Abstract
BACKROUND & AIMS
DNA structural lesions are prevalent in sporadic colorectal cancer, so we proposed that gene variants that predispose to DNA double-strand breaks (DSBs) would be found in patients with familial colorectal carcinomas of an undefined genetic basis (UFCRC).
METHODS
We collected primary T cells from 25 patients with UFCRC and matched patients without colorectal cancer (controls) and assayed for DSBs. We performed exome sequence analyses of germline DNA from 20 patients with UFCRC and 5 undiagnosed patients with polyposis. The prevalence of identified variants in genes linked to DNA integrity was compared to that of individuals without a family history of cancer. The effects of representative variants found to be associated with UFCRC was confirmed in functional assays with HCT116 cells.
RESULTS
Primary T cells from most patients with UFCRC had increased levels of the DSB marker γH2AX following treatment with DNA damaging agents, compared to T cells from controls (P<.001). Exome sequence analysis identified a mean 1.4 rare variants/patient that were predicted to disrupt functions of genes relevant to DSBs. Controls (from public databases) had a much lower frequency of variants in the same genes (P<.001). Knockdown of representative variant genes in HCT116 CRC cells increased γH2AX. Detailed analysis of immortalized patient-derived B cells, which contained variants in the Werner syndrome, RecQ helicase-like gene (WRN, encoding T705I), and excision repair cross-complementation group 6 (ERCC6, encoding N180Y), revealed reduced levels of these proteins and increased DSBs, compared to B cells from controls. This phenotype was rescued by exogenous expression of WRN or ERCC6. Direct analysis of the recombinant variant proteins confirmed defective enzymatic activities.
CONCLUSIONS
These results provide evidence that defects in suppression of DSBs underlie some cases of UFCRC; these can be identified by assays of circulating lymphocytes. We specifically associated UFCRC with variants in WRN and ERCC6 that reduce capacity for repair of DNA DSBs. These observations could lead to a simple screening strategy for UFCRC, and provide insight into the pathogenic mechanisms of colorectal carcinogenesis.
Keywords: colon cancer, hereditary cancer, genomic instability, tumorigenesis
INTRODUCTION
Familial colorectal carcinoma (FCRC) is characterized by early disease onset and/or occurrence of CRC in multiple family members. Several FCRC syndromes have been linked with specific germline defects: Familial Adenomatous Polyposis coli with the Wnt pathway gene adenomatous polyposis coli (APC), Lynch syndrome with a group of mismatch repair (MMR) genes (most commonly MLH1, MSH2, MSH6, and PMS2), and MutY-H polyposis with the eponymous base excision repair gene1. However, most FCRC remains genetically undefined (UFCRC), accounting for some 20% of CRC in the United States.
Clinical guidelines advise starting CRC screening in UFCRC families at earlier ages and, depending on family history, more frequent intervals2. While beneficial, this strategy is inefficient. Family members who are not genetically predisposed are subjected to unnecessary costs and morbidity, while some of those actually at risk may be under-screened. Intensive genome wide association studies have sought to identify additional genes underlying UFCRC. These studies have yielded only moderate associations at multiple genome locations, implying dauntingly complex genetics3–8. No common molecular defect has been recognized.
Most sCRCs exhibit chromosomal instability, yet its molecular basis has remained ill-defined9,10. In a few instances, somatic mutations have been found in genes involved in mitosis or mitotic checkpoints11. Recent studies suggest that replicative stress, rather than mitotic defects, may underlie chromosomal instability in many sCRCs12. One FCRC family was described in which a germline BUBR1 variant perturbed genome stability in peripheral blood lymphocytes (PBLs)13, suggesting that PBLs, the cells most readily obtained from patients, might reveal defects in other UFCRC patients. We hypothesized that genetic defects causing constitutional genome instability underlie a major fraction of UFCRC, and can be detected by biological assays in PBLs. Validation of this hypothesis would suggest strategies to improve screening for CRC.
RESULTS
Constitutional defects in suppression of DNA DSBs in UFCRC patients
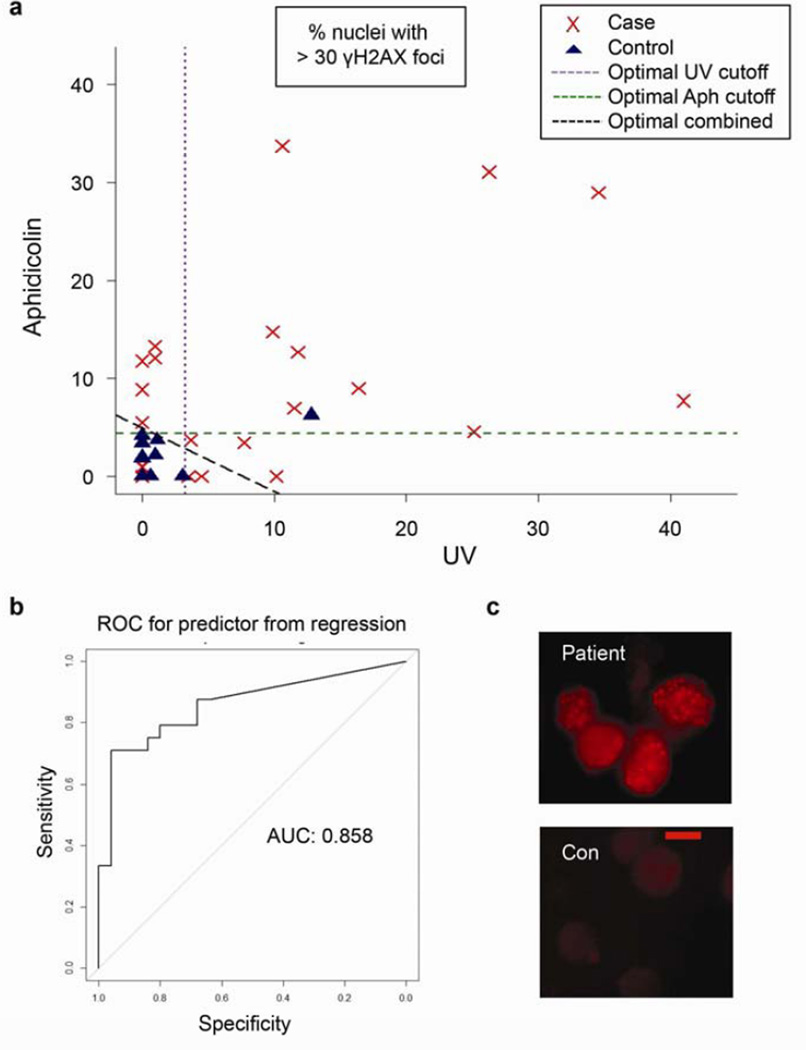
Using patient PBLs, we directly tested the hypothesis that germline mutations caused constitutional cellular defects in suppression of DSBs. For this purpose, we identified 25 CRC patients who developed the disease before age 50 and/or had at least one family member with CRC, and tested negative for defined FCRC syndromes (Table 1, Supplementary Tables 1 and 2). We also examined 5 patients who developed marked polyposis (10 or more polyps) by age 50 and similarly tested negative for known FCRC syndromes (Table 2, Supplementary Table 3). We cultured T-cells, the cell type that is most readily cultured from PBLs, with and without treatment with the DNA polymerase inhibitor aphidicolin or ultraviolet light (UV). Immunofluorescence was used to assay the formation of DSB-associated γ(phospho)-H2AX foci (Figure 1). The UFCRC samples exhibited markedly higher γH2AX levels than the controls (Figure 1A, P < 0.001). The mean γH2AX scores for aphidicolin and UV treatments were each significantly greater than the controls (P = 0.001 and P = 0.0005, respectively), and together, these results yielded an area under the ROC curve of 0.85, indicating strong discrimination between the two groups (Figure 1B). Without DNA damaging treatments, 4 patients also exhibited greater γH2AX staining than any control and 6 patients had greater γH2AX staining than all but one control (Table 1).
