Abstract
Palatogenesis involves the initiation, growth, morphogenesis, and fusion of the primary and secondary palatal shelves from initially separate facial prominences during embryogenesis to form the intact palate separating the oral cavity from the nostrils. The palatal shelves consist mainly of cranial neural crest-derived mesenchyme cells covered under a simple embryonic epithelium. Growth and patterning of the palatal shelves are controlled by reciprocal epithelial-mesenchymal interactions regulated by multiple signaling pathways and transcription factors. During palatal shelf outgrowth, the embryonic epithelium develops a “teflon” coat consisting of a single, continuous layer of periderm cells that prevents the facial prominences and palatal shelves from forming aberrant inter-epithelial adhesions. Palatal fusion involves not only spatiotemporally-regulated disruption of the periderm but also dynamic cellular and molecular processes that result in adhesion and intercalation of the palatal medial edge epithelia to form an inter-shelf epithelial seam, and subsequent dissolution of the epithelial seam to form the intact roof of the oral cavity. The complexity of regulation of these morphogenetic processes is reflected by the common occurrence of cleft palate in humans. This review will summarize major recent advances and discuss major remaining gaps in the understanding of cellular and molecular mechanisms controlling palatogenesis.
Keywords: cleft palate, fusion, morphogenesis, palate development, periderm, signaling, mouse
1. Introduction
Cleft lip and/or cleft palate are among the most common birth defects in humans, occurring at a frequency of about 1 in 500 - 2500 live birth (Vanderas, 1987; Schutte and Murray, 1999; Gorlin et al., 2001; Dixon et al., 2011; Mangold et al., 2011). Clinically, cleft lip with or without cleft palate (CL/P) is a unilateral or bilateral gap between the philtrum and the lateral upper lip, often extending through the upper jaw into the nostril and accompanied by cleft of the secondary palate – the roof of the oral cavity. Cleft palate (CP) also frequently occurs without cleft lip. Both CL/P and CP appear in syndromic and nonsyndromic forms. The Online Mendelian Inheritance in Man (OMIM) database (www.ncbi.nlm.nih.gov/omim/) lists over 500 syndromic disorders in which CL/P or CP is a feature. The genetic basis of some of these syndromes has been identified through positional or candidate gene cloning (reviewed in Dixon et al., 2011). Approximately 70% of CL/P and 50% CP cases are nonsyndromic, for which the etiology and pathogenesis are complex and poorly understood (Dixon et al., 2011; Mangold et al., 2012; Rahimov et al., 2012).
The major reason why CL/P and CP are common birth defects is because development of the midface involves highly coordinated growth and morphogenesis of initially separate promordia. In humans, development of the face begins in the fourth week of embryogenesis with migrating neural crest cells that combine with mesodermal and ectodermal cells to establish the facial primordia, which initially consist of five separate parts surrounding the primitive mouth, the stomodeum (reviewed by Jiang et al., 2006; Dixon et al., 2011; Levi et al., 2011). At the rostral midline is the frontonasal prominence, which is populated by mesenchymal cells derived from the forebrain and midbrain neural crest cells. The stomodeum is bound laterally by a pair of maxillary processes and caudally by a pair of mandibular processes, which are populated by neural crest cells derived from the rostral region of the hindbrain. By the end of the fourth week, nasal pits form bilaterally in the lower part of the frontonasal prominence and extend into the stomodeum such that the frontonasal prominence is divided distally into paired medial and lateral nasal processes. Subsequently, rapid growth of the maxillary processes results in the medial nasal processes merging with each other, forming the intermaxillary segment. The lateral nasal processes fuse with the maxillary processes to form the lateral nose and cheek region. The intact upper lip forms by the maxillary processes fusing medially with the medial nasal processes (Jiang et al., 2006).
Development of the palate begins during the fifth week of human embryogenesis and is divided into two regions: the primary and secondary palates (reviewed by Dixon et al., 2011; Levi et al., 2011; Bush and Jiang, 2012). The primary palate arises from the intermaxillary segment and contains the philtrum, the incisors and a small part of the hard palate situated anterior to the incisive foramen (located immediately behind the incisor gum). The secondary palate includes both the hard and soft palate posterior of the incisive foramen. Development of the secondary palate begins during the sixth week of human embryogenesis with the initiation of two outgrowths, termed palatal shelves, from the oral side of the maxillary processes. The palatal shelves initially grow downward on both sides of the developing tongue. During the eighth week, as the head expands and the tongue moves downward, the bilateral palatal shelves elevate to a horizontal position above the dorsum of the tongue and merge with each other at the midline. In addition, the palatal shelves also fuse anteriorly with the primary palate and dorsally with the nasal septum, both of which are derived from the medial nasal processes. These fusion processes are complete by the twelfth week of gestation.
The dynamic morphogenetic processes giving rise to the midface are often disturbed by genetic or environmental insults. For example, cleft lip with cleft palate may result from disturbances in proliferation, migration and survival of the neural crest cells, or fusion between the medial nasal and maxillary processes, whereas cleft palate without cleft lip may result from disruption of palatal shelf growth, elevation or fusion. Moreover, since palate development occurs concurrently with significant growth and expansion of the whole craniofacial complex, malformation of structures in the vicinity of the palatal shelves, such as the tongue and mandible, sometimes hinders palatal shelf elevation or contact, resulting in cleft palate (reviewed by Chai and Maxson, 2006; Bush and Jiang, 2012; Dotto, 2012). In addition to extensive human genetic studies that have led to identification of the gene mutations responsible for many cleft palate syndromes (reviewed in Dixon et al., 2011), genetic and developmental biology studies in model animal systems, particularly the laboratory mice in which the palatal growth and morphogenetic processes are remarkably similar to that in humans (Bush et al., 2012) (Figure 1), in the last 25 years have provided significant insights into the cellular and molecular processes underlying palatogenesis. Since the studies of the roles of various signaling pathways in controlling palate development have been extensively reviewed recently (Bush and Jiang, 2012; Cobourne and Green, 2012; He and Chen et al., 2012; Parada and Chai, 2012; Stanier and Pauws, 2012; Smith et al., 2013; Lane and Kaartinen, 2014), we focus this article on integrating recent studies of the molecular pathways in palate growth and patterning as well as on highlighting new advances in the cellular and molecular mechanisms regulating epithelial adhesion and palate fusion.
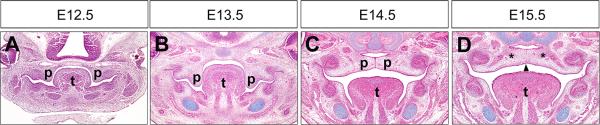
Figure 1.
Palate development in mice. (A-D) Representative HE-stained coronal sections through the middle of the anterior-posterior axis of the developing palatal shelves at E12.5 (A), E13.5 (B), E14.5 (C), and E15.5 (D). The palatal shelves grow vertically from the oral side of the maxillary processes from E11.5 to past E13.5 (A and B). By E14.5, the palatal shelves have elevated to the horizontal position above the tongue, adhered to each other at the midline and formed the midline epithelial seam (MES) (C). At around E15.5, the MES is disintegrated to allow confluence of the palatal mesenchyme (D). Arrowhead in D point to the disintegrating MES. Asteroids in D mark the sites of ossification to form the palatal bone. p, palatal shelf; t, tongue.
2. Regulation of palatal shelf growth and patterning
The central player in the regulation of palatal shelf growth appears to be sonic hedgehog (Shh) signaling. Shh is expressed throughout the early oral epithelium prior to palatal shelf outgrowth (Rice et al., 2006). Mice with tissue-specific inactivation of Smoothened (Smo), which encodes the obligate transducer of hedgehog signaling, in the cranial neural crest cells showed complete agenesis of the secondary palate (Jeong et al., 2004). Studies of mice with tissue-specific inactivation of Smo in the early palatal mesenchyme or of Shh in the epithelium confirmed that Shh signaling is critical for palatal shelf growth (Rice et al., 2004; Lan and Jiang, 2009). Shh signaling is required for activation and/or maintenance of expression of several transcription factors, including Foxf1, Foxf2, and Osr2, in the developing palatal mesenchyme (Lan and Jiang, 2009). Although whether Foxf1 and Foxf2 play primary roles in palatal shelf growth remains to be elucidated, Osr2 has been shown to function as an intrinsic regulator of palatal mesenchyme cell proliferation (Lan et al., 2004). In addition, Shh signaling regulates expression of cell cycle regulators Cyclin D1 and Cyclin D2 in the palatal mesenchyme (Lan and Jiang, 2009).
