Abstract
DNA oxidative lesions are widely considered as a potential risk factor for colorectal cancer development. The aim of this work was to determine the role of the efficiency of base excision repair, both in lymphocytes and in epithelial tissue, in patients with CRC and healthy subjects. SNPs were identified within genes responsible for steps following glycosylase action in BER, and patients and healthy subjects were genotyped. A radioisotopic BER assay was used for assessing repair efficiency and TaqMan for genotyping. Decreased BER activity was observed in lymphocyte extract from CRC patients and in cancer tissue extract, compared to healthy subjects. In addition, polymorphisms of EXO1, LIG3, and PolB may modulate the risk of colorectal cancer by decreasing (PolB) or increasing (LIG3 and EXO1) the chance of malignant transformation.
1. Introduction
Colorectal cancer (CRC) is a common neoplasia in both men and women and is ranked as the second most common type of cancer. The causes of colorectal cancer have not yet been established and its incidence is known to be increasing, with approximately 1.4 million new cases diagnosed each year [1]. CRC occurs mainly in three specific forms: sporadic form, which accounts for about 80% of all cases, a familial form, which represents about 15%, and inherited forms, observed in 5% of all cases, which include familial adenomatous polyposis (FAP), and hereditary nonpolyposis colorectal cancer (HNPCC) [2]. Despite the cause of most colorectal cancers being environmental factors, studies show that individual predispositions for developing this cancer may depend on mutations of certain genes, including those involved in the process of DNA repair. Several DNA repair mechanisms have evolved to protect the genome from DNA damage caused by endogenous or environmental factors, which if unrepaired could lead to the initiation of carcinogenesis. The efficiency of DNA repair varies between individuals, and the reasons for this should be sought in polymorphisms within the DNA repair genes. An increasing number of DNA repair gene polymorphisms are being correlated with increased risk of cancer occurrence. Although an irrefutable link has already been established between colorectal cancer and the presence of mutations in mismatch repair (MMR) genes [3], other polymorphisms of DNA repair genes (BER and NER) are undergoing investigation for a potential influence on CRC.
Base excision repair (BER) is DNA repair system that operates on small lesions such as oxidized or reduced bases. A single damaged base is removed by base-specific DNA glycosylases. The abasic site is then rebuilt by endonuclease action, removal of the sugar residue, DNA synthesis using the other strand as a template, and then ligation. The molecules involved with the process include Exonuclease 1, which cleaves the nucleotides from the end of DNA strand, DNA polymerase beta, which is involved in gap filling, and DNA ligase 3, which seals interruptions in the phosphodiester backbone of duplex DNA [4]. Polymorphisms in the genes encoding these proteins are suspected to influence the efficiency of the whole BER process and thus modulate the risk of CRC.
The present study has two major aims. The first is to evaluate the influence of the presence of polymorphisms within the tested genes with an elevated risk of colorectal cancer. The second is to make an in vitro assessment of the efficiency of restoring DNA continuity via the BER pathway. As all protein products of the genes chosen for SNP screening are involved in the BER stages directly following glycosylase action, the BER assay was adjusted to measure only the gap-filling step, when PolB, EXO3, and LIG3 play crucial roles.
2. Materials and Methods
DNA for genotyping was isolated from lymphocytes of the peripheral blood. The blood samples were taken from 235 unrelated patients hospitalized in the Military Medical Academy University Teaching Hospital-Central Veterans' Hospital in Lodz. Each patient had histopathologically confirmed colorectal cancer. The studied group included 137 men and 98 women (average age 61 years ± 8 years). The stage of the tumors was established according to TNM scale. The control group included 240 individuals not diagnosed with cancer and with ages corresponding to the age of the studied group (p < 0.05). Permission to conduct research was granted by the bioethics committee of the Medical University of Lodz.
DNA isolation was carried out with a commercial kit QIAamp DNA Blood Mini Kit for isolation of high-molecular-weight DNA (Qiagen).
The occurrence of polymorphic variants of 242Pro/Arg of PolB gene, 780Arg/His of LIG3 gene, and 589Glu/Lis of EXO1 gene was studied with TaqMan technique. Briefly, 25 μL of reaction mixture was used for analysis, containing 1 μL of genomic DNA solution, 1 μL of probes designed specifically for each polymorphism, 13 μL of premix with polymerase, and 10 μL of water. The PCR reaction was performed in a Stratagene Mx3005P Real Time PCR Thermocycler. The RS numbers for polymorphisms and thermal conditions of reaction are shown in Table 1. For 10% of the randomly selected samples, genotyping was repeated to confirm reproducibility. Cases and controls were genotyped randomly and researchers were blinded to the case/control status during genotyping.
Table 1.
The refSNP and thermal conditions used in the PCR reaction.
| Gene | PolB | LIG3 | EXO1 |
|---|---|---|---|
| Polymorphism | 242Pro/Arg | 780Arg/His | 589Lys/Glu |
|
| |||
| refSNP | 3136797 | 3136025 | 1047840 |
|
| |||
| Thermal conditions | (1) 95°C—10 min | ||
| (2) 92°C—15 sec | |||
| (3) 60°C—1 min | |||
| (4) Step 2 and 3—45x | |||
|
| |||
| Dyes | ROX, HEX, and FAM | ||
|
| |||
| Ref. dye | ROX | ||
BER efficiency was evaluated according to Matsumoto et al. [5] with some minor modifications to improve the preparation of synthetic lesion site. A plasmid construct with radioactively labeled single-strand breaks was incubated with protein extract isolated from peripheral blood lymphocytes and slices of cancerous tissue removed during surgical procedures. Repair capability was assessed by densitometric analysis of DNA fragments which had been electrophoretically separated and depicted on X-ray film: these fragments vary with regard to length and can identify repaired or unrepaired fractions. The course of procedure is outlined in Figure 1.
Figure 1.
