Scholar]
- 8.Dustin ML, Depoil D. New insights into the T cell synapse from single molecule techniques. Nat Rev Immunol. 2011;11:672–684. doi: 10.1038/nri3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levental I, Grzybek M, Simons K. Greasing their way: lipid modifications determine protein association with membrane rafts. Biochemistry. 2010;49:6305–6316. doi: 10.1021/bi100882y. [DOI] [PubMed] [Google Scholar]
- 10.Lillemeier BF, Mortelmaier MA, Forstner MB, Huppa JB, Groves JT, Davis MM. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. 2010;11:90–96. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choudhuri K, Wiseman D, Brown MH, Gould K, van der Merwe PA. T-cell receptor triggering is critically dependent on the dimensions of its peptide-MHC ligand. Nature. 2005;436:578–582. doi: 10.1038/nature03843. [DOI] [PubMed] [Google Scholar]
- 12.Varma R, Campi G, Yokosuka T, Saito T, Dustin ML. T cell receptor-proximal signals are sustained in peripheral microclusters and terminated in the central supramolecular activation cluster. Immunity. 2006;25:117–127. doi: 10.1016/j.immuni.2006.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim W, Barhoumi R, McMurray DN, Chapkin RS. Dietary fish oil and DHA down-regulate antigen-activated CD4+ T-cells while promoting the formation of liquid-ordered mesodomains. Br J Nutr. 2013:1–7. doi: 10.1017/S0007114513002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim W, Fan YY, Barhoumi R, Smith R, McMurray DN, Chapkin RS. n-3 polyunsaturated fatty acids suppress the localization and activation of signaling proteins at the immunological synapse in murine CD4+ T cells by affecting lipid raft formation. J Immunol. 2008;181:6236–6243. doi: 10.4049/jimmunol.181.9.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zech T, Ejsing CS, Gaus K, de Wet B, Shevchenko A, Simons K, Harder T. Accumulation of raft lipids in T-cell plasma membrane domains engaged in TCR signalling. EMBO J. 2009;28:466–476. doi: 10.1038/emboj.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belluzzi A, Brignola C, Campieri M, Pera A, Boschi S, Miglioli M. Effect of an enteric-coated fish-oil preparation on relapses in Crohn's disease. N Engl J Med. 1996;334:1557–1560. doi: 10.1056/NEJM199606133342401. [DOI] [PubMed] [Google Scholar]
- 17.Farzaneh-Far R, Harris WS, Garg S, Na B, Whooley MA. Inverse association of erythrocyte n-3 fatty acid levels with inflammatory biomarkers in patients with stable coronary artery disease: The Heart and Soul Study. Atherosclerosis. 2009;205:538–543. doi: 10.1016/j.atherosclerosis.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uchiyama K, Nakamura M, Odahara S, Koido S, Katahira K, Shiraishi H, Ohkusa T, Fujise K, Tajiri H. N-3 polyunsaturated fatty acid diet therapy for patients with inflammatory bowel disease. Inflamm Bowel Dis. 2010;16:1696–1707. doi: 10.1002/ibd.21251. [DOI] [PubMed] [Google Scholar]
- 19.Fan YY, Ly LH, Barhoumi R, McMurray DN, Chapkin RS. Dietary docosahexaenoic acid suppresses T cell protein kinase C theta lipid raft recruitment and IL-2 production. J Immunol. 2004;173:6151–6160. doi: 10.4049/jimmunol.173.10.6151. [DOI] [PubMed] [Google Scholar]
- 20.Fan YY, McMurray DN, Ly LH, Chapkin RS. Dietary (n-3) polyunsaturated fatty acids remodel mouse T-cell lipid rafts. J Nutr. 2003;133:1913–1920. doi: 10.1093/jn/133.6.1913. [DOI] [PubMed] [Google Scholar]
