Abstract
We previously developed replication-competent reporter HIV-1 (referred to herein as LucR.T2A reporter viruses), utilizing a “ribosome skipping” T2A peptide strategy to link Renilla luciferase (LucR) with Nef expression. The demonstrated utility for HIV-1 vaccine and transmission study applications included measurement of neutralizing antibody (NAb) activity in vaccine sera, improved cell-mediated virus inhibition assays, such as T cell-mediated virus inhibition and antibody-dependent cell-mediated cytotoxicity (ADCC) assays, and humanized mouse models. Herein, we extend our prior work and introduce reporter virus technology for applications that require fully functional Nef. We demonstrate that in CD4+ T cells productively infected with LucR.T2A reporter viruses, T2A peptide-driven Nef expression and function, such as down-regulation of surface CD4 and MHC-I, were impaired. We overcame this limitation of LucR.T2A reporter viruses and achieved physiological Nef expression and function by engineering novel LucR reporter HIV-1 comprising 11 different internal ribosome entry site (IRES) elements chosen for size and relative activity. A range of Nef expression was observed in 293T cells transfected with the different LucR.IRES reporter virus constructs. Iteratively, we identified IRES reporter genomes that expressed Nef closest to physiological levels and produced virus with infectivity, titers, and replication kinetics similar to nonreporter viruses. Our results demonstrated that LucR reporter activity was stable over multiple replication cycles in peripheral blood mononuclear cells (PBMCs). Furthermore, we analyzed Nef functionality, i.e., down-modulation of MHC-I and CD4, following infection of T cell lines and PBMCs. Unlike LucR.T2A reporter virus, one of the redesigned LucR.IRES reporter viruses [containing the modified encephalomyocarditis virus (EMCV) 6ATR IRES element, “6ATRi”] demonstrated Nef expression and function similar to parental “nonreporter” virus. In a previously validated (nef-independent) T cell-based NAb neutralization assay, LucR.6ATRi reporter virus performed indistinguishably from LucR.T2A reporter virus. In summary, reporter viruses comprising the “6ATRi” element promise to augment HIV-1 vaccine and transmission research approaches requiring a sensitive reporter readout combined with wild-type Nef function.
Introduction
Development of a successful vaccine against HIV-1 requires well-defined tools to determine the breadth of the immune response including elicitation of neutralizing antibody (NAb), T cell-mediated virus inhibition, and antibody-dependent cell-mediated cytotoxicity (ADCC). Several strategies and approaches are employed in existing immunomonitoring assays, including reporter cell lines and HIV-1 reporter viruses (reviewed in Polonis et al.,1 Polonis et al.,2 and Ochsenbauer and Kappes3). Depending on the molecular strategy for reporter gene expression from proviral constructs, viruses may either require Env pseudotyping for single-round infection only or possess replication capacity.
We previously reported on novel replication-competent reporter viruses for studying HIV-1 transmission and correlates of protection-encompassing proviral infectious molecular clones (IMC) of transmitter/founder (T/F) HIV-14–6 and several forms of reporter virus derivatives expressing genes such as enhanced green fluorescent protein (EGFP)7 and Renilla luciferase (LucR),4,8 which underpin new immune monitoring assays and augment the performance of existing assays for various vaccine discovery approaches.3,4,8–26
Among formerly described replication-competent HIV-1 reporter vectors were those designed with a bicistronic EGFP-IRES-nef cassette in place of nef, in which the EGFP gene is downstream of env, and nef is under translational control of an internal ribosome entry site (IRES) from encephalomyocarditis virus (EMCV).3,27–31 However, we found that these EMCV IRES-containing reporter viruses vastly overexpress Nef8 and exhibit both poor replication and stability of the reporter gene (personal observations and Brown et al.29). To overcome these challenges, we explored replication-competent HIV-1 reporter viruses that utilized the small “ribosome skipping” T2A peptide (18 amino acids) to mediate Nef expression in frame with, and release from (via ribosome skipping of the last peptide bond at the C-terminus of the T2A peptide32,33), LucR.8 Furthermore, the molecular strategy was designed to facilitate expression of heterologous env genes in an isogenic (NL4-3-derived) proviral backbone, collectively referred to as Env-IMC-LucR.T2A, or simply LucR.T2A reporter viruses. As previously reported,3,8 this approach enables sensitive detection of infection, as measured by reporter gene expression, in assays that require replication-competent HIV-1, including those utilizing peripheral blood mononuclear cells (PBMCs) or other primary cells and nonreporter T cell lines.
In contrast to the EMCV IRES-containing reporter viruses, LucR.T2A reporter viruses had demonstrated replication kinetics similar to parental “nonreporter” viruses, as well as stable expression of the LucR reporter gene over several replication cycles.8 In this regard, the LucR.T2A reporter virus technology offers several critical advantages and has found wide application, including enabling of a novel, highly sensitive T cell-based assay (referred to as the A3R5/Env-IMC-LucR neutralization assay)9,10 for measuring NAb activity in vaccine sera from the RV144 and Vax003 HIV-1 vaccine trials in Thailand.11,34,35 The reporter viruses have also underpinned the development of novel CD8+ T cell virus inhibition assays (VIA),12,19 ADCC assays,13–15 and a humanized mouse model of HIV-1 transmission.16
The majority of current applications do not require functional Nef expression; however, the LucR.T2A strategy may be a limitation in certain applications such as correlates of protection discovery that require “whole genome” T/F reporter IMC. While the LucR.T2A strategy was conceived to ensure Nef expression, the approach incorporates a Pro residue to the N-terminus of Nef [followed by two additional residues (Ser-Arg) included from translation of a six-nucleotide (nt) sequence encoding an XbaI restriction site engineered into the vector's design] proximal to the native Met start codon. This modification likely interferes with N-terminal myristoylation of Nef,36,37 thereby altering or impairing Nef function and/or expression in infected cells.38
With the goal of improving upon the LucR.T2A strategy for additional applications in the HIV-1 transmission and immune monitoring fields, we revisited the IRES-containing reporter virus approach. We postulated that unlike T2A, an IRES-based strategy could produce authentic Nef protein. Given the shortcomings for earlier IRES-containing reporter viruses, we investigated the utility of several different IRES elements engineered to overcome limitations exhibited by the EMCV IRES used in these previous reporter HIV-1. Our goal was to achieve physiological levels of Nef expression and function as well as stable reporter gene expression over multiple replication cycles. In total, we generated 11 LucR.IRES reporter viruses comprising well-defined IRES elements from insect viruses and human hepatitis C virus (HCV), as well as various derivations of the EMCV IRES.
Herein, we show that compared to the LucR.T2A reporter virus, all 11 of the IRES reporter viruses were similarly infectious and expressed LucR. Following proviral DNA transfection of 293T cells, Nef protein was not detected from insect IRES, whereas Nef expression levels from HCV IRES and some of the derivative EMCV IRES elements were lower than those observed with parental nonreporter viruses. In contrast, Nef expression levels from several other EMCV IRES elements were comparable to or exceeded those of matched nonreporter HIV-1 strains. Three of the LucR.IRES reporter viruses with the most physiological Nef levels were tested for replication kinetics and genetic stability of the LucR reporter gene in pooled PBMCs. From this evaluation, we determined that the LucR.6ATRi reporter virus (containing the 6ATR derivative EMCV IRES element) displayed a favorable phenotype, similar to LucR.T2A reporter viruses. Importantly, and in contrast to LucR.T2A reporter viruses, we observed levels of Nef-mediated down-modulation of MHC-I and CD4 following infection of T cell lines and PBMCs with LucR.6ATRi reporter viruses that were similar to those seen with nonreporter viruses. When tested against a panel of eight broadly NAb (bNAb) in the A3R5/Env-IMC-LucR neutralization assay, the LucR.6ATRi reporter virus yielded results indistinguishable from the LucR.T2A reporter virus that was used in assay standardization.10
Our findings suggest that reporter viruses comprising the EMCV-derived 6ATR IRES element (collectively referred to as Env-IMC-6ATRi) will be useful virological tools for augmenting HIV vaccine correlates of protection discovery efforts, with an emphasis on applications for which fully functional Nef is desirable or required.
Materials and Methods
Cells
All cells were propagated at 37°C in a humidified atmosphere of 5% CO2. The HEK 293T/17 cell line (CRL-11268) was obtained from the American Type Culture Collection (ATCC, Manassas, VA). We previously generated the TZM-bl dual-reporter cell line,39 which is also available through the NIH AIDS Reagent Program, Division of AIDS NIAID, NIH (catalog no. 8129). TZM-bl and 293T cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), penicillin (100 U/ml), streptomycin (100 μg/ml), l-glutamine (2 mM), and HEPES (25 mM). The A3R5.7 cell line was obtained from Drs. Jerome Kim and Robert McLinden at the U.S. Medical HIV Research Program (MHRP). The A3R5.7 cell line is derived from a human CD4+ CXCR4+ lymphoblastoid cell line (CEM/A3.01)40 and is engineered to express CCR5.9 A3R5.7 was maintained in RPMI 1640 growth medium supplemented with 10% FBS, penicillin (100 U/ml), streptomycin (100 μg/ml), l-glutamine (2 mM), HEPES (25 mM), and geneticin G-418 sulfate (1 mg/ml) (Life Technologies, Carlsbad, CA). G-418-free growth medium was used for A3R5.7 experiments. The J2574-R5 cell line was kindly provided by Dr. Olaf Kutsch. J2574-R5 is a Jurkat-derived cell line, previously engineered by stable transduction with an HIV-1 LTR-GFP reporter cassette,41,42 followed by introduction of constitutive CCR5 expression.
Human primary PBMCs were provided by Tom Denny (Duke University) from the CT-VIMC (OPP1032325), part of the CAVD, funded by the Bill & Melinda Gates Foundation. PBMCs were thawed and placed for 48 h into “PBMC culture medium” [i.e., RPMI 1640 growth medium supplemented with 20% FBS, l-glutamine (2 mM), penicillin (100 U/ml), streptomycin (100 μg/ml), HEPES (25 mM), and interleukin-2 (IL-2; 30 U/ml) (Roche, Indianapolis, IN)] and supplemented with phytohemagglutinin (PHA)-P (5 μg/ml) (Sigma-Aldrich, St. Louis, MO). Following PHA-P stimulation, the medium was removed, and cells were placed into fresh “PBMC culture medium” and used immediately.
Proviral plasmid construction: IRES constructs
Previously, we described the construction of a LucR-expressing, replication-competent HIV-1 proviral DNA (designated pNL-LucR.T2A), as well as several derivative LucR.T2A proviruses in which the ectodomain of gp160/Env is encoded by heterologous HIV-1 strain env sequences, including pNL-LucR.T2A-BaL.ecto.8 In the present study, we modified this approach by replacing the bicistronic LucR.T2A-nef fragment in pNL-LucR.T2A-BaL.ecto with a panel of bicistronic LucR.IRES-nef cassettes of different lengths. Detailed cloning schema and methods are available upon request and are summarized hereafter and in Supplementary Fig. S1 (Supplementary Data are available online at www.liebertpub.com/aid). Briefly, 11 different NheI-Kozak-LucR-spacer-IRES cassettes were inserted into the pNL-LucR.T2A-BaL.ecto backbone in place of pNL4-3 nt 8786, between the env stop codon (nt 8783–8785) and the nef start codon (8787–8789) (positions are based on the pNL4-3 backbone; GenBank ID: M19921.1). “LucR” corresponds to the entire Renilla reniformis luciferase reporter gene, including the stop codon, from phRL-CMV (Promega, Madison, WI) (nt 1068–2003; GenBank ID: AF362549). “Spacer” refers to one of two different 26-nt spacers (Spacer A: atcgatgccgccaccatggacaattg; Spacer B: atcgatgccgccaccatggagggtta).
“IRES” elements were amplified from several sources. The Israeli acute paralysis virus (IAPV) intergenic region (IGR) IRES (IAPVi) was amplified from a plasmid that was generously provided by Dr. Sunnie R. Thompson (UAB).43,44 The region amplified corresponds to viral genome nt 6399–6617 (GenBank ID: NC_009025) but with the following compensatory mutations: CA→TC (nt 6566–6567) and ATG→TGA (nt 6612–6614) (termed IAPVuga/uc in Hertz et al.43). The cricket paralysis virus (CrPV) IGR IRES (CrPVi) was amplified from a plasmid generously provided by Dr. Sunnie R. Thompson (UAB).43,45 The region amplified corresponds to viral genome nt 6027–6216 (GenBank ID: AF218039). The HCV 5′-UTR (untranslated region) IRES (HCVi) was also amplified from a plasmid generously provided by Dr. Sunnie R. Thompson (UAB).45,46 The region amplified corresponds to viral genome nt 18–341 (GenBank ID: AB691953). The EMCV IRES (GenBank ID: NC_001479) was amplified from the plasmids pIRES-EGFP (Clontech, Mountain View, CA) and pNL-ENG1-IRES-BaL.ecto (also termed pNLENG1i-BaL.ecto or simply “NLENG1i”).7,27,28
These two EMCV IRES elements differ in that the pNLENG1i-BaL.ecto contains the “wild-type” (A)6 (or “6A”) bifurcation loop and pIRES-EGFP contains the (A)7 (or “7A”) bifurcation loop, which leads to known differences in translation efficacy.47 From each vector, two regions were amplified corresponding to viral genome nt 258–833 and nt 399–833, for “full-length” and “truncated” (TR) forms,47,48 respectively. In addition, all four forms were also amplified to include a spacer or “attenuator” at the 3′ end of the IRES, just before the ATG codon of nef, and are referred to as (att)enuated. The 29-nt attenuator (tctagactcacaaccccagaaacagacat) corresponds to an XbaI restriction site (tctaga) fused to nt 2692–2716 of the mouse β-globin major 5′-UTR (GenBank ID: J00413).49 Overall, eight different EMCV IRES elements were created, referred to as 6Ai, 6ATRi, 7Ai, 7ATRi, att6Ai, att6ATRi, att7Ai, and att7ATRi.
