Abstract
Integrin receptor activation initiates the formation of integrin adhesion complexes (IACs) at the cell membrane that transduce adhesion-dependent signals to control a multitude of cellular functions. Proteomic analyses of isolated IACs have revealed an unanticipated molecular complexity; however, a global view of the consensus composition and dynamics of IACs is currently lacking. Here, we have integrated several IAC proteomes and generated a 2,412-protein integrin adhesome. Analysis of this dataset reveals the functional diversity of proteins in IACs and establishes a consensus adhesome of 60 proteins. The consensus adhesome likely represents a core cell adhesion machinery, centred around four axes comprising ILK-PINCH-kindlin, FAK-paxillin, talin-vinculin and α-actinin-zyxin-VASP, and includes underappreciated IAC components such as Rsu-1 and caldesmon. Proteomic quantification of IAC assembly and disassembly detailed the compositional dynamics of the core cell adhesion machinery. The definition of this consensus view of integrin adhesome components provides a resource for the research community.
Cellular adhesion to the extracellular matrix (ECM) is essential for a multicellular existence. Cell-surface integrin adhesion receptors engage the cytoskeleton and transduce signals that control cell morphology, migration, survival and differentiation in a wide range of developmental, homeostatic and disease processes1. The interactions of integrin cytoplasmic domains with cytoskeletal, adaptor and signalling molecules are central to regulation of integrin-mediated functions2,3. The complex multimolecular structures that form the connection between integrins and the actin cytoskeleton (termed IACs) contain over 200 reported components4-6. IAC proteins have been characterised as either “intrinsic” components, which localise directly to IACs, or “associated” components, which are effectors of intrinsic molecules6. Despite their apparent complexity, IACs are highly dynamic, turning over on a timescale of minutes.
IACs, like other membrane-receptor-associated signalling complexes, have been refractory to proteomic analysis due to their lability and inaccessibility7. Recent approaches to isolate IACs and analyse their molecular composition using mass spectrometry have been performed in multiple cell types under various conditions8-17. These datasets are necessarily context-dependent (e.g. cell-type- or integrin-heterodimer-specific) and generally represent steady-state cell adhesion. Consequently, an integrative, systems-wide description of IAC composition and dynamics is lacking.
To enable a systems-level analysis of IACs, we characterised their composition in mouse fibroblasts and computationally integrated these data with previously reported IAC proteomes from additional cell types11,13-16. Bioinformatic analyses allowed us to define the functional IAC landscape, from which we identified a robustly detected core “consensus” adhesome, which consisted of both well-characterised and underappreciated components. Using this consensus adhesome in combination with proteomic quantification of IAC assembly and disassembly revealed distinct temporal profiles of protein recruitment. Together with identification of IAC components dependent on myosin-II-mediated tension, these results detailed the compositional dynamics and maturation of the core cell adhesion machinery.
RESULTS
An experimentally defined integrin meta-adhesome
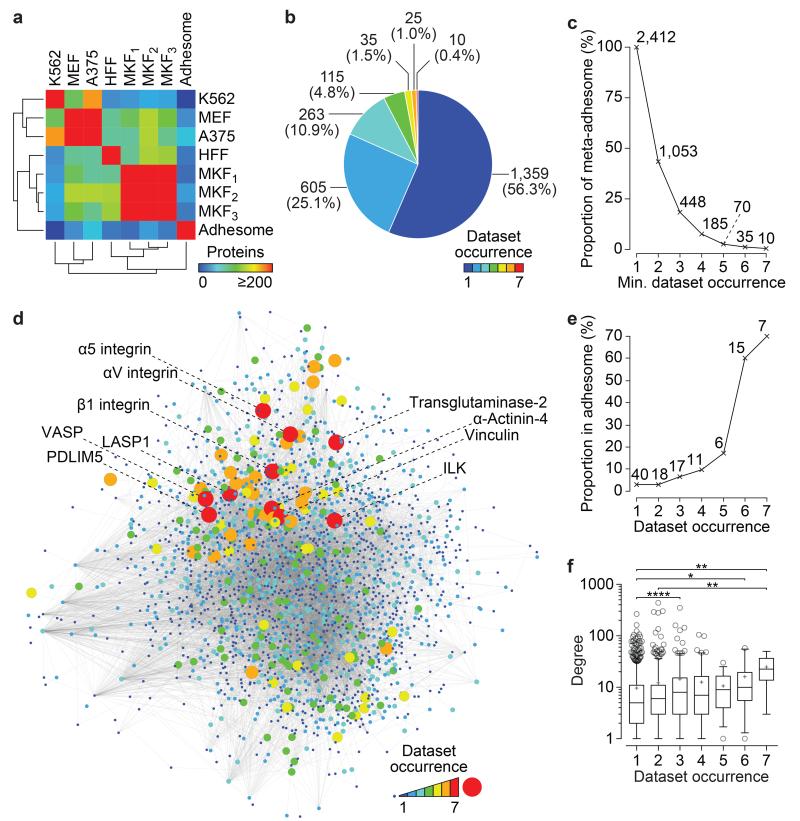
To obtain a global overview of IAC composition, we constructed a database from multiple mass spectrometry (MS)-based proteomics reports. All six published quantitative proteomic datasets detailing the composition of IACs induced by the canonical ligand fibronectin (FN)11,13-16 were assembled. To distinguish and reduce non-specific proteins from the data, we required the proteomic analyses to employ a negative-control ligand, which excluded several published datasets10,12,17, and we included only those proteins that were at least two-fold enriched over their respective controls (Supplementary Table 1; see Methods for details). The assembled datasets were generated in multiple laboratories using a variety of methodologies and from a range of cell types from different lineages: human malignant melanoma (A375), human foreskin fibroblast (HFF), human chronic myelogenous leukaemia (K562) and mouse kidney fibroblast (MKF) cells (Supplementary Table 1). In addition, to expand the number of controlled datasets in the database, we generated a seventh dataset of FN-induced IACs purified from mouse embryonic fibroblast (MEF) cells using published protocols18,19. A total of 1,461 proteins were identified (≥99% confidence) in IACs from MEF cells, of which 674 proteins were at least two-fold enriched to FN-induced IACs over the negative control (transferrin) (Supplementary Table 2), which is of a similar scale to other IAC proteomes (Supplementary Fig. 1, Supplementary Table 1). FN-enriched MEF proteins were integrated with FN-enriched proteins from the six assembled IAC datasets. The resulting experimentally defined database contained 2,412 proteins observed in at least one IAC proteome and was termed the “meta-adhesome” (Supplementary Table 3).
Comparative analyses identified cell-type-, negative-control- and biochemical-isolation-methodology-specific variations in IAC composition (Fig. 1a, Supplementary Fig. 1). Individual IAC proteomes contained hundreds of proteins (602 ± 250, mean ± s.d.; range, 314–1,023) and identified up to a third of literature-curated adhesome4 components (20.9 ± 7.1%, mean ± s.d.; range, 9.1–32.3%) (Fig. 1a, Supplementary Fig. 1c). This variation is likely to result from the context under which the IACs were observed20. Over half of the proteins in the meta-adhesome (1,359; 56.3%) were identified uniquely in a single dataset (Fig. 1b). These proteins represent low-abundance or context-specific adhesome components, or those difficult to detect by MS. The number of proteins identified in the meta-adhesome decreased exponentially as the stringency in dataset number increased (Fig. 1b,c). Four hundred and forty-eight proteins were detected in at least three datasets (Fig. 1c), more than the 63 proteins previously found in common between three published IAC proteomes21. Only 10 proteins were enriched in all seven datasets (labelled in Fig. 1d). We hypothesised that a restricted set of robustly detected proteins may represent a context-independent core of IAC components20. Indeed, the proportion of identified proteins that were literature-curated adhesome4 components increased with dataset occurrence (Fig. 1e, Supplementary Fig. 1a), suggesting that robustly detected proteins are more likely to represent canonical adhesion proteins.
Figure 1.
Overlap and comparison of IAC proteomes in the meta-adhesome. (a) Pairwise overlaps of FN-enriched proteins identified in the seven proteomic datasets and the literature-curated adhesome4 are displayed as a hierarchically clustered heatmap. K562, human chronic myelogenous leukaemia cells11; MEF, mouse embryonic fibroblast cells (this study); A375, human malignant melanoma cells14; HFF, human foreskin fibroblast cells13; MKF1, mouse kidney fibroblast cells15; MKF2 and MKF3, mouse kidney fibroblast cells16. Details of the proteomic datasets are provided in Supplementary Table 1. (b) The number of proteomic datasets in which proteins in the meta-adhesome are identified (dataset occurrence) is displayed as a pie chart. Numbers of proteins identified are indicated for each segment (proportions of the meta-adhesome are shown in parentheses). (c) Line graph showing the cumulative proportion of the meta-adhesome in at least x proteomic datasets, where x is the minimum (min.) dataset occurrence category. Numbers of proteins identified are indicated for each data point. (d) Protein-protein interaction network model of the meta-adhesome. The 2,412 meta-adhesome proteins were mapped onto a curated database of reported protein-protein interactions. The largest connected graph component is displayed, comprising 11,430 interactions (grey lines; edges) between 2,035 proteins (circles; nodes). Node size and colour are proportional to the number of proteomic datasets in which a protein was identified. Locations of proteins identified in all seven datasets are indicated. (e) Line graph showing the proportion of identified proteins that are in the literature-curated adhesome. Numbers of literature-curated adhesome proteins identified are indicated for each data point. (f) The number of reported protein-protein interactions (degree) for each protein is plotted according to the number of proteomic datasets in which it was identified. Box-and-whisker plot shows the median (line), mean (plus sign), 25th and 75th percentiles (box) and 5th and 95th percentiles (whiskers) (n = 1,117, 518, 238, 102, 33, 25 and 10 mapped proteins identified in 1–7 datasets, respectively, with degree ≥ 1). *P < 0.05, **P < 0.01, ****P < 0.0001; Kruskal–Wallis test with Dunn’s post hoc correction (see Supplementary Table 15 for statistics source data).
