Abstract
Parasitic lice (order Phthiraptera) infest birds and mammals. The typical animal mitochondrial (mt) genome organization, which consists of a single chromosome with 37 genes, was found in chewing lice in the suborders Amblycera and Ischnocera. The sucking lice (suborder Anoplura) known, however, have fragmented mt genomes with 9–20 minichromosomes. We sequenced the mt genome of the elephant louse, Haematomyzus elephantis – the first species of chewing lice investigated from the suborder Rhynchophthirina. We identified 33 mt genes in the elephant louse, which were on 10 minichromosomes. Each minichromosome is 3.5–4.2 kb in size and has 2–6 genes. Phylogenetic analyses of mt genome sequences confirm that the elephant louse is more closely related to sucking lice than to the chewing lice in the Amblycera and Ischnocera. Our results indicate that mt genome fragmentation is shared by the suborders Anoplura and Rhynchophthirina. Nine of the 10 mt minichromosomes of the elephant louse differ from those of the sucking lice (Anoplura) known in gene content and gene arrangement, indicating that distinct mt karyotypes have evolved in Anoplura and Rhynchophthirina since they diverged ~92 million years ago.
The parasitic lice (order Phthiraptera) have ~5,000 described species in four suborders. The sucking lice, which feed exclusively on the blood of eutherian mammals, are in the suborder Anoplura1. The chewing lice, which infest both birds and mammals and feed mainly on feathers, dead skin, blood or secretions, are in three other suborders: Amblycera, Ischnocera and Rhynchophthirina2. The mitochondrial (mt) genomes of the wallaby louse, Heterodoxus macropus (Amblycera), and four species in the suborder Ischnocera, Campanulotes bidentatus (small pigeon louse), Bothriometopus macrocnemis (screamer louse), Coloceras sp. (pigeon feather louse) and Ibidoecus bisignatus (ibis head louse), have been sequenced3,4,5,6. Like most other bilateral animals, these five species of chewing lice all have the typical mt chromosomes with 37 genes, 14,670 to 15,564 bp in size6,7. In contrast, all of the sucking lice that have been sequenced (10 species/subspecies in total) have fragmented mt genomes and there is no evidence for the presence of the typical mt chromosome in these lice. The human body louse, Pediculus humanus, and the human head louse, Pe. h. capitis, have their 37 mt genes on 20 minichromosomes8,9. The 37 mt genes of the rat lice, Polyplax asiatica and Po. spinulosa, are on 11 minichromosomes10, whereas those of the pig lice, Haematopinus suis and Haematopinus apri, and the horse louse, Haematopinus asini, are on nine minichromosomes11,12. The 34 mt genes identified in the human pubic louse, Pthirus pubis, are on 14 minichromosomes9; the 34 mt genes identified in the rat louse, Hoplopleura kitti, are on 11 minichromosomes; and the 28 mt genes identified in the mouse louse, Ho. akanezumi, are on 10 minichromosomes13.
Despite being classified as chewing lice based on mouth parts14, the suborder Rhynchophthirina is phylogenetically more closely related to the sucking lice (Anoplura) than to the other two suborders of the chewing lice15,16,17,18,19,20. Rhynchophthirina has one family, Haematomyzidae, one genus, Haematomyzus, and three species: Haematomyzus elephantis (elephant louse), Haematomyzus hopkinsi (African wart-hog louse) and Haematomyzus porci (African red-river hog louse)2. We sequenced the mt genome of the elephant louse, H. elephantis, and identified 33 mt genes in this louse; these genes were on 10 minichromosomes. Our results indicate that mt genome fragmentation is shared by parasitic lice in the suborders Anoplura and Rhynchophthirina. Further, nine of the 10 mt minichromosomes of the elephant louse differ from those of the sucking lice known in gene content and gene arrangement, indicating that distinct mt karyotypes have evolved between Anoplura and Rhynchophthirina since they diverged ~92 million years ago20,21.
Results
The mitochondrial genome of the elephant louse, Haematomyzus elephantis
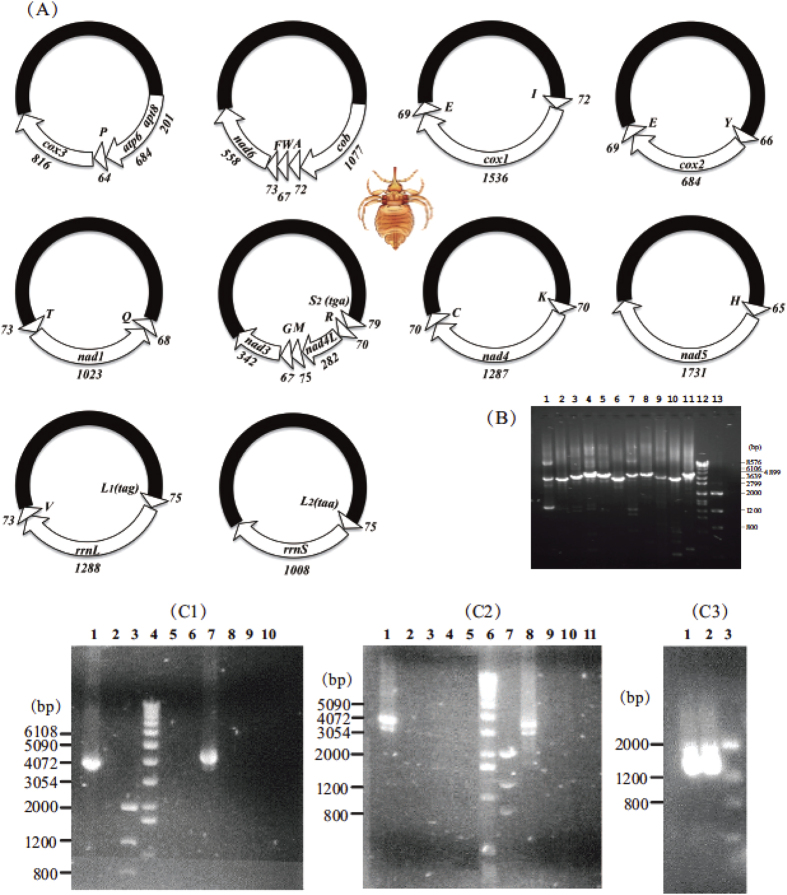
We obtained 14,760 and 1,137,360 sequence-reads from the amplicons of the mt genome of the elephant louse, H. elephantis, by Roche 454 sequencing and Illumina Hiseq sequencing, respectively (Table 1, Table 2). Roche sequence-reads are 100–611 bp in size (mean 344 bp, standard deviation 108); Illumina sequence-reads are all 90 bp in size. We assembled these sequence-reads into contigs and identified 33 of the 37 mt genes typical of bilateral animals. These genes are on 10 minichromosomes; each minichromosome is 3.5–4.2 kb in size and consists of a coding region and a non-coding region (NCR) in a circular organization (Fig. 1A,B). The coding regions have 2–6 genes each and vary in size from 857 bp to 1,879 bp (Table 1; Fig. 1A). All of the mt genes of the elephant louse have the same orientation of transcription relative to the