Abstract
A central hurdle in developing small interfering RNAs (siRNAs) as therapeutics is the inefficiency of their delivery across the plasma and endosomal membranes to the cytosol, where they interact with the RNA interference machinery. With the aim of improving endosomal release, a poorly understood and inefficient process, we studied the uptake and cytosolic release of siRNAs, formulated in lipoplexes or lipid nanoparticles, by live-cell imaging and correlated it with knockdown of a target GFP reporter. siRNA release occurred invariably from maturing endosomes within ~5–15 min of endocytosis. Cytosolic galectins immediately recognized the damaged endosome and targeted it for autophagy. However, inhibiting autophagy did not enhance cytosolic siRNA release. Gene knockdown occurred within a few hours of release and required <2,000 copies of cytosolic siRNAs. The ability to detect cytosolic release of siRNAs and understand how it is regulated will facilitate the development of rational strategies for improving the cytosolic delivery of candidate drugs.
Cytosolic delivery is the major obstacle to siRNA drug development. Cationic lipids1, used for in vitro transfection, form positively charged heterogeneous complexes with nucleic acids, called lipoplexes2. However, because of their size, charge and toxicity, they are not suitable for in vivo use. Smaller (50–100 nm) homogeneous lipid nanoparticles (LNP), formed by mixing siRNAs with PEGylated and cationic lipids and cholesterol, are the furthest advanced in clinical studies3–5. These LNPs are ionizable (neutral at physiological pH, but protonated in endosomes), which facilitates fusion of their lipids with the endosomal membrane and enables cytosolic RNA delivery. LNPs carrying transthyretin siRNAs cause durable gene knockdown in the liver (>80% knockdown lasting weeks after one injection6) with manageable toxicity. These are currently being evaluated in phase 3 clinical trials to treat familial amyloidotic polyneuropathy. LNPs are trapped in the liver and generally cause effective gene knockdown only in that organ. Both lipoplexes and LNPs are taken up by endocytosis, but most of their cargo accumulates in late endosomes and lysosomes, where they are not active7–9.
Figuring out how to improve cytosolic release is hampered by a lack of tools to detect the endosomal escape of nucleic acids. Previous microscopy studies of endocytosed lipoplexes or LNPs either have not directly visualized cytosolic release8,9 or have detected a gradual increase of RNA-oligonucleotide cargo in the cytosol without clearly linking it to knockdown or mechanism7,10.
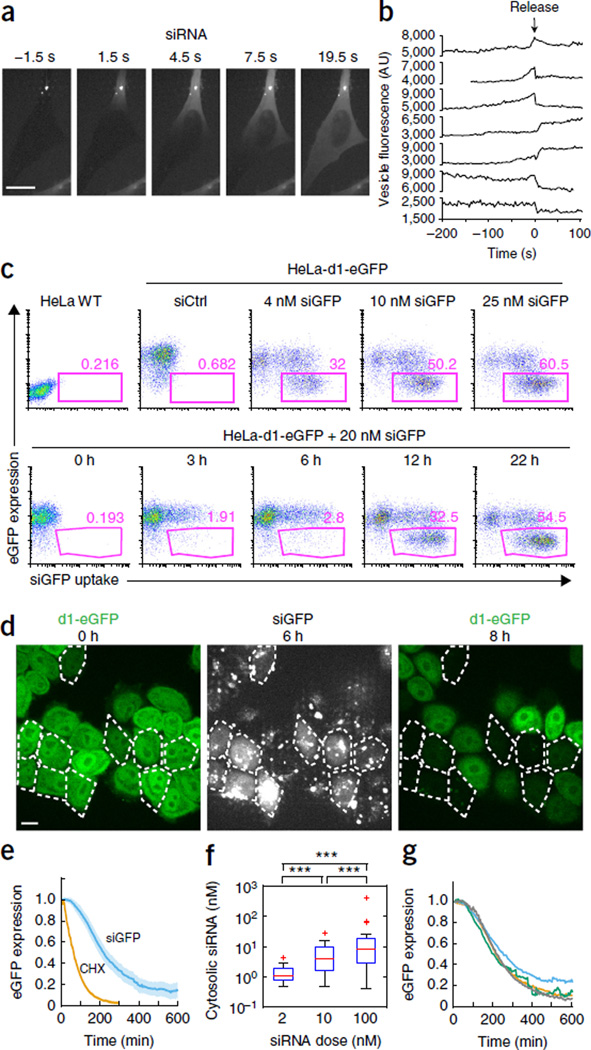
Visualizing endosomal release in live cells is challenging because small amounts of released siRNAs must be detected simultaneously with intensely fluorescent endosomes that are densely packed with lipoplexed siRNA. To handle the large dynamic range, we developed an imaging approach similar to the high-dynamic-range (HDR) technique used in digital cameras. Cells were imaged with two different exposure settings using a spinning-disk microscope equipped with an electron-multiplying charge-coupled device (EMCCD) camera. Multiple planes encompassing most of the cellular volume were acquired with short exposure times and a dynamic range adjusted to the bright structures within the cells (the intact lipoplexes and vesicles). Then a single plane in the lower third of the cell was captured with a long exposure time, intentionally overexposing bright areas to detect the weakly fluorescent siRNA signal in the cytosol (Supplementary Fig. 1). Using this technique, we observed sudden cytosolic release of Alexa Fluor 647–labeled siRNAs (siRNA-AF647) that originated from intensely fluorescent lipoplex-containing vesicles (Fig. 1a and Supplementary Movie 1). The released siRNAs rapidly diffused and filled the entire cytosol within 10–20 s, suggesting that free siRNAs, rather than intact lipoplexes, escaped into the cytosol. Although cytosolic release was detected in a single plane, the method was sensitive enough to detect release events that occurred outside that plane. Typically between one and five release events were observed per cell over several hours. The fluorescence intensity of the releasing particle usually increased gradually 1–2 min before release and then suddenly dropped, concurrently with detection of the cytosolic signal (Fig. 1b). The releasing vesicle’s fluorescence was reduced by only a fraction of its intensity and did not decline further with time. Thus only some cargo was released, and the leaky vesicle did not continue to release its cargo. Therefore, the membrane of the releasing endosome did not rupture. Because fluorophores in close proximity are self-quenched, we interpreted the initial increase in fluorescence as a sign of partial disintegration of the lipoplexes that resulted in dequenching. The sudden drop in fluorescence reflected the actual release event and coincided with a sudden increase in cytosolic siRNA fluorescence adjacent to the releasing vesicle (Supplementary Fig. 2). siRNA release coincided, in many cases, with small cytosolic Ca2+ transients, of variable magnitude as measured using the Fluo-4 Ca2+ sensor, which originated from the damaged endosome (Supplementary Fig. 3). Laser illumination during imaging did not affect the probability of cytosolic release of the siRNA (Supplementary Fig. 4).
Figure 1.