Proviral plasmid construction: Nef mutants
A series of Nef mutants (see Fig. 1A) were created in the pNL-BaL.ecto,8 pNL-LucR.T2A-BaL.ecto,8 and pNL-LucR.6ATRi-BaL.ecto (described above) proviral genomes using standard cloning techniques. To briefly summarize these methods, the NefG2A mutant is based on that reported in Welker et al. in which the Gly-to-Ala change at amino acid position 2 (G2A) resulted in an inability to N-myristoylate (Δmyr) the N-terminus of Nef.38 The NefKR mutant [mutation of six basic amino acid residues important for Nef association with the membrane (Lys and Arg to Val or Ala, see Fig. 1A)] is based on the equally named mutant reported in Bentham et al.50 The NefSTOP mutant was created by combining the G2A mutation with the mutation of the fourth and fifth codons to stop codons. The presence of the T2A motif followed by an XbaI site adds three extra amino acids (Pro-Ser-Arg, or “PSR”) to the N-terminus of Nef (T2A-Nef). Therefore, two additional mutants were created, T2APSD-Nef and T2APSS-Nef, to mutate the basic Arg (R) residue to Asp (D) or Ser (S), respectively. Furthermore, several of the mutations were combined to produce additional functional Nef mutants (see Fig. 1A). Detailed cloning schema and methods are available upon request.
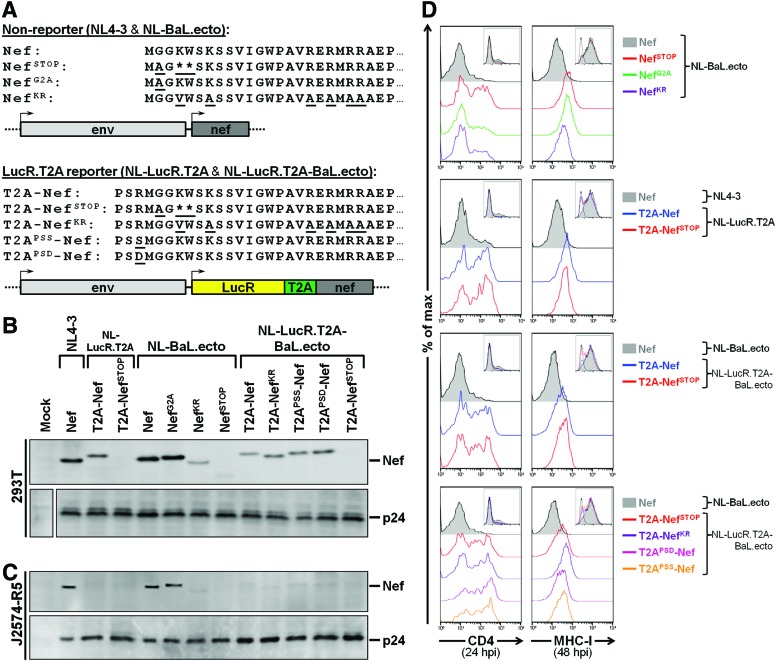
FIG. 1.
Decreased expression and functional impairment of Nef in J2574-R5 cells infected with LucR.T2A reporter viruses compared to nonreporter viruses. (A) Schematic representation and N-terminal amino acid sequence detail of Nef in nonreporter (NL4-3 and NL-BaL.ecto) and LucR.T2A reporter (NL-LucR.T2A and NL-LucR.T2A-BaL.ecto) constructs and the various mutations of nef that were incorporated into either background. Mutations that abrogate either myristoylation (NefG2A), basic residue-mediated membrane association (NefKR), or expression (NefSTOP) of Nef are indicated. Additionally, for LucR.T2A reporter viruses, the presence of the T2A motif followed by an XbaI site incorporates three extra amino acids (Pro-Ser-Arg or “PSR”) to the N-terminus of Nef (T2A-Nef). Therefore two additional mutants were created, T2APSD-Nef and T2APSS-Nef, to mutate the basic Arg (R) residue to an Asp (D) or Ser (S), respectively. (B) Western blot evaluation of Nef levels in 293T cells 48 h after transfection with LucR.T2A reporter and nonreporter proviral DNA. Replica blots were prepared in parallel and probed with anti-p24 (Gag) to demonstrate equivalent transfection efficiencies. (C, D) Evaluation of expression (C) and function (D) of Nef in a Jurkat-derived T cell line, J2574-R5, after infection with 293T transfection-derived LucR.T2A reporter and nonreporter viruses prepared in (B). J2574-R5 cells express GFP upon HIV-1 infection, which was exploited to normalize the number of infected cells analyzed by Western blot, and for gating on HIV-1-infected (GFP+) cells for flow cytometric analysis. (C) Western blot evaluation of Nef levels in J2574-R5 cells 48 hpi with anti-Nef monoclonal antibody (mAb) clone EH1. Replica blots were prepared in parallel and probed with anti-p24 (Gag) to demonstrate loading of equivalent numbers of infected cells. (D) Flow cytometric evaluation of Nef function, as measured by CD4 (left panels) and MHC-I (right panels) down-modulation on infected (GFP+) cells at 24 and 48 hpi, respectively. Levels of CD4 and MHC-I down-regulation were compared to nonreporter viruses (NL4-3 or NL-BaL.ecto) encoding wild-type Nef (filled histograms). Insets illustrate relative numbers of cells (y-axis) that are GFP+ (x-axis) and analyzed in each panel. Color images available online at www.liebertpub.com/aid
Generation of virus stocks and determination of virus infectivity
293T transfection-derived virus stocks were produced by transfection of proviral DNA using FuGENE 6 according to the manufacturer's protocol (Promega) as previously described.4,8 Medium was changed after 24 h, and viral supernatants were harvested 72 h posttransfection, clarified at 300×g for 12 min, and frozen at −80°C. Virus stocks were analyzed for HIV-1 p24 antigen concentration by ELISA (PerkinElmer, Groningen, The Netherlands) as indicated and were titered on the TZM-bl reporter cell line by enumeration of beta-galactosidase (β-gal)-stained colonies [TZM-bl infectious units (IU)] as previously described.39 The titered virus stocks were further analyzed for infectivity and LucR gene expression in the TZM-bl and A3R5.7 cell lines. Briefly, flat- or round-bottom 96-well plates were seeded at 1×104 TZM-bl cells per well 1 day prior to infection or at 9×104 A3R5.7 cells per well on the day of infection, respectively.
On the day of infection, virus was serially diluted starting with a normalized TZM-bl IU input multiplicity of infection (MOI) of 1. The linear range of the LucR readout was examined by infecting TZM-bl cells in triplicate at an MOI equivalent to 0.01, 0.05, 0.1, 0.5, and 1.0 TZM-bl IU (based on β-gal enumeration) in the presence of DEAE-dextran (10 μg/ml). At 48 h postinfection (hpi), 50 μl of 5×Passive Lysis Buffer (PLB; Promega) was added (for a final concentration of 1×) directly to each well containing cells. Samples were frozen at −80°C until use. Samples were brought to room temperature and 20 μl of each sample was analyzed for provirally encoded LucR [Renilla Luciferase Assay System (Promega; catalog no. E2810)] and cell-encoded firefly luciferase (LucF) [as a measure of virus infectivity, irrespective of LucR reporter gene expression; Luciferase Assay System (Promega; catalog no. E4530)] using a Victor X Light Luminometer with single-channel injector (PerkinElmer). Similarly, the linear range of the LucR expression after infection of (nonreporter) A3R5.7 cells with normalized and serially diluted Env-IMC-LucR.T2A and Env-IMC-LucR.IRES viruses [in the presence of DEAE-dextran (5 μg/ml)] was determined at 48 hpi, using the Renilla Luciferase Assay System, including its 5× Renilla Luciferase Assay Lysis Buffer (Promega).
Replication kinetics in PBMCs and determination of LucR stability
PBMCs (2×107) from four different donors were thawed and individually stimulated with PHA-P (5 μg/ml) and IL-2 (30 U/ml) for 48 h, as described above. Cells were then combined (8×107 PBMCs in total) and depleted of CD8+ cells using MACS CD8 MicroBeads according to the manufacturer's protocol (Miltenyi Biotec Inc., Auburn, CA). Following depletion, 2×106 cells were infected with 1×106 TZM-bl IU (MOI of 0.05) of each virus to be analyzed, with and without the protease inhibitor indinavir sulfate (1 μM; obtained through the NIH AIDS Reagent Program, Division of AIDS NIAID, NIH). The cells were incubated with virus in screw-cap Eppendorf tubes in a total volume of 1 ml PBMC culture medium and rotated for 4 h at 37°C. The cells were then washed twice with prewarmed “PBMC culture medium” and transferred to a 48-well plate for the duration of the experiment.
Aliquots of supernatants from each sample were taken at the indicated time points and stored at −80°C until all of the samples were collectively analyzed by quantitative HIV-1 p24 ELISA (PerkinElmer). Aliquots of cells were also removed, lysed with 5× Renilla Luciferase Assay Lysis Buffer (Promega), and stored for LucR analysis, as above, at the time supernatants were collected (equivalent cell numbers were collected at all time points). When applicable, indinavir was readded at day 3, and medium removed for sampling was replenished throughout the experimental time course.
Replicate aliquots of the same supernatant samples were also used to determine the stability of LucR expression over several replication cycles, essentially as previously described.8 Briefly, the supernatants collected on days 5, 9, and 16, as well as the cryopreserved 293T transfection-derived virus stocks, were used to infect TZM-bl cells in triplicate with three 5-fold dilutions. The cells were infected in the presence of DEAE-dextran (40 μg/ml) for 4 h, then the medium was replaced with prewarmed cell culture medium. After a 48-h incubation period, the cells were lysed with 5× PLB (Promega) and analyzed for both LucF and LucR activity, as above. Overall infectivity was measured by cell-encoded LucF activity while virus-encoded LucR activity was determined to assess the relative viral reporter gene expression. Values from those input dilutions yielding relative light unit (RLU) values within the linear range were then used to calculate the ratios of LucR to LucF.
Infection of cells for the determination of Nef, CD4, and MHC-I expression
J2574-R5 cells (1×106) were resuspended in vials with 500 μl of medium and 500 μl of each virus. Cells and virus were brought to a final volume of 1 ml in the presence of DEAE-dextran (5 μg/ml) and rotated at 37°C for 4 h. Cells were then transferred to a 24-well plate at which time the final volume was brought up to 2 ml with fresh medium, keeping the concentration of DEAE-dextran at 5 μg/ml, and then incubated overnight at 37°C. At 18–24 hpi, a portion of the infected cells was analyzed for CD4 expression by flow cytometry (see below), and at 48 hpi the remaining infected cells were analyzed for MHC-I expression and prepared for Western blot analysis (see below).
For A3R5.7 and PBMCs, 5×106 cells were resuspended in vials with 500 μl of their respective medium and 500 μl of each virus (or uninfected controls), rotating them at 37°C for 4 h to promote infection [DEAE-dextran (5 μg/ml) was included for A3R5.7 cells]. The contents of the vials were then transferred into T-12.5-cm2 flasks, at which point the final volume was brought up to 2 ml with fresh medium, and then incubated overnight at 37°C. The next morning 2 ml of fresh medium was added to each flask. At 72 hpi, 1 ml of medium was removed and replaced with 2 ml of fresh medium. The day after (∼96 hpi) the cells were counted, and aliquots from each flask were taken to determine, by flow cytometry, the number of infected cells by intracellular p24 staining, as well as MHC-I down-modulation (see below). Samples were also prepared for Western blot (see below).
Flow cytometric analysis for the number of infected cells and CD4 and MHC-I expression levels
To examine Nef functionality, infected cells were identified by flow cytometry by either GFP expression (for J2574-R5) or intracellular p24 staining with FITC-conjugated mouse anti-HIV-1 p24 (clone KC57; Beckman Coulter, Brea, CA) monoclonal antibody (mAb) (for A3R5.7 and PBMCs). Within these gates (i.e., positivity for GFP or FITC), levels of cell surface MHC-I and CD4 were determined using the following mouse mAbs for flow cytometry: APC-conjugated antihuman CD4 (clone L200) and APC-conjugated antihuman MHC-I (clone G46-2.6, recognizing a monomorphic epitope on HLA-A, HLA-B, and HLA-C) purchased from BD Biosciences, Pharmingen (San Diego, CA). Cells were assayed using a BD FACSCalibur flow cytometer and the resulting data were analyzed with FlowJo software (TreeStar Inc., Ashland, OR).
Western blot analysis
Provirally transfected 293T cells were washed with phosphate-buffered saline (PBS) and lysed with Laemmli sample buffer (Sigma-Aldrich) 60–72 h posttransfection. When indicated, virions were concentrated from the same six-well plate cultures (three wells per sample, ∼7.5 ml) by layering cell-free supernatants over a 20% sucrose cushion before ultracentrifugation at 25,000 rpm (∼110,000×g) for 2 h (Beckman Optima XL-90 polyallomer tubes; Beckman Coulter). Supernatants were removed and virion pellets were lysed directly in 150 μl Laemmli sample buffer. Cell lysates and virion lysates were sonicated for 40 s and then heated for 10 min at 95°C and frozen at −20°C until use.
J2574-R5 cells were infected as above. At 48 hpi, cells were washed with PBS, counted, pelleted, and then lysed with a low volume of Laemmli sample buffer. The percentage and total number of infected cells (i.e., GFP+ cells) were determined by flow cytometric analysis of an aliquot from each sample (see above), and the volume of Laemmli sample buffer was adjusted accordingly to achieve 1×104 infected cells per μl of lysate. Similarly, the percentage and total number of infected A3R5.7 or PBMCs were determined at 96 hpi via intracellular p24 staining by flow cytometric analysis (see above). Based on the available total number of cells and the percentage of infection determined by flow cytometry, aliquots of cells were lysed with a volume of Laemmli sample buffer in order to achieve a final concentration of 1×104 infected cells per μl of lysate. J2574-R5, A3R5.6, and PBMC cell lysates were sonicated for 40 s and then heated for 10 min at 95°C and frozen at −20°C until use.
