Abstract
Purpose
Well-established laboratory mouse lines are important in creating genetically engineered knockout mouse models; however, these routinely used inbred strains are prone to spontaneous and deleterious mutations. One of these strains, the commonly used C57BL/6N (B6N), was discovered to carry a point mutation in the Crumbs homolog 1 (Crb1rd8) gene, which codes for a developmental protein involved in tight junction formation at the outer limiting membrane (OLM). This mutation disrupts photoreceptor polarity and leads to retinal degeneration. It was hypothesized that the G-protein receptor kinase 1 knockouts (Grk1−/−), which were based on the B6N strain, would exhibit abnormal morphological phenotypes in their offspring not related to GRK1’s major phosphorylation function. The hypothesis was tested by examining Grk1−/− with or without the Crb1rd8 mutation.
Methods
The mice strains tested were C57BL/6J (B6J), B6N, and Grk1−/− on either a B6J (Grk1−/−;B6J) or B6N background (Grk1−/−;B6N) and were verified with PCR genotype analysis for Grk1−/− and Crb rd8. The mice were bred and raised in complete darkness until 1 or 3 months of age and then exposed to 1,000 lux light for 24 h, followed by processing for immunohistochemistry (IHC) analysis on the retinal structure to investigate the morphological effects of light exposure. Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) was performed to detect photoreceptor apoptosis.
Results
The microanatomy of the retinal sections revealed disorganization of the outer nuclear layer (ONL) in the B6N and Grk1−/−;B6N mice and a significant decrease in the thickness of the ONL in the 3-month-old Grk1−/−;B6N mice. The adherens-junction-associated protein, Zona occludens-1 (ZO-1), formed a continuous line at the OLM in the 1- and 3-month-old control B6J and Grk1−/−;B6J mice. In contrast, the B6N and Grk1−/−;B6N retinas showed discontinuous and fragmented staining for ZO-1 at the OLM at both ages. After the mice were exposed to light, TUNEL analysis showed a significant increase in photoreceptor cell death in the Grk1−/−;B6J and Grk1−/−;B6N retinas versus either the B6J or B6N retinas at 1 and 3 months of age and a small significant difference between the Grk1−/−;B6J and Grk1−/−;B6N retinas at 1 month. In addition, glial fibrillary acidic protein (GFAP) expression was enhanced in the Grk1−/−;B6J and Grk1−/−;B6N retinas at 1 and 3 months. Occasional sprouting processes of rod bipolar cells were detected in the B6N and Grk1−/−;B6N retinas, but sprouting was not detected in the B6J or Grk1−/−;B6J retinas at either age.
Conclusions
The B6N strain background exhibited abnormal phenotypes in the Grk1−/−;B6N retina. This study demonstrates that the B6N background can influence the phenotype of a genetic mouse knockout and introduces potential visual functional consequences of the Crb1 mutation.
Introduction
G-protein receptor kinase 1 (GRK1) was the first identified member of a superfamily of seven proteins and was initially discovered for its essential role in light-activated rhodopsin phosphorylation in rod photoreceptors [1]. GRK1 is also expressed in the cone photoreceptors of the vertebrate retina and is responsible for the phosphorylation of light-activated mouse M- and S-opsin [2-4]. Phosphorylation of these photopigments is an essential first step in the deactivation of the phototransduction pathway, followed by the binding of the visual arrestin (ARR1 or ARR4) [5,6]. GRK1 is also found in human photoreceptors, along with GRK7, and both are critical for opsin phosphorylation [7,8]. Genetic defects in either GRK1 or ARR1 lead to Oguchi’s disease, a form of congenital stationary night blindness associated with retinitis pigmentosa (RP) [9-11]. Mouse Grk1−/− retinas have profoundly longer electroretinography (ERG) recorded recovery times in the rods and cones in response to flashes of light and undergo significant light-dependent degeneration over time, similar to human patients diagnosed with Oguchi’s disease [12-14]. The Grk1−/− model is therefore an important diagnostic research tool in understanding the underlying etiology of Oguchi’s disease and other forms of RP.
In 2012, Mattapallil and colleagues discovered that the B6N sub-strain of mice, which we discovered was used to create the original Grk1 knockout [3,13], carried a spontaneous rd8 point mutation of the Crumbs homolog 1 gene (Crb1), a mouse ortholog of Drosophila Crumbs [15,16]. Crb1 encodes a transmembrane protein that is highly expressed in the murine eye and central nervous system [15]. In the retina, this protein localizes primarily to the subapical region (SAR) of the Müller glial cells and, to a lesser extent, to the SAR of the photoreceptors, where the protein complexes with other proteins, including the protein associated with Lin-7 (Pals), the Pals1 associated tight junction protein (Patj), and the membrane protein, palmitoylated 5 (Mpp5), to form tight junctions at the SAR and to maintain the integrity of the external limiting membrane [17-19]. The loss of Crb1 leads to significant retinal degeneration that is worsened with exposure to light [19,20]. In humans, the loss of CRB1 leads to Leber congenital amaurosis (LCA8), a progressive degenerative disease that causes severe visual impairment at birth [21,22]. Because the loss of Crb1 results in severe retinal degeneration, this discovery had serious implications for investigators who use B6N mice for retinal degeneration studies [16].
In addition, the severity of the Crb1 knockout phenotypes varies and depends on additional genetic and epigenetic factors [19,23]. The severity of the degeneration in B6N varies from retina to retina, and the degeneration of B6N is much less severe than in the Crb1rd8/rd8 or Crb1−/− retinas [23,24]. The B6N background also does not influence every phenotype equally. The Crbrd8 mutation combined with a chemokine ligand gene (Ccl2) and a fractalkine receptor gene (Cx3cr1) double knockout (DKO) results in a severely degenerated retina, whereas the Crbrd8 mutation combined with a heterozygous mutation in the C1q-tumor necrosis factor-related protein-5 (Ctrp5+/−) gene did not reveal any detectable early onset retinal changes [24,25]. A strain related to B6N, the B6J strain, completely lacks the Crb1rd8 point mutation and the retinal degenerative phenotype [16].
In the current study, the vendor lines of the B6J and B6N, Grk1−/−;B6J, and Grk1−/−;B6N retinas were compared and contrasted, first, to determine whether some of the published phenotypes for Grk1−/− might be caused by the B6N background and, second, to see how the rd8 phenotype of the B6N mouse can contribute to the Grk1−/− degeneration phenotype. Using immunohistochemistry (IHC) techniques with specific retinal antibodies and terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) analysis to determine cell death, retina morphology and apoptosis counts were observed to determine the level of degeneration that occurred in each retina. In addition, changes in the Müller cells and the outer limiting membrane (OLM) potentially associated with retinal degeneration were also studied. Finally, signs of neural remodeling in the rod bipolar cells and the horizontal cells in response to photoreceptor degeneration were investigated.
Methods
Animals
All mice were bred and reared in total darkness or under red light and examined at 1 and 3 months of age. Controls included B6J or B6N (the original breeders for both strains purchased from Jackson Laboratories, Bar Harbor, ME). The Grk1−/−Crb1rd8/rd8 (Grk1−/−;B6N) mice were initially generously provided by Dr. Ching-Kang Chen (Baylor College of Medicine) [3] and crossed with B6J to obtain Grk1−/−Crb1+/−. These mice were backcrossed to obtain Grk1−/−Crb1+/+ (Grk1−/−;B6J). The animals were treated in accordance with the regulations of the Veterinary Authority of the University of Southern California with an approved animal protocol that complied with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Genotype analysis
To verify the genotype of the Grk1 and Crb1 knockout mice, PCR was used to amplify wild-type (WT) and knockout genes. Genomic DNA was extracted from the mouse tails using a mixture of direct PCR tail lysis buffer and proteinase K (Viagen Biotech, Inc., Los Angeles, CA) [26]. GoTaq Green Master Mix (Promega, Madison, WI) was used for PCR along with the appropriate primer sets. For Grk1, the following primer pairs were used: +Grk1 WT/RK-KA5: 5ʹ-AGG AGA GCC TGC TTT ATG TGA GAA CCG-3ʹ; -Grk1 WT/RK-WT4: 5ʹ-TAG CAC CTT TAA GCT TGT GTA TGG TG-3ʹ; +Grk1 KO/RK-KA5: 5ʹ-AGG AGA GCC TGC TTT ATG TGA GAA CCG-3ʹ; and -Grk1 KO/Neo 4: 5ʹ-CCT GCG TGC AAT CCA TCT TGT TCA ATG-3ʹ. For Crb1, the following primers were used [19]: +Crb1 mF1 WT: 5ʹ-GTG AAG ACA GCT ACA GTT CTG ATC-3ʹ; +Crb1 mF2 3481 SNP (rd8): 5ʹ-GCC CCT GTT TGC ATG GAG GAA ACT TGG AAG ACA GCT ACA GTT CTT CTG-3ʹ; and -Crb1 mR: 5ʹ- GCC CCA TTT GCA CAC TGA TGA C-3ʹ. For the forward Crb1 rd8 primer, the amount of primer used in the PCR reaction was halved. All genes were amplified using a thermocycler (Bio-Rad, Hercules, CA). The Grk1 WT and KO genes were amplified by heating the reaction mix to 94 °C for 3 min for initial denaturation, followed by 35 cycles of 94 °C for 1 min for denaturation, 60 °C for 1 min for annealing, and 72 °C for 1 min for extension, followed by 72 °C for 8 min for final extension. PCR produced products for Grk1 WT that were 280 base pairs (bp) long and products for Grk1 KO that were 540 bp long. Crb1 WT and rd8 genes were amplified by heating the reaction mix to 94 °C for 5 min for initial denaturation, followed by 35 cycles of 94 °C for 30 sec for denaturation, 65 °C for 30 sec for annealing, and 72 °C for 30 sec for extension, followed by 72 °C for 7 min for final extension. Crb1 WT products were 220 bp long, while Crb1 rd8 products were 244 bp long. PCR products were analyzed by gel electrophoresis on a 1.5% agarose gel in tris base, acetic acid, and Ethylenediaminetetraacetic acid (TAE) buffer with ethidium bromide at 100V for 30 min. Gels were imaged in a Gel Doc system (Bio-Rad) using Image Lab software (Bio-Rad).