Sudden endosomal release and cytosolic dispersion of lipoplexed siRNA trigger gene knockdown. (a) Images taken every 3 s of HeLa cells incubated with siRNA-AF647 lipoplexes. Single-plane images of the siRNA-AF647 signal were captured with long exposure settings (300 ms) to detect the faint cytosolic siRNA-AF647 signal. Scale bar, 10 µm. (b) siRNA-AF647 fluorescence intensity traces of releasing endosomes from maximum z-projection images of short exposures (30 ms) of the entire cell. (c) Top, flow cytometry analysis of eGFP expression 24 h after incubation of HeLa-d1-eGFP cells with different amounts of siGFP-AF647 lipoplexes (final siGFP concentrations indicated). Untreated HeLa WT cells and HeLa-d1-eGFP cells incubated with nonfluorescent siCtrl lipoplexes were analyzed as controls. Bottom, flow cytometry analysis of eGFP expression in HeLa-d1-eGFP cells incubated with 20 nM siGFP-AF647 lipoplexes for the indicated amounts of time. (d) HeLa-d1-eGFP cells were incubated with siGFP-Cy3 lipoplexes and imaged every 3 min. eGFP fluorescence levels at the start (t = 0 h) and end of the experiment (t = 8 h) are shown in the left and right images, respectively. Cells in which a cytosolic release event occurred during the first 6 h are outlined (middle panel, siGFP-Cy3 signal). Scale bar, 10 µm. (e) eGFP expression (relative to expression at t = 0) in HeLa-d1-eGFP cells after cytosolic release of siGFP-AF647 (siGFP, blue line; number of cells, n = 31) or after inhibition of translation (cycloheximide treatment, CHX, orange line; number of cells, n = 59). Shaded areas are 95% confidence intervals (in the case of CHX, the confidence interval is largely covered by the line). (f) Cytosolic siGFP-AF647 concentration after endosomal release in cells incubated with lipoplexes formed with different amounts of siRNA (2 nM, n = 19; 10 nM, n = 34; 100 nM, n = 27; ***P < 0.001, two-tailed Mann-Whitney U test). Box indicates 25–75 percentiles and whiskers indicate the entire data range excluding outliers (‘+’). (g) Relative eGFP expression of cells exhibiting different levels of cytosolic siGFP-AF647 release. Average cytosolic siRNA concentration within each group of cells: green, 170 nM (n = 7); blue, 15 nM, (n = 8); orange, 4 nM, (n = 8); gray, 1.6 nM, (n = 8). Data in all panels are representative of experiments performed at least three times.
To correlate release events with target gene knockdown, we first used flow cytometry to study lipoplex-mediated gene knockdown in HeLa cells that were stably expressing destabilized eGFP (HeLa-d1-eGFP)11 with an ~1-h half-life (t1/2). HeLa-d1-eGFP cells were incubated with dilutions of lipoplexes containing fluorescent siRNAs targeting eGFP (siGFP-AF647). Although most cells internalized siGFP-AF647 lipoplexes, eGFP knockdown was an all-or-nothing phenomenon at all lipoplex concentrations tested (Fig. 1c), which suggested that endosomal release was a discrete limiting step of knockdown. Although more cells showed knockdown at higher siRNA concentrations, the extent of knockdown was dose independent.
Next we simultaneously monitored eGFP fluorescence and cytosolic siRNA delivery over several hours in HeLa-d1-eGFP cells incubated with fluorescent siGFP lipoplexes (Fig. 1d and Supplementary Movie 2). In cells that showed siGFP-AF647 endosomal release, eGFP fluorescence began to decrease ~60 min after release and had a t1/2 of ~115 min (Fig. 1e). In control cells treated with cycloheximide, which stopped protein synthesis, eGFP fluorescence began to decline after a delay of ~18 min, with a t1/2 of ~45 min after cycloheximide treatment. Thus, cytosolic delivery of siRNAs caused rapid knockdown. Although we could not exclude the possibility that there were small amounts of siRNA leakage that we could not detect, the all-or-nothing knockdown response together with the synchronization of knockdown following the first release event, and its rapidity, suggest that the release events observed here are the dominant pathway for lipoplex-mediated siRNA cytosolic delivery.
To estimate how many cytosolic copies of the siRNAs are needed for complete knockdown, the siRNA-AF647 concentration in the cytosol just after release was estimated by comparing the cytosolic fluorescence of cells with a reference curve for the fluorescence intensities of droplet preparations spanning a range of siRNA-AF647 concentrations (measured with the same imaging setup, linear range: <1–1,000 nM, R2 = 0.9992). Lipoplexes containing more siRNAs (using a fixed amount of lipid) released progressively more cytosolic siRNAs (Fig. 1f). However, knockdown kinetics were indistinguishable between cells with widely different cytosolic concentrations of siGFP (Fig. 1g). Assuming an average cell volume of 2,000 fl, the lowest quartile of cells contained on average ~2,000 cytosolic siRNAs, while the highest quartile had ~200,000 cytosolic molecules. This calculation suggests that <2,000 cytosolic siRNAs/cell caused maximal knockdown, in agreement with previous estimates7,12,13. In comparison, HeLa-d1-eGFP cells contained 1,326 ± 63 (mean ± s.d.) eGFP mRNA molecules/cell by qRT-PCR analysis.
