Abstract
L-Glutamine (Gln) functions physiologically to balance tissue requirements of carbon and nitrogen. It has been proposed that in cancer cells undergoing aerobic glycolysis, accelerated anabolism is sustained by Gln-derived carbons, which replenish the tricarboxylic acid (TCA) cycle (anaplerosis). However, it is shown here that in glioblastoma (GBM) cells, almost half of the Gln-derived glutamate (Glu) is secreted and does not enter the TCA cycle and, that inhibiting glutaminolysis does not affect proliferation. Moreover, Gln-starved cells are not rescued by TCA cycle replenishment. Instead, the conversion of Glu to Gln by Glutamine Synthetase (GS) (cataplerosis) confers Gln prototrophy, and fuels de novo purine biosynthesis. In both orthotopic GBM models and in patients, 13C-glucose tracing showed that GS produces Gln from TCA cycle-derived carbons. Finally, while it is contributed only marginally by the circulation, the Gln required for the growth of GBM tumours is either autonomously synthesized by GS-positive glioma cells, or supplied by astrocytes.
INTRODUCTION
Gln and Glu constitute a metabolic hub in cellular physiology. An increased demand for Gln by transformed cells has been recognized by biochemists for almost a century and, it has been linked to its role as an abundant circulating respiratory fuel1. Notably, Gln carbons can support anabolism through entering the TCA cycle via glutaminolysis. Only specific tumour types display Gln-dependency2-9 whereby its genetic and metabolic basis remains debatable. In certain cancer models, the inhibition of glutaminase (GLS), which deaminates Gln to Glu, reduces proliferation and tumorigenicity10. Conversely, GLS2 can be induced by the tumour suppressor p5311, and in human hepatocellular carcinoma, β-catenin increases the expression of GS, which catalyses the reversed GLS reaction12. Originally, tuning of the Gln-Glu cycle was observed in the central nervous system13 where Glu is the most abundant neurotransmitter14. Unlike astrocytes, glioma cells can release neuro-exitotoxic amounts of Glu, potentially promoting tumorigenesis15. Gln-addiction has been proposed as a mark of GBM, the most aggressive glioma4. Here, we dissected the differential metabolic roles of Gln-derived carbon and nitrogen atoms in sustaining anabolism and growth in six human established GBM cell lines, in primary GBM stem-like cells, and in normal astrocytes. Additionally, Gln-related metabolism was investigated in both primary orthotopic murine xenografts and in GBM patients, leading to the identification of a GBM-astrocyte metabolic crosstalk.
RESULTS
Gln starvation reduces GBM cell proliferation unsystematically
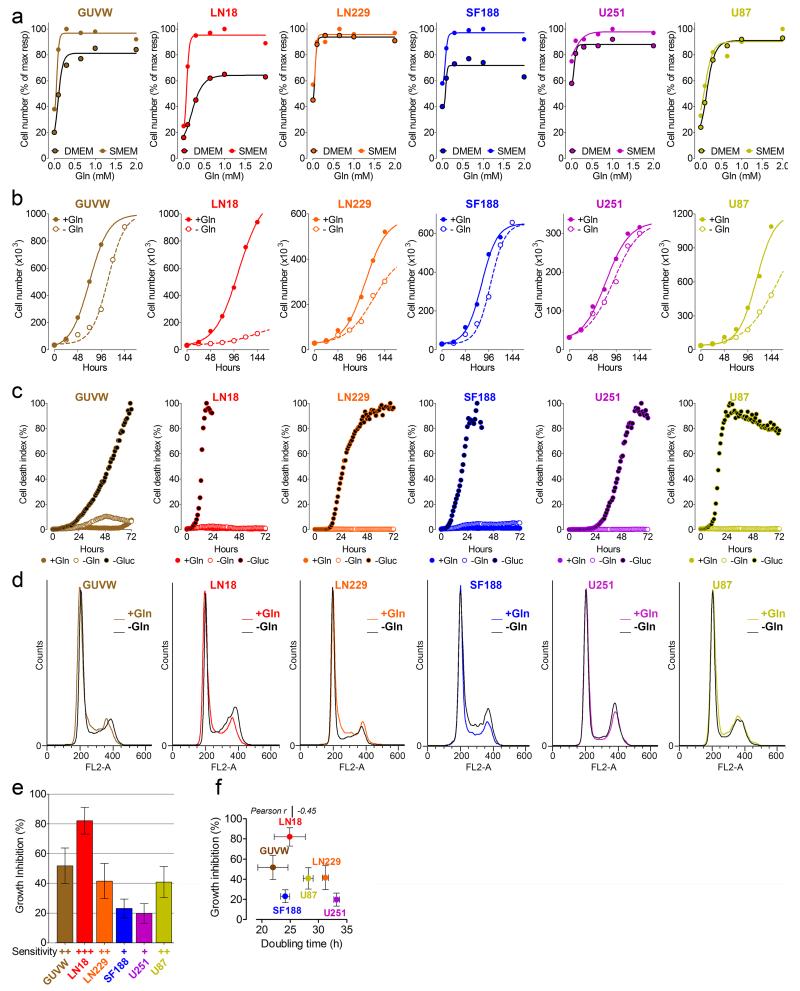
To explore their growth response to different nutrient supplies, GBM cells were incubated either in DMEM containing supra-physiological concentrations of glucose and lacking some of the non-essential amino acids, or in a newly-formulated SMEM, containing nutrient concentrations comparable to human serum (Supplementary Table 1). Both media were supplemented with various concentrations of Gln (Fig. 1a). In serum-like medium, all cell lines grew comparably to or faster than cells cultured in DMEM. In both media, the minimal Gln concentration required for maximal growth was below 0.65 mM, hereafter used as the control concentration. In the absence of Gln, cells grew faster in SMEM, demonstrating that medium formulation affects the response to Gln deprivation. Gln starvation hindered proliferation to different extents (Fig. 1b and Supplementary Fig. 1a) without inducing cell death, contrary to previous reports3,4, and to their response to glucose withdrawal (Fig. 1c). DNA flow-cytometry analysis showed that Gln starvation did not cause cell cycle arrest at any particular phase (Fig. 1d and Supplementary Fig. 1b). Overall, Gln withdrawal resulted in cell line-specific growth inhibition, ranging from 20% for U251 and SF188, to 80% for LN18 cells (Fig. 1e), independently of the initial proliferation rate (Fig. 1f).
Figure 1.
Gln starvation reduces GBM cell proliferation. (a) Dose-response curves for cell lines incubated for 3 days in DMEM or SMEM with the indicated concentrations of Gln. (b) Cells were incubated for the indicated times in SMEM +/− Gln. (c) Cells were incubated for 72 hours in SMEM +/− Gln or Glucose as indicated. Each dot represents the number of dead cells (with plasma membrane integrity loss) normalized over a confluence index. The resulting cell death index was assessed every hour. (d) Cell cycle distribution of cell lines incubated for 3 days +/− Gln. Mean of 3 independent experiments is shown in Supplementary Figure 1b. (e) Growth inhibition caused by Gln starvation. Mean ± S.E.M. n=3 independent experiments. (f) Scatter plot of the doubling time obtained for the cell lines in Gln-fed conditions, in relation to growth inhibition caused by Gln starvation. Mean ± S.E.M. n=3 independent experiments. (a-d) Data derive from one experiment performed twice (a, b, c), or three times (d). Raw data of independent repeats are provided in the statistics source data Supplementary Table 5.
Gln-based anaplerosis is not essential for the proliferation of GBM cell lines
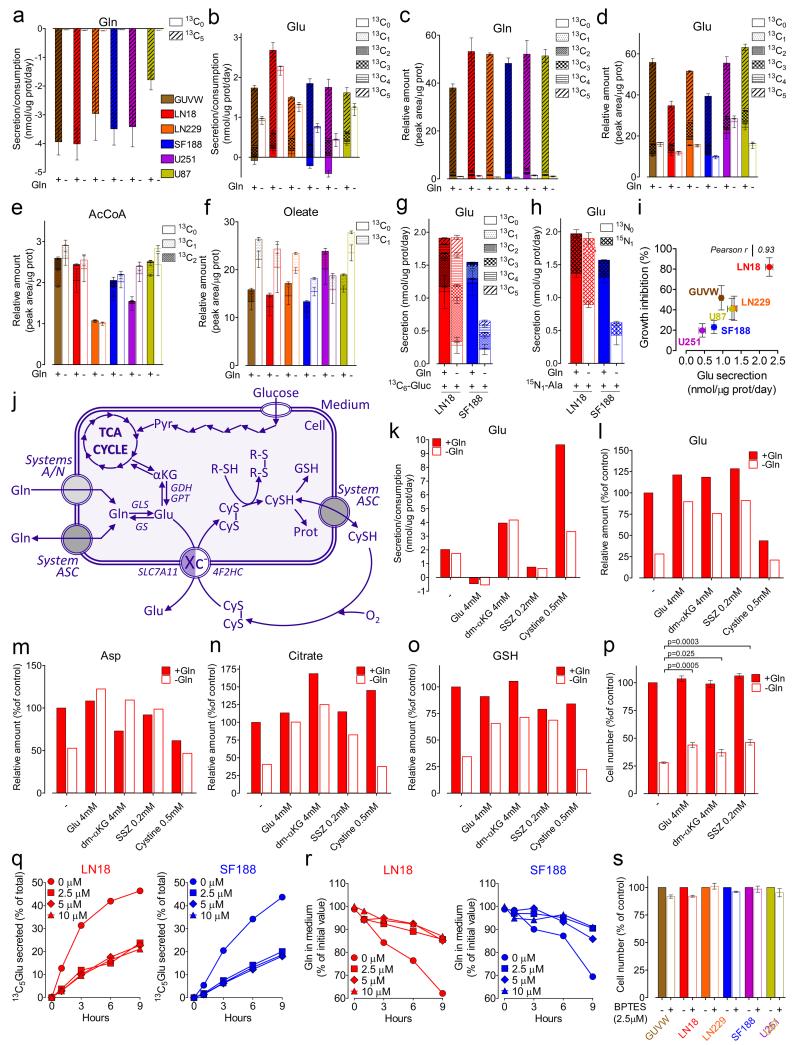
To investigate cellular metabolic alterations upon Gln starvation, the exchange rate of metabolites between cells and medium was analysed by HPLC-MS. Gln was the second most consumed nutrient by all cell lines (Supplementary Fig. 2 and Supplementary Table 2). However, no clear relationship emerged between Gln consumption and Gln dependency (Fig. 2a and Supplementary Fig.3a). In contrast, all cell lines showed a net secretion of Glu despite its presence in the medium (Fig. 2b). Tracing 13C5-labeled Gln revealed that 38 ± 8% to 60 ± 19% (GUVW and U87 cells, respectively) of the Gln consumed was deamidated and secreted as 13C5-Glu (Fig. 2a-b). Unexpectedly, even Gln-starved cells discharged Glu (Fig. 2b, empty bars). Here, intracellular Gln was almost exhausted (Fig. 2c) and Glu concentration fell by more than 50% (Fig. 2d) yet, neither acetyl-CoA (Fig. 2e) nor oleate (Fig. 2f) levels were reduced, re-affirming that Gln does not sustain fatty acids biosynthesis under normoxic condition16-20. Consistently, in all cell lines, less than 15% of the citrate was derived from reductive carboxylation (13C5-Citrate; Supplementary Fig. 3b), and labelled acetyl-CoA and oleate were barely detectable (Fig. 2e-f).