To investigate the organisation of proteins in the meta-adhesome, we performed interaction network analysis (Fig. 1d). Proteins detected in few IAC datasets exhibited lower network connectivity in general, whereas proteins detected in all seven datasets exhibited the potential to exert greater control over the interactions of other proteins in the complex, as determined by network topology (Fig. 1f, Supplementary Fig. 2). The high number of proteins identified in the meta-adhesome, together with their interconnected network of potential interactions, indicates that IACs, and the flow of information that they relay, are highly complex. Furthermore, it suggests that even the literature-curated adhesome4 underestimates this complexity and that heterogeneity in IAC composition exists between experimental contexts even when cells are exposed to very similar extracellular microenvironments and ligands.
Functional analysis of the integrin meta-adhesome
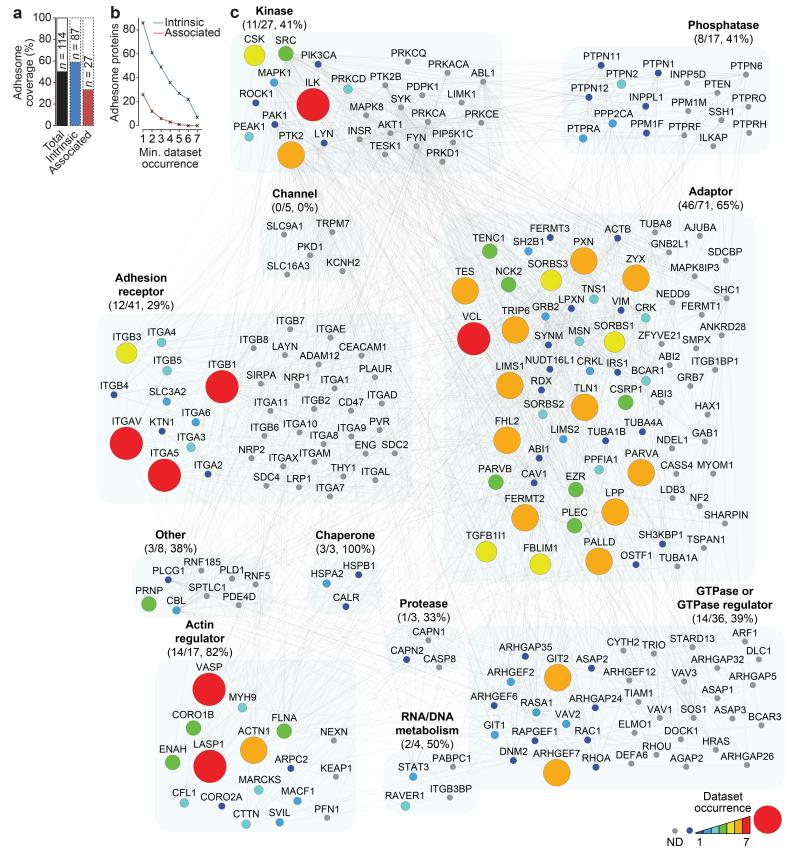
To visualise proteins identified in the meta-adhesome in the context of the literature-curated adhesome4, meta-adhesome proteins were mapped onto adhesome functional categories. In total, 114 (49%) adhesome components were detected across all datasets (Fig. 2a), with almost half (56) detected in three or more datasets (Fig. 2b). The functional categories with the highest coverage in the meta-adhesome were adaptors (46; 65%), actin regulators (14; 82%) and chaperones (3; 100%) (Fig. 2c). GTPases, phosphatases, kinases, channels and adhesion receptors were less well represented. Notably, the receptors most robustly detected were the prominent FN-binding α5β1 and αVβ3 integrins, which confirms the specificity of FN-induced IACs incorporated in the meta-adhesome. The 114 FN-specific adhesome components comprised 87 “intrinsic” and 27 “associated” proteins (Fig. 2a). These data likely reflect the ability of IAC isolation methods to stabilise and identify structural adhesome molecules, such as adaptors and actin regulators. Associated proteins were generally enriched in fewer datasets compared with intrinsic proteins (Fig. 2b), which may be due to the low stoichiometry, context-specificity or highly dynamic and labile nature of associated proteins, such as adhesion-related enzymes, within IACs.
Figure 2.
Meta-adhesome coverage of the literature-curated adhesome. (a) The proportion of the literature-curated adhesome4 identified in the meta-adhesome is plotted as a percentage bar chart. Proportions of the total literature-curated adhesome (black), intrinsic adhesome components (blue) and associated adhesome components (red) are shown. Numbers of identified proteins are indicated. (b) Line graph showing the cumulative number of literature-curated adhesome proteins identified in at least x proteomic datasets, where x is the minimum (min.) dataset occurrence category. Data for intrinsic (blue) and associated (red) adhesome components are shown. (c) Protein-protein interaction network of the literature-curated adhesome proteins identified in the meta-adhesome. Node size and colour are proportional to the number of proteomic datasets in which a protein was identified; ND, not detected (grey node). Nodes are clustered according to literature-curated adhesome functions; numbers (meta-adhesome/literature-curated adhesome total) and proportions of each functional category identified in the meta-adhesome are indicated in parentheses. Nodes are labelled with gene names for clarity (see Supplementary Table 3 for details).
Characterisation of a consensus integrin adhesome
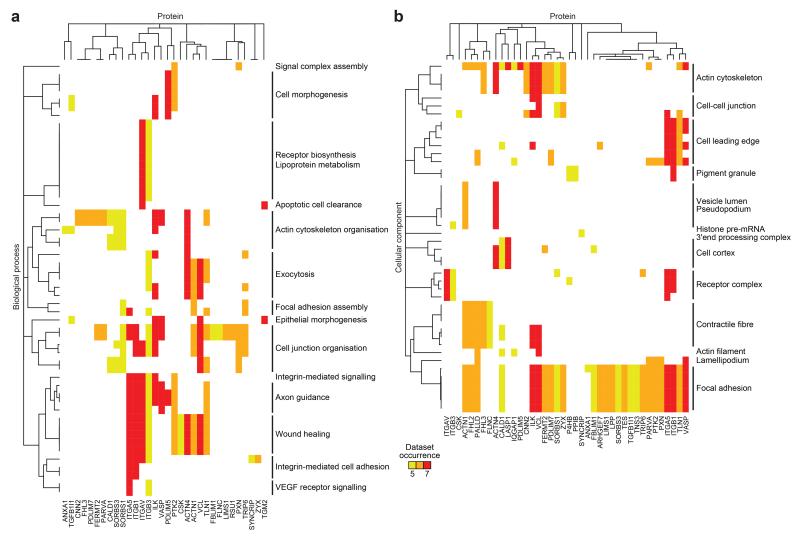
The meta-adhesome provides a resource detailing global IAC composition from multiple cell types and experimental designs. Proteins with diverse cellular functions were detected in the meta-adhesome, but the most robustly detected proteins were overrepresented for numerous adhesion-related functions (Supplementary Fig. 3). To identify the core set of IAC components, and thereby aid the identification of key nodes controlling adhesive functions20, we examined proteins identified in at least five datasets (excluding ECM components), which resulted in a consensus integrin adhesome comprising 60 proteins (Supplementary Table 4). Pathways regulating adhesion-related functions were the most significantly enriched in the consensus adhesome (Fig. 3, Supplementary Table 5), and there was overrepresentation of actin-binding domains and, most significantly, LIM domains, which have been shown previously to be involved in force recognition at adhesion sites12,15,21-24 (Supplementary Tables 4, 5). Nine consensus adhesome genes (15%) had links to inherited diseases (Supplementary Table 4), including seven also identified in a recent report4 and two others (α-actinin-4 and cyclophilin B) associated with glomerular disease and bone disorders, which have previously been linked to adhesome genes4.
Figure 3.
Functional enrichment map of the consensus integrin adhesome. (a,b) Overrepresented biological process (a) and cellular component (b) terms from proteins identified in the consensus adhesome were hierarchically clustered according to proteomic dataset occurrence. This identified clusters of similarly detected proteins associated with a similar set of functional terms. Related terms are summarised (black bars). Proteins are labelled with gene names for clarity (see Supplementary Table 4 for details).