The developing palatal shelves exhibit both morphological and molecular heterogeneity along both the anteroposterior and oronasal axes (reviewed by Hilliard et al., 2005; Bush and Jiang, 2012; Smith et al., 2013; Figure 3). Whereas Shh is initially expressed throughout the palatal epithelium at the onset of palatal outgrowth (Rice et al., 2004), its expression becomes highly restricted to the palatal rugae, the periodic epithelial ridges that form in species-specific patterns on the oral side of the palatal epithelium, in the anterior region and in the sensory papilla in the posterior region of the palate (Pantalacci et al., 2008; Welsh and O'Brien, 2009; Baek et al., 2011; Figure 2). Whereas the periodic formation of rugae during palate development appears to be regulated by a Turing-type reaction-diffusion mechanism (Economou et al., 2012), the restriction of Shh expression to the oral side of the palatal epithelium correlates with differential expression of Fgf10 and Fgf7 along the oronasal axis of the palatal mesenchyme, with Fgf10 preferentially expressed on the oral side and Fgf7 preferentially on the nasal side (Rice et al, 2004; Veistinen et al., 2009). In palatal explant culture assays, Fgf10 induced, whereas Fgf7 repressed, Shh expression in the palatal epithelium (Rice et al., 2004; Han et al., 2009). Moreover, application of function-neutralizing antibody against Fgf7 caused increased Shh expression in the palatal epithelium, suggesting that Fgf7 normally competes with Fgf10 and represses Shh expression in the nasal side of the palatal epithelium (Han et al., 2009). Expression of Fgf7 in the palatal mesenchyme is maintained by the Dlx5 transcription factor whereas Fgf10 and Shh act in a positive feedback loop to regulate each other's expression (Rice et al., 2004; Han et al., 2009; Lan and Jiang, 2009) (Figure 3). Interestingly, exogenous Shh protein inhibited Fgf7 expression in palatal mesenchyme explants (Han et al., 2009), suggesting that Shh signaling plays an active role in maintaining the oronasal asymmetry of the palatal shelves through differential regulation of expression of Fgf10 and Fgf7 in the mesenchyme (Figure 3). In addition, maintenance of Shh expression in the most anterior rugae and in the posterior palatal epithelium requires the functions of the Msx1 and Pax9 transcription factors, respectively, in the palatal mesenchyme (Zhang et al., 2002; Zhou et al., 2013). Msx1 expression is restricted to the anterior region of the developing palatal mesenchyme and regulates anterior palatal mesenchyme proliferation through activation of expression of Bmp4, which in turn signals to the epithelium to maintain Shh expression (Zhang et al., 2002). Whereas Bmp4 expression in the anterior palate is dependent on Msx1 function, Bmp4 is also expressed in a posterior region of the developing palatal shelves where Msx1 is normally not expressed (Zhou et al., 2013). The expression of Bmp4 in the posterior palate depends on Pax9 function for its maintenance (Zhou et al., 2013). Mice lacking Pax9 function exhibit complete penetrance of cleft palate and palatal shelf growth defects (Peters et al., 1998; Zhou et al., 2011; 2013). In Pax9−/− mutant mouse embryos, expression of Fgf10 and Osr2 in the developing palatal mesenchyme as well as expression of Shh in the palatal epithelium were also reduced in comparison with wildtype littermates. Zhou et al. (2013) showed that Fgf10 expression was also reduced in the Osr2−/− palatal mesenchyme and that expression of Osr2 from a Pax9Osr2KI knockin allele restored Fgf10 expression in the palatal mesenchyme and partly rescued posterior palate morphogenesis in the Pax9 mutant embryos although the reductions in Shh expression in the palatal epithelium and in Bmp4 expression in the posterior palatal mesenchyme were not rescued (Zhou et al., 2013). Together, these studies suggest that Pax9 regulates the Osr2-Fgf10 and the Bmp4-Shh pathways independently during palate development, with Pax9 and the Shh signaling converging on the regulation of the Osr2 transcription factor (Figure 3).
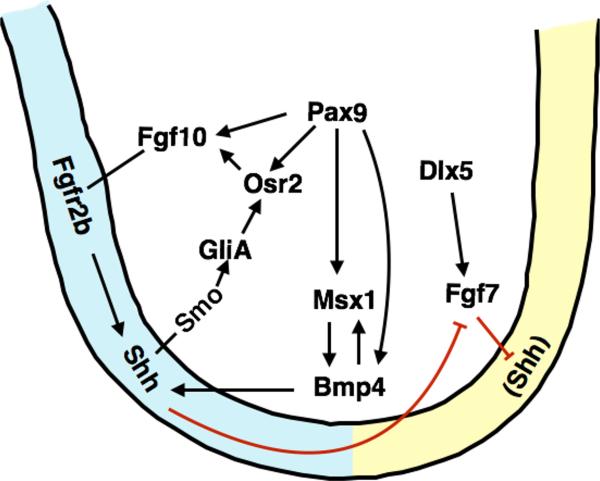
Figure 3.
Molecular regulation of palatal shelf growth and oronasal patterning. Schematic diagram of a coronal section of the developing palatal shelf, with the oral side to the left. Black arrows indicate positive regulation whereas red lines indicate repression. Shh is expressed in the palatal epithelium and positively regulates Fgf10 expression through the Osr2 transcription factor in the palatal mesenchyme. Fgf10 signals through the Fgfr2b receptor to maintain Shh expression in the palatal epithelium. Dlx5 activates Fgf7 expression in the palatal mesenchyme. Fgf7 and Shh represses the expression of each other such that Fgf7 expression is restricted in the nasal side of the palatal mesenchyme and Shh in the oral side of the palatal epithelium. Msx1 maintains Shh expression through Bmp4 in the anterior palate whereas Pax9 acts upstream of Bmp4, Fgf10, Msx1, and Osr2 in the palatal mesenchyme.
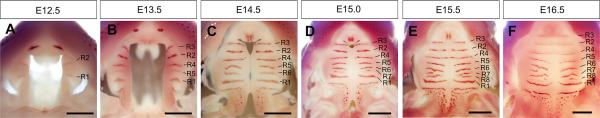
Figure 2.
Whole mount oral view of growth and patterning of the developing palatal shelves in mice. Patterns of Shh mRNA expression were detected by whole mount in situ hybridization. R1 – R8 mark the Shh-expressing palatal rugae in the sequence of their formation from E12.5 (A) to E16.5 (F). R1 marks the anterior-posterior boundary (all panels are orientied with the tip of the snout pointing up). The patchy Shh expression in the posterior palatal mark the sensory papilla buds. Scale bar, 500 μm.
In addition to Msx1 and Pax9, several other transcription factors, including short stature homeobox-2 (Shox2), BarH-like homeobox-1 (Barx1), meningioma-1 (Mn1), and T-box factor 22 (Tbx22), exhibit differential expression along the anteroposterior axis (Figure 4). Shox2 is expressed throughout the anterior half but completely absent from the posterior half of the palatal mesenchyme, with the anterior-posterior boundary corresponding exactly to the first formed palatal rugae (Yu et al., 2005; Li and Ding, 2007; Pantalacci et al., 2008; Welsh and O'Brien, 2009, Figure 4). Expression of Tbx22 is largely restricted to the posterior palatal mesenchyme, whereas expression of Barx1 and Mn1 shows preference in the posterior palatal mesenchyme, with expression expanded into the anterior palate domain during palatal shelf growth (Liu et al., 2008; Welsh and O'Brien, 2009). These patterns of expression during palatal development correlate with palatal developmental defects in several mutant mice, with Shox2 specifically required for anterior palatal growth whereas Mn1 acts upstream of Tbx22 to regulate posterior palatal growth (Yu et al., 2005; Liu et al., 2008; Pauws et al., 2009).
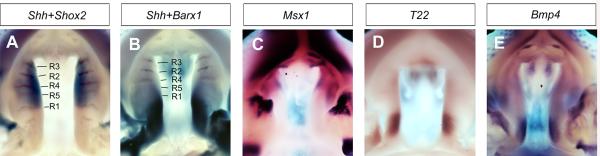
Figure 4.
Differential patterns of gene expression along the anterior-posterior axis of the developing palatal shelves at E13.5. (A) Dual color in situ hybridization detection of Shh (brown) and Shox2 (blue) mRNA expression. Shh is expressed in the palatal rugae (marked as R1 – R5 in the sequence of their formation), whereas Shox2 mRNAs are restricted to the anterior half of the developing palatal shelves. R1 coincides with the posterior boundary of Shox2 mRNA expression. (B) Dual color in situ hybridization detection of Shh (brown) and Barx1 (blue) mRNA expression. Barx1 is mainly expressed in the posterior half of the developing palatal shelves. (C) whole mount in situ hybridization detection of Msx1 (purple) mRNA expression. Msx1 mRNAs were detected in the anterior-most quarter of the developing palatal shelves. (D) Tbx22 mRNA expression is highly restricted in the posterior half of the developing palatal shelves. (E) Bmp4 mRNA expression was detected in the anterior and posterior subregions of the developing palatal shelves.
