Abstract
Notch and EGFR signaling pathways play important roles in photoreceptor differentiation during Drosophila eye development. Notch signaling induces Enhancer of Split (E(spl)) proteins to repress atonal (ato) expression and restrict R8 photoreceptor cell fate. The R8 precursors express rhomboid (rho), which is required for the release of active EGFR ligand to activate EGFR signaling in surrounding cells for the subsequent stepwise recruitment. However, it is not clear about the mechanisms of transcriptional regulation of rho and how the lateral inhibition of Notch signaling and rho expression are coordinated. In this study, we show that inactivation of Groucho (Gro), an evolutionally conserved transcriptional corepressor, inhibits Ato upregulation, delays R8 determination, and promotes differentiation of R2-5 type of neurons. We demonstrate that these phenotypes are caused by a combination of the loss of Notch-mediated lateral inhibition and the precocious activation of EGFR signaling due to deregulated rho expression. Blocking EGFR signaling by Pnt-RNAi in conjunction with Gro-inactivation leads to lateral inhibition defects with deregulated Ato expression and R8 differentiation. We further show that inactivation of E(spl), which are the Gro binding transcription factors, causes deregulated rho expression and extra R8 cells within and posterior to the morphogenetic furrow (MF), and that E(spl) mediates the binding of Gro to the regulatory regions of both rho and ato genes in eye disc cells. Our results suggest that Gro inhibits rho expression in undifferentiated cells and represses the expression of both ato and rho in non-R8 precursors during initiation of photoreceptor differentiation in an E(spl)-dependent manner. The latter function of Gro provides novel insights into the mechanism that coordinates R8 specification with the restriction of initial rho expression to developing R8 cells.
Keywords: Groucho, Su(H), E(spl), Rhomboid, EGFR, Drosophila photoreceptor differentiation
Introduction
The development of multiple organs in Drosophila such as the eye, the adult femoral chordotonal organ, and the embryonic chordotonal stretch receptor involves the initial selection of neuronal precursors by lateral inhibition and the stepwise recruitment of additional neurons through EGFR signaling. Proper recruitment requires locally activated EGFR signaling, which is achieved by restricting the expression of rhomboid (rho), the rate-limiting component of EGFR signaling (Freeman, 1997; Freeman, 2008; Shilo, 2005), to the initially selected neuronal precursors. How the selection of the initial neurons by lateral inhibition and the restriction of rho expression are coordinated is not known.
During Drosophila eye development, a morphogenetic furrow (MF) initiates at the posterior tip of the early 3rd instar eye disc and moves anteriorly. Cells in the MF are arrested in the G1 phase of the cell cycle and initiate photoreceptor differentiation. Photoreceptor differentiation in the Drosophila developing eye initiates with upregulation of the proneural protein Atonal (Ato), which is progressively restricted in the MF to clusters of cells and eventually to single R8 precursors, which also express the sensory marker Senseless (Sens) (Frankfort and Mardon, 2002) (Fig. 1A). Enhancer of Split (E(spl)) proteins, which are HES (Hairy/Enhancer of Split) family of transcription factors induced by Notch signaling activation, are required to mediate lateral inhibition and specification of the R8 precursors (Li and Baker, 2001; Turki-Judeh and Courey, 2012). Although R8 cell fates do not require EGFR, activation of EGFR signaling in cells surrounding the R8 precursors is required for the recruitment and differentiation of all the subsequent photoreceptors and accessory cells in each ommatidia (Freeman, 1997). Interestingly, precocious activation of EGFR signaling in eye discs suppresses Ato expression and R8 cell fates (Chen and Chien, 1999; Dominguez et al., 1998), indicating EGFR activity needs to be precisely regulated to achieve its specific functions during eye development.
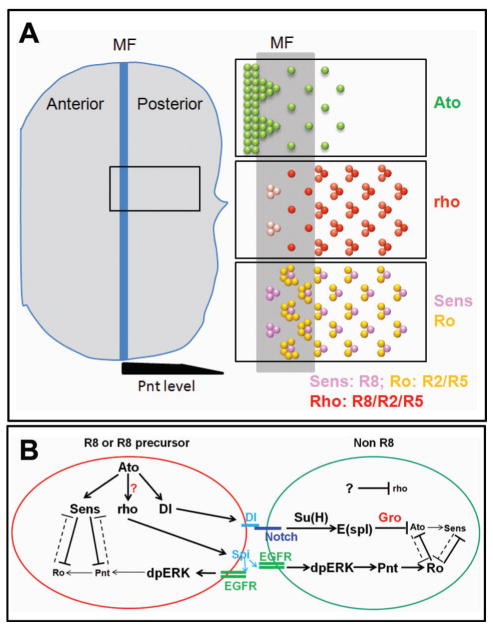
Figure 1.
Schemes of cell fate determination in developing Drosophila eye discs. (A) Diagram of Drosophila late 3rd-instar eye disc. A whole eye disc is on the left side and the enlarged gene expression patterns are on the right side. Ato is initially expressed in most cells just anterior to the MF and is progressively restricted to the intermediate groups and eventually to the single R8 cells in the MF and several rows posterior to the MF. rho expression starts in intermediate groups very weakly and its expression is upregulated and restricted to the R8 precursors and R8 cells. Later, relative weak expression of rho is observed in R2/R5 cells. Sens is expressed in R8 and R8 precursors. Ro is broadly expressed in the cells surrounding R8 precursors in the MF, and then restricted to R2/R5. Pnt, the EGFR nuclear effector, is highly expressed posterior to the MF but decreased in the more posterior area, which is shown at the bottom of the eye disc. (B) A diagram of gene networks regulating R8 and non R8 photoreceptor specification. R8 has high levels of Ato, which positively regulates Sens and the Notch ligand Dl. Sens inhibits Pnt functions in R8 to decrease Pnt targets including Ro. Dl in R8 binds Notch receptor in non R8 to activate Notch signaling and upregulate E(spl). E(spl) proteins suppress R8 markers including Ato and Sens by interacting with Gro. rho cleaves Spitz to activate EGFR signaling in R8 cells and surrounding non-R8 cells. EGFR signaling upregulates Ro expression in non R8 cells through Pnt. The regulation of rho is the subject of this study. Previous results suggest that Ato positively regulates rho, but the molecular mechanisms of rho transcriptional regulation have been largely unknown.
Precise regulation of EGFR signaling depends on restricted expression of positive regulators and negative feedback regulators (Shilo, 2005). Most of the components in the Drosophila EGFR pathway, including ligand (Spitz), receptor (EGFR), and intracellular components (Ras, Raf, MAPK, etc.), are expressed broadly. The temporal and spatial specific activation of EGFR signaling depend on the restricted expression of rhomboid family of membrane serine proteases (Freeman et al., 1992; Lee et al., 2001; Urban et al., 2001), which are required to process and activate Spitz in the presence of the membrane chaperone Star. In eye discs, rho and another Rhomboid family protein Roughoid (Ru) function redundantly. Inactivation of both Ru and rho showed phenotypes similar to complete loss of EGFR signaling (Wasserman et al., 2000).
rho is expressed dynamically during development and has important roles in regulating EGFR activities in different organs at different development stages. During Drosophila eye development, rho expression is first detected in the developing R8 precursors (Freeman et al., 1992; Spencer et al., 1998) (Fig. 1A). The Spitz molecules released from R8 precursors activate EGFR signaling in surrounding cells and induce them to differentiate into R2, R5, R3, R4 and later R1, R6 and R7 (Roignant and Treisman, 2009) (Fig. 1B). Similar to photoreceptor differentiation in eye discs, development of the adult femoral chordotonal organ and the embryonic chordotonal stretch receptor also involve the initial selection of the “primary” sense organ precursor (SOP) by lateral inhibition involving Notch signaling. Furthermore, these “primary” SOPs also express rho and release EGFR ligand, which is required for the activation of EGFR signaling in adjacent ectodermal cells and the recruitment of additional chordotonal SOPs (Lage et al., 1997; Okabe and Okano, 1997; zur Lage and Jarman, 1999). However, little is known of the mechanism by which the initial selection of neuronal precursors is linked with their expression of rho (Fig. 1B). While there are some studies implicating the involvement of Ato and Daughterless (Da) in rho expression in eye discs (Baonza et al., 2001; Chen and Chien, 1999; Spencer et al., 1998) and in the chordotonal organ of the leg discs (zur Lage and Jarman, 1999), the expression pattern of rho and ato are only partially overlapped. Since EGFR signaling is critical for photoreceptor recruitment and differentiation, elucidating the mechanisms by which rho expression is controlled can potentially provide novel insights into the coordinated process of photoreceptor determination and recruitment.