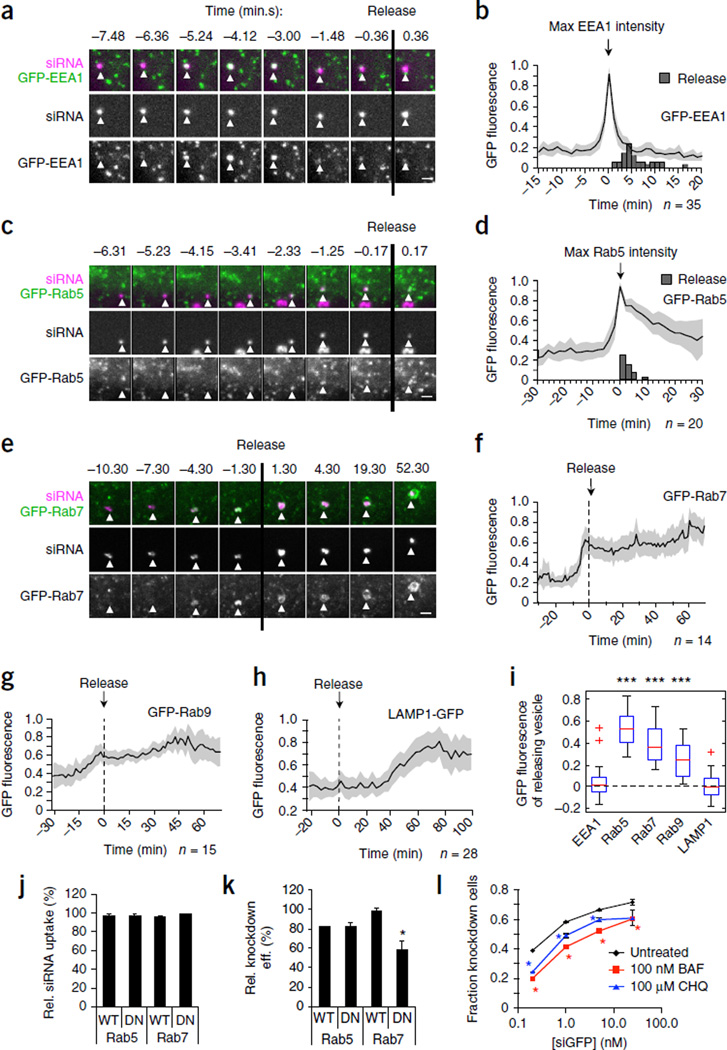
Because we could identify the releasing endosome by its change in siRNA fluorescence (Fig. 1b), we next characterized the stage in endosomal maturation at which release occurred. HeLa cells expressing GFP-tagged endosomal marker proteins were incubated with fluorescent lipoplexes to identify the properties of the releasing vesicle (Fig. 2). Lipoplex-containing vesicles associated with the early endosome marker EEA1 (ref. 14) before release, but became EEA1− just before release (Fig. 2a,b and Supplementary Movie 3). Release occurred 5.5 min (median, 4–9 min, 25–75 percentile, n = 35) after maximal GFP-EEA1 intensity. In GFP-Rab5–expressing HeLa cells, the releasing endosome also became Rab5+ a few minutes before release (Fig. 2c,d and Supplementary Movie 4). Release occurred about 3 min (median, 1–3.5 min, 25–75 percentiles, n = 20) after GFP-Rab5 intensity reached its maximum, when endosomes are normally still Rab5+. In many cases the lateral mobility of lipoplex-containing vesicles increased sharply around the time of GFP-Rab5 recruitment, indicating maturation into a mobile endosome (Supplementary Movie 4 and data not shown). Thus, the majority of release events occurred about 5–10 min after lipoplexes were taken up into the cell. Endosomes undergo a Rab5 to Rab7 conversion during maturation to late endosomes15,16. GFP-Rab7 was recruited to lipoplex-containing endosomes a few minutes before release and remained associated after release (Fig. 2e,f and Supplementary Movie 5). By contrast, the late endosomal (Rab9) and lysosomal (LAMP1) markers were just being recruited or completely absent, respectively, at the time of release (Fig. 2g,h). The vesicle remnants became LAMP1+ 40–50 min after release, when they became lysosomes.
Figure 2.
Lipoplexed siRNA is released from maturing endososomes. (a–i) HeLa cells expressing the indicated GFP-tagged endosomal markers were incubated with siRNA-AF647 lipoplexes and imaged over time. (a,c,e) Details of the releasing endosomes (white triangles) before and after the release event (designated t = 0 min, vertical black lines). Scale bars, 2 µm. (b,d,f–h) Average GFP fluorescence intensities (normalized to 0–1 as described in Online Methods) of releasing endosomes before and after release. Individual traces are aligned with t = 0 set as the maximal GFP-EEA1 signal (b), maximal GFP-Rab5 signal (d) or the time of release (f–h). Shaded areas are 95% confidence intervals. Gray bars indicate time of release (b,d). Number of release events monitored (n) is indicated. (i) Background-corrected GFP fluorescence intensity of releasing endosomes at the time of release in cells expressing GFP-EEA1 (number of cells, n = 33), GFP-Rab5 (n = 16), GFP-Rab7 (n = 13), GFP-Rab9 (n = 13) or LAMP1-GFP (n = 22) (results pooled from two to four independent experiments per condition; ***P < 0.001, Wilcoxon signed-rank test, median ≠ 0). Box indicates 25–75 percentiles and whiskers indicate the entire data range excluding outliers (‘+’). (j,k) Flow cytometry analysis of siGFP-AF647 lipoplex uptake (j) and eGFP knockdown (k) in HeLa-d1-eGFP cells expressing dsRed-tagged WT or DN Rab5 or Rab7, relative to untransfected dsRed− cells in the same sample. (l) Proportion of HeLa-d1-eGFP cells showing eGFP knockdown by flow cytometry upon incubation with siGFP lipoplexes in the presence of bafilomycin A1 (BAF) or chloroquine (CHQ). In j–l, data are mean ± s.d. of three independent experiments. (*P < 0.05, two-tailed Student’s t-test).
Thus lipoplexed siRNAs are released from EEA1−Rab5+Rab7+Rab9±Lamp1− maturing endosomes (Fig. 2i) during a narrow time window within 5–15 min of uptake. The endoplasmic reticulum (ER) was not recruited to the releasing endosome (Supplementary Fig. 5). Previous studies have shown that delivered siRNAs begin to be loaded into Ago2-RISC complexes, which localize near the ER13,17, after 30 min, but we did not study these later events.
We next asked whether endosomal maturation contributes to siRNA release using HeLa-d1-eGFP cells overexpressing either wild-type (WT) or dominant-negative (DN) Rab5 or Rab7. Neither Rab construct affected lipoplex uptake (Fig. 2j). However, DN Rab7, but not DN Rab5, impaired knockdown (Fig. 2k), suggesting that Rab7 activity promotes endosomal siRNA escape. Given its central role in early endosomal maturation16, it is unclear why inhibiting Rab5 did not affect uptake and knockdown. It is possible that lipoplexes are routed through a Rab5-independent pathway or that compensatory mechanisms are triggered by Rab5 inhibition.
We next studied whether inhibiting endosomal acidification (with the V-ATPase inhibitor bafilomycin A1 or with chloroquine) affected knockdown. Both drugs reduced knockdown (Fig. 2l). Thus, endosome maturation enhances siRNA knockdown, presumably because it promotes release.
Transfection reagents induce autophagy18,19. siRNA escape, which presumably results from a damaged endosomal membrane, could trigger macroautophagy of the releasing endosome. HeLa cells transfected with tandem-fluorescent LC3 (tfLC3) (ref. 20), in which LC3 is fused to a pH-sensitive eGFP and a pH-insensitive mRFP, were used to detect autophagy. Within a few minutes of endosomal release, LC3 was recruited to the damaged endosome (Fig. 3a–c). A characteristic cup-shaped membrane engulfed and sealed off the damaged vesicle (Supplementary Movie 6). About 30–50 min later, the GFP-LC3 signal disappeared, whereas the mRFP-LC3 signal was retained (Fig. 3d), which indicated entry of the endosome into an acidic lysosomal compartment. This timing was consistent with LAMP1 detection on endosomes ~40–50 min after siRNA release (Fig. 2h). Thus, endosomal escape triggers macroautophagic isolation of the releasing endosome.
Figure 3.