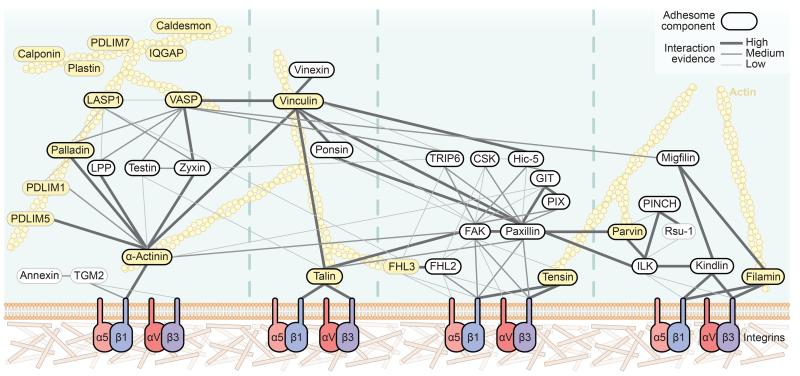
To validate further the consensus integrin adhesome network, interactions between proteins were scored according to the level of supporting experimental evidence (Supplementary Table 6). The resulting interaction network contained many known IAC components, including 31 literature-curated adhesome members4 (black borders, Fig. 4; Supplementary Table 4). The most connected proteins were literature-curated adhesome4 components (proposed interactions; FAK, 15; β1 integrin, 13; paxillin, 12), which may be because these proteins have been studied more extensively than others25. An emergent property of the consensus adhesome network was that it broadly clustered into four theoretical modules based on currently known signalling axes and links from integrins to actin reported in the literature (Fig. 4). The first module contained α-actinin and zyxin family members. The second module contained vinculin, talin and the vinculin-binding proteins vinexin and ponsin. Vinculin contained the highest number of high-evidence interactions (seven) and associated with many proteins in the third module containing FAK and paxillin. The final module consisted of two submodules connected via a kindlin-ILK interaction. In addition, non-consensus meta-adhesome proteins are known to interact with consensus components (Supplementary Table 7) and are therefore also likely to be involved in regulating the integrin-actin connection.
Figure 4.
Curated network model of the consensus integrin adhesome. Protein-protein interaction network of the consensus adhesome. Interactions were manually validated and scored (high, medium, low) according to the level of experimental evidence for that interaction, shown by the thickness and saturation of the grey edges (see Supplementary Table 6). Thick black node border indicates literature-curated adhesome4 protein. Yellow node indicates actin-binding protein. The specific isoforms and subunits of proteins identified are detailed in Supplementary Table 4 by gene name. For clarity, α-actinin is depicted as one node, even though two α-actinin isoforms (α-actinin-1 and -4) were identified. Actin is depicted for illustrative purposes but was not present in the consensus adhesome. The network comprised 41 proteins with 92 interactions, excluding actin binding. Unconnected components or components with only one low-evidence interaction are not shown in the network; proteins unconnected to the main network were ALYREF, BRIX1, DDX18, DDX27, DIMT1, DNAJB1, FAU, FEN1, H1FX, HP1BP3, LIMD1, MRTO4, POLDIP3, RPL23A, SIPA1 and SYNCRIP; proteins connected to the network with a single low-evidence interaction were P4HB and PPIB.
Interestingly, the consensus adhesome contained 29 proteins that were not members of the literature-curated adhesome4, some of which likely represent underappreciated FN-mediated IAC components (grey borders, Fig. 4; Supplementary Table 4). Six of these proteins (Rsu-1, PDLIM1, PDLIM5, FHL3 and a transglutaminase-2-annexin A1 complex) interact with canonical IAC components, while five (caldesmon, calponin, IQGAP, PDLIM7 and plastin) were not connected to other consensus components but bind actin, suggesting that they were isolated as peripheral IAC components or that their connections to other consensus proteins are uncharacterised (Fig. 4, Supplementary Table 6). The remaining proteins in the consensus adhesome were unconnected to the network, were not known to associate with actin, had unknown function or were involved in non-adhesion-related functions (Supplementary Table 4). The fact that these unconnected proteins were identified here using the same methods used to detect many known adhesion-related proteins increases confidence in their involvement in the core adhesion machinery, but their contributions to adhesive functions remain to be elucidated, and it remains possible that their association is non-specific.
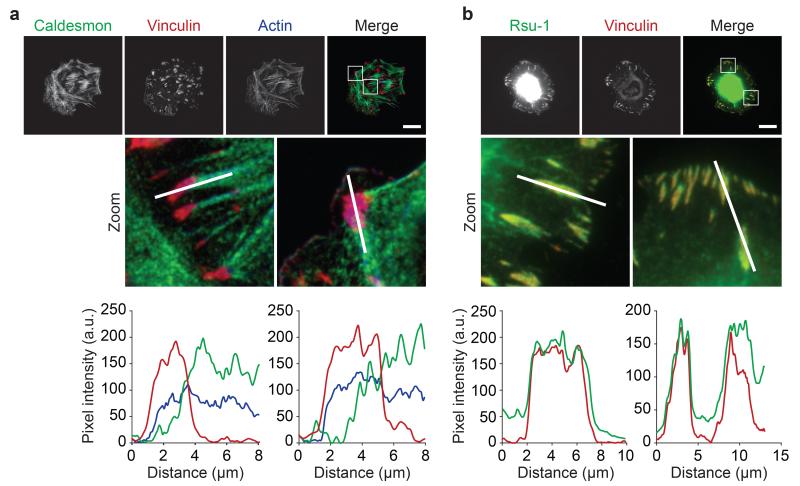
To verify that underappreciated proteins identified in the consensus adhesome localise to IACs, we visualised by immunofluorescence two consensus adhesome proteins that are not literature-curated adhesome4 members (Fig. 5). Caldesmon26 localised to actin within vinculin-positive areas (Fig. 5a; Mander’s overlap coefficient27 (MOC) = 0.51 ± 0.19, mean ± s.d.). Rsu-1, which has been reported to associate with IACs in other cell types28, co-localised with vinculin (Fig. 5b; MOC = 0.98 ± 0.03, mean ± s.d.). These data confirm IAC localisation of caldesmon and Rsu-1 and suggest that underappreciated consensus adhesome proteins may participate in regulating the integrin-actin connection.
Figure 5.
Caldesmon and Rsu-1 localisation in IACs. (a,b) U2OS cells were spread on FN for 2 h and visualised using antibodies against caldesmon (green) (a) and Rsu-1 (green) (b). IACs were visualised by immunofluorescence staining for vinculin (red) and the actin cytoskeleton was visualised by staining with fluorophore-conjugated phalloidin (blue). Graphs show fluorescence intensity values for each channel across line segments in corresponding zoomed areas above each graph. In addition, colocalisation with vinculin-positive areas was quantified for caldesmon (MOC27 = 0.51 ± 0.19) and Rsu-1 (MOC = 0.98 ± 0.03). Values are mean ± s.d. (n = 20 cells from one independent experiment; see Supplementary Table 15 for source data). Scale bars, 20 μm.
Maturation state of the consensus adhesome
To evaluate further the function of the consensus adhesome, we compared consensus adhesome proteins identified in individual IAC proteomes, demonstrating that they were distributed relatively evenly between datasets (range, 26–59 proteins) (Supplementary Fig. 4). The dataset generated from K562 cells using FN-coated beads identified the lowest number of consensus components, with a high proportion of the absent proteins containing LIM domains. As it has been shown previously that LIM-domain proteins are force-sensitive12,15, this suggests that the K562 dataset likely represents a higher proportion of immature IAC structures that form before applied myosin-II-generated cytoskeletal forces. To explore this issue, we analysed changes in the IAC proteome in MEF cells upon myosin II inhibition (Supplementary Table 8). Taken together with two related published datasets of myosin-II-dependent IAC composition12,15, these data support the view that LIM-domain-containing proteins are recruited to more mature IACs under myosin-II-generated tension (Supplementary Table 9). In addition, one published dataset generated from human fibroblasts without a negative control ligand condition12 identified a similar scale of consensus adhesome components to other IAC proteomes (42/60; Supplementary Table 9, Supplementary Fig. 4), further exemplifying the utility of the consensus adhesome as a filter for other datasets. In contrast, the consensus adhesome showed little overlap with datasets that represent other types of IAC29-31, such as podosomes and invadopodia (Supplementary Table 10), which suggests that the consensus adhesome does not represent these structures. In summary, these data suggest that the consensus adhesome contains commonly identified IAC molecules from both nascent and mature IACs.
Temporal dynamics of the consensus integrin adhesome
The consensus adhesome provides a comprehensive view of commonly identified, steady-state IAC composition. To identify the temporal dynamics of IACs, we characterised their composition during assembly and disassembly32 (Supplementary Tables 11, 12). Analysis of meta-adhesome proteins identified in the temporal IAC profiles revealed distinct dynamics of proteins involved in specific functional processes (Supplementary Figs. 5, 6, Supplementary Tables 13, 14). Proteins involved in membrane organisation, which may localise to the plasma membrane to coordinate morphological changes during cell spreading, increased during IAC assembly. Proteins involved in cytoskeletal or adhesive functions were generally more abundant later in IAC assembly and decreased during IAC disassembly. Both consensus and non-consensus adhesome components co-clustered in different groups, which suggests that their combined contributions are involved in IAC dynamics. For example, vimentin and myosin II co-clustered with consensus adhesome molecules that bind actin during IAC disassembly (Supplementary Fig. 6). Moreover, Rac1 and Lyn co-clustered with other consensus proteins and were abundant early and late in IAC assembly (Supplementary Fig. 5). In contrast, proteins involved in RNA processing and translation peaked early during IAC assembly and increased during IAC disassembly, suggesting a reciprocal temporal relationship between these cellular processes at IACs.