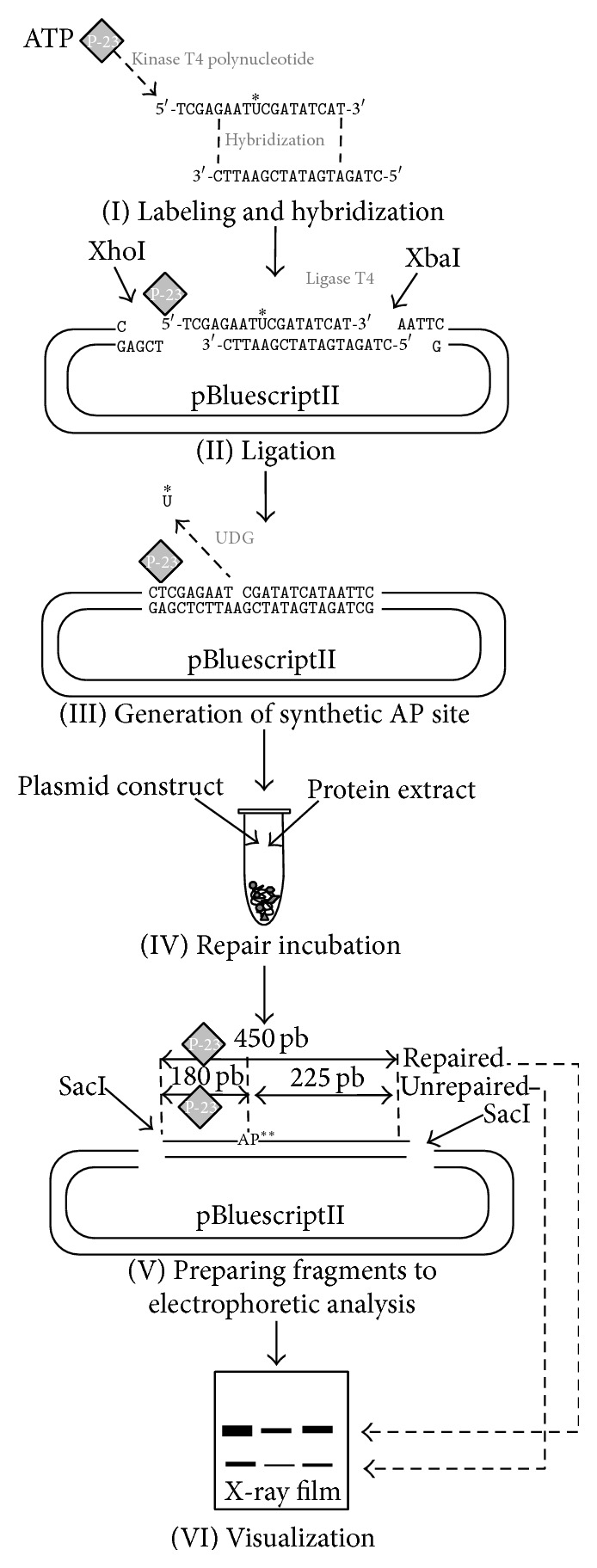
(I) A uracil-containing oligonucleotide was subjected to action of polynucleotide kinase to attach a radioactive phosphate group from [γ-32P] ATP. It was hybridized with a second oligonucleotide whose sequence was adjusted to obtain sticky ends referring to XhoI and XbaI digestion site. (II) The short DNA fragment prepared in stage I was cloned into a pBluescriptII plasmid. (III) Uracil-DNA glycosylase was utilized to remove uracil and, as consequence, create a single gap in DNA to act as a synthetic lesion. (IV) A plasmid with single AP site constituted a substrate for the protein extract in 90-minute repair incubation. (V) Two SacI recognition sites of the pBluescriptII plasmid were used to excise 450 pb-long fragment covering the lesion site and radioactive label for analysis on 8% urea/acrylamide gel. (VI) Interpretation of outcomes was based on detection of two bands. The full-length 450 pb fragment reflects restored DNA fraction, whereas presence of short 180 pb fraction indicates the amount of unrepaired DNA. ∗U: uracil; ∗∗AP: apurinic/apyrimidinic.
The blood donors were a 79-year-old man with histopathologically confirmed adenocarcinoma and a cancer-free woman of the same age. A colorectal cancer tumor had been removed from the 86-year-old female with cecum carcinoma. Control tissue samples of the colon were taken from patients with primary inguinal incarcerate hernia from the macroscopically unchanged tissue during the operation. All individuals enrolled in this experiment were hospitalized in the Military Medical Academy University Teaching Hospital-Central Veterans' Hospital, Lodz.
2.1. Details of the BER Assay Procedure
2.1.1. Preparation of DNA Substrate
The Vector, a pBSII plasmid, was multiplied in E. coli DH5α and isolated by Qiagen Maxiprep and underwent double digestion with 2.5 U of XbaI and 2.5 U XhoII fast digest enzymes for 1 hour (Fast digest, ThermoScientific, Rochester, USA). SAP (ThermoScientific, Rochester, USA) enzyme was applied to avoid self-ligation. Insert 5′-TCGAGAATUCGATATCAT-3′ was labeled in kinase reaction 2 U T4 kinase polynucleotide (thermo) with 2 μL of [γ-32P] ATP (6000 μCi) whereas the second oligo (5′-CTTAAGCTATAGTAGGATC-3′) was incubated under the same conditions but with unlabeled ATP. Equal amounts of the two oligonucleotides were mixed and annealed through heating to 95°C before being left to slowly cool down. A 1 : 5 vector : insert molar ratio (established previously) was applied to allow ligation with 1 U of T4 ligase (ThermoScientific, Rochester, USA) overnight. The construct was purified by elution (GenJet Maxi prep kit, ThermoScientific) from 1% agarose gel.
Preparation of whole-cell protein extract is as follows: Minute Total Protein Extraction Kit (Invent) was used to isolate proteins from peripheral blood lymphocytes or tissue slices. All protein samples were adjusted to 2 μg/mL.