Table 1.
uFCRC Patient High Quality Variants, DSB Phenotypes, Polyp Histories, and Family Histories.
| Patient ID # |
HQ Variants | γH2AX + | Flow Cytometry |
Polyps (No or type) |
Family History of Neoplasia |
|---|---|---|---|---|---|
| 111797 | CHAF1B | Aph, UV | ↑S | No | Sis/CRC@41; M/CRC@72; MA/Ovar@63; MGF/Gastric@46; MGM/Panc@72 |
| 118294 | SHPRH | Aph, Campt | ↑S | No | F/pols@72; M/ut@71; M/BCC@76; MC/Laryn@51; MGF/Panc@69; PA/Br&Skin@33; PGF/Esoph@51 |
| 120713 (Pt1) | WRN; ERCC6 | No RX, Aph, Campt, UV, Etop | ↑G2/M | No | F/Blad; F&M/BCC@40s; Bro/BCC@50; Sis/CRC@48; PC1xR/Br@50; PC1xR/Ovar@40s |
| 122517 | POLD1; MCM10; MPG; SKA1 | Aph, Campt | Nl | mult | Sis/pols@37; F/CRC@41; PA/CRC@27; PGM/CRC@62; PGF/IntestSarc@31; PGF Sis/Sig@62 |
| 123000 | SPAG5 BRD4 | Aph, UV | NT | No | Bro/pols@64; Bro/pols@67; MU/CRC@60s; MA/CRC@60s; MC/CRC@68; F/CRC@79 |
| 123006 | MASTL | UV | ↑S | No | No |
| 123163 | DYNC1H1 CHAF1A | Nl | Nl | No | MA/Melan@45; A/Br@61; A/CRC@49; A/pols@58; Neph/pols@33; GM/CRC@63; GA/lymph@73; 4 1stCs1xR/CRC; GA/Uter@52; GA/Br@80,CRC; C/CRC@55 |
| 123320 | MSH2 | UV | ↑G2/M | No | No |
| 124744 | FANCL | Nl | Nl | No | Sis/pols@51; Bro/pols@56; Sis/pols@41; Sis/pol@39; Bro/pols@36; N/pols@27; M/Adeno@67; MA/CRC@51,pols@56; MGF/pols@47; F/CRC in pols@56; PA/CRC&pols@47; PC/CRC@58 |
| 125380 | [POLK]* | No Rx, Aph, UV | ↑S, G2/M | No | MGA/Esoph; MGA/Gastric; MCA/CRC; F/Pros@63; F/pols@67; F/Panc@70; PU/pols |
| 125659 | EEPD1 | UV | Nl | No | M/pol@70s; MU/Panc@53; PGF/GI@89; PGM/Liver@63 |
| 126780 | ERCC2 | No Rx, Aph | Nl | No | M/Panc@60; MU/Gastric@60; MU/unknown@50; F/CRC@60; F/pols |
| 126875 | ANAPC10 | No Rx, Aph | Aph: ↓S ↑G2/M | No | S/Bileduct@41, MU/CRC@50s; PU/CRC@50s; MSH2 mutation in PA/Gastric@60s; pt – for MSH2 |
| 127581 | CTC1 | No Rx, Aph | Aph: ↑G2/M | No | M/CRC@58; PGF/CRC@58 |
| 129338 | None | Nl | ↑S, G2/M | mult@50s | MU/pols&CRC@79; Son/Blad@50s; Son/pols@50s; MU/pols@75; MC/Br@43; MU/pols&CRC@63; MC/pols@60s; MA/pols@75; MGM/Panc@65; MA/CRC@60s&Endom&Ovar; Son/CRC |
| 132231 | C19Orf40 | Nl | Nl | 2 adv | Bro/pols@50 |
| 132406 | DDX11*; LIG1; | Aph, UV | NT | 1 ser | Bro/CRC&pols@40,55; M/pols@50s&CRC@72 |
| 132667 | LIG3 | Aph, UV | NT | >5 serr adv@<50 | Bro/CRC@44; MU/Canc@70s; MU/Stom/Esopha@50s, MA/Panc@70s; MC/Brain@57; MC/CRC@45; MC/CRC@52; MU/Canc@85; MC/Lung@52 |
| 133012 | POLD3 | No Rx, Aph | Nl | 3 adv | M/pols@60s; P1/2Sis/pols@32; PU/pols@50 |
| 134321 | None | Nl | Nl | 1 adv | NA |
HQ Variant: variant with GO annotation of DNA replication, DNA repair, checkpoint, mitotic, or mitosis; reported in < 1/100 exomes; and scoring > 0.95 on the current PolyPhen2 program and (+) by 2/3: SIFT, Provean, and MutationAssessor programs. γH2AX+: conditions under which the patient's cells showed statistically greater staining than the control or controls: 1) higher than the optimized discriminator line for aphidicolin or UV treatment (Fig 2); 2) higher than all controls for the No Rx condition; or 3) statistically greater than its specific matched controls (Pt1 and Pt2). Flow Cytometry: replicative phases in which the % of patient’s cells was > 20% higher than the controls. Polyps: polyps in the patients - known number and nature are listed (mult: >3, adv: advanced (> 1 cm, high grade dysplasia, or villous histology), ser: serrated adenomas. NT: not tested, Nl: normal. Family history of neoplasia: F: father, M: mother, Sis: sister, Bro: brother, MA: maternal aunt, MU: maternal uncle, PU: paternal uncle, MC: maternal cousin etc./organ site@age at diagnosis, Pols: polyps.
The POLK variant was predicted to be dysfunctional by PP2 and MutationAssessor but narrowly not by SIFT, and hence is technically not HQV (see main text).
The DDX11 variant sequence was also found in 2 other strong homology regions on chr12 and was flagged with low quality metrics in ExAC.
Table 2.
High Quality Variants in Polyposis Patients
| Patient ID No. | HQ Variant | Personal History of Cancer and Polyps |
Family History of Neoplasia |
|---|---|---|---|
| 120200 | ERCC3* | >20 adenomas@50 | No |
| 126784 | None | 10 adenomas in 20s+ | M/Br@44 and 51; MA/Br@45; MGM/Br@84; PGM/Br@73; PGF/Pros@70 |
| 130170 | DCLRE1A ERCC4 SPC24 BMP7 | >20 adenomas@47 | Sis/pols&CRC@52; MA/CRC@50; MGM/Lymph@55; MC/Pols |
| 130924 | SHPRH NCAPD3 ESPL1 TEX14 | Hodgkins@23 50–100 adenomas@50 | Sis/Thyr@40; F/CRC@78; PU/UnknownCancer@70s; PGM/Br@70s; MA/Panc@50 |
| 133486 | RFC2 MIS18BP1 | 11 adenomas@33+ | B/Pols@47and @60, PA/pols@82, PU/CRC&Pols@70 |
HQ Variant: variant with GO annotation of DNA replication, DNA repair, checkpoint, mitotic, or mitosis; reported in < 1/100 exomes; and scoring > 0.95 on the current PolyPhen2 program and deleterious by 2/3 other programs: SIFT, Provean, and MutationAssessor. Polyps: polyps in the patients - known number are listed. Family history of neoplasia: F: father, M: mother, Sis: sister, Br: brother, MA: maternal aunt, MU: maternal uncle, PU: paternal uncle, MC: maternal cousin etc./organ site@age at diagnosis, Pols: polyps.