Preparation of mouse retinal eyecup
Mice were exposed to 24 h of 1000 lux light and then immediately euthanized by intra-peritoneal injection of Euthasol (40 mg/kg body weight, Virbac, Fort Worth, TX). The eyes were enucleated (n=3 animals per group) immediately prior to complete asphyxiation, which was followed by cervical dislocation. The anterior segment of each eye was removed, and the remaining eyecup was fixed by immersion in 4% paraformaldehyde in 0.1 M PBS (1X; 150 mM NaCl, 8.4 mM Na2HPO4·2H2O, 1.9 mM NaH2PO4·2H2O, pH 7.4), pH 7.4, for 1 h. Following fixation, the retinas were transferred to 30% sucrose in PBS for 24 h at 4 °C. For the frozen cryostat sections, the lens of each eyecup was removed, and the eyecups were embedded in optimal cutting temperature (OCT) embedding medium (Tissue-Tek, Elkhart, IN) and then snap frozen in liquid nitrogen. Each block was subsequently sectioned through the optic nerve along the vertical meridian and collected on Superfrost Plus glass slides (VWR, Visalia, CA) 10 µm thick for TUNEL or 20 µm thick for IHC (three sections per slide).
Immunohistochemistry
IHC was performed as previously published [27,28]. Briefly, the 20 µm retinal sections were incubated with blocking buffer (10% ChemiBLOCKER, Millipore, Temecula, CA; 0.5% Triton X-100 in PBS) and incubated in 4 °C overnight with the following primary antibodies: rabbit polyclonal anti-glial fibrillary acidic protein (GFAP, dilution 1:1,000, Sigma-Adrich Corp., St. Louis, MO; #4546), rabbit polyclonal anti-protein kinase C alpha (PKC-α, dilution 1:1,000, Santa Cruz Biotechnology, Santa Cruz, CA; #sc-208), rabbit polyclonal anti-calbindin (dilution 1:1,000, Millipore, Temecula, CA; #AB1778), rabbit polyclonal anti-recoverin (dilution 1:1500, Millipore #AB5585), mouse monoclonal antibodies anti-zona occludens-1 (ZO-1, dilution 1:500, Life Technologies #339111, Grand Island, NY), mouse monoclonal anti-post-synaptic density 95 kDa (PSD-95, dilution 1:500, Affinity Bioreagents #MA1–046, Golden, CO), or mouse monoclonal anti-glutamine synthetase (GS, dilution 1:1,000, Millipore #MAB302). A list of all primary antibodies used is included in Table 1. All antibodies were diluted in PBS. After incubation, the slides were washed three times in PBS for 5 min. The slides were then incubated with either Alexa Fluor 488 donkey anti-rabbit immunoglobulin G (IgG; dilution 1:500, #R37118) or Alexa Fluor 488 donkey anti-mouse IgG (1:500; Life Technologies #A-21202, Grand Island, NY) for 1 h at room temperature. Slides were mounted using mounting medium with 4',6-diamidino-2-phenylindole (DAPI; Vectashield, Vector Laboratories) and covered with a glass coverslip.
Table 1. Primary antibodies.
| Target protein | Abbreviation | Source | Host | Dilution |
|---|---|---|---|---|
|
Glial fibrillary acidic protein |
GFAP |
Sigma Aldrich (#4546) |
Rabbit |
1:1000 |
|
Protein kinase C alpha |
PKCα |
Santa Cruz (#sc-208) |
Rabbit |
1:1000 |
|
Calbindin |
Calbindin |
Millipore (#AB1778) |
Rabbit |
1:1000 |
|
Recoverin |
Recoverin |
Millipore (#AB5585) |
Rabbit |
1:1500 |
|
Zona occludens-1 |
ZO-1 |
Life Technologies (#339111) |
Mouse |
1:500 |
|
Post synaptic density 95 |
PSD-95 |
Affinity Bioreagents (#MA1–046) |
Mouse |
1:500 |
| Glutamine synthetase | GS | Milipore (#MAB302) | Mouse | 1:1000 |
Information list of all the primary antibodies used in this study for immunohistochemistry.
For dual immunological stains, the slides were incubated sequentially using the following mixtures: PSD-95 with anti-PKC-α, PSD-95 with calbindin, and GS with recoverin [3,29,30]. The following secondary antibodies were used: Alexa Fluor 488 donkey anti-rabbit, Alexa Fluor 488 donkey anti-mouse, Alexa Fluor 568 goat anti-rabbit, or Alexa Fluor 568 goat anti-mouse, according to their corresponding primary antibody (dilution 1:500 for all). Dual-stained slides were mounted as described. All slides were imaged on an LSM 710 confocal microscope, using Zen 2010 software to capture the images (Zeiss, Thornwood, NY).
TUNEL assay
Apoptosis in retinas was analyzed using a fluorometric TUNEL assay kit as described previously (Promega) [31]. The 10 µm sections were used to measure the apoptotic cells. Apoptotic nuclei were labeled with fluorescein conjugated dUTP according to the manufacturer’s instructions. Using a Leica DMR fluorescent microscope (Leica Microsystems, Buffalo Grove, IL), TUNEL counts were collected from a 200 µm wide region of the superior and inferior retina at 200 µm intervals, beginning 200 µm from the optic disc (OD).
TUNEL counts were collected from at least three retinas from separate animals for each genotype for 1- and 3- month-old animals. The results were plotted as a spider plot with distance from the OD as the x-axis and TUNEL-positive nuclei per 0.01 mm2 (area of 200 µm retina length × 50 µm average thickness) as the y-axis. Points located 0.4 mm to 1.2 mm away from the OD were considered central to the OD, while points located 1.6 mm and farther were considered peripheral. Plot points were analyzed for statistical significance using two-way analysis of variance (ANOVA) with Tukey’s correction for repeated measures (Prism, GraphPad, La Jolla, CA) [28].
Retinal morphology
The photoreceptor nuclei of multiple frozen sections were stained using a monomeric cyanine nucleic acid stain (TO-PRO-3 Iodide; 1:1,000; Invitrogen, Carlsbad, CA) and incubated for 5 min at room temperature [32]. The sections were washed three times in PBS for 5 min and then mounted with mounting medium with DAPI and a glass coverslip. The sections were imaged on a Leica DMR fluorescent microscope or an LSM 510 confocal microscope and captured using Zen 2009 software (Zeiss). Similar to the TUNEL counts, retina thickness was measured at 400 µm intervals, beginning 200 µm from the optic disc. Three measurements of the ONL thickness were taken at each section, spaced approximately 50 µm apart, which were then averaged for each section [32]. The most peripheral region was not counted due to the variability of the section length. Layer thickness measurements were collected from at least three retinas from separate animals for each genotype for 1- and 3-month-old animals. These data were plotted on a Spider plot with the center point representing the OD. Statistical analysis of the data was performed with two-way ANOVA with Tukey’s correction for repeated measures (Prism) [32].
Results
Crb1rd8 mutation is found in Grk1−/− animals
The presence of the Crb1rd8 mutation was detected in the mouse colonies using PCR to amplify the truncated Crb1 gene. The Grk1−/− animals showed a positive result for truncated Crb1, but they were negative for the wild-type Crb1 allele. The Grk1−/− mice were crossed with the B6J animals to produce heterozygous Grk1+/−Crb+/rd8 animals, which were backcrossed again to produce homozygous Grk1−/−;B6J and Grk1−/−;B6N mice. To confirm the results of the backcross, the mice were tested with both sets of Crb1 and Grk1 primers. All B6J and Grk1−/−;B6J animals were negative for the truncated Crb1 gene, but each animal was positive for the wild-type Crb1 allele. The B6N and Grk1−/−;B6N animals were positive for the truncated Crb1 gene mutation, but they were negative for the wild-type Crb1 allele. The B6N and B6J mice were positive for wild-type Grk1 and negative for the Grk1 knockout, while the Grk1−/−;B6J and Grk1−/−;B6N mice were negative for wild-type Grk1 and positive for the Grk1 knockout (data not shown).