2.1.2. Repair Assay
A 100 ng/reaction of unaltered native pBSII (load control) was mixed with prelabeled plasmid (1000 cpm/sample) carrying an AP site generated by digestion with 1 U of UDG glycosylase (ThermoScientific) for 4 h. This mixture was made up to a total volume of 15 μL by adding the reagents 0.4 μL of 1 M HEPES-KOH, pH 7.5, 0.2 μL of 1 M MgCl2, 0.5 μL of 3 M KCl, 0.2 μL of 0.1 M DTT, 1 μL of 0.1 M ATP, and 0.4 μL of 1 mM dNTP and adjusted to a final volume with H2O. The reaction was launched by adding 5 μL (10 μg) protein extract. Repair incubation was carried out in a thermocycler (Bio-Rad) and took 90 min at 25°C. The reaction was stopped by adding of 6 μL of 2% SDS to each tube. Thereupon, 2 μL of 1 mg/mL proteinase K and 2 μL of 0.2 mg/mL carrier tRNA were added to each tube and incubated at 37°C for 30 min, and then treated with 150 μL stop solution (10 mM Tris-HCl, pH 7.5, 300 mM sodium acetate, 10 mM EDTA pH 8.0, 0.5% SDS). DNA was recovered by phenol/chloroform extraction (1 : 1) and overnight ethanol precipitation. A 450 pb length DNA fragment was excised from plasmid by 1 U of SacI enzyme (ThermoScientific, Rochester, USA) incubated at 37°C for 1 h. To allow the repaired and unrepaired fractions to be differentiated, the remaining unrepaired AP site was treated with 1 U of AP-recognizing endonuclease IV for 1 h. All samples were run on 8% urea-containing polyacrylamide gel for 3 h in 120 V. The accurate electrophoresis was preceded by 1 h preelectrophoresis with loading buffer.
2.1.3. Visualization
The bands were detected by autoradiography. The gels were dried and stored at −20°C with X-ray film for 2 h, 6 h, or overnight exposure. Bromidium ethidium staining was used to visualize load control. Optical density quantification of bends was performed with GeneTools software (Invitrogen).
3. Results
The genotyping results indicate that the Lys/Glu genotype of the EXO1 gene (Table 2) may increase the risk of colorectal cancer (OR = 1.672 (1.109–2.519), p = 0.014). The investigated PolB gene polymorphism was not found to increase the risk of CRC; however, our analysis suggests that occurrence of Arg allele may have a protective effect, since it decreases the risk of colorectal cancer (OR = 0.772 (0.601–0.994), p = 0.044) as shown in Table 3. The 780Arg/His polymorphism of the LIG3 gene was found to contribute to an increase in the risk of CRC (OR = 1.570 (1.109–2.224), p = 0.011) (Table 4).
Table 2.
The distribution of genotypes, allele frequencies, and the analysis of the odds ratio (OR) for 589Lys/Glu polymorphism of EXO1 gene in patients with colorectal cancer (CRC) and the control group.
| Genotype/allele | Patients n = 309 | Controls n = 304∗ | OR (95% CI) | p |
|---|---|---|---|---|
| Lys/Lys | 57 | 69 | 1 (ref.) | — |
| Lys/Glu | 203 | 147 | 1.672 (1.109–2.519) | 0.014 |
| Glu/Glu | 49 | 88 | 0.674 (0.411–1.106) | 0.118 |
| Lys | 317 | 285 | 1 (ref.) | — |
| Glu | 301 | 323 | 0.838 (0.670–1.048) | 0.121 |
∗Genotype distribution in Hardy-Weinberg equilibrium; χ 2 = 0.612.
Table 3.
The distribution of genotypes, allele frequencies, and the analysis of the odds ratio (OR) for 242Pro/Arg polymorphism of PolB gene in patients with colorectal cancer (CRC) and the control group.
| Genotype/allele | Patients n = 303 | Controls n = 302∗ | OR (95% CI) | p |
|---|---|---|---|---|
| Pro/Pro | 147 | 121 | 1 (ref.) | — |
| Pro/Arg | 123 | 142 | 0.713 (0.507–1.003) | 0.052 |
| Arg/Arg | 33 | 39 | 0.697 (0.413–1.174) | 0.174 |
| Pro | 417 | 384 | 1 (ref.) | — |
| Arg | 189 | 220 | 0.772 (0.601–0.994) | 0.044 |
∗Genotype distribution in Hardy-Weinberg equilibrium; χ 2 = 0.791.
Table 4.
The distribution of genotypes, allele frequencies, and the analysis of the odds ratio (OR) for 780Arg/His polymorphism of LIG3 gene in patients with colorectal cancer (CRC) and the control group.
| Genotype/allele | Patients n = 310 | Controls n = 305∗ | OR (95% CI) | p |
|---|---|---|---|---|
| Arg/Arg | 101 | 121 | 1 (ref.) | — |
| Arg/His | 173 | 132 | 1.570 (1.109–2.224) | 0.011 |
| His/His | 36 | 52 | 0.829 (0.503–1.368) | 0.462 |
| Arg | 375 | 374 | 1 (ref.) | — |
| His | 245 | 236 | 1.035 (0.823–1.302) | 0.764 |
∗Genotype distribution in Hardy-Weinberg equilibrium; χ 2 = 0.125.
In order to investigate the interaction of the polymorphisms of the studied genes and to evaluate their mutual influence on the risk of colorectal cancer, gene-gene interactions were analyzed. The simultaneous occurrence of the Lys/Glu genotype of the EXO1 gene and the Pro/Pro genotype of the PolB gene was found to possibly increase the risk of colorectal cancer (OR = 2.265 (1.193–4.301), p = 0.011) (Table 5). In case of gene-gene interactions between 589Lys/Glu EXO1 SNP and 780Arg/His LIG3 SNP, the simultaneous occurrence of Lys/Glu and Arg/His genotypes may increase risk of colorectal cancer (OR = 1.970 (1.041–3.731), p = 0.036) while concomitant presence of Glu/Glu and Arg/Arg genotypes may decrease the risk (OR = 0.402 (0.178–0.906), p = 0.026) (Table 6). Finally, the analysis of gene-gene interactions for 242Pro/Arg PolB gene and 780Arg/His LIG3 gene indicated that the cooccurrence of genotypes Pro/Pro and Arg/His may increase the risk of CRC (OR = 2.154 (1.265–3.667), p = 0.004) (Table 7).
Table 5.