NCRs, except for trnT, nad1 and trnQ, which form a cluster and have an opposite orientation of transcription to that of other genes (Fig. 1A; see Fig. 1 legend for the full name of each mt gene). With the exception of trnE, all of the mt genes we identified in the elephant louse were found in only one type of minichromosome. trnE gene was found in two types of minichromosomes in the elephant louse: one in the trnI-cox1-trnE minichromosome and the other in the trnY-cox2-trnE minichromosome (Note: minichromosomes are named after the genes they contain hereafter). Further, the two copies of trnE gene have identical sequences to each other. We did not find trnD, trnN, trnS1(tct) and nad2 genes in the sequence-reads of the elephant louse generated by Roche sequencing, nor Illumina sequencing. A possible explanation is that the primer pair USFB1567–ELR, which we used to amplify the coding regions of the mt minichromosomes of the elephant louse, are not conserved in the minichromosome(s) that contain trnD, trnN, trnS1(tct) and nad2 genes; these minichromosome(s) were thus not amplified by the PCR with USFB1567–ELR.
Table 1. The mitochondrial minichromosomes of the elephant louse, Haematomyzus elephantis, identified by Roche GS FLX (454) sequencing.
| Gene content and gene arrangement in each minichromosome | Size of coding region (bp) | Number of Roche sequence reads | Coverage (fold) |
|
|---|---|---|---|---|
| Range | Mean | |||
| atp8-atp6-trnP-cox3* | 1754 | 410 | 14–109 | 74 |
| cob-trnA-trnW-trnF-nad6 | 1879 | 215 | 6–70 | 38 |
| trnI-cox1-trnE | 1707 | 1157 | 77–340 | 208 |
| trnY-cox2-trnE | 857 | 678 | 23–299 | 127 |
| trnT-nad1-trnQ | 1143 | 4041 | 308–1701 | 983 |
| trnS2(tga)-trnR-nad4L-trnM-trnG-nad3 | 968 | 1384 | 102–558 | 336 |
| trnK-nad4-trnC | 1528 | 38 | 1–20 | 8 |
| trnH-nad5 | 1795 | 455 | 28–197 | 71 |
| trnL2(taa)-rrnS | 961 | 5125 | 352–2165 | 1310 |
| trnL1(tag)-rrnL-trnV | 1436 | 1257 | 50–497 | 263 |
*See Fig. 1 legend for the full names of mt genes.
Table 2. Illumina sequencing of PCR amplicons of four near full-length mitochondrial minichromosomes of the elephant louse, Haematomyzus elephantis.
| Target minichromosome | Primer pair used# | Size of amplicon (bp) | Number of Illumina sequence reads | Coverage (fold) |
|
|---|---|---|---|---|---|
| Range | Mean | ||||
| trnI-cox1-trnE* | YSC1F(2)-YSC1R(2) | 4026 | 114037 | 807–8319 | 2558 |
| trnT-nad1-trnQ | YSN1F-YSN1R | 3600 | 176234 | 828–16628 | 4388 |
| trnK-nad4-trnC | YSN4F-YSN4R | 3875 | 278107 | 1068–19823 | 6462 |
| trnH-nad5 | YSN5F(2)-YSN5R(2) | 4153 | 158711 | 1258–9427 | 3453 |
| trnH-nad5 | YSHF-YSHR | 4150 | 410271 | 2487–25650 | 8921 |
Figure 1.
(A) Mitochondrial minichromosomes of the elephant louse, Haematomyzus elephantis. Arrows indicate protein-coding and rRNA genes: cox1-3 for cytochrome c oxidase subunits 1–3, cob for cytochrome b, nad1-5 and nad4L for NADH dehydrogenase subunits 1–5 and 4L, rrnS and rrnL for small and large ribosome RNA subunits. Triangles indicate tRNA genes (labeled with single-letter abbreviations of their corresponding amino acids). Numbers near each gene indicate gene length in bp. Non-coding regions are in black. Drawing of elephant louse was by Hu Li. (B) Verification of mitochondrial minichromosomes of H. elephantis by PCR. Lanes 1–11: amplicons from minichromosomes T-nad1-Q, S2(tga)-R-nad4L-M-G-nad3, K-nad4-C, H-nad5, I-cox1-E, Y-cox2-E, atp8-atp6-P-cox3, cob-A-W-F-nad6, L1(tag)-rrnL-V, L2(taa)-rrnS, and H-nad5 (genes from which PCR primers were designed are in bold). Lanes 12-13: DNA Molecular Weight Marker VII (Roche) and Low DNA Mass Ladder (Life Technologies). (C) PCR amplicons with elephant-louse-specific primers (see also Table S5). (C1) Lane 1: primer pair EL16SF-EL16SR, which amplified trnL1(tag)-rrnL-trnV minichromosome in near full length. Lanes 2, 5 and 6: negative controls for EL16SF-LX16SR that had no forward primer, no reverse primer and no DNA template respectively. Lanes 3 and 4: Low Mass Ladder (LML) and DNA Molecular Weigh Marker X (DMWMX). Lane 7: primer pair ELC1F-ELC1R, which amplified trnI-cox1-trnE minichromosome in near full length. Lanes 8, 9 and 10: negative controls for ELC1F-ELC1R that had no forward primer, no reverse primer and no DNA template respectively. (C2) Lane 1: primer pair ELC2F-ELC2R1, which amplified trnY-cox2-trnE minichromosome in near full length. Lanes 2, 3 and 4: negative controls for ELC2F-ELC2R1 that had no forward primer, no reverse primer and no DNA template respectively. Lanes 6 and 7: DMWMX and LML. Lane 8: primer pair EL12SF1-EL12SR, which amplified trnL2(taa)-rrnS minichromosome in near full length. Lanes 9, 10 and 11: negative controls for EL12SF1-EL12SR that had no forward primer, no reverse primer and no DNA template respectively. (C3) Lanes 1 and 2: amplicons produced by the primer pair USFB1567-ELR from the full-length coding regions of all mitochondrial minichromosomes. Lane 3: LML.