Retinal morphology and thickness
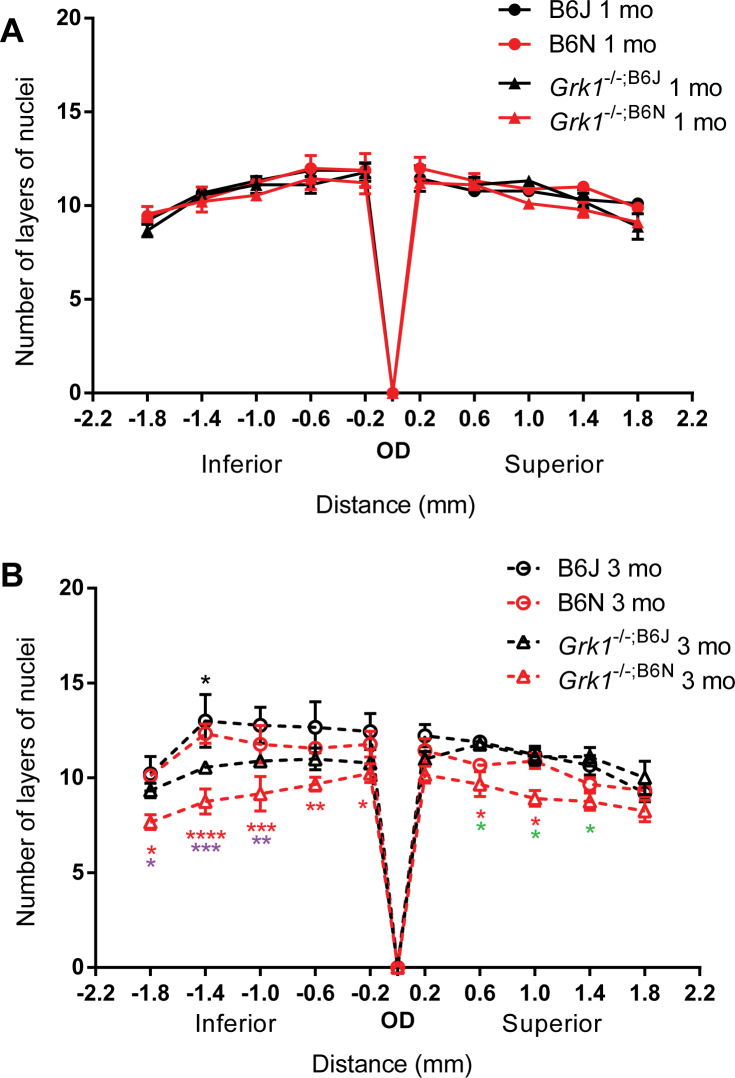
The ONL thickness of the light-exposed retinas was examined by counting the number of layers of nuclei in the TO-PRO-3 stained vertical sections (Figure 1 and Figure 2), which is an established procedure for detecting photoreceptor degeneration [32,33]. Previously published papers have shown that Grk1−/− and Crb1−/− retinas undergo light-dependent degeneration, with Grk1−/− mice exhibiting rapid retinal degeneration within 24 h of continuous light exposure (450 lux) [13]. Retinas without CRB1 expression also undergo photoreceptor degeneration following 72 h of continuous brighter light exposure (3,000 lux) starting at 3 months of age, but the degeneration is limited to the nasal inferior quadrant of the retina [20]. Two-way ANOVA analysis of the number of layers of nuclei revealed that at 1 month, the genotype contributes significantly to the variation between groups, but post hoc comparisons showed no significant differences between any of the retinas studied (F(3,88)=3.896, p=0.0115, n=3 for all groups; Figure 1A). At 3 months, the thickness of of the Grk1−/−;B6N ONL was significantly decreased in most regions of the retina compared to the other genotypes (two-way ANOVA main effect of genotype F(3,99)=29.84, p<0.0001, n=3 for all groups; Figure 1B). Compared to the B6J mice, the thickness of the Grk1−/−;B6N ONL was significantly decreased in all points in the inferior retina, as well as 0.6 and 1.0 mm superior from the OD (Figure 1B, black asterisk, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). When compared to the B6N ONL, the thickness of the Grk1−/−;B6N ONL was significantly decreased at −1.0, −1.4, and −1.8 mm inferior from the OD (Figure 1B, purple asterisk, *p<0.05, **p<0.01, ***p<0.001). When compared to the thickness of the Grk1−/−;B6J ONL, the thickness of the Grk1−/−;B6N ONL was significantly decreased at 0.6, 1.0, and 1.4 superior from the OD (Figure 1B, green asterisk, *p<0.05). The thickness of the Grk1−/−;B6J ONL was also significantly decreased compared to that of the B6J ONL at −1.4 mm of the inferior retina (Figure 1B, *p<0.05).
Figure 1.
Outer nuclear layer thickness. The nuclei of the ONL were stained and visualized using TO-PRO-3, and the layers of the nuclei were counted for 1-month-old (A) and 3-month-old (B) retinas at the labeled distances from the OD. At 1 month (A), there are no significant changes in layer thickness between any of the groups. The Grk1−/−;B6N layers are significantly reduced at 3 months compared to the other retinas (B), and the Grk1−/−;B6J layers are reduced at one point −1.4 mm from the OD (B). The black asterisk denotes the Grk1−/−;B6J layers compared to the B6J layers, the red asterisk denotes the Grk1−/−;B6N layers compared to the B6J layers, the purple asterisk denotes the Grk1−/−;B6N layers compared to the B6N layers, and the green asterisk denotes the Grk1−/−;B6N layers compared to the Grk1−/−;B6J layers (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). ONL=outer nuclear layer; OD=optic disc.
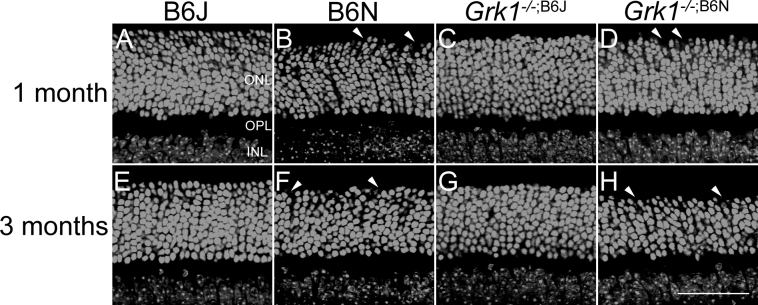
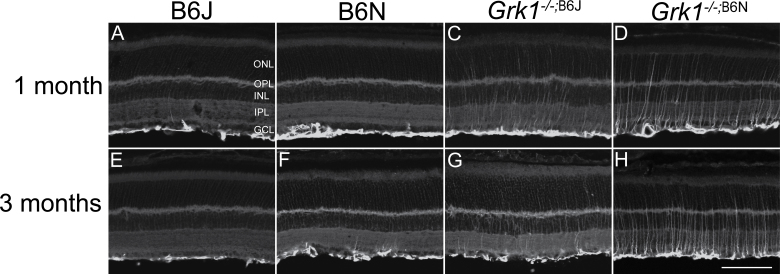
Figure 2.
Microanatomy and outer nuclear layer thickness. Retinal nuclei of the ONL stained with TO-PRO-3 and imaged for 1-month-old (A–D) and 3-month-old (E–H) animals. The B6J and Grk1−/−;B6J retinas have well-organized outer nuclear layers while the B6N (B and F) and Grk1−/−;B6N (D and H) retinas show disorganization in the outer portion of the ONL, with holes forming at the outer edge of the ONL (arrowheads; scale bar, 50 µm). ONL=outer nuclear layer; OPL=outer plexiform layer; INL=inner nuclear layer.
In addition to degeneration, retinal disorganization in the B6N background retinas was investigated (Figure 2). The B6J and Grk1−/−;B6J retinas had well-defined ONLs with a clearly delineated outer edge (Figure 2A,C,E,G). However, the B6N and Grk1−/−;B6N retinas showed retinal disorganization in the proximal ONL (Figure 2B,D,F,H). This disorganization was observed in the inferior and superior regions of the retina (Figure 3). Overall, the nuclear stains showed that retinal disorganization was apparent at 1 and 3 months in the B6N and Grk1−/−;B6N retinas and that the Grk1−/−;B6N retinas degenerated significantly by 3 months of age compared to each of the other genotypes.
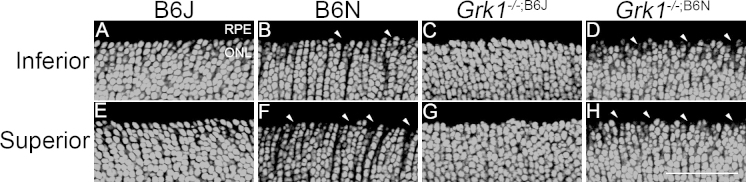
Figure 3.
Comparison of nuclear organization between inferior (A-D) and superior (E-H) regions of 1 month retinas. Retinas were stained with To-Pro3 to visualize nuclear layers. B6J (A, E) and Grk1-/-;B6J (C, G) ONLs are organized in both the superior and inferior regions of the retina. B6N (B, F) and Grk1-/-;B6N (D, H) retinas exhibit disorganization and formation of holes (arrowheads) in the outer portion of the ONL in both the inferior and superior retinas. (Scale bar, 50 µm). ONL=outer nuclear layer; RPE=retinal pigment epithelium.