In the current study, we show that Gro is required for the repression of rho in the developing eye discs and that loss of Gro causes neuronal differentiation defects as a consequence of both deregulated EGFR signaling and loss of Notch-mediated lateral inhibition. We show that the E(spl) proteins recruit Gro to the ato and rho regulatory regions to mediate repression of both ato and rho in non-R8 cells within and posterior to MF. These results provide novel insights into the mechanisms that coordinate R8 specification with the restriction of rho expression to allow precise regulation of EGFR activities.
Results
Gro inactivation induces neurogenic phenotypes but inhibits Ato expression and delays R8 determination
Gro is a transcriptional co-repressor and performs its functions through interacting with a number of transcription factors including E(spl) proteins (Turki-Judeh and Courey, 2012). Consistent with the functions of E(spl) proteins in lateral inhibition, a previous study showed that loss of Gro led to neurogenic phenotypes in Drosophila eye discs (Chanut et al., 2000). However, the exact function of Gro in eye development is not well characterized.
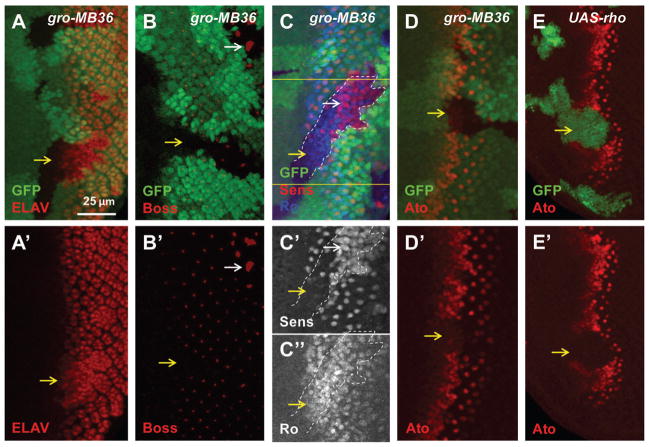
We analyzed the developmental defects caused by loss of Gro with several markers labeling different cell types. Similar to the previous report, most of the gro mutant or RNAi cells posterior to MF expressed ELAV, the pan-neural marker (Fig. 2A, S1A). The neurogenic phenotypes of Su(H) or E(spl) mutant cells are caused by loss of Notch signaling-mediated lateral inhibition, which leads to ectopic Ato expression and extra R8 specification as shown by the R8 marker Senseless (Sens) (Li and Baker, 2001). Unexpectedly, inactivation of Gro inhibited Ato upregulation within and anterior to the MF (Fig. 2D) and delayed the initiation of R8 differentiation as shown by decreased expression of R8-specific markers, Boss and Sens, close to the MF (Fig. 2B, 2C–C′, yellow arrows). Increased R8 differentiation was observed only in more posterior eye disc regions as shown by the large clusters of Boss-expressing cells (Fig. 2B, white arrow) and the extensive Sens-positive cells (Fig. 2C–C′, white arrow). These data show that Gro inactivation initially inhibits R8 differentiation but eventually extra R8 cells are differentiated at later stages.
Figure 2.
Gro inactivation causes photoreceptor differentiation defects in eye discs. The orientation of eye discs in all images is dorsal up and posterior to right (same in other figures). (A–D) Mutant mosaic clones are marked by absence of GFP. (E) overexpression clones are marked with GFP. Yellow arrows point to mutant clones in the MF. Most of the gro mutant cells posterior to the MF have ELAV expression (A). gro mutant cells close to the MF lack Boss (R8 marker), while some gro mutant cells away from the MF (white arrow) have extra Boss signal (B). gro mutant cells close to the MF have extra Ro expression (R2/R5 marker) and a lack of Sens (marker of R8 precursor and R8) expression, while some gro mutant cells away from the MF have extra Sens expression (C–C″). Ato expression is downregulated in gro mutant clones (D). Ectopic EGFR activity induced by rho overexpression reduces Ato levels (E).
To characterize the neuronal cell types that were differentiated in gro mutant clones, we further examined the expression of Rough (Ro), which is mainly found in R2/R5 cells and is upregulated in response to EGFR signaling activity. Significantly increased Ro-positive cells were observed in Gro-inactivated cells close to MF (Fig. 2C, 2C″, S1B). These results show that Gro inactivation delays R8 differentiation but promotes R2/5 photoreceptor neuron differentiation.
Blocking EGFR signaling in Gro-inactivated cells induces lateral inhibition defects
Since Sens expression is directly regulated by proneural factor Ato (Pepple et al., 2008), and since EGFR signaling activity upregulates Ro and downregulates Ato (Dokucu et al., 1996; Dominguez et al., 1998) (also see Fig. 1B), the observed effects of Gro inactivation on Ato and Ro suggest Gro may affect EGFR signaling. Indeed, ectopic EGFR activity induced by rho expression also blocked Ato upregulation within and anterior to the MF (Fig. 2E).
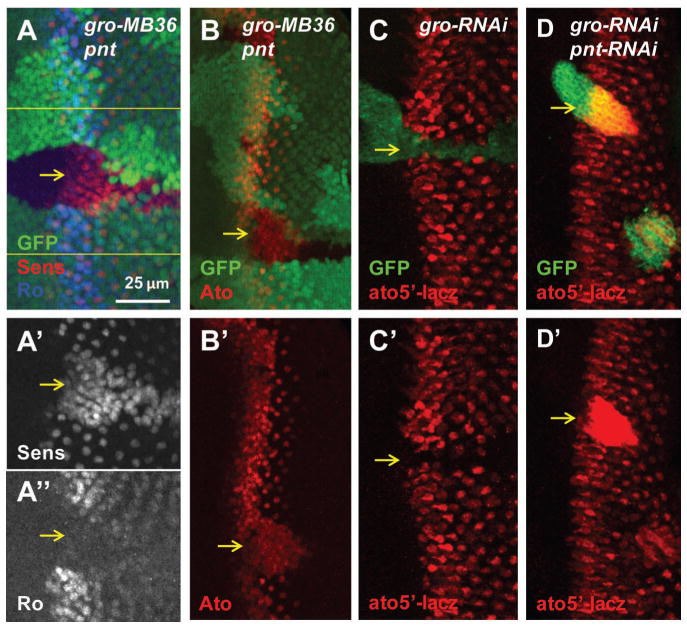
To determine whether the neurogenic defects of Gro-inactivated cells are potentially mediated by a combination of upregulated EGFR signaling and loss of Notch-mediated lateral inhibition, we tested the effect of blocking EGFR signaling on Ato expression and R8 differentiation. Indeed, blocking EGFR signaling by inactivation of Pointed (Pnt), the nuclear effector of the EGFR pathway, led to inhibited Ro expression and increased R8 differentiation (shown by Sens) in the gro pnt double inactivated cells (Fig. 3A–A″, Fig. S1C). Similarly, blocking EGFR signaling by Pnt inactivation led to increased Ato protein in the MF (Fig. 3B). We further tested whether Gro inhibited ato expression at the transcriptional level by using an ato5′-lacz reporter, which contains enhancers regulating ato expression in the intermediate groups and individual R8 precursors (Sun et al., 1998). Inactivation of Gro by RNAi inhibited ato5′-lacz reporter expression (Fig. 3C), while ato5′-lacZ reporter was upregulated in the gro pnt double inactivated cells (Fig. 3D).