Figure 2.
The effects of Glu secretion, and GLS inhibition on GBM cells growth and metabolism. (a-b) Cells were incubated for 24h +/− 13C5-Gln. Secretion (positive bars) and consumption (negative bars) rates of Gln and Glu isotopologues are shown. Mean ± S.E.M. n=3 independent experiments. (c-f) Cells were incubated as in (a-b) and the levels of intracellular Gln, Glu, Acetyl CoA, and oleate isotopologues are shown. Mean ± S.E.M. n=3 independent experiments. (g-h) LN18 and SF188 cells were incubated for 24h +/− Gln in media where glucose (g) or alanine (h) where fully replaced by 13C6-glucose or 15N1-alanine respectively. The isotopologues distributions of Glu released in the medium are shown. Mean ± S.E.M. n=3 independent experiments. (i) Scatter plot of Glu secretion observed in the absence of Gln, in relation to the growth inhibition caused by Gln starvation. Mean ± S.E.M. n=3 independent experiments. (j) A schematic representation of the activity in the context of Glu metabolism. Adapted from Bannai et al.40 (k) LN18 cells were incubated for 24h +/− Gln in media supplemented or not with Glu, α-ketoglutarate dimethylester (dm-αKG), sulfasalazine (SSZ), or cystine, at the indicated concentrations, and the secretion/consumption rates of Glu are shown. (l-o) LN18 cells were incubated as in (k) and the intracellular levels of Glu (l), aspartate (m), citrate (n), and reduced form of glutathione (o) are shown as % of untreated control. (p) LN18 cells were incubated for 72h as described for (k). Cell number is shown as % of untreated control. Mean ± S.E.M. n=4 independent experiments. p values refer to a two-tailed t test for unpaired samples. (q-r) Cells were pre-incubated in medium with 0, 2.5, 5, 10, 15, 30μM BPTES for 3h. At t=0 medium was replaced with one containing 13C5-Gln. The abundance of 13C5-Glu (q) or 13C5-Gln (r) in the medium was monitored over time. In all conditions cells were exposed to 0.3% DMSO. (s) Cells were incubated in medium +/− 2.5μM BPTES for 72h, and counted. DMSO was 0.3% in all conditions. Mean ± S.E.M. n=3 independent experiments. (k, l, m, n, o, q, r) Data derive from one experiment performed once (k, l, m, n, o), or twice (q, r). Raw data of independent repeats are provided in the statistics source data Supplementary Table 5.
To identify the carbon source for Glu synthesis under Gln starvation, Gln-deprived LN18 and SF188 cells were incubated with 13C6-glucose. In both cases the contribution of glucose carbons to Glu was markedly increased (Fig. 2g). Accordingly, alanine consumption was increased (Supplementary Fig. 2, inset), providing the nitrogen required for Glu production (Fig. 2h). Moreover, a strong direct correlation between Glu efflux and growth inhibition was observed (Fig. 2i), suggesting that upon Gln withdrawal, Glu efflux limits the intracellular Glu available for reactions essential for growth.
To test whether Glu efflux indeed limited both its availability and the proliferation of Gln-starved cells, LN18 cells were incubated with sulfasalazine, an inhibitor of , a Glu/Cystine antiporter (Fig. 2j) which is active in glioma cells15, or with 4 mM Glu, largely exceeding the Ki for 21. In both conditions Glu release was largely inhibited (Fig. 2k). Consistently, the activation of by increasing the extracellular concentrations of cystine, boosted Glu efflux (Fig. 2k), firmly associating with the escape of Glu from GBM cells. Inhibiting prevented the drop in intracellular Glu, aspartate, and citrate, caused by Gln withdrawal (Fig. 2l-n). The level of glutathione, an alternative metabolic fate for Glu, was also substantially decreased upon Gln starvation, and significantly replenished by Glu and sulfasalazine (Fig. 2o). However, the drop in glutathione upon Gln starvation was not accompanied by its increased oxidation (Supplementary Fig. 3c), indicating that Gln delimits Glu availability for glutathione biosynthesis without causing oxidative stress. Furthermore, adding the membrane permeable dimethylester of α-ketoglutarate (dm-αKG) to Gln-starved cells doubled the Glu efflux (Fig. 2k) and replenished Glu, aspartate, citrate and glutathione intracellular pools (Fig. 2l-o). Overall, in the absence of Gln, inhibition or dm-αKG supplementation restored the cellular Glu, aspartate, citrate and glutathione levels to that of non-starved cells. However, maintaining the levels of these metabolites only moderately rescued the proliferation of Gln starved cells (Fig. 2p). Together with the observations that, under Gln starvation, intracellular oleate was unaffected (Fig. 2f), and glucose-dependent Glu production increased (Fig. 2g-h), these results imply that the contribution of Gln to growth is largely independent on anaplerosis.
To evaluate this directly, BPTES, a GLS inhibitor, was employed. The kinetics of Gln-derived Glu secretion from LN18 and SF188 cells showed that BPTES inhibited GLS activity (Fig. 2q). Accordingly, the rate of Gln consumption was reduced (Fig. 2r). Nevertheless, at the minimum effective concentration, BPTES did not affect the growth of the GBM lines (Fig. 2s and Supplementary Fig. 3d-i) confirming that here, GLS and Gln-based anaplerosis are dispensable for maximal growth.
Glutamine Synthetase sustains purine availability and cell growth under Gln starvation
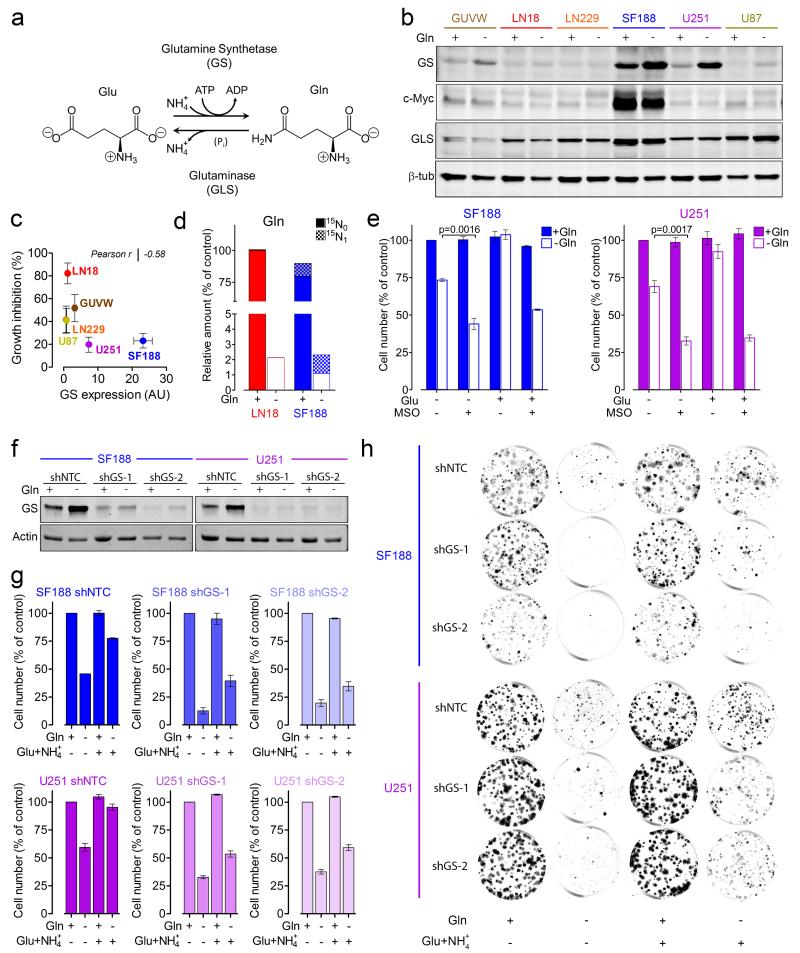
Next, a genome-scale constraint-based metabolic modelling approach was employed22, searching for reactions that become essential when Gln is removed from the SMEM medium. According to the model, GS was the only enzyme essential for sustaining biomass production after Gln starvation (Supplementary Note).