Whereas the differential gene expression patterns and mutant mouse phenotypes clearly demonstrate regionalized control of palatal growth along the anteroposterior axis, the molecular mechanism setting up the anteroposterior patterns of the developing palatal shelves remains to be elucidated. Hilliard et al. (2005) showed that exogenous Bmp4 induced Msx1 expression in the anterior but not posterior palatal mesenchyme in explant culture assays, which is probably due to the fact that Bmpr1a is preferentially expressed in the anterior palate (Baek et al., 2011). Yu et al. (2005) showed that anterior palatal epithelium was able to induce Shox2 expression in posterior palatal mesenchyme in recombinant explant culture assays (Yu et al., 2005). Together, these results indicate that both the palatal epithelium and mesenchyme are patterned along the anteroposterior axis.
3. Proper periderm differentiation is critical for normal palatogenesis
During early embryogenesis, the ectoderm consists initially of a single layer of undifferentiated cuboidal epithelial cells covering the entire embryo, including lining the primitive oral cavity (Richardson et al., 2014). As embryonic development proceeds, the embryonic skin undergoes a series of defined stratification and differentiation events to generate the mature epidermis. The first stratification event produces the periderm, a single layer of flattened epithelial cells covering the embryonic epithelia during subsequent developmental stages until shortly before birth (Holbrook and Odland, 1975; Sanes et al., 1986; M'Boneko and Merker, 1988; Polakowska et al., 1994; Richardson et al., 2014). Several functions have been proposed for the periderm, including protection from the environment (Hayward, 1983), regulation of underlying mesenchyme (Scott et al., 1987), and contribution to cornified evelope formation (Akiyama et al., 1999). Whereas all of these functions have yet to be proven, a series of mouse genetic studies and human disease phenotypes have shown a critical role for the periderm in craniofacial and palate development, as discussed below.
Through characterization of the cellular mechanisms underlying cleft palate pathogenesis in the Jag2−/− mutant mice, we first reported a critical role for the oral periderm in preventing aberrant oral epithelial adhesions that interfered with palatal shelf elevation (Jiang et al., 1998a; Casey et al., 2006; Richardson et al., 2009). We showed that Jag2 is expressed in the developing oral epithelium and activates the Notch1 receptor during periderm differentiation. In the Jag2−/− mutant embryos, although oral epithelial stratification occurred, the resultant suprabasal cells did not have the flattened morphology characteristic of normal periderm cells and the mutant oral epithelia was disorganized and subsequently showed multiple intraoral inter-epithelial adhesions, including adhesion between the maxillary and mandibular oral epithelia as well as adhesion of the palatal shelves to the developing tongue (Jiang et al., 1998a; Casey et al., 2006) (Figure 5A). The correlation of aberrant oral epithelial adhesions with the disruption in oral periderm differentiation in the Jag2−/− mutant mice suggests that periderm normally provides a non-sticking barrier for the formation of the oral cavity. The specific activation and nuclear localization of intracellular domain of Notch1 in the periderm cells in wildtype embryos and the dramatic reduction of nuclear accumulation of Notch1 in the suprabasal oral epithelium in Jag2−/− mutant embryos suggest that Jag2-Notch1 signaling plays a critical role in periderm cell differentiation.
Figure 5.
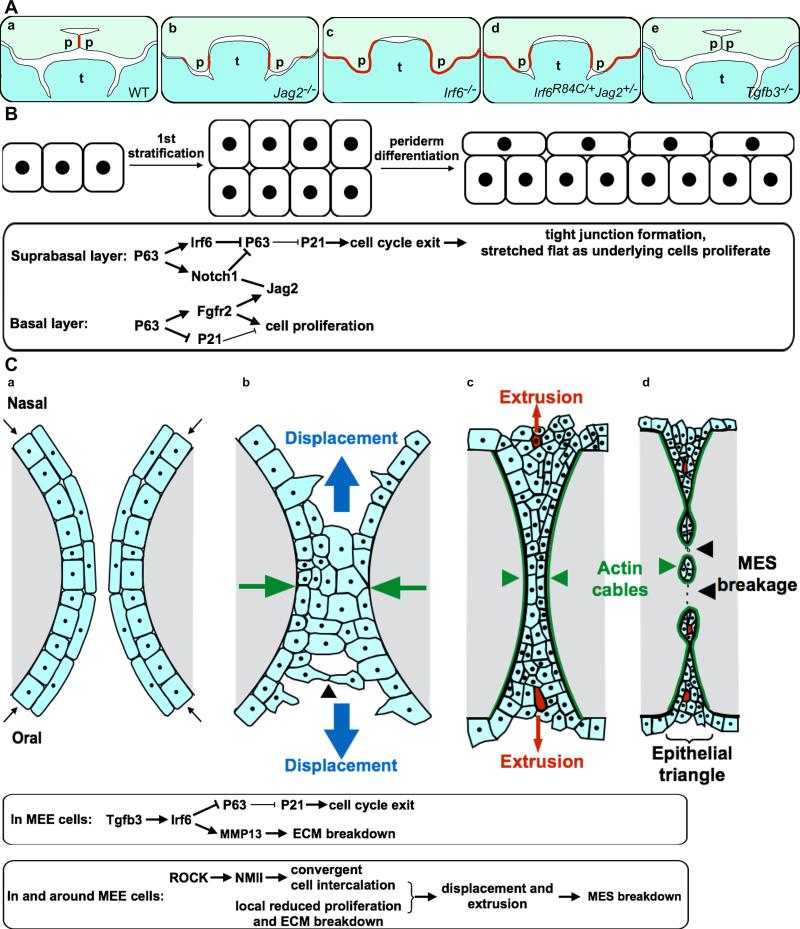
Periderm differentiation plays a critical role in palatogenesis. (A) Schematic diagrams depicting palatal shelf adhesion at the onset of palatal fusion in wildtype mouse embryos (a), palate-tongue and maxillary-mandibular adhesions in the Jag2−/− mutant embryos (b), extensive intra-oral epithelial adhesions in Irf6−/− mutant embryos (c), palate-tongue, palate-mandible, and maxillary-mandibular adhesions in the Irf6R84C/+Jag2+/− compound heterozygous mouse embryos (d), and failure of palatal adhesion in Tgfb3−/− mutant mouse embryos. Red line marks sites of inter-epithelial adhesion. p, palatal shelf; t, tongue. (B) Schematic diagram of 1st epithelial stratification and periderm differentiation, with the molecular and cellular pathways in the suprabasal and basal cells outlined in the rectangular box below. (C) Schematic diagram of coronal sections of palatal shelves depicting cell convergence and extrusion during palatal adhesion and fusion, with the molecular and cellular processes regulating MES breakdown outlined in the rectangular boxed below. The cell convergence and extrusion diagrams are modified from Kim et al. (2015). Palatal epithelial cells are in ligh blue color and the palatal mesenchyme in gray. The paired palatal shelves are each covered with a single layer of periderm cells prior to inter-palatal adhesion (a). At the onset of palatal fusion, periderm cells and basal MEE cells extend protrusions and converge toward (green arrows) the midline (b), with some cells in the multicellular seam displaced toward the oral or nasal side. As the MES forms, cellular rosettes form and cells at the center of the rosettes are squeezed out by extrusion (marked in red) (c). The actomyosin cables pull the MES apart and surround the epithelial islands, continuing to extrude cells from cellular rosettes (d). Palatal MES breakdown is driven by both the nonmyosin II mediated cell convergence and extrusion as well as by Tgfb3-Irf6 regulated local cell cycle arrest and ECM breakdown.
Aberrant oral epithelial adhesions have been associated with several congenital ectodermal disorders, including Van der Woude syndrome (VWS) and popliteral pterygium syndrome (PPS), which are also characterized by varying degrees cleft lip, cleft palate (Van der Woude, 1954; Kondo et al., 2002; Richardson et al., 2006). Mutations in the IRF6 gene have been shown to underlie both syndromes (Kondo et al., 2002). Gene expression studies in mice showed that Irf6 is highly specifically expressed during periderm cell differentiation (Knight et al., 2006; Richardson et al., 2009; 2014). Mouse embryos deficient in Irf6 as well as mouse embryos homozygous for the PPS mutation Irf6R84C showed severe intraoral epithelial adhesions, including maxillary-mandibular and palatal-tongue adhesions that caused cleft palate by blocking palatal shelf elevation (Richardson et al., 2006; Ingraham et al., 2006) (Figure 5A). More recently, Richardson et al. (2014) carefully examined periderm development, using multiple molecular markers, in wildtype and Irf6R84C/R84C mutant mouse embryos and demonstrated that Irf6 function is required for normal periderm differentiation. Furthermore, two other mutant mouse strains, Ikka−/− and SfnEr/Er, which exhibit remarkably similar phenotypes with the Irf6−/− mutant mice, also lack periderm differentiation (Richardson et al., 2014). Furthermore, genetic ablation of periderm cells during embryogenesis caused aberrant intraoral epithelial adhesions, which led to cleft palate in the severely affected embryos (Richardson et al., 2014). Together, these results indicate that proper periderm formation prevents pathological inter-epithelial adhesions and is critical for orofacial and palatal morphogenesis.