Endosomal release triggers autophagy. (a) HeLa cells expressing tfLC3 were incubated with siRNA-AF647 lipoplexes and imaged every 3 min. Whole cell (top row; scale bar, 10 µm) and detailed (bottom four rows; scale bar, 2 µm) time-lapse images of cells exhibiting release events. White triangles indicate releasing endosome. (b–d) Average GFP-LC3 (b) and mRFP-LC3 (c) fluorescence intensities of releasing endosomes, and their ratio (d). Time of release = 0 min, shaded areas indicate 95% confidence interval, and number of recorded events (n) is shown. (e–h) Images of HeLa cells expressing YFP-tagged galectin, incubated with siRNA-AF647 lipoplexes. Gal1, galectin-1; Gal3, galectin-3; Gal4, galectin-4; Gal8, galectin-8; Gal9, galectin-9. (e) Detailed images of releasing endosome before (top row) and after (bottom row) release. Scale bar, 2 µm. Fluorescence intensities of YFP–galectin-3 (f), YFP–galectin-8 (g) and YFP–galectin-9 (h) from individual releasing endosomes (relative to cytosolic fluorescence intensity, time of release = 0 s). (i) YFP–galectin-8–expressing HeLa cells were incubated with siRNA-AF647 lipoplexes and imaged every 1 min. White triangles indicate releasing endosome. Scale bar, 10 µm. (j) Quantification of GFP-LC3 recruitment to releasing endosomes 15 min after release from HeLa cells cotransfected with the tfLC3 expression plasmid and either control (siCtrl; number of release events, n = 28), galectin-8 (n = 19), NDP52 (n = 26) or galectin-3 siRNA (n = 17), which were incubated with siRNA-AF647 lipoplexes and imaged every 1 min (results pooled from two to four independent experiments per condition; **P < 0.01, ***P < 0.001, two-tailed Mann-Whitney U test). (k) Analysis of target gene knockdown in (j) by qRT-PCR. Data are mean ± s.d. of three independent experiments. (l) HeLa-d1-eGFP cells, transfected with the indicated siRNAs 3 d earlier, were incubated with siGFP lipoplexes for 12 h and analyzed for GFP expression by flow cytometry. Data are mean ± s.d. of technical replicates (untreated cells: n = 2; 0.1 nM and 0.4 nM siGFP treatment: n = 3) and representative of two independent experiments.
We then asked how autophagy is activated. Cytosolic carbohydrate-sensing galectins are recruited to endosomes damaged by cytosolic invasion by bacteria12,21, when endosomal glycans become exposed to the cytosol. To determine whether galectin recruitment might trigger autophagy, we used HeLa cells to express YFP-tagged versions of all the cytosolic galectins and found that they stained the cytoplasm diffusely under basal conditions (Fig. 3e). In lipoplex-treated cells, galectin-8 and galectin-9 were strongly recruited to the releasing endosome within seconds, galectin-3 was weakly recruited, and galectin-1 and galectin-4 were barely recruited (Fig. 3f–h). These differences may reflect differences in carbohydrate recognition by the galectins, which are not well characterized. In a few cases, galectins were detected on the vesicle 1–2 min before we detected cytosolic siRNAs, presumably due to limited or transient endosomal damage and release that was below our limit of detection. For most of the release events, galectins were recruited within 5–10 s of cytosolic siRNA detection. As galectin-8 was uniformly cytosolic in undisturbed cells, galectin-8 recruitment could be used to identify the siRNA-releasing endosomes (Fig. 3i and Supplementary Movie 7).
The autophagic double membrane around a releasing endosome might limit further siRNA release. Indeed, a recent report suggests that lipoplex-mediated plasmid DNA delivery is restricted by autophagy19. During microbial invasion3,12, activation of autophagy occurs by a noncanonical pathway that depends on galectin-8 and the adaptor NDP52. Knockdown of either galectin-8 or NDP52 severely impaired LC3 recruitment to damaged endosomes, whereas galectin-3 knockdown had no effect (Fig. 3j,k). (We were unable to knock down galectin-9.) However, neither galectin-8 nor NDP52 knockdown enhanced target gene knockdown (Fig. 3l). Thus, something other than autophagy limits siRNA release. siRNA and plasmid DNA release might be differentially sensitive to autophagy because siRNAs diffuse more rapidly than larger plasmid DNAs.
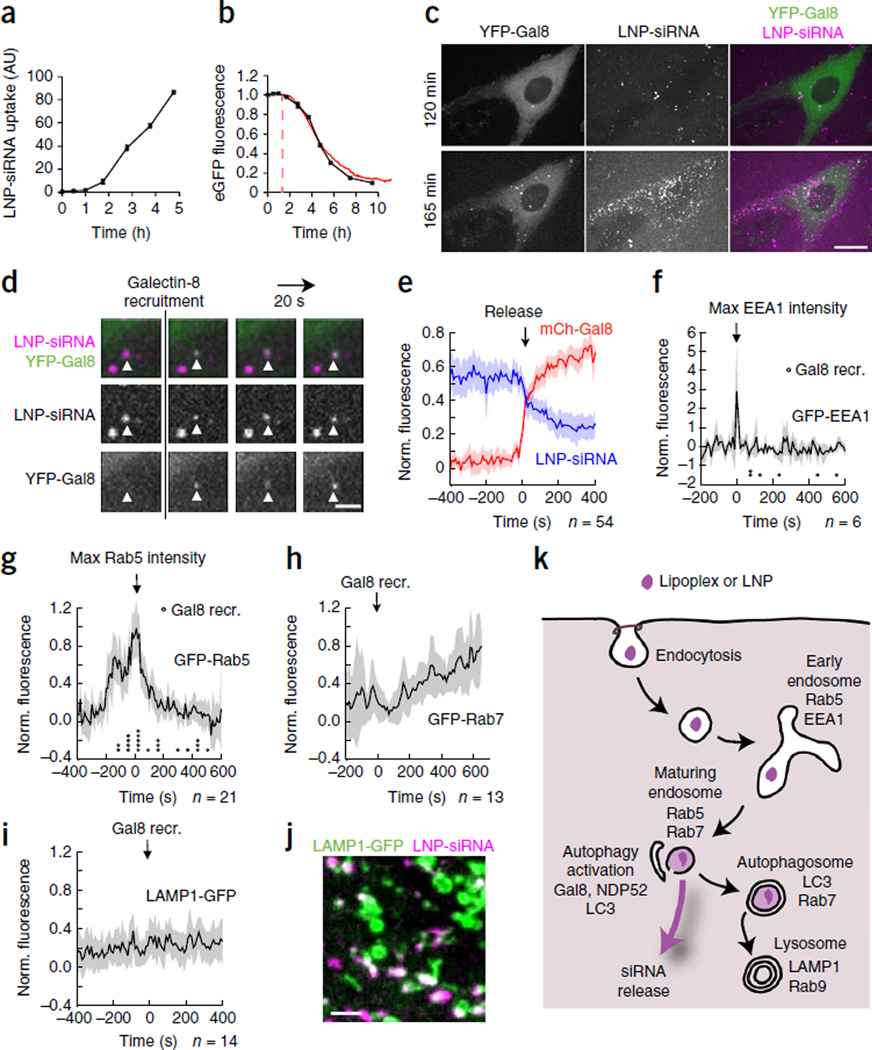
To determine whether LNP-formulated siRNAs traffic like lipoplexed siRNA, we studied LNPs that contained the ionizable lipid L319 (a biodegradable derivative of DLin-MC3-DMA), distearoylphosphatidylcholine, cholesterol, 1,2-dimyristoyl-sn-glycerol and methoxypolyethylene glycol22, which resemble LNPs currently in clinical trials6,23. These small LNPs, which carry much less siRNA than the larger lipoplexes, were efficiently internalized (Fig. 4a). Each cell took up more than 100 LNPs over several hours. After a lag phase of 1–2 h, which corresponded to a lag in endocytosis, the kinetics of LNP-mediated knockdown closely tracked those of lipoplex- mediated knockdown (Fig. 4b). Thus, knockdown occurred rapidly after endocytosis. However, like other groups7,8, we could not detect siRNA release from LNPs, presumably because of their smaller cargo. For those particles whose fluorescence intensity changed, we could not reliably assess whether this was due to endosomal escape or to acquisition-related errors from bleaching, statistical fluctuations or movement relative to the imaging plane. However, we could use galectin-8 recruitment as a surrogate for endosomal release. Indeed, when HeLa cells expressing YFP–galectin-8 were incubated with