Abstract
Aim
To investigate the cost of antibiotic resistance versus the potential for resistant clones to adapt in maintaining polymorphism for resistance.
Materials & methods
Experimental evolution of Escherichia coli carrying different resistance alleles was performed under an environment devoid of antibiotics and evolutionary parameters estimated from their frequencies along time.
Results & conclusion
Costly resistance mutations were found to coexist with lower cost resistances for hundreds of generations, contrary to the hypothesis that the cost of a resistance dictates its extinction. Estimated evolutionary parameters for the different resistance backgrounds suggest a higher adaptive potential of clones with costly antibiotic resistance mutations, overriding their initial cost of resistance and allowing their maintenance in the absence of drugs.
Keywords: antibiotic resistance, clonal interference, epistasis, evolvability, experimental evolution
Bacterial populations can acquire antibiotic resistance (AR) as a result of transfer and acquisition of new genetic material between individuals of the same or different species but also by chromosomal DNA mutations, which alter existing bacterial proteins. One landmark example of this second process is provided by Mycobacterium tuberculosis (the etiologic agent of tuberculosis in humans) which, albeit incapable of horizontal gene transfer, can display ‘total drug resistance’ [1]. This kind of resistance is typically acquired by the sequential accumulation of mutations that alter the cellular target for the drug action. During this process extensive clonal competition has also been observed [2]. Understanding how these AR determinants disseminate and are maintained in bacterial populations is, therefore, of paramount importance.
Mutations that confer spontaneous AR can occur at relatively high rates [3]. For instance, rifampicin (Rif) resistance occurs spontaneously at frequencies that can be higher than 10−8 in wild isolates of E. coli [4]. In fact, the levels of resistance in pathogenic populations continue to rise at an alarming rate [5,6] having reached the same significance as any other virulence factor [7]. However, in the absence of the drug, AR mutations typically bear a cost [8–10]. This cost depends on the specific resistance allele [11], on the environment [12,13] and on the genomic background where the mutation happens to arise [14,15]. Nevertheless, suppressive mutations that mitigate this cost can occur either in the presence or absence of antibiotics. In the absence of drugs, one can expect that sensitive bacteria will sweep through the population, driving the AR mutant to extinction. However, more often than not, resistant strains have been observed to acquire additional beneficial mutations that reduce the costs of resistance without loss of resistance, thus preventing the elimination of resistance alleles [16]. These compensatory mutations are common in clinical isolates [17,18] and hinder the possibility of reverting the resistance mutation, due to their epistatic nature [19]. Since the probability of a compensatory mutation tends to be much higher than that of reversion for several resistance mutations [10,20–23], it is likely that resistance alleles may take long periods of time to be eliminated from populations.
Many mutations that confer AR occur in essential genes and are likely to result in pleiotropic effects altering several bacterial traits. Important examples include resistance to Rif and streptomycin (Str) [24,25]. Rif is one of the frontline antituberculosis drugs [26]. The co-occurrence of resistance to this antibiotic and isoniazid typically classifies M. tuberculosis as multidrug-resistant. These are the two most commonly used and effective drugs for the treatment of tuberculosis. The main genetic target for Rif resistance is rpoB, which codes for the β subunit of the RNA polymerase. Besides typically decreasing the rate of transcription and consequently the growth rate, rpoB resistance mutations are probably some of the most pleiotropic among AR mutations. Their effects can range from regulation of competence, sporulation and germination in Bacillus subtilis [27], temperature and phage sensitivity in E. coli [28], growth advantage in stationary phase in E. coli and Salmonella enterica [29], increased antibiotic resistance in Staphylococcus aureus [30,31], among others (see [26] for a review). rpoA and rpoC, as well as additional mutations in rpoB, have been pointed as targets for compensatory mutation relieving the deleterious effects of rpoB Rif resistance mutations in M. tuberculosis, S. enterica and E. coli [17,32–34]. Another frequent resistance is Str, occurring through mutations in the rpsL gene, which codes for the S12 subunit of the ribosome compromising translational speed and accuracy. Compensatory mutations for Str resistance have been observed in Salmonella thyphimurium, and some of the target genes include rpsL, rpsD, rpsE and rplS [12,35–36], which encode the ribosomal subunits S12, S4, S5 and L19. Recent analysis of 161 genomes of M. tuberculosis with a broad range of resistance profiles, revealed seven possible additional targets for compensatory mutations to Str resistance [37].
Given such broad targets for increasing the fitness of AR mutants we may expect that these resistances change the evolvability of bacteria, through altering the range of beneficial mutations that can be accessed by a given resistance genotype. Here we use the term evolvability as ‘the genome’s ability to produce adaptive variants’ [38]. In this sense, a costly AR mutant may be able to effectively compete with a less costly resistance allele (i.e., with a higher selective coefficient when in direct competition) if the evolvability of the former is higher. The conditions for this to happen will depend on the distribution of effects of beneficial mutations (DEBM), as well as on the rate of beneficial mutations available to each genotype [22,39,40].
Most studies addressing the competitive fitness effects associated with AR, consider resistant strains competing with the ancestral sensitive strain. However, in many natural environments the frequency of resistance can be very high and competition between different resistant alleles may be a common event [2,37,41–45]. For instance in [2], a monoclonal M. tuberculosis infection was followed and both clonal sweeps and the coexistence of different resistant mutants were observed in the dynamics of the population. Competition between different resistant strains is also likely to occur whenever there is spatial heterogeneity, with different areas posing different selective pressures [2,46,47]. Furthermore, bacteria with multiple resistance alleles are also commonly segregating in natural populations [48–50]. This competitive context might, therefore, play a crucial role in the maintenance of antibiotic resistance.
Here we study the process of fitness recovery mimicking an environment with different resistance backgrounds competing at high frequency. We use an experimental evolution approach [51] to test the ability of clones with costly AR alleles to coexist or even outcompete clones with less costly resistances. In a drug-free environment, the differences in the fitness costs of resistance alleles should determine their probability of extinction. Contrary to this simple expectation, we observe the maintenance of costly resistance alleles over hundreds of generations even when their fitness impairment should predict fast extinction. We infer that differences in adaptive potential for each AR mutant exist and suggest that these can explain the observed outcomes in the evolution of resistance.
Materials & methods
Bacterial strains & growth conditions
All bacterial strains used in this study were derived from Escherichia coli K12 MG1655 and have in common the following genotype: galK::yfp/cfp cmR (pKD3), δlacIZYA. The yellow (yfp) and cyan (cfp) alleles were integrated at the galK locus under the control of the lac promoter and were constructed by P1 transduction [52] of yfp/cfp inserts from previously constructed strains [53]. In these strains the fluorescence marker is constitutively expressed. In this common backbone, different antibiotic resistance mutations were introduced by P1 transduction. The donor strains were spontaneous antibiotic-resistant mutants previously obtained by plating the sensitive bacteria in Luria-Bertani (LB) supplemented with agar and 100 mg/ml of either streptomycin or rifampicin [14]. A total of eight strains were constructed: K43TStr-YFP, S531FRif-YFP, H526YRif-YFP, H526DRif-YFP and K43TStr-CFP, S531FRif-CFP, H526YRif-CFP, H526DRif-CFP, such that the same resistance allele was introduced in the two fluorescence backgrounds. K43TStr confers resistance to streptomycin and all the other amino acid changes confer resistance to rifampicin. In order to confirm the identity of the mutations transferred by P1 transduction, the antibiotic resistance target gene (rpoB for rifampicin or rpsL for streptomycin) was amplified and sequenced using the following primers: for the relevant fragment of the rpoB gene, 5′-CGTCGTATCCGTTCCGTTGG-3′ and 5′-TTCACCCGGATAACATCTCGTC-3′ and for the rpsL gene, 5′-ATGATGGCGGGATCGTTG-3′ and 5′-CTTCCAGTTCAGATTTACC-3′. Each resistant clone was grown from a single colony in LB medium supplemented with the respective antibiotic at 37°C with aeration and stored in 15% glycerol at −80°C.
Fitness assays & test for frequency-dependent selection
The fitness costs of antibiotic resistance mutations were first measured in competition against a sensitive reference strain (Supplementary Table S1). The reference strain carried a yfp allele if the resistant strain carried the cfp allele (and vice versa). Competitions were done after acclimatization, where each bacterial strain was grown in the same environment of the competition: in a 96-well plate with 150 μl of LB per well at 37°C with aeration. Acclimatization consisted of two consecutive passages where 5 μl from the first 24 h grown culture were used to inoculate a new plate for another 24 h. Competitions were performed by inoculating approximately 105 cells of both competitor and reference strain in LB medium and allowed to grow for 24 h (~9 generations). The initial and final ratios of both strains were determined by Flow Cytometry. Fitness effects of the resistance mutations were estimated as the slope between 0 and 24 h of the ln(f(NR)/(1-f(NR)), where f(NR) is the frequency of resistant bacteria in the population or one of the reference resistance backgrounds, in the case of the competitions of the evolved clones.