Before use, all samples were reheated at 95°C for 5 min and the proteins resolved by denaturing SDS-PAGE (10% resolving gel) and transferred to a nitrocellulose membrane. Membranes were blocked with 5% nonfat dried milk in Tris-buffered saline with Tween 20 (TBST) or SuperBlock T20 (PBS) Blocking Buffer (Thermo Scientific, Rockford, IL) and then incubated with the appropriate primary antibodies at 4°C overnight. Mouse mAb to LucR (clone 5B11.2; Chemicon International, Temecula, CA) was used at a 1:1000 dilution. Polyclonal rabbit HIV-1 Nef antiserum (obtained from the NIH AIDS Reagent Program, Division of AIDS NIAID, NIH, contributed by Ronald Swanstrom, catalog no. 2949, Lot# 10-070932)51 was used at 1:1000 dilution (Fig. 2B). Mouse mAb to HIV-1 Nef (clone EH1; kindly provided by Dr. James Hoxie at the University of Pennsylvania), directed against the 13 C-terminal residues of SF-2 Nef and reactive against NL4-3 Nef, was used at a 1:1000 dilution (Figs. 1, 4, and 5). Mouse mAb to HIV-1 p24 (Gag) (also known as “ARF”) was prepared from the HIV-1 p24 hybridoma (clone 183-H12-5C; obtained from the NIH AIDS Reagent Program, Division of AIDS NIAID, NIH, contributed by Drs. Bruce Chesebro and Hardy Chen, catalog no. 1513)52 and used at a 1:1000 dilution. Membranes were probed with horseradish peroxidase (HRP)-conjugated goat polyclonal secondary antibodies to mouse IgG or rabbit IgG (SouthernBiotech, Birmingham, AL) and developed with Immobilon Western Chemiluminescent HRP Substrate (Millipore, Billerica, MA) or Amersham ECL Western Blotting Detection Reagents (GE Healthcare UK Limited, Little Chalfont, Buckinghamshire, UK). PageRuler Prestained Protein Ladder (Thermo Scientific) was used to estimate the molecular weight of bands.
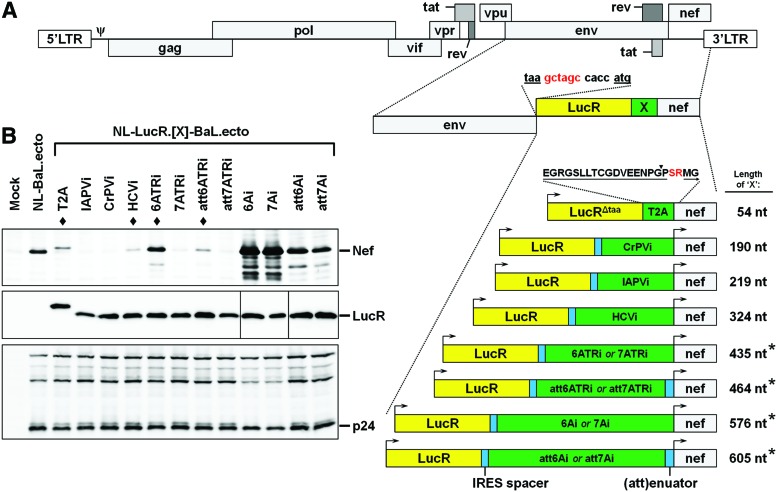
FIG. 2.
Derivation of a panel of novel LucR.IRES reporter viruses. (A) Schematic representation of the insertion of various LucR.IRES elements into the NL4-3 or NL-Env.ecto (e.g., NL-BaL.ecto) backbones (also see Supplementary Fig. S1). Eleven LucR.IRES elements were inserted between the NL4-3 env and nef genes. The nucleotide (nt) sequence is shown for the junction between the stop codon (taa) of env and the start codon (atg) of LucR, which contains an NheI restriction site (gctagc) and Kozak sequence (ccacc). Each LucR.IRES contains a 26 nt “IRES spacer” between the LucR gene and internal ribosome entry site (IRES) element. For the encephalomyocarditis virus (EMCV) IRES, two regions were amplified corresponding to “full-length” and “truncated” (TR) forms (where the 3′ nucleotide sequences are identical). Each of these EMCV IRES elements was also designed to contain either the “wild-type” (A)6 (i.e., “6A”) or (A)7 (i.e., “7A”) bifurcation loop, which corresponds to known differences in translation efficacy. In addition, all four forms were also engineered to include a 29 nt spacer or “(att)enuator” at the 3′ end of the IRES, just before the start codon of nef, resulting in eight EMCV IRES variants (6Ai, 7Ai, 6ATRi, 7ATRi, att6Ai, att7Ai, att6ATRi, and att7ATRi). LucR.T2A (described elsewhere8) is also shown for comparison. The LucR gene (excluding stop codon) and 18 amino acid T2A sequence (underlined; “EGR…PGP”) were fused in-frame. The closed arrowhead (▼) indicates the cotranslational cleavage point between the penultimate (Gly, G) and last amino acid (Pro, P) of T2A. The first two amino acids (→; Met-Gly or “MG”) of Nef are also depicted, as well as the translated XbaI restriction site (Ser-Arg or “SR”) that was introduced by design (see Fig. 1A). Cleavage of the LucR.T2A-Nef bicistron results in the N-terminus of Nef containing three additional amino acids preceding the Met (Pro-Ser-Arg “PSR”). The length of each element (“X”) is shown to the right. The asterisk (*) indicates that the element is 1 nt longer in the 7A variant than in the 6A variant. (B) Western blot evaluation of Nef (upper panel; polyclonal rabbit HIV-1 Nef antiserum) and LucR (middle panel) levels in 293T cells 48 h after transfection with nonreporter (NL-BaL.ecto), LucR.T2A reporter (NL-LucR.T2A-BaL.ecto), and the 11 LucR.IRES reporter proviral DNA. Replica blots were prepared in parallel and probed with anti-p24 (Gag) (lower panel) to demonstrate equivalent transfection efficiencies. The LucR.T2A reporter and three LucR.IRES reporter constructs chosen for further analysis are denoted by a closed diamond (♦). (A, B) IAPV, Israeli acute paralysis virus; CrPV, cricket paralysis virus; HCV, hepatitis C virus; i, IRES. Color images available online at www.liebertpub.com/aid
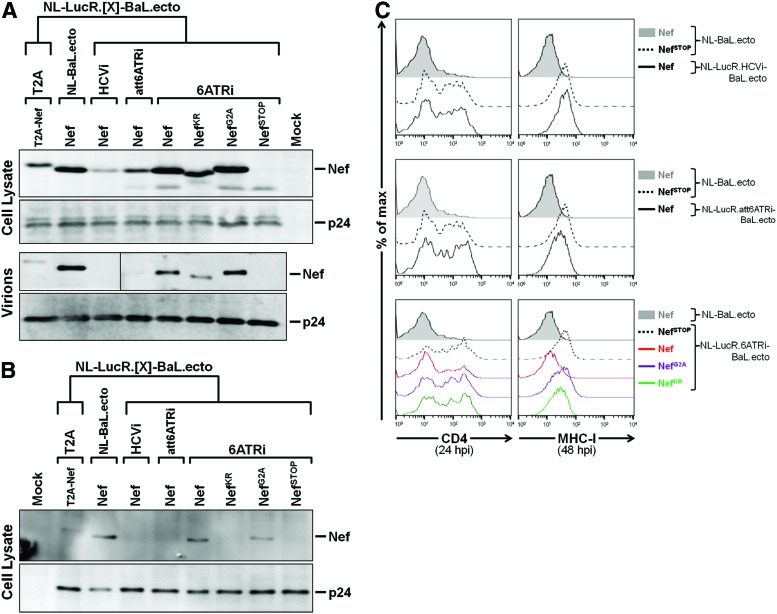
FIG. 4.
Similar expression and function of Nef in J2574-R5 cells infected with LucR.6ATRi reporter or nonreporter viruses. (A) Western blot evaluation of Nef levels (with anti-Nef mAb clone EH1) in 293T cells (top) and virions (bottom) 48 hpi with LucR.T2A reporter (NL-LucR.T2A-BaL.ecto), LucR.HCVi reporter (NL-LucR.HCVi-BaL.ecto), LucR.att6ATRi reporter (NL-LucR.att6ATRi-BaL.ecto), LucR.6ATRi reporter (NL-LucR.6ATRi-BaL.ecto), and nonreporter (NL-BaL.ecto) proviral DNA. Mutations that abrogate either myristoylation (NefG2A), basic residue-mediated membrane association (NefKR), or expression (NefSTOP) of Nef were incorporated into the LucR.6ATRi reporter background and were also included. Unaltered (wild-type) Nef is indicated as “Nef.” Replica blots were prepared in parallel and probed with anti-p24 (Gag) to demonstrate equivalent transfection efficiencies and virion production. (B, C) Evaluation of expression (B) and function (C) of Nef in J2574-R5 after infection with 293T transfection-derived reporter and nonreporter viruses prepared in (A). As in Fig. 1, samples were normalized to the number of infected cells before analysis by Western blot. (B) Western blot evaluation of Nef levels in J2574-R5 cells 48 hpi. Replica blots were prepared in parallel and probed with anti-p24 (Gag) to demonstrate loading of equivalent numbers of infected cells. (C) Flow cytometric evaluation of Nef function, as measured by CD4 (left panels) and MHC-I (right panels) down-modulation on infected (GFP+) J2574-R5 cells at 24 and 48 hpi, respectively. Levels of CD4 and MHC-I down-regulation for NL-LucR.HCVi-BaL.ecto, NL-LucR.att6ATRi-BaL.ecto, and NL-LucR.6ATRi-BaL.ecto reporter viruses were compared to nonreporter virus (NL-BaL.ecto), containing wild-type Nef (filled histograms), and either NL-BaL.ecto or NL-LucR.6ATRi-BaL.ecto containing the abrogated Nef expression mutation (NefSTOP) (dashed histogram). Color images available online at www.liebertpub.com/aid
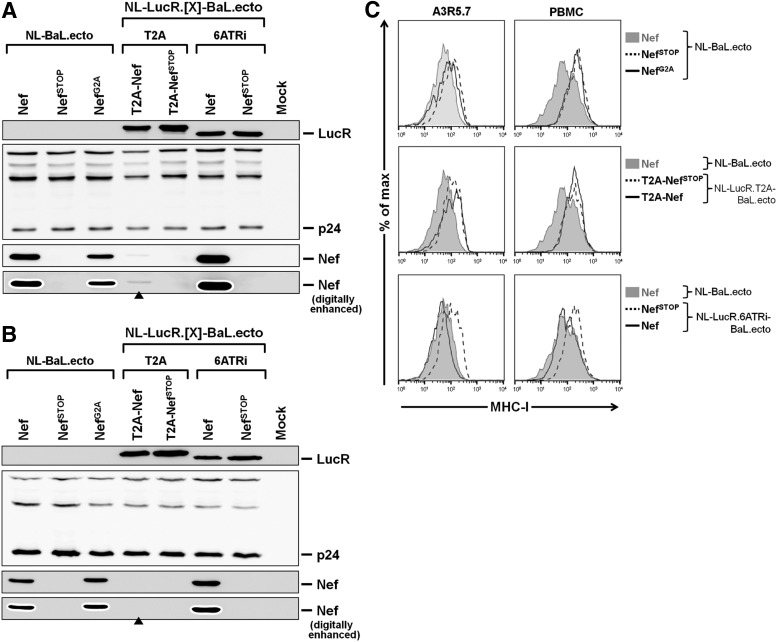
FIG. 5.
Nef expression and function are preserved in A3R5.7 cells and PBMCs infected with LucR.6ATRi reporter virus. (A–C) Evaluation of expression (A, B) and function (C) of Nef in the T cell line A3R5.7 and PBMCs after infection with 293T transfection-derived (TD) LucR.T2A reporter (NL-LucR.T2A-BaL.ecto), LucR.6ATRi reporter (NL-LucR.6ATRi-BaL.ecto), and nonreporter (NL-BaL.ecto) viruses. Mutations that abrogate Nef expression (NefSTOP) were incorporated into each background. An abrogated Nef myristoylation mutant (NefG2A) was also included in the NL-BaL.ecto background. Unaltered nonmutated (wild-type) Nef is indicated as “Nef.” A3R5.7 cells or PBMCs infected with HIV-1 were identified by intracellular p24 staining in order to normalize the number of infected cells analyzed by Western blot, and for gating on HIV-1-infected (p24+) cells for flow cytometric analysis. (A, B) Western blot evaluation of Nef (lower panels, with anti-Nef mAb clone EH1) and LucR (upper panel) levels in A3R5.7 cells (A) or PBMCs (B) 96 hpi with TD LucR.T2A or LucR.6ATRi reporter and nonreporter viruses. For the evaluation of Nef, original and digitally enhanced images are presented in parallel to illustrate significantly reduced (A) or absent (B) Nef levels (see closed arrowheads, ▲). Replica blots were prepared in parallel and probed with anti-p24 (Gag) (middle panel) to demonstrate loading of equivalent numbers of infected cells. (C) Flow cytometric evaluation of Nef function, as measured by MHC-I down-modulation on infected (p24+) A3R5.7 cells (left panels) or PBMCs (right panels) 96 hpi. Levels of MHC-I down-regulation for NL-LucR.T2A-BaL.ecto and NL-LucR.6ATRi-BaL.ecto were compared to both nonreporter virus (NL-BaL.ecto), containing wild-type Nef (filled histograms), and either NL-LucR.T2A-BaL.ecto or NL-LucR.6ATRi-BaL.ecto containing the abrogated Nef expression mutation (NefSTOP) (dashed histogram). Additionally, levels of MHC-I down-regulation for nonreporter virus encoding the NefG2A mutation were compared to nonreporter virus containing wild-type Nef (filled histograms) and the NefSTOP mutation (dashed histogram).