Together, GS and GLS control Gln homeostasis by catalysing opposite reactions (Fig. 2j and 3a). The mRNA levels of GS and GLS in all cell lines revealed no pattern of Gln dependency (Supplementary Fig. 4a). While it has been proposed that c-Myc determines Gln addiction by increasing GLS expression3,23,24, MYC-induced lung tumours were shown to increase GS expression2. In line with this, SF188 cells, harboring MYC amplification25, and expressing high levels of c-Myc (Fig. 3b and supplementary Fig. 4a), showed the highest levels of GS mRNA and protein (Fig. 3b and Supplementary Fig. 4a). In most cell lines, GS protein levels rose upon Gln deprivation (Fig. 3b). Furthermore, GS protein levels tend to increase in cells with decreased sensitivity to Gln withdrawal (Fig. 3c). Nevertheless, GS did not match the residual low amount of Gln found in Gln-starved cells (Fig. 2c). To investigate this apparent discrepancy, the metabolic flux via GS was assessed by the incorporation of 15N-labeled ammonia () into Gln in LN18 and SF188 cells, which display low and high levels of GS, respectively. Significant levels of 15N-labeled Gln were indeed detected in SF188 but not in LN18 cells (Fig. 3d). Next, SF188 and U251 cells, displaying high levels of GS and low sensitivity to Gln withdrawal, were incubated with L-methionine sulfoximine (MSO), a selective irreversible inhibitor of GS. MSO sensitized cells to Gln starvation and, further, abolished the protective effect of Glu supplementation (Fig. 3e). To complement this approach, GS expression was stably silenced in these cells by two shRNA sequences (Fig. 3f). Upon Gln starvation cell proliferation (Fig. 3g) as well as colony formation (Fig. 3h and Supplementary Fig. 4b) was lowered by GS silencing. Supplementation with the GS substrates Glu and ammonia, rescued Gln-deprived control cells more effectively than GS-silenced cells.
Figure 3.
GS sustains cell growth during Gln starvation. (a) GS and GLS catalysed reactions. (b) Cells were incubated for 24h +/− Gln and protein expression was assessed. Unprocessed scans of western blots are shown in Supplementary Figure 8. (c) Scatter plot of GS protein expression observed in Gln-fed condition (Arbitrary Units, AU) in relation to the growth inhibition caused by Gln starvation. Mean ± S.E.M. n=3 independent experiments. (d) LN18 and SF188 cells were incubated for 24h +/− Gln in the presence of 0.8 mM . The intracellular levels of Gln isotopologues are shown as % of 15N0-Gln in LN18 cells. Data derive from one experiment performed twice. Raw data of independent repeats are provided in the statistics source data Supplementary Table 5. (e) SF188 and U251 cells were incubated for 72h +/− Gln in medium supplemented with 4mM Glu and 1mM MSO as indicated. Cells numbers are shown as % of untreated control. Mean ± S.E.M. n=3 independent experiments. (f) SF188 and U251 cells stably expressing a non-targeting control shRNA (shNTC) or two sequences targeting GS (shGS-1 and shGS-2) were incubated for 24h +/− Gln. (g) Cells were incubated for 72h +/− Gln in medium supplemented with 4mM Glu, and 0.8mM , as indicated. Cell number is shown as % of the respective Gln-fed control. Mean ± S.E.M. n=3 independent experiments. (h) Cells were incubated for 12-17 days +/− Gln in medium supplemented with 4mM Glu, and 0.8mM as indicated. Colonies obtained in representative wells are shown. n=4 independent experiments, quantified as shown in Supplementary Figure 4b.
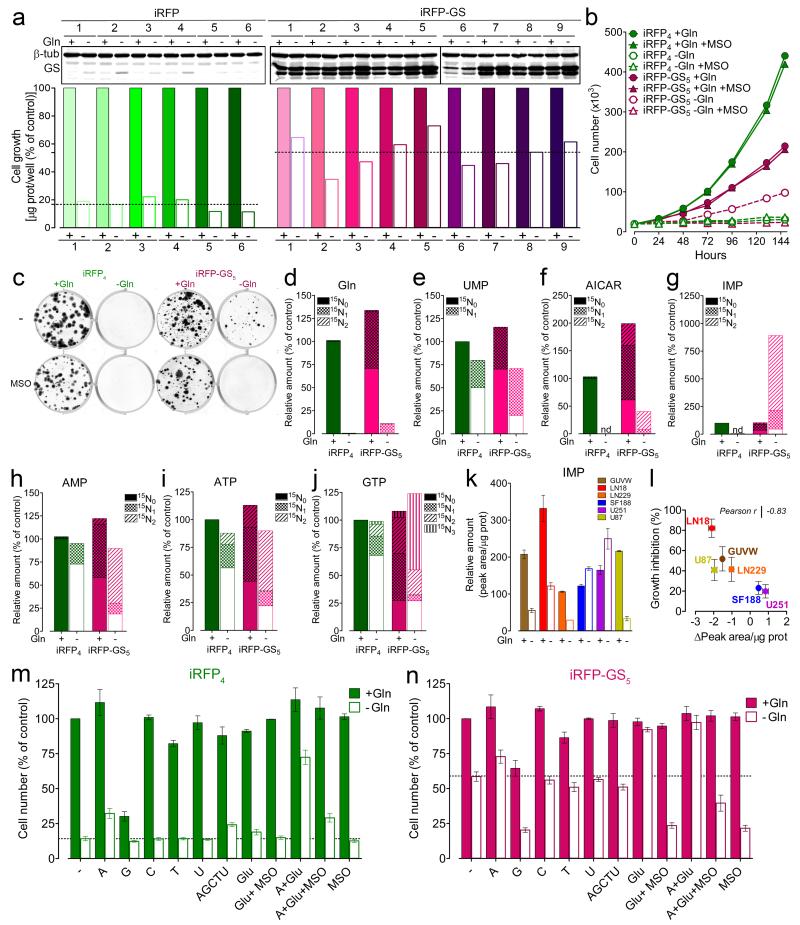
To corroborate the causal link between Gln biosynthesis and Gln-dependency, GS was overexpressed in LN18 cells, which display low GS levels and high sensitivity to Gln deprivation. To this end, LN18-derived clones stably expressing infra-Red Fluorescent Protein only (iRFP)26,27 or iRFP and GS were established. To eliminate intrinsic clonal variability, GS expression and its effect on growth under Gln starvation were evaluated in multiple clones (6 iRFP and 9 iRFP-GS). After five days of starvation, growth of iRFP control cells reached, on average, 16% ± 5 % of control Gln-fed cells, while iRFP-GS clones reached on average, 54% ± 12 % (Fig. 4a). Under Gln-supplementation, the iRFP-GS5 clone proliferated slower than iRFP controls. This was not rectified by GS inhibition with MSO (Fig. 5b), consistent with a reported non-metabolic, anti-proliferative role of GS28. Nevertheless, iRFP-GS5, but not control iRFP4 cells, proliferated and formed colonies in Gln-free medium and this growth advantage was blocked by MSO (Fig. 4b-c). These results imply that under Gln starvation the amidation of Glu via GS sustains cell growth. In line with this, when supplemented with 15N1-ammonia, GS-expressing cells displayed 15N incorporation into ~50% of the total Gln pool, even when Gln fed (Fig 4d). When incubated with 15N1-ammonia without Gln, residual intracellular Gln was higher in iRFP-GS5 cells compared to control iRFP4 cells, and produced entirely by GS as judged by 15N incorporation (Fig. 4d).
Figure 4.
GS activity regulates cell growth and purine availability under Gln starvation. (a) Top: LN18 clones stably expressing iRFP or iRFP-GS were incubated +/− Gln for 5 days and protein expression assessed. Unprocessed scans of western blots are shown in Supplementary Figure 8. Bottom: For each clone growth was determined from the protein amount and referred as % of respective control. Dashed lines show the mean % values obtained - Gln. (b) iRFP4 and iRFP-GS5 cells were incubated for the indicated times in medium +/− Gln and MSO (1mM), and counted. (c) iRFP4 and iRFP-GS5 cells were incubated +/− Gln and MSO (1mM) for 21 days, and colonies in representative wells are shown. Data derive from one experiment performed twice. (d-j) iRFP4 and iRFP-GS5 cells were incubated +/− Gln for 24h in medium supplemented with 0.8 mM . The intracellular isotopologues of Gln, UMP, AICAR, IMP, AMP, ATP, and GTP are shown as % of values obtained for the 15N0 metabolites in iRFP4 cells in the presence of Gln. (k) Cell lines were incubated +/− Gln for 24h and the relative amount of intracellular IMP is shown. Mean ± S.E.M. n=3 independent experiments. (l) Scatter plot of the changes in intracellular IMP levels in relation to the growth inhibition caused by Gln starvation. Mean ± S.E.M. n=3 independent experiments. (m-n) iRFP4 (m) and iRFP-GS5 (n) cells were incubated for 72h +/− Gln in medium containing adenosine (A) guanosine (G) cytidine (C) thymidine (T) uridine (U), each at 0.2mM, or in combination (AGCTU) at 0.2 mM each, Glu (4mM), MSO (1mM), as indicated. Cells numbers are shown as % of untreated control. Dashed lines show percentage values obtained in the absence of Gln without any further supplementation. Mean ± S.E.M. n=3 independent experiments. (a, b, d, e, f, g, h, i, j) Data derive from one experiment performed once (a, b) or twice (d-j). Raw data of independent repeats are provided in the statistics source data Supplementary Table 5.
Figure 5.