siRNA-AF647 LNPs, multiple galectin-8 foci appeared progressively after an initial lag phase of ~1.5–2 h (Fig. 4c and Supplementary Movie 8). Galectin-8 foci only formed on LNP-fluorescent vesicles (Fig. 4d and Supplementary Movie 9), but only a small fraction of internalized LNPs (~7%, 62 of 862 LNP-containing vesicles in 10 cells) recruited galectin-8, suggesting that most internalized LNPs did not release their cargo. Galectin-8 recruitment coincided with a sudden reduction of siRNA-AF647 fluorescence, thereby confirming that galectin recruitment could be used to identify releasing vesicles (Fig. 4e). Some individual LNP traces increased fluorescence just before release (data not shown), indicating possible disintegration and dequenching as observed for lipoplexes. Vesicular fluorescence after release was reduced by ~50%, suggesting that about half of the siRNAs in the LNPs were released during each event. Thus, ~3.5% of internalized siRNAs in LNPs were delivered into the cytosol.
Figure 4.
Release of LNP siRNAs from maturing endosomes recruits galectin-8. (a,b) Flow cytometric detection of LNP uptake (a) and GFP fluorescence (b, black line) in HeLa-d1-eGFP cells incubated with LNP–siGFP-AF647. In b, knockdown by lipoplex siGFP-AF647 is superimposed in red (from Fig. 1e), but shifted 1.5 h to account for the slower internalization of LNPs. n = 2; data are mean ± s.d.; representative of two independent experiments. (c) YFP–galectin-8–expressing HeLa cells were incubated with LNP–siRNA-AF647 and imaged every 20 s. Scale bar, 10 µm. After 120 min, few release events had occurred; by 165 min, multiple release events had occurred. (d) Detail of time-lapse images of LNP-containing vesicles (white triangles) recruiting YFP–galectin-8. Scale bar, 2 µm. (e) HeLa cells expressing mCherry–galectin-8 were incubated with LNP–siRNA-AF647 and imaged every 10 s, and fluorescence intensities of vesicles that became mCherry–galectin-8+ were quantified (time of positive mCherry–galectin-8 signal = 0). Blue line, average normalized LNP–siRNA-AF647 fluorescence; red line, mCherry–galectin-8 fluorescence. (f–j) HeLa cells expressing mCherry–galectin-8 and the indicated GFP-tagged endosomal markers were incubated with LNP–siRNA-AF647 and imaged every 10 s. Average normalized GFP fluorescence intensities of LNP vesicles that became mCherry–galectin-8+ are shown. Traces are shifted in time with t = 0 set as maximum GFP-EEA1 signal (f), maximum GFP-Rab5 signal (g) or time of mCherry–galectin-8 recruitment (h,i). Circles in f and g indicate time of release. Shaded areas are 95% confidence intervals. The number of release events (n) analyzed is indicated. (j) Colocalization of LNP–siRNA-AF647 with LAMP1-GFP 4 h after LNP addition. Scale bar, 2 µm. (k) Model of intracellular trafficking of siRNAs delivered by internalized lipoplexes or LNPs.
To identify in which endosomal compartment release occurred, we incubated HeLa cells expressing mCherry–galectin-8 and GFP-tagged endosomal markers with siRNA-AF647 LNPs and used galectin-8 recruitment to time cytosolic release. Galectin-8 recruitment occurred after EEA1 fluorescence disappeared and both before and after Rab5 fluorescence reached its peak (Fig. 4f,g and Supplementary Fig. 6). Most galectin-8 recruitment occurred before vesicles became Rab7+ (Fig. 4h) and always before they became LAMP1+ (Fig. 4i). Unreleased siRNAs accumulated in LAMP1+ lysosomes from which there was no further release (Fig. 4j). Like lipoplexes, LNP vesicles were also targeted for autophagy, and a subset became LC3+ (Supplementary Fig. 7). Thus, LNP siRNAs were released from EEA1−Rab5+Rab7±LAMP1− maturing endosomes over a narrow ~10-min time window, which was somewhat earlier than that for lipoplexed siRNAs, which were released from Rab7+ endosomes (many of which had acquired Rab9). The slight difference in the stage of endosomal maturation at which lipoplexes and LNPs release RNA may reflect differences in their pH sensitivities and physical characteristics.
Here we developed an imaging method to visualize endosomal escape of siRNAs from lipoplexes and LNPs. For both, cytosolic release occurred from maturing endosomes (Fig. 4k) and triggered the autophagic isolation of the releasing endosome. A previous mathematical model also suggested that LNP siRNAs are released early in the endosomal pathway7. It is worth noting that LNPs containing ionizable lipids with pKas in the range of the maturing endosome pH (6–7 (ref. 24) or 6.2–6.5 (ref. 25)) knock down gene expression most efficiently.
siRNA release occurred during a narrow ‘window of opportunity’. No release occurred from late endosomes or lysosomes. We do not know what limits release from these compartments. Although autophagic isolation and subsequent fusion with the lysosome would be expected to terminate release, inhibiting autophagy did not enhance knockdown or increase release. This suggests that other factors that act before autophagy is complete—such as changes in cholesterol, lipid and protein composition that accompany endosomal maturation—might regulate release.
To our knowledge, this is the first study to show that nonmicrobial endosomal damage triggers autophagy through a galectin-8–dependent pathway. As glycoproteins and glycolipids are typically excluded from the cytosol, these glycan sensors might act as alarms to inform the cell about cytosolic invasion. However, we did not detect innate immune or inflammasome activation in lipoplex- or LNP-treated cells (data not shown).