How do Irf6 and the Jag2-Notch1 pathways regulate periderm differentiation? Recent studies suggest that both Irf6 and Notch signaling are activated by and negatively feedback to regulate the transcription factor p63 to drive periderm differentiation. P63 is expressed in the earliest embryonic epithelial and continue to be expressed in basal undifferentiated epithelial cells but downregulated upon periderm differentiation (Richardson et al., 2009; 2014). The p63 gene is transcribed using two alternative promoters, producing two major classes of protein isoforms that different in the N-terminal amino acid sequences (King and Weinberg, 2007; Crum and McKeon, 2010; Romano et al., 2012). Mice deficient in all p63 isoforms and mice with specific inactivation of ΔNp63, the predominant isoform in epidermal cells, exhibit a thin, undifferentiated epidermis with significantly reduced level of expression of Irf6 in oral epithelial cells (Mills et al., 1999; Yang et al., 1999; Moretti et al., 2010; Thomason et al., 2010). Moreover, the p63 protein binds enhancer sequences upstream of the Irf6 gene and can activate reporter gene expression driven by the Irf6 gene sequences, suggesting that p63 directly activates Irf6 expression during early epithelial development. However, upon periderm differentiation, Irf6 is strongly expressed but p63 is down-regulated in the periderm cells (Richardson et al., 2009; Richardson et al., 2014). Moretti et al. (2010) showed that IRF6 promotes proteasome-mediated degradation of the ΔNp63 protein in human keratinocytes, suggesting that Irf6 negatively regulates p63 during periderm differentiation. Moreover, whereas p63 has previously been shown to positively regulate Jag1 and Jag2 expression (Sasaki et al., 2002; Candi et al., 2007), a recent study suggests that active Notch signaling represses p63 expression in the developing surface ectoderm (Tadeu and Horsley, 2013). Furthermore, although Irf6 and Jag2 are not required for the expression of each other in the developing oral epithelium, Irf6R84C/+;Jag2+/− compound heterozygous mice exhibited similar persistent expression of p63 in stratified and aberrantly adhering oral epithelium (Richardson et al., 2009) (Figure 5A), suggesting that Irf6 and Jag2-Notch signaling act synergistically to downregulate p63 during periderm differentiation. P63 has been shown to repress transcription of p21 in vitro and in vivo (Westfall et al., 2003; Laurikkala et al., 2006; Welsh and O'Brien, 2009). Thus, the Irf6 and Jag2-Notch signaling pathways drive periderm differentiation through indirect activation of p21 mediated cell cycle exit, with continued proliferation of the basal epithelial cells and new tight junctions that form between the suprabasal cells (Yoshida et al., 2012) contributing to stretching and flattening the periderm cells (Figure 5B).
The functions of many other genes, including Grhl3, Ikka, and Sfn, are required for periderm differentiation. Mutations in GRHL3 have been associated with a subset of VWS patients with no linkage to IRF6 (Peyrard-Janvid et al., 2014). Grhl3−/− mice also exhibit aberrant inter-epithelial adhesions function (Peyrard-Janvid et al., 2014). Studies in zebrafish showed that Grhl3 is specifically expressed in periderm cells and its expression is dependent on Irf6 (de la Garza et al., 2013). Ikka−/− and SfnEr/Er mice exhibit failure of periderm differentiation similar to Irf6−/− mice, but they do not appear to be regulated by Irf6 and it is not clear yet how they regulate periderm differentiation (Richardson et al., 2014). Mice lacking Fgf10 or its epithelial receptor Fgfr2b also exhibit aberrant palate-mandible or palate-tongue adhesions (Alappat et al., 2005; Rice et al., 2004). Alappat et al. (2005) showed that Fgf10−/− embryos exhibit loss of Jag2 expression in the oral and palatal epithelium, suggesting that Fgf10 signals through Fgfr2b to maintain Jag2 expression in the oral epithelium. Recently, Ferone et al. (2012) showed that Fgfr2 is a direct target of p63 in the developing epithelium (Ferone et al., 2012). Together with previous studies showing upregulation of Jag2 expression by p63 and the loss of periderm integrity in Jag2−/− mice (Jiang et al., 1998a; Sasaki et al., 2002; Casey et al., 2006; Candi et al., 2007), these studies suggest that p63 indirectly regulates Jag2 expression in the basal epithelial cells through Fgfr2b and that Jag2 signals to the suprabasal cells to maintain the integrity of the periderm (Dotto, 2012) (Figure 5B).
4. Palatal fusion: formation and dissolution of inter-shelf epithelial seam by cell convergence and extrusion
In order for the paired palatal shelves to initiate adhesion and fusion after they have elevated to the horizontal position above the dorsum of the developing tongue, the periderm covering the medial edge of the shelves has to be disrupted (Yoshida et al., 2012). Classic embryological as well as transmission and scanning electron microscopy studies showed that, just prior to inter-shelf adhesion, the periderm cells at the medial edge change shape dramatically by elongating and contracting and their nuclei become pyknotic (Waterman and Meller, 1974; Fitchett and Hay, 1989; Griffith and Hay, 1992; Yano et al., 1996). Similar apoptotic events occur in the periderm cells of the edge epithelia of medial nasal and maxillary prominences prior to lip fusion (Sun et al., 2000). The desquamation of the periderm cells enables the apposing pair of palatal shelves to adhere to each other at the midline, with the adhered medial edge epithelial layers (MEE) subsequently undergoing intercalation to form the midline epithelial seam (MES) (Yoshida et al. 2012).
Although the cellular processes of palatal fusion have been extensively studied, the exact mechanisms regulating palatal periderm desquamation and MES dissolution are still incompletely understood (Sun et al., 2000; Cuervo and Covarrubias, 2004; Bush and Jiang, 2012; Hu et al., 2015; Kim et al., 2015). Historically, three distinct but non-exclusive cellular mechanisms driving MES dissolution, including epithelial-mesenchyme transformation, apoptotic cell death, and migration of MES cells into the oral and nasal sides of the palatal epithelia, were proposed and supported by experimental evidence from palatal culture studies (Sun et al., 2000; Cuervo and Covarrubias, 2004; reviewed by Dudas et al., 2007; Gritli-Linde, 2007; Nawshad, 2008; Bush and Jiang, 2012). With the development of Cre/loxP-mediated genetic lineage labeling methods, the fate of the MES cells during palatal fusion in mouse embryos have been analyzed by several laboratories using different epithelial Cre-driver transgenic mice (Vaziri Sani et al., 2005; Xu et al., 2006; Jin and Ding, 2006a). Since no genetically labeled derivatives of the MES are found in the post-fusion palatal mesenchyme, results from these studies mostly favored the apoptosis hypothesis for MES dissolution although MES migration was also detected (Bush and Jiang, 2012)(reviewed by Bush and Jiang, 2012). However, direct analysis of the requirement for important apoptosis regulators in palatal fusion have been inconclusive (Cecconi et al., 1998; Cuervo et al., 2002; Takahara et al., 2004; Jin and Ding, 2006b). Most recently, Kim et al. (2015) examined cell behavior during palatal fusion using a combination of genetic lineage labeling, tissue-specific gene inactivation, and live imaging. These studies reveal an essential role for actomyosin contractility-driven convergence and cell intercalation in the formation of MES and subsequent cell displacement and extrusion during MES breakdown (Figure 5C). Whereas previously an argument against cell death as the major mechanism for MES dissolution suggested that extensive cell death of the MES cells would weaken the fusion site and lead to the bilateral palatal shelves been pulled apart (Sun et al., 2000), Kim et al. (2015) demonstrate by using live imaging the process of MES cell extrusion, during which converging MES cells form rosettes and the cells in the center of these rosettes are squeezed out by multicellular actin cables. Immunodetection of active Caspase-3 and E-cadherin of E14.5 mouse embryonic palatal tissues showed that about half of the apoptotic cells in the MES were part of the cellular rosettes. Thus, apoptosis in the MES, at least during early stages of MES breakdown, does not involve simultaneous death of neighboring cells that might weaken the fusion site but rather through extrusion of the apoptotic cell by its neighbors. Previous studies showed that an epithelial cell destined for apoptosis initiates signaling to its neighbors to form a contractile ring of actin and myosin prior to procaspase activation (Rosenblatt et al., 2001). Furthermore, pharmacological inhibition of stretch-activated ion channels blocked palatal fusion in explant culture, indicating that live cell extrusion due to crowding, in addition to apoptosis, also plays a key role in MES dissolution. Together with a series of genetic and pharmacological experiments demonstrating the requirement for Rho kinase- and myosin light chain kinase-mediated activation of non-muscle myosin-II in MES cell intercalation and displacement during palatal fusion, these studies reveal the mechanistic connections of all previously observed cellular behaviors during palatal fusion, including extensive epithelial filapodia and cell shape changes at the beginning of palatal shelve adhesion, cell displacement and collective cell migration in the oronasal directions during MES formation and breakdown, and apoptotic cell death, to actomyosin-driven MES cell convergence (Figure 5C).