- 21.Fan YY, Kim W, Callaway E, Smith R, Jia Q, Zhou L, McMurray DN, Chapkin RS. fat-1 transgene expression prevents cell culture-induced loss of membrane n-3 fatty acids in activated CD4+ T-cells. Prostaglandins Leukot Essent Fatty Acids. 2008;79:209–214. doi: 10.1016/j.plefa.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang JX, Wang J, Wu L, Kang ZB. Transgenic mice: fat-1 mice convert n-6 to n-3 fatty acids. Nature. 2004;427:504. doi: 10.1038/427504a. [DOI] [PubMed] [Google Scholar]
- 23.Hou TY, Monk JM, Fan YY, Barhoumi R, Chen YQ, Rivera GM, McMurray DN, Chapkin RS. n-3 polyunsaturated fatty acids suppress phosphatidylinositol 4,5-bisphosphate-dependent actin remodelling during CD4+ T-cell activation. Biochem J. 2012;443:27–37. doi: 10.1042/BJ20111589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yog R, Barhoumi R, McMurray DN, Chapkin RS. n-3 polyunsaturated fatty acids suppress mitochondrial translocation to the immunologic synapse and modulate calcium signaling in T cells. J Immunol. 2010;184:5865–5873. doi: 10.4049/jimmunol.0904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rentero C, Zech T, Quinn CM, Engelhardt K, Williamson D, Grewal T, Jessup W, Harder T, Gaus K. Functional implications of plasma membrane condensation for T cell activation. PLoS One. 2008;3:e2262. doi: 10.1371/journal.pone.0002262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mihailescu M, Soubias O, Worcester D, White SH, Gawrisch K. Structure and dynamics of cholesterol-containing polyunsaturated lipid membranes studied by neutron diffraction and NMR. J Membr Biol. 2011;239:63–71. doi: 10.1007/s00232-010-9326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaikh SR, Dumaual AC, Castillo A, LoCascio D, Siddiqui RA, Stillwell W, Wassall SR. Oleic and docosahexaenoic acid differentially phase separate from lipid raft molecules: a comparative NMR, DSC, AFM, and detergent extraction study. Biophys J. 2004;87:1752–1766. doi: 10.1529/biophysj.104.044552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams JA, Batten SE, Harris M, Rockett BD, Shaikh SR, Stillwell W, Wassall SR. Docosahexaenoic and eicosapentaenoic acids segregate differently between raft and nonraft domains. Biophys J. 2012;103:228–237. doi: 10.1016/j.bpj.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaikh SR, Kinnun JJ, Leng X, Williams JA, Wassall SR. How polyunsaturated fatty acids modify molecular organization in membranes: insight from NMR studies of model systems. Biochim Biophys Acta. 2015;1848:211–219. doi: 10.1016/j.bbamem.2014.04.020. [DOI] [PubMed] [Google Scholar]
- 30.Shaikh SR. Biophysical and biochemical mechanisms by which dietary N-3 polyunsaturated fatty acids from fish oil disrupt membrane lipid rafts. J Nutr Biochem. 2012;23:101–105. doi: 10.1016/j.jnutbio.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turk HF, Chapkin RS. Membrane lipid raft organization is uniquely modified by n-3 polyunsaturated fatty acids. Prostaglandins Leukot Essent Fatty Acids. 2013;88:43–47. doi: 10.1016/j.plefa.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chapkin RS, Wang N, Fan YY, Lupton JR, Prior IA. Docosahexaenoic acid alters the size and distribution of cell surface microdomains. Biochim Biophys Acta. 2008;1778:466–471. doi: 10.1016/j.bbamem.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonilla DL, Fan YY, Chapkin RS, McMurray DN. Transgenic mice enriched in omega-3 fatty acids are more susceptible to pulmonary tuberculosis: impaired resistance to tuberculosis in fat-1 mice. J Infect Dis. 2010;201:399–408. doi: 10.1086/650344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaki SP, Barhoumi R, Berginski ME, Sreenivasappa H, Trache A, Gomez SM, Rivera GM. Nck enables directional cell migration through the coordination of polarized membrane protrusion with adhesion dynamics. J Cell Sci. 2013;126:1637–1649. doi: 10.1242/jcs.119610. [DOI] [PubMed] [Google Scholar]