It will be interesting to see whether the imaging methods developed here can be applied to follow the cytosolic delivery of endocytosed nucleic acids and other large molecules, such as antibodies, by other delivery approaches. These include siRNA delivery strategies, such as conjugates to N-acetylgalactosamine (GalNAc)3 or aptamers26, which have shown in vivo efficacy. Because GalNAc-conjugates are monomeric and are not delivered as particles, detecting their release may be challenging unless they are released in packets, their endosomal escape also activates galectin sensors, or more sensitive single-molecule imaging methods are developed. In this study, the detection limit for cytosolic siRNA was ~0.5 nM.
The molecular basis of endosomal release is not well understood. The ability to detect both the released contents and the releasing endosome at the very moment of release (by assaying galectin recruitment) should provide a valuable tool to characterize the process and the molecules involved. It could provide insights into how to modulate intracellular sorting or enable screening of endosome-destabilizing compounds. It should also help in the design and optimization of delivery strategies that can overcome the endosomal-release bottleneck that thwarts effective intracellular therapeutics for siRNAs and other large-molecule drugs.
ONLINE METHODS
Cell culture
HeLa cells (American Type Culture Collection (ATCC)) and HeLa cells stably expressing destabilized d1-eGFP11 (HeLa-d1-eGFP27) were cultured in DMEM (Gibco) supplemented with 10% FCS, 2 mM glutamine, 2 mM HEPES pH 7.5, 100 U/ml penicillin, 100 mg/ml streptomycin and 50 µM β-mercaptoethanol. Cells were verified to be free of mycoplasma contamination.
Plasmids
Plasmids encoding tfLC3 (ref. 20), WT and DN dsRed-Rab5 (ref. 28) and dsRed-Rab7 (ref. 29) were provided by Addgene. Plasmids encoding C-terminal eGFP-tagged human Rab5, Rab7, Rab9 and EEA1 and N-terminal eGFP-tagged human LAMP1 were provided by the Kirchhausen lab. Cells expressing the ER-GFP marker were transduced with CellLight ER-GFP, BacMam 2.0 (Life Technologies). Plasmids encoding YFP-tagged galectins and mCherry-tagged galectin-8 were kind gifts of F. Randow12.
siRNAs and LNP formulation
The following siRNA sequences were used: Custom-synthesized Alexa Fluor 647– or Cy3-labeled siRNAs targeting eGFP: sense: 5′-GGC UAC GUC CAG GAG CGC Atts-AF647/Cy3-3′, antisense: 5′-UGC GCU CCU GGA CGU AGC Ctst-3′, where ‘s’ indicates a phosphorothioate linkage and lowercase denotes a deoxynucleotide. Silencer Select (Ambion, Life Technologies) siRNA targeting NDP52: sense: 5′-GGA GGA GCU AGA AAC CCU Att-3′, antisense: 5′-UAG GGU UUC UAG CUC CUC Ctg-3′; LGALS8: sense: 5′-CUG UCG UCG UUA AAG GAG Att-3′, antisense: 5′-UCU CCU UUA ACG ACG ACA Gtt-3′; LGALS3: sense: 5′-CGG UGA AGC CCA AUG CAA Att-3′, antisense: 5′-UUU GCA UUG GGC UUC ACC Ctg-3′; Negative control #2 siRNA (siCtrl): strand 1: 5′-UCG UAA GUA AGC GCA ACC Ctt-3′, strand 2: 5′-GGG UUG CGC UUA CUU ACG Att-3′.
LNPs were prepared using ionizable lipid L319, distearoylphosphatidylcholine (DSPC), cholesterol and PEG-DMG at a molar ratio of 55:10:32.5:2.5 (L319:DSPC:cholesterol:PEG- DMG) according to the procedure in ref. 22.
Spinning-disk confocal microscopy image acquisition
Images were acquired with a spinning disk confocal head (Yokogawa) coupled to a fully motorized epifluorescence microscope (Axio Observer) equipped with a 63× lens (Plan Apochromat, 1.4 numerical aperture (NA), Carl Zeiss) and a 40× lens (Plan Apochromat, 1.2 NA, Carl Zeiss). Three 50-mW solid-state lasers (491, 561 and 660 nm; cobalt laser), coupled to the spinning head through an acoustic-optical tunable filter (AOTF), were used as light source. The imaging system operates under control of SlideBook 5.0 and an EMCCD camera (Quant-EM, Hamamatsu) was used to acquire images.
HDR live-cell imaging of lipoplex uptake
Cells transfected with fluorescent fusion proteins were transfected 72–96 h before imaging, with an Amaxa Nucleofector (Lonza) according to the manufacturer’s instructions for HeLa cells (Kit R). Cells were plated on 25-mm poly-l-lysine–coated coverslips 12–24 h before imaging. Coverslips were transferred to a sample holder (20/20 Technology, Inc.) and 900 µl DMEM without phenol red, supplemented with 10% FCS, 2 mM glutamine, 20 mM HEPES was added. The sample holder was then put in the microscope within an environmental chamber, maintained at 37 °C, 5% CO2 and 100% humidity, containing the objective lenses. siRNA-AF647 or siRNA-Cy3 and Lipofectamine 2000 was precomplexed for 15 min at room temperature in OptiMEM (Gibco) (100:4:96, pmol:µl:µl, siRNA:Lipofectamine 2000:OptiMEM), to obtain a 1000 nM lipoplex solution. Cells were then selected for subsequent imaging. For nucleofected cells, low transgene expressing cells were chosen. Then, 100 µl siRNA-solution was added (100 nM final siRNA concentration), and image acquisition started immediately. During acquisition, normally 14 z-planes, 0.7 µm apart were acquired with 30-ms exposures of the siRNA-AF647 channel and ~100-ms exposures in the GFP channel (for endocytic marker–expressing cells). After all z-planes were acquired, a single long exposure (generally 300 ms) in the lower third of the cell was acquired in the siRNA-AF647 channel (Supplementary Fig. 1). When permitted by frame rate, multiple positions were imaged in parallel. For visualization purposes, the display lookup table (LUT) is skewed to enhance weak signals in the images (i.e., cytosolic siRNA).
In experiments with varying siRNA to transfection lipid ratios, the Lipofectamine 2000 amount was kept constant (4 µl) while either 2, 10 or 100 pmol siRNA-AF647 was complexed in 96 µl OptiMEM, and added to cells in 900 µl DMEM supplemented as above, to obtain a 2, 10 or 100 nM final siRNA concentration.
Calcium imaging
HeLa cells were preloaded with the Fluo-4 acetoxymethyl calcium indicator (Life Technologies) according to the manufacturer’s instructions. 100 nM siRNA-AF647 lipoplexes were added to the cells and imaging of a single plane was started immediately using a Zeiss LSM 710 confocal microscope with a 40× objective (Plan Apochromat, 1.3 NA, Carl Zeiss). The maximal field of view was acquired, intentionally overexposing the siRNA channel to detect the cytosolic siRNA signal.