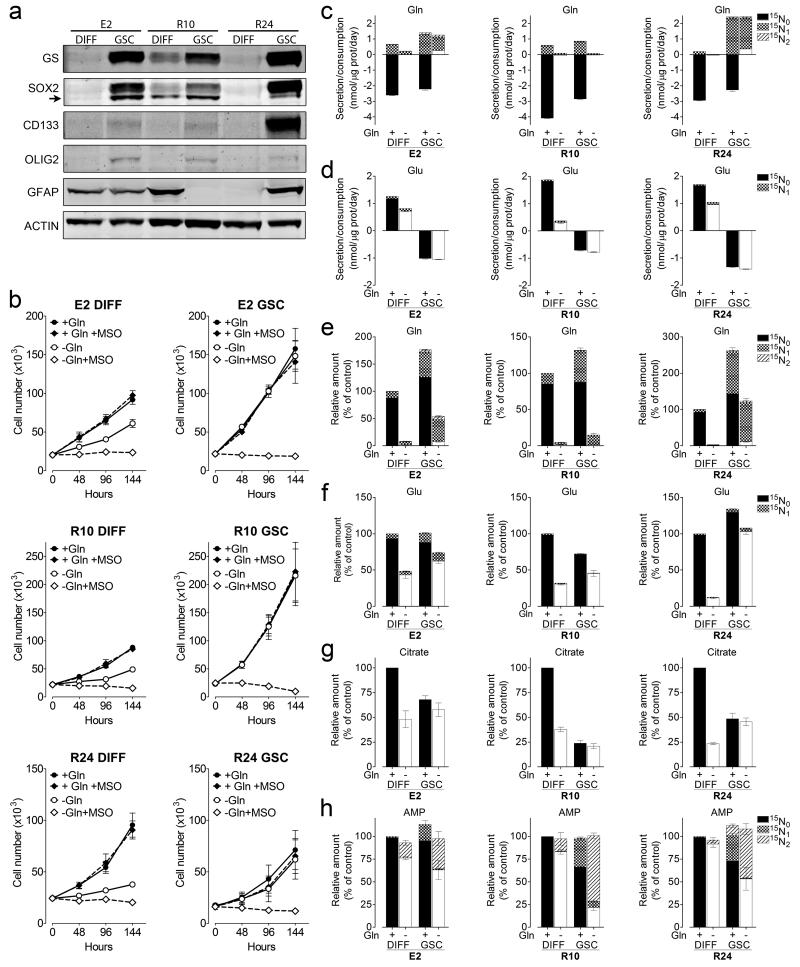
Gln metabolism in differentiated (DIFF) and GBM Stem-like (GSC) primary human GBM cells. (a) Protein expression was assessed in E2, R10 and R24 cells maintained in DMEM/F-12 and supplemented as described in the Methods section. Arrow points to SOX2 specific band. A representative experiment repeated twice is shown. Unprocessed scans of western blots are shown in Supplementary Figure 8. (b) Cells were incubated in SMEM supplemented as described in the Methods section, +/− 0.65mM Gln and 1mM MSO as indicated. Mean ± S.E.M. n=3 independent experiments. (c-d) The exchange rates of Gln (c) and Glu (d) isotopologues in cells incubated for 24h in SMEM +/− 0.65mM Gln supplemented with of 0.8mM . Mean ± S.E.M. n=3 independent experiments. (e-h) The Intracellular content of Gln (e), Glu (f), citrate (g), and AMP (h) isotopologues in cells incubated as in (c-d). Mean ± S.E.M. n=3 independent experiments.
To explore the essentiality and metabolic fate of de novo synthesized Gln under Gln starvation, we employed Flux Imbalance Analysis29. In silico, nucleotide biosynthesis delimited cell growth, with a higher weighted cost for purine biosynthesis (Supplementary Note). Indeed, regardless of GS status, Gln removal only marginally affected the levels of the pyrimidine nucleotides uridine monophosphate (UMP) (Fig. 4e). Moreover, the contribution of GS-derived 15N1-Gln to 15N1-UMP showed that the low GS-activity in iRFP4 cells could maintain UMP production. Similarly, the biosynthesis of UDP-N-acetylglucosamine, an intermediate of hexosamine biosynthesis, which requires Gln-derived nitrogen, was also sustained in iRFP4 cells during Gln starvation (Supplementary Fig. 4c). By contrast, 5-Aminoimidazole-4-carboxamide ribotide (AICAR), a purine precursor for inosine monophosphate (IMP), dropped to undetectable levels in Gln-starved iRFP4 cells (Fig. 4f). The ammonia-derived AICAR (15N2-isotopologue) in Gln-starved iRFP-GS5 cells demonstrates that GS contributes the two Gln nitrogen atoms required for its biosynthesis (Fig. 4f). Thus, 15N2-IMP accumulated in Gln-deprived iRFP-GS5 cells, but not in iRFP4 cells (Fig. 4g). The IMP found in iRFP-GS5 starved cells corroborates the reported inhibitory effect of Gln deprivation on IMP dehydrogenase30, and suggests that under these conditions over-activity of GS exceeds the rate of IMP conversion to AMP and GMP. Indeed, AMP levels were not significantly affected by either Gln presence or GS overexpression (Fig. 4h). Moreover, ATP and GTP levels, indices of the cell’s bioenergetics state, were comparable between iRFP-GS5 and iRFP4 starved cells (Fig. 4i-j). Conversely the fractions of 15N labelled AMP, ATP, and GTP found in iRFP-GS5 cells exceeded those of iRFP4 cells, demonstrating that, under Gln starvation, GS sustains de novo biosynthesis of purine nucleotides.
Notably, the reduction in IMP caused by Gln starvation in the six GBM cell lines correlated with Gln-dependency (Fig. 4k-l). Accordingly, adenosine, but not guanosine or pyrimidine nucleosides, partially restored Gln-independent growth of iRFP4 cells (Fig. 4m). Furthermore, the combined addition of adenosine and Glu compensated for the lack of exogenous Gln in iRFP4 cells (Fig. 4m), and Glu alone completely restored proliferation of iRFP-GS5 starved cells (Fig. 4n). In both lines, MSO prevented the Glu rescue, confirming that under Gln starvation Glu availability determines Gln production rather than anaplerosis. The effect of adenosine on Gln-starved cells was MSO-independent (Fig. 4m-n), since it supported proliferation downstream of GS.
Primary human GBM stem-like cells are self-sufficient for Gln requirements
The clinical relevance of studying established glioma cell lines has been questioned due to their inability to form tumours that recapitulate human pathology. Therefore, we utilized three primary patient-derived GBM cell lines (E2, R10, R24), generating paired populations of differentiated cells (DIFF) and glioma stem-like cells (GSC)31. Stem cell markers such as CD133, Olig2 and Sox2, were predominantly expressed in GSC but not DIFF, however the astrocytic marker glial fibrillary acidic protein (GFAP), was not consistently associated with the DIFF population (Fig. 5a). The expression of GS was markedly higher in all GSC compared to DIFF (Fig. 5a) and while DIFF proliferation was attenuated in the absence of Gln, GSC grew independently of Gln supplementation (Fig. 5b). Once more, the growth of Gln-starved DIFF and GSC was abolished by MSO (Fig. 5b).
Next, the exchange rates (Fig. 5c-d) and intracellular composition of metabolites (Fig. 5e-h) were analysed in these primary Gln-starved or control cells, in the presence of 15N1-ammonia. The net Gln consumption was consistently higher in DIFF compared to GSC, whereby R24-GSC demonstrated no net Gln consumption (Fig. 5c). As the intracellular 15N1-Gln fraction shows, GSC have higher GS activity compared to DIFF and sustain higher residual Gln levels upon starvation (Fig. 5e). The differences in Glu exchange rates were also striking: GSC exhibited a net uptake of Glu, while the reverse occurred in DIFF (Fig 5d). Also, upon Gln withdrawal, intracellular levels of citrate (Fig. 5g) decreased in DIFF, but remained unaltered in GSC, showing that Gln-derived anaplerosis was redundant to the stem-like population. Finally ammonia-derived 15N incorporation into purine nucleotides (15N2-AMP) under Gln starvation was greater in GSC compared to the paired DIFF.
Human GBM tumours rely on in situ de novo Gln synthesis
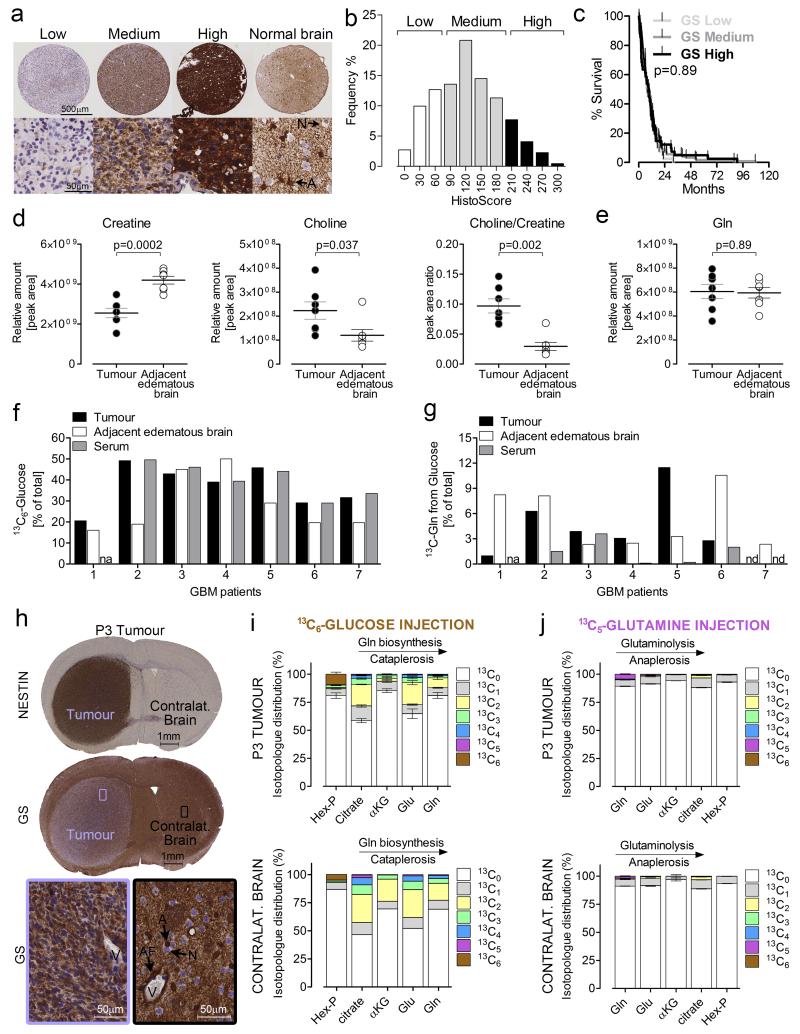
As shown above, GS activity, largely determining Gln dependency, varies between established and primary human GBM cells. Similarly, tissue microarray (TMA) analysis showed that GS expression varies between human GBM patients (n=209), resembling a Gaussian distribution ranging from tumours with low GS levels, comparable to neurons (25% of patients), to high-expression tumours comparable to astrocytes (15%) (Fig 6a-b). However, GS expression did not predict patient median survival (Fig. 6c). Of 20 biopsies, from which core TMA were sampled, 5 showed substantial intra-tumoral GS immunostaining heterogeneity (3 examples are reported in Supplementary Fig. 5). Most GBMs either showed GS uniformity, or a mosaic infiltration of GS-positive cells, suggesting autonomous intra-tumoral Gln biosynthetic capacity. To assess this hypothesis in human tumours, seven GBM patients were injected with 13C6-glucose prior to surgery, and metabolites were extracted from the resected tumours and their edematous margins. The metabolic analysis reliably discriminated between tumour and adjacent tissues by using the choline to creatine ratio, a parameter for classifying brain tumours by MR spectroscopy32 (Fig. 6d). No significant difference in Gln content was observed between tumour and adjacent tissues (Fig. 6e). At the time of resection, 13C6-glucose enrichment in serums ranged between 16% and 50% (Fig. 6f, and Supplementary Fig. 6a). Glucose-derived 13C-Gln was detected in 6/7 tumours and in 7/7 adjacent edematous tissues with an enrichment ranging between 1% and 12% (Fig. 6g). In 3/5 patients the fraction of glucose-derived Gln in the tumour was higher than in the serum sample, and so not in equilibrium with the circulating Gln, suggesting that the tumour Gln pool is synthesized in situ and/or provided by adjacent normal brain.