Figure 3.
Repression of EGFR activity restores R8 cell fate in gro mutant cells in the MF. (A–B) Mutant mosaic clones are marked by the absence of GFP. (C–D) RNAi clones are marked with GFP. Yellow arrows point to mutant cells close to the MF. gro pnt mutant cells lack Ro and have extra Sens expression (A–A″). Ato is expressed in most of the gro pnt mutant cells (B). ato5′-lacz is suppressed by gro-RNAi (C). ato5′-lacz is expressed strongly in gro-pnt-RNAi clones (D).
These results suggest that loss of Gro causes both increased EGFR signaling as well as lateral inhibition defects and that blocking EGFR signaling allows the typical lateral inhibition defects to be observed.
Loss of Gro upregulates EGFR activity by deregulating rho expression
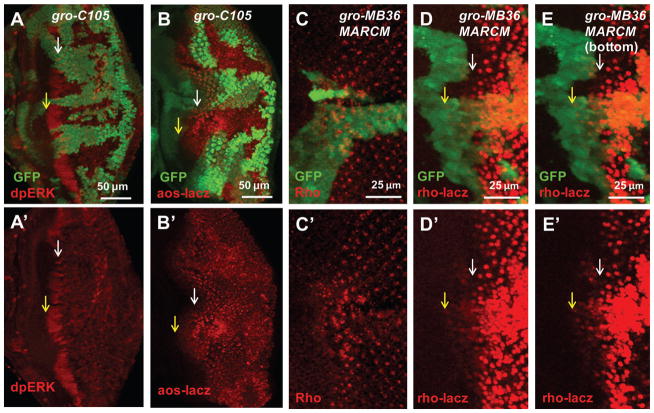
To directly characterize whether Gro inactivation affects EGFR activity, we used two readouts to examine EGFR signaling activity: the phosphorylated ERK (pERK) and enhancer trap expression of the EGFR target gene argos (aos-lacz). In WT cells, relatively high levels of pERK were detected in distinct clusters of the “intermediate groups” in the MF (Fig. 4A, white arrow). Interestingly, in gro mutant clones, expanded and higher levels of pERK were detected near the MF (Fig. 4A, yellow arrow). Argos is a target as well as a negative feedback regulator of EGFR signaling expressed in the posterior eye disc where EGFR activity is high. Precocious aos-lacz expression was detected in gro mutant cells near the MF (Fig. 4B, compare yellow and white arrows) and increased aos-lacz expression was detected in gro mutant cells posterior to the MF. Taken together, these results show that loss of Gro leads to increased EGFR signaling activity.
Figure 4.
Loss of Gro induces ectopic EGFR activity and rho expression. (A–B) Mutant mosaic clones are marked by the absence of GFP. (C–E) MARCM clones are marked with GFP. Yellow arrows point to mutant clones and white arrows point to WT cells. dpERK is upregulated in gro mutant clones in the MF (A). aos-lacz expression is increased in gro mutant clones posterior to the MF and ectopically expressed in clones in the MF (B). Comparing to the WT cells, loss of Gro induces both anterior and posterior ectopic rho protein levels (C). Transcription of rho is indicated by expression of rho-lacz. rho-lacz is expressed in R8 precursors in the MF and R8, R2 and R5 photoreceptor cells posterior to MF in WT cells, and gro mutation induces both anterior and posterior ectopic rho-lacz expression (D). In the bottom layers of the disc, the ectopic rho-lacz expression induced by gro mutation anterior to the MF is more obvious (E).
Since rho is a rate-limiting component of the EGFR pathway (Wasserman et al., 2000), we checked whether it was upregulated in gro mutant cells. rho expression starts in the R8 precursors in the MF and is later restricted mainly to the developing R8 and weakly to the R2 and R5 photoreceptor cells posterior to the MF (Freeman et al., 1992; Spencer et al., 1998). Interestingly, precocious rho protein expression in the more anterior region and increased rho protein levels within and posterior to the MF were detected in gro mutant clones (Fig. 4C). The X-81 rho-lacz enhancer trap can be used to detect rho expression (Freeman et al., 1992). Low levels of precocious rho expression in the more anterior region and deregulated rho expression in and posterior to the MF were observed in gro mutant or gro-RNAi cells (Fig. 4D–E, Fig. 5A). Furthermore, the deregulated induction of rho expression by loss of Gro is not limited to the eye discs. rho-lacz expression was also detected in the developing chordotonal SOP region of leg discs (Fig. S2A, yellow arrow), and gro-RNAi significantly expanded this rho-lacz positive region (Fig. S2B, yellow arrow). These results show that Gro is required for the repression of rho expression in multiple developing tissues.
Figure 5.
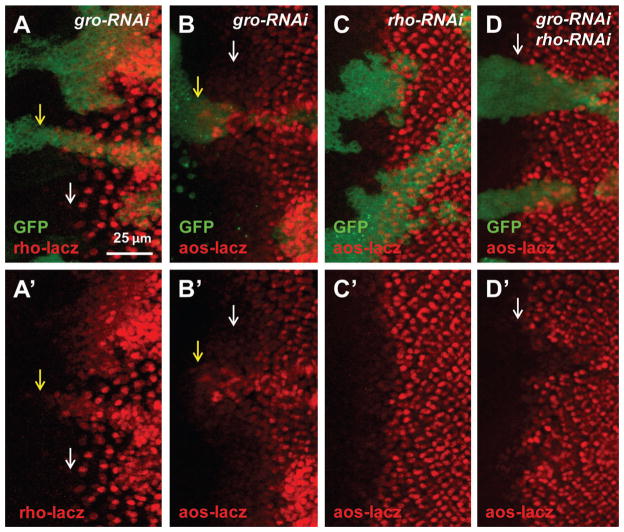
Ectopic EGFR activity induced by loss of Gro is rho dependent. (A–D) RNAi clones are marked with GFP. Yellow arrows point to mutant or RNAi clones and white arrows point to WT cells. Similar to gro mutation, gro-RNAi also induces ectopic rho-lacz and aos-lacz expression around the MF (A–B). rho-RNAi does not change aos-lacz expression (C). gro-rho-RNAi cells do not show ectopic aos-lacz expression in the MF (D).
To determine whether increased rho expression contributes to increased EGFR signaling in Gro-inactivated cells, we tested the effect of knocking down rho in conjunction with gro. Similar to gro mutation, gro-RNAi induced ectopic EGFR activity as shown by increased aos-lacz both in the MF and posterior to the MF (Fig. 5B). While rho-RNAi itself did not cause obvious changes to aos-lacz expression, rho-RNAi significantly decreased the ectopic aos-lacz expression induced by gro-RNAi (Fig. 5C–D). Taken together, these data suggest that the increased EGFR activity in Gro inactivated cells largely depends on rho expression.