A3R5/Env-IMC-LucR neutralization assay
To analyze neutralization of HIV-1 infection in A3R5.7 cells, we followed the standardized protocol essentially as described.10 Briefly, appropriate volumes of 293T transfection-derived virus (NL-LucR.T2A-BaL.ecto or NL-LucR.6ATRi-BaL.ecto) that would yield approximately 1.5×105 LucR RLU under the sampling conditions were added to each well of a 96-well plate with different bNAbs (see below) diluted in a series of eight 3-fold dilutions from starting concentrations of either 25 or 5 μg/ml. After incubation for 45–90 min at 37°C in 96-well plates, 9×104 A3R5.7 cells (in 100 μl) were added to each well in a final volume of 250 μl. After incubation for 96 h at 37°C, 125 μl of medium was removed from each well and 30 μl of 5× Renilla Luciferase Assay Lysis Buffer (Promega) was added (for a final concentration of 1×) directly to each well containing cells. Samples were frozen at −80°C until use. At that point, samples were brought to room temperature and 20 μl of each sample was analyzed for LucR activity, as described above. Percent neutralization was calculated as previously described.8
The 2F5, 4E10, IgG1b12, PG9, PG16, and 2G12 bNAbs were obtained from Polymun GmbH (Vienna, Austria). CH01 and VRC-CH31 were manufactured by Catalent (Madison, WI) with permission from the Center for HIV/AIDS Vaccine Immunology (CHAVI). 2F5 and 4E10 are directed against the HIV-1 gp41 membrane-proximal external region (MPER).53,54 IgG1b12 (b12) and VRC-CH31 are directed against the HIV-1 gp120 CD4-binding site55,56; CH01, PG9, and PG16 are directed against the HIV-1 gp120 V1-V2 conformational epitope56–59; and 2G12 is directed against a distinct HIV-1 gp120 epitope.60
Results
Nef expression and function in LucR.T2A reporter viruses
Nef is a 27-kDa myristoylated protein that is not essential for HIV-1 replication in cell culture (reviewed in Foster and Garcia61). In vivo, the maintenance of high viral loads and the progression to AIDS appear to require functional Nef.62,63 Among its many functions, Nef has been shown to down-regulate CD4 and MHC-I, modulate signal transduction pathways, and enhance viral infectivity (reviewed in Foster and Garcia61). Down-modulation of CD4 and MHC-I as well as incorporation of Nef into virus particles require Nef association with the plasma membrane.38,50,64 Membrane localization is mediated by complex processes involving features within the N-terminal anchor region of Nef: N-terminal myristoylation and a cluster of basic amino acids.38,50
Our previously described LucR.T2A reporter virus strategy entailed expression of LucR and Nef from a bicistronic mRNA in which translation of Nef is linked in-frame to LucR with the T2A peptide, which results in ribosome skipping of the last peptide bond at the C-terminus of the T2A peptide that leaves the last T2A-encoded amino acid residue (Pro) to be added to the N-terminus of Nef. In addition, two additional residues (Ser-Arg) following the Pro are introduced from translation of a 6-nt sequence encoding an XbaI restriction site engineered in the vector's design. Thus, the LucR.T2A strategy may lead to impairment of this modified Nef's (“T2A-Nef”) functions and/or viral incorporation secondary to loss of the N-terminal myristoylation signal (i.e., N-terminal Gly)36, 37 (Fig. 1A). We have previously shown that for LucR.T2A reporter viruses (NL-LucR.T2A, NL-LucR.T2A-BaL.ecto, and NL-LucR.T2A-SF162.ecto), levels of Nef expression in 293T cells following transfection of proviral DNA were somewhat lower than in parental nonreporter viruses (NL4-3, NL-BaL.ecto, and NL-SF162.ecto)8; however, Nef function in T cells infected with these reporter viruses has not been analyzed.
Here, we further evaluated the expression and function of T2A-Nef in transfected 293T cells and in infected CD4+ T cell lines and PBMCs. We compared the LucR.T2A reporter viruses, NL-LucR.T2A and NL-LucR.T2A-BaL.ecto, with their parental nonreporter viruses, NL4-3 and NL-BaL.ecto, respectively. The R5-tropic virus expressing the BaL reference env was chosen as a stand-in for the large panel (n>120) of Env-IMC-LucR.T2A encoding env genes from heterologous HIV-1 strains4,8,11,17 (also Jones et al., unpublished observations). Additional nef mutations were analyzed (Fig. 1A) in both LucR.T2A reporter (NL-LucR.T2A-BaL.ecto) and nonreporter (NL-BaL.ecto) backbones, including those that abrogate either myristoylation (NefG2A), membrane association (NefKR), or expression (NefSTOP) of Nef. As noted above, the T2A motif followed by an XbaI site added three extra amino acids (Pro-Ser-Arg) to the N-terminus of Nef (T2A-Nef). Therefore, two additional mutants were created, T2APSD-Nef and T2APSS-Nef, mutating the basic Arg (R) residue to either Asp (D) or Ser (S), in order to evaluate (1) whether the basic residue in T2A-Nef may, at least in part, functionally compensate for the loss of myristoylation through provision of an additional residue available for membrane association, and (2) whether removal of this additional charged residue would affect Nef function in the T2A context.
First, we compared Nef expression in 293T cells transfected with LucR.T2A reporter (NL-LucR.T2A and NL-LucR.T2A-BaL.ecto) and nonreporter (NL4-3 and NL-BaL.ecto) constructs by Western blot (Fig. 1B). Consistent with previous results,8 T2A-mediated Nef expression was reduced compared to Nef expression from matched nonreporter constructs and the slightly higher molecular weight of T2A-Nef (Fig. 1B) was consistent with the three additional C-terminal amino acids (Fig. 1A). Replica blots probed with anti-p24 (Gag) mAb demonstrated proteolytic processing and similar levels of Gag expression (Fig. 1B). The NefG2A mutation did not adversely affect the Nef expression level in the context of NL-BaL.ecto virus; however, the NefKR mutation reproducibly reduced Nef expression, consistent with the findings of Bentham et al.50 None of the mutations analyzed in the LucR.T2A background had further adverse effect on the expression levels of Nef (Fig. 1B). As expected, Nef expression was undetectable for constructs containing NefSTOP (Fig. 1B).
We next evaluated Nef expression and function, as measured by down-modulation of CD4 (at 24 h) and MHC-I (at 48 h), in infected J2574-R5 cells (Fig. 1C and D). These Jurkat-derived cells express GFP upon HIV-1 infection, which we exploited to normalize the number of infected cells analyzed by Western blot, and for gating on HIV-1-infected (GFP+) cells using flow cytometric analysis. The levels of wild-type Nef versus Nef mutants (i.e., NefG2A and NefKR) expressed from nonreporter viruses (NL-BaL.ecto) in J2574-R5 cells at 48 hpi (Fig. 1C) were similar to those observed in 293T cells following transfection of the respective proviral DNAs (Fig. 1B). Unexpectedly and importantly, the same level of T2A-Nef expression observed in the context of 293T cells transfected with proviral DNA (NL-LucR.T2A and NL-LucR.T2A-BaL.ecto) (Fig. 1B) was not exhibited upon productive infection of J2574-R5 cells with these LucR.T2A reporter viruses (Fig. 1C). Rather, infection of J2574-R5 cells with LucR.T2A reporter viruses resulted in severely reduced Nef levels compared to cells infected with matched, wild-type nef-encoding nonreporter viruses (Fig. 1C).
The effect of Nef on CD4 surface expression is best assessed early in infection since it will otherwise become increasingly obscured by down-modulation mediated by Vpu and Env. We assessed CD4 down-modulation at 24 hpi, when GFP expression in infected cells reached levels sufficient for reliable gating (Fig. 1D, insets). At this time point, infection with nonreporter viruses (NL4-3 and NL-BaL.ecto) encoding wild-type Nef resulted in nearly complete loss of surface CD4 expression. CD4 down-regulation was significantly impaired for NL-BaL.ecto encoding NefSTOP virus (Fig. 1D, left upper panel). As expected, abrogating Nef myristoylation (NefG2A) did not affect Nef-dependent CD4 down-modulation, whereas abrogating Nef membrane association (NefKR) resulted in an intermediate phenotype (Fig. 1D, left upper panel), despite the low level of NefKR observed by Western blot analysis (Fig. 1C). However, and in line with the nearly complete absence of detectable T2A-Nef in J2574-R5 cells infected with LucR.T2A reporter viruses by Western blot (Fig. 1C), Nef-dependent down-regulation of CD4 was not observed with T2A-Nef, T2A-NefSTOP, or with any of the T2A-Nef mutants (i.e., KR, PSD, or PSS) (Fig. 1D, left middle and lower panels). Interestingly, we observed an overall lesser degree of non-Nef-mediated (i.e., Vpu-mediated) CD4 down-regulation for LucR.T2A reporter viruses containing T2A-NefPSD and T2A-NefPSS (Fig. 1D, left lower panel); however, this may be related to very low infection levels, and thus possibly an earlier stage of the replication cycle, at the time of analysis.
We also assessed Nef effects on surface MHC-I in infected J2574-R5 cells at 48 hpi. As expected, infection with nonreporter viruses (NL4-3 and NL-BaL.ecto) expressing wild-type Nef resulted in lower MHC-I surface levels than infection with viruses lacking detectable Nef expression (i.e., NefSTOP) (Fig. 1D, right upper and middle panels) and similar (only slightly lower) MHC-I surface levels as seen in mock-infected control cells (see Fig. 6, left upper panel). Down-modulation of MHC-I was either completely inhibited by abrogating Nef myristoylation (NefG2A) or impaired by mutations that affect Nef membrane association (NefKR) (Fig. 1D, right upper panel). Down-modulation of surface MHC-I expression was also not observed on J2574-R5 cells infected with LucR.T2A reporter viruses (NL-LucR.T2A and NL-LucR.T2A-BaL.ecto) encoding T2A-Nef or its mutants (Fig. 1D, right middle and lower panels). This result was again consistent with the very low or undetectable levels of Nef in these infected cells (Fig. 1C). Thus, surface expression of MHC-I in LucR.T2A reporter virus-infected cells was unlike that of nonreporter virus-infected cells expressing wild-type Nef; rather, it was similar to that detected on cells infected with nonreporter viruses with altered (NefG2A or NefKR) or abrogated (NefSTOP) expression of Nef (Fig. 1D).
FIG. 6.
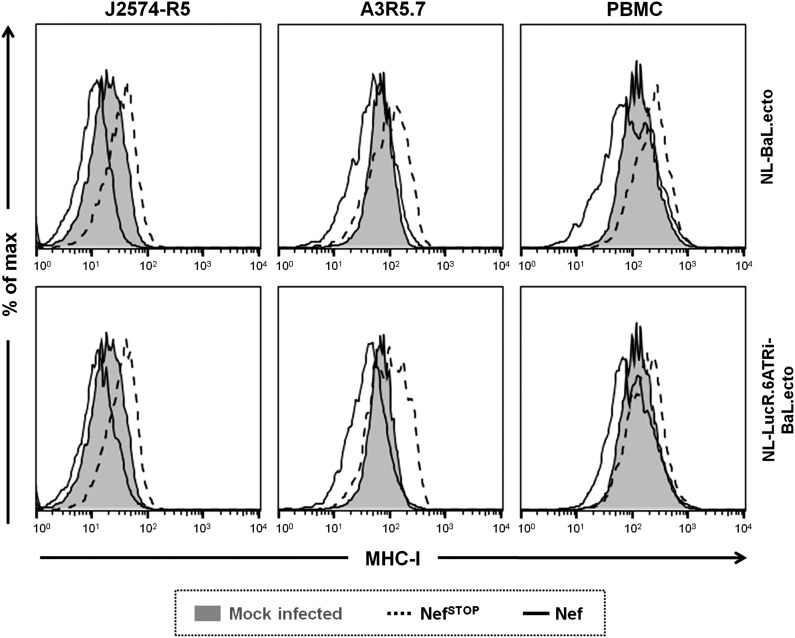
Compared to virus-naive cells, MHC-I levels increase or decrease after infection of T cells with viruses harboring abrogated or functional Nef expression, respectively. The level of MHC-I expression (HLA-A/B/C) was analyzed by flow cytometry in T cell lines, J2573-R5 and A3R5.7, and pooled donor PBMCs depleted of CD8+ cells with 293T transfection-derived nonreporter (NL-BaL.ecto) and LucR.6ATRi reporter (NL-LucR.6ATRi-BaL.ecto) viruses encoding either unaltered functional Nef (solid histogram) or NefSTOP (dashed histogram). Cells from virus-naive (mock-infected) cultures were included to provide normal MHC-I levels (filled histograms). A3R5.7 cells or PBMCs productively infected with HIV-1 were identified by intracellular p24 staining at 96 hpi (as in Fig. 5), and productively infected J2574-R5 cells were identified by GFP expression at 48 hpi (as in Fig. 1). Data from a representative experiment are shown.
Derivation of novel LucR.IRES reporter viruses
Given that both Nef expression and function, as measured by down-modulation of CD4 and MHC-I, were impaired in the context of LucR.T2A reporter virus infection of Jurkat T cells (Fig. 1), as well as A3R5.7 cells and PBMCs (see below, Figs. 5 and 6), we investigated alternative reporter virus strategies that utilized an IRES bicistronic reporter system. We and others have found the performance of previous EMCV IRES HIV-1 reporter viruses (e.g., NLENG1i)3,27–29 to be suboptimal with respect to replication, stability of the EGFP reporter gene (personal observations), and supraphysiological overexpression of Nef.8 Therefore, we examined several different IRES elements, including eight iterations of an EMCV IRES element similar to that found in the previous NLENG1i virus strategy. These elements were engineered in place of the T2A element within the LucR.T2A reporter virus (Fig. 2A).