We sequenced the NCRs of four mt minichromosomes of the elephant louse in full length. The NCRs are 2,324, 2,457, 2,348 and 2,359 bp respectively for trnI-cox1-trnE, trnT-nad1-trnQ, trnK-nad4-trnC and trnH-nad5 minichromosomes, and account for 58%, 68%, 61% and 57% of the total size of each minichromosome (Fig. S1; Alignment S1). The NCR of T-nad1-Q minichromosome has a 99-bp sequence at the 3′ end, which is not seen in the NCRs of other minichromosomes. Otherwise, the NCRs of the four minichromosomes have high sequence similarity (>97%) to each other. Contrary to those of most animals, the NCRs of the elephant louse have higher GC content (thus lower AT content) than the coding regions: 40% vs 38% for trnI-cox1-trnE minichromosome, 39% vs 34% for trnT-nad1-trnQ minichromosome, 40% vs 34% for trnK-nad4-trnC minichromosome, and 41% vs 35% for trnH-nad5 minichromosome. Whether or not the high GC content in the NCRs has any functional significance to the fragmented mt genome of the elephant louse remains to be investigated. There are tandem-repeat units in the NCRs of the elephant louse, 225 and 346 bp in length, respectively. The 225-bp unit repeated four times whereas the 346-bp unit repeated twice; the different copies of each unit have ~85% identity to each other (Fig. 2). We also sequenced the NCRs of the other six minichromosomes of the elephant louse except for the GC-rich parts and the tandem-repeat parts using a Sanger sequencing platform (Fig. S2; Alignment S2). The exact length of the NCRs of these six minichromosomes was unknown. The full-length NCR sequences shown in Fig. S1 were the consensus sequences obtained from the assembly of the Illumina sequence-reads. The high coverage of the Illumina sequence-reads also revealed numerous heteroplasmic sites in these NCRs of the elephant louse (Table S2–S5), which is consistent with that observed in human lice and pig lice8,9,11,22.
Figure 2. Schematic illustration of the non-coding regions of the mitochondrial minichromosomes of the elephant louse, Haematomyzus elephantis.
USFB1567 and ELR are the primers used to amplify the coding regions of all mitochondrial minichromosomes. A 346-bp unit (in green) repeated twice and a 225-bp unit (in yellow) repeated 4 times; the sequence similarity between/among the repeat copies of each unit is ~85%. A 99-bp sequence (in red) is present only in T-nad1-Q minichromosome.
Concerted evolution between the two mitochondrial tRNA genes for leucine in the elephant louse and the sucking lice
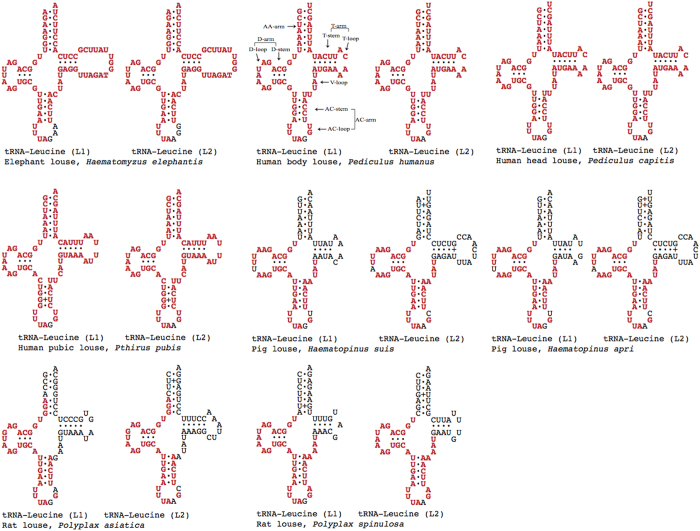
It is common for animals to have two tRNA genes for leucine, trnL1(tag) and trnL2(taa), in their mt genomes23. The two tRNA genes for leucine of the elephant louse have identical sequences except for three nucleotides starting from the third anti-codon position (Fig. 3). Near identical sequences between the two trnL genes were also observed in the human body louse, Pe. h. humanus, the head louse, Pe. h. capitis, and the pubic louse Pthirus pubis8,9. In the pig lice, Haemapinus suis and Haemapinus apri, and the rat lice, Polyplax asiatica and Po. spinulosa, the two trnL genes have identical or near identical sequences at the D-arm, the anti-codon arm and the V-loop but differ at the amino-acid arm and the T-arm (Fig. 3)10,11.
Figure 3. Inferred secondary structures of the mitochondrial tRNAs for leucine of the elephant louse, three human lice, two pig lice and two rat lice.
Shared identical sequences between the two tRNA genes of each species are in red.