This variant does not technically fit the HQV definition used in this study, but is of interest due to frameshift resulting in early truncation. In addition this patient harbors another TT to AA change (c.113_114delinsAA). This variant is not represented in ExAC.
Figure 1. Elevated γH2AX in T-cells from patients with UFCRC.
(a) Primary T-cells from 25 UFCRC patients and 25 age- and sex-matched controls were stimulated by PHA and IL-2, treated with aphidicolin or UV, and stained for nuclear γH2AX foci. The percent of cells with ≥ 30 nuclear foci is depicted. Cases: red ‘x’s, controls: blue triangles. Dashed lines: statistically optimal cutoff points to discriminate between samples with high and low γH2AX levels for each treatment or the two tests combined, as indicated. Using the combination (diagonal line), 18/25 patients exhibited high γH2AX levels versus 1/25 controls (P < 0.001). A total of 14,368 cells were scored, with a minimum of 100 and mean of 143/condition (b) The area under the curve gauges the ability of the combined γH2AX scores for aphidicolin and UV to discriminate between patients and controls. (c) Representative images of γH2AX foci in a patient and control (con). Scale bar = 5 um.
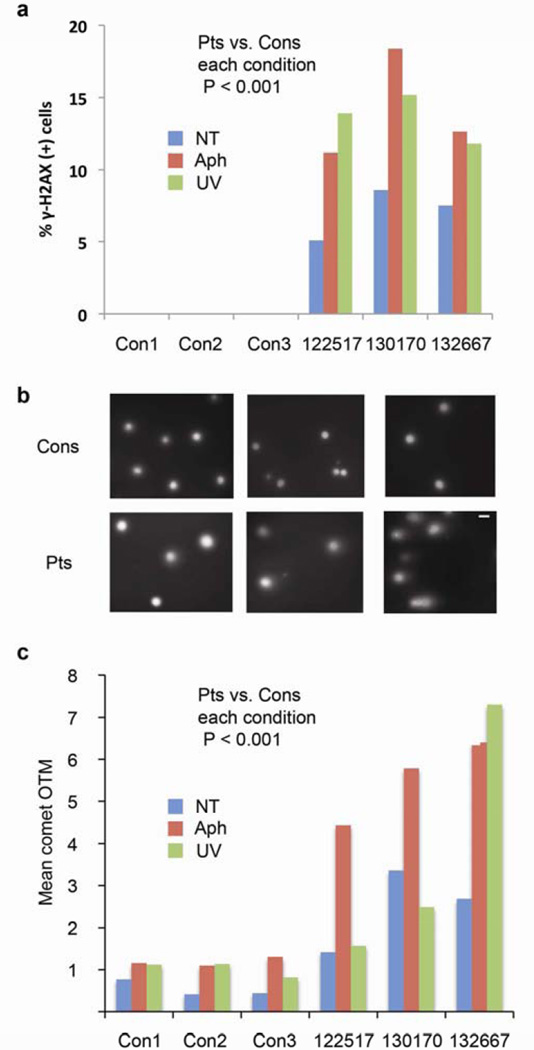
Next, we subjected T cells from 2 UFCRC patients, a polyposis patient, and 3 controls to parallel analysis of γH2AX foci and nuclear ‘comets’. The latter assay measures the migration of broken DNA fragments from nuclei upon application of electric current. All three patients showed greater γH2AX and larger comets than the three controls under all conditions (Figure 2). The presence of this phenotype in a polyposis patient suggested that it is not a secondary effect of malignancy or its treatment. Further supporting this idea, we compared pretreatment PBLs from UFCRC patients and controls to PBLs from 10 sporadic cases of CRC (sCRC). Baseline values for γH2AX in untreated PBLs showed no statistically significant differences between patients versus matched controls for UFCRC or sCRC, although UFCRC trended higher (Supplementary Figure S1a). UV treatment did not strongly induce γH2AX in PBLs from sCRC patients but did in PBLs from UFCRC (P = 0.04, Supplementary Figure S1b). Interestingly, sCRC PBLs did respond to aphidicolin treatment, although not as strongly as UFCRC PBLs, representing a partial phenotype.
Figure 2. Increased γH2AX and nuclear comets in UFCRC patients and a polyposis patient.
(a) T-cells from 2 UFCRC patients (122517 and 132667) and an undiagnosed polyposis patient (130170) were cultured, treated, and scored for γH2AX as described in Fig 1. NT, not treated; Aph, aphidicolin; UV, ultraviolet irradiation. (b) Images of nuclear comet ‘olive tail moments’ (OTM). Scale bar = 10 um. (c) Quantitation of OTM for the specimens in (a) provides a second measurement of DSB.
Defects in DNA damage responses might either cause or be augmented by changes in cell cycle progression, e.g. through cell cycle checkpoints. Flow cytometry was used to screen for altered cell cycle compartmentalization (Table 1). A subset (9/17 analyzed) of patients exhibited modestly increased S or G2/M fractions, but these differences did not achieve statistical significance. There was also no statistically significant correlation (or substantial trend) of increased basal or induced γH2AX levels with age, tumor location, smoking, or history of chemotherapy (Supplementary Table 4).
Exome sequencing of UFCRC patients reveals variants in DNA DSB repair genes
To identify inherited defects in pathways that suppress DSBs, we subjected germline (PBL) DNA from 20 UFCRC and 5 polyposis patients to exome sequencing. More than 83% of target sequence was covered at least 20-fold. Rare missense variants were flagged in a set of 1155 genes that had Gene Ontology (GO) terms including DNA replication, DNA repair, checkpoint, mitosis, or mitotic, and/or which had been identified in a genome-wide siRNA screen to strongly suppress γH2AX (top 500 genes)14. High quality variants (HQVs) were defined stringently as those predicted to disrupt protein dysfunction by scores > 0.95 in PolyPhen2 (PP2), the most established prediction algorithm15, and by at least two of three other protein prediction programs: Provean16, SIFT17, and MutationAssessor18. All HQVs were confirmed by examining the exome reads for sequence quality and variant representation in the Exome Aggregation Consortium (ExAC) and by direct Sanger sequencing.