Four well-characterized IRES elements from the viruses, IAPV, CrPV, HCV, and EMCV (Fig. 2A and Supplementary Fig. S1), were chosen based on a number of characteristics, with particular attention given to the size and previously reported translational activity levels. The 5′-UTR IRES from HCV was of moderate size (324 nt) and activity.65 In contrast, the smaller IGR IRES elements from the insect viruses IAPV and CrPV have evolved to function in a bicistronic manner (as opposed to the 5′-UTR),66,67 similar to the design of our reporter virus technology concept. In addition, these were the smallest of the elements (219 and 190 nt, respectively) tested, and we predicted that they would have the least negative impact on reporter gene retention and stability.
The EMCV IRES is among the best described IRES elements. To overcome limitations observed in the context of NLENG1i-based HIV-1 strains, we engineered the EMCV IRES with eight different variations that have been described in the literature. The first variant was based on the “wild-type” (A)6 (or “6A”) versus (A)7 (or “7A”; found in pIRES-EGFP) bifurcation loop, which leads to known differences in translation efficacy.47 Given the relatively large size of the preferred “consensus” EMCV IRES found in a number of commercial plasmids, we derived both “full-length” and minimal, or “truncated” (TR), forms (576 and 435 nt, respectively).47 All four of these variants were configured to utilize the preferred native “ATG” in EMCV as the start codon for Nef. Lastly, each of the above variants was also designed to include a 29-nt spacer or “attenuator”49 at the 3′ end of the IRES (in place of the native EMCV “ATG” codon), just before the ATG codon of Nef; these are referred to as the “attenuated” (att) variants (as inclusion of the spacer is expected to result in reduced Nef expression compared to EMCV IRES without it).47 Overall, eight different EMCV IRES elements were created and designated as described in Materials and Methods (e.g., “6Ai” indicates the full-length EMCV IRES with the 6A bifurcation loop, and “att7ATRi” indicates the truncated EMCV IRES with the 7A bifurcation loop and attenuator spacer) (see also Supplementary Fig. S1).
Infectivity of 293T transfection-derived LucR.IRES reporter viruses
The 11 different 293T transfection-derived LucR.IRES reporter viruses showed infectious titers (IU/ml) and relative infectivity (IU/ng p24) similar to that of LucR.T2A reporter (NL-LucR.T2A-BaL.ecto) and nonreporter (NL-BaL.ecto) viruses in TZM-bl cells (data not shown). Furthermore, for each virus, linear responses of both cell-encoded LucF and virus-encoded LucR activities were observed up to an MOI of 0.5 following infection of TZM-bl cells (Supplementary Fig. S2A and B). As expected, infection with LucR.IRES reporter viruses resulted in nearly identical LucF values compared to cognate LucR.T2A reporter and nonreporter viruses, indicating similar cell-encoded LTR transactivation in the TZM-bl cell line (Supplementary Fig. S2A). Among all LucR.IRES reporter viruses, RLU values of LucR activity fell within a 3-fold boundary of RLU values obtained with the LucR.T2A reporter virus (Supplementary Fig. S2B). In A3R5.7 cells (which together with LucR.T2A reporter viruses have found application for sensitive detection of neutralizing immune responses9,10), linear responses of LucR activity were also observed for all LucR.IRES reporter viruses, which were once again similar to and within the 3-fold range of LucR.T2A reporter virus RLU values (Supplementary Fig. S2C).
Nef expression in 293T cells following transfection with LucR.IRES reporter proviral DNA
Expression of Nef in 293T cells transfected with the panel of LucR.IRES reporter constructs was compared to that from cognate LucR.T2A reporter (NL-LucR.T2A-BaL.ecto) and nonreporter (NL-BaL.ecto) constructs (Fig. 2B). We did not detect IAPVi-mediated and CrPVi-mediated expression of Nef. HCVi-mediated Nef expression was detectable but lower than T2A-mediated expression, which, as seen before (Fig. 1B and Edmonds et al.8), was reduced compared to nonreporter virus (Fig. 2B, upper panel). As hypothesized, we observed a range of Nef expression levels from the eight EMCV IRES variants (Fig. 2B, upper panel). Supraphysiological levels of Nef expression were seen with the LucR.6Ai and LucR.7Ai reporter constructs, comparable to expression levels detected for the NLENG1i reporter construct (data not shown and Edmonds et al.8); as expected, the “7A” variants generally demonstrated reduced expression compared to their “6A” counterparts (i.e., 6Ai vs. 7Ai, 6ATRi vs. 7ATRi, att6Ai vs. att7Ai, and att6ATRi vs. att7ATRi); the shorter “TR” variants resulted in lower Nef levels than the “full-length” versions (i.e., 6Ai vs. 6ATRi, 7Ai vs. 7ATRi, att6Ai vs. att6ATRi, and att7Ai vs. att7ATRi), and “attenuated” (att) variants resulted in lower Nef expression than their nonattenuated counterparts (i.e., att6Ai vs. 6Ai, att7Ai vs. 7Ai, att6ATRi vs. 6ATRi, and att7ATRi vs. 7ATRi). Furthermore, products of Nef proteolytic cleavage by HIV-1 protease, a process of unknown relevance that predominantly occurs in virions,68 are also detectable in provirally transfected 293T cells69 (and O. Fackler, personal communication), and become more prominent with increasing, and especially supraphysiological levels of Nef expression. As before, the proteolytic processing of Gag was normal and equal across samples, but supraphysiological Nef levels seem to have a moderate effect on Gag processing (Fig. 2B, lower panel). Equivalent expression levels of LucR were seen for the LucR.T2A and all LucR.IRES reporter constructs (Fig. 2B, middle panel).
Based on Nef expression levels most closely matching those of nonreporter or LucR.T2A reporter viruses, and avoiding supraphysiological Nef levels, we selected a subset of three LucR.IRES reporter viruses to evaluate further: LucR.HCVi (NL-LucR.HCVi-BaL.ecto), LucR.6ATRi (NL-LucR.6ATRi-BaL.ecto), and LucR.att6ATRi (NL-LucR.att6ATRi-BaL.ecto).
Replication kinetics and stability of LucR expression for LucR.IRES reporter viruses in PBMCs
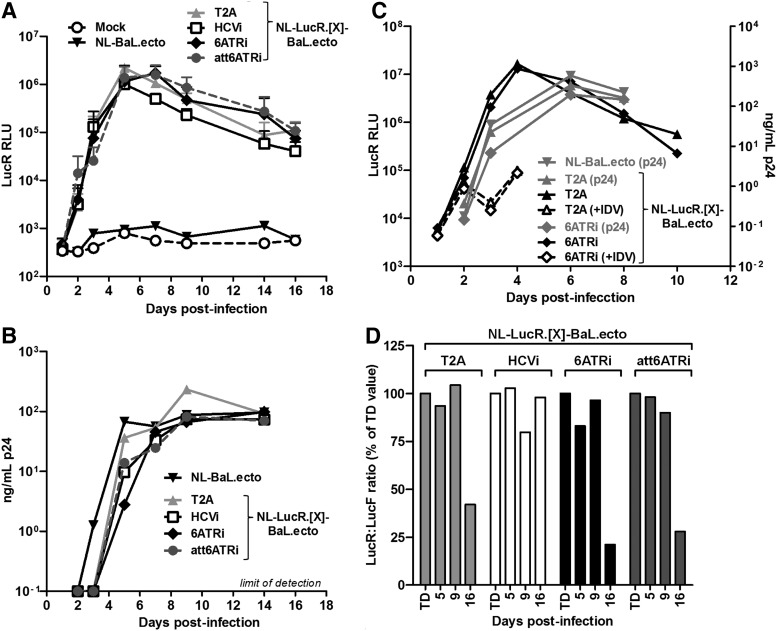
The addition of exogenous sequence into the HIV-1 genome can have negative effects on viral replication, presenting as severely delayed replication kinetics and/or rapid selection of viruses defective for the expression of the exogenous reporter gene.70–72 Thus, we tested the selected subset of LucR.IRES reporter viruses for replication fitness by measuring p24 antigen production and LucR activity over time after infection at a low MOI of PHA-P-stimulated pooled donor PBMCs depleted of CD8+ cells (Fig. 3). Measurement of p24 antigen production (Fig. 3B) confirmed that the LucR.T2A reporter virus (NL-LucR.T2A-BaL.ecto) was replication competent with slightly delayed replication kinetics compared to the nonreporter virus (NL-BaL.ecto), in line with our previous results.8 The three tested LucR.IRES reporter viruses were replication competent similar to the LucR.T2A virus (Fig. 3B). The kinetics of LucR activity (Fig. 3A), as a sensitive and robust measure of proviral gene expression and viral replication, were also highly similar in PBMCs infected with LucR.T2A reporter virus and all three LucR.IRES reporter viruses. In an independent experiment, inclusion of the protease inhibitor, idinavir (IDV), which restricts infection to a single round, validated that the ∼100-fold increase in LucR activity in the absence of IDV beyond day 2 postinfection was attributable to virus replication and spread (Fig. 3C).
FIG. 3.
Replication kinetics and stability of Renilla luciferase (LucR) expression for LucR.IRES reporter viruses in peripheral blood mononuclear cells (PBMCs). (A, B) Replication kinetics for a subset of LucR.IRES reporter viruses, LucR.HCVi (NL-LucR.HCVi-BaL.ecto), LucR.att6ATRi (NL-LucR.att6ATRi-BaL.ecto), and LucR.6ATRi (NL-LucR.6ATRi-BaL.ecto), was compared to cognate LucR.T2A reporter (NL-LucR.T2A-BaL.ecto) and nonreporter (NL-BaL.ecto) viruses. Replication kinetics was measured by LucR activity (A) and p24 antigen production (B) over time after infection at a low MOI (MOI of 0.05 TZM-bl IU, corresponding to an approximate MOI of 0.001 PBMC IU) of phytohemagglutinin (PHA)-P-stimulated pooled donor PBMCs depleted of CD8+ cells. In (B), the limit of p24 detection (100 pg/ml) is indicated. (C) PBMCs were infected with LucR.T2A and LucR.6ATRi reporter viruses, as above, but in the absence (solid lines) and presence (dashed lines) of the protease inhibitor, indinavir (IDV), and viral replication was monitored by LucR expression (left y-axis, black lines) and p24 (right y-axis, gray lines). p24 production for nonreporter virus is also shown for comparison. (D) To assess the stability of LucR expression from LucR.T2A and LucR.IRES reporter viral genomes over multiple cycles of virus replication, we collected culture supernatants from the infected PBMC cultures at days 5, 9, and 16. TZM-bl cells were then infected with the PBMC infection-derived, as well as the 293T transfection-derived (TD), virus supernatants, as detailed in Materials and Methods. The stability of LucR expression relative to infectivity was calculated as the ratio of virus-encoded LucR RLU to cell-encoded LucF RLU (which represents a measure of the overall infectivity of the respective virus stocks at each time point) and compared to the LucR:LucF ratio obtained with TD supernatants.
Next, we examined whether LucR expression from the viral genome was stably maintained over multiple cycles of virus replication, essentially as previously described.8 For this, 293T transfection-derived virus and supernatants collected at days 5, 9, and 16 from the infected PBMC cultures described above were used to infect TZM-bl cells as detailed in Materials and Methods. Since activity of cell-encoded LucF represents a measure of the overall infectivity of the respective virus stocks at each time point, we calculated the ratio of virus-encoded LucR RLU to LucF RLU for PBMC supernatants and compared it to the LucR:LucF ratio obtained with transfection-derived virus. As illustrated for a representative experiment in Fig. 3D, nearly full retention of LucR activity was observed over the 16-day course of replication of the LucR.HCVi reporter virus. Reduction of LucR activity was seen for LucR.6ATRi and LucR.att6ATRi reporter viruses as of day 16, similar to what was observed for the LucR.T2A reporter virus (NL-LucR.T2A-BaL.ecto) here and previously,8 as well as with two additional LucR.T2A reporter virus constructs with different env genes.12
Nef expression and function in J2574-R5 T cells infected with LucR.IRES reporter viruses
Given that the results for replication kinetics and LucR reporter gene stability in PBMCs were similar for LucR.T2A virus as compared to the subset of three LucR.IRES viruses, we further compared all of them with respect to Nef virion incorporation and Nef expression and function in T cells. In addition, since we had observed the most physiological Nef levels in 293T cells for the LucR.6ATRi reporter virus, we also introduced into this construct the functional nef mutations described above (see Fig. 1A) that abrogate myristoylation (NefG2A), membrane association (NefKR), or expression (NefSTOP) of Nef protein to use as controls.
Nef levels in 293T cells following transfection with LucR.T2A (NL-LucR.T2A-BaL.ecto), LucR.HCVi (NL-LucR.HCVi-BaL.ecto), LucR.6ATRi (NL-LucR.6ATRi-BaL.ecto), and LucR.att6ATRi (NL-LucR.att6ATRi-BaL.ecto) reporter constructs (Fig. 4A) were consistent with earlier results (Figs. 1B and 2B). The nef mutations, when analyzed in the context of the LucR.6ATRi reporter backbone, had effects on expression (Fig. 4A) similar to those observed when analyzed in the nonreporter (NL-BaL.ecto) background (Fig. 1B). Incorporation of unaltered Nef and T2A-Nef into virions was reduced disproportionally for those virus constructs that resulted in lower-than-wild-type cellular Nef expression levels (Fig. 4A). In this regard, Nef was substantially decreased in the LucR.T2A reporter virions compared with nonreporter virions, and Nef was absent or significantly decreased in virions for LucR.HCVi and LucR.att6ATRi reporter viruses, respectively (Fig. 4A).