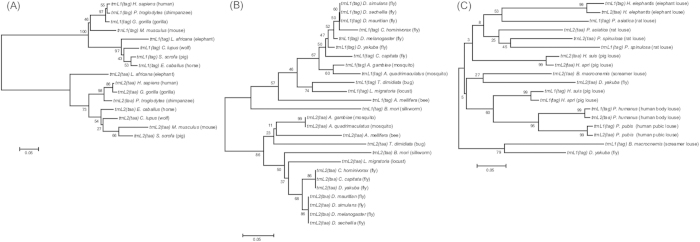
We used distance-based neighbor-joining (NJ) method to analyze the sequences of trnL1(tag) and trnL2(taa) genes. The NJ tree showed that these two genes evolved independently in a wide range of species of mammals and insects that have the typical single-chromosome mt genome organization. As would be expected for any pair of homologous genes, trnL1(tag) of a given species was more closely related to trnL1(tag) of other species than it is to trnL2(taa) of its own, and vice versa (Fig. 4A,B; Alignment S3 and S4). In the elephant lice and the sucking lice that have fragmented mt genomes, however, both independent evolution and concerted evolution were observed between the two tRNA genes for leucine (Fig. 4C; Alignment S5). In the two species of pig lice, Haematopinus suis and Haematopinus apri, both trnL genes evolved independently: trnL1(tag) genes of the two species are most closely related to each other whereas trnL2(tag) genes of the two species are most closely related. In the elephant louse, the human lice and the rat louse, Po. spinulosa, however, the two trnL genes evolved in concert: trnL1(tag) of each species is more closely related to trnL2(taa) of the same species than to trnL1(tag) of other species, and vice versa.
Figure 4. Independent evolution of the two mitochondrial (mt) tRNA genes for leucine in mammals (A) and insects (B), and a mixture of both independent and concerted evolution of these two genes in parasitic lice (C).
Trees were constructed using neighbor-joining method with MEGA 5.2.2 (Tamura et al. 2011). Bootstrap support values (%) are indicated on branches. All of the mammals and insects in (A) and (B) and the fly and the screamer louse in (C) have the typical, single-chromosome mt genomes of bilateral animals. The elephant louse and the sucking lice (Anoplura) in (C) have fragmented mt genomes with multiple minichromosomes. The human head louse, Pe. h. capitis, was not included in (C) because it has identical sequences with the human body louse, Pe. h. humanus, for trnL1 and trnL2 genes.
The complexity in the evolution of the two trnL genes in the elephant louse and the sucking lice has apparently been driven by recombination events between these genes8,9,24. Depending on the frequency of recombination between trnL1(tag) and trnL2(taa), different patterns of evolution can be observed. Concerted evolution of these two genes would be expected over long term or between distantly related species whereas independent evolution would be expected over short term (between two recombination events) or between closely related species, such as the two species of pig lice. Also, if recombination occurs only between parts of the genes, then concerted evolution would be expected only for the recombinant parts whereas the other parts of the genes will remain to evolve independently from each other.
Phylogenetic relationship of the elephant louse to other parasitic lice, barklice and booklice inferred with mitochondrial genome sequences and gene rearrangements
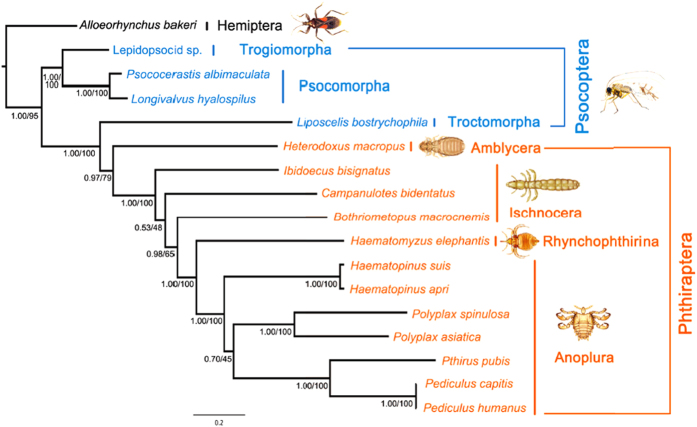
The elephant louse, Haematomyzus elephantis, together with the African wart-hog louse, Haematomyzus hopkinsi, and the African red-river hog louse, Haematomyzus porci, differ markedly from other parasitic lice in morphology and thus are in a suborder of their own, Rhyncophthirina14. We used maximum likelihood (ML) and Bayesian inference (BI) methods and generated eight phylogenetic trees in total with concatenated sequences of: 1) 11 mt protein-coding genes (Alignment S6); 2) the first and second codon positions of the 11 mt protein-coding genes (Alignment S7); 3) the 11 mt protein-coding genes and the two mt rRNA genes (Alignment S8); and 4) the first and second codon positions of the 11 mt protein-coding genes and the two mt rRNA genes (Alignment S9; also see Method below). In all of the eight trees, the elephant louse (suborder Rhyncophthirina) was placed as the sister group to the seven species of sucking lice (suborder Anoplura) (Fig. 5; Fig. S3). The monophyly of Anoplura and the sister-group relationship between Anoplura and Rhyncophthirina (represented by the elephant louse in the current study) were strongly supported in all of the trees with bootstrap support values (BSV) 97–100% and posterior probabilities (PP) 1. The suborder Ischnocera was paraphyletic: one of its species, Bothriometopus macrocnemis (screamer louse), was more closely related to Anoplura + Rhyncophthirina than to the other two ischnoceran species (BSV 62–88%, PP 0.88–1). The monophyly of the parasitic lice (order Phthiraptera) was supported in six of the eight trees (PCGRNA_ML, PCGRNA_BA, PCG12_ML, PCG12_BA, PCG12RNA_ML, PCG12RNA_BA; BSV 72–79%, PP 0.97–1). The monophyly of the barklice and booklice (order Psocoptera) was strongly rejected in all of the eight trees as the booklouse Liposcelis bostrychophila was more closely related to the parasitic lice than to the barklice (BSV 100%, PP 1). The sister-group relationship between Anoplura and Rhyncophthirina was also supported by four shared characters of mt gene arrangement: L1-rrnL, L2-rrnS, T-nad1-Q and rrnL-V (Table 3). These characters are derived for insects and present only in the elephant louse and the sucking lice, not in other parasitic lice, nor in other insects.
Figure 5. Phylogenetic relationship of the elephant louse to other parasitic lice (Phthiraptera), booklice and barklice (Psocoptera).
The tree was constructed using Bayesian and maximum likelihood methods with concatenated sequences of the first and second codon positions of 11 mitochondrial (mt) protein-coding genes and two mt rRNA genes (6,850 nucleotides). Posterior probability and bootstrap support value (%) for each grouping were indicated near the branch nodes. Trees were rooted with the true bug, Alloeorhynchus bakeri. Drawing of insects was by Hu Li.