What are the molecular mechanisms underlying the spatiotemporal regulation of MEE fusion competence and the fusion process? Embryological and genetic studies have demonstrated an essential role for Tgfb3 signaling regulating palatal fusion. During mouse palate development, Tgfb3 expression is specifically activated in the MEE by E13 (Fitzpatrick et al., 1990; Pelton et al., 1990). Mice lacking Tgfb3 function and mice with epithelium-specific deletion of either Tgfbr1 or Tgfbr2 exhibit cleft palate due to failure of palatal fusion (Kaartinen et al., 1995; Proetzel et al., 1995; Dudas et al., 2006; Xu et al., 2006). In Tgfb3−/− embryos, although the palatal shelves grow and elevate normally, approach and contact each other at the midline, appropriate adhesion does not occur. Even when placed in direct contact with each other in explant culture, the Tgfb3−/− mutant palatal shelves adhere poorly or not at all (Kaartinen et al., 1997). Recent studies demonstrated that periderm cells persisted as an intact single layer covering the palatal shelves in Tgfb3−/− mutant embryos (Wu et al., 2013; Hu et al., 2015). Remarkably, whereas initial differentiation of the periderm cells requires Irf6 function, recent studies suggest that Irf6 is also required to mediate Tgfb3-induced palatal fusion. This second important function of Irf6 in palate development is brought about by Tgfb3 activation of its expression in the basal layer of the MEE cells just prior to palatal fusion (Fakhouri et al., 2012; Iwata et al., 2013). Tgfb3−/− as well as Tgfbr2fl/fl;K14-Cre mutant embryos show significantly reduced Irf6 expression in MEE cells (Knight et al., 2006; Iwata et al., 2013). Transgenic expression of Irf6 in the basal epithelial layer driven by the keratin-14 gene promoter was able to rescue palatal fusion in Tgfbr2fl/fl;K14-Cre mutant mice (Iwata et al., 2013). Iwata et al. (2013) suggest that Irf6 might employ the same downstream molecular cascade, involving repression of p63 and indirectly activating p21 as in periderm differentiation, to regulate palatal MES degeneration (Morreti et al., 2010; Iwata et al., 2013). Whereas the up-regulation of p21 likely contributes to MEE cell cycle exit, whether Irf6 contributes to Tgfb3 regulation of periderm disruption remains to be investigated. On the other hand, Irf6 is required downstream of Tgfb3 for MES breakdown since Irf6R84C/R84C mutant embryos showed persistence of the aberrantly adhered MEE and tongue epithelial cells whereas the aberrant palate-tongue epithelial seam was disintegrated in Jag2−/− mutant embryos (Casey et al., 2006; Richardson et al., 2009). Like Tgfb3, Irf6 function is also required for the expression of Mmp13 in MEE cells (Blavier et al., 2001; Richardson et al., 2009). Thus, Irf6 might also mediate Tgfb3 induced MES disintegration through the up-regulation of Mmp13 and other factors involved in degradation of the basal lamina (Figure 5C).
Upon activation of the receptor complexes, Tgfβ signaling activates receptor activated Smads, including Smad2 and Smad3, which then partner with the common mediator Smad4 to regulate expression of downstream target genes (Shi and Massague, 2003). Receptor Smads can also bind to Trim33, a chromatin reader (He et al., 2006). In addition, Tgfβ signaling can activate Smad-independent pathways, including the Tgfβ-activated kinase-1 (Tak1) and p38 MAPK kinase cascade (Derynck and Zhang, 2003). Studies of mice with epithelium-specific inactivation of Smad4, Tak1, and Trim33, show that all three pathways act partly redundantly to mediate Tgfβ3 signaling in palatal MES breakdown (Xu et al., 2008; Lane et al., 2015).
Mice with K14-Cre mediated deletion of Tgfbr1 or Tgfbr2, or Tgfb3, exhibit milder palatal fusion defects than the Tgfb3−/− germ line knockout mice, such that inter-palatal shelf adhesion occurred initially but the MES failed to disintegrate in the K14-Cre conditional mutant mouse strains (Kaartinen et al., 1995; Proetzel et al., 1995; Dudas et al., 2006; Xu et al., 2006; Lane et al., 2014). This has been attributed to lack of Cre activity in the palatal periderm cells in K14-Cre embryos (Lane et al., 2014). The milder phenotypes in the K14-Cre conditional mutants than in the Tgfb3 null mice indicate that direct effects of Tgfb3 signaling on the periderm are critical for disruption of periderm integrity and initiation of inter-shelf adhesion. Whereas a recent report suggested that Tgfb3 regulates periderm removal through repression of ΔNp63 (Hu et al., 2015), it is likely that Tgfb3 affects another pathway in the periderm cells for their desquamation since ΔNp63 is already down-regulated during periderm differentiation, as discussed above. On the other hand, some other transcription factors expressed in the MEE, in particular Runx1 and Snail1, have been shown required for palatal fusion (Murray et al., 2007; Charoenchaikorn et al., 2009). Charoenchaikorn et al. (2009) showed that Runx1−/− mutant embryos with a transgenic rescue of hematopoietic defects exhibit defects in MEE cell behavior and cleft anterior palate. Whether Runx1 deficiency affected expression of Tgfb3 in the MEE was characterized, however. Whereas Snail1−/− embryos die during early embryogenesis and Snail2−/− mice exhibit an incomplete penetrance of cleft palate, all Snail1+/−Snail2−/− compound mutant mice had cleft palate (Jiang et al., 1998b; Carver et al., 2001; Murray et al., 2007). Although the palatal shelves grow and elevated normally and made contact at the midline, they remained covered with periderm cells and failed to form the MES in Snail1+/−Snail2−/− compound mutant embryos. Expression of Tgfb3 was not affected in the Snail1+/−Snail2−/− mutant MEE, indicating that these transcription factors regulate inter-palatal shelf adhesion downstream of, or in parallel to, Tgfb3 signaling. Since the Snail family transcription factors have been implicated in epithelial-mesenchymal transition by directly repressing the expression of components of epithelial cell adhesive complexes (Nieto, 2002), it is possible that Snail1 and Snail2 act downstream of or in corporation with Tgfb3 signaling to loosen adhesion between the MEE periderm cells at the onset of palatal fusion. Further studies are necessary to delineate these molecular relationships and how they relate to activation of the actomyosin contractility-driven cell convergence behavior during MES formation.
5. Concluding remarks
Nearly 30 years ago, the introduction of the gene targeting technology for manipulating the mouse genome via homologous recombination revolutionized mouse genetics and propelled the laboratory mouse to the forefront of biomedical research (reviewed by Capecchi, 2005). Since then, mice carrying each of thousands of constitutive or conditional alleles have been generated and analyzed, which have provided much of the current understanding of mammalian biology and mechanisms of human diseases. Much of the progress in our understanding of the cellular and molecular mechanisms of palatogenesis is also due to the widespread application of sophisticated genetic manipulations in mice and detailed morphological and molecular analysis of mutant mouse models (Bush and Jiang, 2012). Whereas the major signaling pathways governing palatogenesis have been well studied genetically, the underlying biochemical mechanisms remain largely uncharacterized. Moreover, as whole exome and whole genome sequencing approaches are increasingly effectively applied for human disease gene discovery (Newman and Black, 2014; Tabor et al., 2014; The Deciphering Developmental Disorders Study, 2015), it can be expected that the number of genes associated with craniofacial and palate defects will increase rapidly. The major challenge is to integrate new discoveries into the understanding of the gene regulatory networks controlling craniofacial and palate development. By combining high-throughput molecular and biochemical approaches, such as chromatin immunoprecipitation and high throughput sequencing (ChIP-seq), whole transcriptome RNA-seq based gene expression profiling, and mass spectrometry with bioinformatics analysis will help generate gene regulatory and protein-protein interaction network models that can be further tested genetically and biochemically to provide new insights into the mechanisms of palate development. Moreover, the recent adaptation of the versatile CRISPR-Cas9 system for genome editing has once again revolutionized genetics and developmental biology research and enabled unprecedented access to investigating the detailed functions of proteins, genes, RNAs, and other elements as well as cellular and molecular processes in the endogenous context (Doudna and Charpentier, 2014; Harrison et al., 2014). Together, these new technological tools will help rapidly advance our understanding of the mechanisms of palate development and lead to development of new and better treatment or prevention strategies for cleft palate birth defects.
Acknowledgements
We apologize to many colleagues whose work we could not discuss in this article due to space limitations. We thank Yang Gao for contributing data used in Figure 4A and 4B. We thank Jeff Bush for sharing the PLoS Biology publication on cell convergence and extrusion during palate fusion prior to online access. Palate development research in the Lan and Jiang laboratories was supported by the National Institutes of Health National Institute of Dental and Craniofacial Research (NIH/NIDCR) grants R03DE023864 (Y.L.) and R01DE013681 (R.J.).