The distribution of genotypes and the analysis of the odds ratio (OR) for gene-gene interactions: 589Lys/Glu EXO1 and 242Pro/Arg PolB in patients with colorectal cancer (CRC) and the control group.
| Genotype | Patients n = 302 | Controls n = 302 | OR (95% CI) | p |
|---|---|---|---|---|
| Lys/Lys-Pro/Pro | 24 | 28 | 1 (ref.) | — |
| Lys/Lys-Pro/Arg | 20 | 26 | 0.897 (0.404–1.994) | 0.791 |
| Lys/Lys-Arg/Arg | 11 | 15 | 0.856 (0.331–2.212) | 0.752 |
| Lys/Glu-Pro/Pro | 99 | 51 | 2.265 (1.193–4.301) | 0.011 |
| Lys/Glu-Pro/Arg | 81 | 72 | 1.313 (0.698–2.467) | 0.396 |
| Lys/Glu-Arg/Arg | 19 | 23 | 0.964 (0.426–2.180) | 0.920 |
| Glu/Glu-Pro/Pro | 23 | 42 | 0.639 (0.303–1.346) | 0.238 |
| Glu/Glu-Pro/Arg | 22 | 44 | 0.583 (0.276–1.232) | 0.156 |
| Glu/Glu-Arg/Arg | 3 | 1 | — | — |
Table 6.
The distribution of genotypes and the analysis of the odds ratio (OR) for gene-gene interactions: 589Lys/Glu EXO1 and 780Arg/His LIG3 in patients with colorectal cancer (CRC) and the control group.
| Genotype | Patients n = 302 | Controls n = 302 | OR (95% CI) | p |
|---|---|---|---|---|
| Lys/Lys-Arg/Arg | 21 | 27 | 1 (ref.) | — |
| Lys/Lys-Arg/His | 31 | 27 | 1.476 (0.684–3.185) | 0.320 |
| Lys/Lys-His/His | 3 | 15 | — | — |
| Lys/Glu-Arg/Arg | 64 | 46 | 1.789 (0.902–3.547) | 0.094 |
| Lys/Glu-Arg/His | 118 | 77 | 1.970 (1.041–3.731) | 0.036 |
| Lys/Glu-His/His | 17 | 23 | 0.950 (0.407–2.218) | 0.920 |
| Glu/Glu-Arg/Arg | 15 | 48 | 0.402 (0.178–0.906) | 0.026 |
| Glu/Glu-Arg/His | 18 | 26 | 0.890 (0.389–2.038) | 0.777 |
| Glu/Glu-His/His | 15 | 13 | 1.484 (0.582–3.784) | 0.409 |
Table 7.
The distribution of genotypes and the analysis of the odds ratio (OR) for gene-gene interactions: 242Pro/Arg PolB and 780Arg/His LIG3 in patients with colorectal cancer (CRC) and the control group.
| Genotype | Patients n = 302 | Controls n = 302 | OR (95% CI) | p |
|---|---|---|---|---|
| Pro/Pro-Arg/Arg | 52 | 56 | 1 (ref.) | — |
| Pro/Pro-Arg/His | 82 | 41 | 2.154 (1.265–3.667) | 0.004 |
| Pro/Pro-His/His | 12 | 24 | 0.539 (0.245–1.185) | 0.121 |
| Pro/Arg-Arg/Arg | 38 | 48 | 0.853 (0.483–1.506) | 0.584 |
| Pro/Arg-Arg/His | 66 | 73 | 0.974 (0.589–1.611) | 0.920 |
| Pro/Arg-His/His | 19 | 21 | 0.974 (0.471–2.015) | 1.000 |
| Arg/Arg-Arg/Arg | 9 | 15 | 0.646 (0.261–1.603) | 0.343 |
| Arg/Arg-Arg/His | 20 | 17 | 1.267 (0.599–2.679) | 0.538 |
| Arg/Arg-His/His | 4 | 7 | — | — |
In general, optical density detection of particular DNA bands revealed higher BER repair efficiency among cancer-free individuals than CRC patients in both lymphocytes and colon tissue samples. The percentage ratio of repaired to damaged fractions was found to be 89.67%/10.32% in lymphocytes taken from healthy subjects and 70.5%/29.5% in those of CRC patients. Examination of the ability of tissue protein extract to perform BER indicated a significantly greater repair level in normal tissue (68.11%/31.89%) than CRC tissue (58.36%/41.64%). The results of the BER assay analysis are presented in Figure 2.
Figure 2.
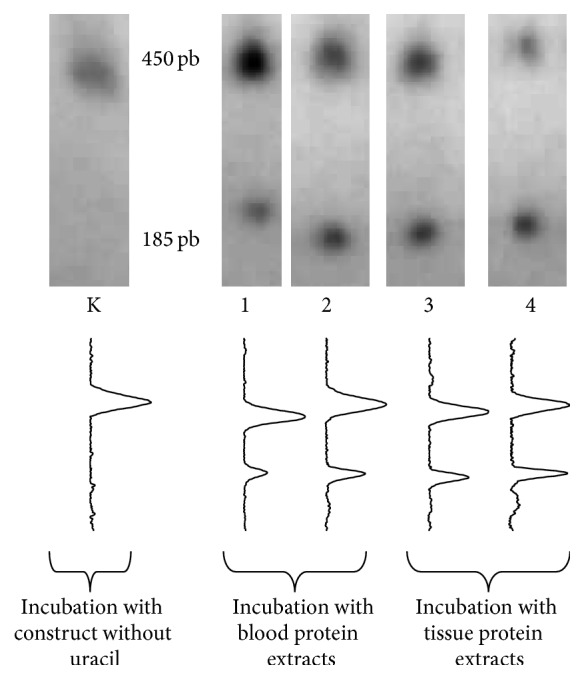
A comparison of BER activity in the lymphocytes and tissue of CRC patients and healthy controls. Each electropherogram shows two fractions of DNA: 450 pb repaired and 185 pb unrepaired. Lanes 1-2 indicate lymphocyte BER efficiency while lanes 3-4 refer to BER in tissue. Samples are presented in the following order: K: positive control; DNA substrate did not contain uracil and so reflects 100% of repair; 1: healthy control; 2: colorectal cancer; 3: unchanged colon tissue; 4: colorectal cancer.