Table 3. The four derived characters of mitochondrial gene arrangement shared exclusively by the elephant louse and the sucking lice.
| Species of insects | Order/suborder | L1-rrnL | L2-rrnS | T-nad1-Q | rrnL-V |
|---|---|---|---|---|---|
| Haematomyzus elephantis (elephant louse) | Phthiraptera/Rhynchophthirina | + | + | + | + |
| Pediculus humanus capitis (human head louse) | Phthiraptera/Anoplura | + | + | — | — |
| Pediculus humanus humanus (human body louse) | Phthiraptera/Anoplura | + | + | — | — |
| Pthirus pubis (human pubic louse) | Phthiraptera/Anoplura | — | + | — | — |
| Polyplax asiatica (rat louse) | Phthiraptera/Anoplura | + | — | — | + |
| Polyplax spinulosa (rat louse) | Phthiraptera/Anoplura | — | — | — | + |
| Haematopinus suis (domestic pig louse) | Phthiraptera/Anoplura | + | — | + | — |
| Haemapinus apri (wild pig louse) | Phthiraptera/Anoplura | + | — | + | — |
Note: “+” is for “presence”; “-” is for “absence”.
Discussion
The sister-group relationship between the suborder Anoplura (sucking lice) and the suborder Rhynchophthirina (elephant louse, wart-hog louse and red-river hog louse) indicated above by mt genome sequence analyses and derived gene-arrangement characters is consistent with the phylogeny inferred previously by morphological and molecular analyses15,16,17,18,19,20. Furthermore, Light et al.20 and Smith et al.21 dated the most recent common ancestor (MRCA) of Anoplura and Rhynchophthirina to be ~92 million years old. The species from Anoplura and Rhynchophthirina investigated to date all have fragmented minichromosomes. Thus, the most plausible explanation is that fragmented mt genomes were already present in the MRCA of Anoplura and Rhynchophthirina, and all species of these two suborders retained this novel mt genome organization (Fig. 6). Cameron et al. reported three minicircles with six mt genes in a Damalinia species (Trichodectidae, Ischnocera)6, indicating that fragmented mt genomes may be present in parasitic lice outside Anoplura and Rhynchophthirina. The exact origin of fragmented mt genomes in parasitic lice remains to be determined with more and wider sampling of species from the suborders Ischnocera and Amblycera.
Figure 6. Comparison of mitochondrial gene arrangement between the elephant louse, Haematomyzus elephantis, and other parasitic lice.
The phylogenetic tree is consistent with that shown in Fig. 5. The black dot on the tree indicates the most recent common ancestor (MRCA) shared by species of the two suborders, Rhynchophthirina and Anoplura. nad5, rrnS and rrnL genes of the elephant louse and sucking lice are highlighted with asterisks; each of these genes has its own minichromosomes, not shared with other protein-coding genes or rRNA genes. trnT, nad1 and trnQ genes are highlighted in bold red. The ancestral gene-arrangements of insects retained in the elephant louse are in bold green; the derived gene-arrangements of insects present in the elephant louse and shared by other parasitic lice are in bold black. Hyphens link neighboring genes on the same minichromosome; commas separate minichromosomes.
What did the fragmented mitochondrial genome of the MRCA of Anoplura and Rhynchophthirina look like? How different was it from those of the elephant louse and the sucking lice we observed today? For the convenience and accuracy of communication, we find it helpful to introduce the expression (and concept) of “mitochondrial karyotype” for the elephant louse and the sucking lice whose mt genomes contain multiple minichromosomes. Analogous to “nuclear karyotype”, by “mitochondrial karyotype” we mean: 1) the number of mt minichromosomes, 2) the topology of minichromosomes (i.e. linear or circular), and 3) the gene content and gene arrangement of each minichromosome. With the data available now, we cannot re-construct yet the ancestral mt karyotype of Anoplura and Rhyncophthirina. However, we can infer that several common features that the elephant louse shares with the sucking lice were likely present in the ancestral mt karyotype of Anoplura and Rhyncophthirina (Fig. 6). These features include: 1) the two rRNA genes, rrnS and rrnL, and nad5 gene were on their own minichromosomes, likely with tRNA genes downstream and/or upstream but not with any other protein-coding or rRNA genes; 2) three genes, trnT, nad1 and trnQ, were in a cluster and had the opposite orientation of transcription relative to that of other mt genes, with subsequent rearrangement of these genes in Pediculus species, Pthirus pubis and Polyplax species; 3) three ancestral gene-arrangement characters of insects were retained: atp8-atp6, trnG-nad3 and trnH-nad5; and 4) six derived gene-arrangement characters to insects were present: trnP-cox3, trnF-nad6, trnI-cox1, trnY-cox2, trnK-nad4 and trnL1-rrnL-trnV.
The number of mt minichromosomes known varies from 9 in the pig lice, Haemapinus suis and Haemapinus apri, and the horse louse, Haemapinus asini, to 20 in the human head louse, Pe. h. capitis, and the human body louse, Pe. h. humanus8,9,11,12. The elephant louse, Haematomyzus elephantis, has 10 minichromosomes (with 4 mt genes not identified yet). The rat lice, Po. asiatica and Po. spinulosa, have 11 minichromosomes10 and the human pubic louse, Pt. pubis, has 14 minichromosomes9 (3 genes not identified; Fig. 6). The substantial variation in the number of mt minichromosomes among the elephant louse and the sucking lice indicates that the process of mt genome fragmentation is likely a continuous process. One possibility is that the MRCA of Anoplura and Rhyncophthirina had less minichromosomes than what we have seen today in the species investigated from these two suborders. Fragmentation since the MRCA may have occurred at different rates in different lineages of the Anoplura and Rhyncophthirina and thus generated the mt minichromosome diversity we see today. We speculate that lineage-specific factors may have a role in generating the variation in the extent and the rate of mt genome fragmentation. Dong et al. noticed a casual, inverse link between the length of life cycle of the sucking lice and the extent of mitochondrial genome fragmentation10. Among the sucking lice investigated to date, the pig lice have the longest life cycle (29–48 days depending on weather) and the least fragmented mt genome. In contrast, the human head lice and the body lice have the shortest life cycle (13–20 days) and the most fragmented mt genomes. The rat lice, Po. spinulosa, have both the length of life cycle (25–28 days) and the extent of mt genome fragmentation in between the pig lice and the human head and body lice. Furthermore, the human pubic lice have both the length of life cycle (16–25 days) and the extent of mt genome fragmentation in between the rat lice and the human head and body lice. This link can be tested further with data from more sucking lice and other eukaryotes that have fragmented mt genomes10. In theory, if mt genome fragmentation was a continuous process and each life cycle contributed equally to the fragmentation, then species with shorter life cycles would be expected to have more fragmented mt genomes than species with longer life cycles.