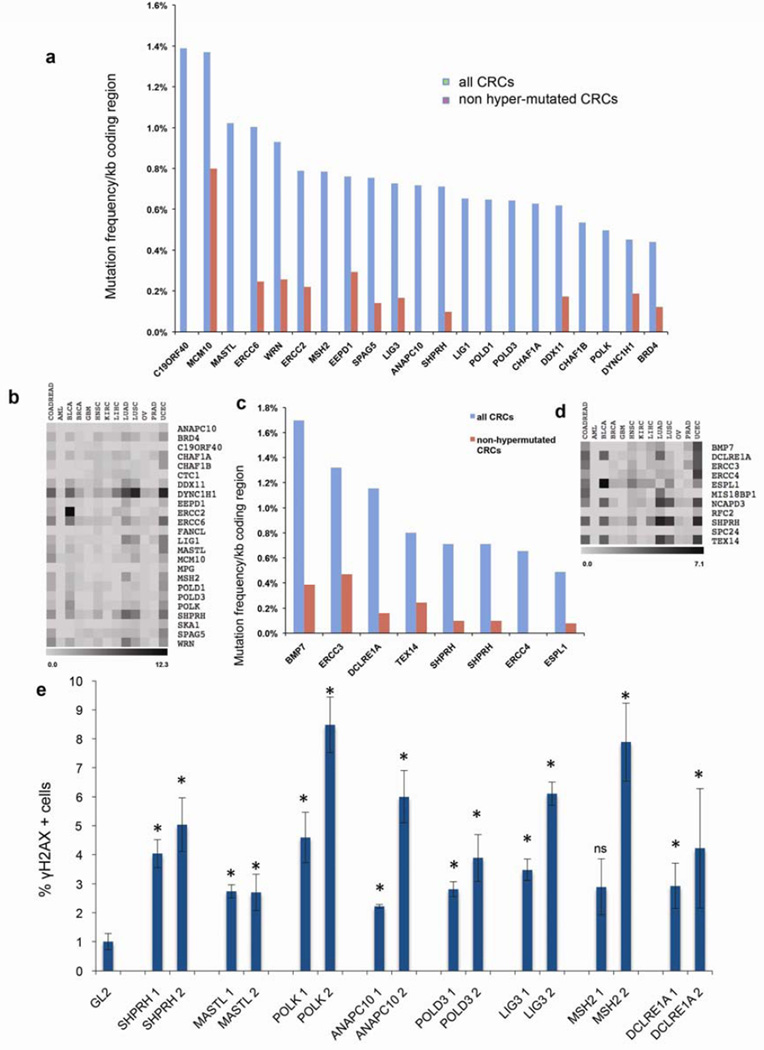
HQVs were found in 17/20 patients with UFCRC (Table 1, Supplementary Tables 1 and 2) and 4/5 patients with undiagnosed polyposis (Table 2, Supplementary Table 3), yielding a total of 35 variants in 25 patients (1.4/patient). Supplementary Table S1 includes the frequency of these alleles detected in non-cancer prone controls as reported in the Exome Variant Server (EVS), while Supplementary Tables S5 and S6 presents the frequency of these variants by ethnicity using data from EVS and ExAC. Overall, these data show the variants present in the UFCRC are present at a very low frequency or not detected in controls. In addition, analysis of exomes from an independent ITMI (Inova Translational Medicine Institute) cohort of 1508 controls who denied a personal or family history of CRC indicated a 7-fold lower frequency of HQVs in this gene set (Supplementary Table 7 lists filtration of HQVs in the ITMI set, 0.2/control, P < 0.001), supporting a relationship between the identified genes and UFCRC. We also investigated whether mutations in these genes characterized sporadic cases of CRC (hypermutated and non-mutated) and other common cancers represented in The Cancer Genome Atlas (TCGA) (Figure 3A–D). While mutations were identified in some genes at an appreciable frequency in both hypermutated and non-hypermutated CRC, and following correction for coding length, comparison to the total rate of mutations in non-DSB genes indicated no statistically significant enrichment (p =0.41). These data suggested a preferential role for mutation in the DSB genes in predisposition to UFCRC.
Figure 3. Some variant Genes are mutated in sCRC and suppress DSBs in CRC cells.
(a) TCGA CRC somatic mutation frequencies in genes that exhibited HQVs in the UFCRC patients (a) or polyposis patients (c), normalized to coding region length (in kb). Abbreviations (cBioportal): COADREAD, Colorectal Adenocarcinoma; AML, Acute Myeloid Leukemia; BLCA, Bladder Urothelial Carcinoma; BRCA, Breast Carcinoma; GBM, Glioblastoma Multiforme; HNSC, Head and Neck Squamous Cell Carcinoma; KIRC, Kidney Renal Clear Cell Carcinoma; LIHC, Liver Hepatocellular Carcinoma; UAD, Lung Adenocarcinoma; LUSC, Lung Squamous Cell Carcinoma; OV, Ovarian Serous Cystadenocarcinoma; PRAD, Prostate Adenocarcinoma; UCEC, Uterine Carcinoma. (b) ‘Heat map’ of somatic mutation frequencies in the gene sets from (a), in several major tumor types. Grayscale ranges from white (no mutations) to black (the highest possible number, as indicated, expressed in %) (c, d) Same analysis as for (a, b), performed for polyposis patients. (e) siRNA depletion of genes identified as variant in UFCRC and polyposis patients elevates γH2AX in HCT116 cells. Cells were transfected with the designated siRNAs (two per gene) and scored for γH2AX. *, P < 0.05 versus GL2; ns, not significant.
Overall variant patterns and risk of neoplasia
Compared to HQVs in all GO-flagged relevant genes, the HQVs were enriched for nucleotide excision repair (NER) genes (N = 5, P = 0.0015). Using those with a minimum threshold of PP2 > 0.95, HQVs were enriched in NER (P = 0.0014), Fanconi’s anemia (FA) (P = 0.015), and DNA polymerase (P = 0.025) proteins. FA is a rare syndrome associated with bone marrow failure, genome instability, cancer, and cellular sensitivities to mitomycin C and UV. Patients 118294 and 130924 harbored the same rare R1184C variant in the DNA helicase SHPRH. This change alters an amino acid that is stringently conserved as a basic residue and was suggested by molecular modeling to directly mediate DNA binding (Supplementary Figure S2). Variants were also seen in the helicases WRN (Pt 120713) and DDX11 (Pt 132406), and the helicase linker MCM10 (Pt 122517). Although most of the UFCRC patients we studied were young (Supplementary Table 4) and had no other neoplasm, their blood relatives often manifested other cancers (Table 1). These findings are consistent with an inherited constitutional genomic instability with predisposition to CRC and potentially other tumor types.
Knockdown of variant genes induces γH2AX in HCT116 cells
To test whether the involved genes were needed to suppress DNA DSBs in CRC cells, we used siRNA to deplete 9 of these genes in HCT116 cells. For each gene, knockdown resulted in increased γH2AX foci relative to control knockdowns (Figure 3E), supporting a role for these genes in maintaining genomic stability in CRC cells. To emulate a heterozygous state, and determine whether haploinsufficiency was adequate to induce DSB phenotypes, for a number of genes we repeated experiments using very low siRNA concentrations, resulting in only ~40–60% protein depletion (Supplementary Figure S3). We observed similar results in these knockdowns, supporting the idea that loss of function of a single allele of each gene might impair the DNA damage response.