4. Discussion
All cells in the human body are permanently exposed to the negative effects of reactive oxygen species. Virtually all kinds of cell components, including proteins, lipids, and nucleic acids, can be targets for attack by ROS, which may interfere with the proper functioning of cellular biochemical processes. Oxidative stress has been confirmed to play a role in carcinogenesis by a number of previous studies [6]. Oxidative damage to DNA has significant mutagenic potential, and excessive accumulation of damage to DNA leads to cell necrosis or apoptosis, the most abundant types of lesion being 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG) and 4,6-diamino-5-formamidopyrimidine (FapyA) and 8-oxo-7,8-dihydroguanine (8-oxoG).
By virtue of its high stability and relative simplicity of detection, 8-oxoG is a vital biomarker of DNA oxidative damage [7]. Hence, colorectal cancer was extensively examined in terms of 8-oxoG presence. An analysis performed on a Spanish population indicated a twofold higher level of 8-oxoG in colorectal tumors than in normal mucosa [8]. In other studies, immunohistochemical tests complemented by high-performance liquid chromatography (HPLC) have revealed considerable higher levels of 8-oxoG in colorectal carcinoma than nontumorous colon epithelial cells [9]. Furthermore, the same team reports the presence of an elevated 8-oxoG level accompanied by 8-oxoG lyase overexpression [10]. Increased levels of 8-oxoG have been observed in CRC patient lymphocytes [11] and plasma [12] compared to those of healthy controls.
These high levels create a very challenging environment for base excision repair processes, which have to operate with high efficiency to maintain genome integrity. Omission of oxidized forms of guanine may lead to incorrect base-pairing, resulting in G:C to T:A transversion mutation [13, 14]. However, no evaluation of BER system activity can be performed purely on the basis of 8-oxoG level. In addition, the majority of previous studies tests have been based on the total 8-oxoG content including unbound 8-oxoG and that bound to DNA. A high level of free 8-oxoG might occur as consequence of efficient performance of the BER initial step, when the damaged bases are being recognized and excised by glycosylase. Conversely, higher numbers of damaged bases remaining in a DNA-associated form may indicate that BER activity is insufficient to cope with repairs.
A more precise tool to evaluate the level of DNA single-strand breaks and repair capacity is the comet assay. An alkaline version of the comet assay used in a previous investigation indicated statistically significant differences in repair efficiency between CRC patients and healthy subjects. After a 240-minute repair incubation, the level of single-strand DNA breaks was significantly diminished in lymphocytes from a cancer-free control group in comparison to CRC subjects. A similar difference was observed in a comparative analysis of cells from normal colon mucosa tissue and a CRC tumor [15]. In the course of a comet assay, cells are incubated intravitally after hydrogen peroxide treatment, whereby the BER process, consisting of the excision of the damaged base and restoration of the DNA sequence, can be tracked holistically.
Glycosylase activity in initial stages of BER is an issue that has been the focus of a great degree of research interest. Thus far, eukaryotic cells have been found to possess several glycosylases such as NEIL1-3, UNG, NTH1, MUTYH, APE1, and OGG1 [16–19]. OGG1 is the primary BER enzyme capable of cleaving N-glycosyl bond between the sugar component and 8-oxoG. Studies based on a mouse model with OGG1−/− knock-out revealed this deficiency to have minor or even marginal importance in pathogenesis and cancer frequency [20]. MUTYH has a unique ability to remove normal adenines misincorporated opposite to 8-oxoG. Similar to OGG1, studies based on biallelic MUTYH mutation implied no significant increase in sensitivity to oxidative stress [21]. Surprisingly, an additive effect has been observed in mice with the double mutation OGG1−/− and MUTYH−/−, where higher tumor appearance frequencies have been noted [22].
Both OGG1 and MUTYH have numerous polymorphic variants which are being eagerly examined in the context of carcinogenesis. The common polymorphisms of the OGG1 gene, S326 C and R46Q, have been found to slightly decrease the activity of the enzyme [23, 24]. Regarding population screening, definitely more attention has been paid to the screening of S326 C, especially its involvement in lung cancer development. However, several investigations summarized in a meta-analysis do not reveal any linkage with lung cancer [25]. In contrast, certain variants of MUTYH, a polymorphism-rich gene, have been shown to elevate the risk of CRC 28-fold [26]. A great deal of current research into genetic variation of proteins has focused on the XRCC1 gene. XRCC1 is an important protein due to its participation in the recruitment of the other BER proteins, making it a binder of all stages [27]. However, several large scale meta-analyses display contradictory conclusion about its role in carcinogenesis. To be specific, while the Arg194Trp polymorphism was found to have a protective effect on tobacco smoking with regard to cancer risk [28], it was found to have no such role for other examined cancers [25, 29, 30].
As the literature shows no consensus on role of the early stages of BER, the present study addresses the gap-filling stage. The present study is so far the only one aimed to evaluate the effectiveness of BER in CRC. Undoubtedly, although our findings show an interesting trend, they should be treated with great caution. As a BER deficiency can be observed in lymphocytes from a CRC individual, it can be inferred that the native repair system is also deficient, which may result in a slow, gradual accumulation of damage that, at some critical moment, may contribute to the development of cancer.
It is important to determine whether some difficult to exclude factors can interfere with the result. To minimize this risk of appearance of additional undesirable damaging agents, the primary inclusion criteria for the BER assay were place of residence (the same city), the subject not taking medication, including cancer therapy for CRC, and the lack of any smoking addiction or alcohol abuse. However, it is difficult to predict the influence of other significant factors such as ionizing radiation, UV light, diet, or stress associated with everyday situations. There is some risk that any of these factors could put BER on standby, while it is forced to repair more cellular proteins which had been produced as a response to greater exposure [31]. In the follow-up phase of the experiment, the tumor cell extract demonstrated a similar reduction of BER activity in comparison to normal tissue.
However, it is unclear whether this reduced repair ability is innate and this phenotype is maintained after tumorigenesis, as differences could emerge due to the presence of mutations which were nested during malignant transformation. In addition, weak BER capacity may help exacerbate the genotoxic effect of ROS, allowing malignancy to progress. Chan et al. report that the presence of hypoxia in CRC provokes changes in BER [32]. Other reports note that some characteristics of colon tissue factor may induce oxidative stress: an increased amount of free radicals may occur as result of diet rich in red meat [33] or with low calcium or vitamin D levels [34]. What is more, bacteria living in the intestine can also be an important extracellular source of ROS which promotes increased DNA damage in colonic epithelial cells [35]. The available evidence seems to suggest that BER has a possible impact on both the development and progression of CRC.