References
- Akiyama M, Smith LT, Yoneda K, Holbrook KA, Hohl D, Shimizu H. Periderm cells form cornified cell envelope in their regression process during human epidermal development. J Invest Dermatol. 1999;112:903–909. doi: 10.1046/j.1523-1747.1999.00592.x. [DOI] [PubMed] [Google Scholar]
- Alappat SR, Zhang Z, Suzuki K, Zhang X, Liu H, Jiang R, Yamada G, Chen Y. The cellular and molecular etiology of the cleft secondary palate in Fgf10 mutant mice. Dev Biol. 2005;277:102–113. doi: 10.1016/j.ydbio.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Baek JA, Lan Y, Liu H, Maltby KM, Mishina Y, Jiang R. Bmpr1a signaling plays critical roles in palatal shelf growth and palatal bone formation. Dev Biol. 2011;350:520–531. doi: 10.1016/j.ydbio.2010.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blavier L, Lazaryev A, Groffen J, Heisterkamp N, DeClerck YA, Kaartinen V. TGF-beta3-induced palatogenesis requires matrix metalloproteinases. Mol Biol Cell. 2001;12:1457–1466. doi: 10.1091/mbc.12.5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush JO, Jiang R. Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development. 2012;139:231–243. doi: 10.1242/dev.067082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Rufini A, Terrinoni A, Giamboi-Miraglia A, Lena AM, Mantovani R, Knight R, Melino G. DeltaNp63 regulates thymic development through enhanced expression of FgfR2 and Jag2. Proc Natl Acad Sci U S A. 2007;104:11999–12004. doi: 10.1073/pnas.0703458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21:8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey LM, Lan Y, Cho ES, Maltby KM, Gridley T, Jiang R. Jag2-Notch1 signaling regulates oral epithelial differentiation and palate development. Dev Dyn. 2006;235:1830–1844. doi: 10.1002/dvdy.20821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi F, Alvarez-Bolado G, Meyer BI, Roth KA, Gruss P. Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell. 1998;94:727–737. doi: 10.1016/s0092-8674(00)81732-8. [DOI] [PubMed] [Google Scholar]
- Chai Y, Maxson RE., Jr. Recent advances in craniofacial morphogenesis. Dev Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- Charoenchaikorn K, Yokomizo T, Rice DP, Honjo T, Matsuzaki K, Shintaku Y, Imai Y, Wakamatsu A, Takahashi S, Ito Y, et al. Runx1 is involved in the fusion of the primary and the secondary palatal shelves. Dev Biol. 2009;326:392–402. doi: 10.1016/j.ydbio.2008.10.018. [DOI] [PubMed] [Google Scholar]
- Cobourne MT, Green JB. Hedgehog signalling in development of the secondary palate. Front Oral Biol. 2012;16:52–59. doi: 10.1159/000337543. [DOI] [PubMed] [Google Scholar]
- Crum CP, McKeon FD. p63 in epithelial survival, germ cell surveillance, and neoplasia. Annu Rev Pathol. 2010;5:349–371. doi: 10.1146/annurev-pathol-121808-102117. [DOI] [PubMed] [Google Scholar]
- Cuervo R, Covarrubias L. Death is the major fate of medial edge epithelial cells and the cause of basal lamina degradation during palatogenesis. Development. 2004;131:15–24. doi: 10.1242/dev.00907. [DOI] [PubMed] [Google Scholar]
- Cuervo R, Valencia C, Chandraratna RA, Covarrubias L. Programmed cell death is required for palate shelf fusion and is regulated by retinoic acid. Dev Biol. 2002;245:145–156. doi: 10.1006/dbio.2002.0620. [DOI] [PubMed] [Google Scholar]
- de la Garza G, Schleiffarth JR, Dunnwald M, Mankad A, Weirather JL, Bonde G, Butcher S, Mansour TA, Kousa YA, Fukazawa CF, et al. Interferon regulatory factor 6 promotes differentiation of the periderm by activating expression of Grainyhead-like 3. J Invest Dermatol. 2013;133:68–77. doi: 10.1038/jid.2012.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12:167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto GP. p63 and FGFR: when development meets proliferation. EMBO Mol Med. 2012;4:165–167. doi: 10.1002/emmm.201200202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Dudas M, Li WY, Kim J, Yang A, Kaartinen V. Palatal fusion - where do the midline cells go? A review on cleft palate, a major human birth defect. Acta Histochem. 2007;109:1–14. doi: 10.1016/j.acthis.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Dudas M, Kim J, Li WY, Nagy A, Larsson J, Karlsson S, Chai Y, Kaartinen V. Epithelial and ectomesenchymal role of the type I TGF-beta receptor ALK5 during facial morphogenesis and palatal fusion. Dev Biol. 2006;296:298–314. doi: 10.1016/j.ydbio.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou AD, Ohazama A, Porntaveetus T, Sharpe PT, Kondo S, Basson MA, Gritli-Linde A, Cobourne MT, Green JB. Periodic stripe formation by a Turing mechanism operating at growth zones in the mammalian palate. Nat Genet. 2012;44:348–351. doi: 10.1038/ng.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhouri WD, Rhea L, Du T, Sweezer E, Morrison H, Fitzpatrick D, Yang B, Dunnwald M, Schutte BC. MCS9.7 enhancer activity is highly, but not completely, associated with expression of Irf6 and p63. Dev Dyn. 2012;241:340–349. doi: 10.1002/dvdy.22786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferone G, Thomason HA, Antonini D, De Rosa L, Hu B, Gemei M, Zhou H, Ambrosio R, Rice DP, Acampora D, et al. Mutant p63 causes defective expansion of ectodermal progenitor cells and impaired FGF signalling in AEC syndrome. EMBO Mol Med. 2012;4:192–205. doi: 10.1002/emmm.201100199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitchett JE, Hay ED. Medial edge epithelium transforms to mesenchyme after embryonic palatal shelves fuse. Dev Biol. 1989;131:455–474. doi: 10.1016/s0012-1606(89)80017-x. [DOI] [PubMed] [Google Scholar]
- The Deciphering Developmental Disorders Study Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2014;519:223–228. doi: 10.1038/nature14135. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick DR, Denhez F, Kondaiah P, Akhurst RJ. Differential expression of TGF beta isoforms in murine palatogenesis. Development. 1990;109:585–595. doi: 10.1242/dev.109.3.585. [DOI] [PubMed] [Google Scholar]
- Gorlin RJ, Cohen MM, Hennekam RCM. Syndromes of the Head and Neck. Oxford University Press; Oxford, New York: 2001. [Google Scholar]
- Griffith CM, Hay ED. Epithelial-mesenchymal transformation during palatal fusion: carboxyfluorescein traces cells at light and electron microscopic levels. Development. 1992;116:1087–1099. doi: 10.1242/dev.116.4.1087. [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A. Molecular control of secondary palate development. Dev Biol. 2007;301:309–326. doi: 10.1016/j.ydbio.2006.07.042. [DOI] [PubMed] [Google Scholar]
- Han J, Mayo J, Xu X, Li J, Bringas P, Jr., Maas RL, Rubenstein JL, Chai Y. Indirect modulation of Shh signaling by Dlx5 affects the oral-nasal patterning of palate and rescues cleft palate in Msx1-null mice. Development. 2009;136:4225–4233. doi: 10.1242/dev.036723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Jenkins BV, O'Connor-Giles KM, Wildonger J. A CRISPR view of development. Genes Dev. 2014;28:1859–1872. doi: 10.1101/gad.248252.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward AF. The permeability of the epithelium of the skin of fetal rats demonstrated with a lanthanum-containing solution. J Anat. 1983;136:379–388. [PMC free article] [PubMed] [Google Scholar]
- He F, Chen Y. Wnt signaling in lip and palate development. Front Oral Biol. 2012;16:81–90. doi: 10.1159/000337619. [DOI] [PubMed] [Google Scholar]
- He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massague J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- Hilliard SA, Yu L, Gu S, Zhang Z, Chen YP. Regional regulation of palatal growth and patterning along the anterior-posterior axis in mice. J Anat. 2005;207:655–667. doi: 10.1111/j.1469-7580.2005.00474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook KA, Odland GF. The fine structure of developing human epidermis: light, scanning, and transmission electron microscopy of the periderm. J Invest Dermatol. 1975;65:16–38. doi: 10.1111/1523-1747.ep12598029. [DOI] [PubMed] [Google Scholar]
- Hu L, Liu J, Li Z, Ozturk F, Gurumurthy C, Romano RA, Sinha S, Nawshad A. TGFbeta3 Regulates Periderm Removal Through DeltaNp63 in the Developing Palate. J Cell Physiol. 2015;230:1212–1225. doi: 10.1002/jcp.24856. [DOI] [PubMed] [Google Scholar]
- Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, Malik MI, Dunnwald M, Goudy SL, Lovett M, et al. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6). Nat Genet. 2006;38:1335–1340. doi: 10.1083/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata J, Suzuki A, Pelikan RC, Ho TV, Sanchez-Lara PA, Urata M, Dixon MJ, Chai Y. Smad4-Irf6 genetic interaction and TGFbeta-mediated IRF6 signaling cascade are crucial for palatal fusion in mice. Development. 2013;140:1220–1230. doi: 10.1242/dev.089615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Mao J, Tenzen T, Kottmann AH, McMahon AP. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 2004;18:937–951. doi: 10.1101/gad.1190304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Bush JO, Lidral AC. Development of the upper lip: morphogenetic and molecular mechanisms. Dev Dyn. 2006;235:1152–1166. doi: 10.1002/dvdy.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Lan Y, Chapman HD, Shawber C, Norton CR, Serreze DV, Weinmaster G, Gridley T. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 1998a;12:1046–1057. doi: 10.1101/gad.12.7.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Lan Y, Norton CR, Sundberg JP, Gridley T. The Slug gene is not essential for mesoderm or neural crest development in mice. Dev Biol. 1998b;198:277–285. [PubMed] [Google Scholar]
- Jin JZ, Ding J. Analysis of cell migration, transdifferentiation and apoptosis during mouse secondary palate fusion. Development. 2006a;133:3341–3347. doi: 10.1242/dev.02520. [DOI] [PubMed] [Google Scholar]
- Jin JZ, Ding J. Analysis of Meox-2 mutant mice reveals a novel postfusion-based cleft palate. Dev Dyn. 2006b;235:539–546. doi: 10.1002/dvdy.20641. [DOI] [PubMed] [Google Scholar]
- Kaartinen V, Cui XM, Heisterkamp N, Groffen J, Shuler CF. Transforming growth factor-beta3 regulates transdifferentiation of medial edge epithelium during palatal fusion and associated degradation of the basement membrane. Dev Dyn. 1997;209:255–260. doi: 10.1002/(SICI)1097-0177(199707)209:3<255::AID-AJA1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Kaartinen V, Voncken JW, Shuler C, Warburton D, Bu D, Heisterkamp N, Groffen J. Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial-mesenchymal interaction. Nat Genet. 1995;11:415–421. doi: 10.1038/ng1295-415. [DOI] [PubMed] [Google Scholar]
- King KE, Weinberg WC. p63: defining roles in morphogenesis, homeostasis, and neoplasia of the epidermis. Mol Carcinog. 2007;46:716–724. doi: 10.1002/mc.20337. [DOI] [PubMed] [Google Scholar]
- Kim S, Lewis AE, Singh V, Ma X, Adelstein R, Bush JO. Convergence and extrusion are required for normal fusion of the mammalian secondary palate. PLoS Biol. 2015 doi: 10.1371/journal.pbio.1002122. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight AS, Schutte BC, Jiang R, Dixon MJ. Developmental expression analysis of the mouse and chick orthologues of IRF6: the gene mutated in Van der Woude syndrome. Dev Dyn. 2006;235:1441–1447. doi: 10.1002/dvdy.20598. [DOI] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, Howard E, de Lima RL, Daack-Hirsch S, Sander A, et al. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet. 2002;32:285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Jiang R. Sonic hedgehog signaling regulates reciprocal epithelial-mesenchymal interactions controlling palatal outgrowth. Development. 2009;136:1387–1396. doi: 10.1242/dev.028167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Ovitt CE, Cho ES, Maltby KM, Wang Q, Jiang R. Odd-skipped related 2 (Osr2) encodes a key intrinsic regulator of secondary palate growth and morphogenesis. Development. 2004;131:3207–3216. doi: 10.1242/dev.01175. [DOI] [PubMed] [Google Scholar]
- Lane J, Kaartinen V. Signaling networks in palate development. Wiley Interdiscip Rev Syst Biol Med. 2014;6:271–278. doi: 10.1002/wsbm.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane J, Yumoto K, Pisano J, Azhar M, Thomas PS, Kaartinen V. Control elements targeting Tgfb3 expression to the palatal epithelium are located intergenically and in introns of the upstream Ift43 gene. Front Physiol. 2014;5:258. doi: 10.3389/fphys.2014.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane J, Yumoto K, Azhar M, Ninomiya-Tsuji J, Inagaki M, Hu Y, Deng CX, Kim J, Mishina Y, Kaartinen V. Tak1, Smad4 and Trim33 redundantly mediate TGF-beta3 signaling during palate development. Dev Biol. 2015;398:231–241. doi: 10.1016/j.ydbio.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurikkala J, Mikkola ML, James M, Tummers M, Mills AA, Thesleff I. p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development. 2006;133:1553–1563. doi: 10.1242/dev.02325. [DOI] [PubMed] [Google Scholar]
- Levi B, Brugman S, Wong VW, Grova M, Longaker MT, Wan DC. Palatogenesis: engineering, pathways and pathologies. Organogenesis. 2011;7:242–254. doi: 10.4161/org.7.4.17926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Ding J. Gene expression analysis reveals that formation of the mouse anterior secondary palate involves recruitment of cells from the posterior side. Int J Dev Biol. 2007;51:167–172. doi: 10.1387/ijdb.062212ql. [DOI] [PubMed] [Google Scholar]
- Liu W, Lan Y, Pauws E, Meester-Smoor MA, Stanier P, Zwarthoff EC, Jiang R. The Mn1 transcription factor acts upstream of Tbx22 and preferentially regulates posterior palate growth in mice. Development. 2008;135:3959–3968. doi: 10.1242/dev.025304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M'Boneko V, Merker HJ. Development and morphology of the periderm of mouse embryos (days 9-12 of gestation). Acta Anat (Basel) 1988;133:325–336. doi: 10.1159/000146662. [DOI] [PubMed] [Google Scholar]
- Mangold E, Ludwig KU, Nothen MM. Breakthroughs in the genetics of orofacial clefting. Trends Mol Med. 2011;17:725–733. doi: 10.1016/j.molmed.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Murray SA, Oram KF, Gridley T. Multiple functions of Snail family genes during palate development in mice. Development. 2007;134:1789–1797. doi: 10.1242/dev.02837. [DOI] [PubMed] [Google Scholar]
- Moretti F, Marinari B, Lo Iacono N, Botti E, Giunta A, Spallone G, Garaffo G, Vernersson-Lindahl E, Merlo G, Mills AA, et al. A regulatory feedback loop involving p63 and IRF6 links the pathogenesis of 2 genetically different human ectodermal dysplasias. J Clin Invest. 2010;120:1570–1577. doi: 10.1172/JCI40267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawshad A. Palatal seam disintegration: to die or not to die? that is no longer the question. Dev Dyn. 2008;237:2643–2656. doi: 10.1002/dvdy.21599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman WG, Black GC. Delivery of a clinical genomics service. Genes (Basel) 2014;5:1001–1017. doi: 10.3390/genes5041001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- Pantalacci S, Prochazka J, Martin A, Rothova M, Lambert A, Bernard L, Charles C, Viriot L, Peterkova R, Laudet V. Patterning of palatal rugae through sequential addition reveals an anterior/posterior boundary in palatal development. BMC Dev Biol. 2008;8:116. doi: 10.1186/1471-213X-8-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada C, Chai Y. Roles of BMP signaling pathway in lip and palate development. Front Oral Biol. 2012;16:60–70. doi: 10.1159/000337617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauws E, Hoshino A, Bentley L, Prajapati S, Keller C, Hammond P, Martinez-Barbera JP, Moore GE, Stanier P. Tbx22null mice have a submucous cleft palate due to reduced palatal bone formation and also display ankyloglossia and choanal atresia phenotypes. Hum Mol Genet. 2009;18:4171–4179. doi: 10.1093/hmg/ddp368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelton RW, Hogan BL, Miller DA, Moses HL. Differential expression of genes encoding TGFs beta 1, beta 2, and beta 3 during murine palate formation. Dev Biol. 1990;141:456–460. doi: 10.1016/0012-1606(90)90401-4. [DOI] [PubMed] [Google Scholar]
- Peters H, Neubuser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrard-Janvid M, Leslie EJ, Kousa YA, Smith TL, Dunnwald M, Magnusson M, Lentz BA, Unneberg P, Fransson I, Koillinen HK, et al. Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development. Am J Hum Genet. 2014;94:23–32. doi: 10.1016/j.ajhg.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakowska RR, Piacentini M, Bartlett R, Goldsmith LA, Haake AR. Apoptosis in human skin development: morphogenesis, periderm, and stem cells. Dev Dyn. 1994;199:176–188. doi: 10.1002/aja.1001990303. [DOI] [PubMed] [Google Scholar]