Experimental validation of variants in patient 120713
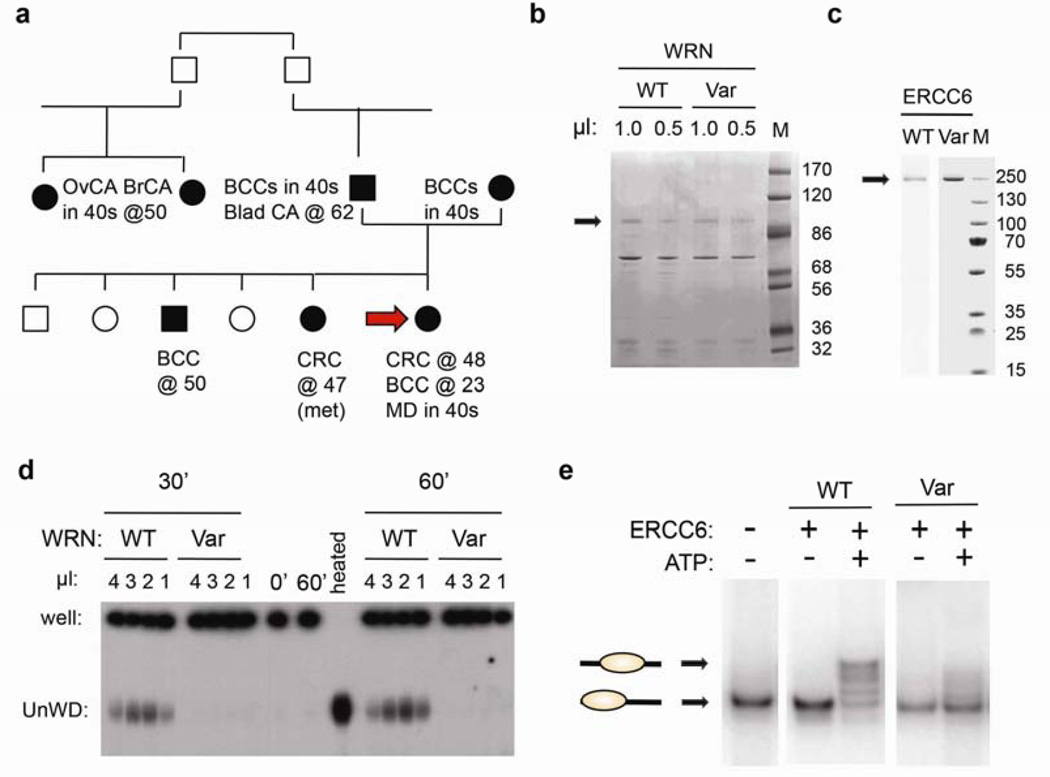
We selected patient 120713, referred to hereafter as Pt1, for detailed analysis. This patient exhibited moderately elevated γH2AX levels in response to aphidicolin and UV, typical of the UFCRC patients, and had HQVs in the WRN and ERCC6 genes (Table 1, Supplementary Table 1). The personal and family history of Pt1 revealed a predisposition to cancer consistent with WRN and ERCC6 dysfunction. Germline homozygous null mutations cause Werner’s syndrome19, characterized by chromosomal instability and cancer. WRN heterozygotes manifest subtle genome instability20. WRN is also mutant in 4% of sCRCs and other major cancers9 (Figure 3). CRC was diagnosed in Pt1 at 48 and a sister at 47, suggesting an inherited predisposition to CRC (Figure 4A).
Figure 4. Variants of WRN and ERCC6 identified in Pt1 exhibit defective enzymatic activity.
(a) Pt1 pedigree with ages of diagnosis. Carcinomas: basal cell: BCC, bladder: Blad CA, ovarian: OvCA, breast: BrCA. Macular degeneration: MD. Arrow: Pt1. (b) Comparable preparations of recombinant wild type (WT) and variant (Var) Xenopus WRN helicase domains (assessed by Coomassie blue staining of the designated extract volumes. M: molecular weight markers, in KD. (c) Defective helicase activity of the WRN T705I variant. Persistently basepaired DNA remains in the well, while unwound (UnWD) oligonucleotide migrates into the gel. (−) controls: No added helicase (0’ or 60’), (+) control: heat denaturation of DNA (heated). Similar results were seen in three independent experiments using two independent preparations. (d) Comparable preparations of recombinant wild type (WT) and variant (Var) ERCC6 helicase domains, as in (b). (e) ERCC6 N180Y variant is defective in chromatin remodeling. End positioned nucleosomes with a single 91-bp DNA overhang (lower band) used as substrate. WT ERCC6 generated nucleosomes at different translational positions in an ATP-dependent manner (upper bands). Similar results were seen in three independent experiments.
Homozygous germline mutations in ERCC6 account for the majority of cases of Cockayne syndrome, which are characterized by developmental deficiency and sun sensitivity21. ERCC6 knockout mice are prone to UV-induced skin tumors22 and ERCC6 somatic mutations are seen in CRC and other major carcinoma types21,9 (Figure 3). Basal cell carcinoma was diagnosed in Pt1 at 23, in a brother at 50, and in both parents in their 40s. This tumor is uncommon before 50, rare before 25, UV-associated, and familial23. Pt1 also developed early onset macular degeneration, a UV-associated disorder that is also often familial and controversially associated with ERCC622. Her father developed bladder cancer; sporadic bladder tumors are known to harbor WRN and ERCC6 mutations (Figure 3B). Two paternal cousins developed early breast and ovarian cancer, respectively (Figure 4A). Thus, Pt1’s clinical history and pedigree raise the possibility of genetic defects combining in Pt1 to yield a strong constitutional defect in repair of UV-associated and spontaneous DNA damage, with CRC predisposition. The identified WRN and ERCC6 variants are excellent candidates to account for this condition.
To evaluate the importance of WRN and ERCC6 in the phenotype of patient 1, we first used siRNA studies to confirm the need for both genes to suppress DSBs in CRC cells (Supplementary Figure S4). We next directly assessed the activity of the detected variants, using standard in vitro assays that have been established for both WRN and ERCC6. The WRN helicase unwinds branched DNA and fosters end resection, for recombination-based repair. The Pt1 WRN variant T705I represents a non-conservative change in a highly conserved residue of the helicase domain (Supplementary Table 1). We introduced the homologous WRN T705I variant (T646I in Xenopus) into the Xenopus WRN helicase domain, which is active in vitro17. The variant protein was devoid of helicase activity (Figure 4B, D).
ERCC6 is an ATP-dependent chromatin remodeling protein, essential for NER. The Pt1 N180Y variant is a novel, non-conservative change in a stretch of highly conserved residues that do not define a motif but are predicted to be functionally important (Supplementary Figure S5). We introduced the ERCC6 N180Y variant into human ERCC6. Native ERCC6 translocated nucleosomes in vitro in an ATP-dependent manner (Figure 4C, E). In contrast, the variant protein showed much reduced activity.
Pt1 primary lymphocytes exhibited an elevated G2/M fraction (Table 1)). In analysis of metaphase spreads, Pt1 cells exhibited an increased number of sporadic chromosomal gains (4/50 or 8% (each a different chromosome) versus 1.3% in three controls, Supplementary Figure S6A). These data provided further evidence for genomic instability in Pt1. We assayed primary T-cells from Pt1 and her matched control, with and without treatment with additional agents known to induce DSBs, and scored γH2AX foci using a more sensitive scale than before. Pt1 cells showed greater γH2AX foci under each condition (Supplementary Figure S6B; each P < 0.001, except UV: P = 0.006). Similar results were seen using an independent blood draw and confocal microscopy, to confirm that the foci were nuclear (Supplementary Figure S7). Pt1 cells were particularly sensitive to camptothecin (Supplementary Figure S6B), the canonical drug to which WRN cells are sensitive20. These results support a genomic instability phenotype in Pt1 due to defective suppression of DNA DSBs and defects in WRN and/or ERCC6.