- 35.Rivera GM, Vasilescu D, Papayannopoulos V, Lim WA, Mayer BJ. A reciprocal interdependence between Nck and PI(4,5)P(2) promotes localized N-WASp-mediated actin polymerization in living cells. Mol Cell. 2009;36:525–535. doi: 10.1016/j.molcel.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Switzer KC, Fan YY, Wang N, McMurray DN, Chapkin RS. Dietary n-3 polyunsaturated fatty acids promote activation-induced cell death in Th1-polarized murine CD4+ T-cells. J Lipid Res. 2004;45:1482–1492. doi: 10.1194/jlr.M400028-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallace PK, Muirhead KA. Cell tracking 2007: a proliferation of probes and applications. Immunol Invest. 2007;36:527–561. doi: 10.1080/08820130701812584. [DOI] [PubMed] [Google Scholar]
- 38.Byrum JN, Van Komen JS, Rodgers W. CD28 Sensitizes TCR Ca2+ Signaling during Ag-Independent Polarization of Plasma Membrane Rafts. J Immunol. 2013 doi: 10.4049/jimmunol.1300485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chichili GR, Westmuckett AD, Rodgers W. T cell signal regulation by the actin cytoskeleton. J Biol Chem. 2010;285:14737–14746. doi: 10.1074/jbc.M109.097311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodgers W. Making membranes green: construction and characterization of GFP-fusion proteins targeted to discrete plasma membrane domains. Biotechniques. 2002;32:1044–1046. 1048, 1050–1041. doi: 10.2144/02325st05. [DOI] [PubMed] [Google Scholar]
- 41.Chichili GR, Rodgers W. Clustering of membrane raft proteins by the actin cytoskeleton. J Biol Chem. 2007;282:36682–36691. doi: 10.1074/jbc.M702959200. [DOI] [PubMed] [Google Scholar]
- 42.Koushik SV, Chen H, Thaler C, Puhl HL, 3rd, Vogel SS. Cerulean, Venus, and VenusY67C FRET reference standards. Biophys J. 2006;91:L99–L101. doi: 10.1529/biophysj.106.096206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brzostowski JA, Meckel T, Hong J, Chen A, Jin T. Imaging protein-protein interactions by Forster resonance energy transfer (FRET) microscopy in live cells. Curr Protoc Protein Sci. 2009;19:19.15. doi: 10.1002/0471140864.ps1905s56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khakh BS, Fisher JA, Nashmi R, Bowser DN, Lester HA. An angstrom scale interaction between plasma membrane ATP-gated P2X2 and alpha4beta2 nicotinic channels measured with fluorescence resonance energy transfer and total internal reflection fluorescence microscopy. J Neurosci. 2005;25:6911–6920. doi: 10.1523/JNEUROSCI.0561-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pralle A, Keller P, Florin EL, Simons K, Horber JK. Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J Cell Biol. 2000;148:997–1008. doi: 10.1083/jcb.148.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varnai P, Lin X, Lee SB, Tuymetova G, Bondeva T, Spat A, Rhee SG, Hajnoczky G, Balla T. Inositol lipid binding and membrane localization of isolated pleckstrin homology (PH) domains. Studies on the PH domains of phospholipase C delta 1 and p130. J Biol Chem. 2002;277:27412–27422. doi: 10.1074/jbc.M109672200. [DOI] [PubMed] [Google Scholar]