E(spl) and Gro are required for the repression of rho within and posterior to the MF in eye discs
Drosophila E(spl) proteins belong to the HES family of transcription factors, which have a C-terminal WRPW motif that interacts with Gro (Turki-Judeh and Courey, 2012). The E(spl) proteins are induced by Notch signaling through the transcription factor Su(H) and mediate Notch signaling-induced lateral inhibition to restrict Ato expression to R8 precursors during photoreceptor differentiation (Li and Baker, 2001; Pepple et al., 2008; Voas and Rebay, 2004) (Fig. 1B). Since rho expression starts in the R8 precursor in the MF and is restricted mainly to the developing R8 neurons behind the MF (Freeman et al., 1992; Spencer et al., 1998) (Fig. 1A), it is possible that the E(spl)/Gro repressor complex may mediate the repression of both ato and rho to coordinate the restriction of R8 differentiation with the expression of rho in the developing R8 neurons.
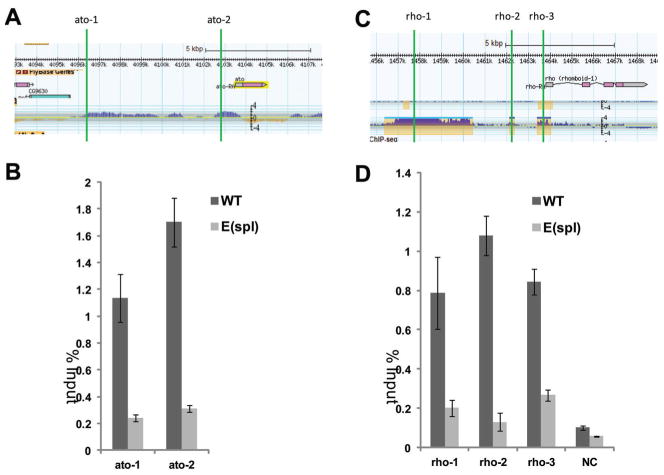
To investigate whether E(spl) may recruit Gro to coordinate the repression of ato and rho, we determined the binding of Gro to the ato and rho regulatory regions in WT and E(spl) mutant eye discs by Chromatin Immunoprecipitation (ChIP). ModENCODE data show that Gro binds to both the promoter and 5′ distal regions of ato gene (Fig. 6A) (Negre et al., 2011) (Fig. 6A). We designed oligos to detect Gro binding to the ato 5′ enhancer and promoter region (ato-1 and ato-2). As expected, significant binding of Gro to ato-1 and ato-2 regions were observed in WT eye discs as determined by ChIP (Fig. 6B). We generated eye discs that contained mostly the E(spl) mutant cells by inducing E(spl) mutant clones (all seven linked E(spl) genes are deleted in this E(spl) mutant allele) in a Minute background to determine the effect of removing E(spl) proteins on Gro binding to the ato regulatory regions. As shown in Fig. 6B, binding of Gro to the ato regulatory regions was significantly decreased in eye discs comprised mostly of E(spl) mutant cells. These results indicate that E(spl) proteins recruit Gro to the ato regulatory regions.
Figure 6.
Gro binding to the regulatory regions of ato and rho genes depends on E(spl) in eye discs. (A, C) diagrams of ChIP-chip or ChIP-seq on the ato or rho gene from modENCODE, and the regions for ChIP are marked with green lines. Gro ChIP analysis indicates significant binding of Gro to the regulatory regions of ato (ato-1, ato-2) and rho (rho-1, rho-2, rho-3) in comparison to a negative control region (NC) in cells from WT eye discs. However the Gro binding activities on these regions are much weaker in the cells from E(spl) mutant eye discs (B and D).
We used the same approach to determine the effect of removing E(spl) on the binding of Gro to the rho regulatory regions. We designed oligos to detect Gro binding to three Gro positive regions upstream of the rho gene (rho-1, rho-2, and rho-3) and one negative Gro binding region (NC) based on the ModENCODE data (Fig. 6C) (Negre et al., 2011). Consistent with the ModENCODE data, significant binding of Gro to regions rho-1, rho-2, and rho-3 were observed in WT eye discs while little binding of Gro was observed to the NC region (Fig. 6D). In contrast, in eye discs that contain mostly the E(spl) mutant cells, Gro binding to the rho-1, rho-2, and rho-3 regions was significantly decreased (Fig. 6D). These results indicate that E(spl) proteins are required for the recruitment of Gro to the rho regulatory regions. Taken together, all these data support the notion that the Gro/E(spl) complex directly represses rho and ato expression.
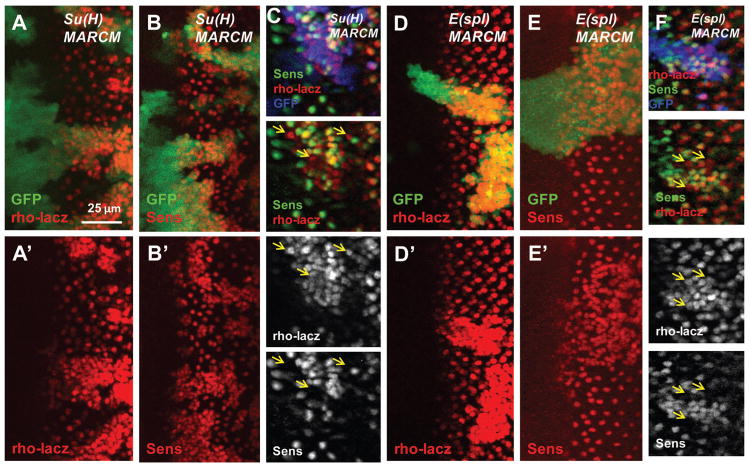
To further determine whether E(spl) proteins regulate rho expression, we examined rho-lacz expression in Su(H) or E(spl) mutant clones. Most of the cells in the Su(H) or E(spl) mutant clones had deregulated rho-lacz expression within and posterior to the MF (Fig. 7A, D). Consistent with previous studies, loss of Su(H) or E(spl) also induced ectopic Sens-positive R8 precursors or neurons (Fig. 7B, E). Double staining of rho-lacZ and Sens revealed the presence of rho-lacz positive but Sens-negative cells in both the Su(H) and the E(spl) mutant clones (yellow arrows in Fig. 7C, F). Therefore, E(spl) proteins can regulate rho expression independently of their regulation of R8 cell fate. These results suggest that E(spl) proteins coordinately repress rho and ato expression within and posterior to the MF by recruiting Gro.
Figure 7.
Some Su(H) or E(spl) mutant cells are rho-lacz positive and Sens negative. (A–F) MARCM clones are marked with GFP (GFP is colored blue in C and F). Su(H) or E(spl) mutation induces posterior ectopic rho-lacz expression (A, D) and ectopic Sens expression (B, E). Some cells in E(spl) or Su(H) mutant clones have ectopic rho-lacz expression but no Sens expression (C, F, indicated by yellow arrows).
It should be pointed out that in contrast to Gro-inactivation, precocious rho-lacz expression in the more anterior region was not observed in Su(H) or E(spl) mutant clones (Fig. 7A, D, compare with Fig. 4D–E). The difference between gro and E(spl) in inducing precocious rho expression is likely due to the presence of additional factors that function to recruit Gro to repress rho in the anterior regions.