Figure 6.
Gln metabolism in GBM patients and primary orthotopic xenografts. (a) GBM tissue microarray. GS immuno-staining of representative tissue cores at low and high magnification (top and bottom respectively). A: Astrocyte, N: Neuron. (b) Frequency distribution of GBM patients (n=209) divided according to their histoscore for GS, and categorized as low, medium, and high. Normal astrocytes were used as a reference for defining maximal immunoreactivity. (c) Kaplan Meier curves for GBM patients divided into low, medium, and high GS expression. p value refers to a log-rank (Mantel-Cox) test. (d-e) creatine, choline, choline to creatine ratio (d), and Gln (e) levels in tumor tissue and adjacent edematous brain of GBM patients injected with 13C6-glucose before surgical intervention. n= 7 patients, p values refer to a two-tailed t test for paired samples. (f-g) 13C6-glucose (f), and 13C Gln (g) enrichment in serum at time of tumour resection, in tumor tissue, and in adjacent edematous tissue. Gln isotopologues incorporating one or more 13C-atoms, over the total amount of Gln detected (% of total) are shown. na: not available, nd: not detectable. Values were corrected for the natural abundance of 13C. (h) Coronal section of human P3 GBM xenograft grown in brain of immunocompromised mice, and stained for human nestin and GS. Lower panels are magnification of the respective framed regions. A: astrocytes, AF: astrocytic end-feet, N: neuron, V: blood vessel. (i, j) Isotopologue distribution of metabolites (Hexose phosphates, citrate, α-KG, Glu, Gln) obtained in mice orthotopically xenografted with human P3 GBM, and injected in the tail vain with a bolus of 13C6-Glucose (i) or 13C5-Gln (j). Tissues were sampled 22min after injection. The values are mean ± S.E.M. n=3 mice for all conditions, except for contralateral brain of mice injected with glucose, where 2 mice were used. (i, j) Raw data of independent repeats are provided in the statistics source data Supplementary Table 5.
To complement the analysis in patients, mice were orthotopically transplanted with a GS positive human GBM (P3, Fig. 6h) and injected with 13C-labelled glucose or Gln ~20 minutes prior to tissue extraction. An enrichment of 44 ± 3% of 13C6-glucose was found in the blood at the time of tissue sampling. The intracellular hexoses phosphate pool derived from 13C6-glucose was ~10% and 5% in tumour and contralateral brain tissue, respectively (Fig. 6i). Concomitantly, ~10% and 15% of the total Gln was labelled (13C2) from 13C6-glucose in tumours and contralateral brain, respectively (Fig. 6i), consistent with GS activity in those tissues. After 13C5-Gln injection, the enrichment in circulating 13C5-Gln at the time of tissue sampling was 17 ± 1%. Isotopologue distribution analysis showed an enrichment in 13C5-Gln of <5% in both tumour and contralateral brain tissues (Fig. 6j), while in the liver, 13C5-Gln presented 12% of the total (Supplementary Fig. 6e). Products of glutaminolysis, such as Glu and α-ketoglutarate, were labelled below 1% in both tumour and brain tissues but ~5% in liver (Fig. 6j and Supplementary Fig. 6e). These results suggest slow kinetics both for Gln uptake from the blood, and for glutaminolysis in GBM and brain tissues, compared to liver. Similar results were obtained upon constant carotid artery 13C5-Gln infusion: within two hours, circulating 13C5-Gln levels plateaued at ~20% enrichment, with an overall ~70% increase in steady-state levels of Gln (Supplementary Fig. 6f). Here too, 13C5-Gln and glutaminolysis products were scarce in tumour and contralateral brain (Supplementary Fig. 6h-i) indicating that physiologically, circulating Gln does not significantly supply Gln to the brain or the tumour within it.
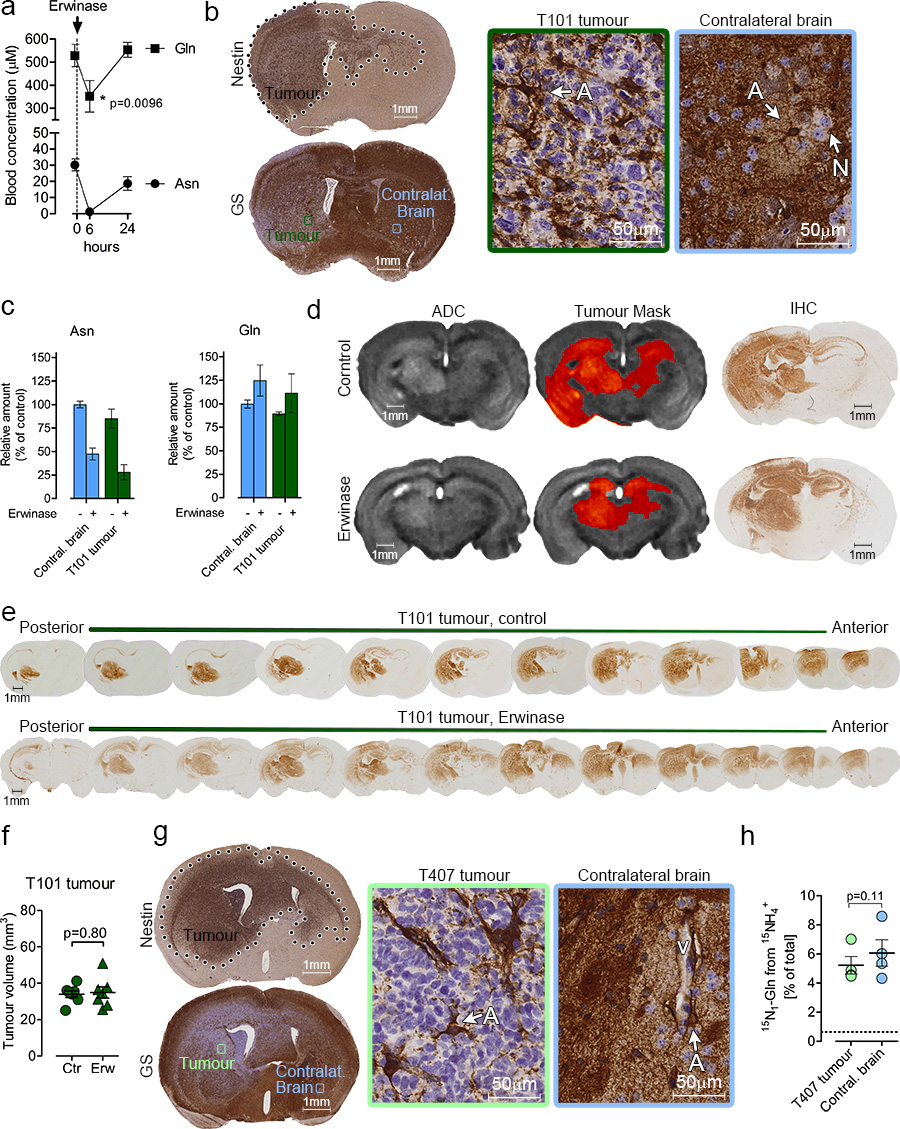
Next, Erwinase, an enzyme that temporarily depletes circulating asparagine (Asn) and significantly reduces Gln9,33 (Fig 7a), was injected daily (five times per week) into mice bearing GS-negative orthotopic GBM xenografts (T101, Fig. 7b). Erwinase reduced intra-tumoral and intracerebral Asn but not Gln (Fig. 7c). The diffuse morphology of GS-negative tumours impaired Ex vivo MRI analysis volumetric quantification (Fig. 7d). Histological reconstruction to assess tumour burden (Fig. 7e) revealed no significant differences between control and Erwinase treated groups (Fig. 7f). Altogether, these results indicate that Gln is not provided to GBM by the blood and so, the proximity of GS-positive astrocytes and GS-negative glioma cells (Fig. 7b and 7g as representatives) suggest that astrocytes may be a source of Gln. Indeed, when mice bearing GS-negative GBM xenografts (T407, Fig 7g) were infused with 15N1-ammonia into the carotid artery for 4 hours, the fraction of 15N-Gln was ~5% in both tumour and contralateral brain tissues (Fig 7h) indicating that they are a secluded, autonomous compartment for Gln biosynthesis and utilization, where GS expressing cells supply Gln to GS-negative ones.
Figure 7.