Despite the common gene-arrangement features above, nine of the 10 mt minichromosomes of the elephant louse differ from those of the sucking lice known in gene content and gene arrangement; only trnH-nad5 minichromosome is also seen in Po. asiatica, the louse of greater bandicoot rat (Fig. 6). Apparently, distinct mt karyotypes have evolved between Anoplura and Rhynchophthirina since they diverged. Distinct mt karyotypes have evolved within the sucking lice too. The human lice, pig lice, the horse louse and rat lice all have their unique mt karyotypes not seen in other lice8,9,10,11,12. Animal mt genomes are usually extremely stable in terms of gene content, gene arrangement and genome organization23,25. The mt genomes of the elephant louse and the sucking lice apparently evolved at a much higher rate than in other insects and animals. What caused the fragmentation of mt genomes in the sucking lice and the elephant louse? Why are the mt karyotypes of these lice so variable? We cannot address these questions yet with the data available now. Parasitic lice do display features not usually seen in other insects or animals. For instance, a recent study showed that DNA substitution rate in the parasitic lice of humans and chimpanzees was on average 14 times higher than in their hosts26. We expect further studies on parasitic lice and related insects will be able to reveal the evolutionary forces and/or the cellular or biochemical mechanisms that drove the fragmentation of mt genomes in these insects.
Methods
Sample collection, DNA extraction and amplification of mitochondrial genome
Specimens of the elephant louse, Haematomyzus elephantis, were collected from elephants in Chitwan National Park, Nepal. Specimens were stored in 100% ethanol at −20 °C. Total DNA was extracted from individual specimens with DNeasy Blood & Tissue kit (QIAGEN).
Fragments of mt cox1, cox2, rrnS and rrnL genes were amplified by PCR with primer pairs mtd6–mtd11 (659 bp), mtd16–mtd18 (299 bp), 12SA–12SB (487 bp) and mtd32m–mtd34 (544 bp), respectively (Table S6). These four pairs of primers target conserved sequence motifs in the mt genome of the elephant louse. The amplicons of these fragments were sequenced with AB3730xl 96-capillary sequencers at the AGRF (Australian Genome Research Facilities). Four pairs of elephant-louse-specific primers, ELC1F–ELC1R, ELC2F–ELC2R1, EL12SF1–EL12SR and EL16SF–LX16SR, were designed from cox1, cox2, rrnS and rrnL genes; the forward and reverse primer in each pair are close to each other with 78, 7, 2 and 8 bp in between respectively (Table S6). PCRs with these four pairs of elephant-louse-specific primers amplified in near full-length the four minichromosomes that contain cox1, cox2, rrnS and rrnL genes respectively (Fig. 1C). The amplicons from these four minichromosomes were cloned into JM109 Competent Cells with pGEM-T Easy Vector System (Promega) and sequenced with AB3730xl 96-capillary sequencers at the AGRF using a primer-walking strategy (Table S6). Sequences of the non-coding regions of these four minichromosomes near the 5′ end and the 3′ end of the coding regions were obtained and aligned with ClustalX27; a forward primer USFB1567 and a reverse primer ELR were designed from the conserved sequence motives (Table S6). The PCR with USFB1567–ELR produced a mixture of amplicons, 1.2–2.0 kb in size (Fig. 1C), expected from the coding regions of all mt minichromosomes of the elephant louse; these amplicons were sequenced with next-generation platforms (see below). The PCR amplification strategy used in this study was developed from our observations in previous studies that each mt minichromosome of a sucking louse species has a distinct coding region and a well-conserved non-coding region13.
Elongase Enzyme (Life Technologies) was used in short PCRs to amplify fragments of cox1, cox2, rrnS and rrnL genes; TaKaRa Ex and LA Taq were used subsequently in long PCR amplifications, following the manufacturers’ instructions. PCR cycling conditions were: 94 °C for 1 min, 35 cycles of 98 °C for 10 sec, 50–65 °C (depending on primers) for 30 sec, and 72 °C for 1–4 min (depending on target size, ~1 min/kb), followed by 72 °C for 2–8 min. Negative controls were executed with each PCR experiment to detect DNA contamination and false positive amplicons. PCR amplicons were checked with agarose-gel electrophoresis. The sizes of PCR amplicons were estimated by comparison with molecular markers. PCR amplicons used for cloning and sequencing were purified with Wizard SV Gel/PCR Clean-up System (Promega).
Next-generation sequencing, assembly and verification of mitochondrial minichromosomes
Purified PCR amplicons generated above with the primer pair USFB1567–ELR from the coding regions of mt minichromosomes of the elephant louse were sequenced with Roche GS FLX (454) platform at the AGRF. We chose this platform because of its long reads relative to other platforms, which was an advantage for de novo assembly. Sequence-reads were assembled de novo into contigs with Geneious 6.1.228. The parameters for sequence assembly were: 1) minimum overlap 100 bp; 2) minimum overlap identity 90%; 3) maximum gaps per read 10%; and 4) maximum gap size 10 bp. tRNA genes were identified using tRNAscan-SE and ARWEN based on their secondary structures (Fig. S4)29,30. Protein-coding genes and rRNA genes were identified by BLAST searches of NCBI database31. Identical sequences shared between mt genes were identified with Wordmatch32. BLAST searches did not find significant matches to nad6 gene; this gene was identified by comparison of the hydrophilicity profile of its putative protein NAD6 and the conserved amino acid sequences of NAD6 with those of other animals (Figs S5, 6; Alignment S10).