To assess the impact of the variants on expression of the cognate genes in Pt1 cells, we generated two independent Epstein Barr Virus-transformed B-cell lines from Pt1 as well as a matched control. Immunoblotting (IB) showed that protein levels of WRN and ERCC6 were lower in the patient-derived lines than the control lines (Supplementary Figure S8). These results suggested that the variants also result in a net reduced stability of the variant proteins and/or reduced formation of the native transcript in vivo, beyond the potential direct effects on protein function suggested by the in vitro analysis.
These Pt1 cell lines recapitulated the genomic instability phenotypes seen in Pt1 primary cells. The Pt1 cell lines exhibited higher γH2AX levels than control lines, at baseline and after treatment with aphidicolin (Supplementary Figure S9A), and showed higher levels of p53 and P-ATM (Supplementary Figure S9B), both associated with DSBs, and greater comet sizes (Figure 5C–D; each P < 0.001) than control lines. Finally, overexpression of either WRN or ERCC6 (Figure 5A, B) markedly reduced γH2AX levels (Figure 5A, B) and resulted in smaller comets (Figure 5C–D; each comparison versus empty vector P < 0.0001, except as noted) in Pt1 cells, indicating that dysfunction of both WRN and ERCC6 contributed to genomic instability in these cells.
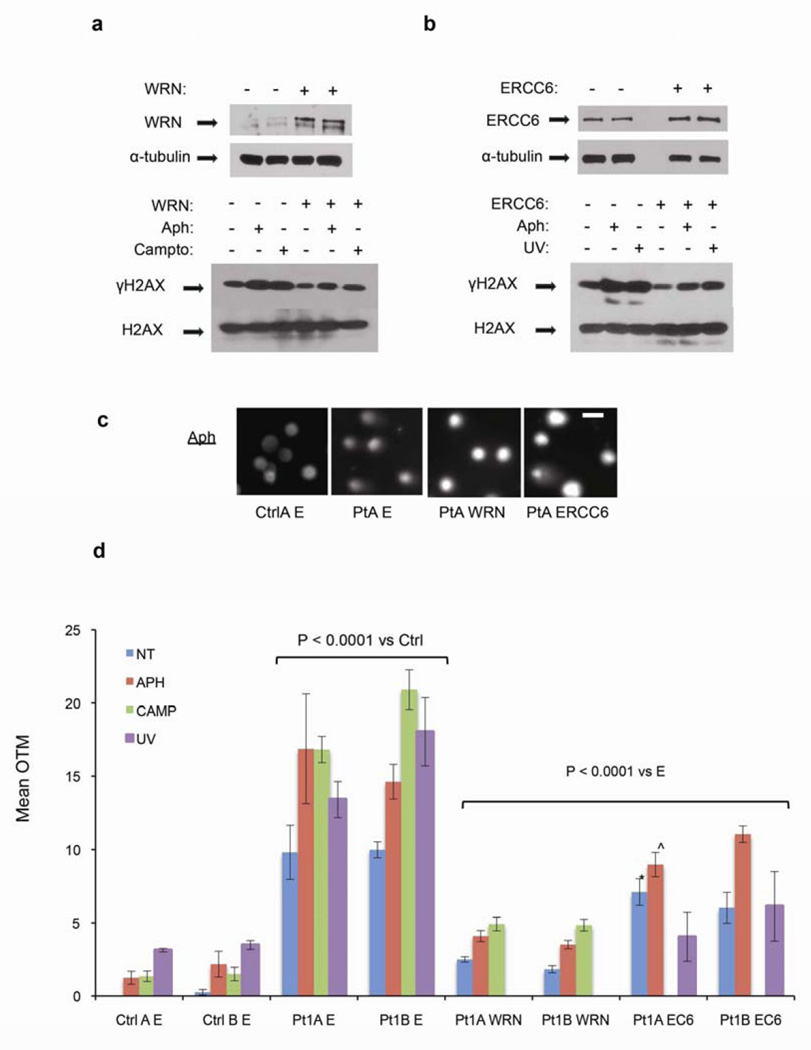
Figure 5. Pt1-derived PBL cell lines exhibit excess DNA DSBs that are suppressed by exogenous expression of WRN or ERCC6.
Two independently derived cell lines (A, B) from the matched control and Pt1, respectively, were transfected with empty vector (−) or vector expressing wild type WRN or ERCC6 and were left untreated (NT) or treated with aphidicolin, camptothecin, or UV, as indicated. (a, b) Top: Representative IB of the exogenous proteins in WRN- and ERCC6-transfected cells (+), referenced to α-tubulin loading control. Bottom: Representative γH2AX levels referenced to total H2AX. (c, d) Nuclear comets/olive tail moments (OTM) for the specimens analyzed in (a, b) are shown. (c) Representative comets from aphidicolin-treated cells from matched control (Ctrl) or Patient (Pt) transfected with empty vector (E) or vector expressing the indicated genes (WRN, ERCC6). Scale bar = 10 um. (d) OTM was elevated in Pt1 cells relative to control lines in each condition (P < 0.0001), and WRN or ERCC6 (EC6) expression reduced these scores (P < 0.0001 for each comparison versus the empty vector (E), except *, P = 0.04 (*), or ^, P = 0.001 (^)). A total of 1915 comets were scored, a mean of 68 and minimum of 34/condition. Similar results were seen in a two independent experiments.
DISCUSSION
We present evidence here that many UFCRC patients exhibit constitutional defects in suppression of DNA DSBs. Primary lymphocytes from the UFCRC patients exhibited higher γH2AX levels than sCRC and non-cancer control patients, at baseline and in response to treatments that augment DSBs, confirming constitutional predispositions to DSBs and suggesting potential for a non-invasive and low cost screening strategy that might augment current CRC screening approaches. Exome sequencing revealed HQVs in relevant genes, including the NER and FA pathways, DNA polymerases, and DNA helicases. Large control populations exhibited much lower prevalence of HQVs in the same genes. Although some of the genes with HQVs are mutated at detectable frequencies in sporadic tumors, no specific enrichment for mutation of these genes was seen in sCRC tumors. Together, these data support a preferential mutation in our gene set in UFCRC. siRNA studies showed that compromise of many of these genes was sufficient to erode genome stability in CRC cells. Detailed study of Pt1 implicated WRN and ERCC6 variants in the phenotype.
Beyond the WRN and ERCC6 genes studied in detail here, 4 HQVs mapped at or near chromosomal sites linked previously to CRC risk (Supplementary Table 1, ‘Chr band’ column, bold). Patient 122517 was found to harbor the POLD1 S478N variant recently described in several British families with FCRC and polyposis24. This variant, in the proofreading domain, appears to increase base substitution errors. Of note, however, CRCs in the British patients also exhibited chromosomal instability, not seen in other base substitution syndromes, such as Lynch. Patient 122517 also harbored HQVs in MCM10, MPG, and SKA1. Among the genes analyzed, MCM10 was one of the most frequently somatically mutated in CRC (Figure 3). Thus, the POLD1 S478N variant, alone or in conjunction with other variants, may also augment DSBs. The only HQV in Pt 133012 is in POLD3. POLD3 was recently linked to CRC susceptibility by genome-wide association studies of non-coding polymorphisms5 and implicated in DNA break-induced replication25. The variant in pt 133012 is the first POLD3 coding variant reported in FCRC. While this manuscript was under review, FA gene variants were described in several FCRC families26. These data strengthen genetic linkage of the DSB phenotype to CRC risk.