H526YRif and H526DRif strains were tested for negative frequency-dependent selection in the same conditions as described above, and the number of cells was measured using the Flow Cytometer BD LSR Fortessa (BD Biosciences), at different initial ratios of the two strains: 100:1, 10:1, 1:1, 1:10 and 1:100 (H526YRif:H526DRif).
Long-term propagation of resistant populations
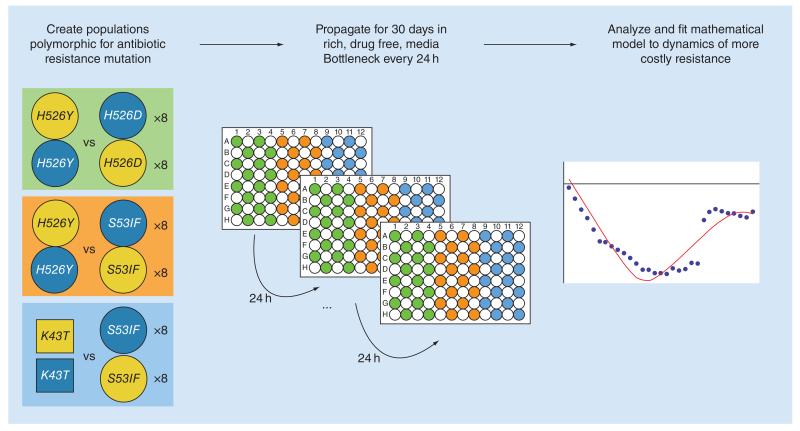
Prior to the start of the long-term competitions, acclimatization of the bacterial strains was performed in plates with a checkered arrangement (one plate for YFP strains, another for CFP strains). Each well was inoculated with an independent starting sample, from a frozen culture, into 150 μl of LB medium, to maximize the probability of sampling a large pool of beneficial mutations. We note that it is possible that beneficial mutations occur during the acclimatization period, since the rate of mutations which compensate for the cost of resistance can be very high [22]. After 24 h, 5 μl of the grown cultures where inoculated into fresh LB media and after 48 h the numbers of bacteria were measured. Appropriate dilutions were done to achieve the required initial ratio (1:1) of YFP and CFP strains for the long-term evolution of competing clones. This evolution was performed in the same conditions as the fitness assays, with daily passages of about approximately 105 bacteria for approximately 280 generations (30 days). Samples of the evolving populations were frozen every day, from which the relative abundance of each resistance was followed by measuring the frequencies of their linked fluorescent alleles by Flow Cytometry. The following three pairs of mutants were studied: K43TStr vs S531FRif, H526YRif versus H526DRif and H526YRif versus S531FRif. For each pairwise competition of mutants, 16 replicas were performed: half where one of the resistances, say R1, was linked to the yfp background and the other half where it was linked with the cfp background. Pairwise combinations of mutants (R1-YPF vs R2-CFP and R1-CFP vs R2-YFP) were settled in a checkerboard arrangement (Figure 1), where half of the wells were filled solely with LB to control for external contamination.
Figure 1. Testing the evolvability of different antibiotic resistances.
First, polymorphic populations are created by mixing, in a 1:1 ratio, clones with two different antibiotic resistance mutations. All clones are isogenic, except for this different resistance mutation and a neutral fluorescent marker. 16 replicate populations are followed for each competition, where half of these replicates have one of the resistances linked with one of the neutral markers, while the other is linked to the other marker. Three different competitive scenarios were studied, between Rifampicin mutations (circles) and between Rifampicin and Streptomycin (squares). Next, populations are passaged in rich media in 96-well plates, organized in a checkered layout and separated by control wells (with media but without bacteria). Every 24 h, all populations are passaged serially into a new 96-well plate with fresh media, during 30 days. The frequencies of the neutral markers (and hence the resistance alleles) are analyzed to unravel possible differences in the evolutionary parameters according to the resistance background.
Estimation of relative parameters of beneficial mutations
The dynamics of a given costly resistance allele (R1) when competing with another resistant clone (R2), with a different fitness cost, were analyzed under a simple model of positive selection, that assumes the occurrence of new beneficial alleles which are sweeping towards fixation. We first fitted the simplest possible model (Model 1), which can allow for a costly resistance allele to be maintained for hundreds of generations despite its initial cost. In this model we assume an initial population composed of two distinct genotypes, with different resistances and initial fitnesses, wR1 and wR2. The initial frequency of the more costly genotype, R20, is taken from a uniform distribution within the interval R2Experimental0 ± 0.1, while that of the other genotype is R10 = 1-R20. At generation TN a new genotype, with fitness wN, is assumed to have arisen and reached a frequency N0 = 0.001. From that time onwards it is assumed to change in frequency deterministically towards fixation. Therefore, we modeled selection deterministically in discrete time with two genotypes for t < TN, and three for t > TN. We estimated, by maximum likelihood, the parameters R20, TN and wN that best fit the observed values of ln(M/(1-M)), where M is the measured frequency of a fluorescent marker, at several time points, assuming a normal distribution for measurement error (with average 0 and standard deviation 0.2). The search for the set of parameters that maximize the likelihood of the data in the space of the possible parameters was performed using the Nelder–Mead method, as implemented in Mathematica 8.0 [54], with 100 iterations and repeated for 100 realizations with different initial starting combinations of parameter values. While Model 1 could provide a reasonable fit for some of the replicate experimental lines, for others it did not. We, therefore, fitted the next simplest model (Model 2), which assumes that two beneficial mutants (N1 and N2), one for each background and fluorescent marker (with fitnesses WN1 and WN2), emerged at times (TN1 and TN2). We selected the model with the lower AIC (Akaike Information Criteria) to test if the different resistances would have distinct evolvabilities. Specifically, for each pair of competing resistances and each experimental evolved population, we asked if the times of appearance of beneficial mutations or the effects of accumulated beneficial mutations were significantly different. If the genetic target for acquiring beneficial mutations is larger for resistance background R2 than for R1, but the effects of the compensatory mutations are similar, then we expect the time of appearance of mutations to be smaller in R2 than in R1. If the target is similar but the effects depend on the background we expect to observe a significant difference between the effects accumulated in one resistance background versus the other. The method used here is a discrete adaptation of existing methodologies to infer evolutionary parameters [55], with the added difference of allowing for different initial fitness values in competing genetic backgrounds. Code for these simulations is available upon request. The inference process and summary relative effects are shown for a single line, as an example, in Supplementary Figure S1.
Whole genome sequencing of H526YRif & S531FRif evolved clones
A single clone of each resistance (H526YRif and S531FRif) from each evolved population was isolated in LB agar plates with rifampicin at the end of the evolution experiment. In six populations the frequency of clones from the S531FRif background was very low and no resistant clone was sampled. DNA was extracted from each sampled clone, and then pooled according to resistance background. Paired-end sequencing using Illumina MiSeq Benchtop Sequencer, with mean coverage of 207× was performed. The resulting reads were trimmed at a Phred quality score of 99.9%, and were then aligned using Escherichia coli K12 MG1655 (NC_000913.2) as the reference genome. Mutation prediction was done using version 0.23 of BRESEQ pipeline [56], with polymorphism detection on. Other settings were used as default except for: requirement of a minimum coverage of three reads on each strand per polymorphism; polymorphism predictions occurring in homopolymers of length >3 were discarded; polymorphism predictions with significant (p < 0.05) strand or base quality score bias were discarded.
Results
Pairwise competitions between different AR alleles were performed in an antibiotic-free environment for approximately 280 generations. Each strain carries a neutral fluorescence marker. Three pairs of resistance mutations were studied: two pairs involving three different Rif resistance alleles, and one pair involving a Str and a Rif resistance allele (Figure 1). The costs of each resistance allele measured against a sensitive reference strain of E. coli are shown in Supplementary Table S1. The cost of resistance in relation to the sensitive is higher for the S531FRif mutation (0.1 ± 0.01) and the K43TRif mutation (0.09 ± 0.03), followed by H526YRif (0.07 ± 0.01) and H526DRif (0.06 ± 0.02) mutations. We then inferred the fitness cost of the same resistance mutations but this time in the presence of another resistance. Whether a resistance allele was more or less costly was estimated from the change in frequency of the fluorescent markers, linked to the resistances, during the first 3 days of evolution where the pairs of AR mutants were competing (Figures 2, 3 & 4, and also Supplementary Figure S2 for each of the replicates identified by its neutral marker). We assume that during this short period new beneficial mutations have not yet arisen, or are at a frequency too low to interfere with the relative fitness differences that both clones may have.