Light-exposed Grk1−/−;B6N retinas experience increased degeneration
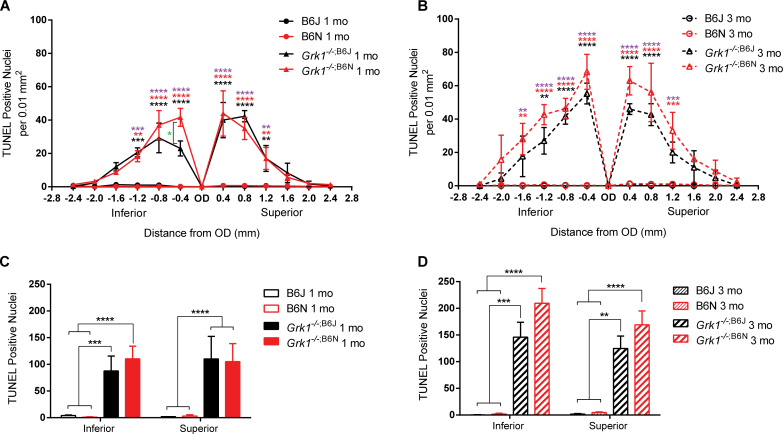
To determine whether the Grk1−/− retina undergoes degeneration independent of the B6N background, a TUNEL assay was performed to quantify the number of apoptotic cells for each genotype [13]. At 1 and 3 months, lower TUNEL counts were observed in the B6J control and B6N mice, and significantly increased TUNEL counts were observed in the Grk1−/−;B6J and Grk1−/−;B6N mice. TUNEL counts for the 1-month-old Grk1−/−;B6J and Grk1−/−;B6N retinas were significantly elevated at −0.4, −0.8, and −1.2 mm inferior from the OD and 0.4, 0.8, and 1.2 mm superior from the OD (two-way ANOVA main effect of genotype F(3,117)=88.97, p<0.0001, n=3 for all groups; Figure 4A, **p<0.01, ***p<0.001, ****p<0.0001, black asterisk, Grk1−/−;B6J compared to B6J and B6N; red asterisk, Grk1−/−;B6N compared to B6J; purple asterisk, Grk1−/−;B6N compared to B6N). TUNEL counts in the 1-month-old Grk1−/−;B6N retina also increased compared to the 1-month-old Grk1−/−;B6J at −0.4 mm inferior from the OD (Figure 4A; *p<0.05).
Figure 4.
Superior and inferior retina TUNEL analysis. A and B are representative plots of terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL)-positive nuclei at defined points in the retina at (A) 1 and (B) 3 months. Mice were exposed to 24 h of 1,000 lux light and were immediately euthanized. TUNEL staining was performed on 10 µm thick sections, and counts were taken from a 0.01 mm2 area every 0.4 µm from the OD. The Grk1−/−;B6J (black asterisk) and Grk1−/−;B6N (red asterisk) retinas have significantly increased levels of TUNEL-positive nuclei compared to the B6J retinas. The purple asterisk shows Grk1−/−;B6N significance compared to that of B6N. The level of significance of the Grk1−/−;B6J retinas compared to the B6N retinas is identical in comparison to the B6J retinas (A). Grk1−/−;B6N TUNEL is significantly increased against Grk1−/−;B6J at one point in the inferior retina (A, −0.4 mm, green asterisk). C and D are total TUNEL-positive counts in the inferior and superior retina at (C) 1 and (D) 3 months. The Grk1−/−;B6J and Grk1−/−;B6N retinas are significantly increased compared to the B6J and B6N retinas (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). OD=optic disc.
For the 3-month-old retinas, the TUNEL counts were also elevated at −0.4, −0.8, and −1.2 mm inferior from the OD and at 0.4 and 0.8 mm superior from the OD in the Grk1−/−;B6J and Grk1−/−;B6N retinas compared to the B6J and B6N retinas (two-way ANOVA main effect of genotype F(3,104)=94.70, p<0.0001, n=3 for all groups; Figure 4B, **p<0.01, ****p<0.0001). The TUNEL counts for the 1-month-old Grk1−/−;B6N retinas were also significantly increased compared to the B6J and B6N retinas at two points that were not significant in the Grk1−/−;B6J retina; one of those points lay in the peripheral inferior retina (Figure 4B at −1.6 mm and 1.2 mm, **p<0.01 and ***p<0.001, respectively).
Summation of the TUNEL counts of the inferior and superior retinas revealed consistently lower TUNEL counts in the 1- and 3-month-old B6J and B6N retinas with significantly increased TUNEL counts in the 1-month-old (two-way ANOVA main effect of genotype F(3,18)=45.75, p<0.0001, n=3 for all groups; Figure 4C) and 3-month-old (two-way ANOVA main effect of genotype F(3,16)=52.04, p<0.0001, n=3 for all groups; Figure 4D) Grk1−/−;B6J and Grk1−/−;B6N retinas. The increases in the TUNEL count for the Grk1−/−;B6N mice were extremely significant (****p<0.0001) when compared with the B6J and B6N mice. The TUNEL counts for the Grk1−/−;B6J mice were also highly significant compared to the B6J and B6N mice in the 1-month-old superior retinas (****p<0.0001) but were less significant for the 1- and 3-month-old inferior retinas (***p<0.001) and the 3-month-old superior retinas (**p<0.01). Overall, the TUNEL counts revealed that the light-exposed Grk1-/- retinas of both backgrounds experienced significant apoptosis in the ONL, while the B6J control retinas and the B6N retinas did not. The TUNEL counts also suggested that the Grk1−/−;B6N retinas experienced slightly elevated apoptosis compared to the Grk1−/−;B6J retinas, notably in the peripheral regions of the 3-month-old retinas.
GFAP upregulation in the Grk1−/−;B6N retina
Because of the decrease in the ONL thickness and the elevated TUNEL counts in the Grk1−/−;B6N retina, we explored potential mechanisms for resolving the observed increases in apoptosis and degeneration. Induced expression of the cytoskeletal fibrillary component, GFAP, in Müller cells is a typical glial reaction associated with an inflammatory response connected to photoreceptor degeneration [34-36]. This glial reaction was visualized using an antibody directed against GFAP. In the B6J retinas, immunoreactive-labeled GFAP at 1 and 3 months revealed no GFAP upregulation, and expression was limited to the nerve fiber layer (NFL; Figure 5A,E). The B6N retinas had similar GFAP expression but revealed occasional GFAP upregulation into the inner plexiform layer (IPL; Figure 5B,F). The Grk1−/−;B6J and Grk1−/−;B6N retinas showed GFAP upregulation throughout the layers of the retina, but the Grk1−/−;B6N retinas showed a more prominent activation of GFAP (Figure 5C–D,G–H). These results indicate that the Grk1−/−;B6N retinas experienced greater environmental stress and injury in response to light than the Grk1−/−;B6J retinas.
Figure 5.
Immunohistochemistry analysis of retinal GFAP. Vertical sections were immunologically stained with the primary antibody for glial fibrillary acidic protein (GFAP) (1:1,000) followed with the secondary antibody Alexa Fluor donkey anti-rabbit 488 (1:500) for 1-month-old (A–D) and 3-month-old (E–H) retinas. A and E: Immunoreactive GFAP is restricted to the innermost layer of the retina in the B6J retinas. B and F: GFAP is predominantly restricted to the nerve fiber layer (NFL) but with occasional fibrils extending into the IPL (C and G). GFAP is moderately upregulated in the processes extended into the INL and some into the ONL. D and H: Widespread GFAP upregulation in the processes extending into the ONL. Images were taken using a 20X objective lens (scale bar, 50 µm). ONL=outer nuclear layer; OPL=outer plexiform layer; INL=inner nuclear layer; IPL=inner plexiform layer; GCL=ganglion cell layer.
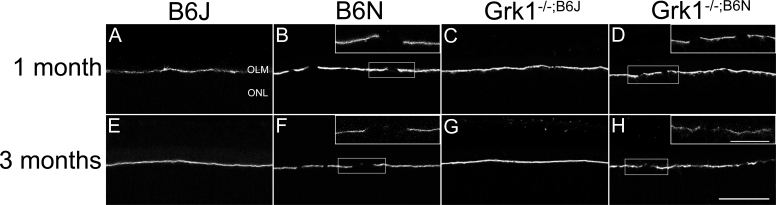
Disruptions of the OLM in the B6N and Grk1−/−;B6N retinas
The ZO-1 at the OLM is normally present between the photoreceptor inner segments and the apical processes of Müller cells [37-40]. A previous study showed that mutation of Crb1 prevents tight junctions from forming at the subapical region in photoreceptor cells, causing retinal disorganization and OLM disruption in B6N and Crb1−/− retinas [19]. Because of the nuclear disorganization observed in the ONL of the B6N and Grk1−/−;B6N retinas (Figure 2), we hypothesized that the OLM was also disrupted in the Grk1−/−;B6N retinas but not the Grk1−/−;B6J retinas and that the disruption of the OLM contributed to the increase in degeneration. ZO-1 formed a continuous barrier at the OLM in the B6J and Grk1−/−;B6J retinas (Figure 6A,C,E,G). In contrast, the B6N and Grk1−/−;B6N retinas showed discontinuous and fragmented staining for ZO-1 at the OLM in the inferior and superior retina (Figure 2B,D,F,H). This staining confirmed the hypothesis that Grk1−/−;B6N retinas, but not Grk1−/−;B6J retinas, contain disrupted OLMs.