Quantification of lipoplex associated fluorescence intensity
The fluorescence intensity of siRNA-releasing endosomes was analyzed from confocal microscopy images using SlideBook 5.0 software. A segmentation mask was created using the siRNA-AF647 signal in maximum projection time-lapse images (short exposures). The identified particles were tracked over time following the center of intensity. Releasing endosomes were identified manually as follows: 1) cells exhibiting sudden increases in cytosolic siRNA-AF647 fluorescence were identified from long exposure images; 2) the siRNA-AF647 fluorescence intensity of all identified particles in that cell was examined; and 3) if one, and only one, particle within that cell changed significantly in fluorescence intensity, this particle was identified as the releasing particle, otherwise, the endosomal escape event was discarded in the analysis. Fluorescence intensity traces were then shifted in time with t = 0 for all release events, or in some cases, t = 0 at the maximal endocytic marker fluorescence intensity. Endosomal marker fluorescence intensity values were normalized to 0–1 for every trace, with 0 being the background (autofluorescence) intensity as determined in untransfected cells, and 1 being the highest intensity value in each trace to compensate for variations in expression levels. To compare the relative abundance of the different GFP-labeled endocytic markers at the time of release, background-corrected, normalized GFP fluorescence intensities of the releasing endosomes (average of the frame before and after release) was calculated.
Flow cytometry
HeLa or HeLa-d1-eGFP cells were plated 24 h before start of incubation. For experiments with a fixed ratio of siRNA to cationic lipid, a 1,000-nM lipoplex solution was mixed (100:4:96, pmol:µl:µl, siRNA:Lipofectamine 2000:OptiMEM). Aliquots of this solution were added to cells in fresh DMEM, with 10% FCS, to obtain the desired final siRNA concentration. After incubation, cells were washed once with PBS, followed by a wash in CellScrub Wash Buffer (Genlantis) and one more PBS wash. Cells were then removed from plates by trypsin digestion, resuspended and washed twice with FACS buffer (PBS, 0.5% BSA, 10 mM HEPES). Fluorescence was analyzed on a FACSCanto (BD) or FACSAria (BD) cell analyzer using FlowJo software (Tree Star).
For LNP incubations, cells were incubated with LNPs dissolved in DMEM, 10% FCS and 1.5 µg/ml recombinant apolipoprotein E3 (ApoE3, R&D Systems). After incubation, cells were washed once with PBS, trypsinized and analyzed by flow cytometry as described above.
Live-cell eGFP expression measurements
To assess the half-life of destabilized d1-eGFP, HeLa-d1-eGFP cells were plated 24 h before imaging. After transfer to the microscope, 10 µg/ml cycloheximide (Sigma) in DMSO was added to the cells and six positions were imaged in parallel every 3 min for 5 h with a 40× objective. Control cells receiving only DMSO were imaged in a separate acquisition. In the acquired images, cells were tracked over time using an in-house–developed tracking algorithm implemented in the open-source CellProfiler (MIT) software package. Relative changes in eGFP expression were quantified by measuring the average signal intensity of every tracked cell (normalized to 1 at t = 0 for cycloheximide-treated cells and to 1 throughout the acquisition for the untreated cells). To assess the kinetics of siRNA-mediated knockdown of eGFP, HeLa-d1-eGFP cells were plated 24 h before imaging. Coverslips were transferred to a sample holder and 900 µl ice-cold DMEM (with 10% FCS) was added followed by 100 µl siGFP-AF647 lipoplex solution (100:4:96, pmol:µl:µl, siRNA:Lipofectamine 2000:OptiMEM). Cells were then incubated for 1 h at 4 °C in the dark and finally transferred to fresh medium at 37 °C and imaged using spinning-disk confocal microscopy for 12 h with a 40× objective. This procedure ensured that the majority of cells had either one or no release event. Relative changes to eGFP fluorescence levels, in cells experiencing cytosolic release events, was quantified as described above. t = 0 was set to time of release for each cell and the intensity level of nonreleasing cells was set to 1 throughout the acquisition.
Intracellular siRNA concentration estimation
In time-lapse images acquired during lipoplex–siRNA-AF647 incubations, the cytosolic siRNA-AF647 fluorescence after release was quantified as the difference in median cellular siRNA-AF647 fluorescence before and after release. Thus, the fluorescence increase of the ‘typical’ cytosolic pixel was quantified; this was practically unaffected by the fluorescence of bright vesicles and intact lipoplexes. This cytosolic siRNA-AF647 fluorescence was then converted to an estimated cytosolic siRNA concentration through the use of a reference curve. An siRNA-AF647 reference concentration curve was established by measuring four-fold dilutions of siRNA-AF647 between 1 and 1,000 nM in a cytosol-mimicking buffer (15 g/l BSA, 125 mM KCl, 4 mM KH2PO4, 14 mM NaCl, 1 mM MgCl2, 20 mM HEPES, pH 7.2). An ~10-µm high liquid column of these solutions were imaged (in triplicate) using the same imaging conditions as in the live-cell eGFP measurements. Images were corrected for background fluorescence and uneven illumination, and the median fluorescence of the entire frame was then calculated.
Interference with endosomal maturation
HeLa-d1-eGFP cells were nucleofected with dsRed-tagged Rab constructs (WT and DN) and plated for flow cytometry analysis. One day later the cells were put in fresh DMEM (with 10% FCS), and siGFP-AF647 lipoplexes (10 nM final siGFP-AF647 concentration) were added. Cells were analyzed by flow cytometry 24 h later.
For drug treatments, HeLa-d1-eGFP cells were plated for flow cytometry analysis, and one day later fresh DMEM (with 10% FCS) containing 100 µM chloroquine (Sigma) or 100 nM bafilomycin A1 (Sigma) was added. After 1 h, one-tenth the volume of siGFP-AF647 lipoplex solution was added (0.2–25 nM final siGFP-AF647 concentration). Cells were analyzed by flow cytometry 24 h later.
siRNA treatment
HeLa cells were co-nucleofected with tfLC3 and 30 pmol siRNA, followed by a second nucleofection with 30 pmol siRNA the day after. The following day, cells were plated and 24 h later the cells were incubated with 100 nM siRNA-AF647 lipoplexes and imaged using spinning-disk confocal microscopy. The GFP-LC3 signal of releasing endosomes was quantified 15 min after release.
For flow cytometry experiments, HeLa-d1-eGFP cells were nucleofected with 30 pmol siRNA, followed by a second nucleofection with 30 pmol siRNA the day after. For ‘mock’ samples, cells were treated identically except for omission of the siRNA. Two days later, the cells were put in fresh medium and one-tenth the volume of lipoplex solution (0.1–0.4:4:96, pmol:µl:µl, siRNA:Lipofectamine 2000:OptiMEM) was added. Cells were incubated for 12 h and analyzed by flow cytometry as described above.