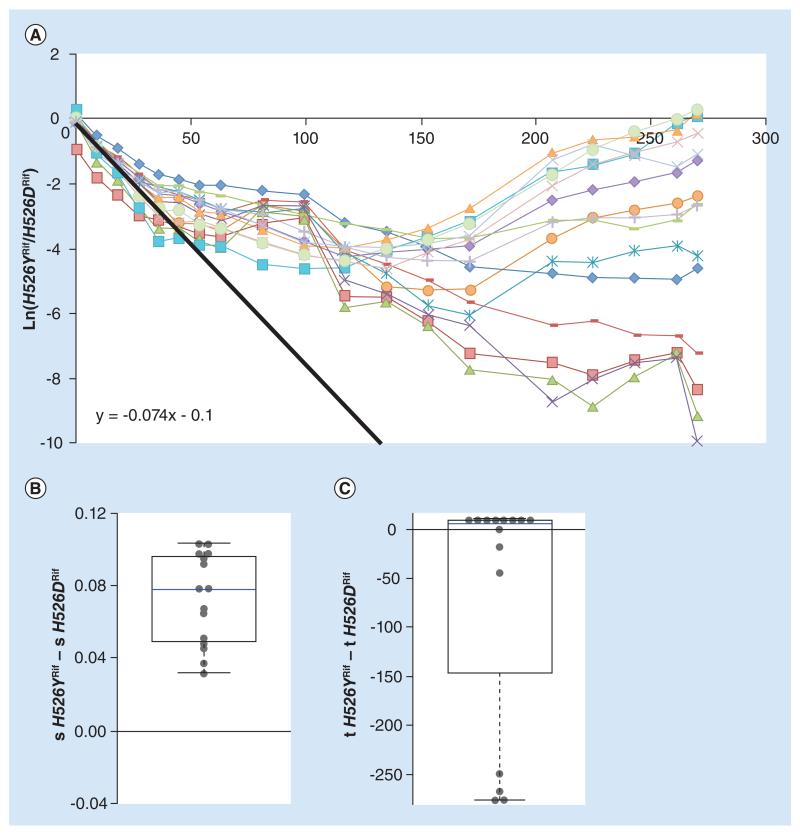
Figure 2. High evolvability of costly H526YRif allows its long-term maintenance.
(A) Long-term dynamics, for 30 days (280 generations) of evolution in 15 replicates of a population composed of resistance strain H562YRif and resistance strain H526DRif. Shown are the dynamics for the H526YRif background. The slope of the black line represents the initial difference in fitness between the resistances. (B) Whisker-box shows the relative fitness differences inferred for new beneficial mutations between the two resistant backgrounds, with H526YRif background as a reference. (C) Whisker-box shows the relative differences in time of appearance inferred for new beneficial mutations between the two resistance backgrounds.
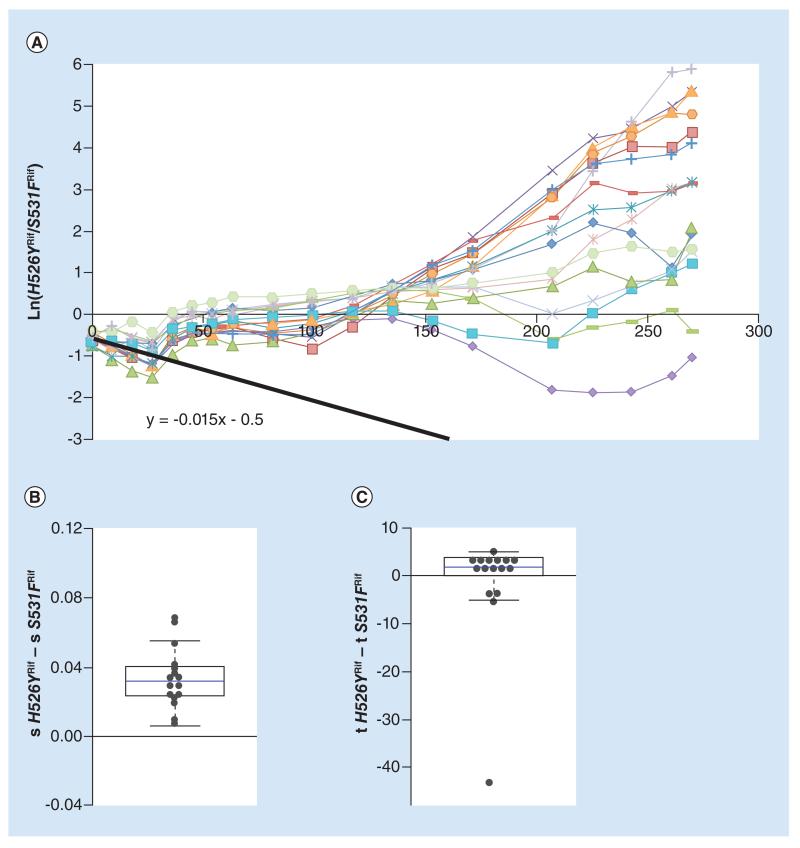
Figure 3. Different evolvabilities between Rifampicin resistance alleles.
(A) Long-term dynamics, for 30 days (280 generations) of evolution in 16 replicates of a population composed of resistance strain H562YRif and resistance strain S531FRif. Shown are the dynamics for the H526YRif background. The slope of the black line represents the initial difference in fitness between the resistances. (B) Whisker-box shows the relative fitness differences inferred for new beneficial mutations between the two resistant backgrounds, with H526YRif background as a reference. (C) Whisker-box shows the relative differences in time of appearance inferred for new beneficial mutations between the two resistance backgrounds.
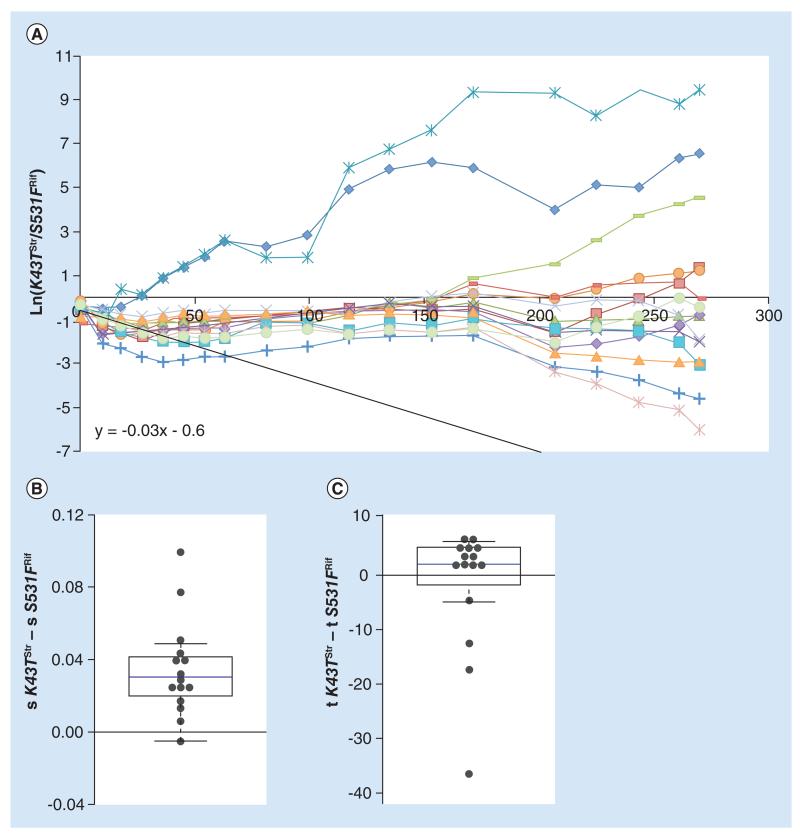
Figure 4. Differences in evolvability between Rifampicin and Streptomycin resistance alleles.
(A) Long-term dynamics, for 30 days (280 generations) of evolution in 14 replicates of a population composed of resistance strain K43TStr and resistance strain S531FRif. Shown are the dynamics for the K43T background. The slope of the black line represents the initial difference in fitness between the resistances. (B) Whisker-box shows the relative fitness differences inferred for new beneficial mutations between the two resistant backgrounds, with K43TStr background as a reference. (C) Whisker-box shows the relative differences in time of appearance inferred for new beneficial mutations between the two resistance backgrounds.