In contrast, 6ATRi-mediated expression of Nef led to similar (to slightly reduced) levels in virions, as compared with the nonreporter virus (Fig. 4A). Abrogation of Nef myristoylation (NefG2A) alone did not seem to affect Nef incorporation into virions produced from 293T cells, while the NefKR mutant resulted in impaired packaging (Fig. 4A). Probing with anti-p24 (Gag) mAb demonstrated that the proteolytic processing of Gag was normal and that similar amounts of each of the viruses had been produced and analyzed (Fig. 4A). In summary, the Nef virion incorporation phenotype of LucR.6ATRi reporter virus was essentially the same as that of wild-type nonreporter virus.
Similar to earlier results (Fig. 1C), the level of Nef detected in J2574-R5 cells infected with the 293T transfection-derived LucR.T2A reporter virus was significantly less compared to that detected for nonreporter virus (NL-BaL.ecto) (Fig. 4B). Interestingly, in contrast to 293T cell lysates, Nef was nearly absent in J2574-R5 cells infected with the LucR.HCVi and LucR.att6ATRi reporter viruses. On the other hand, in J2574-R5 cells infected with the LucR.6ATRi reporter virus, Nef was expressed at levels similar to wild-type nonreporter virus, with the NefG2A mutation imparting only moderate effects on levels of Nef, as would be expected (Fig. 4B). Of note, in cells infected with the LucR.6ATRi reporter virus, the degree of MHC-I down-modulation (Fig. 4C) mediated by Nef essentially reached levels obtained following infection with nonreporter virus, as predicted based on the expression level of this unaltered Nef. As expected, in cells infected with LucR.6ATRi reporter viruses harboring nef functional mutations, MHC-I down-modulation was abrogated (NefG2A or NefSTOP) or impaired (NefKR) (Fig. 4C). Consistent with the near absence of detectable Nef by Western blot, we did not observe down-modulation of MHC-I expression in J2574-R5 cells infected with the LucR.HCVi reporter virus (Fig. 4C).
Despite very low Nef levels in cells infected with the LucR.att6ATRi reporter virus (Fig. 4B), MHC-I expression was partially down-modulated (Fig. 4C). The effect of Nef expressed from the various LucR.IRES proviruses on CD4 down-regulation was analyzed at 24 hpi (Fig. 4C), as described above. As seen for MHC-I, in cells infected with the LucR.6ATRi reporter virus, the magnitude of CD4 down-modulation (Fig. 4C) mediated by Nef was similar to that observed following infection with nonreporter virus. We attribute the small amount of remaining surface CD4 expression to the observation that induced GFP levels in cells infected with the LucR.6ATRi reporter virus lagged slightly behind those of nonreporter virus (NL-BaL.ecto)-infected cells (data not shown), possibly indicating a slightly earlier stage of infection for some reporter virus-infected cells, and thus a lesser degree of non-Nef-mediated (e.g., Vpu-mediated) CD4 down-modulation. By 48 hpi, a difference in GFP levels in infected cells was no longer observed.
Taken together, our findings suggest that the absence of Nef-mediated CD4 and MHC-I down-regulation in cells infected with LucR.T2A reporter viruses may be a consequence of low Nef levels in combination with defective Nef function (i.e., the presumed lack of myristoylation). Importantly, results obtained with the LucR.6ATRi reporter virus indicated that this novel strategy succeeded in overcoming the limitation of LucR.T2A reporter constructs with regard to Nef function while preserving several other important features (e.g., replication capacity, reporter stability, and sensitive detection of infection), which have made the LucR.T2A reporter viruses such valuable tools for a number of applications.
Nef expression and function in A3R5.7 and PBMCs infected with LucR.IRES reporter viruses
To further investigate the utility of the LucR.6ATRi reporter approach, we proceeded with analyzing Nef function after infection of A3R5.7 cells (a representative CCR5+ T cell line chosen for its role in HIV-1 NAb neutralization assays)9–11,17 and PBMCs (as a model of primary HIV-1 target cells). We infected both cell types with a subset of 293T transfection-derived LucR.T2A and LucR.6ATRi reporter viruses with and without functional nef mutations and assessed Nef expression and MHC-I down-modulation at 96 hpi (Figs. 5 and 6). Staining for intracellular HIV-1 p24 allowed us to quantify the number of infected cells by flow cytometry in order to normalize samples for Western blot and to gate on HIV-infected (p24+) cells for flow cytometric analysis of MHC-I surface expression.
As seen in J2574-R5-infected cells (Fig. 4B), the levels of Nef in A3R5.7 cells and PBMCs infected with LucR.6ATRi reporter virus (NL-LucR.6ATRi-BaL.ecto) were similar to nonreporter virus (NL-BaL.ecto) (Fig. 5A and B). However, Nef expression from the LucR.T2A reporter virus (NL-LucR.T2A-BaL.ecto) was significantly reduced in A3R5.7 (Fig. 5A, see arrowhead) and undetectable in PBMCs (Fig. 5B, see arrowhead). The consistent lack of detectable Nef expression mediated by the T2A molecular strategy in infected T cells is noteworthy, but further investigation was outside the scope of the current study.
In agreement with Nef expression data, and illustrated in Fig. 5C, we did not observe down-regulation of MHC-I expression on A3R5.7 cells or PBMCs that were infected with LucR.T2A reporter viruses (Fig. 5C, middle panels) or with any of the viruses with abrogated Nef (NefSTOP) expression. Importantly, in both A3R5.7 cells and PBMCs infected with the LucR.6ATRi reporter virus, MHC-I expression levels were similar to those in cells infected with nonreporter virus encoding wild-type Nef (Fig. 5C), as we had seen in J2574-R5 cells (Fig. 4C).
Interestingly, we observed in PBMCs, as well as in the two T cell lines tested, that infection with “nef-minus” (NefSTOP) or functionally Nef-defective (NefG2A or NefKR) viruses reproducibly led to an increase in MHC-I levels compared to “mock-infected” cells in a parallel culture [with the increase in mean fluorescence intensity (MFI) ranging from 1.3- to 1.8-fold]. Infection by cognate viruses expressing fully functional Nef (i.e., NL-BaL.ecto and NL-LucR.6ATRi-BaL.ecto) led to a modest degree of down-regulation of MHC-I compared to mock-infected cells (to a range of 0.56- to 0.75-fold). Thus, the effect of Nef on MHC-I was most apparent when comparing MFI between cells productively infected with virus encoding wild-type Nef versus virus encoding NefSTOP (Fig. 6). Of note, when we gated on uninfected (i.e., p24− or GFP−) cells in a virus-exposed culture harboring productively infected cells, MHC-I levels were elevated compared to virus-naive or “mock-infected” cultures, irrespective of the nef genotype (data not shown).
Comparison of LucR.T2A and LucR.6ATRi reporter viruses in the A3R5/Env-IMC-LucR neutralization assay
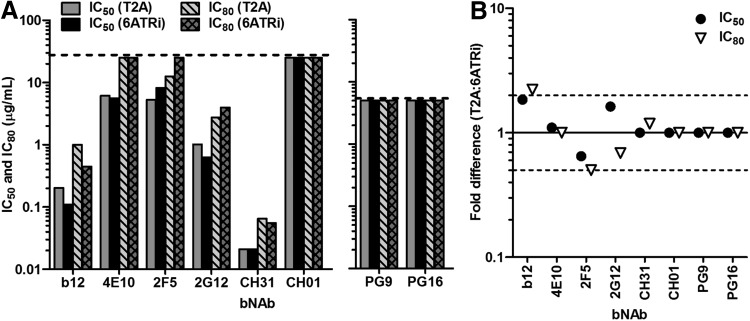
We previously developed a large panel of LucR.T2A reporter viruses with env genes from seven clades17 (also Jones et al., unpublished observations) that enables a number of immune monitoring approaches (e.g., of vaccine sera11) including the recently formally validated A3R5/Env-IMC-LucR neutralization assay.9,10 While this, or other T cell-based-NAb assays, does not require functional Nef, we wanted to ensure that the new LucR.6ATRi reporter virus (also collectively referred to as “Env-IMC-LucR.6ATRi”) performed similarly (“noninferior”) to its counterpart, the LucR.T2A reporter virus (i.e., “Env-IMC-LucR.T2A”). We recently conducted a carefully controlled neutralization tier phenotyping study of over 120 Env-IMC-LucR.T2A viruses (Jones et al., unpublished observations), including NL-LucR.T2A-BaL.ecto, which allowed us to test the performance of NL-LucR.6ATRi-BaL.ecto in parallel. Both viruses were similarly neutralized by the eight different bNAbs tested, according to IC50 and IC80 data (Fig. 7A). Neutralization curves for NL-LucR.T2A-BaL.ecto and NL-LucR.6ATRi-BaL.ecto essentially overlapped. The curve-based (Fig. 7A) and point-based (data not shown) IC50 and IC80 values for each respective bNAb/virus combination were closely matched. Importantly, the fold-difference between NL-LucR.T2A-BaL.ecto and NL-LucR.6ATRi-BaL.ecto IC50 values for each bNAb was very low (Fig. 7B, 2-fold range indicated), and well within the 3-fold range that was previously defined as acceptable for interexperimental variation of IC50 values for the same virus in the A3R5/Env-IMC-LucR neutralization assay.10
FIG. 7.
Equivalent performance of Env-IMC-LucR.T2A and Env-IMC-LucR.6ATRi in the A3R5/Env-IMC-LucR neutralization assay. Inhibition of NL-LucR.T2A-BaL.ecto and NL-LucR.6ATRi-BaL.ecto by the IgG1b12 (b12), 2G12, 2F5, 4E10, CH31, CH01, PG9, and PG16 bNAbs was determined on A3R5.7 cells, following the recently validated neutralization assay protocol, and utilizing reduction of LucR activity as a readout to determine 50% and 80% inhibitory concentrations (IC50 and IC80, respectively). (A) Curve-based IC50 and IC80 values obtained for each of the bNAbs against LucR.T2A and LucR.6ATRi reporter viruses. Dashed lines indicate the starting concentrations of each bNAb (5 μg/ml for PG9 and PG16; 25 μg/ml for all others), i.e., maximum achievable IC50 or IC80 concentration for the assay. (B) Fold-difference (ratio) between NL-LucR.T2A-BaL.ecto and NL-LucR.IRES-BaL.ecto (T2A:6ATRi) IC50 (or IC80) values for each bNAb. Dashed lines indicate a 2-fold range of IC50 values (note: a 3-fold range for interassay and intraassay variation was previously demonstrated to be acceptable and indicative of equivalent IC50 values in the validated A3R5/Env-IMC-LucR neutralization assay10).
Discussion
Previously we developed and described the replication-competent Env-IMC-LucR.T2A reporter virus technology8 that has demonstrated widespread use in a variety of applications including HIV-1 transmission studies,4,73 humanized mouse models,16 and several immunological assays,9,10,12–15,17,18,20 including measurement of NAb activity in sera from vaccinees participating in the RV144 trial.11 We also demonstrated the expression of Nef from the LucR.T2A-nef cassette comprised within LucR.T2A reporter virus after transfection of 293T cells with proviral DNA.8 However, we observed that T2A-mediated expression of Nef was consistently lower than Nef expression levels associated with nonreporter proviral DNA in transfected cells, and we thus predicted that this would lead to lower or altered Nef function. We also postulated that T2A-mediated expression of Nef may interfere with N-terminal myristoylation of Nef,36,37 thus altering or impairing certain Nef phenotypes and functions in infected cells.38
Mutational analysis by Welker et al. revealed that both myristoylation and an N-terminal cluster of basic amino acids were required for virion incorporation of Nef in infected CD4+ T cells.38 Herein, we now demonstrate that incorporation of Nef into virions produced in 293T cells was reduced for the LucR.T2A reporter virus to a degree greater than that expected from the reduced cellular expression levels alone, suggesting that T2A-Nef is indeed myristoylation impaired. Interestingly, the deleterious effects of the NefG2A myristoylation and NefKR mutants on virion incorporation of Nef in infected T cells described by Welker et al. were less pronounced in provirally transfected 293T cells.38 Interestingly and somewhat surprisingly, we observed that in T cell lines and PBMC-derived primary CD4+ T cells infected with LucR.T2A-containing viruses, detectable T2A-Nef protein was markedly decreased or absent, respectively, despite significant T2A-Nef detection in provirally transfected 293T cells. Consequently, Nef function, as measured by down-regulation of CD4 and MHC-I, was impaired accordingly in T cells infected with LucR.T2A reporter viruses.
It is not immediately clear what factors contributed to these phenomena. Our findings may indicate significant differences in gene expression from proviral plasmids following transfection compared to integrated provirus in infected cells, including alternative splicing or altered early-versus-late gene expression. They may reflect decreased mRNA stability, decreased protein stability (possibly due to N-terminal Nef modification as a result of T2A ribosome skipping), or decreased T2A ribosome skipping efficiency. Of note, however, we found strong LucR protein expression in T cell lines and PBMCs infected with LucR.T2A reporter viruses, and we did not observe abnormally large-sized LucR and/or Nef protein products by Western blot, as we would have expected in the case of diminished T2A ribosome skipping efficiency in primary cells. Instability of the likely nonmyristolyated form of Nef (T2A-Nef) in cells infected with the LucR.T2A reporter virus was also unlikely, as the NefG2A myristoylation mutant expressed from nonreporter virus produced normal amounts of Nef protein in transfected 293T cells and infected J2574-R5 T cells.