A similar concept has recently been presented by Stanczyk et al., who, by using a similar methodology to the present study, report significantly lower BER efficiency in lymphocytes taken from children suffering from childhood acute lymphoblastic leukemia in comparison to healthy controls [36]. Although CRC and leukemia are virtually incomparable due to their totally different natures, the BER system was found to play a crucial role in both and may also be involved in the pathogenesis of several other diseases.
It is undeniable that polymorphisms of DNA repair systems participate in the carcinogenesis process, as mentioned before in the first part of Section 4. Extensive studies suggest that these polymorphisms play a role in almost all types of cancer [37–40], including colorectal cancer [29, 41]. The genetic polymorphisms of MMR system appear to participate in the pathogenesis of hereditary nonpolyposis colorectal cancer [42, 43], and a growing body of evidence suggests their involvement in the BER system [44, 45], but reports concerning the NER system are inconclusive, with some confirming the link [46] and others denying it [47].
The present paper examines the impact of polymorphisms of PolB, LIG3, and EXO1 of the BER repair system on the modulation of the risk of colon cancer. The genes were selected on the basis that the products of these three genes do not have glycosylase activity, thus avoiding any negative impact on the first part of the experiment. All three proteins are involved in stages of BER directly following the glycosylase action. Therefore, BER assay was adjusted to measure only the gap-filling step where PolB, EXO1, and LIG3 play crucial roles. The protective effect of the Arg allele for the 242Pro/Arg gene polymorphism of PolB demonstrated in our work (Table 3) has been shown in previous publications [46]. In addition, the 242Pro/Pro genotype of the PolB gene in combination with the genotype 780Arg/His of LIG3 gene increases the risk of CRC (Table 7), and the risk is much higher than in case of the 780Arg/His SNP of LIG3 (OR = 2.154; 1.265–3.667, p = 0.004 versus OR = 1.570; 1.109–2.224, p = 0.011) (Table 4). This clearly shows the important role of gene-gene interactions in modulating the risk of malignant transformation, which has been confirmed in many other publications [48, 49].
Our finding that the 589Lys/Glu SNP of EXO1 is associated with an increased risk of CRC is contrary to those of Akbari et al. [50]. However, it should be noted that the previous study was performed on an Iranian population, while our tests were carried out on a Polish population. Ethnic group has been repeatedly demonstrated to have a significant impact on the modulation of the risk of particular diseases [51, 52]. Yamamoto et al. [53] suggest that the potential impact of polymorphism 589Lys/Glu on increased risk of carcinogenesis may depend on the presence of cigarette smoking by the patient. Again, however, these studies concern a Japanese population, which may exert an influence on the results.
In our opinion, it is important to note interaction of polymorphisms 589Lys/Glu of the EXO1 gene and 780Arg/His of the LIG3 gene (Table 6), whose coexistence increases the risk of CRC compared to the presence of polymorphism 589Lys/Glu itself (OR = 1.970; 1.041–3.731, p = 0.036 versus OR = 1.570; 1.109–2.224, p = 0.011). As no extant publications describe the influence of the 780Arg/His polymorphism of LIG3 on the risk of CRC, our own findings in this regard showing an elevated risk (Table 4) are significant. In addition, attention should be once again directed to the mentioned earlier gene-gene interaction of LIG3 with the 589Lys/Glu polymorphism of the EXO1 gene. Furthermore, not only does the potential protective effect of the Arg allele of the 242Pro/Arg PolB SNP and the increased CRC risk associated with the 780Arg/His SNP of LIG3 and 589Lys/Glu SNP of EXO1 merit attention, but also, more importantly, the modulation of risk induced by gene-gene interactions identified in this study can significantly affect individual predisposition to the development of cancer.
5. Conclusions
Decreased BER activity may play a crucial role in the pathogenesis, development, and progression of colorectal cancer, as the activity of BER is distinctly reduced in lymphocytes and cancer tissue from CRC individuals. In addition, the genotyping of SNPs which have so far not been thought to be associated with CRC (LIG3, PolB, and EXO1) suggests that potential BER dysfunction may lay not only in its first steps, but equally or at even greater level in the gap-filling events. We believe that our results are promising, yet further studies are needed on this subject to establish a link between a given polymorphism and its phenotypic effect in the modulation of BER activity and thus its impact on carcinogenesis.