The relationships between Ato, Da, and Gro on rho expression in developing eye discs
To further investigate the transcriptional mechanisms of rho in eye discs, we analyzed the relationships between Gro and potential transcriptional activators. Previous studies showed that the bHLH transcription factor Ato, which forms a heterodimer with another bHLH transcription factor Da, is essential for EGFR activation when the MF initiated in early L3 eye discs (Baonza et al., 2001; Chen and Chien, 1999; Jarman et al., 1993). In addition, ectopic expression of Ato using sevenless-Gal4 induces ectopic rho expression (Baonza et al., 2001). Recent studies suggest that Da may also form homodimers to activate transcription (Bhattacharya and Baker, 2011; Tanaka-Matakatsu et al., 2014). The ChIP-chip data from flybase show that Da binds to several sites upstream of rho gene (Fig. S3). These observations suggest that Da-containing complexes such as Ato/Da or Da/Da may regulate rho transcription directly. Here, we studied the relationships between Ato, Da and Gro on rho expression using mutations and RNAi knockdown.
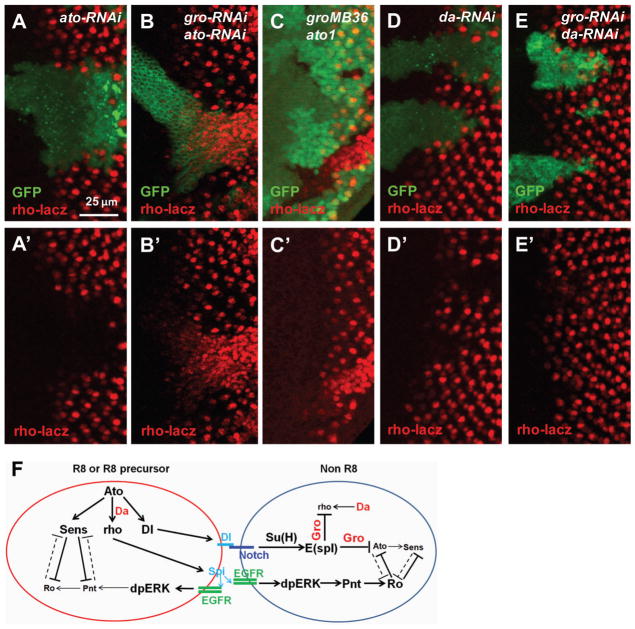
Consistent with the published results showing that Ato positively regulates rho, ato-RNAi strongly inhibited rho-lacz expression (Fig. 8A). However, high levels of rho-lacz expression was still detected in ato-gro-RNAi or gro ato mutant clones (Fig. 8B, C). Therefore Ato is required for rho expression in WT cells but is dispensable when Gro is inactivated. These results are consistent with the observation that ato expression was inhibited in gro mutant clones (Fig. 2D) and support the model that Gro represses rho directly.
Figure 8.
rho expression regulated by Ato, Da, and Gro in eye discs. (A–E) RNAi clones are marked with GFP in all images. ato-RNAi efficiently suppresses rho-lacz expression (A). ato-gro-RNAi clones have high levels of ectopic rho-lacz expression(B). ato gro mutant clones also have high levels of ectopic rho-lacz expression (C). da-RNAi efficiently suppresses rho-lacz expression (D). da-gro-double RNAi still suppresses rho-lacz expression (E). Model of Gro in regulating ato and rho transcription in R8 and non-R8 cells (F). The diagram is based on Fig. 1B and incorporates the results of this study about Gro/E(spl) repress both rho and ato and Da is essential for rho expression (shown in red).
In contrast to the effect of inactivating Ato, da-RNAi efficiently suppressed rho-lacz expression in both WT and gro-RNAi cells (Fig. 8D, E). Therefore Da is necessary for activation of rho expression even in the absence of Gro. It is possible that Da homodimer or heterodimers with other bHLH proteins can also regulate rho transcription.
Discussion
In this study, we show that Gro plays critical roles in repressing rho expression in eye discs and that E(spl) proteins are required to recruit Gro to both the ato and the rho regulatory regions to mediate their transcriptional repression in developing photoreceptor cells. Besides the mechanisms of rho transcriptional regulation, our studies provide novel insights into how the regulation of R8 selection is coordinated with the restriction of rho expression during eye development. This mechanism is likely also relevant to other settings where Notch signaling is used to select the initial founder cells and localized EGFR signaling is used for subsequent stepwise recruitment. Furthermore this study reveals that the neuronal differentiation defects observed in gro mutant clones are mediated by a combination of a loss of Notch-mediated lateral inhibition and the precocious activation of EGFR signaling.
E(spl)/Gro coordinates selection of the initial neuron and the restriction of rho expression to the selected neurons for stepwise recruitment
Photoreceptor differentiation in eye disc is initiated with the selection of R8 precursors by lateral inhibition via Notch signaling followed by stepwise recruitment of additional neurons by EGFR signaling. Proper stepwise recruitment requires localized EGFR signaling achieved by restricting rho expression to the initially selected neurons to activate EGFR signaling in the surrounding cells. Our results suggest that the E(spl) and Gro proteins are directly involved in restricting rho expression to the initially selected R8 precursors.
E(spl) proteins have WRPW motifs that can interact with Gro (Paroush et al., 1994; Turki-Judeh and Courey, 2012). The expression of E(spl) genes are regulated by the Notch signaling pathway and require the transcription factor Su(H) (Jennings et al., 1994; Li and Baker, 2001). Our results show that E(spl) proteins, which are highly expressed in non-R8 cells by Notch signaling, are also directly involved in the recruitment of Gro to the rho regulatory region to repress rho expression in non-R8 cells (Fig. 8F, non R8). This is supported by the observation that gro, E(spl) or Su(H) mutant clones show deregulated rho expression in posterior eye discs independently of R8 determination and that loss of E(spl) blocks recruitment of Gro protein to the rho regulatory region. The model that E(spl) proteins recruit Gro to repress rho expression during photoreceptor differentiation is consistent with the fact that most non-R8 cells posterior to the MF show little rho expression. Furthermore, EGFR activity in non-R8 cells can repress Ato expression through Pnt (Fig. 8F, non-R8). The low level of rho expression in R2 and R5 cells has two possible explanations: 1) high EGFR signaling in the R2-5 cells negatively regulates the repression by Gro (Hasson et al., 2005; Helman et al., 2011); or 2) specific factors in R2/R5 may contribute to the activation of rho expression (Freeman et al., 1992). In R8 cells of WT eye discs, the lack of E(spl) to recruit Gro contributes to the high rho expression (Fig. 8F, R8). Consistent with this, most of the cells in E(spl) or Su(H) mutant clones also have high level rho expression. In addition, high levels of Sens in R8 cells inhibits Pnt, and Ato can positively regulate rho expression (Fig. 8F, R8).
It is of interest to note that the development of the adult femoral chordotonal organ and the embryonic chordotonal stretch receptor also involve the initial selection of “primary” SOPs by lateral inhibition and the recruitment of additional neurons by these SOPs through the release of EGFR ligands (Lage et al., 1997; Okabe and Okano, 1997; zur Lage and Jarman, 1999). Therefore it is tempting to speculate that the E(spl)/Gro complex is also involved in coordinating the selection of the primary SOPs with the restriction of rho expression in these cells in chordotonal organ development. Consistent with this idea, we found that inactivation of Gro alone significantly increased the area of rho expression in femoral chordotonal organs in leg discs. These observations suggest that E(spl)/Gro may coordinate the selection of initial neuronal precursors with the restriction of rho expression in the selected cells in multiple organs.