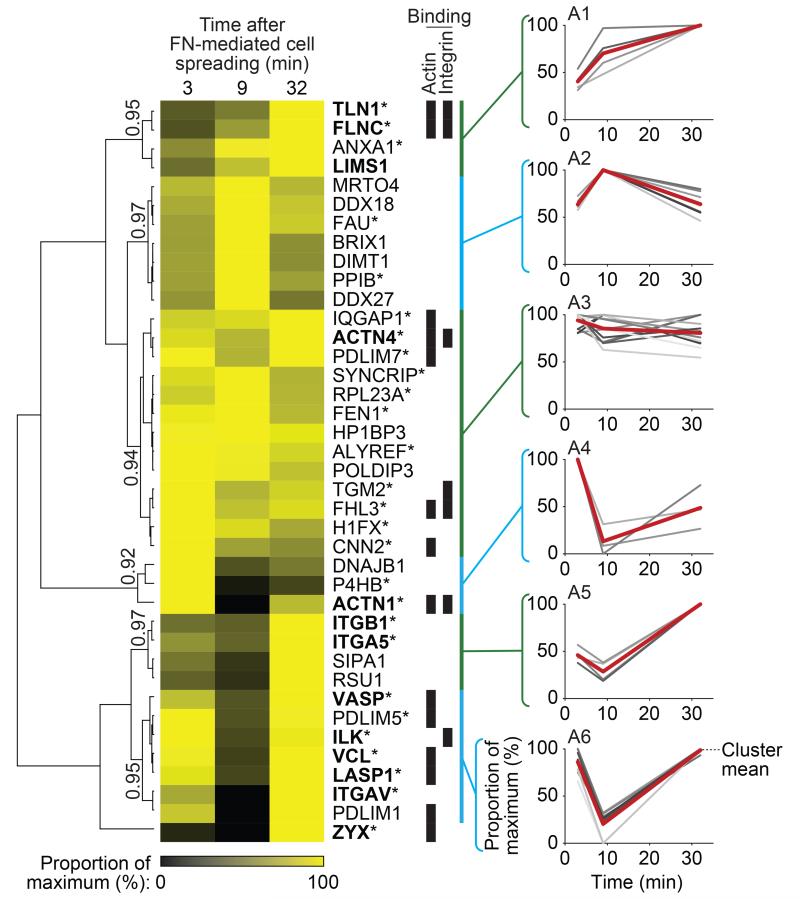
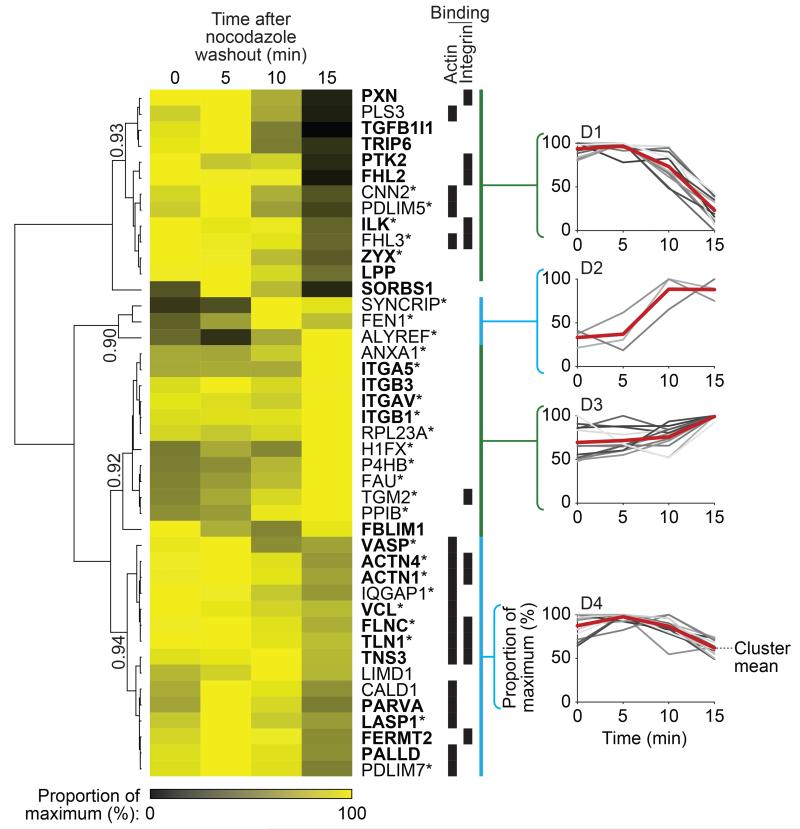
To examine the core adhesion machinery, hierarchical clustering revealed that different consensus adhesome components display distinct dynamics (Figs. 6, 7). β1, α5 and αV integrins reached maximum abundance by 30 minutes in this system. Integrins were relatively stable throughout IAC disassembly, and this was also the case for other cell-surface molecules (e.g. annexin A1, transglutaminase-2 and the CD98 heavy chain (SLC3A2)). Most consensus components, although distributed in different clusters (Fig. 6), were detected in high abundance late in IAC assembly here, indicating distinct dynamics of protein recruitment. Integrin-binding proteins decreased during IAC disassembly but with different kinetics (clusters D1, D4; Fig. 7). Most of the adaptors in the consensus adhesome were almost completely absent from IACs after 15 minutes (cluster D1, Fig. 7), while 13 of the 17 actin-binding proteins, five of which were integrin-binding, decreased in abundance less rapidly (cluster D4, Fig. 7). These data suggest that adaptor proteins located between actin and integrins are lost earlier and at a faster rate than actin-binding proteins and that the integrin-actin linkage is disrupted late during IAC disassembly.
Figure 6.
Temporal profiling of the consensus adhesome during IAC assembly. IACs were isolated from K562 cells in biological duplicate after 3, 9 and 32 min incubation with FN-coated beads and analysed by MS (data are from 2 independent experiments; see Supplementary Table 11). Throughout IAC assembly, 39 of the 60 consensus adhesome proteins were identified and were analysed by unsupervised hierarchical clustering, revealing distinct temporal profiles of protein recruitment to IACs. Six clusters, labelled A1–6, were chosen on the basis of a Pearson correlation threshold greater than 0.9 and are indicated by blue and green bars. Clusters are shown alongside corresponding profile plots, with the mean temporal profile for each cluster indicated by a red line. Quantitative heat map displays mean spectral counts as a proportion of the maximum spectral count for each given protein. Proteins are labelled with gene names for clarity. Proteins also identified during IAC disassembly (Fig. 7, Supplementary Table 12) are indicated by an asterisk. Literature-curated adhesome4 proteins and their isoforms are in bold. Proteins able to bind actin or integrin are indicated by black bars.
Figure 7.
Temporal profiling of the consensus adhesome during IAC disassembly. IACs were isolated from U2OS cells in biological triplicate upon nocodazole removal and 5, 10 and 15 min after nocodazole washout to examine changes in IAC composition throughout IAC disruption32. Isolated IACs at each time point were analysed by MS (data are from 3 independent experiments; see Supplementary Table 12). Throughout IAC disassembly, 43 of the 60 consensus adhesome proteins were identified and were analysed by unsupervised hierarchical clustering, revealing distinct temporal profiles of protein dissociation from IACs. Four clusters, labelled D1–4, were chosen on the basis of a Pearson correlation threshold greater than 0.9 and are indicated by blue and green bars. Clusters are shown alongside corresponding profile plots, with the mean temporal profile for each cluster indicated by a red line. Quantitative heat map displays mean spectral counts as a proportion of the maximum spectral count for each given protein. Proteins are labelled with gene names for clarity. Proteins also identified during IAC assembly (Fig. 6, Supplementary Table 11) are indicated by an asterisk. Literature-curated adhesome4 proteins and their isoforms are in bold. Proteins able to bind actin or integrin are indicated by black bars.
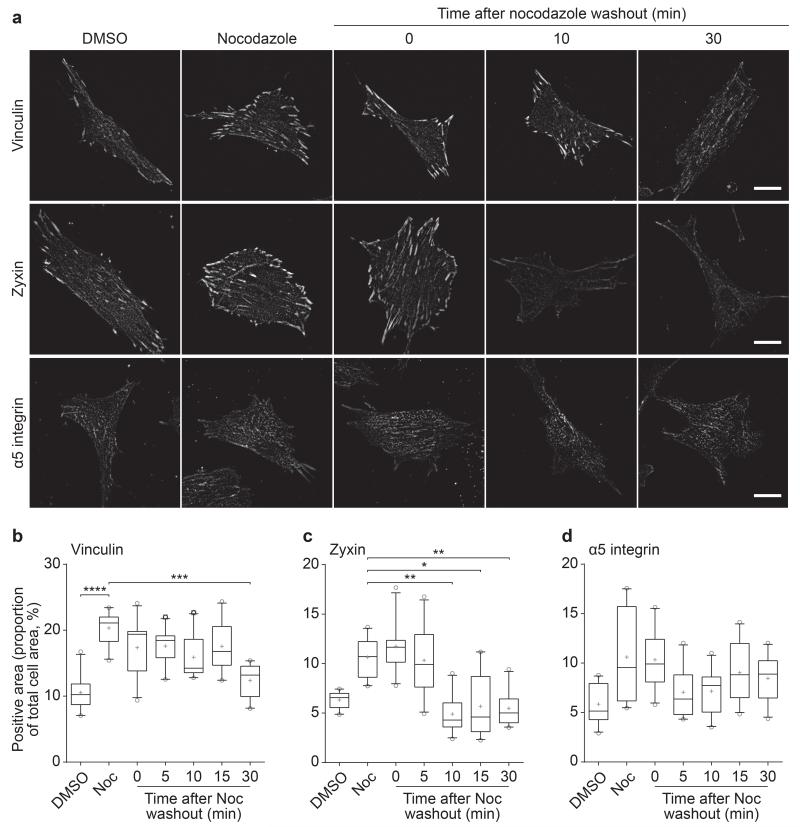
To confirm the temporal differences in IAC components revealed by MS, IAC proteins were visualised during nocodazole washout32. Upon nocodazole washout, the area of the ventral cell surface covered by α5 or β1 integrin did not change (Fig. 8, Supplementary Fig. 7). In support of the different rates of loss of IAC components, the decrease in vinculin (30 min; Fig. 8) was delayed compared with the loss of zyxin (10 min; Fig. 8) and other adhesion molecules (phospho-FAKY397, 10 min; paxillin and phospho-paxillinY118, 15 min; Supplementary Fig. 7). These data validate the findings obtained using MS that indicate different adhesion molecules display distinct temporal profiles during IAC disassembly.