For each pairwise evolution experiment we query whether different AR mutants show distinct adaptive potential. For that, we inferred the fitness effects of new beneficial mutations that could explain the observed long-term frequency dynamics of the AR alleles. We allowed for one or two beneficial mutations of different effects to emerge at different times, in either of the backgrounds (see Methods). We sought to infer the combination of parameters (time of emergence, effect of the beneficial mutation, and the initial frequency of one of the resistance alleles) that provided the best fit to the observed evolutionary dynamics. We started by fitting a model where a single new beneficial mutation (with effect SN, at time TN) escapes stochastic loss in the background with lower initial fitness (Model 1). We estimated the value of SN and TN that best fits the dynamics of each resistance, under this model. We then used a model assuming that two beneficial mutations had increased in frequency, one in each background, and fitted the parameters of this second model (TN1, SN1, and TN2, SN2) to the dynamics observed. The lines shown in Supplementary Figure S3 represent the model that best fits the data, and the inferred parameters are presented in Tables 1, 2 & 3.
Table 1.
Evolutionary parameters estimated for the competition between strains H526Y and H526D.
| Initial H526Y frequency | H526D (R1) | H526Y (R2) | |||
|---|---|---|---|---|---|
| W | T | W | T | ||
| Population 1 | 0.538 | 1.576 | 11 | 1.552 | 15 |
| Population 2 | 0.382 | 1.293 | 11 | 1.263 | 22 |
| Population 3 | 0.488 | 1.387 | 41 | 1.348 | 51 |
| Population 4 | 0.468 | 1.483 | 26 | 1.44 | 32 |
| Population 5 | 0.413 | 1.421 | 21 | 1.409 | 30 |
| Population 6 | 0.476 | 1.474 | 31 | 1.478 | 42 |
| Population 7 | 0.513 | 1.218 | 11 | 1.19 | 19 |
| Population 8 | 0.493 | 1.776 | 18 | 1.767 | 23 |
| Population 9 | 0.557 | 1.017 | 3 | ||
| Population 10 | 0.461 | 1.028 | 29 | ||
| Population 11 | 0.581 | 1.023 | 3 | ||
| Population 12 | 0.58 | 1.002 | 46 | 1.02 | 3 |
| Population 13 | 0.523 | 1.023 | 14 | ||
| Population 14 | 0.55 | 1.001 | 38 | 1.028 | 23 |
| Population 15 | 0.59 | 1.668 | 37 | 1.671 | 45 |
W stands for the fitness of the emerging haplotype and T its time of appearance. Initial Freq stands for the inferred initial frequency of the H526Y background. In the cases where only one of the backgrounds has acquired a mutation, the inferred parameters are in bold for the background where no mutation was inferred.
Table 2.
Evolutionary parameters estimated for the competition between strains H526Y and S531F.
| Initial H526Y frequency | S531F (R1) | H526Y (R2) | |||
|---|---|---|---|---|---|
| W | T | W | T | ||
| Population 1 | 0.369 | 1.459 | 15 | 1.472 | 17 |
| Population 2 | 0.458 | 1.041 | 46 | 1.051 | 3 |
| Population 3 | 0.277 | 1.436 | 15 | 1.45 | 19 |
| Population 4 | 0.472 | 1.034 | 7 | 1.054 | 4 |
| Population 5 | 0.286 | 1.529 | 8 | 1.554 | 12 |
| Population 6 | 0.454 | 1.585 | 55 | 1.637 | 60 |
| Population 7 | 0.381 | 1.615 | 36 | 1.655 | 40 |
| Population 8 | 0.325 | 1.465 | 11 | 1.491 | 15 |
| Population 9 | 0.473 | 1.199 | 19 | 1.194 | 14 |
| Population 10 | 0.378 | 1.163 | 14 | 1.154 | 10 |
| Population 11 | 0.356 | 1.595 | 17 | 1.601 | 19 |
| Population 12 | 0.483 | 1.028 | 3 | 1.049 | 4 |
| Population 13 | 0.37 | 1.344 | 11 | 1.349 | 12 |
| Population 14 | 0.444 | 1.542 | 9 | 1.562 | 12 |
| Population 15 | 0.446 | 1.505 | 16 | 1.514 | 17 |
| Population 16 | 0.468 | 1.647 | 55 | 1.7 | 59 |
Table 3.
Evolutionary parameters estimated for the competition between strains K43T and S531F.
| Initial K43T frequency | S531F (R1) | K43T (R2) | |||
|---|---|---|---|---|---|
| W | T | W | T | ||
| Population 1 | 0.505 | 1.207 | 16 | 1.188 | 3 |
| Population 2 | 0.319 | 1.349 | 20 | 1.359 | 26 |
| Population 3 | 0.299 | 1.546 | 18 | 1.547 | 21 |
| Population 4 | 0.349 | 1.26 | 16 | 1.253 | 18 |
| Population 5 | 0.319 | 1.669 | 4 | 1.737 | 5 |
| Population 6 | 0.359 | 1.457 | 4 | 1.474 | 10 |
| Population 7 | 0.333 | 1.088 | 52 | 1.06 | 34 |
| Population 8 | 0.256 | 1.325 | 7 | 1.334 | 12 |
| Population 9 | 0.43 | 1.687 | 37 | 1.732 | 42 |
| Population 10 | 0.335 | 1.097 | 3 | 1.09 | 4 |
| Population 11 | 0.357 | 1.537 | 50 | 1.529 | 53 |
| Population 12 | 0.388 | 1.16 | 11 | 1.144 | 6 |
| Population 13 | 0.41 | 1.195 | 5 | 1.193 | 7 |
| Population 14 | 0.379 | 1.095 | 43 | 1.058 | 6 |
| Population 15 | 0.329 | 1.663 | 25 | 1.672 | 30 |
Potential for adaptation causes the maintenance of high cost resistance
We first studied the fate of two AR mutations, conferring resistance to the same antibiotic – Rif – and altering the same amino acid in the β subunit of RNA polymerase: H526YRif and H526DRif. While the costs of these resistances are not statistically different when measured against a sensitive strain, their relative fitness cost differs when they compete. From the initial generations of competition between these two strains, a fitness difference of 0.074 was inferred, with strain H526YRif being highly detrimental in this competitive environment (Figure 2A, black solid line). The cost carried by strain H562YRif leads to the rapid decline in its frequency, detected in the first tens of generations. With such a relative fitness cost, it should go extinct in 100 generations (as indicated by the black line). Strikingly though, H526YRif is clearly able to resist extinction in the majority of the replicate lines that were evolved. Four of the lines show a frequency too low to be reliably detected by flow cytometry. However, plating these populations at generation 280 allows observing the presence of fluorescent colonies of the H526YRif background at extremely low frequencies. This lack of extinction can be expected under two different scenarios: negative frequency-dependent selection and/or an ability to access higher effect beneficial mutations. We, therefore, tested for a possible signature of negative frequency-dependent selection, that is, advantage from rarity of the H526YRif mutation when competing against H526DRif. In competitions between these two backgrounds, starting with different frequencies of the H526YRif allele, no advantage from rarity can be detected (Supplementary Figure S4), so frequency-dependent selection is unlikely to be responsible for the observed lack of extinction of this mutation. On the other hand, analysis of the long-term frequency dynamics revealed a significant difference between the fitness effects of the mutations inferred to have emerged, with its median difference (sH526YRif – sH526DRif) deviating from 0 (p < 0.001, Wilcoxon Signed Rank Test) (Figure 2B). The mean difference in fitness effects between these two genetic backgrounds was 0.07 (Table 1 & Figure 2B), with H526YRif background accumulating stronger effect mutations. Regarding the times of appearance of new beneficial mutations, no significant difference was detected between the backgrounds (p = 0.41, Wilcoxon Signed Rank Test) (Figure 2C). When considering these results together, there is a strong indication that the mean effect of beneficial or compensatory mutations for H526YRif is higher, thus qualifying the strain with the H526YRif mutation as more evolvable than the strain with the H526DRif mutation. Such higher evolvability can explain the avoidance of extinction of background H526YRif, contrary to the a priori expectation based on its lower initial fitness.
Higher evolvability of H526YRif upheld in competition with an alternative resistance
We then studied the ability of the H562YRif background to outcompete a mutant, bearing a resistance mutation in a different amino acid, S531FRif. Resistance to the antibiotic rifampicin is now provided by mutations causing different amino acid substitutions in the β subunit of RNA polymerase. Relative to the sensitive strain, H526YRif is estimated to impose a smaller mean fitness cost than S531FRif (see Supplementary Table S1), although the difference is not significant. The relative fitness difference between strains H562YRif and S531FRif inferred from their direct competition is 0.015 (Figure 3A, black solid line). All lines initially decrease in frequency, in accordance with a cost of H526YRif, but in all replicates this tendency is inverted. After generation 30, the frequency of each of the resistance background stabilizes at 50% in all populations. After generation 125, additional changes in frequency can be detected. This is expected if other arising beneficial mutations increase in frequency in one or both backgrounds. In some replicate lines one of the resistance alleles starts to increase in frequency but later decreases. This is likely the result of clonal interference [57], with multiple beneficial mutations competing among them. There were more replicates in which H526YRif increased in frequency than expected, even under the assumption that the initial fitness difference between the backgrounds would be negligible. At generation 280, 14 out of 16 lines have a frequency of this allele above that expected by chance (p = 0.004, Binomial two-sided test), which is even more striking considering that its initial frequency is below 50% in most of the lines. This deviation suggests again differences in the adaptive potential between the resistance backgrounds.