In this regard, decreased translation initiation of Nef following ribosome skipping of the last peptide bond at the C-terminus of T2A (resulting in an imbalance of LucR to Nef products) may play a role.33 Nevertheless, future studies will be required to fully understand the mechanism(s) for the impaired performance of the T2A approach in the context of infected cells. Such mechanistic studies, while intriguing, were outside of the scope of this project, which aimed at overcoming this unexpected limitation.
In an effort to provide an alternative reporter virus technology for applications that require functional Nef, we revisited an IRES-based approach and analyzed several different IRES elements engineered to replace the T2A in the LucR.T2A reporter virus. These comprised a panel of 11 novel HIV-1 reporter constructs, collectively referred to as LucR.IRES reporter HIV-1 or “Env-IMC-LucR.IRES,” analogous to the terminology “Env-IMC-LucR.T2A” we established elsewhere.4,8,11,12,17 In general, the titer and infectivity for 293T transfection-derived Env-IMC-LucR.IRES were similar to Env-IMC-LucR.T2A reporter and nonreporter (parental) viruses, and all of them expressed similar levels of LucR. As intended, we observed a range of Nef expression in 293T cells following transfection with LucR.IRES constructs.
Several constructs resulted in Nef levels lower than that observed for wild-type Nef, and interestingly, these LucR.IRES reporter viruses resulted in an even greater reduction of Nef levels after infection of T cell lines (J2574-R5 and A3R5.7) and PBMCs. Importantly, infection of T cell lines and PBMCs with one of the other constructs, the LucR.6ATRi reporter virus, resulted in physiological levels of Nef expression. Furthermore, and in contrast to the LucR.T2A reporter virus, T cell lines and PBMCs infected with LucR.6ATRi HIV-1 exhibited patterns of CD4 and MHC-I down-modulation similar to those detected on cells infected with nonreporter viruses. Thus, the LucR.6ATRi reporter virus behaved in a manner similar to the nonreporter (parental) virus with regard to Nef expression and function.
Replication kinetics of the LucR.6ATRi reporter HIV-1 were only slightly delayed when compared to nonreporter virus in PBMCs, as we previously observed for LucR.T2A reporter viruses.8 Thus, in comparison to the much greater replication delays we had seen with the “classic” EMCV IRES-containing reporter viruses, like the GFP-expressing NLENG1i HIV-1 (personal observations), the new LucR.6ATRi reporter approach performed well. In part, this may be attributable to the smaller size of the encoded 6ATRi element [435 nt versus 576 nt for the “wild-type” EMCV IRES element (6Ai), Supplementary Fig. S1]. In addition, the relative stability of LucR reporter gene expression over several rounds of replication in PBMCs, a hallmark of LucR.T2A reporter viruses,8 was similarly achieved with LucR.6ATRi reporter virus in the current study. Interestingly, among the various LucR.IRES reporter viruses analyzed, the one comprising the LucR.HCVi cassette best retained LucR activity over time; however, due to the virtual absence of Nef expression and function in infected cells, this desirable characteristic was irrelevant except for supporting the notion that smaller exogenous gene insertions are better tolerated.
Lastly, in an effort to demonstrate that functionality of the LucR.6ATRi reporter virus was “noninferior” to the LucR.T2A reporter virus in already established applications that do not require functional Nef, we compared the two reporter viruses in the recently validated A3R5/Env-IMC-LucR neutralization assay.10 Indeed, both NL-LucR.T2A-BaL.ecto and NL-LucR.6ATRi-BaL.ecto viruses were similarly neutralized by the eight different bNAbs tested, resulting in essentially indistinguishable IC50 and IC80 values for each. Several studies assessing the performance of the LucR.6ATRi reporter virus in applications that desire or require wild-type Nef have been initiated. In particular, efforts toward further optimization of our recently described CD8+ T cell virus inhibition assay (LucR VIA),12 which include evaluating the effects of functional, 6ATRi-driven Nef expression on VIA performance in the context of different, VIA-relevant proviral IMC backbones, are ongoing and will be reported elsewhere.
The majority of published studies assessing the effect of nef mutations on Nef function, including CD4 and MHC-I down-regulation, have been performed in the context of cells transfected or transduced with nef-expression plasmids. Here, we assessed the modulation of MHC-I surface levels in three types of T cells productively infected with HIV-1 expressing Nef or several functional mutants. Of note, we observed that infection with nonreporter and LucR.6ATRi reporter viruses with abrogated Nef expression (NefSTOP) resulted in a slight increase in MHC-I expression on infected cells (identified as either GFP+ or HIV-1 p24+) by flow cytometry with anti-HLA-A/B/C compared to control (“mock infected”) cells (illustrated in Fig. 6). Interestingly, and similar to others,74,75 we did not observe any difference in MHC-I (HLA-A/B/C) levels between the cells infected (HIV-1+) with Nef-deficient virus and the cells that remained uninfected (HIV-1−) within the same culture. Rather, we found it was the entire virus-exposed cell population that showed slightly elevated MHC-I expression (data not shown) compared to the mock-infected cells. We suggest that this increase in MHC-I on uninfected cells within a Nef-deficient virus-infected culture is attributable to a bystander effect, possibly from the activity of interferon-gamma (IFN-γ) produced in response to HIV-1 infection, since IFN-γ has been described as up-regulating MHC-I (reviewed in Roff et al.76).
In contrast, we demonstrated that infection with matched viruses expressing functional Nef resulted in a clear downward shift in MHC-I expression on infected cells compared to uninfected cells within the same culture. Thus, importantly, in the context of productively HIV-1-infected T cells, down-regulation of MHC-I by Nef was most pronounced when compared to the cells infected with Nef-deficient virus or with uninfected bystander cells in the same culture. MHC-I down-regulation was less apparent when compared to virus-unexposed (mock infected) cultures of PBMCs as well as two CD4+ T cell lines. Interestingly, similar findings were reported by Brown et al. in infected PBMCs (7 days postinfection) and primary monocyte-derived macrophages (10–13 days postinfection),29 and to a lesser degree by Kirchhoff and colleagues in PBMCs77 but not Jurkat cells.75
Our flow cytometric analysis of endogenous MHC-I following HIV-1 infection (like analyses done by others75) utilized a pan-reactive antibody that did not distinguish between HLA-A, HLA-B, and HLA-C. However, Nef has been described as selectively down-modulating HLA-A and HLA-B but not HLA-C (although initial data for HLA-C were somewhat contradictory,74,78 it has since been demonstrated that Nef does not directly affect HLA-C and HLA-E31,79). Thus, we assume that our observation is also limited to HLA-A and HLA-B. It may be biologically meaningful that Nef counteracts the up-regulation of MHC-I by the IFN-γ response to viral infection, and consequently the recognition by cytotoxic T lymphocytes (CTL),74 while avoiding down-regulation below normal MHC-I levels. Even though (the Nef-unaffected) HLA-C and HLA-E have been described as protective of the NK cell response,31 avoiding lower-than-normal HLA-A and HLA-B levels may contribute to preventing NK cell recognition (reviewed in Othman and Yusof80). Possibly, the remaining MHC-I does not “flag” the cell as infected through presentation of HIV-1-derived peptides since Tat, among its many functions, inhibits the generation of such peptides.81
Cohen et al. have posed the question of how anti-HIV CTL are generated in vivo in light of MHC-I down-regulation.31 They proposed that epitope density on infected cells may be sufficient for CTL activation but not for lysis; or, alternatively, HIV epitopes may instead be presented in association with MHC-I by uninfected antigen-presenting cells (APCs).31 The characterization we conducted herein of novel IRES encoding reporter viruses engineered to provide wild-type Nef function provided an unexpected glimpse at the remarkable complexity of lentiviral manipulation of the host cell. It also highlights that for applications that delve into questions of pathogenesis, or for immune-monitoring assays that rely on cell interactions as in the LucR VIA12,19 that measures CD8+ T cell responses to infected CD4+ T cells, it may be highly relevant to utilize a sensitive reporter virus technology that retains expression and function of all viral genes.
Our work further highlights that subtle genetic differences in the EMCV IRES sequence result in a broad range of expression levels of the downstream gene. Our observations are in line with Bochkov et al. who performed a similar, but more extensive, comparison of EMCV IRES elements in an alternative system.47 Indeed, insufficient sequence details regarding the use of the “EMCV IRES” are overwhelmingly present throughout the literature; it could be that the “inefficiency” attributed to EMCV IRES-mediated expression may be related to the use of suboptimal and/or attenuated versions of the EMCV IRES. In this regard, further optimization of the CrPV, IAPV, and HCV IRES elements utilized in our panel of constructs could lead to significant gains in IRES-mediated translation of downstream nef. For example, in nature the CrPV IGR IRES initiates translation of its polyprotein from a GCU (Ala) codon rather than from an AUG (Met) codon. In the design of LucR.CrPV proviral construct, we chose to utilize a Met codon to initiate translation of nef in order to preserve the native N-terminus of the Nef protein; however, translation from an Ala codon is more efficient.82 Nevertheless, we are in the process of transferring the 6ATRi-nef approach, as well as select alternative IRES elements, into different strains of T/F HIV-1 to define their performance in the context of different proviral backbones.
We predict that 6ATRi-nef will work well but will test this extensively before starting to build a new generation of a T/F HIV-1-based reporter virus panel comprising a variety of reporter genes. In particular, it may be worthwhile to explore whether the performance of the HCVi elements83 within the LucR.IRES reporter system can be improved upon since its small size (∼325 nt) may be particularly beneficial in proviral backbones that are transcriptionally less robust than that derived from NL4-3, so long as the problem of substantially reduced Nef expression could be overcome. Despite the favorable results obtained here, there may still be limitations on utilizing the truncated 435 nt EMCV IRES (6ATRi) compared with the much smaller T2A element (54 nt). In this regard, synthetic IRES technology (some as small as 9 nt) may offer an alternative to the larger EMCV IRES.49,84,85 However, initial strategies to utilize synthetic IRES elements in our reporter virus system were not pursued due to the low translational activity associated with these elements. On the other hand, concatemerized versions of these small elements were found to enhance IRES activity to levels equivalent to or greater than that of EMCV49,84; however, the repetitive nature of these elements may be selected against in a replicating virus.
In summary, our iterative characterization of the redesigned reporter viruses with modified IRES elements resulted in an improved reporter virus concept that balances favorable attributes regarding replication capacity and kinetics, robustness and stability of sensitive reporter gene expression, and wild-type-like Nef expression and function. Thus, the Env-IMC-LucR.6ATRi approach promises to provide a useful virological tool for augmenting HIV transmission and pathogenesis-related investigations and HIV-1 vaccine immune monitoring efforts, including the measurement of parameters such as the generation of NAb, ADCC, and CD8+ T cell virus inhibition assays and other novel approaches emphasizing primary cell and tissue models.