Figure 8.
Changes in consensus adhesome components during IAC disassembly. (a) HFF cells treated with DMSO, 10 μM nocodazole or after nocodazole removal at different times were stained for vinculin, zyxin and α5 integrin. Representative images are shown. Scale bars, 20 μm. (b–d) Quantification of images in a. Vinculin, zyxin and α5 integrin levels were quantified as a proportion of total cell area. Box-and-whisker plot shows the median (line), mean (plus sign), 25th and 75th percentiles (box) and 5th and 95th percentiles (whiskers) (n = 10 cells per condition from one independent experiment). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; Kruskal–Wallis test with Dunn’s post hoc correction (comparisons with the nocodazole treatment group are shown; see Supplementary Table 15 for statistics source data).
DISCUSSION
Here, we performed extensive analyses of IAC proteomes, resulting in an experimentally defined meta-adhesome of 2,412 proteins. An emergent property of the meta-adhesome was the identification of a consensus adhesome comprising core adhesion machinery robustly detected in IAC proteomes. The proteomic datasets used here provide a global description of IACs in the context of FN-mediated adhesion. Analysis of the meta-adhesome overcomes the heterogeneity encountered when studying individual datasets from different laboratories and cell types. The heterogeneity between IAC proteomes collected from cells exposed to similar microenvironments and the increased number of proteins identified in the meta-adhesome compared with the literature-curated adhesome4 highlights an unanticipated complexity in IAC composition. Evidence for IAC localisation of 118 adhesome proteins (51%; 64 intrinsic, 54 associated) that were not detected in the meta-adhesome may be context-dependent or may need re-examining. One outstanding question is how the consensus adhesome differs for other ECM ligands, such as laminin or collagen, or integrin heterodimers or cell types. Increasing the numbers of proteomic datasets of IACs induced by alternative ECM ligands and cell types to those previously investigated would help clarify this view.
To identify core adhesion machinery involved in adhesive function, we defined a consensus adhesome of 60 proteins commonly identified in FN-induced IAC proteomes that incorporated negative controls, which enabled us to threshold the identification of canonical and underappreciated IAC proteins objectively. Many actin-binding proteins were identified in the consensus adhesome, which may represent a specific subset of actin-binding proteins that localise in IACs at the ends of actin fibres, which was shown to be the case for caldesmon. Importantly, not all cellular actin-binding proteins were identified by these analyses, indicating that the IAC isolation strategies allow the separation and characterisation of a functionally distinct pool of actin and associated proteins. Most (46; 90%) candidate IAC proteins common to three proteomic datasets highlighted in a recent analysis21 were not present in the consensus adhesome, but some protein isoforms exhibit cell-type-specific expression33-35 and related isoforms, and additional non-canonical IAC components, were identified. With the exception of signal-induced proliferation-associated 1 (SIPA1)36 and LIM domains containing 1 (LIMD1)37, we found no evidence supporting the involvement of the unconnected consensus adhesome proteins (Fig. 4, legend) in IACs or adhesive function. Some of these proteins have functional roles related to RNA processing and translation (Supplementary Table 4) and therefore may be involved in localised protein synthesis, which is supported by IAC localisation of translation machinery and β-actin mRNA38-42. They may be co-purifying contaminants from the IAC isolation process, which is supported by their identification in the contaminant repository for affinity purification–MS data (CRAPome)43. However, comparative analysis of IAC proteomes with the CRAPome is risky, as many canonical IAC proteins and cytoskeletal components (e.g. talin and β1 integrin) occur in the CRAPome. Conversely, some well-characterised IAC components were not enriched in all seven datasets (e.g. β3 integrin, FAK, kindlin, paxillin and talin) or were observed in the meta-adhesome but not the consensus adhesome (e.g. p130Cas and Src family kinases). These omissions may be due to cell-type-specific expression, cell-type-specific IAC maturation, protein abundance at IACs, preferential use of β1 integrin or non-specific detection in negative controls. Additional examination of the phosphoproteome14 and stoichiometry44 of IACs will provide deeper coverage of IAC composition and further insights into their relative functions in adhesion signalling.
The consensus adhesome contained many evolutionarily conserved proteins across multiple species and whose genetic depletion causes dramatic defects in integrin-mediated adhesion45-49, indicating that consensus adhesome proteins form an essential contribution to integrin function. Analysis of interactions between consensus adhesome molecules resulted in the identification of four interconnected axes that link integrins to actin. Proteomics methods are complementary to super-resolution microscopy approaches used to study IACs20. Indeed, the four axes that form the integrin-actin structural connection defined in this study support the vertical Z-plane model of IACs50 where talin spans IACs, FAK and paxillin are in an integrin-proximal signalling layer and α-actinin, zyxin and VASP are localised distal to integrins near actin50. The association of α-actinin with β1 integrin51,52 occurs in early adhesions and is lost during maturation44,53. Applied cytoskeletal force could induce α-actinin-integrin dissociation, allowing distal localisation of α-actinin and potentially associated actin-binding and LIM-domain proteins from the membrane. Interactions with other consensus adhesome proteins may maintain the localisation of α-actinin and associated molecules in IACs. Important next steps will be to determine the dynamics54 and nanoscale localisation50 of other consensus adhesome proteins using super-resolution microscopy.
IACs are highly dynamic structures that can be characterised depending on their size, localisation and maturation state55,56. Current MS-based approaches to analyse IACs result in the combined analysis of these heterogeneous IAC structures (which are compositionally different from podosomes and invadopodia) from a cell population to give a compositional snapshot at a particular time point. To demonstrate how the meta-adhesome and consensus adhesome can be used by the research community, we generated time-course datasets during IAC assembly and disassembly. By filtering the acquired datasets using these adhesomes, we found that adhesion molecules are recruited to, and disassembled from, IACs with distinct kinetics, suggesting that these processes are differentially regulated and not simply reciprocal events. In support of studies showing hierarchical IAC formation44,55,56, α-actinin was abundant early during IAC assembly, whereas zyxin was most abundant later. Most IAC molecules decreased in abundance during disassembly with different kinetic rates, and adaptor proteins were lost from IACs earlier than actin-binding proteins, suggesting that adaptor proteins may be primary targets for disassembly. Therefore, as demonstrated here, we propose that the meta-adhesome and consensus adhesome can be used for removal of non-specific components from future analyses of IAC composition by MS, thus contextualising and streamlining identification of candidate adhesion molecules for follow-up studies.
In summary, the data presented in this study provide a systems-wide analysis of FN-induced IAC composition, detail a comprehensive reductionist view of an experimentally defined integrin adhesome and catalogue the first global characterisation of IAC dynamics during the initial phases of assembly and disassembly.
METHODS
Reagents
FN, PDL, transferrin, blebbistatin and nocodazole were from Sigma-Aldrich. Monoclonal antibodies used for immunofluorescence were mouse anti-vinculin (hVIN-1, V9131, Sigma-Aldrich; 1:400), rabbit anti-phospho-FAKY397 (141-9, 44-625G, Invitrogen; 1:200), mouse anti-paxillin (349/Paxillin, 610052, BD Biosciences; 1:400), mouse anti-zyxin (ZZ001, 39-6000, Thermo Fisher Scientific; 1:400), rat anti-α5 integrin (mAb11, provided by K. M. Yamada, NIH, Bethesda, MD, USA; 1:200), rat anti-β1 integrin (9EG7, provided by D. Vestweber, University of Münster, Münster, Germany; 1:200) and rabbit anti-caldesmon-1 (D5C8D, 12503, Cell Signaling Technology; 1:400). Polyclonal antibodies used for immunofluorescence were rabbit anti-phospho-paxillinY118 (44-722G, Invitrogen; 1:200) and rabbit anti-Rsu-1 (provided by M. L. Cutler, University of Manitoba, Winnipeg, MB, Canada; 1:500). Secondary antibodies were from Jackson Immunoresearch and Alexa Fluor 647-conjugated phalloidin was from Invitrogen.