We tested for possible differences in the adaptive potential of each strain by inferring the time of appearance and fitness effects of new beneficial mutations that could explain the changes in frequency of the neutral markers (Figure 3B & C). The mean value for differences in fitness between the two genetic backgrounds was 0.034 (see Figure 3B & Table 2). The median of the distribution of relative effects deviates significantly from 0 (p < 0.001, Wilcoxon Signed Rank test) (Figure 3B). The times of appearance of the new inferred beneficial mutations, however, did not appear to be significantly different between the genetic backgrounds (p = 0.3, Wilcoxon Signed Rank test) (Figure 3C). The overrepresentation of the H526YRif resistance background in the evolving populations, together with this analysis, indicates that H526YRif has a higher evolvability across different competitive contexts and, therefore, can be easily maintained.
Potential for adaptation drives the fate of resistances to different antibiotics
Differences in evolvability are expected to be larger amongst resistances affecting different genes than between alleles from the same gene. This is so because the target for beneficial mutations is expected to be more similar between mutations affecting the same function than between mutations impairing different traits. To query if the costs or the evolvabilities are determinant to the competition of resistances to distinct drugs, we studied the fate of the Str resistance allele (K43TStr) when in competition with the Rif resistance allele (S531FRif). K43TStr is estimated to impose a cost to the sensitive strain of approximately 0.09, which is not statistically different from the cost imposed by S531FRif. However, upon competition between these two strains, K43TStr shows a disadvantage of around 0.03 relative to S531FRif (Figure 4A, black solid line). This is observed in the initial 25 generations, where K43TStr decreases in frequency in most replicate competitions. The long-term evolutionary dynamics, however, depart from those observed in the previous studied cases. In Figure 4A, a higher variation in the outcome of which resistance wins the competition emerges in this pair of competing resistances. In the vast majority of the replicate lines the K43TStr mutation survived extinction for the duration of the experiment, contrary to what would have been expected from its initial relative fitness cost. In most of the lines, K43TStr is kept at a stable frequency of around 30% and in three lines it rises in frequency, sweeping to majority status (above 99% frequency) by generation 280. Contrary to expectations, extinction (frequency below 1%) of the more costly allele was only observed in two of the populations. Most of the lines where the K43TStr resistance was kept at a stable but low frequency at the middle time points had different outcomes, with some rising and others decreasing in frequency. Overall, at the end of the 280 generations, the background with the K43TStr allele reached a frequency higher than 50% in 6 out of 15 evolved replicate lines. The inference of evolutionary parameters performed for the long-term dynamics indicates that there is a significant difference in the strength of mutations acquired by each background (p = 0.014, Wilcoxon Signed Rank test) (Figure 4B). K43TStr acquires beneficial mutations of a stronger effect than does the background S531FRif (mean difference 0.03, Table 3). No significant difference was detected for the times of appearance of beneficial mutations between both backgrounds (p = 0.6, Wilcoxon Signed Rank Test) (Figure 4C). The differences inferred from the dynamics in Figure 4A, therefore, depart from the expectation of a non-epistatic model of beneficial mutations, where both backgrounds would access mutations of similar effect. Extinction might have been avoided for this Str resistance mutation by its ability to accumulate stronger effect mutations when competing with the Rif resistant strain.
Genetic characterization & fitness determination of evolved clones isolated from the long-term competition between H526YRif & S531FRif
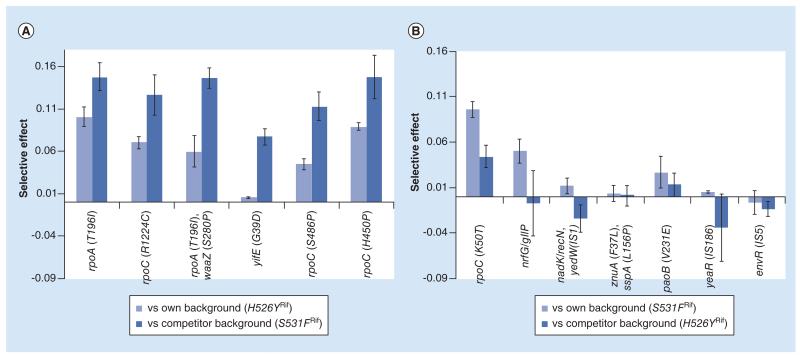
In the three cases of long-term competitions studied, the differences in evolvability inferred resulted from differences in the effects of the beneficial mutations acquired. We inferred that H526YRif could acquire higher effect mutations, relative to those emerging in S531FRif clones, even though these two different alleles cause very similar fitness costs. To gain further insight into the mutations acquired by the H526YRif background in this competitive context, we performed whole genome sequencing (see Methods for a description of the procedure) of a sample of H526YRif and of S531FRif clones, after these two lineages had evolved in competition (Figure 3). Table 4 shows the mutations identified in each of the evolved backgrounds. We could identify 12 targets (in 10 distinct genes or intergenic regions) for possible beneficial (and compensatory) mutations in H526YRif (all single point mutations), and 10 targets (all in different genes/intergenic regions) in S531FRif (7 of them were single point mutations, the other 3 involved transpositions of insertion sequence elements). Mutations in the H526YRif background were observed at a higher frequency compared with the ones observed in S531FRif. This suggests that the mutations appearing in the H526YRif background have stronger effects, since they are acquired by different replicate evolving populations, pointing to a rapid fixation due to their beneficial effect. To enquire if this is indeed the case we performed competition assays between each individual clone and the ancestral strains. Since the whole genome sequencing was performed with a mixture of clones (see ‘Methods’ section), we targeted sequencing each individual clone to gain access to the haplotypic composition of each clone. For the clones carrying the H526YRif mutation, we targeted the most prevalent mutations detected (see Table 4). Fifteen out of the 16 H526YRif clones carried at least one mutation in the target genes (Supplementary Table 2) and one clone was found to have two mutations, one single point mutation in rpoA and another in waaZ. The amino acid change T196I in rpoA was present in eight independently evolved clones. Since all of them have the same genetic background (H526YRif YFP), this suggests that this mutation could already have been present prior to the long-term evolution experiment. All S531FRif clones, except one, carried one of the previously identified mutations (Supplementary Table 2) and rpoC was also a target for beneficial mutations in this RifR background. We then measured the competitive fitness of each of these evolved clones to directly assay their fitness advantage. In order to increase in frequency during the long-term propagation, these mutants had to outcompete their ancestral and also the competitor with a different resistance background. Figure 5 shows that when competing either against their respective ancestral or the other resistance clone, the selective effects of the evolved H526YRif clones are stronger than the selective effects of the mutations acquired by the S531FRif background. For the latter background, the mutations are advantageous against their ancestral, supporting their increase to detectable frequencies, but most are neutral or even deleterious versus the opposite H526YRif background. Remarkably, mutations in rpoA and rpoC provided the strongest competitive fitness advantages. Overall, these results therefore provide further support for the previously identified higher evolvability of the H526YRif background and strongly suggest that the H526YRif resistance may be easily maintained in populations.
Table 4.