Figure 6.
Immunohistochemistry analysis of retinal ZO-1. Primary antibody to ZO-1 (1:1,000) followed by secondary antibody Alexa Fluor donkey anti-rabbit 488 (1:500) identifies immunohistochemical staining in mouse retina sections at 1 month (A–D) and 3 months (E–H) localized to the OLM. A, C, E, G: The OLM in the B6J background retinas is unbroken. B, D, F, H: The OLM in the B6N background retinas is discontinuous. The top-right corners show higher magnification images. Lower magnification images were taken with a 20X objective lens (scale bar, 50 µm); higher magnification images were taken at 100X objective (scale bar, 20 µm). OLM=outer limiting membrane; ONL=outer nuclear layer.
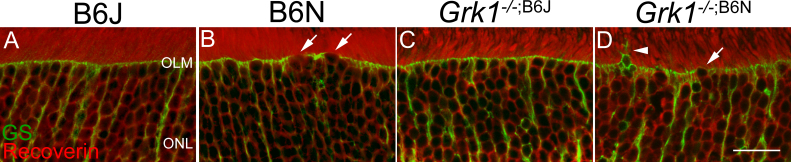
A previous study showed that after retinal damage, Müller cells processes extend toward the sub-retinal space and wrap around the cones [41]. Thus, whether apical processes of Müller cells might be similarly changed in B6N retinas where the OLM is weakened was examined. Vertical sections immunolabeled for glutamine synthetase, an established marker for Müller cells, and recoverin, a marker for photoreceptors [42], are shown in Figure 7. In the B6J and Grk1−/−;B6J retinas, the apical processes of the Müller cells formed a clear boundary line at the OLM (Figure 7A,C). In contrast, the B6N and Grk1−/−;B6N retinas showed weakening of this boundary at the OLM, similar to the gaps observed in the ZO-1 staining (Figure 5, Figure 7B,D). However, the migration of cell bodies into the sub-retinal space was not observed. In addition to a weaker OLM, extensions of the Müller cell processes beyond the OLM were observed in the Grk1−/−;B6N retinas (Figure 7B,D, arrows denote gaps; the arrowhead denotes processes). These extensions, however, did not appear to wrap around specific photoreceptors. These results indicate that weakening of the OLM is associated with changes in the Müller cells in a degenerating retina.
Figure 7.
Immunological dual staining of GS and recoverin. Primary antibody to glutamine synthetase (GS; 1:1,000) and recoverin (1:1,000) followed with secondary antibody Alexa Fluor goat anti-mouse 488 (1:500) and Alexa Fluor donkey anti-rabbit 568 (1:500), respectively, for immunohistochemical staining. A and B: Photoreceptors (red) are healthy and organized in the B6J and B6N retinas. C and D: Photoreceptors show disorganization and gaps between the outer segments in the Grk1−/−;B6J and Grk1−/−;B6N retinas. A and C: The OLM (green) is intact in the B6J and Grk1−/−;B6J retinas. B and D: The OLM disruption is apparent in GS staining in the B6N and Grk1−/−;B6N retinas (arrows). Nuclei and Müller cell projections extend toward the sub-retinal space where the OLM is disrupted (D, arrowhead). Images were taken at 2X zoom on a 40X objective lens (scale bar, 20 µm). NFL=nerve fiber layer; OLM=outer limiting membrane; ONL=outer nuclear layer.
Remodeling of second-order neurons
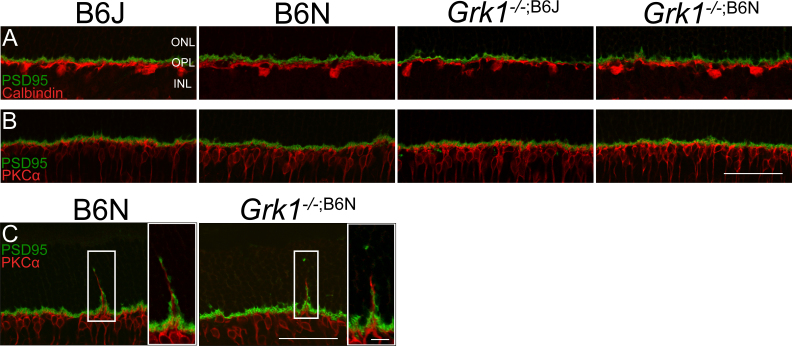
Changes in the wiring of the second-order neurons of the retina are a common effect of photoreceptor degeneration [43,44]. Since extensive apoptosis was observed in the Grk1−/− retinas and significant thinning of Grk1−/−;B6N, the study was extended to determine if the degeneration led to changes in the horizontal and bipolar cell synapses. Retinas were labeled with either PKC-α, a marker for rod bipolar cells, or calbindin, a marker for horizontal cells, and colabeled with postsynaptic density (PSD-95), a specific marker for terminal of rods and cones in the mammalian retina [42,45]. In most retinas, apical dendrites of the rod bipolar cells and horizontal cells are confined to the OPL, where the photoreceptor terminals are present (Figure 8A,B). However, apical dendrites of rod bipolar cells extending into the ONL were occasionally observed in the B6N and Grk1−/−;B6N retinas, appearing in close apposition to the immunoreactive PSD-95 staining (Figure 8C). These results suggest that the retinal rewiring at 1 and 3 months is due to the B6N background alone. Although not prevalent, occasional sprouting dendrites from horizontal cells were also apparent in the B6N background retinas (data not shown).
Figure 8.
Immunohistochemistry analysis of the inner retina with PSD-95, calbindin, and PKC-α. A: Primary antibodies for PSD-95 (1:500) and calbindin (1:1,000) were followed by Alexa Fluor donkey anti-mouse 488 (1:500) and Alexa Fluor goat anti-rabbit 568 (1:500), respectively, for dual localization immunoreactive stain. The synapses (green) between the photoreceptor and horizontal cells (red) were normal for all genotypes. B: Primary antibodies to PSD-95 (1:500) and PKC-α (1:1,000) followed by Alexa Fluor donkey anti-mouse 488 (1:500) and Alexa Fluor goat anti-rabbit 568 (1:500), respectively, for the dual localization immunoreactive stain. The synapses (green) between the photoreceptor and bipolar cells (red) for most retinas were normal for all genotypes. C: Occasionally, bipolar cells in the B6N background retinas appear to form ectopic synapses in the outer nuclear layer. Images taken with a 40X objective lens at 2X zoom (scale bar, 50 µm). Magnified images taken with a 63X objective lens (scale bar, 10 µm). ONL=outer nuclear layer; OPL=outer plexiform layer; INL=inner nuclear layer.
Discussion
Based on the retinal morphological characterization and the immunohistochemical analysis, the experiments demonstrated that although previous characterizations of the Grk1−/− retina are valid, the Grk1−/−;B6N retina clearly has retinal phenotypes that are distinct from those documented in the Grk1−/−;B6J retina. These phenotypic differences are relatively mild with incremental changes but, as a whole, indicate an exacerbation of the Grk1−/− phenotype that is linked to the Crb1 point mutation known to be present in the B6N background.
The most widely known Grk1−/− phenotypes should still be considered accurate for the Grk1−/− mouse. TUNEL analysis has confirmed that light-induced apoptosis [13] was significantly increased in the Grk1−/−;B6J retinas compared to the B6J retinas while the B6N retinas did not experience an increase in the TUNEL counts compared to the B6J retinas. The TUNEL results for the Grk1−/−;B6N retina appeared much more similar to the Grk1−/−;B6J retina than the B6N retina. Grk1−/−, rather than B6N, is therefore the most likely cause of the increased TUNEL counts in the Grk1−/−;B6N retina. Although not covered in this study, it is likely that the other widely recognized phenotype of Grk1−/−, delayed ERG recovery times and photoresponses [12,13], is also caused by the Grk1−/− mutation and not the B6N background. Previous studies on Crb1rd8 retinas revealed that ERG amplitudes for Crb1rd8 are generally not significantly different from those for wild-type retinas [46]. However, ERG data for Grk1−/− retinas revealed significantly reduced a-wave and b-wave responses as well as profoundly slowed recovery of cone-driven responses, which is linked to Grk1’s role in the phosphorylation of rhodopsin [12,13].
However, the Grk1−/−;B6N retinas showed exacerbated degeneration compared to the Grk1−/−;B6J retinas in response to light. At 1 month, the difference in degeneration between the Grk1−/−;B6J and Grk1−/−;B6N retinas was minimal, with apoptotic nuclei significant at only one point in the inferior retina (Figure 4A). This significance could be linked to the tendency for CRB1-deficient retinas to exhibit degeneration in the inferior nasal quadrant of the retina [19,20,47]. However, at 3 months, the difference between the degeneration in the Grk1−/−;B6J and Grk1−/−;B6N retinas was significant and spread beyond the inferior region of the retina. The TUNEL counts for the Grk1−/−;B6N retinas showed a tendency to have more significantly increased apoptosis counts in the retina compared to the B6J retinas, whereas the Grk1−/−;B6J retinas had fewer points of significance (Figure 4B). The thickness of the 3-month-old Grk1−/−;B6N retinas also significantly decreased in the superior and inferior regions of the retina, not just the inferior region (Figure 2B). GFAP upregulation—a common sign of retinal injury and degeneration [35,48]—was likewise observed along the entire retina in the Grk1−/−;B6N retinas at 1 and 3 months. These data suggest that the B6N background potentiates the degenerative phenotype of this and other retinal degeneration mouse models and that this effect is ubiquitous throughout the retina, instead of being limited to the previously characterized CRB1-deficient patterns of degeneration.