RNA from transfected and control cells was collected in TRIzol (Life Technologies) and extracted according to the manufacturer’s protocol. Total RNA (1 µg) was converted to cDNA using SuperScript III Reverse Transcriptase (Life Technologies). For qRT-PCR, 20 µl reactions containing SsoFast EvaGreen mastermix (Bio-Rad), appropriate primers and template cDNAs made from 10 ng RNA were amplified on a Bio-Rad CFX 96 Thermal Cycler. qRT-PCR data were normalized to GAPDH or ACTB and to the expression level in siCtrl-treated cells..
LNP microscopy
HeLa cells were nucleofected with YFP–galectin-8 or conucleofected with expression plasmids for mCherry–galectin-8 and GFP-tagged endosomal markers. Cells were prepared for microscopy 24 h later. LNP–siRNA-AF647 (50 nM) in DMEM, 10% FCS and 1.5 µg/ml ApoE3 was added to the cells. The cells were then incubated for 1.5–2 h at 37 °C before spinning- disk confocal imaging was started. To limit photobleaching and toxicity, only the lower half of the cells were imaged (8 z-planes, 0.5 µm apart) at low illumination intensities for up to 45 min with frame rates of 10–30 s. Then new cells were selected and imaged up to a maximum of ~4 h after adding LNPs.
LNP image analysis
The fluorescence intensity of LNP–siRNA-AF647–releasing endosomes was analyzed from maximum z-projection confocal microscopy images using SlideBook 5.0 software. Galectin-8–targeted LNPs were first identified. The targeted LNP was then tracked manually from the first frame of appearance with a circular mask (diameter of 5 pixels). Local background fluorescence intensities were obtained from a donut-shaped mask around the particle mask. Local background–corrected fluorescence intensities of all galectin+ LNPs were then analyzed using a Matlab algorithm to identify the first frame of galectin positivity in an unbiased manner (above a defined threshold). Fluorescence intensity traces were then shifted in time with t = 0, for the time of galectin-8 recruitment, or in some cases the maximal endocytic marker fluorescence intensity (along a moving-average window). siRNA-AF647 and mCherry–galectin-8 traces were normalized to 0–1 for every trace, with 0 being the background and 1 the highest value in each trace. Background-corrected endocytic marker (GFP) traces were normalized to the average cellular GFP fluorescence.
To estimate the fraction of galectin-8+ LNP vesicles, vesicles were counted and scored manually after 165 min of LNP incubation.
Statistical analysis
Data are expressed as mean ± s.d. (s.d.). Shaded areas in intensity tracings are estimated 95% confidence values of the calculated average, defined as the s.e.m. multiplied by 1.96.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank E. Marino for maintaining the Imaging Resource used in this study and K. Ketman and N. Barteneva for technical assistance. This work was supported by US National Institutes of Health (NIH) grants AI090671 and CA139444 (J.L.), the Swedish Research Council (A.W.), NIH grant GM075252 (T.K.) and New England Regional Center of Excellence in Biodefense and Emerging Infectious Disease, Core Imaging Facility grant AI057159 (T.K.).
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
A.W. and J.L. conceived the study, designed the experiments and wrote the manuscript. T.K. provided advice. A.W. performed most of the experiments. A.A., P.H., R.T. and X.L. performed some of the experiments. K.C. and M.M. provided LNPs and advice for their use.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Felgner PL, et al. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elouahabi A, Ruysschaert J-M. Formation and intracellular trafficking of lipoplexes and polyplexes. Mol. Ther. 2005;11:336–347. doi: 10.1016/j.ymthe.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 4.Zimmermann TS, et al. RNAi-mediated gene silencing in nonhuman primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 5.Semple SC, et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 6.Coelho T, et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 2013;369:819–829. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- 7.Gilleron J, et al. Image-based analysis of lipid nanoparticle–mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 8.Sahay G, et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 2013;31:653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Payne CK, Jones SA, Chen C, Zhuang X. Internalization and trafficking of cell surface proteoglycans and proteoglycan-binding ligands. Traffic. 2007;8:389–401. doi: 10.1111/j.1600-0854.2007.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.ur Rehman Z, Hoekstra D, Zuhorn IS. Mechanism of polyplex- and lipoplex-mediated delivery of nucleic acids: real-time visualization of transient membrane destabilization without endosomal lysis. ACS Nano. 2013;7:3767–3777. doi: 10.1021/nn3049494. [DOI] [PubMed] [Google Scholar]
- 11.Li X, et al. Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- 12.Thurston TLM, Wandel MP, von Muhlinen N, Foeglein Á, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stalder L, et al. The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing. EMBO J. 2013;32:1115–1127. doi: 10.1038/emboj.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mu FT, et al. EEA1 is a conserved α-helical peripheral membrane protein flanked by cysteine “fingers” and contains a calmodulin-binding IQ motif. J. Biol. Chem. 1995;270:13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- 15.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poteryaev D, Datta S, Ackema K, Zerial M, Spang A. Identification of the switch in early-to-late endosome transition. Cell. 2010;141:497–508. doi: 10.1016/j.cell.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 17.Nechaev S, et al. Intracellular processing of immunostimulatory CpG–siRNA: Toll-like receptor 9 facilitates siRNA dicing and endosomal escape. J. Control. Release. 2013;170:307–315. doi: 10.1016/j.jconrel.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, et al. Autophagy induced by calcium phosphate precipitates targets damaged endosomes. J. Biol. Chem. 2014;289:11162–11174. doi: 10.1074/jbc.M113.531855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts R, et al. Autophagy and formation of tubulovesicular autophagosomes provide a barrier against nonviral gene delivery. Autophagy. 2013;9:667–682. doi: 10.4161/auto.23877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 21.Paz I, et al. Galectin-3, a marker for vacuole lysis by invasive pathogens. Cell. Microbiol. 2010;12:530–544. doi: 10.1111/j.1462-5822.2009.01415.x. [DOI] [PubMed] [Google Scholar]
- 22.Maier MA, et al. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akinc A, et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alabi CA, et al. Multiparametric approach for the evaluation of lipid nanoparticles for siRNA delivery. Proc. Natl. Acad. Sci. USA. 2013;110:12881–12886. doi: 10.1073/pnas.1306529110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jayaraman M, et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed. Engl. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNamara JO, et al. Cell type–specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
References
- 27.Stewart SA, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma DK, et al. Glycosphingolipids internalized via caveolar-related endocytosis rapidly merge with the clathrin pathway in early endosomes and form microdomains for recycling. J. Biol. Chem. 2003;278:7564–7572. doi: 10.1074/jbc.M210457200. [DOI] [PubMed] [Google Scholar]
- 29.Choudhury A, et al. Rab proteins mediate Golgi transport of caveola-internalized glycosphingolipids and correct lipid trafficking in Niemann-Pick C cells. J. Clin. Invest. 2002;109:1541–1550. doi: 10.1172/JCI15420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.