Cell culture
K562 cells (provided by M. E. Hemler, Dana-Farber Cancer Institute, Boston, MA, USA) were cultured in RPMI 1640 medium supplemented with 10% (v/v) foetal calf serum (FCS; Lonza Bioscience) and 2 mM L-glutamine. Telomerase-immortalised HFF (provided by K. Clark, University of Leicester, Leicester, UK), conditionally immortalised MEF (generated in-house, see ref. 57), A375-SM (provided by I. J. Fidler, MD Anderson Cancer Center, Houston, TX, USA) and osteosarcoma (U2OS; purchased from Sigma Aldrich, 92022711) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) FCS and 2 mM L-glutamine. MEF cells were supplemented with interferon-γ (Sigma-Aldrich). All cells were maintained at 37 °C in a humidified 5% (v/v) CO2 atmosphere, except for MEF cells, which were maintained at 33 °C. All cell lines were frequently tested for mycoplasma and were negative. Cell lines were not authenticated and are not listed in the database of commonly misidentified cell lines maintained by ICLAC (http://iclac.org) and NCBI Biosample (http://www.ncbi.nlm.nih.gov/biosample).
IAC isolation
IACs were isolated using a similar approach to the ligand affinity purification method described previously18. For isolation of IACs from MEF cells, cells were resuspended in DMEM supplemented with 25 mM HEPES (Sigma-Aldrich) and incubated in suspension for 20 min at 37 °C to down-regulate ECM adhesion signalling events. Cells were spread on tissue culture dishes coated with 10 μg/ml FN or transferrin for 120 min at 37 °C, 8% (v/v) CO2, in the presence or absence of 50 μM blebbistatin. Cells were incubated with the membrane-permeable cross-linker dimethyl-3, 3′-dithiobispropionimidate (DTBP; Sigma-Aldrich; 3 mM, 30 min), washed twice with PBS, and DTBP was quenched using 1 M Tris-HCl (pH 8, 10 min), after which cells were washed twice with PBS and incubated in PBS at 4 °C. Cell bodies were removed by a combination of cell lysis in ice-cold extraction buffer [20 mM NH4OH, 0.5% (w/v) Triton X-100] and sonication for 1 min (VibraCell VCX 500; Sonics & Materials). Protein complexes left bound to the substrate were washed five times with PBS, recovered by scraping in 100 μl recovery solution [125 mM Tris-HCl (pH 6.8), 1% (w/v) SDS, 15% (v/v) β-mercaptoethanol] and incubated at 70 °C for 10 min (2 biological replicates).
For analysis of IAC assembly, the ligand affinity purification method described previously11,18 was adapted to enable isolation of newly formed IACs. To examine IAC assembly during the early stages of cell adhesion, complexes were isolated 3, 9 and 32 min after initial cell attachment to FN. Paramagnetic beads (4.5 μm-diameter; M-450 Dynabeads; Life Technologies) coated with FN were incubated with K562 cells in DMEM supplemented with 0.2% (w/v) BSA and 25 mM HEPES at 70 rpm for either 1, 7 or 30 min at 37 °C. Bead-bound cells were incubated with DTBP (10 mM, 2 min) and DTBP was quenched with 20 mM Tris-HCl (pH 8.5). Bead-bound cells were washed with CSK buffer [10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (pH 6.8), 50 mM NaCl, 150 mM sucrose, 3 mM MgCl2, and 1 mM MnCl2] supplemented with 20 mM Tris-HCl (pH 8.5) and 2 mM Na3VO4, and were lysed in CSK+ buffer [CSK buffer supplemented with 0.5% (w/v) Triton X-100, leupeptin (10 μg/ml), aprotinin (10 μg/ml), 0.5 mM 4-(2-aminoethyl)benzenesulfonylfluoride hydrochloride (AEBSF), and 2 mM Na3VO4] for 30 min on ice with sonication (VibraCell VCX 500). Isolated IACs were eluted from beads with reducing sample buffer [50 mM Tris-HCl (pH 6.8), 10% (w/v) glycerol, 4% (w/v) SDS, 0.004% (w/v) bromophenol blue, and 8% (v/v) β-mercaptoethanol] and were separated from beads with a magnet. IACs isolated at each time point were analysed by quantitative MS (2 biological replicates).
To examine IAC disassembly, complexes were isolated upon microtubule-induced disassembly, and at 5, 10 and 15 min after nocodazole removal. U2OS cells plated on FN-coated dishes were serum-starved for 16 h, treated with 10 μM nocodazole for 4 h, washed three times with DMEM and incubated for appropriate times after nocodazole removal at 37 °C, 5% (v/v) CO2 (ref. 32). Cells were incubated with DTBP (6 mM, 3 min), and DTBP was quenched with 1 M Tris-HCl (pH 8.5), followed by sonication for 2.5 min (VibraCell VCX 500) to lyse cells. Isolated IACs were washed and collected using the same method used to generate the MEF dataset. IACs isolated at each time point were analysed by quantitative MS (3 biological replicates).
Immunoblotting
Proteins were resolved by SDS-PAGE and transferred to nitrocellulose membrane (Whatman). Membranes were washed and incubated with antibodies as described previously11. Briefly, membranes were blocked with blocking buffer (Sigma-Aldrich) for 1 h and were incubated with appropriate primary antibodies diluted in blocking buffer overnight at 4 °C. After three 5-min washes, membranes were incubated with secondary antibodies diluted in blocking buffer for 45 min in the dark. Secondary antibodies used were donkey Alexa Fluor 680-conjugated anti-goat IgG or anti-mouse IgG (Life Technologies) and donkey IRDye 800-conjugated anti-mouse IgG (Rockland Immunochemicals). Membranes were washed in the dark and scanned using the Odyssey infrared imaging system (LI-COR).
MS data acquisition
Following SDS-PAGE, gel lanes were sliced and subjected to in-gel digestion with trypsin58 with modifications11. Peptide samples were analysed by liquid chromatography (LC)-tandem MS using a nanoACQUITY UltraPerformance LC system (Waters) coupled online to an LTQ Velos mass spectrometer (Thermo Fisher Scientific) or using an UltiMate 3000 Rapid Separation LC system (Thermo Fisher Scientific) coupled online to an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific). Peptides were concentrated and desalted on a Symmetry C18 preparative column (20 mm × 180 μm, 5-μm particle size; Waters) and separated on a bridged ethyl hybrid C18 analytical column (250 mm × 75 μm, 1.7-μm particle size; Waters) using a 45-min linear gradient from 1% to 25% or 8% to 33% (v/v) acetonitrile in 0.1% (v/v) formic acid at a flow rate of 200 nl/min. Peptides were selected for fragmentation automatically by data-dependent analysis.
MS data analysis
MS data were searched using an in-house Mascot server (version 2.2.03; Matrix Science)59 as described previously9. Mass tolerances for precursor and fragment ions were 0.4 Da and 0.5 Da, respectively, for LTQ Velos data or 5 ppm and 0.5 Da, respectively, for Orbitrap Elite data. Data were validated in Scaffold (version 3.00.06; Proteome Software) using a threshold of identification of at least 90% probability at the peptide level, assignment of at least two unique, validated peptides, and at least 99% probability at the protein level. These acceptance criteria resulted in an estimated protein false discovery rate of ≤0.1% for all datasets. MS data were quantified as described previously9. Briefly, relative protein abundance was calculated using the unweighted spectral count of a given protein normalised to the total number of spectra observed in that sample and to the molecular weight of that protein (termed normalised spectral count). Final results were reported as mean normalised spectral counts of biological replicate isolations. Only proteins with a spectral count of at least four were used for further analysis. For the IAC assembly and disassembly datasets, data were reported as mean spectral counts as a proportion of the maximum spectral count for each given protein.
Meta-adhesome construction
The assembled IAC datasets (Supplementary Table 1) were filtered to include only proteins from cells spread on FN and the corresponding negative control in the absence of perturbation. To reduce the identification of non-specific IAC components, only those proteins enriched in FN-induced IACs compared to the negative control, with relative abundance satisfying log2(FN/control) ≥ 1, were assembled into the meta-adhesome database (Supplementary Table 3). Two datasets were included from Schiller et al. (2013)16 for cells expressing αV and β1 integrins (α5β1, αVβ3 and αVβ5 integrins; cells spread for 45 min (MKF2) and 90 min (MKF3)), excluding proteins uniquely identified in cells expressing only either β1- or αV-class integrins as integrin heterodimer-specific adhesomes were not presented in this version of the meta-adhesome.
Consensus adhesome construction
Proteins enriched in at least five proteomic datasets in the meta-adhesome database were incorporated into the consensus adhesome. ECM or secreted proteins (COL1A1, COL1A2, COL5A2, COL6A1, COL6A2, FGG, FN1, PCOLCE, PRSS23, SERPINE1) were excluded since, although relevant to adhesion biology, we sought to focus on intracellular components of IACs. All isoforms of literature-curated adhesome4 members were classified as adhesome molecules. Functional information was adapted from the HUGO Gene Nomenclature Committee (HGNC) database61, protein domain information was assigned from InterPro62 and disease annotations were extracted from the Online Mendelian Inheritance in Man database (http://www.omim.org).
Hierarchical clustering and principal component analyses
Proteins or datasets were hierarchically clustered on the basis of uncentred Pearson correlation using Cluster 3.0 (C Clustering Library, version 1.50)63 and visualised using Java TreeView (version 1.1.5)64. Binary data were clustered on the basis of Jaccard distance and visualised using R (version 3.1.0). Distances between hits were computed using a complete-linkage matrix in all cases. Additional heatmaps were visualised using MultiExperiment Viewer (version 4.8.1)65. Principal component analysis was performed using MATLAB (version R2012a; MathWorks).