One possible reason for this spread of degeneration could be due to the disruption of the OLM. OLM integrity is critical for cell survival in various retinal disease models and is maintained until the most advanced stages of degeneration [49]. An intact OLM is also important in the regulation of the size of the apical villi of Müller cells [17,47] and the coupling of the photoreceptors to the Müller glial cells by the adherens junction (AJ) complex [19,20]. Loss of Crb1 leads to disruption of the OLM complexes at the SAR and the AJ [17,20]. In the present study, OLM disruption was observed alongside the extension of apical Müller cell processes in the Grk1−/−;B6N retinas (Figure 7C). These apical processes were likely caused by the weakening of the junctions between the photoreceptors and the Müller cells, but these processes were not observed in the B6N retinas (Figure 7B). This suggests that the disruption of the OLM is necessary, but not sufficient, for the sprouting of these apical processes beyond the OLM, and that the light-induced degeneration caused by the loss of Grk1 in the Grk1−/−;B6N retina is a second contributor to the growth of these processes. Since Müller cells play a role in preventing and decreasing the severity of photoreceptor degeneration [50], the loss of Müller cell interaction with photoreceptors caused by a disrupted OLM in the Grk1−/−;B6N retina could also enhance photoreceptor death following exposure to light. The OLM appears to be an important element of retinal health, and additional studies on OLM disruption in degenerative disease could explore the effects of increased light exposure on Müller cell health and morphology.
An interesting mechanism related to the Grk1−/−;B6N retina is the role of abnormally stable rhodopsin-arrestin complexes in causing photoreceptor degeneration [51-53]. These complexes are involved in retinal degeneration and are proposed to be the main cause of degeneration in mammalian retinas that lack CRB1 [53]. Since rhodopsin in Grk1−/− retinas cannot be phosphorylated by GRK1, these rhodopsin-arrestin complexes would be unable to form in Grk1−/−;B6N mice. If these complexes are the underlying mechanism causing retinal degeneration in Crb1−/− and Crbrd8/rd8 mice, a possible reason why a greater increase in the number of apoptotic cells in the Grk1−/−;B6N retina is not observed could be due to the failure of these complexes to form. Future studies on the presence of these complexes in Crb1−/− and Crb1rd8/rd8 retinas would help elucidate whether this is the case.
Downstream of the photoreceptors, changes were also detected in the second-order neurons—the bipolar and horizontal cells—in the B6N and Grk1−/−;B6N retinas. While normal bipolar and horizontal cell morphology and wiring were the typical phenotype for all genotypes, the B6N and Grk1−/−;B6N retinas displayed occasional neurite extensions from rod bipolar cells into the ONL (Figure 8B,D). PSD-95 immunoreactive staining also showed that these neurite extensions maintained or developed synapses with photoreceptors inside the ONL. Retinal plasticity and neurite growth are associated with retinal degenerative disease models, such as RP and age-related macular degeneration (AMD) [54-57]. This neurite growth and plasticity usually occur in three phases: photoreceptor stress, photoreceptor cell death, and extensive retinal remodeling [56,57]. In degenerative disease, such as AMD, neurite sprouting and growth follow photoreceptor death and the retraction of photoreceptor axons into the ONL [54]. Interestingly, neurite growth was observed in the B6N retinas, which experienced minimal photoreceptor apoptosis, and not in the Grk1−/−;B6J retinas, which experienced significant photoreceptor apoptosis. Additionally, the Grk1−/−;B6N mice did not exhibit an increase in retinal rewiring compared to the B6N mice. Therefore, the process of cell death itself does not induce second-order neuron rewiring but other factors in the B6N retina. The disruption of the CRB1 complex at the OLM and the loss of photoreceptor polarity may result in a release of trophic factors that can influence the beginnings of neurite growth by bipolar and horizontal cells [54]. Since Crb1−/− retinas are known to degenerate significantly with age [20], it is likely that B6N and Grk1−/−;B6N mice beyond 3 months of age would exhibit more extensive neurite sprouting and remodeling of synapses.
The Crb1rd8 mutation of the B6N mouse contributes to an increased degenerative phenotype in the retina, but the influence of the B6N background is not limited to the eye. In 2013, Kumar and collaborators discovered a mutation in the B6N mouse in the cytoplasmic fragile X mental retardation protein (FMRP) interacting protein 2 that was not found in the B6J mouse [58]. This mutation sensitized the B6N mice’s responses to cocaine, whereas the B6J mice had a normal response. Researchers are becoming increasingly aware that background strain differences between mice can contribute significantly to the phenotypes and results of an experiment [23,25,59,60]. Of particular interest is the use of the B6N mouse as the background model for the International Knockout Mouse Consortium/Knockout Mouse Project (IKMC/KOMP), a historic initiative to create null alleles for every gene in the mouse genome [61,62]. Knockout mouse lines for every gene would undoubtedly prove to be an immense resource for investigators. However, if mutations in the B6N background alter the phenotypes of these knockout mice, the resulting characterization of these knockouts could be potentially misleading. Comparative analyses of polymorphisms between B6N and other common strains such as the B6J [23,59,63,64] enable researchers to be aware and use caution when interpreting these genotypic and phenotypic differences.
Acknowledgments
This work was supported, in part, by grant awards from the NIH-National Eye Institute R01EY015851 (CMC), P30EY03040 (Doheny Eye Institute, NEI Core Grant), VSoE Research Innovation Fund (EJL), and Research to Prevent Blindness (USC Ophthalmology), and the Mary D. Allen Foundation (CMC, JSP), Dorie Miller (JSP), William Hansen Sandberg Memorial Foundation (JSP), and Tony Gray Foundation (JSP). Dr. Craft is the inaugural Doheny Eye Institute’s Mary D. Allen Chair in Vision Research. We also acknowledge members of our Mary D. Allen Laboratory for Vision Research, including Bruce Brown for animal assistance, Erik Haw for genotype analysis, Ernesto Barron for help with confocal imaging, Dr. Janise Deming for critical reading and suggestions of the manuscript, and USC undergraduate student, Daphne Derose, for technical assistance with immunohistochemistry.
References
- 1.Shichi H, Somers RL. Light-dependent phosphorylation of rhodopsin. Purification and properties of rhodopsin kinase. J Biol Chem. 1978;253:7040–6. [PubMed] [Google Scholar]
- 2.Weiss ER, Ducceschi MH, Horner TJ, Li A, Craft CM, Osawa S. Species-specific differences in expression of G-protein-coupled receptor kinase (GRK) 7 and GRK1 in mammalian cone photoreceptor cells: implications for cone cell phototransduction. J Neurosci. 2001;21:9175–84. doi: 10.1523/JNEUROSCI.21-23-09175.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu X, Brown B, Li A, Mears AJ, Swaroop A, Craft CM. GRK1-dependent phosphorylation of S and M opsins and their binding to cone arrestin during cone phototransduction in the mouse retina. J Neurosci. 2003;23:6152–60. doi: 10.1523/JNEUROSCI.23-14-06152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nikonov SS, Daniele LL, Zhu X, Craft CM, Swaroop A, Pugh EN., Jr Photoreceptors of Nrl −/− mice coexpress functional S- and M-cone opsins having distinct inactivation mechanisms. J Gen Physiol. 2005;125:287–304. doi: 10.1085/jgp.200409208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kühn H, Hall SW, Wilden U. Light-induced binding of 48-kDa protein to photoreceptor membranes is highly enhanced by phosphorylation of rhodopsin. FEBS Lett. 1984;176:473–8. doi: 10.1016/0014-5793(84)81221-1. [DOI] [PubMed] [Google Scholar]