Glutamine supply for GBM tumours with low GS expression. (a) Gln and Asn levels were measured by HPLC-MS in peripheral blood samples obtained at indicated time points from immunocompromised mice intraperitoneally injected with Erwinase (5U/gr of body weight). Mean ± S.E.M. n=5 mice. p values refer to a two-tailed t test for paired samples. (b) Coronal section of human T101 GBM xenograft grown in brain of immunocompromised mice, and stained for human nestin and GS. Left panels are magnification of the respective framed regions. A: astrocytes, N: neuron. (c) Immunocompromised mice were orthotopically implanted with T101 GBM tumours and treated with Erwinase for 6 weeks as described in the Methods section. Asn and Gln were assessed in the tumour and contralateral brain 6h after the last Erwinase injection. Mean ± S.E.M. n=3 mice. (d) MRI-based Apparent Diffusion Coefficient (ADC) maps of T101 GBM tumours treated with Erwinase as described in (c). The tumour mask has been manually delineated to highlight the tumour region. IHC staining of brain sections corresponding to the MRI scans are shown. T101 tumours were stained with an anti-human EGFR antibody. (e) Two representative series of coronal sections of T101 brain xenografts, were stained for human EGFR. Mice were treated with Erwinase as in (c). (f) Volumes of T101 orthotopic tumours obtained thorough quantitative imaging of EGFR-stained serial sections of brains. Mice were treated with Erwinase as in (c). Mean ± S.E.M. n=7 mice. p value refer to a two-tailed t test for unpaired samples. (g) Coronal section of human T407 GBM xenograft grown in brain of immunocompromised mice, and stained for human nestin and GS. Left panels are magnification of the respective framed regions. A: astrocytes, V: blood vessel. (h) 15N1-Gln enrichment in T407 GBM tumours and in contralateral brains, after a 4h intracarotid infusion with . The dashed lines correspond to the natural abundance of 15N1-Gln. Mean ± S.E.M. n=4 mice. p value refer to a two-tailed t test for paired samples.
Gln-starved GBM cells feed on astrocyte-derived Gln
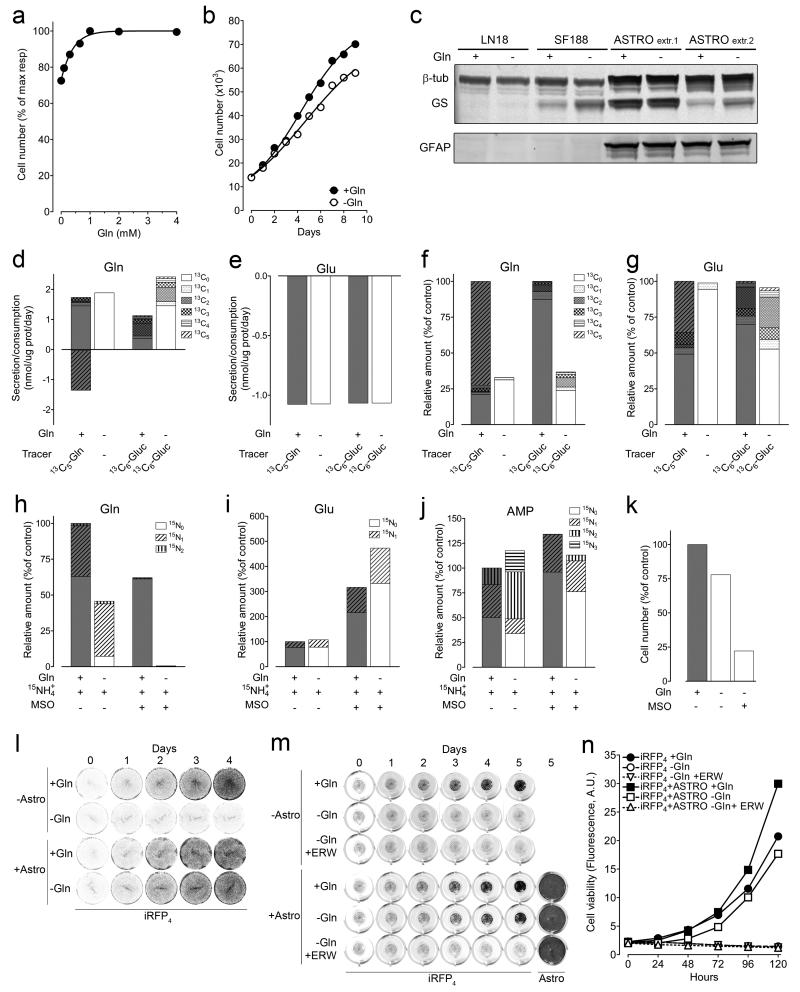
To assess this potential interaction, rat primary cortical astrocytes were cultured and their Gln requirement and metabolism investigated. Similar to GBM cells, the minimal Gln concentration required for maximal astrocyte growth was ~0.65 mM (Fig. 8a). Nonetheless, astrocyte proliferation was barely affected by Gln deprivation (Fig. 8b). As observed in the human TMA (Fig. 6a), GS protein levels in astrocytes and in the highest expressing GBM cells were comparable (Fig. 8c). However, only astrocytes demonstrated no net Gln consumption but rather, rapid Glu uptake (Fig. 8d-e), in line with the expression of Excitatory Amino Acids Transporters (EAAT) in this cell type34. Under Gln starvation, Glu consumption was unaffected and paralleled by an equimolar net Gln efflux (Fig. 8d-e). The absence of Gln in the medium reduced intracellular Gln, but not Glu (Fig. 8f-g). Moreover, 13C6-glucose tracing showed that only 30-40% of both intracellular Glu and Gln (Fig. 8f-g) were glucose-derived. Astrocytes maintained ~30% of the control level of intracellular Gln under Gln starvation (Fig. 8f and 8h), fitting with high GS expression. Gln maintenance depended on GS activity, as seen from both 15N1-ammonia tracing and GS inhibition by MSO (Fig. 8h). Moreover, GS inhibition dramatically elevated the intracellular amounts of its substrate, Glu (Fig. 8i), without changing the steady state levels of the Gln-product, AMP (Fig. 8j). However, combined Gln withdrawal and GS inhibition significantly reduced the labelled fraction of AMP derived from de novo synthesis (15N2 and 15N3; Fig. 8j), and hindered proliferation (Fig. 8k).
Figure 8.
Astrocytes feed with Gln GBM cells. (a, b) Astrocytes were incubated in SMEM for 6 days with 0, 0.1, 0.3, 0.65, 1, 2, 4mM Gln (a) or for the indicated times with 0 and 0.65mM Gln (b). (c) Astrocytes derived from two independent extractions, and cell lines, were incubated for 3 days +/− Gln and protein expression assessed. Unprocessed scans of western blots are shown in Supplementary Figure 8. (d-j) Astrocytes were incubated in SMEM for 24h in the presence of 5.56mM Glucose (13C6 or 13C0), 0.65mM Gln (13C5 or 13C0), 0.8mM 15NH4+, and 1mM MSO as indicated. Secretion/consumption rates (positive/negative bars respectively) are shown for Gln (d) and Glu (e). Intracellular levels of Gln (f) and Glu (g) isotopologues are reported as % of control (total of isotopologues in Gln fed conditions). The intracellular isotopologues of Gln (h), Glu (i), AMP (j) are shown as % of control (total isotopologues in the presence of Gln and 15NH4. (k) Astrocytes were incubated for 6 days +/− 0.65mM Gln, and 1mM MSO and counted. (l) Astrocytes were grown to confluence in multi-well plates. iRFP4 cells were seeded in wells +/− astrocytes, and +/− Gln. The fluorescence of iRFP4 cells in representative wells is shown. The experiment was performed twice with comparable results. (m) Astrocytes were grown to confluence in multi-well plates. iRFP4 cells seeded in transwell inserts were co-cultured +/− astrocytes, +/−Gln, and +/− Erwinase (Erw) as indicated. Fluorescence of iRFP4 cells in representative inserts is shown. At day 5 astrocytes were stained with sulphorodamine-B and the fluorescence of representative wells is shown. (n) Quantification of the iRFP4 fluorescence as described for (m). (a, b, d, e, f, g, h, i, j, k, n) Data derive from one experiment performed twice. Raw data of independent repeats are provided in the statistics source data Supplementary Table 5.
Finally, in co-culture, astrocytes enabled the proliferation of GS-negative LN18 iRFP4 cells without Gln supplementation (Fig. 8l). Moreover, transwell co-culturing of these cells showed that the factor conveying growth was diffusible (Fig. 8m-n). The addition of Erwinase, which depletes both Asn and Gln, prevented the rescue of Gln-deprived cells by astrocytes (Fig. 8m-n). Since Asn was present in the media during all Gln-starvation experiments, and since astrocytes consume Asn but produce and secrete Gln (Fig. 8d and Supplementary Fig. 6k), these results designate astrocyte-derived Gln as the growth-supporting factor for Gln-starved GBM cells.
DISCUSSION
Gln plays multiple metabolic and non-metabolic roles. Consequently, the dependency of cancer cells on Gln is difficult to discern. Nevertheless, it was demonstrated here that the Gln requirement in GBM goes beyond anaplerosis (Fig 2q-s). We identified two alternative metabolic determinants for Gln sensitivity: 1) Glu release via the antiporter and 2) GS-dependent conversion of Glu to Gln. GS and seemingly compete for cytoplasmic Glu which, during Gln starvation, becomes limiting (Fig. 2d,l). Nevertheless, the rescuing effect achieved by maintaining intracellular Glu through inhibition depends on GS activity (Fig. 3e, h, g and Fig. 4m-n). These results demonstrate that upon Gln starvation, Glu conversion to Gln constitutes a critically limiting reaction required for growth. This metabolic trait applies to established GBM lines, and to naïve primary cells. Indeed, when primary GBM cells were maintained in a stem-like state, GS expression was dramatically increased (Fig. 5a) and Glu was taken-up rather than released (Fig. 5d). Both responses enable growth of glioma stem-like cells independent of extracellular Gln (Fig. 5b). The fate of newly-synthesized Gln in both established and primary GBM cells was followed by 15N1-ammonia tracing, identifying AMP biosynthesis pathway as a significant player in Gln dependency (Fig. 4 d-j and Fig. 5h).