Outbound primers were designed from the coding regions of each mt minichromosome to verify the size and the circular organization of the minichromosomes revealed above by sequence assembly (Table S1). The two primers in each pair are either immediately next to each other or are 2–35 bp apart; these primers amplify each minichromosome in full or near-full length if that minichromosome has a circular organization. PCR set-up and cycling conditions were the same as described above.
To obtain full-length non-coding region sequences, we sequenced, in a separate batch, the amplicons from four full-length mt minichromosomes of the elephant louse with Illumina Hiseq 2000 platform at the BGI (Beijing Genomics Institute). We chose this platform because of its high coverage and accuracy relative to other platforms. Illumina sequence-reads were assembled de novo with Geneious 7.0.528; the assembly parameters were: 1) minimum overlap 70 bp; 2) minimum overlap identity 90%; 3) maximum gaps per read 5%; and 4) maximum gap size 5 bp. We also sequenced the non-coding regions of the other six minichromosomes except for the GC-rich parts and the tandem-repeat parts using a Sanger sequencing platform. The Sanger sequence-reads were assembled using the full-length NCR sequences generated above as references with the parameters: 1) minimum overlap 200 bp; 2) minimum overlap identity 95%; 3) maximum gaps per read 5%; and 4) maximum gap size 5 bp. Heteroplasmic sites in NCRs were called with the “Find Variations/ SNPs” function in Geneious 7.0.5 with the parameters “Minimum Coverage” 50, and “Minimum Variant Frequency” 10%.
Phylogenetic analyses of mitochondrial genome sequences and tRNA-leucine gene sequences
We inferred the phylogenetic relationship of the elephant louse to 15 other parasitic lice (Phthiraptera), barklice and booklice (Psocoptera) with concatenated sequences of: 1) 11 mt protein-coding genes (nad2 and nad4 excluded; 8,040 nucleotides, PCG hereafter); 2) the first and second codon positions of the 11 mt protein-coding genes (5,360 nucleotides, PCG12 hereafter); 3) the 11 mt protein-coding genes and the two mt rRNA genes (9,530 nucleotides, PCGRNA hereafter); and 4) the first and second codon positions of the 11 mt protein-coding genes and the two mt rRNA genes (6,850 nucleotides, PCG12RNA hereafter) (Table S7)38,39,40,41. Sequences of mt protein-coding and rRNA genes were aligned individually with MUSCLE (gap open: -2.9; gap extend; 0; hydrophobicity multiplier: 1.2; clustering methods: UPGMB; diagonals min length: 24)33. nad2 and nad4 were excluded because nad2 was not identified in the elephant louse whereas nad4 was not identified in the human pubic louse9. Segments of identical sequences (26–127 bp long) shared between different mt genes in the sucking lice8,9,11 were also excluded to ensure only homologous regions of the mt genes were aligned and used in subsequent analyses. Putative amino acid sequences were used to guide nucleotide sequence alignments for the protein-coding genes. Alignments of individual genes were concatenated after removing poorly aligned sites using Gblocks 0.91 (minimum number of sequences for a conserved position: 11; minimum number of sequences for a flanking position: 11; maximum number of contiguous nonconserved positions: 8; minimum length of an initial block: 5; minimum length of a block: 5; allowed gap positions: with half)34. The concatenated alignment was used in maximum likelihood (ML) and Bayesian analyses with RAxML 7.0.3 and MrBayes 3.2.135,36. Bootstrap values for node support in ML trees were obtained by heuristic searches of 1,000 resampled datasets. For Bayesian analyses, two simultaneous runs of 10 million generations were conducted; trees were sampled every 1,000 generations with the first 25% discarded as burn-in. Stationarity was considered to be reached when the average standard deviation of split frequencies was below 0.01.
We analyzed the sequences of the two mt tRNA genes for leucine, trnL1(tag) and trnL2(taa), with distanced-based neighbor-joining (NJ) method to compare the evolution patterns of these two genes in the elephant louse, the sucking lice and other insects and mammals. The sequences of trnL1(tag) and trnL2(taa) were retrieved from GenBank for sucking lice and other insects and mammals, and were aligned according to their secondary structures. NJ trees were constructed with MEGA 5.2.237. P-distance model was used and 5,000 replicates of bootstrap resampling were run.
Additional Information
Accession codes: The nucleotide sequence of the mt genome of the elephant louse reported in this manuscript has been deposited in GenBank under accession numbers KF933032-41. http://www.nature.com/srep
How to cite this article: Shao, R. et al. Fragmented mitochondrial genomes in two suborders of parasitic lice of eutherian mammals (Anoplura and Rhynchophthirina, Insecta). Sci. Rep. 5, 17389; doi: 10.1038/srep17389 (2015).
Supplementary Material
Acknowledgments
We thank Gary Ah-kee, Kate Herd and Haoyu Xiong for assistance in the lab. We acknowledge funding support by the Australian Research Council (DP120100240 to RS and SCB), Australia-China Science & Research Fund (ACSRF00980 to RS), China Postdoctoral Science Foundation (2013M540167 and 2014T70145 to HL) and Beijing Natural Science Foundation (6144027 to HL).
Footnotes
Author Contributions R.S. and S.C.B. designed the research. S.S.P. collected elephant lice. R.S. S.S. and Y.S. performed the research. R.S. and S.C.B. contributed reagents and materials. R.S., H.L. and Y.S. analyzed the data. R.S. and Y.S. wrote the manuscript. All authors have read and approved the final manuscript.