- 6.Wilden U, Hall SW, Kuhn H. Phosphodiesterase activation by photoexcited rhodopsin is quenched when rhodopsin is phosphorylated and binds the intrinsic 48-kDa protein of rod outer segments. Proc Natl Acad Sci USA. 1986;83:1174–8. doi: 10.1073/pnas.83.5.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao X, Huang J, Khani SC, Palczewski K. Molecular forms of human rhodopsin kinase (GRK1). J Biol Chem. 1998;273:5124–31. doi: 10.1074/jbc.273.9.5124. [DOI] [PubMed] [Google Scholar]
- 8.Cideciyan AV, Jacobson SG, Gupta N, Osawa S, Locke KG, Weiss ER, Wright AF, Birch DG, Milam AH. Cone Deactivation Kinetics and GRK1/GRK7 Expression in Enhanced S Cone Syndrome Caused by Mutations inNR2E3. Invest Ophthalmol Vis Sci. 2003;44:1268. doi: 10.1167/iovs.02-0494. [DOI] [PubMed] [Google Scholar]
- 9.Fuchs S, Nakazawa M, Maw M, Tamai M, Oguchi Y, Gal A. A homozygous 1-base pair deletion in the arrestin gene is a frequent cause of Oguchi disease in Japanese. Nat Genet. 1995;10:360–2. doi: 10.1038/ng0795-360. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto S, Sippel KC, Berson EL, Dryja TP. Defects in the rhodopsin kinase gene in the Oguchi form of stationary night blindness. Nat Genet. 1997;15:175–8. doi: 10.1038/ng0297-175. [DOI] [PubMed] [Google Scholar]
- 11.Cideciyan AV, Zhao X, Nielsen L, Khani SC, Jacobson SG, Palczewski K. Null mutation in the rhodopsin kinase gene slows recovery kinetics of rod and cone phototransduction in man. Proc Natl Acad Sci USA. 1998;95:328–33. doi: 10.1073/pnas.95.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyubarsky AL, Chen C, Simon MI, Pugh EN., Jr Mice lacking G-protein receptor kinase 1 have profoundly slowed recovery of cone-driven retinal responses. J Neurosci. 2000;20:2209–17. doi: 10.1523/JNEUROSCI.20-06-02209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen CK, Burns ME, Spencer M, Niemi GA, Chen J, Hurley JB, Baylor DA, Simon MI. Abnormal photoresponses and light-induced apoptosis in rods lacking rhodopsin kinase. Proc Natl Acad Sci USA. 1999;96:3718–22. doi: 10.1073/pnas.96.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cideciyan AV, Hood DC, Huang Y, Banin E, Li ZY, Stone EM, Milam AH, Jacobson SG. Disease sequence from mutant rhodopsin allele to rod and cone photoreceptor degeneration in man. Proc Natl Acad Sci USA. 1998;95:7103–8. doi: 10.1073/pnas.95.12.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.den Hollander AI, Ghiani M, de Kok YJ, Wijnholds J, Ballabio A, Cremers FP, Broccoli V. Isolation of Crb1, a mouse homologue of Drosophila crumbs, and analysis of its expression pattern in eye and brain. Mech Dev. 2002;110:203–7. doi: 10.1016/s0925-4773(01)00568-8. [DOI] [PubMed] [Google Scholar]
- 16.Mattapallil MJ, Wawrousek EF, Chan CC, Zhao H, Roychoudhury J, Ferguson TA, Caspi RR. The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes. Invest Ophthalmol Vis Sci. 2012;53:2921–7. doi: 10.1167/iovs.12-9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Rossum AG, Aartsen WM, Meuleman J, Klooster J, Malysheva A, Versteeg I, Arsanto JP, Le Bivic A, Wijnholds J. Pals1/Mpp5 is required for correct localization of Crb1 at the subapical region in polarized Muller glia cells. Hum Mol Genet. 2006;15:2659–72. doi: 10.1093/hmg/ddl194. [DOI] [PubMed] [Google Scholar]
- 18.Richard M, Roepman R, Aartsen WM, van Rossum AG, den Hollander AI, Knust E, Wijnholds J, Cremers FP. Towards understanding CRUMBS function in retinal dystrophies. Hum Mol Genet. 2006;15(Spec No 2):R235–43. doi: 10.1093/hmg/ddl195. [DOI] [PubMed] [Google Scholar]
- 19.Mehalow AK, Kameya S, Smith RS, Hawes NL, Denegre JM, Young JA, Bechtold L, Haider NB, Tepass U, Heckenlively JR, Chang B, Naggert JK, Nishina PM. CRB1 is essential for external limiting membrane integrity and photoreceptor morphogenesis in the mammalian retina. Hum Mol Genet. 2003;12:2179–89. doi: 10.1093/hmg/ddg232. [DOI] [PubMed] [Google Scholar]
- 20.van de Pavert SA, Kantardzhieva A, Malysheva A, Meuleman J, Versteeg I, Levelt C, Klooster J, Geiger S, Seeliger MW, Rashbass P, Le Bivic A, Wijnholds J. Crumbs homologue 1 is required for maintenance of photoreceptor cell polarization and adhesion during light exposure. J Cell Sci. 2004;117:4169–77. doi: 10.1242/jcs.01301. [DOI] [PubMed] [Google Scholar]
- 21.den Hollander AI, Heckenlively JR, van den Born LI, de Kok YJM, van der Velde-Visser SD, Kellner U, Jurklies B, van Schooneveld MJ, Blankenagel A, Rohrschneider K, Wissinger B, Cruysberg JRM, Deutman AF, Brunner HG, Apfelstedt-Sylla E, Hoyng CB, Cremers FPM. Leber Congenital Amaurosis and Retinitis Pigmentosa with Coats-like Exudative Vasculopathy Are Associated with Mutations in the Crumbs Homologue 1 (CRB1) Gene. Am J Hum Genet. 2001;69:198–203. doi: 10.1086/321263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lotery AJ, Jacobson SG, Fishman GA, Weleber RG, Fulton AB, Namperumalsamy P, Heon E, Levin AV, Grover S, Rosenow JR, Kopp KK, Sheffield VC, Stone EM. Mutations in the CRB1 gene cause Leber congenital amaurosis. Arch Ophthalmol. 2001;119:415–20. doi: 10.1001/archopht.119.3.415. [DOI] [PubMed] [Google Scholar]
- 23.Luhmann UF, Carvalho LS, Holthaus SM, Cowing JA, Greenaway S, Chu CJ, Herrmann P, Smith AJ, Munro PM, Potter P, Bainbridge JW, Ali RR. The severity of retinal pathology in homozygous Crb1rd8/rd8 mice is dependent on additional genetic factors. Hum Mol Genet. 2015;24:128–41. doi: 10.1093/hmg/ddu424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu XK, Wang Y, Ardeljan D, Tuo J, Chan CC. Controversial view of a genetically altered mouse model of focal retinal degeneration. Bioengineered. 2013;4:130–5. doi: 10.4161/bioe.22949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sahu B, Chavali VR, Alapati A, Suk J, Bartsch DU, Jablonski MM, Ayyagari R. Presence of rd8 mutation does not alter the ocular phenotype of late-onset retinal degeneration mouse model. Mol Vis. 2015;21:273–84. [PMC free article] [PubMed] [Google Scholar]
- 26.Yetemian RM, Craft CM. Characterization of the pituitary tumor transforming gene 1 knockout mouse retina. Neurochem Res. 2011;36:636–44. doi: 10.1007/s11064-010-0334-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu X, Li A, Brown B, Weiss ER, Osawa S, Craft CM. Mouse cone arrestin expression pattern: light induced translocation in cone photoreceptors. Mol Vis. 2002;8:462–71. [PubMed] [Google Scholar]
- 28.Yetemian RM, Brown BM, Craft CM. Neovascularization, enhanced inflammatory response, and age-related cone dystrophy in the Nrl−/−Grk1−/− mouse retina. Invest Ophthalmol Vis Sci. 2010;51:6196–206. doi: 10.1167/iovs.10-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu X, Craft CM. Modulation of CRX transactivation activity by phosducin isoforms. Mol Cell Biol. 2000;20:5216–26. doi: 10.1128/mcb.20.14.5216-5226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hombrebueno JR, Tsai MM, Kim HL, De Juan J, Grzywacz NM, Lee EJ. Morphological changes of short-wavelength cones in the developing S334ter-3 transgenic rat. Brain Res. 2010;1321:60–6. doi: 10.1016/j.brainres.2010.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li A, Zhu X, Brown B, Craft CM. Gene expression networks underlying retinoic acid-induced differentiation of human retinoblastoma cells. Invest Ophthalmol Vis Sci. 2003;44:996–1007. doi: 10.1167/iovs.02-0434. [DOI] [PubMed] [Google Scholar]
- 32.Brown BM, Ramirez T, Rife L, Craft CM. Visual Arrestin 1 contributes to cone photoreceptor survival and light adaptation. Invest Ophthalmol Vis Sci. 2010;51:2372–80. doi: 10.1167/iovs.09-4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J, Simon MI, Matthes MT, Yasumura D, LaVail MM. Increased susceptibility to light damage in an arrestin knockout mouse model of Oguchi disease (stationary night blindness). Invest Ophthalmol Vis Sci. 1999;40:2978–82. [PubMed] [Google Scholar]
- 34.Bignami A, Raju T, Dahl D. Localization of vimentin, the nonspecific intermediate filament protein, in embryonal glia and in early differentiating neurons. In vivo and in vitro immunofluorescence study of the rat embryo with vimentin and neurofilament antisera. Dev Biol. 1982;91:286–95. doi: 10.1016/0012-1606(82)90035-5. [DOI] [PubMed] [Google Scholar]
- 35.Lewis GP, Fisher SK. Up-regulation of glial fibrillary acidic protein in response to retinal injury: its potential role in glial remodeling and a comparison to vimentin expression. Int Rev Cytol. 2003;230:263–90. doi: 10.1016/s0074-7696(03)30005-1. [DOI] [PubMed] [Google Scholar]