GS is found in the majority of human GBM (Fig. 6a-b), its expression associated with poor prognosis35 though this is not supported by our TMA study. However, we show that GS expression varies greatly between tumours, ranging from negative, comparable to GS expression in neurons, to high, as in normal astrocytes. This variation accords with a Gln-rich tumour microenvironment, which alleviates the need to synthesize Gln. Nevertheless, in agreement with previous reports36,37, it is shown here that most human GBM, as well as GS-proficient orthotopic GBM xenografts, withdraw carbons from the TCA cycle (cataplerosis) to synthesize Gln via GS. When compared to the liver, GBM are inclined towards net Gln synthesis, rather than glutaminolysis (Fig. 6i and Supplementary Fig. 6d). Accordingly, the circulation provides minimal amounts of Gln to normal brain38 and GBM (Fig 6l and Supplementary Fig. 6f, h, i). Moreover, a marked decrease in circulating Gln levels did not affect tumour growth (Fig 7a-h) and, GBM expressing low GS had levels of ammonia-derived Gln comparable to contralateral brain (Fig. 7j). The stability of Gln levels may explain the heterogeneity in GS expression between patients (Fig 6b) and within some tumours (Supplementary Fig. 5): GS-positive astrocytes and/or GBM cells (potentially GSC) excrete Gln that supports the growth of GS-negative GBM cells. Indeed, primary astrocytes in culture retain the metabolic traits of Glu uptake, GS-dependent Gln synthesis and Gln secretion, and support the proliferation of GS-negative, Gln-auxotrophic GBM cells (Fig 8l-n). Thus, this brain-to-tumour metabolic communication portrays a scenario in which Gln, provided by astrocytes, feeds GS-negative cancer cells (Fig. 3g). Indeed, GS-positive cellular protrusions of astrocytes surround GS-negative GBM cells in orthotopic xenografts and human tumours (Fig 7b, g and Supplementary Fig. 5p). This ‘parasitic’ behaviour of cancer cells could divert the physiological Gln-Glu cycle in the brain and, indeed, an increase in epileptic seizures has been reported for patients with GBM expressing low GS39.
METHODS
Cell cultures
The human glioblastoma cell lines MOG-G-UVW, LN-18, LN-229, SF-188, U-251 MG, U-87 MG (hereafter referred to as GUVW, LN18, LN229, SF188, U251, U87, respectively) were obtained from the following sources: GUVW: frozen stock originally isolated by Dr Ian Freshney at the University of Glasgow; LN18 and LN229: ATCC; SF188: Brain Tumour Tissue Bank University of California San Francisco; U251 and U87 were kindly given by Prof Kevin Ryan and Prof Anthony Chalmers, respectively. The cell lines were authenticated using Promega GenePrint 10 system (STR multiplex assay that amplifies 9 tetranucleotide repeat loci and the Amelogenin gender determining marker). No cell line used in the manuscript has been found in the ICLAC database of commonly misidentified cell lines. Cortical Astrocytes were generated from the cortex of P7 Sprague-Dawley rat pups (unclassified gender). Briefly both cortices were removed from each pup cleared of meninges and enzymatically dissociated and purified as previously described. Tissue from 2 pups was added to each 75cm2 tissue culture poly-L-Lysine coated flask (13μg/ml, Sigma). Astrocytes prepared in this way were over 95% pure as assessed by immunoreactivity to glial fibrillary acidic protein41.
All the cell lines tested negative for mycoplasma with MycoAlert™ when tested with Mycoplasma Detection Kit (Lonza). Cells were maintained in DMEM (Invitrogen 21969-035) supplemented with 2 mM Glutamine and 10% FBS. All the experiments were performed on cells seeded and incubated for at least 24h in a serum-like medium, SMEM, formulated as reported in Supplementary Table 1, and supplemented with 10% FBS, dialyzed against a semi-permeable cellulose membrane (3.5kDa MW cutoff).
For co-culture experiments 30,000 astrocytes /well were plated in a 24 well plate and allowed to grow to confluence, then LN18-derived clone stably expressing infrared fluorescent protein, iRFP4, was plated 25,000 cells/well or 5000 iRFP4 cells/transwell (Costar, CLS3413) were seeded in complete medium for direct or transwell co-culture experiments, respectively. The day after iRFP4 cell cultures were extensively washed with PBS, a transwell was inserted in wells containing astrocytes, and medium was replaced with the conditions indicated in Figure 8l, n. Half of the medium volume was renewed daily for the whole duration of the experiments. Growth was assessed by measuring fluorescence with a Licor Odyssey scanner and quantified Image Studio 2.0 software.
E2, R10 and R24 human glioblastoma cells were obtained and cultured as previously reported with minor modifications31. In brief differentiated (DIFF) cells were maintained in advanced DMEM/ F-12 (Gibco) supplemented with 10% FBS and 2mM L-Gln, while glioma stem-like cells (GSC) were maintained in advanced DMEM/F-12 supplemented with B27 (1%, Invitrogen), N2 (0.5%, Invitrogen), 4 μg/ml heparin, 20 ng/ml fibroblast growth factor 2 (bFGF, Sigma), 20 ng/ml epidermal growth factor (EGF, Sigma), 1mM pyruvate, and 2mM L-Glutamine. For all the experiments stem cells were seeded and incubated in SMEM supplemented as described above, while DIFF cells were seeded and incubated in advanced DMEM/F-12 supplemented with 10% dialyzed FBS. 0.65mM Glutamine was added as indicated in figures.
Cell number assessment
Cells were plated at a density of 20000 cells/well in 24 well plates. Cell number was assessed at the specified experimental time as follows: each well was washed twice with PBS, trypsinized, cells were re-suspended in a Casyton solution and counted with a Casy cell counter. The Doubling Time (DT) was obtained using the Exponential Growth equation with GraphPad prism 5.0. The % of growth inhibition was calculated according to the following equation:
Cell death assessment
Cells were plated at a density of 15000 cells/well in 12 well plates. After 2 days cells were placed in medium with or without glucose and Gln, and supplemented with CellTox™ Green Dye (1:1000, Promega, G8731). Cells were incubated in an IncuCyteTM FLR (EssenBioScience) for 72h, during which time bright field and fluorescence pictures were taken every 15 minutes. For each time point a cell death index was calculated by dividing the number of dead cells on the confluency index.
Cell cycle analysis
Culture media and cells were collected, centrifuged and washed with PBS. Cells were then fixed with cold methanol and stained with a PBS solution containing propidium idodide (100μg/ml) and RNAase A (250 μg/ml). DNA nuclear content was measured by using a Beckton Dickinson FACScan flow cytometer and data analyzed with the FlowJo software.
Tissue Microarray (TMA) and immunohistochemistry (IHC)
The collection of human biopsy tissue was approved by the regional ethical committee at Haukeland University Hospital, Bergen, Norway (REK 013.09). All patients gave a written informed consent for tumour biopsy collection and signed a declaration permitting the use of their biopsy specimens for research. Core tissue of glioblastoma cancer tissue (n=209) were identified by a pathologist, and collected from representative areas of tumours derived from patients’ surgical resection. Tissue cores were collected in triplicate for each patient. Follow-up details including date and cause of death were collected. For IHC analysis of the TMA, and mouse brain slices, the following procedure was followed. Epitope retrieval was performed at 98°C for 15’ in citrate buffer, pH 6. The immunostaining was performed using an EnVision System-HRP DAB (Dako, K4006). The cores were incubated over night with primary antibody. Cores were counterstained with haematoxylin, dehydrated and coverslipped. The staining was quantified using a weighted histoscore method with a final value range 0–300. The intensity of the immunostaining observed for normal astrocytes was classified as 300 and used as reference. Low- medium- and high-Glutamine Synthetase (GS) expressing groups were defined as reported in Figure 6b. For TMA and IHC the following antibodies were diluted in a solution of 5% BSA in TBS-tween and employed at the specified dilutions: purified mouse anti-human monoclonal GS antibody (BD transduction laboratories, Cat. 610518, clone 6 at a final concentration of 250ng/ml); mouse monoclonal anti-human nestin antibody (Abcam ab6320, clone 3k1, 1:500); mouse monoclonal anti-human EGFR antibody (Dako 7239 clone E30, 1:200, 1h at room temperature). For tumour volume quantification by IHC the mouse brain was paraffin embedded and entirely sliced into sections of 8μm. Two sections were loaded on each slide, and one slide in every 10 was stained for EGFR. Digital images were acquired with a Leica SCN400 Slide scanner, loaded into Matlab, calibrated and pixels positive for the staining were identified and used to quantify tumour volume. All IHC staining shown are representative of three or more sections.
Metabolic study in GBM patients
Before undergoing surgical tumour resection, seven GBM patients were infused for 15 minutes with 20 gr of 13C6-Glucose. During the operation tissue fragments from the core and peripheral region of the tumour were stereotactically sampled (~200 mg for each region). On average the specimens were snap-frozen 47 minutes after the end of the glucose injection (range 30-80 minutes). Peripheral blood samples were also collected at the same time of tissue sampling. Sub-fragments of frozen tissues (15-25 mg) were further dissected from peripheral and core regions of the tumour, and from edematous adjacent tissue. The samples were extracted on dry ice and processed as described below for human orthotopic GBM xenografts.
Informed consent was obtained from all human subjects involved in the study, the heavy isotope administration and the collection of tissue from surgical resections was approved by the regional ethical committee at Haukeland University Hospital, Bergen, Norway (REK 2010/130-2).