Acknowledgments
This work has been supported by Umed in Lodz Grants 502-03/5-108-05/502-54-158 and 502-03/5-108-05/502-54-144 and by Polish Ministry of Science and Higher Education Grants N402422138 and NN403250340.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Giordano A. I., Domènech I., Torres A., et al. Results in the surgical treatment of giant acoustic neuromas. Acta Otorrinolaringologica Espanola. 2012;63(3):194–199. doi: 10.1016/j.otorri.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Power D. G., Gloglowski E., Lipkin S. M. Clinical genetics of hereditary colorectal cancer. Hematology/Oncology Clinics of North America. 2010;24(5):837–859. doi: 10.1016/j.hoc.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Papadopoulos N., Lindblom A. Molecular basis of HNPCC: mutations of MMR genes. Human Mutation. 1997;10(2):89–99. doi: 10.1002/(SICI)1098-1004(1997)10:2<89::AID-HUMU1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 4.Wood R. D., Mitchell M., Sgouros J., Lindahl T. Human DNA repair genes. Science. 2001;291(5507):1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 5.Matsumoto Y., Kim K., Bogenhagen D. F. Proliferating cell nuclear antigen-dependent abasic site repair in Xenopus laevis oocytes: an alternative pathway of base excision DNA repair. Molecular and Cellular Biology. 1994;14(9):6187–6197. doi: 10.1128/mcb.14.9.6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klaunig J. E., Kamendulis L. M. The role of oxidative stress in carcinogenesis. Annual Review of Pharmacology and Toxicology. 2004;44:239–267. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- 7.Valavanidis A., Vlachogianni T., Fiotakis C. 8-hydroxy-2′ -deoxyguanosine (8-OHdG): a critical biomarker of oxidative stress and carcinogenesis. Journal of Environmental Science and Health Part C: Environmental Carcinogenesis and Ecotoxicology Reviews. 2009;27(2):120–139. doi: 10.1080/10590500902885684. [DOI] [PubMed] [Google Scholar]
- 8.Oliva M. R., Ripoll F., Muñiz P., et al. Genetic alterations and oxidative metabolism in sporadic colorectal tumors from a Spanish community. Molecular Carcinogenesis. 1997;18(4):232–243. doi: 10.1002/(SICI)1098-2744(199704)18:4<232::AID-MC7>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 9.Kondo S., Toyokuni S., Iwasa Y., et al. Persistent oxidative stress in human colorectal carcinoma, but not in adenoma. Free Radical Biology and Medicine. 1999;27(3-4):401–410. doi: 10.1016/s0891-5849(99)00087-8. [DOI] [PubMed] [Google Scholar]
- 10.Kondo S., Toyokuni S., Tanaka T., et al. Overexpression of the hOGG1 gene and high 8-hydroxy-2′-deoxyguanosine (8-OHdG) lyase activity in human colorectal carcinoma: regulation mechanism of the 8-OHdG level in DNA. Clinical Cancer Research. 2000;6(4):1394–1400. [PubMed] [Google Scholar]
- 11.Gackowski D., Banaszkiewicz Z., Rozalski R., Jawien A., Olinski R. Persistent oxidative stress in colorectal carcinoma patients. International Journal of Cancer. 2002;101(4):395–397. doi: 10.1002/ijc.10610. [DOI] [PubMed] [Google Scholar]
- 12.Sato T., Takeda H., Otake S., et al. Increased plasma levels of 8-hydroxydeoxyguanosine are associated with development of colorectal tumors. Journal of Clinical Biochemistry and Nutrition. 2010;47(1):59–63. doi: 10.3164/jcbn.10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neeley W. L., Essigmann J. M. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chemical Research in Toxicology. 2006;19(4):491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 14.Grollman A. P., Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends in Genetics. 1993;9(7):246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 15.Przybylowska K., Kabzinski J., Sygut A., Dziki L., Dziki A., Majsterek I. An association selected polymorphisms of XRCC1, OGG1 and MUTYH gene and the level of efficiency oxidative DNA damage repair with a risk of colorectal cancer. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2013;745-746:6–15. doi: 10.1016/j.mrfmmm.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 16.Krokan H. E., Standal R., Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochemical Journal. 1997;325(1):1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hazra T. K., Izumi T., Kow Y. W., Mitra S. The discovery of a new family of mammalian enzymes for repair of oxidatively damaged DNA, and its physiological implications. Carcinogenesis. 2003;24(2):155–157. doi: 10.1093/carcin/24.2.155. [DOI] [PubMed] [Google Scholar]
- 18.McCullough A. K., Dodson M. L., Lloyd R. S. Initiation of base excision repair: glycosylase mechanisms and structures. Annual Review of Biochemistry. 1999;68:255–285. doi: 10.1146/annurev.biochem.68.1.255. [DOI] [PubMed] [Google Scholar]
- 19.Krokan H. E., Bjørås M. Base excision repair. Cold Spring Harbor Perspectives in Biology. 2013;5(4) doi: 10.1101/cshperspect.a012583.a012583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arai T., Kelly V. P., Minowa O., Noda T., Nishimura S. The study using wild-type and Ogg1 knockout mice exposed to potassium bromate shows no tumor induction despite an extensive accumulation of 8-hydroxyguanine in kidney DNA. Toxicology. 2006;221(2-3):179–186. doi: 10.1016/j.tox.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 21.Hirano S., Tominaga Y., Ichinoe A., et al. Mutator phenotype of MUTYH-null mouse embryonic stem cells. The Journal of Biological Chemistry. 2003;278(40):38121–38124. doi: 10.1074/jbc.c300316200. [DOI] [PubMed] [Google Scholar]
- 22.Xie Y., Yang H., Cunanan C., et al. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Research. 2004;64(9):3096–3102. doi: 10.1158/0008-5472.can-03-3834. [DOI] [PubMed] [Google Scholar]
- 23.Dherin C., Radicella J. P., Dizdaroglu M., Boiteux S. Excision of oxidatively damaged DNA bases by the human α-hOgg1 protein and the polymorphic α-hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Research. 1999;27(20):4001–4007. doi: 10.1093/nar/27.20.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill J. W., Evans M. K. Dimerization and opposite base-dependent catalytic impairment of polymorphic S326C OGG1 glycosylase. Nucleic Acids Research. 2006;34(5):1620–1632. doi: 10.1093/nar/gkl060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiyohara C., Takayama K., Nakanishi Y. Association of genetic polymorphisms in the base excision repair pathway with lung cancer risk: a meta-analysis. Lung Cancer. 2006;54(3):267–283. doi: 10.1016/j.lungcan.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 26.Theodoratou E., Campbell H., Tenesa A., et al. A large-scale meta-analysis to refine colorectal cancer risk estimates associated with MUTYH variants. British Journal of Cancer. 2010;103(12):1875–1884. doi: 10.1038/sj.bjc.6605966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campalans A., Marsin S., Nakabeppu Y., O'Connor T. R., Boiteux S., Radicella J. P. XRCC1 interactions with multiple DNA glycosylases: a model for its recruitment to base excision repair. DNA Repair. 2005;4(7):826–835. doi: 10.1016/j.dnarep.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 28.Hu Z., Ma H., Chen F., Wei Q., Shen H. XRCC1 polymorphisms and cancer risk: a meta-analysis of 38 case-control studies. Cancer Epidemiology Biomarkers and Prevention. 2005;14(7):1810–1818. doi: 10.1158/1055-9965.epi-04-0793. [DOI] [PubMed] [Google Scholar]