Interaction network analyses
Interaction network analysis was performed using Cytoscape (version 3.0.2)66. Enriched proteins were mapped onto a merged human interactome consisting of physical protein-protein interactions as described previously9. Graph clustering was performed using the yFiles Organic algorithm implemented in Cytoscape. Topological parameters were computed from undirected graphs, excluding self-interactions, using NetworkAnalyzer67.
For the consensus adhesome, evidence for protein-protein interactions was manually verified and scored. Low-evidence interactions included those based on a single publication or on coprecipitation or yeast two-hybrid studies. Medium-evidence interactions were based on data from multiple sources, or a single source if there were phosphorylation or peptide binding data. High-evidence interactions were based on structural evidence of direct binding between two proteins, such as X-ray crystallography or nuclear magnetic resonance, or confirmation using a wide variety of techniques. Experimental evidence and source publications are detailed in Supplementary Table 6.
Functional enrichment analyses
Functional enrichment analysis was performed using DAVID (version 6.7)68. Keywords with fold enrichment ≥ 1.5, Bonferroni-corrected P value < 0.05, EASE score (modified Fisher’s exact test) < 0.05 and at least two proteins per keyword were considered significantly overrepresented.
For generation of functional enrichment maps, overrepresentation of gene ontology terms was calculated using High-Throughput GoMiner69. One thousand randomisations were performed and data were thresholded for a 5% false discovery rate. Overrepresented terms with ≥5 and ≤500 assigned proteins were reported. Dataset occurrence was mapped onto proteins assigned to each overrepresented term, and the data matrix was subjected to hierarchical clustering analysis as described above.
Immunofluorescence microscopy
To confirm localisation of Rsu-1 and caldesmon at IACs, U2OS cells were spread on FN-coated dishes (MatTek) for 2 h at 37 °C, 8% (v/v) CO2. To validate MS data of IAC disassembly, HFF cells were treated with nocodazole and nocodazole was washed out as described previously32. Cells were washed with PBS, fixed with −20 °C methanol or 4% (w/v) paraformaldehyde for 7 min at room temperature and permeabilised with 0.5% (v/v) Triton X-100 for 10 min. Permeabilised cells were washed three times with PBS before incubation with appropriate primary antibodies diluted in 2% (w/v) BSA in PBS for 1 h. Vinculin, zyxin and α5 integrin were analysed, as each protein displayed different IAC disassembly dynamics (Fig. 7). In addition, phospho-paxillinY118, paxillin, phospho-FAKY397 and β1 integrin were visualised to test whether proteins within the same cluster displayed similar disassembly dynamics (Supplementary Fig. 7). Cells were washed three times with PBS and incubated with appropriate secondary antibodies diluted in 2% (w/v) BSA in PBS for 30 min in the dark. Stained cells were washed once in PBS, twice in water and stored in water at 4 °C until imaging. Images were acquired on a Delta Vision RT (Applied Precision) restoration microscope using a 60×/1.42 Plan Apo objective and the Sedat filter set (Chroma 89000). Images were collected with a Z optical spacing of 0.2 μm, five images per stack, using a Coolsnap HQ camera (Photometrics) and Softworx software (Applied Precision). To assess colocalisation of Rsu-1 and caldesmon with vinculin, cells were also imaged using a spinning-disk confocal inverted microscope (Marianas; 3i). Images were collected with a Z optical spacing of 0.2 μm, three images per stack, using a 63×/1.4 Plan Apochromat objective and SlideBook 6.0 software (3i).
Image analysis and quantification
Maximum intensity projections of raw images were generated and background filtered (rolling ball, 10-pixel radius) using ImageJ (version 1.48o)70. Areas containing positive staining of IAC proteins were measured and normalised to total cell area. Box-and-whisker plots were generated using Prism (version 6.04; GraphPad). To quantify Rsu-1 and caldesmon colocalisation with vinculin, images were individually band-pass filtered (A trous wavelet, linear 3 × 3 filter, keeping scales 2–8) using custom software written in Python and NumPy to create a mask of vinculin-positive adhesion structures. Colocalisation analysis was performed using the ImageJ plugins Coloc 2, with the mask as a region of interest to calculate MOC27, and Plot_Multicolor (version 4.3) to plot line profiles. Figures were assembled using Illustrator (Adobe).
Statistics and reproducibility of experiments
Statistical significance was calculated using Kruskal–Wallis tests (non-parametric, one-way ANOVA) with Dunn’s post hoc correction as indicated in the figure legends, with P < 0.05 considered statistically significant (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). No statistical method was used to predetermine sample size. Statistical analyses were carried out using Prism software. MS data were obtained from two independent experiments for the MEF and IAC assembly datasets and from three independent experiments for the IAC disassembly dataset. Immunofluorescence quantification is based on data obtained from one independent experiment from at least 10 cells, and representative images are shown.
Data deposition and accession numbers
MS data were deposited in ProteomeXchange (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository60 with the primary accession identifiers PXD000018 (DOI: 10.6019/PXD000018; MEF dataset), PXD002159 (DOI: 10.6019/PXD002159; IAC assembly dataset) and PXD002129 (DOI: 10.6019/PXD002129; IAC disassembly dataset). Details of all identified proteins are provided in Supplementary Tables 2 and 8 for the MEF dataset, Supplementary Table 11 for the IAC assembly dataset and Supplementary Table 12 for the IAC disassembly dataset.
Previously published MS datasets that were re-analysed here are available in the PRIDE database (http://www.ebi.ac.uk/pride) with the referenced accession identifiers 9985–9987 (K562 dataset)11 and in ProteomeXchange with the identifiers PXD001578 (DOI: 10.6019/PXD001578; A375 dataset)14 and PXD001183 (DOI: 10.6019/PXD001183; HFF dataset)13. MS data from the MKF1 dataset15 or MKF2 and MKF3 datasets16 are not available in an online repository.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. E. Craig for technical assistance, J. N. Selley for bioinformatic support, M. Manca for preliminary data analysis, P. March, S. Marsden and E. Zindy for assistance with microscopy, the PRIDE team for assistance with MS data deposition and G. Jacquemet, M. C. Jones and A. P. Mould for discussions. We are grateful to K. Clark, M. L. Cutler, I. J. Fidler, M. E. Hemler, D. Vestweber, J. A. Wilkins and K. M. Yamada for reagents. This work was supported by the Wellcome Trust (grant 092015 to M.J.H.), a Wellcome Trust Institutional Strategic Support Fund award (grant 097820 to the University of Manchester) and a Biotechnology and Biological Sciences Research Council studentship from the Systems Biology Doctoral Training Centre (to E.R.H). The mass spectrometers and microscopes used in this study were purchased with grants from the Biotechnology and Biological Sciences Research Council, Wellcome Trust and the University of Manchester Strategic Fund. Data are available via ProteomeXchange with identifiers PXD000018, PXD002159 and PXD002129.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 2.Liu S, Calderwood DA, Ginsberg MH. Integrin cytoplasmic domain-binding proteins. J. Cell Sci. 2000;113:3563–3571. doi: 10.1242/jcs.113.20.3563. [DOI] [PubMed] [Google Scholar]
- 3.Morse EM, Brahme NN, Calderwood DA. Integrin cytoplasmic tail interactions. Biochemistry. 2014;53:810–820. doi: 10.1021/bi401596q. (Mosc.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winograd-Katz SE, Fässler R, Geiger B, Legate KR. The integrin adhesome: from genes and proteins to human disease. Nat. Rev. Mol. Cell Biol. 2014;15:273–288. doi: 10.1038/nrm3769. [DOI] [PubMed] [Google Scholar]
- 5.Zaidel-Bar R, Geiger B. The switchable integrin adhesome. J. Cell Sci. 2010;123:1385–1388. doi: 10.1242/jcs.066183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat. Cell Biol. 2007;9:858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byron A, Humphries JD, Bass MD, Knight D, Humphries MJ. Proteomic analysis of integrin adhesion complexes. Sci. Signal. 2011;(pt2) doi: 10.1126/scisignal.2001827. [DOI] [PubMed] [Google Scholar]
- 8.Byron A, Humphries JD, Craig SE, Knight D, Humphries MJ. Proteomic analysis of α4β1 integrin adhesion complexes reveals α-subunit-dependent protein recruitment. Proteomics. 2012;12:2107–2114. doi: 10.1002/pmic.201100487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byron A, et al. A proteomic approach reveals integrin activation state-dependent control of microtubule cortical targeting. Nat. Commun. 2015;6:6135. doi: 10.1038/ncomms7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang I-H, et al. GEF-H1 controls focal adhesion signaling that regulates mesenchymal stem cell lineage commitment. J. Cell Sci. 2014;127:4186–4200. doi: 10.1242/jcs.150227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Humphries JD, et al. Proteomic analysis of integrin-associated complexes identifies RCC2 as a dual regulator of Rac1 and Arf6. Sci. Signal. 2009;2:ra51. doi: 10.1126/scisignal.2000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo J-C, Han X, Hsiao C-T, Yates JR, Waterman CM. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat. Cell Biol. 2011;13:383–393. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng DHJ, Humphries JD, Byron A, Millon-Frémillon A, Humphries MJ. Microtubule-dependent modulation of adhesion complex composition. PLoS ONE. 2014;9:e115213. doi: 10.1371/journal.pone.0115213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson J, et al. Defining the phospho-adhesome through the phosphoproteomic analysis of integrin signalling. Nat. Commun. 2015;6:6265. doi: 10.1038/ncomms7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schiller HB, Friedel CC, Boulegue C, Fässler R. Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 2011;12:259–266. doi: 10.1038/embor.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schiller HB, et al. β1- and αv-class integrins cooperate to regulate myosin II during rigidity sensing of fibronectin-based microenvironments. Nat. Cell Biol. 2013;15:625–636. doi: 10.1038/ncb2747. [DOI] [PubMed] [Google Scholar]