Inactivation of Gro induces precocious rho expression before photoreceptor differentiation
Although loss of E(spl) or Su(H) showed an effect similar to Gro inactivation in inducing strong ectopic rho expression within and posterior to the MF, E(spl) or Su(H) inactivation did not induce obvious precocious rho expression anterior to the MF (Fig. 7A, D), which is distinct from Gro inactivation (Fig. 4D–E). Interestingly, the effects of gro, E(spl), or Su(H) mutations on differentiation are also different: while most E(spl) or Su(H) mutant cells differentiate into R8 with high levels of Ato and Sens, gro mutant cells close to the MF exhibit properties of R2/R5 with high level of Ro and low levels of Ato and Sens. On the other hand, inhibiting EGFR signaling allows high levels of Ato and Sens in gro pnt double mutant cells, which is similar to E(spl) or Su(H) mutant cells. All these data suggest that the precocious rho expression before differentiation initiation in gro mutant cells contributes to the observed effects of Gro on photoreceptor differentiation.
Gro is a corepressor protein that can interact with a number of transcription factors. It is likely that other factors are involved in Gro’s repression of rho in the anterior MF region. Hairy is also a HES protein with a WRPW motif that can interact with Gro, and is specifically expressed anterior to the MF. While loss of Hairy does not cause eye development defects, overexpression of Hairy causes neural differentiation defects (Brown et al., 1991). It is possible that Hairy functions redundantly with other proteins. In addition to Hairy, Su(H), which can also form complex with Gro (Barolo et al., 2002), is also expressed in the MF. Therefore Hairy and Su(H) may recruit Gro to repress rho expression in the anterior MF eye disc regions.
Another interesting phenotype is that Gro inactivation inhibits R8 initiation only in the areas close to the MF and significant numbers of gro mutant cells away from the MF have extra Sens and have lost Ro (Fig. 2C). This is potentially due to the significantly decreased expression of the Pnt protein in the more posterior regions of the eye discs (Boisclair Lachance et al., 2014) (Fig. 1A). Because of the mutual inhibition between Senseless and Ro (Pepple et al., 2008) (Fig. 1B), the absence of lateral inhibition to shut off Sens expression in conjunction with the reduced levels of Pnt may lead to decreased levels of Ro and accumulation of Sens in posterior eye discs.
The activation of rho expression in developing eyes
Ato/Da was implicated in rho expression in eye discs (Baonza et al., 2001; Chen and Chien, 1999). Interestingly, our results show that Ato is necessary for rho transcription in WT cells but dispensable for rho expression in gro-RNAi or gro mutant cells in eye discs. In contrast to the effects of Ato, we found Da to be necessary for rho transcription in both WT and gro-RNAi cells, suggesting that Da contributes to the activation of rho expression without Ato. This is consistent with the recent discoveries that Da homodimers may have important functions (Bhattacharya and Baker, 2011; Tanaka-Matakatsu et al., 2014). Alternatively Da may form heterodimers with other bHLH proteins that function redundantly with Ato/Da to activate rho (Fig. 8F).
Materials and Methods
Drosophila stocks and genetics
Fly stocks used in this study include: groC105 (BL 2124), groMB36 (Jennings et al., 2008), Su(H) Δ47 (BL 51295), E(spl) (BL 52011), pntΔ88 (BL 861), ato1 (BL 25779), gro RNAi (BL 35759), pnt RNAi (BL 31936), ato RNAi (BL 35017), da RNAi (BL 26319), aos-lacz (BL 2513), rho-lacz (Freeman et al., 1992), UAS-rho (Freeman lab).
Main genetic technologies used in this study include: UAS/Gal4 and Flp-out system to induce ectopic expression of protein or RNAi in clones (Brand and Perrimon, 1993; Ni et al., 2008; Pignoni and Zipursky, 1997); FLP/FRT to generate regular loss of function mosaic clones (Xu and Rubin, 1993); MARCM to generate mosaic clones with both mutation and ectopic expression (Lee and Luo, 2001). Drosophila genotypes used in each figures are listed in the supplemental materials.
hsFLP induced regular mosaic or MARCM clones were generated by heat shocking 24–48 hours after egg deposition (AED) larvae at 38°C for 1 hour. hsFLP induced Flp-out clones using Act>CD2>Gal4 were generated by heat shocking 24–48 AED larvae at 34°C for 20 minutes to 1 hour, depending on the clone sizes of each genotype. The imaginal discs were dissected from larvae three days after heat shock. All flies for the experiments were kept at 25°C.
Immunostaining
Immunostaining and imaging were performed with the protocols as described in our previous studies (Gordon et al., 2013; Zhang et al., 2014). Primary antibodies used in this study include: mouse anti-β-Galactosidase (1:100, DSHB), mouse anti-Ro (1:100, DSHB), rabbit anti-Ato (Jarman et al., 1993), Guinea pig anti-Senseless (Nolo et al., 2000), rabbit anti-dpERK (1:200, cell signaling), rabbit anti-rho (1:500) (Sturtevant et al., 1996). Secondary antibodies are from Jackson ImmunoResearch (1:200 to 1:400).
Chromatin Immunoprecipitation (ChIP)
Eye discs of 3rd instar larvae were dissected in ice-cold PBS. Samples were double fixed with 1.5 mM DSP for 40 minutes and 1.8% formaldehyde for 15 minutes (Martinez and Arnosti, 2008). After double fixation, following steps are the same as the protocol described previously (Austin et al., 1999), except that SDS was omitted from the solutions. Sixty pairs of eye discs and 20 ul Gro antibody (concentrated, from Hybridoma Bank) were used for each ChIP assay. Real time RT-PCR (qPCR) was determined using the Thermo Scientific™ DyNAmo™ SYBR™ Green qPCR Kit on an Opticon 2 Real-time PCR detector (BioRad). qPCR with each pair of primers was done in triplicate. The efficiency of Gro binding was shown by the ratio of ChIP DNA and input DNA content determined by qPCR. Primers include: rho-Gchip-1-F, 5′-AAGAACGCGAGTCAGCATTT-3′; rho-Gchip-1-R, 5′-CGTCCTCTGTCCTCCATTTC-3′; rho-Gchip-2-F, 5′-CTGGTCATGGGCATGTAGTG-3′; rho-Gchip-2-R, 5′-TTCAGAGTCCTCGTCCTCGT-3′; rho-Gchip-3-F, 5′-CAGTGAATAACCAGCGACGA-3′; rho-Gchip-3-R, 5′-GCACATCCGGATCTTGTCTT-3′; rho-Gchip-NC-F, 5′-TTCAACGGTGCATGAATGAT-3′; rho-Gchip-NC-R, 5′-ACTAGTCCGAGCGATTGCAG-3′; ato5′-1-F, 5′-TGTGCCCAAAGGAATAATCA-3′; ato5′-1-R, 5′-TCAAGGGTTGGACAAACAAA-3′; ato5′-2-F, 5′-GAGGGCTAAGGTGAAGGTCA-3′; ato5′-2-R, 5′-CAATTGATACGCTTGTTGCC-3′.
Supplementary Material
Figure S1. gro-RNAi or gro-RNAi pnt-RNAi also induce phenotypes similar to the corresponding mutations. gro-RNAi induces extra ELAV expression (A) and extra Ro and decreased Sens in the cells close to the MF but extra Sens in the cells away from the MF (B). Most of the gro-RNAi pnt-RNAi cells have Sens expression (C).
Figure S2. rho-lacz is expressed in the developing chordotonal SOP region of the leg discs (A). gro-RNAi significantly expands the rho-lacz positive region (B, yellow arrow).
Figure S3. Da ChIP-chip diagrams on the rho gene region were downloaded and adapted from Flybase. Da binds to multiple sites in the regulatory regions of the rho gene (brown dashed lines indicate fragments of DNA bound by Da).
Highlights.