- 29.Hung R. J., Hall J., Brennan P., Boffetta P. Genetic polymorphisms in the base excision repair pathway and cancer risk: a huge review. American Journal of Epidemiology. 2005;162(10):925–942. doi: 10.1093/aje/kwi318. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y., Newcomb P. A., Egan K. M., et al. Genetic polymorphisms in base-excision repair pathway genes and risk of breast cancer. Cancer Epidemiology Biomarkers and Prevention. 2006;15(2):353–358. doi: 10.1158/1055-9965.EPI-05-0653. [DOI] [PubMed] [Google Scholar]
- 31.Fry R. C., Begley T. J., Samson L. D. Genome-wide responses to DNA-damaging agents. Annual Review of Microbiology. 2005;59:357–377. doi: 10.1146/annurev.micro.59.031805.133658. [DOI] [PubMed] [Google Scholar]
- 32.Chan N., Ali M., McCallum G. P., et al. Hypoxia provokes base excision repair changes and a repair-deficient, mutator phenotype in colorectal cancer cells. Molecular Cancer Research. 2014;12(10):1407–1415. doi: 10.1158/1541-7786.mcr-14-0246. [DOI] [PubMed] [Google Scholar]
- 33.Angeli J. P. F., Garcia C. C. M., Sena F., et al. Lipid hydroperoxide-induced and hemoglobin-enhanced oxidative damage to colon cancer cells. Free Radical Biology and Medicine. 2011;51(2):503–515. doi: 10.1016/j.freeradbiomed.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 34.Erdelyi I., Levenkova N., Lin E. Y., et al. Western-style diets induce oxidative stress and dysregulate immune responses in the colon in a mouse model of sporadic colon cancer. Journal of Nutrition. 2009;139(11):2072–2078. doi: 10.3945/jn.108.104125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huycke M. M., Abrams V., Moore D. R. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis. 2002;23(3):529–536. doi: 10.1093/carcin/23.3.529. [DOI] [PubMed] [Google Scholar]
- 36.Stanczyk M., Sliwinski T., Trelinska J., et al. Role of base-excision repair in the treatment of childhood acute lymphoblastic leukaemia with 6-mercaptopurine and high doses of methotrexate. Mutation Research—Genetic Toxicology and Environmental Mutagenesis. 2012;741(1-2):13–21. doi: 10.1016/j.mrgentox.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 37.Ali M. F., Meza J. L., Rogan E. G., Chakravarti D. Prevalence of BER gene polymorphisms in sporadic breast cancer. Oncology Reports. 2008;19(4):1033–1038. [PubMed] [Google Scholar]
- 38.Goode E. L., Ulrich C. M., Potter J. D. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiology Biomarkers and Prevention. 2002;11(12):1513–1530. [PubMed] [Google Scholar]
- 39.Leibeling D., Laspe P., Emmert S. Nucleotide excision repair and cancer. Journal of Molecular Histology. 2006;37(5–7):225–238. doi: 10.1007/s10735-006-9041-x. [DOI] [PubMed] [Google Scholar]
- 40.García-Closas M., Malats N., Real F. X., et al. Genetic variation in the nucleotide excision repair pathway and bladder cancer risk. Cancer Epidemiology Biomarkers and Prevention. 2006;15(3):536–542. doi: 10.1158/1055-9965.EPI-05-0749. [DOI] [PubMed] [Google Scholar]
- 41.Farrington S. M., Tenesa A., Barnetson R., et al. Germline susceptibility to colorectal cancer due to base-excision repair gene defects. American Journal of Human Genetics. 2005;77(1):112–119. doi: 10.1086/431213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peltomäki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. Journal of Clinical Oncology. 2003;21(6):1174–1179. doi: 10.1200/jco.2003.04.060. [DOI] [PubMed] [Google Scholar]
- 43.Jäger A. C., Rasmussen M., Bisgaard H. C., Singh K. K., Nielsen F. C., Rasmussen L. J. HNPCC mutations in the human DNA mismatch repair gene hMLH1 influence assembly of hMutLα and hMLH1-hEXO1 complexes. Oncogene. 2001;20(27):3590–3595. doi: 10.1038/sj.onc.1204467. [DOI] [PubMed] [Google Scholar]
- 44.Chow E., Thirlwell C., Macrae F., Lipton L. Colorectal cancer and inherited mutations in base-excision repair. Lancet Oncology. 2004;5(10):600–606. doi: 10.1016/s1470-2045(04)01595-5. [DOI] [PubMed] [Google Scholar]
- 45.Tudek B. Base excision repair modulation as a risk factor for human cancers. Molecular Aspects of Medicine. 2007;28(3-4):258–275. doi: 10.1016/j.mam.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 46.Moreno V., Gemignani F., Landi S., et al. Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clinical Cancer Research. 2006;12(7):2101–2108. doi: 10.1158/1078-0432.ccr-05-1363. [DOI] [PubMed] [Google Scholar]
- 47.Mort R., Mo L., McEwan C., Melton D. W. Lack of involvement of nucleotide excision repair gene polymorphisms in colorectal cancer. British Journal of Cancer. 2003;89(2):333–337. doi: 10.1038/sj.bjc.6601061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ritchie M. D., Hahn L. W., Roodi N., et al. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. American Journal of Human Genetics. 2001;69(1):138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cordell H. J. Detecting gene-gene interactions that underlie human diseases. Nature Reviews Genetics. 2009;10(6):392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akbari Z., Safari-Alighiarloo N., Haghighi M. M., et al. Lack of influence of the SMAD7 gene rs2337107 polymorphism on risk of colorectal cancer in an Iranian population. Asian Pacific Journal of Cancer Prevention. 2014;15(11):4437–4441. doi: 10.7314/apjcp.2014.15.11.4437. [DOI] [PubMed] [Google Scholar]
- 51.Kamangar F., Dores G. M., Anderson W. F. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. Journal of Clinical Oncology. 2006;24(14):2137–2150. doi: 10.1200/jco.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 52.Manuguerra M., Saletta F., Karagas M. R., et al. XRCC3 and XPD/ERCC2 single nucleotide polymorphisms and the risk of cancer: a HuGE review. American Journal of Epidemiology. 2006;164(4):297–302. doi: 10.1093/aje/kwj189. [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto H., Hanafusa H., Ouchida M., et al. Single nucleotide polymorphisms in the EXO1 gene and risk of colorectal cancer in a Japanese population. Carcinogenesis. 2005;26(2):411–416. doi: 10.1093/carcin/bgh335. [DOI] [PubMed] [Google Scholar]