- 47.Day CA, Kenworthy AK. Mechanisms underlying the confined diffusion of cholera toxin B-subunit in intact cell membranes. PLoS One. 2012;7:e34923. doi: 10.1371/journal.pone.0034923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calloway N, Owens T, Corwith K, Rodgers W, Holowka D, Baird B. Stimulated association of STIM1 and Orai1 is regulated by the balance of PtdIns(4,5)P(2) between distinct membrane pools. J Cell Sci. 2011;124:2602–2610. doi: 10.1242/jcs.084178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Litvinov RI, Chen X, Bach TL, Lian L, Petrich BG, Monkley SJ, Kanaho Y, Critchley DR, Sasaki T, Birnbaum MJ, Weisel JW, Hartwig J, Abrams CS. Loss of PIP5KIgamma, unlike other PIP5KI isoforms, impairs the integrity of the membrane cytoskeleton in murine megakaryocytes. J Clin Invest. 2008;118:812–819. doi: 10.1172/JCI34239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Zhao L, Suzuki A, Lian L, Min SH, Wang Z, Litvinov RI, Stalker TJ, Yago T, Klopocki AG, Schmidtke DW, Yin H, Choi JK, McEver RP, Weisel JW, Hartwig JH, Abrams CS. Platelets lacking PIP5KIgamma have normal integrin activation but impaired cytoskeletal-membrane integrity and adhesion. Blood. 2013;121:2743–2752. doi: 10.1182/blood-2012-07-445205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Imai A, Yano K, Kameyama Y, Nozawa Y. Reversible thrombin-induced modification of positional distribution of fatty acids in platelet phospholipids: involvement of deacylation-reacylation. Biochem Biophys Res Commun. 1981;103:1092–1099. doi: 10.1016/0006-291x(81)90920-7. [DOI] [PubMed] [Google Scholar]
- 52.Webb Y, Hermida-Matsumoto L, Resh MD. Inhibition of protein palmitoylation, raft localization, and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J Biol Chem. 2000;275:261–270. doi: 10.1074/jbc.275.1.261. [DOI] [PubMed] [Google Scholar]
- 53.Zeyda M, Staffler G, Horejsi V, Waldhausl W, Stulnig TM. LAT displacement from lipid rafts as a molecular mechanism for the inhibition of T cell signaling by polyunsaturated fatty acids. J Biol Chem. 2002;277:28418–28423. doi: 10.1074/jbc.M203343200. [DOI] [PubMed] [Google Scholar]
- 54.Stulnig TM, Huber J, Leitinger N, Imre EM, Angelisova P, Nowotny P, Waldhausl W. Polyunsaturated eicosapentaenoic acid displaces proteins from membrane rafts by altering raft lipid composition. J Biol Chem. 2001;276:37335–37340. doi: 10.1074/jbc.M106193200. [DOI] [PubMed] [Google Scholar]
- 55.Stulnig TM, Berger M, Sigmund T, Raederstorff D, Stockinger H, Waldhausl W. Polyunsaturated fatty acids inhibit T cell signal transduction by modification of detergent-insoluble membrane domains. J Cell Biol. 1998;143:637–644. doi: 10.1083/jcb.143.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gu Z, Wu J, Wang S, Suburu J, Chen H, Thomas MJ, Shi L, Edwards IJ, Berquin IM, Chen YQ. Polyunsaturated fatty acids affect the localization and signaling of PIP3/AKT in prostate cancer cells. Carcinogenesis. 2013;34:1968–1975. doi: 10.1093/carcin/bgt147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kenworthy AK, Nichols BJ, Remmert CL, Hendrix GM, Kumar M, Zimmerberg J, Lippincott-Schwartz J. Dynamics of putative raft-associated proteins at the cell surface. J Cell Biol. 2004;165:735–746. doi: 10.1083/jcb.200312170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Owen DM, Williamson DJ, Magenau A, Gaus K. Sub-resolution lipid domains exist in the plasma membrane and regulate protein diffusion and distribution. Nat Commun. 2012;3:1256. doi: 10.1038/ncomms2273. [DOI] [PubMed] [Google Scholar]
- 59.Saarikangas J, Zhao H, Lappalainen P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol Rev. 2010;90:259–289. doi: 10.1152/physrev.00036.2009. [DOI] [PubMed] [Google Scholar]