Among the patients with polyposis, Pt 130170 harbored variants in (NER gene) ERCC4, SPC24, and the FA gene DCLRE1A, as well as BMP7. DCLRE1A (DNA cross-link repair 1A) has recently been linked to CRC risk27. BMP receptor mutations have been found in juvenile polyposis28, and BMP2 and BMP4 have been genetically linked to CRC risk29. Patient 123320 exhibited a single HQV in MSH2, even though her CRC showed stable microsatellites and normal expression of mismatch repair proteins. This MSH2 variant has not been linked to Lynch syndrome (Table 1). Beyond its role in MMR, MSH2 has been found to facilitate DSB repair via homologous recombination30,31.
Only one patient with elevated γH2AX did not carry a HQV: patient 125380. However, this patient harbors a POLK variant G403D, which received the strongest possible score from PolyPhen2, was predicted to be dysfunctional by MutationAssessor (Table 1, Supplementary Table 2), and narrowly missed being called deleterious by SIFT. POLK is recruited to DNA structural lesions by SHPRH, which was variant in two other patients (Supp Table 1, Supp Table 3). Thus, we suspect that the POLK variant is responsible for the cellular and clinical phenotype in patient 125380 and that 3 of our 25 patients harbor defective variants in these two directly interacting proteins.
The HQVs we identified appear to confer risk when heterozygous, consistent with most FCRC syndromes and a previously described pedigree with a BUBR1 variant13. Further study will be needed to clarify whether the variants commonly act through haploinsufficiency, dominant negative effects, and/or inter-variant genetic interactions. Our siRNA studies suggest that many of these genes are haploinsufficient. Results from Pt1 imply effects from two HQVs in different genes in a single patient. This situation is relatively novel in hereditary cancer and may help to account for the genetic complexity of UFCRC.
In our study, the majority of UFCRC samples exhibited elevated γH2AX levels and HQVs in genes that suppress DNA DSBs. Our findings suggest that this phenotype might represent the largest molecular class of hereditary CRC defined to date. Classic cytogenetic studies in PBLs from patients with colon and other cancers have noted increased micronuclei, a marker of genomic instability32–34. Moreover, while our manuscript was under review, it was reported that lymphocytes from colon cancer patients exhibit on average greater comet sizes35. While these studies did not distinguish between inherited and somatic causes or identify specific gene defects, they are consistent with our findings and support the notion that functional assays of PBLs may provide non-invasive, low cost tests to assist in diagnosing a predisposition to cancer. Our preliminary data suggesting a mixed response of PBLs from patients with sporadic cancer to a polymerase inhibitor, aphidicolin, versus an agent that directly damaged DNA is also intriguing. Further exploration of this difference in a larger patient cohort is clearly of interest for future work. In sum, study of PBLs offers a new approach to resolve the biological and clinical significance of rare gene variants identified by exome sequencing, and may improve clinical approaches to risk assessment.
METHODS
Patients and controls
The Institutional Review Board approved all work. Patients were seen in the Fox Chase Cancer Center Familial Risk Assessment Program. They were diagnosed with CRC before age 50 and/or had another family member with CRC and tested negative for known FCRC syndromes (see Supplementary Methods). sCRCs were defined as individuals with CRCs without a family history of cancer. Our cohort included patients from ages 40 to 82. Control samples for biological studies for both UFCRC and CRC were collected from individuals who denied a personal or family history of CRC, and were age- and sex-matched to the patients. For the large-scale exome sequencing comparison, controls were drawn from a population-based study in Virginia36 and similarly denied a personal or family history of cancer. The EVS (http://evs.gs.washington.edu/EVS/) and the ExAC website, Cambridge, MA (version 0.3) (http://exac.broadinstitute.org/) were used to assess the frequency of the selected variants in the general population or in a particular ethnic group. The ExAC data set contains information on 60,706 unrelated individuals. In each patient group, European-descent Caucasians were the dominant racial group.
Lymphocyte cell preservation, culture, drug treatments, flow cytometry, and metaphase spreads
PBLs were collected from patients and controls in identical fashion and preserved by standard methods and stimulated with PHA-M and IL-2. At 72 h, cells were left untreated or treated under the following conditions: 20 µM aphidicolin, 100 µM etoposide, 25 µM camptothecin and fixed in paraformaldehyde 2h later or 5J/m2 UV and fixed in 5h later. For flow cytometry, cells were fixed in ethanol and stained with propidium iodide. Metaphase spreads were generated by classical methods (see Supplementary Methods).
Exome sequencing and variant calling
DNA libraries were prepared from 4 µg genomic DNA using NEBNext Ultra DNA library prep Kit from Illumina (New England Biolabs, Beverly, MA) and sequenced on HiSeq2500 (Illumina, San Diego, CA). All variants were analyzed on the current PolyPhen2 website (VAR version)15. To be classified as HQV, a patient variant had to receive a score of at least 0.95 (‘probably damaging’) and at least 2/3 of the following: a SIFT score of ‘damaging’, a Provean score of ‘deleterious’ (< −2.5), and a MutationAssessor score of at least ‘moderately damaging’. HQVs were required to map to the major transcript in the Uniprot or ENTREZ Gene databases. Exome sequencing controls were seen at Inova Fairfax Hospital (see Supplementary Methods). Deleteriousness was predicted as above except that any PP2 score > 0.85 was included, regardless of their presence in a dominant transcript.
Immunofluorescence and biochemistry
For scoring in primary lymphocytes, cells were allowed to attach to poly-d-lysine-coated slides or 96-well plates and stained with anti-γH2AX antibody (#05-636, Millipore, Temecula, CA). Cells on slides were photographed on a Nikon Eclipse E800 microscope and the number of bright foci per nucleus was scored. Cells in 96-well plates were imaged on the ImageXpress Micro automated microscope (Molecular Devices, Sunnyvale, CA) driven by MetaXpress software. Images were analyzed in the Multiwavelength Scoring module of MetaXpress and results were displayed and exported utilizing the AcuityXpress software package (Molecular Devices, Sunnyvale, CA). Confocal microscopy was performed on a BioRad Radiance2000 confocal microscope. For IB for γH2AX, chromatin extracts were prepared from cell nuclei that were disrupted by sonication37.
WRN helicase assays
The helicase domain (amino acids 467–1031) of Xenopus WRN and the T646I variant (homologous to the human WRN T705I variant in Pt1) were subcloned into a pGEX vector in frame with the glutathione-S-transferase (GST) open reading frame. The DNA unwinding assay to detect the helicase activity was performed as previously described17 (see Supplementary Methods).
ERCC6 chromatin remodeling assay
Constructs encoding the Pt1 ERCC6 variant were generated by site-directed mutagenesis (Stratagene)37. ERCC6 and the ERCC6 N180Y variant were C-terminally tagged with the Flag epitope, expressed using insect SF9 culture system, purified by affinity chromatography, and assayed for chromatin remodeling as described18 (see Supplementary Methods).
Comet assays
B-cell lines cells were either left untreated or treated with aphidicolin 20 µM, camptothecin 25 µM, or UV at 8 J/m2 where indicated. The presence of DNA DSBs was assessed by neutral comet assay38. See Supplementary Methods.