Potential compensatory mutations identified in the genomes of the clones evolved in the competition between resistances H526Y and S531F.
| Background | Genome Position | Gene(s) | Mutation | Annotation | Frequency | (%) Function |
|---|---|---|---|---|---|---|
| H526Y | 3,438,465 | rpoA | C→T | T196I | 51.5 | RNA polymerase, alpha subunit |
| 4,184,828 | rpoC | T→C | S486P | 25.5 | ||
| 4,187,042 | rpoC | C→T | R1224C | 6.1 | RNA polymerase, beta prime subunit | |
| 4,184,721 | rpoC | A→C | H450P | 2.9 | ||
| 3,797,382 | waaZ | T→C | S280P | 19.7 | Lipopolysaccharide core biosynthesis protein | |
| 3,946,224 | yifE | G→A | G39D | 12.0 | Conserved protein, UPF0438 family, unknown function | |
| 4,292,389 | nrfG/gltP | A→T | Intergenic | 5.0 | [nrfG] Heme lyase (NrfEFG) for insertion of heme into c552, subunit NrfG/[gltP] glutamate/aspartate: proton symporte | |
| 3,637,091 | pitA | C→T | A476V | 2.6 | Phosphate transporter, low- affinity; tellurite importer | |
| 374,196 | mhpE/mhpT | T→C | Intergenic | 2.4 | 4- hyroxy- 2- oxovalerate/4- hydroxy- 2- oxopentanoic acid aldolase, class I/putative 3- hydroxyphenylpropionic transporter | |
| 3,464,375 | bfr | C→T | R125C | 2.0 | Bacterioferritin, iron storage and detoxification protein | |
| 1,743,180 | ydhQ | T→C | S324S | 3.1 | Hypothetical protein | |
| 3,898,043 | yieL | C→A | T186T | 2.5 | Putative xylanase | |
|
| ||||||
| S531F | 4,183,521 | rpoC | A→C | K50T | 14.8 | RNA polymerase, beta prime subunit |
| 4,292,389 | nrfG/gltP | A→T | Intergenic | 19.6 | [nrfG] Heme lyase (NrfEFG) for insertion of heme into c552, subunit NrfG/[gltP] Glutamate/aspartate:proton symporte | |
| 2,749,808 | nadK/recN | δ1 bp | Intergenic | 18.1 | [nadK] NAD kinase/ [recN] Recombination and repair protein | |
| 1,940,499 | znuA | T→C | F37L | 17.7 | Zinc transporter subunit: periplasmic- binding component of ABC superfamily | |
| 3,374,976 | sspA | T→C | L156P | 12.2 | Stringent starvation protein A | |
| 300,420 | paoB | T→A | V231E | 11.8 | PaoABC aldehyde oxidoreductase, FAD- containing subunit | |
| 2,036,728 | yedW | IS1 Ins | Coding | 9.9 | Putative DNA- binding response regulator in two- component system with | |
| 1,877,853 | yeaR | IS186 Ins | Coding | 8.4 | Hypothetical protein | |
| 3,410,893 | envR | IS5 Ins | Coding | 6.8 | DNA-binding transcriptional regulator | |
| 2,361,326 | yfaY | A→G | V110V | 9.4 | Hypothetical protein | |
Mutations were identified in the genomes of the evolved clones in comparison with a reference genome. Shown are the single nucleotide polymorphisms or IS events identified in either of the backgrounds, and the frequencies at which they were detected. Mutations that occurred between genes (intergenic) are identified as such, otherwise all remaining mutations occurred within the gene indicated. The mutations previously identified to be compensatory are identified with a gray shaded cell.
IS: Insertion sequence.
Figure 5. Differences in selective effects of evolved clones from the competition between H526YRif and S531FRif.
(A) Fitness effects of selected clones evolved from the H526YRif background, when in competition with their ancestor (light bars) or against their other resistant competitor (S531FRif, dark bars). (B) Fitness effects of selected clones evolved from the S531FRif background, when in competition with their ancestor (light bars) or against their other resistant competitor (H526YRif, dark bars).
Discussion
In order to understand the relative role of cost versus evolvability in the maintenance of AR mutations we studied how subpopulations carrying different resistant alleles compete. This system mimics the composition of a population after drug exposure, where the different AR mutants co-exist at relatively high frequencies. This scenario has rarely been studied, although it possibly occurs in natural contexts, as has been observed in strains sampled from the same patient [2,45]. Here we address this situation by following the fate of pairwise combinations of different alleles conferring resistance to the same or different drugs. We observe that the cost of resistance measured in competition against the sensitive bacterium is not always a good predictor of the difference in costs between two resistance mutations, and transitivity between these two fitness measures is not always observed. Therefore, to understand why some resistant alleles are rarely segregating while others are pervasive, it is important to also measure their selective coefficients when in coexistence in addition to measure their costs against the sensitive strain.
In this work we studied two sets of mutations whose fitness costs were barely distinguishable when compared with the sensitive strain. The first set comprised three alleles of the same gene (H526YRif, H526DRif and S531FRif), thus conferring resistance to the same drug; the second group comprised alleles of different genes (S531FRif and K43TStr), thus conferring resistance to different antibiotics. In the first set we found that H526YRif has a very significant cost (0.074) when competing with mutation H526DRif and it is less costly when competing with S531F (0.015), implying that, all else being equal, H526YRif should rapidly go extinct when competing with H526DRif, but do so at a slower pace when in competition with S531FRif. The long-term outcome of these competitions indicated that this does not always occur and the H526YRif allele can be maintained in both cases. The evolvability analysis undertaken suggests that the difference between H526YRif and H526DRif could be attributed to a higher mean effect of beneficial mutations accessible to H526YRif. This result was unexpected, because this pair of mutations involves the same amino acid replacement and the different single point mutations cause a fitness defect of similar magnitude. Given this, one could expect similar evolvabilities between the resistance backgrounds [58]. The outcome of the competition between H526YRif and S531FRif (mutations in different amino acids but again in the same gene) shows a tendency for the former to increase in frequency, and this could also be due to access of the former background to higher effect mutations, when in comparison with the latter. This observation is further supported by the different competitions between H526YRif and S531FRif clones, where competitive fitness assays showed stronger effects of the mutations acquired by H526YRif, as predicted by our theoretical analysis. The two cases of competition between different AR mutations both show a long-term advantage of the H526YRif allele, through its higher mean effect of mutations. Interestingly, H526YRif has been reported to be the second most frequent mutation, in a variety of clinical settings, among the ones that confer Rif resistance in M. tuberculosis, even though it was also estimated that this mutation was the third mutation more costly in the same set [59,60]. This could be explained by the particularly high evolvability of this mutation. In the case of the pair of AR mutants composed of K43TStr and S531FRif, differences in evolvability were expected a priori since these mutations alter genes responsible for different cellular traits. In fact, evolution was less reproducible in this situation, with different replicates following different dynamics. Our results indicate that K43TStr acquired beneficial mutations of stronger effect than S531FRif, suggesting that their DEBM and hence their evolvability are different.
The ability to access specific subsets of beneficial mutations determines how evolvable an organism is. If there were no constraints, then all genotypes, regardless of their composition or competitive context, would be able to adapt in a predictable sequence of mutational events. However, pleiotropy and epistasis may limit the access to new beneficial mutations [14,61–62], imposing different evolutionary outcomes in different genetic backgrounds, environmental conditions and/or competitive contexts. This is particularly relevant in the context of AR, as emergence of resistance mutants is fairly common. The resistant clones would presumably be driven to extinction when competing against less costly strains, in the absence of the drugs. Here we show that it is very likely that such extinction events will not occur and, instead, mutations that buffer the effects of resistance alleles will accumulate [36,49,63–64], allowing their maintenance. Our results indicate that different adaptive abilities and the access to strong effect beneficial mutations depends on the genetic background and the competitive context, determining the long-term fate of a given resistance allele.
Antibiotic-based treatments focus on immediate clinical results, but the adaptive potential of resistant bacteria is subtle [65,66] and we suggest it can affect their long-term fate within a host or between hosts. An interesting recent observation suggests that the effects of mutations conferring resistance to streptomycin tend to be smaller for genotypes that are well adapted to a given environment, relative to genotypes not adapted at all [15]. This observation, along with the one we have made here, indicates that it is relevant to determine the relative roles of cost versus evolvability in other environments of special clinical relevance [67], in order to be better able to predict the evolution of pathogens carrying resistance alleles.
Conclusion
The frequency of antibiotic resistance constitutes an alarming concern for public health. A key factor determining the extinction or maintenance of resistance alleles is the fitness costs they may entail. High cost resistance alleles are expected to rapidly go extinct. However, this may not be an inescapable fate. If the availability of beneficial mutations is dependent on the genetic background, clones with less fit resistance alleles may also have higher evolvability, in other words, a higher potential for adaptation. If so, this will lead to the maintenance of resistances with a higher initial cost in populations. Here we perform competitions between strains of E. coli, which carry resistance alleles of different costs, and estimate the relative differences in their adaptive potential. We demonstrate that costly resistance alleles can coexist with resistance alleles of lower cost for hundreds of generations, suggesting that their adaptive potential can override the initial relative cost of resistance.
Future perspective
Antibiotic resistance poses an ever-increasing danger to public health, and its maintenance is the result of a multitude of processes, which require increasing evaluation. How the costs of resistance depend both on the specific resistant alleles, the environment where the bacteria grow and the ecological context to which they are exposed should lead to a better understanding on how resistance can be reduced or avoided. The ability of resistant bacteria for acquiring compensatory mutations and revert to sensitive state across environments should also be evaluated with the help of increasing powerful genomic technics. This is especially important for bacteria carrying multiple resistances as these are becoming more and more common. Assaying the ability of resistance alleles to emerge and thrive in ecologically relevant contexts, which are very likely to include several resistances competing simultaneously and multiple biotic factors will become crucial for our proper understanding of their long-term pathogenicity.
Supplementary Material
EXECUTIVE SUMMARY.