- 36.Bringmann A, Wiedemann P. Muller glial cells in retinal disease. Ophthalmologica. 2012;227:1–19. doi: 10.1159/000328979. [DOI] [PubMed] [Google Scholar]
- 37.Campbell M, Humphries M, Kennan A, Kenna P, Humphries P, Brankin B. Aberrant retinal tight junction and adherens junction protein expression in an animal model of autosomal dominant Retinitis pigmentosa: the Rho(−/−) mouse. Exp Eye Res. 2006;83:484–92. doi: 10.1016/j.exer.2006.01.032. [DOI] [PubMed] [Google Scholar]
- 38.Paffenholz R, Kuhn C, Grund C, Stehr S, Franke WW. The arm-repeat protein NPRAP (neurojungin) is a constituent of the plaques of the outer limiting zone in the retina, defining a novel type of adhering junction. Exp Cell Res. 1999;250:452–64. doi: 10.1006/excr.1999.4534. [DOI] [PubMed] [Google Scholar]
- 39.Pearson RA, Barber AC, West EL, MacLaren RE, Duran Y, Bainbridge JW, Sowden JC, Ali RR. Targeted disruption of outer limiting membrane junctional proteins (Crb1 and ZO-1) increases integration of transplanted photoreceptor precursors into the adult wild-type and degenerating retina. Cell Transplant. 2010;19:487–503. doi: 10.3727/096368909X486057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tserentsoodol N, Shin BC, Suzuki T, Takata K. Colocalization of tight junction proteins, occludin and ZO-1, and glucose transporter GLUT1 in cells of the blood-ocular barrier in the mouse eye. Histochem Cell Biol. 1998;110:543–51. doi: 10.1007/s004180050316. [DOI] [PubMed] [Google Scholar]
- 41.Lewis GP, Fisher SK. Muller cell outgrowth after retinal detachment: association with cone photoreceptors. Invest Ophthalmol Vis Sci. 2000;41:1542–5. [PubMed] [Google Scholar]
- 42.Haverkamp S, Wassle H. Immunocytochemical analysis of the mouse retina. J Comp Neurol. 2000;424:1–23. [PubMed] [Google Scholar]
- 43.Marc RE, Jones BW. Retinal remodeling in inherited photoreceptor degenerations. Mol Neurobiol. 2003;28:139–47. doi: 10.1385/MN:28:2:139. [DOI] [PubMed] [Google Scholar]
- 44.Jones BW, Marc RE. Retinal remodeling during retinal degeneration. Exp Eye Res. 2005;81:123–37. doi: 10.1016/j.exer.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 45.Koulen P, Fletcher EL, Craven SE, Bredt DS, Wassle H. Immunocytochemical localization of the postsynaptic density protein PSD-95 in the mammalian retina. J Neurosci. 1998;18:10136–49. doi: 10.1523/JNEUROSCI.18-23-10136.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aleman TS, Cideciyan AV, Aguirre GK, Huang WC, Mullins CL, Roman AJ, Sumaroka A, Olivares MB, Tsai FF, Schwartz SB, Vandenberghe LH, Limberis MP, Stone EM, Bell P, Wilson JM, Jacobson SG. Human CRB1-associated retinal degeneration: comparison with the rd8 Crb1-mutant mouse model. Invest Ophthalmol Vis Sci. 2011;52:6898–910. doi: 10.1167/iovs.11-7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van de Pavert SA, Sanz AS, Aartsen WM, Vos RM, Versteeg I, Beck SC, Klooster J, Seeliger MW, Wijnholds J. Crb1 is a determinant of retinal apical Muller glia cell features. Glia. 2007;55:1486–97. doi: 10.1002/glia.20561. [DOI] [PubMed] [Google Scholar]
- 48.Bignami A, Dahl D. The radial glia of Muller in the rat retina and their response to injury. An immunofluorescence study with antibodies to the glial fibrillary acidic (GFA) protein. Exp Eye Res. 1979;28:63–9. doi: 10.1016/0014-4835(79)90106-4. [DOI] [PubMed] [Google Scholar]
- 49.Hippert C, Graca AB, Barber AC, West EL, Smith AJ, Ali RR, Pearson RA. Muller glia activation in response to inherited retinal degeneration is highly varied and disease-specific. PLoS ONE. 2015;10:e0120415. doi: 10.1371/journal.pone.0120415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harada T, Harada C, Nakayama N, Okuyama S, Yoshida K, Kohsaka S, Matsuda H, Wada K. Modification of glial-neuronal cell interactions prevents photoreceptor apoptosis during light-induced retinal degeneration. Neuron. 2000;26:533–41. doi: 10.1016/s0896-6273(00)81185-x. [DOI] [PubMed] [Google Scholar]
- 51.Alloway PG, Howard L, Dolph PJ. The formation of stable rhodopsin-arrestin complexes induces apoptosis and photoreceptor cell degeneration. Neuron. 2000;28:129–38. doi: 10.1016/s0896-6273(00)00091-x. [DOI] [PubMed] [Google Scholar]
- 52.Kiselev A, Socolich M, Vinos J, Hardy RW, Zuker CS, Ranganathan R. A molecular pathway for light-dependent photoreceptor apoptosis in Drosophila. Neuron. 2000;28:139–52. doi: 10.1016/s0896-6273(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 53.Johnson K, Grawe F, Grzeschik N, Knust E. Drosophila crumbs is required to inhibit light-induced photoreceptor degeneration. Curr Biol. 2002;12:1675–80. doi: 10.1016/s0960-9822(02)01180-6. [DOI] [PubMed] [Google Scholar]
- 54.Lewis GP, Linberg KA, Fisher SK. Neurite outgrowth from bipolar and horizontal cells after experimental retinal detachment. Invest Ophthalmol Vis Sci. 1998;39:424–34. [PubMed] [Google Scholar]
- 55.Marc RE, Jones BW, Anderson JR, Kinard K, Marshak DW, Wilson JH, Wensel T, Lucas RJ. Neural reprogramming in retinal degeneration. Invest Ophthalmol Vis Sci. 2007;48:3364–71. doi: 10.1167/iovs.07-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marc RE, Jones BW, Watt CB, Strettoi E. Neural remodeling in retinal degeneration. Prog Retin Eye Res. 2003;22:607–55. doi: 10.1016/s1350-9462(03)00039-9. [DOI] [PubMed] [Google Scholar]
- 57.Jones BW, Watt CB, Frederick JM, Baehr W, Chen CK, Levine EM, Milam AH, Lavail MM, Marc RE. Retinal remodeling triggered by photoreceptor degenerations. J Comp Neurol. 2003;464:1–16. doi: 10.1002/cne.10703. [DOI] [PubMed] [Google Scholar]
- 58.Kumar V, Kim K, Joseph C, Kourrich S, Yoo SH, Huang HC, Vitaterna MH, de Villena FP, Churchill G, Bonci A, Takahashi JS. C57BL/6N mutation in cytoplasmic FMRP interacting protein 2 regulates cocaine response. Science. 2013;342:1508–12. doi: 10.1126/science.1245503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B, Heger A, Agam A, Slater G, Goodson M, Furlotte NA, Eskin E, Nellaker C, Whitley H, Cleak J, Janowitz D, Hernandez-Pliego P, Edwards A, Belgard TG, Oliver PL, McIntyre RE, Bhomra A, Nicod J, Gan X, Yuan W, van der Weyden L, Steward CA, Bala S, Stalker J, Mott R, Durbin R, Jackson IJ, Czechanski A, Guerra-Assuncao JA, Donahue LR, Reinholdt LG, Payseur BA, Ponting CP, Birney E, Flint J, Adams DJ. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–94. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kiselycznyk C, Holmes A. All (C57BL/6) Mice are not Created Equal. Front Neurosci. 2011;5:10. doi: 10.3389/fnins.2011.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT, Grieder FB, Heintz N, Hicks G, Insel TR, Joyner A, Koller BH, Lloyd KC, Magnuson T, Moore MW, Nagy A, Pollock JD, Roses AD, Sands AT, Seed B, Skarnes WC, Snoddy J, Soriano P, Stewart DJ, Stewart F, Stillman B, Varmus H, Varticovski L, Verma IM, Vogt TF, von Melchner H, Witkowski J, Woychik RP, Wurst W, Yancopoulos GD, Young SG, Zambrowicz B. The knockout mouse project. Nat Genet. 2004;36:921–4. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.International Mouse Knockout C Collins FS, Rossant J, Wurst W. A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 63.Mekada K, Abe K, Murakami A, Nakamura S, Nakata H, Moriwaki K, Obata Y, Yoshiki A. Genetic Differences among C57BL/6 Substrains. Exp Anim Tokyo. 2009;58:141–9. doi: 10.1538/expanim.58.141. [DOI] [PubMed] [Google Scholar]
- 64.Simon MM, Greenaway S, White JK, Fuchs H, Gailus-Durner V, Wells S, Sorg T, Wong K, Bedu E, Cartwright EJ, Dacquin R, Djebali S, Estabel J, Graw J, Ingham NJ, Jackson IJ, Lengeling A, Mandillo S, Marvel J, Meziane H, Preitner F, Puk O, Roux M, Adams DJ, Atkins S, Ayadi A, Becker L, Blake A, Brooker D, Cater H, Champy MF, Combe R, Danecek P, di Fenza A, Gates H, Gerdin AK, Golini E, Hancock JM, Hans W, Holter SM, Hough T, Jurdic P, Keane TM, Morgan H, Muller W, Neff F, Nicholson G, Pasche B, Roberson LA, Rozman J, Sanderson M, Santos L, Selloum M, Shannon C, Southwell A, Tocchini-Valentini GP, Vancollie VE, Westerberg H, Wurst W, Zi M, Yalcin B, Ramirez-Solis R, Steel KP, Mallon AM, de Angelis MH, Herault Y, Brown SD. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol. 2013;14:R82. doi: 10.1186/gb-2013-14-7-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]