- 17.Yue J, et al. Microtubules regulate focal adhesion dynamics through MAP4K4. Dev. Cell. 2014;31:572–585. doi: 10.1016/j.devcel.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones MC, et al. Isolation of integrin-based adhesion complexes. Curr. Protoc. Cell Biol. 2015;66:9.8.1–9.8.15. doi: 10.1002/0471143030.cb0908s66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo J-C, Han X, Yates JR, Waterman CM. Isolation of focal adhesion proteins for biochemical and proteomic analysis. Methods Mol. Biol. 2012;757:297–323. doi: 10.1007/978-1-61779-166-6_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humphries JD, Paul NR, Humphries MJ, Morgan MR. Emerging properties of adhesion complexes: what are they and what do they do? Trends Cell Biol. 2015;25:388–397. doi: 10.1016/j.tcb.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Geiger T, Zaidel-Bar R. Opening the floodgates: proteomics and the integrin adhesome. Curr. Opin. Cell Biol. 2012;25:562–568. doi: 10.1016/j.ceb.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Smith MA, Hoffman LM, Beckerle MC. LIM proteins in actin cytoskeleton mechanoresponse. Trends Cell Biol. 2014;24:575–583. doi: 10.1016/j.tcb.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uemura A, Nguyen T-N, Steele AN, Yamada S. The LIM domain of zyxin is sufficient for force-induced accumulation of zyxin during cell migration. Biophys. J. 2011;101:1069–1075. doi: 10.1016/j.bpj.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schiller HB, Fässler R. Mechanosensitivity and compositional dynamics of cell-matrix adhesions. EMBO Rep. 2013;14:509–519. doi: 10.1038/embor.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rolland T, et al. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janovick JA, Natarajan K, Longo F, Conn PM. Caldesmon: a bifunctional (calmodulin and actin) binding protein which regulates stimulated gonadotropin release. Endocrinology. 1991;129:68–74. doi: 10.1210/endo-129-1-68. [DOI] [PubMed] [Google Scholar]
- 27.Manders EMM, Verbeek FJ, Aten JA. Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 28.Dougherty GW, Jose C, Gimona M, Cutler ML. The Rsu-1-PINCH1-ILK complex is regulated by Ras activation in tumor cells. Eur. J. Cell Biol. 2008;87:721–734. doi: 10.1016/j.ejcb.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Attanasio F, Caldieri G, Giacchetti G, van Horssen R, Wieringa B, Buccione R. Novel invadopodia components revealed by differential proteomics analysis. Eur. J. Cell Biol. 2011;90:115–127. doi: 10.1016/j.ejcb.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 30.Cervero P, Himmel M, Krüger M, Linder S. Proteomic analysis of podosome fractions from macrophages reveals similarities to spreading initiation centres. Eur. J. Cell Biol. 2012;91:908–922. doi: 10.1016/j.ejcb.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 31.de Hoog CL, Foster LJ, Mann M. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell. 2004;117:649–662. doi: 10.1016/s0092-8674(04)00456-8. [DOI] [PubMed] [Google Scholar]
- 32.Ezratty EJ, Partridge MA, Gundersen GG. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat. Cell Biol. 2005;7:581–590. doi: 10.1038/ncb1262. [DOI] [PubMed] [Google Scholar]
- 33.Beggs AH, et al. Cloning and characterization of two human skeletal muscle alpha-actinin genes located on chromosomes 1 and 11. J. Biol. Chem. 1992;267:9281–9288. [PubMed] [Google Scholar]
- 34.Bialkowska K, et al. The integrin co-activator Kindlin-3 is expressed and functional in a non-hematopoietic cell, the endothelial cell. J. Biol. Chem. 2010;285:18640–18649. doi: 10.1074/jbc.M109.085746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karaköse E, Schiller HB, Fässler R. The kindlins at a glance. J. Cell Sci. 2010;123:2353–2356. doi: 10.1242/jcs.064600. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, et al. Nuclear SIPA1 activates integrin β1 promoter and promotes invasion of breast cancer cells. Oncogene. 2015;34:1451–1462. doi: 10.1038/onc.2014.36. [DOI] [PubMed] [Google Scholar]
- 37.Bai SW, et al. Identification and characterization of a set of conserved and new regulators of cytoskeletal organization, cell morphology and migration. BMC Biol. 2011;9:54. doi: 10.1186/1741-7007-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buxbaum AR, Wu B, Singer RH. Single β-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science. 2014;343:419–422. doi: 10.1126/science.1242939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chicurel ME, Singer RH, Meyer CJ, Ingber DE. Integrin binding and mechanical tension induce movement of mRNA and ribosomes to focal adhesions. Nature. 1998;392:730–733. doi: 10.1038/33719. [DOI] [PubMed] [Google Scholar]
- 40.Katz ZB, et al. β-Actin mRNA compartmentalization enhances focal adhesion stability and directs cell migration. Genes Dev. 2012;26:1885–1890. doi: 10.1101/gad.190413.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willett M, Pollard HJ, Vlasak M, Morley SJ. Localization of ribosomes and translation initiation factors to talin/beta3-integrin-enriched adhesion complexes in spreading and migrating mammalian cells. Biol. Cell. 2010;102:265–276. doi: 10.1042/BC20090141. [DOI] [PubMed] [Google Scholar]
- 42.Willett M, Brocard M, Davide A, Morley SJ. Translation initiation factors and active sites of protein synthesis co-localize at the leading edge of migrating fibroblasts. Biochem. J. 2011;438:217–227. doi: 10.1042/BJ20110435. [DOI] [PubMed] [Google Scholar]
- 43.Mellacheruvu D, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bachir AI, et al. Integrin-associated complexes form hierarchically with variable stoichiometry in nascent adhesions. Curr. Biol. 2014;24:1845–1853. doi: 10.1016/j.cub.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Etheridge T, et al. The integrin-adhesome is required to maintain muscle structure, mitochondrial ATP production, and movement forces in Caenorhabditis elegans. FASEB J. 2015;29:1235–1246. doi: 10.1096/fj.14-259119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bulgakova NA, Klapholz B, Brown NH. Cell adhesion in Drosophila: versatility of cadherin and integrin complexes during development. Curr. Opin. Cell Biol. 2012;24:702–712. doi: 10.1016/j.ceb.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wickström SA, Lange A, Montanez E, Fässler R. The ILK/PINCH/parvin complex: the kinase is dead, long live the pseudokinase! EMBO J. 2010;29:281–291. doi: 10.1038/emboj.2009.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sebé-Pedrós A, Roger AJ, Lang FB, King N, Ruiz-Trillo I. Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc. Natl Acad. Sci. USA. 2010;107:10142–10147. doi: 10.1073/pnas.1002257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meller J, et al. Emergence and subsequent functional specialization of kindlins during evolution of cell adhesiveness. Mol. Biol. Cell. 2015;26:786–796. doi: 10.1091/mbc.E14-08-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanchanawong P, et al. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kelly DF, Taylor KA. Identification of the beta1-integrin binding site on alpha-actinin by cryoelectron microscopy. J. Struct. Biol. 2005;149:290–302. doi: 10.1016/j.jsb.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 52.Otey CA, Pavalko FM, Burridge K. An interaction between alpha-actinin and the beta 1 integrin subunit in vitro. J. Cell Biol. 1990;111:721–729. doi: 10.1083/jcb.111.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi CK, et al. Actin and α-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 2008;10:1039–1050. doi: 10.1038/ncb1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rossier O, et al. Integrins β1 and β3 exhibit distinct dynamic nanoscale organizations inside focal adhesions. Nat. Cell Biol. 2012;14:1057–1067. doi: 10.1038/ncb2588. [DOI] [PubMed] [Google Scholar]
- 55.Zaidel-Bar R, Cohen M, Addadi L, Geiger B. Hierarchical assembly of cell-matrix adhesion complexes. Biochem. Soc. Trans. 2004;32:416–420. doi: 10.1042/BST0320416. [DOI] [PubMed] [Google Scholar]
- 56.Zaidel-Bar R, Ballestrem C, Kam Z, Geiger B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 2003;116:4605–4613. doi: 10.1242/jcs.00792. [DOI] [PubMed] [Google Scholar]
- 57.Bass MD, Roach KA, Morgan MR, Mostafavi-Pour Z, Schoen T, Muramatsu T, Mayer U, Ballestrem C, Spatz JP, Humphries MJ. Syndecan-4-dependent Rac1 regulation determines directional migration in response to the extracellular matrix. J. Cell Biol. 2007;117:527–538. doi: 10.1083/jcb.200610076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 59.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 60.Vizcaíno JA, et al. The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gray KA, Yates B, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 2015;43:D1079–D1085. doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mitchell A, et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 64.Saldanha AJ. Java Treeview-extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 65.Saeed AI, et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 66.Cline MS, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Assenov Y, Ramírez F, Schelhorn S-E, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 68.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 69.Zeeberg BR, et al. High-Throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID) BMC Bioinformatics. 2005;6:168. doi: 10.1186/1471-2105-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.