Long-term evolution of polymorphic antibiotic resistance populations
In all three pairwise competitions studied, the more costly resistance avoided extinction with high probability.
In the majority of populations the costly resistance did not sweep to fixation.
The second most frequent rifampicin resistance mutation to segregate in natural pathogen populations (H526YRif) shows a high cost but also higher resistance to extinction, across multiple competitive contexts.
Estimation of evolutionary parameters
The distribution of effects for the beneficial mutations depends on the resistance background, with more costly resistance backgrounds acquiring mutations of stronger effects.
Differences in rate of acquisition of beneficial mutations were not detected between the resistance backgrounds.
Sequencing & competitive fitness of evolved populations
Whole genome sequencing of the evolving replicate populations with the H526YRif and S531FRif competing clones revealed that the H526YRif background acquired more mutations at higher frequencies pointing towards its increased adaptive potential.
The number of targets identified for beneficial mutations was not significantly different between the resistance backgrounds, supporting the results from the theoretical analysis.
Competitive fitness assays showed that the mean effect of beneficial mutations is different between these two resistant backgrounds (H526YRif and S531FRif), as inferred from theoretical modeling.
Conclusion
The initial relative difference in fitness costs between resistances is not predictive of their long-tern evolution.
The long-term outcome of competitions between pairs of distinct antibiotic resistance alleles is polymorphism for resistance.
The results indicate that it is crucial to understand the ecological contexts and the adaptive potential of antibiotic resistance mutations in order to make informed clinical decisions regarding the treatment of bacterial infections.
Acknowledgements
The authors thank Guillaume Martin for the useful comments, from which we considered some, and Hajrabibi Ali for the experimental support.
The research leading to these results has received funding from the European Research Council under the European Community’s Seventh Framework Programme (FP7/2007–2013)/ERC grant agreement no 260421 – ECOADAPT, and PTDC/BIA-EVF/114622/2009, financed by Fundação para a Ciência e Tecnologia (FCT).
No writing assistance was utilized in the production of this manuscript.
Footnotes
Data accessibility
The whole genome sequencing data will be available through the NCBI Sequence Read Archive (SRA) database.
Financial & competing interests disclosure
I Gordo acknowledges the salary support of LAO/ITQB & FCT. JM de Sousa acknowledges the scholarship SFRH/BD/89151/2012, from FCT. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Velayati AA, Masjedi MR, Farnia P, et al. Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in Iran. Chest. 2009;136(2):420–425. doi: 10.1378/chest.08-2427. [DOI] [PubMed] [Google Scholar]
- 2.Mariam SH, Werngren J, Aronsson J, Hoffner S, Andersson DI. Dynamics of antibiotic resistant Mycobacterium tuberculosis during long-term infection and antibiotic treatment. PLoS ONE. 2011;6(6):e21147. doi: 10.1371/journal.pone.0021147. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• In a Mycobacterium tuberculosis infection, both clonal sweeps and the coexistence of different resistant mutants were observed in the dynamics of the population.
- 3.Mwangi MM, Kim C, Chung M, et al. Whole-genome sequencing reveals a link between β-lactam resistance and synthetases of the alarmone (p)ppGpp in Staphylococcus aureus. Microb. Drug Res. 2013;19(3):153–159. doi: 10.1089/mdr.2013.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baquero MR, Nilsson AI, del Carmen Turrientes M, et al. Polymorphic mutation frequencies in Escherichia coli: emergence of weak mutators in clinical isolates. J. Bacteriol. 2004;186(16):5538–5542. doi: 10.1128/JB.186.16.5538-5542.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh C. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000;406(6797):775–781. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- 6.Laxminarayan R, Duse A, Wattal C, et al. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013;13(12):1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 7.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010;74(3):417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersson DI, Levin BR. The biological cost of antibiotic resistance. Curr. Opin. Microbiol. 1999;2(5):489–493. doi: 10.1016/s1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
- 9.Lenski RE. Bacterial evolution and the cost of antibiotic resistance. Int. Microbiol. 1998;1(4):265–270. [PubMed] [Google Scholar]
- 10.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 2010;8(4):260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 11.Mariam DH, Mengistu Y, Hoffner SE, Andersson DI. Effect of rpoB mutations conferring rifampin resistance on fitness of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2004;48(4):1289–1294. doi: 10.1128/AAC.48.4.1289-1294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Björkman J, Nagaev I, Berg OG, Hughes D, Andersson DI. Effects of environment on compensatory mutations to ameliorate costs of antibiotic resistance. Science. 2000;287(5457):1479–1482. doi: 10.1126/science.287.5457.1479. [DOI] [PubMed] [Google Scholar]
- 13.Chait R, Craney A, Kishony R. Antibiotic interactions that select against resistance. Nature. 2007;446(7136):668–671. doi: 10.1038/nature05685. [DOI] [PubMed] [Google Scholar]
- 14.Trindade S, Sousa A, Xavier KB, Dionisio F, Ferreira MG, Gordo I. Positive epistasis drives the acquisition of multidrug resistance. PLoS Genet. 2009;5(7):e1000578. doi: 10.1371/journal.pgen.1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angst DC, Hall AR. The cost of antibiotic resistance depends on evolutionary history in Escherichia coli. BMC Evol. Biol. 2013;13(1):1–l. doi: 10.1186/1471-2148-13-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maisnier-Patin S, Andersson DI. Adaptation to the deleterious effects of antimicrobial drug resistance mutations by compensatory evolution. Res. Microbiol. 2004;155(5):360–369. doi: 10.1016/j.resmic.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 17.Comas I, Borrell S, Roetzer A, et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat. Genet. 2012;44(1):106–110. doi: 10.1038/ng.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Vos M, Müller B, Borrell S, et al. Putative compensatory mutations in the rpoC gene of rifampin-resistant Mycobacterium tuberculosis are associated with ongoing transmission. Antimicrob. Agents Chemother. 2013;57(2):827–832. doi: 10.1128/AAC.01541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis BH, Poon AFY, Whitlock MC. Compensatory mutations are repeatable and clustered within proteins. Proc. Biol. Sci. 2009;276(1663):1823–1827. doi: 10.1098/rspb.2008.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levin BR, Perrot V, Walker N. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics. 2000;154(3):985–997. doi: 10.1093/genetics/154.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poon A. The coupon collector and the suppressor mutation: estimating the number of compensatory mutations by maximum likelihood. Genetics. 2005;170(3):1323–1332. doi: 10.1534/genetics.104.037259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sousa A, Magalhaes S, Gordo I. Cost of antibiotic resistance and the geometry of adaptation. Mol. Biol. Evolut. 2012;29(5):1417–1428. doi: 10.1093/molbev/msr302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gifford DR, MacLean RC. Evolutionary reversals of antibiotic resistance in experimental populations of Pseudomonas aeruginosa. Evolution. 2013;67(10):2973–2981. doi: 10.1111/evo.12158. [DOI] [PubMed] [Google Scholar]
- 24.Couturier M, Desmet L, Thomas R. High pleiotropy of streptomycin mutations in Escherichia coli. Biochem. Biophys. Res. Comm. 1964;16(3):244–248. doi: 10.1016/0006-291x(64)90333-x. [DOI] [PubMed] [Google Scholar]
- 25.Romero E, Riva S, Berti M, Fietta AM, Silvestri LG. Pleiotropic effects of a rifampicin-resistant mutation in E. coli. Nature New Biol. 1973;246(155):225–228. doi: 10.1038/newbio246225a0. [DOI] [PubMed] [Google Scholar]
- 26.Koch A, Mizrahi V, Warner DF. The impact of drug resistance on Mycobacterium tuberculosis physiology: what can we learn from rifampicin? Emerg. Microbes Infect. 2014;3(3):e17. doi: 10.1038/emi.2014.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maughan H, Galeano B, Nicholson WL. Novel rpoB mutations conferring rifampin resistance on Bacillus subtilis: global effects on growth, competence, sporulation, and germination. J. Bacteriol. 2004;186(8):2481–2486. doi: 10.1128/JB.186.8.2481-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin DJ, Walter WA, Gross CA. Characterization of the termination phenotypes of rifampicin-resistant mutants. J. Mol. Biol. 1988 doi: 10.1016/0022-2836(88)90455-x. [DOI] [PubMed] [Google Scholar]