Supplementary Material
Acknowledgments
This work was supported by a subcontract to C.O. from the Comprehensive Antibody Vaccine Immune Monitoring Consortium (CA-VIMC) (grant #1032144), which is part of the Collaboration for AIDS Vaccine Discovery (CAVD)/CA-VIMC, funded by the Bill & Melinda Gates Foundation; a subcontract to J.C.K. from the Center for HIV/AIDS Vaccine Immunology-Immunogen Discovery (CHAVI-ID), NIH U01-AI-067854; and facilities of the Virology, Genetic Sequencing, and Flow Cytometry cores of the UAB Center for AIDS Research (CFAR), P30-AI-27767. We would also like to acknowledge Dr. Marla Hertz and Dr. Sunnie Thompson for very helpful discussions and the generous provision of IRES reagents; Dr. James Hoxie for insightful discussion and anti-Nef antibodies; Dr. Alok Mulky and Dr. Qun Dai for technical expertise and useful discussions; and Dr. David Montefiori for his support and discussion.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Polonis VR, Brown BK, Rosa Borges A, et al. : Recent advances in the characterization of HIV-1 neutralization assays for standardized evaluation of the antibody response to infection and vaccination. Virology 2008;375(2):315–320 [DOI] [PubMed] [Google Scholar]
- 2.Polonis VR, Schuitemaker H, Bunnik EM, et al. : Impact of host cell variation on the neutralization of HIV-1 in vitro. Curr Opin HIV AIDS 2009;4(5):400–407 [DOI] [PubMed] [Google Scholar]
- 3.Ochsenbauer C. and Kappes JC: New virologic reagents for neutralizing antibody assays. Curr Opin HIV AIDS 2009;4(5):418–425 [DOI] [PubMed] [Google Scholar]
- 4.Ochsenbauer C, Edmonds TG, Ding H, et al. : Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J Virol 2012;86(5):2715–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parrish NF, Wilen CB, Banks LB, et al. : Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin alpha4beta7. PLoS Pathog 2012;8(5):e1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salazar-Gonzalez JF, Salazar MG, Keele BF, et al. : Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med 2009;206(6):1273–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ochiel DO, Ochsenbauer C, Kappes JC, et al. : Uterine epithelial cell regulation of DC-SIGN expression inhibits transmitted/founder HIV-1 trans infection by immature dendritic cells. PLoS One 2010;5(12):e14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edmonds TG, Ding H, Yuan X, et al. : Replication competent molecular clones of HIV-1 expressing Renilla luciferase facilitate the analysis of antibody inhibition in PBMC. Virology 2010;408(1):1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLinden RJ, Labranche CC, Chenine AL, et al. : Detection of HIV-1 neutralizing antibodies in a human CD4(+)/CXCR4(+)/CCR5(+) T-lymphoblastoid cell assay system. PLoS One 2013;8(11):e77756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarzotti-Kelsoe M, Daniell X, Todd CA, et al. : Optimization and validation of a neutralizing antibody assay for HIV-1 in A3R5 cells. J Immunol Methods 2014;409:147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montefiori DC, Karnasuta C, Huang Y, et al. : Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials. J Infect Dis 2012;206(3):431–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naarding MA, Fernandez N, Kappes JC, et al. : Development of a luciferase based viral inhibition assay to evaluate vaccine induced CD8 T-cell responses. J Immunol Methods 2014;409:161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smalls-Mantey A, Doria-Rose N, Klein R, et al. : Antibody-dependent cellular cytotoxicity against primary HIV-infected CD4+ T cells is directly associated with the magnitude of surface IgG binding. J Virol 2012;86(16):8672–8680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrari G, Pollara J, Kozink D, et al. : An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J Virol 2011;85(14):7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollara J, Hart L, Brewer F, et al. : High-throughput quantitative analysis of HIV-1 and SIV-specific ADCC-mediating antibody responses. Cytometry A 2011;79(8):603–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seay K, Qi X, Zheng JH, et al. : Mice transgenic for CD4-specific human CD4, CCR5 and cyclin T1 expression: A new model for investigating HIV-1 transmission and treatment efficacy. PLoS One 2013;8(5):e63537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.deCamp A, Hraber P, Bailer RT, et al. : Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 2014;88(5):2489–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chenine AL, Wieczorek L, Sanders-Buell E, et al. : Impact of HIV-1 backbone on neutralization sensitivity: Neutralization profiles of heterologous envelope glycoproteins expressed in native subtype C and CRF01_AE backbone. PLoS One 2013;8(11):e76104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slichter CK, Friedrich DP, Smith RJ, et al. : Measuring inhibition of HIV replication by ex vivo CD8(+) T cells. J Immunol Methods 2014;404:71–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Planque SA, Mitsuda Y, Chitsazzadeh V, et al. : Deficient synthesis of class-switched, HIV-neutralizing antibodies to the CD4 binding site and correction by electrophilic gp120 immunogen. AIDS 2014;28(15):2201–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fenton-May AE, Dibben O, Emmerich T, et al. : Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology 2013;10:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu P, Yates NL, Shen X, et al. : Infectious virion capture by HIV-1 gp120-specific IgG from RV144 vaccinees. J Virol 2013;87(14):7828–7836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pace CS, Song R, Ochsenbauer C, et al. : Bispecific antibodies directed to CD4 domain 2 and HIV envelope exhibit exceptional breadth and picomolar potency against HIV-1. Proc Natl Acad Sci USA 2013;110(33):13540–13545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miglietta R, Pastori C, Venuti A, et al. : Synergy in monoclonal antibody neutralization of HIV-1 pseudoviruses and infectious molecular clones. J Transl Med 2014;12(1):346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freel SA, Lamoreaux L, Chattopadhyay PK, et al. : Phenotypic and functional profile of HIV-inhibitory CD8 T cells elicited by natural infection and heterologous prime/boost vaccination. J Virol 2010;84(10):4998–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown BK, Wieczorek L, Kijak G, et al. : The role of natural killer (NK) cells and NK cell receptor polymorphisms in the assessment of HIV-1 neutralization. PLoS One 2012;7(4):e29454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy DN, Aldrovandi GM, Kutsch O, and Shaw GM: Dynamics of HIV-1 recombination in its natural target cells. Proc Natl Acad Sci USA 2004;101(12):4204–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gelderblom HC, Vatakis DN, Burke SA, et al. : Viral complementation allows HIV-1 replication without integration. Retrovirology 2008;5:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown A, Gartner S, Kawano T, et al. : HLA-A2 down-regulation on primary human macrophages infected with an M-tropic EGFP-tagged HIV-1 reporter virus. J Leukoc Biol 2005;78(3):675–685 [DOI] [PubMed] [Google Scholar]
- 30.Mahlknecht U, Deng C, Lu MC, et al. : Resistance to apoptosis in HIV-infected CD4+ T lymphocytes is mediated by macrophages: Role for Nef and immune activation in viral persistence. J Immunol 2000;165(11):6437–6446 [DOI] [PubMed] [Google Scholar]
- 31.Cohen GB, Gandhi RT, Davis DM, et al. : The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 1999;10(6):661–671 [DOI] [PubMed] [Google Scholar]
- 32.de Felipe P: Skipping the co-expression problem: the new 2A “CHYSEL” technology. Genet Vaccines Ther 2004;2(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donnelly ML, Luke G, Mehrotra A, et al. : Analysis of the aphthovirus 2A/2B polyprotein 'cleavage' mechanism indicates not a proteolytic reaction, but a novel translational effect: A putative ribosomal 'skip.' J Gen Virol 2001;82(Pt 5):1013–1025 [DOI] [PubMed] [Google Scholar]
- 34.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, et al. : Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 2009;361(23):2209–2220 [DOI] [PubMed] [Google Scholar]
- 35.Pitisuttithum P, Gilbert P, Gurwith M, et al. : Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis 2006;194(12):1661–1671 [DOI] [PubMed] [Google Scholar]
- 36.Eisenhaber F, Eisenhaber B, Kubina W, et al. : Prediction of lipid posttranslational modifications and localization signals from protein sequences: Big-Pi, NMT and PTS1. Nucleic Acids Res 2003;31(13):3631–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maurer-Stroh S, Eisenhaber B, and Eisenhaber F: N-terminal N-myristoylation of proteins: Refinement of the sequence motif and its taxon-specific differences. J Mol Biol 2002;317(4):523–540 [DOI] [PubMed] [Google Scholar]
- 38.Welker R, Harris M, Cardel B, and Krausslich HG: Virion incorporation of human immunodeficiency virus type 1 Nef is mediated by a bipartite membrane-targeting signal: Analysis of its role in enhancement of viral infectivity. J Virol 1998;72(11):8833–8840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei X, Decker JM, Liu H, et al. : Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 2002;46(6):1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Folks T, Benn S, Rabson A, et al. : Characterization of a continuous T-cell line susceptible to the cytopathic effects of the acquired immunodeficiency syndrome (AIDS)-associated retrovirus. Proc Natl Acad Sci USA 1985;82(13):4539–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duverger A, Wolschendorf F, Zhang M, et al. : An AP-1 binding site in the enhancer/core element of the HIV-1 promoter controls the ability of HIV-1 to establish latent infection. J Virol 2013;87(4):2264–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shishido T, Wolschendorf F, Duverger A, et al. : Selected drugs with reported secondary cell-differentiating capacity prime latent HIV-1 infection for reactivation. J Virol 2012;86(17):9055–9069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hertz MI. and Thompson SR: In vivo functional analysis of the Dicistroviridae intergenic region internal ribosome entry sites. Nucleic Acids Res 2011;39(16):7276–7288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jang CJ, Lo MC, and Jan E: Conserved element of the dicistrovirus IGR IRES that mimics an E-site tRNA/ribosome interaction mediates multiple functions. J Mol Biol 2009;387(1):42–58 [DOI] [PubMed] [Google Scholar]
- 45.Landry DM, Hertz MI, and Thompson SR: RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev 2009;23(23):2753–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hertz MI, Landry DM, Willis AE, et al. : Ribosomal protein S25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Mol Cell Biol 2013;33(5):1016–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bochkov YA. and Palmenberg AC: Translational efficiency of EMCV IRES in bicistronic vectors is dependent upon IRES sequence and gene location. Biotechniques 2006;41(3):283–284, 286, 288 passim [DOI] [PubMed] [Google Scholar]
- 48.Duke GM, Hoffman MA, and Palmenberg AC: Sequence and structural elements that contribute to efficient encephalomyocarditis virus RNA translation. J Virol 1992;66(3):1602–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chappell SA, Edelman GM, and Mauro VP: A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc Natl Acad Sci USA 2000;97(4):1536–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bentham M, Mazaleyrat S, and Harris M: Role of myristoylation and N-terminal basic residues in membrane association of the human immunodeficiency virus type 1 Nef protein. J Gen Virol 2006;87(Pt 3):563–571 [DOI] [PubMed] [Google Scholar]
- 51.Shugars DC, Smith MS, Glueck DH, et al. : Analysis of human immunodeficiency virus type 1 nef gene sequences present in vivo. J Virol 1993;67(8):4639–4650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chesebro B, Wehrly K, Nishio J, and Perryman S: Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: Definition of critical amino acids involved in cell tropism. J Virol 1992;66(11):6547–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muster T, Steindl F, Purtscher M, et al. : A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol 1993;67(11):6642–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zwick MB, Labrijn AF, Wang M, et al. : Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol 2001;75(22):10892–10905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burton DR, Pyati J, Koduri R, et al. : Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 1994;266(5187):1024–1027 [DOI] [PubMed] [Google Scholar]
- 56.Bonsignori M, Montefiori DC, Wu X, et al. : Two distinct broadly neutralizing antibody specificities of different clonal lineages in a single HIV-1-infected donor: Implications for vaccine design. J Virol 2012;86(8):4688–4692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bonsignori M, Hwang KK, Chen X, et al. : Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol 2011;85(19):9998–10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walker LM, Phogat SK, Chan-Hui PY, et al. : Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 2009;326(5950):285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, et al. : Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat Struct Mol Biol 2013;20(7):804–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trkola A, Purtscher M, Muster T, et al. : Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol 1996;70(2):1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Foster JL. and Garcia JV: HIV-1 Nef: At the crossroads. Retrovirology 2008;5:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kestler HW, 3rd, Ringler DJ, Mori K, et al. : Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 1991;65(4):651–662 [DOI] [PubMed] [Google Scholar]
- 63.Kirchhoff F, Greenough TC, Brettler DB, et al. : Brief report: Absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med 1995;332(4):228–232 [DOI] [PubMed] [Google Scholar]
- 64.Fackler OT, Moris A, Tibroni N, et al. : Functional characterization of HIV-1 Nef mutants in the context of viral infection. Virology 2006;351(2):322–339 [DOI] [PubMed] [Google Scholar]
- 65.Borman AM, Le Mercier P, Girard M, and Kean KM: Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res 1997;25(5):925–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jan E: Divergent IRES elements in invertebrates. Virus Res 2006;119(1):16–28 [DOI] [PubMed] [Google Scholar]
- 67.Bonning BC. and Miller WA: Dicistroviruses. Annu Rev Entomol 2010;55:129–150 [DOI] [PubMed] [Google Scholar]
- 68.Piguet V. and Trono D: In A Structure-Function Analysis of the Nef Protein of Primate Lentiviruses (Kuiken CL, Foley B, Hahn B, et al., eds.). Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM, 1999 [Google Scholar]
- 69.Miller MD, Warmerdam MT, Ferrell SS, et al. : Intravirion generation of the C-terminal core domain of HIV-1 Nef by the HIV-1 protease is insufficient to enhance viral infectivity. Virology 1997;234(2):215–225 [DOI] [PubMed] [Google Scholar]
- 70.Muller B, Daecke J, Fackler OT, et al. : Construction and characterization of a fluorescently labeled infectious human immunodeficiency virus type 1 derivative. J Virol 2004;78(19):10803–10813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jamieson BD. and Zack JA: In vivo pathogenesis of a human immunodeficiency virus type 1 reporter virus. J Virol 1998;72(8):6520–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Herbein G, Van Lint C, Lovett JL, and Verdin E: Distinct mechanisms trigger apoptosis in human immunodeficiency virus type 1-infected and in uninfected bystander T lymphocytes. J Virol 1998;72(1):660–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fouda GG, Mahlokozera T, Salazar-Gonzalez JF, et al. : Postnatally-transmitted HIV-1 envelope variants have similar neutralization-sensitivity and function to that of nontransmitted breast milk variants. Retrovirology 2013;10:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Collins KL, Chen BK, Kalams SA, et al. : HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 1998;391(6665):397–401 [DOI] [PubMed] [Google Scholar]
- 75.Specht A, DeGottardi MQ, Schindler M, et al. : Selective downmodulation of HLA-A and -B by Nef alleles from different groups of primate lentiviruses. Virology 2008;373(1):229–237 [DOI] [PubMed] [Google Scholar]
- 76.Roff SR, Noon-Song EN, and Yamamoto JK: The significance of interferon-gamma in HIV-1 pathogenesis, therapy, and prophylaxis. Front Immunol 2014;4:498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schindler M, Rajan D, Specht A, et al. : Association of Nef with p21-activated kinase 2 is dispensable for efficient human immunodeficiency virus type 1 replication and cytopathicity in ex vivo-infected human lymphoid tissue. J Virol 2007;81(23):13005–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Le Gall S, Erdtmann L, Benichou S, et al. : Nef interacts with the mu subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC I molecules. Immunity 1998;8(4):483–495 [DOI] [PubMed] [Google Scholar]
- 79.Williams M, Roeth JF, Kasper MR, et al. : Direct binding of human immunodeficiency virus type 1 Nef to the major histocompatibility complex class I (MHC-I) cytoplasmic tail disrupts MHC-I trafficking. J Virol 2002;76(23):12173–12184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Othman S. and Yusof R: Regulation of MHC class I by viruses. In Histocompatibility (Abdel-Salam BKA-H, ed.). InTech Open Access Publisher, 2012, pp. 133–144 [Google Scholar]
- 81.Gavioli R, Gallerani E, Fortini C, et al. : HIV-1 tat protein modulates the generation of cytotoxic T cell epitopes by modifying proteasome composition and enzymatic activity. J Immunol 2004;173(6):3838–3843 [DOI] [PubMed] [Google Scholar]
- 82.Brodel AK, Sonnabend A, Roberts LO, et al. : IRES-mediated translation of membrane proteins and glycoproteins in eukaryotic cell-free systems. PLoS One 2013;8(12):e82234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fraser CS. and Doudna JA: Structural and mechanistic insights into hepatitis C viral translation initiation. Nat Rev Microbiol 2007;5(1):29–38 [DOI] [PubMed] [Google Scholar]
- 84.Owens GC, Chappell SA, Mauro VP, and Edelman GM: Identification of two short internal ribosome entry sites selected from libraries of random oligonucleotides. Proc Natl Acad Sci USA 2001;98(4):1471–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stoneley M, Paulin FE, Le Quesne JP, et al. : C-Myc 5' untranslated region contains an internal ribosome entry segment. Oncogene 1998;16(3):423–428 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.