- 60.Johnson CM, Chichili GR, Rodgers W. Compartmentalization of phosphatidylinositol 4,5-bisphosphate signaling evidenced using targeted phosphatases. J Biol Chem. 2008;283:29920–29928. doi: 10.1074/jbc.M805921200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chapkin RS, Somers SD, Erickson KL. Dietary manipulation of macrophage phospholipid classes: selective increase of dihomogammalinolenic acid. Lipids. 1988;23:766–770. doi: 10.1007/BF02536219. [DOI] [PubMed] [Google Scholar]
- 62.Damsgaard CT, Frokiaer H, Lauritzen L. The effects of fish oil and high or low linoleic acid intake on fatty acid composition of human peripheral blood mononuclear cells. Br J Nutr. 2008;99:147–154. doi: 10.1017/S0007114507791900. [DOI] [PubMed] [Google Scholar]
- 63.Feskens EJ, Kromhout D. Epidemiologic studies on Eskimos and fish intake. Ann N Y Acad Sci. 1993;683:9–15. doi: 10.1111/j.1749-6632.1993.tb35688.x. [DOI] [PubMed] [Google Scholar]
- 64.Kim W, McMurray DN, Chapkin RS. n-3 polyunsaturated fatty acids--physiological relevance of dose. Prostaglandins Leukot Essent Fatty Acids. 2010;82:155–158. doi: 10.1016/j.plefa.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen G, Raman P, Bhonagiri P, Strawbridge AB, Pattar GR, Elmendorf JS. Protective effect of phosphatidylinositol 4,5-bisphosphate against cortical filamentous actin loss and insulin resistance induced by sustained exposure of 3T3-L1 adipocytes to insulin. J Biol Chem. 2004;279:39705–39709. doi: 10.1074/jbc.C400171200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bhonagiri P, Pattar GR, Habegger KM, McCarthy AM, Tackett L, Elmendorf JS. Evidence coupling increased hexosamine biosynthesis pathway activity to membrane cholesterol toxicity and cortical filamentous actin derangement contributing to cellular insulin resistance. Endocrinology. 2011;152:3373–3384. doi: 10.1210/en.2011-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ingolfsson HI, Thakur P, Herold KF, Hobart EA, Ramsey NB, Periole X, de Jong DH, Zwama M, Yilmaz D, Hall K, Maretzky T, Hemmings HC, Jr, Blobel C, Marrink SJ, Kocer A, Sack JT, Andersen OS. Phytochemicals perturb membranes and promiscuously alter protein function. ACS Chem Biol. 2014;9:1788–1798. doi: 10.1021/cb500086e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bruno MJ, Rusinova R, Gleason NJ, Koeppe RE, 2nd, Andersen OS. Interactions of drugs and amphiphiles with membranes: modulation of lipid bilayer elastic properties by changes in acyl chain unsaturation and protonation. Faraday Discuss. 2013;161:461–480. doi: 10.1039/c2fd20092a. discussion 563-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bruno MJ, Koeppe RE, 2nd, Andersen OS. Docosahexaenoic acid alters bilayer elastic properties. Proc Natl Acad Sci U S A. 2007;104:9638–9643. doi: 10.1073/pnas.0701015104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McFarland CT, Fan YY, Chapkin RS, Weeks BR, McMurray DN. Dietary polyunsaturated fatty acids modulate resistance to Mycobacterium tuberculosis in guinea pigs. J Nutr. 2008;138:2123–2128. doi: 10.3945/jn.108.093740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woodworth HL, McCaskey SJ, Duriancik DM, Clinthorne JF, Langohr IM, Gardner EM, Fenton JI. Dietary fish oil alters T lymphocyte cell populations and exacerbates disease in a mouse model of inflammatory colitis. Cancer Res. 2010;70:7960–7969. doi: 10.1158/0008-5472.CAN-10-1396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.