References
- Durden L. A. & Musser G. G. The mammalian hosts of the sucking lice (Anoplura) of the world: a host-parasite list. Bull. Soc. Vector Ecol. 19, 130–168 (1994). [Google Scholar]
- Johnson K. P. & Clayton D. H. The biology, ecology, and evolution of chewing lice. Pages 449-476 in The chewing lice: world checklist and biological overview. (eds Price R. D., Hellenthal R. A., Palma R. L., Johnson K. P. & Clayton D. H.) Champaign: Illinois Natural History Survey Special Publication (2003). [Google Scholar]
- Shao R., Campbell N. J. H. & Barker S. C. Numerous gene rearrangements in the mitochondrial genome of the wallaby louse, Heterodoxus macropus (Phthiraptera). Mol. Biol. Evol. 18, 858–865 (2001). [DOI] [PubMed] [Google Scholar]
- Covacin C., Shao R. F., Cameron S. & Barker S. C. Extraordinary number of gene rearrangements in the mitochondrial genomes of lice (Phthiraptera: Insecta). Insect Mol. Biol. 15, 63–68 (2006). [DOI] [PubMed] [Google Scholar]
- Cameron S., Johnson K. & Whiting M. The mitochondrial genome of the screamer louse Bothriometopus (Phthiraptera: Ischnocera): effects of extensive gene rearrangements on the evolution of the genome. J. Mol. Evol. 65, 589–604 (2007). [DOI] [PubMed] [Google Scholar]
- Cameron S. L., Yoshizawa K., Mizukoshi A., Whiting M. F. & Johnson K. P. Mitochondrial genome deletions and minicircles are common in lice (Insecta: Phthiraptera). BMC Genomics 12, 394 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajander O. A. et al. Human mtDNA sublimons resemble rearranged mitochondrial genoms found in pathological states. Human Mol. Genet. 9, 2821–2835 (2000). [DOI] [PubMed] [Google Scholar]
- Shao R., Kirkness E. F. & Barker S. C. The single mitochondrial chromosome typical of animals has evolved into 18 minichromosomes in the human body louse, Pediculus humanus. Genome Res. 19, 904–912 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao R., Zhu X. Q., Barker S. C. & Herd K. Evolution of extensively fragmented mitochondrial genomes in the lice of humans. Genome Biol. Evol. 4, 1088–1101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W. G., Song S., Jin D. C., Guo X. G. & Shao R. Fragmented mitochondrial genomes of the rat lice, Polyplax asiatica and Polyplax spinulosa: intra-genus variation in fragmentation pattern and a possible link between the extent of fragmentation and the length of life cycle. BMC Genomics 15, 44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H. W., Barker S. C. & Shao R. F. Substantial variation in the extent of mitochondrial genome fragmentation among blood-sucking lice of mammals. Genome Biol. Evol. 5, 1298–1308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Barker S. C. & Shao R. Variation in mitochondrial minichromosome composition between blood-sucking lice of the genus Haematopinus that infest horses and pigs. Parasit. Vectors 7, 144 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W. G. et al. Fragmented mitochondrial genomes are present in both major clades of the blood-sucking lice (suborder Anoplura): evidence from two Hoplopleura rodent lice (family Hoplopleuridae). BMC Genomics 15, 751 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris G. F. The louse of elephants. Haematomyzus elephantis Piaget (Mallophaga: Haematomyzidae). Parasitol. 23, 112 (1931). [Google Scholar]
- Lyal C. H. C. Phylogeny and classification of the Psocodea, with particular reference to the lice (Psocodea: Phthiraptera). Syst. Entomol. 10, 145–165 (1985). [Google Scholar]
- Barker S. C. Phylogeny and classification, origins, and evolution of host associations of lice. Int. J. Parasitol. 24, 1285–1291 (1994). [DOI] [PubMed] [Google Scholar]
- Barker S. C., Whiting M. F., Johnson K. P. & Murrell A. Phylogeny of the lice (Insecta, Phthiraptera) inferred from small subunit rRNA. Zool. Scr. 32, 407–414 (2003). [Google Scholar]
- Murrell A. & Barker S. C. Multiple origins of parasitism in lice: phylogenetic analysis of SSU rDNA indicates that the Phthiraptera and Psocoptera are not monophyletic. Parasitol. Res. 97, 274–280 (2005). [DOI] [PubMed] [Google Scholar]
- Yoshizawa K. & Johnson K. P. Morphology of male genitalia in lice and their relatives and phylogenetic implications. Syst. Entomol. 31, 350–361 (2006). [Google Scholar]
- Light J., Smith V., Allen J., Durden L. & Reed D. Evolutionary history of mammalian sucking lice (Phthiraptera: Anoplura). BMC Evol. Biol. 10, 292 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith V. S. et al. Multiple lineages of lice pass through the K-Pg boundary. Biol. Lett. 5, 782–785 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H., Barker S. C., Burger T. D., Raoult D. & Shao R. Heteroplasmy in the mitochondrial genomes of human lice and ticks revealed by high throughput sequencing. PLoS ONE 8, e73329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore J. L. Animal mitochondrial genomes. Nucleic Acids Res. 27, 1767–1780 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao R. & Barker S. C. Chimeric mitochondrial minichromosomes of the human body louse, Pediculus humanus: Evidence for homologous and non-homologous recombination. Gene 473, 36–43 (2011). [DOI] [PubMed] [Google Scholar]
- Lavrov D. V. Key transitions in animal evolution: a mitochondrial DNA perspective. Integr. Comp. Biol. 47, 734–743 (2007). [DOI] [PubMed] [Google Scholar]
- Johnson K. P. et al. Rates of genomic divergence in humans, chimpanzees and their lice Proc Biol Sci. 281, 20132174 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- Kearse M. et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M. & Eddy S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D. & Canbäck B. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 24, 172–175 (2008). [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W. & Lipman D. J. Basic local alignment search tool. J Mol Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- Rice P., Longden I. & Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 (2000). [DOI] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000). [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006). [DOI] [PubMed] [Google Scholar]
- Ronquist F. & Huelsenbeck J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003). [DOI] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D. D. et al. The multipartite mitochondrial genome of Liposcelis bostrychophila: insights into the evolution of mitochondrial genomes in bilateral animals. PLoS ONE 7, e33973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. Mitochondrial genomes of two barklice, Psococerastis albimaculata and Longivalvus hyalospilus (Psocoptera: Psocomorpha): contrasting rates in mitochondrial gene rearrangement between major lineages of Psocodea. PLoS ONE 8, e61685 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao R., Dowton M., Murrell A. & Barker S. C. Rates of gene rearrangement and nucleotide substitution are correlated in the mitochondrial genomes of insects. Mol. Biol. Evol. 20, 1612–1619 (2003). [DOI] [PubMed] [Google Scholar]
- Li H. et al. Comparative mitogenomic analysis of damsel bugs representing three tribes in the family Nabidae (Insecta: Hemiptera). PLoS ONE 7, e45925 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.