Supplementary Material
ACKNOWLEDGMENTS
The authors and this work were supported by NCI Core Grant no. P30 CA006927, supporting the BioSample Repository, Cell Culture, Genomics, High Throughput, and Biological Imaging Facilities, the FCCC Institute for Personalized Medicine, an unrestricted gift to the FCCC Dept. of Medicine by the Fuginon Corporation, Temple Genomics Funds, the Gianforte family fund, FCCC institutional funds (to S.A. and G.H.E.), NIH GM084983 (to H.-Y. F), NIH CA63366 (to E.A.G.), and a subsidy of the Russian Government to Kazan Federal University (to I.G.S.). Manali Shah, Sanat Deshpande, and Margret Einarson contributed to the γH2AX scoring; Pam Nakajima, Joann Lutje, and Kerry Campbell to culture and transformation of primary lymphocytes; and Jianming Pei, Zemin Liu, and Joe Testa to scoring of metaphase spreads.
Abbreviations
- UFCRC
undefined familial colorectal cancer
- sCRC
sporadic colorectal cancer
- PBL
peripheral blood lymphocytes
- HQV
high quality variant
- DSB
double strand break
- NER
nucleotide excision repair
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: None. Transcript profiling: None. Writing Assistance: None.
Author contributions: SA, most studies, planning and figure preparation; HY, WRN biochemistry; IC & H-YF: ERCC6 biochemistry; BL, sequencing; XG, EN, IS, DLB, & JGV, variant analysis; YZ, annotation; EH and BLE, statistics; MA, modeling; EN: siRNA; MJH, patients; TJY, EAG, GHE: planning and writing manuscript
REFERENCES
- 1.Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044–2058. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burt RW, et al. Colorectal cancer screening. J Natl Compr Canc Netw. 2013;11:1538–1575. doi: 10.6004/jnccn.2013.0180. [DOI] [PubMed] [Google Scholar]
- 3.Neklason DW, et al. Common familial colorectal cancer linked to chromosome 7q31: a genome-wide analysis. Cancer Res. 2008;68:8993–8997. doi: 10.1158/0008-5472.CAN-08-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neklason DW, et al. Colorectal adenomas and cancer link to chromosome 13q22.1–13q31.3 in a large family with excess colorectal cancer. Journal of medical genetics. 2010;47:692–699. doi: 10.1136/jmg.2009.076091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunlop MG, et al. Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat Genet. 2012;44:770–776. doi: 10.1038/ng.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters U, et al. Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology. 2013;144:799–807. e724. doi: 10.1053/j.gastro.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houlston RS, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet. 2010;42:973–977. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tenesa A, Farrington SM, Prendergast JG, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 11.Cahill DP, et al. Mutations of mitotic checkpoint genes in human cancers [see comments] Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 12.Burrell RA, McClelland SE, Endesfelder D, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rio Frio T, et al. Homozygous BUB1B mutation and susceptibility to gastrointestinal neoplasia. N Engl J Med. 2010;363:2628–2637. doi: 10.1056/NEJMoa1006565. [DOI] [PubMed] [Google Scholar]
- 14.Paulsen RD, Soni DV, Wollman R, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi Y, Sims GE, Murphy S, et al. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 18.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen J, Loeb LA. Unwinding the molecular basis of the Werner syndrome. Mech Ageing Dev. 2001;122:921–944. doi: 10.1016/s0047-6374(01)00248-2. [DOI] [PubMed] [Google Scholar]
- 20.Moser MJ, Kamath-Loeb AS, Jacob JE, et al. WRN helicase expression in Werner syndrome cell lines. Nucleic Acids Res. 2000;28:648–654. doi: 10.1093/nar/28.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho I, Tsai PF, Lake RJ, et al. ATP-dependent chromatin remodeling by Cockayne syndrome protein B and NAP1-like histone chaperones is required for efficient transcription-coupled DNA repair. PLoS Genet. 2013;9:e1003407. doi: 10.1371/journal.pgen.1003407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baas DC, Despriet DD, Gorgels TG, et al. The ERCC6 gene and age-related macular degeneration. PLoS One. 2010;5:e13786. doi: 10.1371/journal.pone.0013786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacquemont C, Simon JA, D'Andrea AD, Taniguchi T. Non-specific chemical inhibition of the Fanconi anemia pathway sensitizes cancer cells to cisplatin. Mol Cancer. 2012;11:26. doi: 10.1186/1476-4598-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palles C, Cazier JB, Howarth KM, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45:136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costantino L, Sotiriou SK, Rantala JK, et al. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segui N, Mina LB, Lazaro C, et al. Germline Mutations in FAN1 Cause Hereditary Colorectal Cancer by Impairing DNA Repair. Gastroenterology. 2015 doi: 10.1053/j.gastro.2015.05.056. [DOI] [PubMed] [Google Scholar]
- 27.Zhang B, Jia WH, Matsuda K, et al. Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nat Genet. 2014;46:533–542. doi: 10.1038/ng.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howe JR, et al. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat Genet. 2001;28:184–187. doi: 10.1038/88919. [DOI] [PubMed] [Google Scholar]
- 29.Tomlinson IP, Carvajal-Carmona LG, et al. Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part of the missing heritability of colorectal cancer. PLoS genetics. 2011;7:e1002105. doi: 10.1371/journal.pgen.1002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith JA, et al. Accurate homologous recombination is a prominent double-strand break repair pathway in mammalian chromosomes and is modulated by mismatch repair protein Msh2. Mol Cell Biol. 2007;27:7816–7827. doi: 10.1128/MCB.00455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang N, Liu X, Li L, Legerski R. Double-strand breaks induce homologous recombinational repair of interstrand cross-links via cooperation of MSH2, ERCC1-XPF, REV3, and the Fanconi anemia pathway. DNA Repair (Amst) 2007;6:1670–1678. doi: 10.1016/j.dnarep.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iarmarcovai G, Ceppi M, Botta A, Orsiere T, Bonassi S. Micronuclei frequency in peripheral blood lymphocytes of cancer patients: a meta-analysis. Mutat Res. 2008;659:274–283. doi: 10.1016/j.mrrev.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 33.Bonassi S, El-Zein R, Bolognesi C, et al. Micronuclei frequency in peripheral blood lymphocytes and cancer risk: evidence from human studies. Mutagenesis. 2011;26:93–100. doi: 10.1093/mutage/geq075. [DOI] [PubMed] [Google Scholar]
- 34.Karaman A, Binici DN, Kabalar ME, Calikusu Z. Micronucleus analysis in patients with colorectal adenocarcinoma and colorectal polyps. World J Gastroenterol. 2008;14:6835–6839. doi: 10.3748/wjg.14.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maffei F, et al. Micronucleus frequency in human peripheral blood lymphocytes as a biomarker for the early detection of colorectal cancer risk. Mutagenesis. 2014 doi: 10.1093/mutage/geu007. [DOI] [PubMed] [Google Scholar]
- 36.Bodian DL, McCutcheon JN, Kothiyal P, et al. Germline variation in cancer-susceptibility genes in a healthy, ancestrally diverse cohort: implications for individual genome sequencing. PLoS One. 2014;9:e94554. doi: 10.1371/journal.pone.0094554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lake RJ, Geyko A, Hemashettar G, et al. UV-induced association of the CSB remodeling protein with chromatin requires ATP-dependent relief of N- terminal autorepression. Mol Cell. 2010;37:235–246. doi: 10.1016/j.molcel.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olive PL, Wlodek D, Banath JP. DNA double-strand breaks measured in individual cells subjected to gel electrophoresis. Cancer Res. 1991;51:4671. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.