Gro inactivation inhibits Ato upregulation and delays R8 differentiation
Gro inactivation promotes the R2-5 photoreceptor differentiation
Blocking EGFR signaling in gro mutant clones reveals lateral inhibition defects
Gro inactivation induces precocious rho expression and EGFR signaling activation
E(spl) recruits Gro to repress both ato and rho in posterior non-R8 cells
Acknowledgments
We thank Andy Vo, Emily Mu and Xun Pei for assistance in immunostaining or fly stock maintenance. We thank Dr. Albert Courey and Dr. Matthew Freeman for providing stocks, and Dr. Hugo Bellen, Dr. Yuh-Nung Jan, Dr. Ethan Bier and Developmental Studies Hybridoma Bank (created by the NICHD of the NIH and maintained at The University of Iowa) for providing antibodies. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. This Study was supported by grants from National Institutes of Health GM074197 and CA149275.
Footnotes
Author Contributions
T.Z. and W.D. designed the experiments. T.Z. did the experiments and collected data presented in this manuscript. T.Z. and W.D. wrote the manuscript.
Drosophila genotypes used in each figures
yw, hsFLP; FRT82B, Ubi-GFP/FRT82B groMB36
yw, hsFLP, Act>CD2>Gal4, UAS-CD8GFP; UAS-rho
yw, hsFLP; FRT82B, Ubi-GFP/FRT82B groMB36 pntΔ88
yw, hsFLP, Act>CD2>Gal4, UAS-CD8GFP; UAS-gro-RNAi/ato5′-lacz
yw, hsFLP, Act>CD2>Gal4, UAS-CD8GFP; UAS-gro-RNAi ato5′-lacz/UAS-pnt-RNAi
yw, eyFLP; FRT82B, Ubi-GFP/FRT82B groC105
yw, hsFLP; FRT82B, Ubi-GFP/aos-lacz, FRT82B groC105
yw, hsFLP; Act>y>Gal4, UAS-GFP; FRT82B, tub-Gal80/rho-lacz FRT82B groMB36
yw, hsFLP, Act>CD2>Gal4, UAS-CD8GFP; UAS-gro-RNAi/rho-lacz
yw, hsFLP, Act>CD2>Gal4, UAS-CD8GFP; UAS-gro-RNAi/aos-lacz
yw, hsFLP, Act>CD2>Gal4, UAS-CD8GFP; UAS-rho-RNAi/aos-lacz
yw, hsFLP, Act>CD2>Gal4, UAS-CD8GFP; UAS-gro-RNAi aos-lacz/UAS-rho-RNAi
yw, eyFLP;FRT82B, Ubi-GFP, rps3/FRT82B
yw, eyFLP;FRT82B, Ubi-GFP, rps3/FRT82B, Df(3R)gro[b32.2] P{ry[+t7.2]=E(spl)E8}3
yw, hsFLP, Act>CD2>Gal4,UAS-CD8GFP; FRT82B, tub-Gal80/ rho-lacz, FRT82B, Df(3R)gro[b32.2] P{ry[+t7.2]=E(spl)E8}3
yw, hsFLP; tub-Gal80, FRT40A/Su(H)Δ47, FRT40A; rho-lacz/Act>y>Gal4, UAS-GFP
yw, hsFLP, Act>CD2>Gal4,UAS-CD8GFP; rho-lacz/UAS-ato-RNAi
yw, hsFLP, Act>CD2>Gal4,UAS-CD8GFP; rho-lacz, UAS-gro-RNAi/UAS-ato-RNAi
yw, hsFLP; FRT82B, Ubi-GFP/ rho-lacz, FRT82B ato1 groMB36
yw, hsFLP, Act>CD2>Gal4,UAS-CD8GFP; rho-lacz/UAS-da-RNAi
yw, hsFLP, Act>CD2>Gal4,UAS-CD8GFP; rho-lacz, UAS-gro-RNAi/UAS-da-RNAi
yw, hsFLP, Act>CD2>Gal4, UAS-CD8GFP; UAS-gro-RNAi
yw, hsFLP, Act>CD2>Gal4, UAS-CD8GFP; UAS-gro-RNAi/UAS-pnt-RNAi
yw, hsFLP, Act>CD2>Gal4, UAS-CD8GFP; UAS-gro-RNAi/rho-lacz
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Austin RJ, Orr-Weaver TL, Bell SP. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev. 1999;13:2639–49. doi: 10.1101/gad.13.20.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baonza A, Casci T, Freeman M. A primary role for the epidermal growth factor receptor in ommatidial spacing in the Drosophila eye. Curr Biol. 2001;11:396–404. doi: 10.1016/s0960-9822(01)00125-7. [DOI] [PubMed] [Google Scholar]
- Barolo S, Stone T, Bang AG, Posakony JW. Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless. Genes Dev. 2002;16:1964–76. doi: 10.1101/gad.987402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Baker NE. A network of broadly expressed HLH genes regulates tissue-specific cell fates. Cell. 2011;147:881–92. doi: 10.1016/j.cell.2011.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisclair Lachance JF, Pelaez N, Cassidy JJ, Webber JL, Rebay I, Carthew RW. A comparative study of Pointed and Yan expression reveals new complexity to the transcriptional networks downstream of receptor tyrosine kinase signaling. Dev Biol. 2014;385:263–78. doi: 10.1016/j.ydbio.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brown NL, Sattler CA, Markey DR, Carroll SB. hairy gene function in the Drosophila eye: normal expression is dispensable but ectopic expression alters cell fates. Development. 1991;113:1245–56. doi: 10.1242/dev.113.4.1245. [DOI] [PubMed] [Google Scholar]
- Chanut F, Luk A, Heberlein U. A screen for dominant modifiers of ro(Dom), a mutation that disrupts morphogenetic furrow progression in Drosophila, identifies groucho and hairless as regulators of atonal expression. Genetics. 2000;156:1203–17. doi: 10.1093/genetics/156.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Chien CT. Negative regulation of atonal in proneural cluster formation of Drosophila R8 photoreceptors. Proc Natl Acad Sci U S A. 1999;96:5055–60. doi: 10.1073/pnas.96.9.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokucu ME, Zipursky SL, Cagan RL. Atonal, rough and the resolution of proneural clusters in the developing Drosophila retina. Development. 1996;122:4139–47. doi: 10.1242/dev.122.12.4139. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Wasserman JD, Freeman M. Multiple functions of the EGF receptor in Drosophila eye development. Curr Biol. 1998;8:1039–48. doi: 10.1016/s0960-9822(98)70441-5. [DOI] [PubMed] [Google Scholar]
- Frankfort BJ, Mardon G. R8 development in the Drosophila eye: a paradigm for neural selection and differentiation. Development. 2002;129:1295–306. doi: 10.1242/dev.129.6.1295. [DOI] [PubMed] [Google Scholar]
- Freeman M. Cell determination strategies in the Drosophila eye. Development. 1997;124:261–70. doi: 10.1242/dev.124.2.261. [DOI] [PubMed] [Google Scholar]
- Freeman M. Rhomboid proteases and their biological functions. Annu Rev Genet. 2008;42:191–210. doi: 10.1146/annurev.genet.42.110807.091628. [DOI] [PubMed] [Google Scholar]
- Freeman M, Kimmel BE, Rubin GM. Identifying targets of the rough homeobox gene of Drosophila: evidence that rhomboid functions in eye development. Development. 1992;116:335–46. doi: 10.1242/dev.116.2.335. [DOI] [PubMed] [Google Scholar]
- Gordon GM, Zhang T, Zhao J, Du W. Deregulated G1-S control and energy stress contribute to the synthetic-lethal interactions between inactivation of RB and TSC1 or TSC2. J Cell Sci. 2013;126:2004–13. doi: 10.1242/jcs.121301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson P, Egoz N, Winkler C, Volohonsky G, Jia S, Dinur T, Volk T, Courey AJ, Paroush Z. EGFR signaling attenuates Groucho-dependent repression to antagonize Notch transcriptional output. Nat Genet. 2005;37:101–5. doi: 10.1038/ng1486. [DOI] [PubMed] [Google Scholar]
- Helman A, Cinnamon E, Mezuman S, Hayouka Z, Von Ohlen T, Orian A, Jimenez G, Paroush Z. Phosphorylation of Groucho mediates RTK feedback inhibition and prolonged pathway target gene expression. Curr Biol. 2011;21:1102–10. doi: 10.1016/j.cub.2011.05.043. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grau Y, Jan LY, Jan YN. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell. 1993;73:1307–21. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- Jennings B, Preiss A, Delidakis C, Bray S. The Notch signalling pathway is required for Enhancer of split bHLH protein expression during neurogenesis in the Drosophila embryo. Development. 1994;120:3537–48. doi: 10.1242/dev.120.12.3537. [DOI] [PubMed] [Google Scholar]