- 29.Wrande M, Roth JR, Hughes D. Accumulation of mutants in ‘aging’ bacterial colonies is due to growth under selection, not stress-induced mutagenesis. Proc. Natl Acad. Sci. USA. 2008;105(33):11863–11868. doi: 10.1073/pnas.0804739105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui L, Isii T, Fukuda M, et al. An rpoB mutation confers dual heteroresistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 2010;54(12):5222–5233. doi: 10.1128/AAC.00437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe Y, Cui L, Katayama Y, Kozue K, Hiramatsu K. Impact of rpoB mutations on reduced vancomycin susceptibility in Staphylococcus aureus. J. Clin. Microbiol. 2011;49(7):2680–2684. doi: 10.1128/JCM.02144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brandis G, Wrande M, Liljas L, Hughes D. Fitness-compensatory mutations in rifampicin-resistant RNA polymerase. Mol. Microbiol. 2012;85(1):142–151. doi: 10.1111/j.1365-2958.2012.08099.x. [DOI] [PubMed] [Google Scholar]
- 33.Brandis G, Hughes D. Genetic characterization of compensatory evolution in strains carrying rpoB Ser531Leu, the rifampicin resistance mutation most frequently found in clinical isolates. J. Antimicrob. Chemother. 2013;68(11):2493–2497. doi: 10.1093/jac/dkt224. [DOI] [PubMed] [Google Scholar]
- 34.Reynolds MG. Compensatory evolution in rifampin-resistant Escherichia coli. Genetics. 2000;156(4):1471–1481. doi: 10.1093/genetics/156.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schrag SJ, Perrot V, Levin BR. Adaptation to the fitness costs of antibiotic resistance in Escherichia coli. Proc. Royal Soc. B Biol. Sci. 1997;264(1386):1287–1291. doi: 10.1098/rspb.1997.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maisnier-Patin S, Berg OG, Liljas L, Andersson DI. Compensatory adaptation to the deleterious effect of antibiotic resistance in Salmonella typhimurium. Mol. Microbiol. 2002;46(2):355–366. doi: 10.1046/j.1365-2958.2002.03173.x. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, Li D, Zhao L, et al. Genome sequencing of 161 Mycobacterium tuberculosis isolates from China identifies genes and intergenic regions associated with drug resistance. Nat. Genet. 2013;45(10):1255–1260. doi: 10.1038/ng.2735. [DOI] [PubMed] [Google Scholar]
- 38.Wagner GP, Altenberg L. Perspective: complex adaptations and the evolution of evolvability. Evolution. 1996:967–976. doi: 10.1111/j.1558-5646.1996.tb02339.x. [DOI] [PubMed] [Google Scholar]
- 39.Handel A, Regoes RR, Antia R. The role of compensatory mutations in the emergence of drug resistance. PLoS Comput. Biol. 2006;2(10):e137. doi: 10.1371/journal.pcbi.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrick JE, Kauth MR, Strelioff CC, Lenski RE. Escherichia coli rpoB mutants have increased evolvability in proportion to their fitness defects. Mol. Biol. Evolut. 2010;27(6):1338–1347. doi: 10.1093/molbev/msq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farhat MR, Shapiro BJ, Kieser KJ, et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat. Genet. 2013;45(10):1183–1189. doi: 10.1038/ng.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MOA, Dantas G. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337(6098):1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nolan CM, Williams DL, Cave MD, et al. Evolution of rifampin resistance in human immunodeficiency virus-associated tuberculosis. Am. J. Respir. Crit. Care Med. 1995;152(3):1067–1071. doi: 10.1164/ajrccm.152.3.7663785. [DOI] [PubMed] [Google Scholar]
- 44.Mwangi MM, Wu SW, Zhou Y, et al. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc. Natl Acad. Sci. USA. 2007;104(22):9451–9456. doi: 10.1073/pnas.0609839104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun G, Luo T, Yang C, et al. Dynamic population changes in Mycobacterium tuberculosis during acquisition and fixation of drug resistance in patients. J. Infect. Dis. 2012;206(11):1724–1733. doi: 10.1093/infdis/jis601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hermsen R, Deris JB, Hwa T. On the rapidity of antibiotic resistance evolution facilitated by a concentration gradient. Proc. Natl Acad. Sci. USA. 2012;109(27):10775–10780. doi: 10.1073/pnas.1117716109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Q, Lambert G, Liao D, et al. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science. 2011;333(6050):1764–1767. doi: 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]
- 48.Wright GD. The antibiotic resistome. Expert Opin. Drug Discov. 2010;5(8):779–788. doi: 10.1517/17460441.2010.497535. [DOI] [PubMed] [Google Scholar]
- 49.Borrell S, Teo Y, Giardina F, et al. Epistasis between antibiotic resistance mutations drives the evolution of extensively drug-resistant tuberculosis. Evolut. Med. Public Health. 2013;2013(1):65–74. doi: 10.1093/emph/eot003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merker M, Kohl TA, Roetzer A, et al. Whole genome sequencing reveals complex evolution patterns of multidrug-resistant Mycobacterium tuberculosis Beijing strains in patients. PLoS ONE. 2013;8(12):e82551. doi: 10.1371/journal.pone.0082551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jansen G, Barbosa C, Schulenburg H. Experimental evolution as an efficient tool to dissect adaptive paths to antibiotic resistance. Drug Resist. Updat. 2014;16(6):96–107. doi: 10.1016/j.drup.2014.02.002. [DOI] [PubMed] [Google Scholar]; • Important discussion on the role of experimental evolution in understanding the long-term outcome of antibiotic resistance.
- 52.Silhavy TJ, Berman ML, Enquist LW. Experiments With Gene Fusions. Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 53.Hegreness M. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science. 2006;311(5767):1615–1617. doi: 10.1126/science.1122469. [DOI] [PubMed] [Google Scholar]
- 54.Wolfram Mathworld™ Nelder-Mead Method. http://mathworld.wolfram.com/Nelder-MeadMethod.html.
- 55.Illingworth CJR, Mustonen V. A method to infer positive selection from marker dynamics in an asexual population. Bioinformatics. 2012;28(6):831–837. doi: 10.1093/bioinformatics/btr722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barrick Lab breseq. http://barricklab.org/twiki/bin/view/Lab/ToolsBacterialGenomeResequencing.
- 57.Sniegowski PD, Gerrish PJ. Beneficial mutations and the dynamics of adaptation in asexual populations. Philos. Trans. Royal Soc. B Biol. Sci. 2010;365(1544):1255–1263. doi: 10.1098/rstb.2009.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fisher RA. The Genetical Theory of Natural Selection. Clarendon Press; Oxford, UK: [Google Scholar]
- 59.Gagneux S, Long CD, Small PM, Van T, Schoolnik GK, Bohannan BJM. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science. 2006;312(5782):1944–1946. doi: 10.1126/science.1124410. [DOI] [PubMed] [Google Scholar]; •• Multiresistant strains are obtained from clinical infections, despite the fact that the laboratory evolved resistance mutations carry a fitness cost.
- 60.Trauner A, Borrell S, Reither K, Gagneux S. Evolution of drug resistance in tuberculosis: recent progress and implications for diagnosis and therapy. Drugs. 2014;74(10):1063–1072. doi: 10.1007/s40265-014-0248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khan AI, Dinh DM, Schneider D, Lenski RE, Cooper TF. Negative epistasis between beneficial mutations in an evolving bacterial population. Science. 2011;332(6034):1193–1196. doi: 10.1126/science.1203801. [DOI] [PubMed] [Google Scholar]
- 62.Woods RJ, Barrick JE, Cooper TF, Shrestha U, Kauth MR, Lenski RE. Second-order selection for evolvability in a large Escherichia coli population. Science. 2011;331(6023):1433–1436. doi: 10.1126/science.1198914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maisnier-Patin S, Paulander W, Pennhag A, Andersson DI. Compensatory evolution reveals functional interactions between ribosomal proteins S12, L14 and L19. J. Mol. Biol. 2007;366(1):207–215. doi: 10.1016/j.jmb.2006.11.047. [DOI] [PubMed] [Google Scholar]
- 64.Hall AR, MacLean RC. Epistasis buffers the fitness effects of rifampicin-resistance mutations in Pseudomonas aeruginosa. Evolution. 2011;65(8):2370–2379. doi: 10.1111/j.1558-5646.2011.01302.x. [DOI] [PubMed] [Google Scholar]; • Characterization of patterns of epistatic interactions for a broad range of rifampicin-resistance mutations.
- 65.Read AF, Day T, Huijben S. The evolution of drug resistance and the curious orthodoxy of aggressive chemotherapy. Proc. Natl Acad. Sci. USA. 2011;108(Suppl. 2):10871–10877. doi: 10.1073/pnas.1100299108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stearns SC. Evolutionary medicine: its scope, interest and potential. Proc. Royal Soc. B Biol. Sci. 2012;279(1746):4305–4321. doi: 10.1098/rspb.2012.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miskinyte M, Sousa A, Ramiro RS, et al. The genetic basis of Escherichia coli pathoadaptation to macrophages. PLoS Pathog. 2013;9(12):e1003802. doi: 10.1371/journal.ppat.1003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.