- Proetzel G, Pawlowski SA, Wiles MV, Yin M, Boivin GP, Howles PN, Ding J, Ferguson MW, Doetschman T. Transforming growth factor-beta 3 is required for secondary palate fusion. Nat Genet. 1995;11:409–414. doi: 10.1038/ng1295-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimov F, Jugessur A, Murray JC. Genetics of nonsyndromic orofacial clefts. Cleft Palate Craniofac J. 2012;49:73–91. doi: 10.1597/10-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice R, Connor E, Rice DP. Expression patterns of Hedgehog signalling pathway members during mouse palate development. Gene Expr Patterns. 2006;6:206–212. doi: 10.1016/j.modgep.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Rice R, Spencer-Dene B, Connor EC, Gritli-Linde A, McMahon AP, Dickson C, Thesleff I, Rice DP. Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J Clin Invest. 2004;113:1692–1700. doi: 10.1172/JCI20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Jiang R, Dixon MJ. Integration of IRF6 and Jagged2 signalling is essential for controlling palatal adhesion and fusion competence. Hum Mol Genet. 2009;18:2632–2642. doi: 10.1093/hmg/ddp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Malhotra S, Hardman MJ, Knowles L, Boot-Handford RP, Shore P, Whitmarsh A, Dixon MJ. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat Genet. 2006;38:1329–1334. doi: 10.1038/ng1894. [DOI] [PubMed] [Google Scholar]
- Richardson RJ, Hammond NL, Coulombe PA, Saloranta C, Nousiainen HO, Salonen R, Berry A, Hanley N, Headon D, Karikoski R, et al. Periderm prevents pathological epithelial adhesions during embryogenesis. J Clin Invest. 2014;124:3891–3900. doi: 10.1172/JCI71946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano RA, Smalley K, Magraw C, Serna VA, Kurita T, Raghavan S, Sinha S. DeltaNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development. 2012;139:772–782. doi: 10.1242/dev.071191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt J, Raff MC, Cramer LP. An epithelial cell destined for apoptosis signals its neighbors to extrude it by an actin- and myosin-dependent mechanism. Curr Biol. 2001;11:1847–1857. doi: 10.1016/s0960-9822(01)00587-5. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Rubenstein JL, Nicolas JF. Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 1986;5:3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Ishida S, Morimoto I, Yamashita T, Kojima T, Kihara C, Tanaka T, Imai K, Nakamura Y, Tokino T. The p53 family member genes are involved in the Notch signal pathway. J Biol Chem. 2002;277:719–724. doi: 10.1074/jbc.M108080200. [DOI] [PubMed] [Google Scholar]
- Schutte BC, Murray JC. The many faces and factors of orofacial clefts. Hum Mol Genet. 1999;8:1853–1859. doi: 10.1093/hmg/8.10.1853. [DOI] [PubMed] [Google Scholar]
- Scott WJ, Jr., Nau H, Wittfoht W, Merker HJ. Ventral duplication of the autopod: chemical induction by methoxyacetic acid in rat embryos. Development. 1987;99:127–136. doi: 10.1242/dev.99.1.127. [DOI] [PubMed] [Google Scholar]
- Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Smith TM, Lozanoff S, Iyyanar PP, Nazarali AJ. Molecular signaling along the anterior-posterior axis of early palate development. Front Physiol. 2012;3:488. doi: 10.3389/fphys.2012.00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier P, Pauws E. Development of the lip and palate: FGF signalling. Front Oral Biol. 2012;16:71–80. doi: 10.1159/000337618. [DOI] [PubMed] [Google Scholar]
- Sun D, Baur S, Hay ED. Epithelial-mesenchymal transformation is the mechanism for fusion of the craniofacial primordia involved in morphogenesis of the chicken lip. Dev Biol. 2000;228:337–349. doi: 10.1006/dbio.2000.9946. [DOI] [PubMed] [Google Scholar]
- Tabor HK, Auer PL, Jamal SM, Chong JX, Yu JH, Gordon AS, Graubert TA, O'Donnell CJ, Rich SS, Nickerson DA, et al. Pathogenic variants for Mendelian and complex traits in exomes of 6,517 European and African Americans: implications for the return of incidental results. Am J Hum Genet. 2014;95:183–193. doi: 10.1016/j.ajhg.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadeu AM, Horsley V. Notch signaling represses p63 expression in the developing surface ectoderm. Development. 2013;140:3777–3786. doi: 10.1242/dev.093948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara S, Takigawa T, Shiota K. Programmed cell death is not a necessary prerequisite for fusion of the fetal mouse palate. Int J Dev Biol. 2004;48:39–46. doi: 10.1387/ijdb.15005573. [DOI] [PubMed] [Google Scholar]
- Thomason HA, Zhou H, Kouwenhoven EN, Dotto GP, Restivo G, Nguyen BC, Little H, Dixon MJ, van Bokhoven H, Dixon J. Cooperation between the transcription factors p63 and IRF6 is essential to prevent cleft palate in mice. J Clin Invest. 2010;120:1561–1569. doi: 10.1172/JCI40266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Woude A. Fistula labii inferioris congenita and its association with cleft lip and palate. Am J Hum Genet. 1954;6:244–256. [PMC free article] [PubMed] [Google Scholar]
- Vanderas AP. Prevalence of craniomandibular dysfunction in children and adolescents: a review. Pediatr Dent. 1987;9:312–316. [PubMed] [Google Scholar]
- Vaziri Sani F, Hallberg K, Harfe BD, McMahon AP, Linde A, Gritli-Linde A. Fate-mapping of the epithelial seam during palatal fusion rules out epithelial-mesenchymal transformation. Dev Biol. 2005;285:490–495. doi: 10.1016/j.ydbio.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Veistinen L, Aberg T, Rice DP. Convergent signalling through Fgfr2 regulates divergent craniofacial morphogenesis. J Exp Zool B Mol Dev Evol. 2009;312B:351–360. doi: 10.1002/jez.b.21276. [DOI] [PubMed] [Google Scholar]
- Waterman RE, Meller SM. Alterations in the epithelial surface of human palatal shelves prior to and during fusion: a scanning electron microscopic study. Anat Rec. 1974;180:111–135. doi: 10.1002/ar.1091800111. [DOI] [PubMed] [Google Scholar]
- Welsh IC, O'Brien TP. Signaling integration in the rugae growth zone directs sequential SHH signaling center formation during the rostral outgrowth of the palate. Dev Biol. 2009;336:53–67. doi: 10.1016/j.ydbio.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall MD, Mays DJ, Sniezek JC, Pietenpol JA. The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol Cell Biol. 2003;23:2264–2276. doi: 10.1128/MCB.23.7.2264-2276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Endo M, Yang BH, Radecki MA, Davis PF, Zoltick PW, Spivak RM, Flake AW, Kirschner RE, Nah HD. Intra amniotic transient transduction of the periderm with a viral vector encoding TGFbeta3 prevents cleft palate in Tgfbeta3(−/−) mouse embryos. Mol Ther. 2013;21:8–17. doi: 10.1038/mt.2012.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Han J, Ito Y, Bringas P, Jr., Urata MM, Chai Y. Cell autonomous requirement for Tgfbr2 in the disappearance of medial edge epithelium during palatal fusion. Dev Biol. 2006;297:238–248. doi: 10.1016/j.ydbio.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Xu X, Han J, Ito Y, Bringas P, Jr., Deng C, Chai Y. Ectodermal Smad4 and p38 MAPK are functionally redundant in mediating TGF-beta/BMP signaling during tooth and palate development. Dev Cell. 2008;15:322–329. doi: 10.1016/j.devcel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Yano H, Ohtsuru A, Ito M, Fuji T, Yamashita S. Involvement of c-Fos proto-oncogene during palatal fusion and interdigital space formation in the rat. Dev Growth Differ. 1996;38:351–357. doi: 10.1046/j.1440-169X.1996.t01-3-00003.x. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Shimono Y, Togashi H, Matsuzaki K, Miyoshi J, Mizoguchi A, Komori T, Takai Y. Periderm cells covering palatal shelves have tight junctions and their desquamation reduces the polarity of palatal shelf epithelial cells in palatogenesis. Genes Cells. 2012;17:455–472. doi: 10.1111/j.1365-2443.2012.01601.x. [DOI] [PubMed] [Google Scholar]
- Yu L, Gu S, Alappat S, Song Y, Yan M, Zhang X, Zhang G, Jiang Y, Zhang Z, Zhang Y, et al. Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate. Development. 2005;132:4397–4406. doi: 10.1242/dev.02013. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Song Y, Zhao X, Zhang X, Fermin C, Chen Y. Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development. 2002;129:4135–4146. doi: 10.1242/dev.129.17.4135. [DOI] [PubMed] [Google Scholar]
- Zhou J, Gao Y, Zhang Z, Zhang Y, Maltby KM, Liu Z, Lan Y, Jiang R. Osr2 acts downstream of Pax9 and interacts with both Msx1 and Pax9 to pattern the tooth developmental field. Dev Biol. 2011;353:344–353. doi: 10.1016/j.ydbio.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Gao Y, Lan Y, Jia S, Jiang R. Pax9 regulates a molecular network involving Bmp4, Fgf10, Shh signaling and the Osr2 transcription factor to control palate morphogenesis. Development. 2013;140:4709–4718. doi: 10.1242/dev.099028. [DOI] [PMC free article] [PubMed] [Google Scholar]