- Jennings BH, Wainwright SM, Ish-Horowicz D. Differential in vivo requirements for oligomerization during Groucho-mediated repression. EMBO Rep. 2008;9:76–83. doi: 10.1038/sj.embor.7401122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage P, Jan YN, Jarman AP. Requirement for EGF receptor signalling in neural recruitment during formation of Drosophila chordotonal sense organ clusters. Curr Biol. 1997;7:166–75. doi: 10.1016/s0960-9822(97)70087-3. [DOI] [PubMed] [Google Scholar]
- Lee JR, Urban S, Garvey CF, Freeman M. Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell. 2001;107:161–71. doi: 10.1016/s0092-8674(01)00526-8. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–4. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Li Y, Baker NE. Proneural enhancement by Notch overcomes Suppressor-of-Hairless repressor function in the developing Drosophila eye. Curr Biol. 2001;11:330–8. doi: 10.1016/s0960-9822(01)00093-8. [DOI] [PubMed] [Google Scholar]
- Martinez CA, Arnosti DN. Spreading of a corepressor linked to action of long-range repressor hairy. Mol Cell Biol. 2008;28:2792–802. doi: 10.1128/MCB.01203-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, Li Z, Ishii H, Spokony RF, Chen J, Hwang L, Cheng C, Auburn RP, Davis MB, Domanus M, Shah PK, Morrison CA, Zieba J, Suchy S, Senderowicz L, Victorsen A, Bild NA, Grundstad AJ, Hanley D, MacAlpine DM, Mannervik M, Venken K, Bellen H, White R, Gerstein M, Russell S, Grossman RL, Ren B, Posakony JW, Kellis M, White KP. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–31. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JQ, Markstein M, Binari R, Pfeiffer B, Liu LP, Villalta C, Booker M, Perkins L, Perrimon N. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat Methods. 2008;5:49–51. doi: 10.1038/nmeth1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolo R, Abbott LA, Bellen HJ. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell. 2000;102:349–62. doi: 10.1016/s0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- Okabe M, Okano H. Two-step induction of chordotonal organ precursors in Drosophila embryogenesis. Development. 1997;124:1045–53. doi: 10.1242/dev.124.5.1045. [DOI] [PubMed] [Google Scholar]
- Paroush Z, Finley RL, Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–15. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Pepple KL, Atkins M, Venken K, Wellnitz K, Harding M, Frankfort B, Mardon G. Two-step selection of a single R8 photoreceptor: a bistable loop between senseless and rough locks in R8 fate. Development. 2008;135:4071–9. doi: 10.1242/dev.028951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignoni F, Zipursky SL. Induction of Drosophila eye development by decapentaplegic. Development. 1997;124:271–8. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- Roignant JY, Treisman JE. Pattern formation in the Drosophila eye disc. Int J Dev Biol. 2009;53:795–804. doi: 10.1387/ijdb.072483jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilo BZ. Regulating the dynamics of EGF receptor signaling in space and time. Development. 2005;132:4017–27. doi: 10.1242/dev.02006. [DOI] [PubMed] [Google Scholar]
- Spencer SA, Powell PA, Miller DT, Cagan RL. Regulation of EGF receptor signaling establishes pattern across the developing Drosophila retina. Development. 1998;125:4777–90. doi: 10.1242/dev.125.23.4777. [DOI] [PubMed] [Google Scholar]
- Sturtevant MA, Roark M, O’Neill JW, Biehs B, Colley N, Bier E. The Drosophila rhomboid protein is concentrated in patches at the apical cell surface. Dev Biol. 1996;174:298–309. doi: 10.1006/dbio.1996.0075. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jan LY, Jan YN. Transcriptional regulation of atonal during development of the Drosophila peripheral nervous system. Development. 1998;125:3731–40. doi: 10.1242/dev.125.18.3731. [DOI] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M, Miller J, Borger D, Tang WJ, Du W. Daughterless homodimer synergizes with Eyeless to induce Atonal expression and retinal neuron differentiation. Dev Biol. 2014;392:256–65. doi: 10.1016/j.ydbio.2014.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M, Xu J, Cheng L, Du W. Regulation of apoptosis of rbf mutant cells during Drosophila development. Dev Biol. 2009;326:347–56. doi: 10.1016/j.ydbio.2008.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turki-Judeh W, Courey AJ. Groucho: a corepressor with instructive roles in development. Curr Top Dev Biol. 2012;98:65–96. doi: 10.1016/B978-0-12-386499-4.00003-3. [DOI] [PubMed] [Google Scholar]
- Urban S, Lee JR, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–82. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- Voas MG, Rebay I. Signal integration during development: insights from the Drosophila eye. Dev Dyn. 2004;229:162–75. doi: 10.1002/dvdy.10449. [DOI] [PubMed] [Google Scholar]
- Wasserman JD, Urban S, Freeman M. A family of rhomboid-like genes: Drosophila rhomboid-1 and roughoid/rhomboid-3 cooperate to activate EGF receptor signaling. Genes Dev. 2000;14:1651–63. [PMC free article] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–37. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Zhang T, Liao Y, Hsu FN, Zhang R, Searle JS, Pei X, Li X, Ryoo HD, Ji JY, Du W. Hyperactivated Wnt signaling induces synthetic lethal interaction with Rb inactivation by elevating TORC1 activities. PLoS Genet. 2014;10:e1004357. doi: 10.1371/journal.pgen.1004357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Lage P, Jarman AP. Antagonism of EGFR and notch signalling in the reiterative recruitment of Drosophila adult chordotonal sense organ precursors. Development. 1999;126:3149–57. doi: 10.1242/dev.126.14.3149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. gro-RNAi or gro-RNAi pnt-RNAi also induce phenotypes similar to the corresponding mutations. gro-RNAi induces extra ELAV expression (A) and extra Ro and decreased Sens in the cells close to the MF but extra Sens in the cells away from the MF (B). Most of the gro-RNAi pnt-RNAi cells have Sens expression (C).
Figure S2. rho-lacz is expressed in the developing chordotonal SOP region of the leg discs (A). gro-RNAi significantly expands the rho-lacz positive region (B, yellow arrow).
Figure S3. Da ChIP-chip diagrams on the rho gene region were downloaded and adapted from Flybase. Da binds to multiple sites in the regulatory regions of the rho gene (brown dashed lines indicate fragments of DNA bound by Da).