Primary orthotopic human GBM xenografts
Patient derived GBM xenografts were generated as previously reported42,43. Briefly, GBM patient-derived spheroids were stereotactically implanted into the right frontal cortex of 8-12 weeks old, male and female, NOD-SCID mice (Charles River, Lyon) using a Hamilton syringe (Hamilton, Reno, USA). After 4-6 weeks P3 GBM tumour was fully established in the brain, and U-13C6-glucose or U-13C5-glutamine (Cambridge Isotope Laboratories, Tewksbury, MA) were injected as a bolus in the tail vein at a dose of 1mg/gr and 0.15mg/gr of body weight respectively. 22 minutes after injection the animals were euthanized, organs were dissected, and tumour and contralateral brain fragments were snap frozen. For intracarotid infusion the mice were kept under anesthesia with ketamine medetomidine isoflurane and underwent surgical procedure to expose the right carotid artery. 4.7μmoles/Kg/min of 15N1 ammonia was infused 0.833μl/min for 4h in NOD SCID mice (Charles River, Lyon) xenografted orthotopically with T407 GBM. 13μmoles/kg/min of 13C5-glutamine were infused 0.125μl/min for 4h in 8-weeks old, female, Swiss Nu/Nu mice (Charles River, Lyon) xenografted orthotopically with the GS-positive T16 GBM. To assess the effects of Erwinase, 8-weeks old, female, Swiss Nu/Nu mice (Charles River, Lyon) were implanted with T101 tumour spheroids (six/mouse). 42 days after implantation mice were randomly assigned to Erwinase treatment or control group. Erwinase was injected intraperitoneal 5U/gr of body weight, five times / week. The treatment continued for 6 weeks. Mice of the control group were intraperitoneally injected with saline solution. Mice were euthanized six hours after the last injection and tumour and contralateral brain fragments were snap frozen for metabolic analysis. Whole brains were immediately fixed in 4% paraformaldehyde for ex vivo MRI. For metabolic analysis, 10-20mg of tissue were extracted with a beads mill (Qiagen, Tissuelyzer) in a solution containing 20% water, 50% Methanol, 30% Acetonitrile (25μl of extraction solution/mg tissue). The extracts were spun down at 16000g for 10 min and supernatants stored −80°C for LC-MS analysis. Human glioblastoma biopsies were obtained from the Neurosurgery Department of the Centre Hospitalier in Luxembourg (CHL). All patients had provided written informed consent, with procedures that were approved for the project (project number: REC-LRNO-20110708) by the National Research Ethics Committee for Luxembourg (CNER). The handling of the animals and the surgical procedures were performed in accordance with the European Directive on animal experimentation (2010/63/EU) and the national regulations of Luxembourg and the local ethical committee (the Animal Welfare Structure (AWS) LIH) approved the protocol (protocol number: LRNO-2014-06).
MRI analysis
At the end of Erwinase treatment the brains of the 20-weeks old Swiss Nu/Nu mice with T101 tumours (7 female mice/group) were harvested and fixed in 4% paraformaldehyde. Ex vivo scans were acquired on a 7T micro-MRI system (Bruker PharmaScan) using a mouse brain volume coil. The T2-weighted MRI protocol used a Fast Spin Echo sequence with TE = 36 ms, TR = 4300 ms, and 63 μm in plane resolution. The Diffusion Weighted Imaging (DWI) sequence used EPI readout with TE=28.5ms, TR=7000ms and b-values ranging from 500 to 6000 s/mm2 in 3 directions. The tumours were independently delineated by two users (A.O. and O.K.) using Paravision 5.0 (Bruker; Ettlingen, Germany).
Immunoblotting
Cells were washed twice with cold PBS and lysed in RIPA buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1mM EGTA, 1% Triton, 2,5mM sodium pyrophosphate, 1mM glycerophosphate) supplemented with 1mM Na3VO4, 1mM NaF and protein inhibitor cocktail. Protein concentration was determined with the bicinchoninic acid assay (Thermo scientific) using BSA as a standard. Equal amounts of protein were mixed with Laemmli buffer 4× (250 mM Tris-HCl, pH 6.8, 8% SDS, 40% glycerol, 0.4 M DTT), warmed at 95°C for 5 min, and loaded on 10% gels for SDS-PAGE. After electrophoretic separation, proteins were blotted onto 0.22 mm nitrocellulose membrane (Millipore), blocked with 5% non-fat milk in TBS-Tween, and incubated at 4°C overnight with the following antibodies: Glutamine Synthetase (BD Transduction Labs, 610517, clone 6, 1:1000), β-tubulin (Sigma Aldrich, T5201, clone AA2, 1:5000), GLS (Abcam, ab60709, 1:1000), c-Myc (Cell Signalling Technology, 5605, clone D84C12, 1:1000) Glial Fibrillary Acidic Protein (Dako, Z0334, 1:10000), SOX2 (Abcam, ab75485, clone 57CT23.3.4, 1:1000), CD133 (Miltenyl Biotec, W6B3C1, clone W6B3C1, 1:1000), Olig2 (R&D system, AF2418, 1:500), Actin (Santa Cruz Biotechnology, sc-1616-R, 1:1000). Membranes were then washed and incubated with secondary donkey anti-rabbit (Licor 926 32213, 1:10000) or donkey anti-mouse (Licor 926 32212, 1:10000). The IR scanning was performed using a Licor Odyssey scanner and acquired using Image Studio 2.0. All western blots shown are representative of two experiments unless otherwise indicated.
qPCR analysis
For qPCR analysis 1 μg of total RNA was isolated from cells with RNeasy Mini Kit (Qiagen) and retro-transcribed into cDNA using M-Mulv Reverse Transcriptase and random hexamers. qPCR reactions were performed using a Fast Sybr Green Master Mix (Applied Biosystems), 0.5 mM primers, ROX dye, and a 25ng of cDNA in a final volume of 20 μl according to the manufacturer’s instructions. The following primers were used MYC_F CACCAGCAGCGACTCTGA, MYC_R GATCCAGACTCTGACCTTTTGC; GS_F TCATCTTGCATCGTGTGTGTG, GS_R CTTCAGACCATTCTCCTCCCG; GLS_F GAAATTCGGAACAAGACTGTG, GLS_R AACTTCGATGTGTCCTTCAC β-actin_F TCCATCATGAAGTGTGACGT, β-actin_R TACTCCTGCTTGCTGATCCAC. The following PCR steps were performed on the 7500 Fast Real-Time PCR System (Life Technologies Corporation), 15 min at 95°C followed by 40 cycles of 10 s at 94°C, 30 s at 55°C, 30 s at 72°C. A 10 min final step at 72°C was performed to visualize the PCR products by means of the melting curves. The expression levels of the indicated genes were calculated using the ΔΔCT method using actin for normalization.
Metabolites Extraction and HPLC-MS
The established cell lines were plated in 6 well plates in complete DMEM from 125000 to 250000 cells / well. After 24h the medium was replaced with SMEM supplemented with 10% dialyzed FBS. Stem-like and differentiated primary glioma stem cells were plated at 150000 cells / well in 6 well plates in SMEM supplemented as described in the cell culture section. When optimal confluence was reached (1-3 days) 3 wells were extracted as described below, and the protein determined (μg prott0). The media in the remaining wells was changed with 2 or 7 ml/well of complete SMEM for the analysis of metabolites present in the medium or in the cells, respectively. U-13C5-Gln (0.65 mM, Cambridge Isotopes), U-13C6-Glucose (5.56mM, Cambridge Isotopes), or 15N1-Ammonia (0.8mM Sigma) were supplemented as indicated in figures. Cells, as well as parallel cell-free wells, were incubated for 24h at 37°C. At the end of the incubation, an aliquot of the media was diluted in a solution containing 20% water, 50% Methanol, and 30% Acetonitrile. The medium derived from wells devoid of cells, was used to estimate the exchange rates of metabolites. For the profiling of intracellular metabolites, monolayers were rapidly washed 3 times with ice-cold PBS and extracted with an aqueous solution of 50% Methanol, and 30% Acetonitrile. Both media and cell extracts were centrifuged 16000g for 10min at 4°C and the supernatants were analyzed by means of HPLC-MS. The extracted cell monolayer was used for protein determination (μg prott24) with a Lowry assay. Aliquots of freshly prepared SMEM, without FBS, and spiked with known concentrations of lactate were processed in parallel and used as a reference for metabolite quantification. The secretion/consumption rate for a specific metabolite (x) was obtained according to the equation where Δ metabolite = [(x)nmol medium with no cell – (x)nmol medium with cells]. For the LC separation of medium samples a ZIC-HILIC (SeQuant) with a guard column (Hichrom) was used. The mobile phase A was a 0.1% formic acid solution in water and mobile phase B was 0.1% formic acid in acetonitrile. The flow rate of mobile phases was kept at 100 μl/min and gradient was as follows: 0 minutes 80% of B, 12 minutes 50% of B, 26 minutes 50% of B, 28 minutes 20% of B, 36 minutes 20% of B and 37 37 to 45 minutes 80% of B. For the separation, cell extracts were injected on a ZIC-pHILIC column with a guard column. Mobile phase C: 20 mM ammonia carbonate plus 0.1% ammonia hydroxide in water. Mobile phase D: acetonitrile. The flow rate was kept at 100 μL/min and gradient as follows: 0 minutes 80% of D, 30 minutes 20% of D, 31 minutes 80% of D and 45 minutes 80% of D. The Exactive Orbitrap mass spectrometer (Thermo Scientific, Waltham, MA, USA) was operated in a polarity switching mode.
Cell transfection and infection
For stable transfection 10 cm dishes of 80% confluent LN18 cells were incubated for 9h with 10 ml of FBS-free transfection medium containing 1μg/ml of DNA (Vector containing P2A-iRFP IRES puro only or GS-P2A-iRFP IRES puro and 2μl/ml of Lipofectamine2000 (Invitrogen). The transfection mixture was than replaced with 10ml of complete DMEM. The following day medium was changed and cells were cultured for 3 weeks in selection medium containing 2μg/ml of puromycin. Fluorescent colonies were visualized with the Licor Odyssey scanner, picked and amplified in selection medium to obtain stable cultures. For the establishment of cell lines expressing shRNA against Glutamine Synthetase (GS) four human unique 29mer shRNA constructs and scrambled negative control non effective shRNA cassette (NTC) in lentiviral GFP vector were employed according to the manufacturer’s instructions (TL312740a: GGTGAGAAAGTCCAGGCCATGTATATCTG; TL312740b: GAATGGTGCAGGCTGCCATACCAACTTCA; TL312740c: TGGTACTGGAGAAGGACTGCGCTGCAAGA; TL312740d: GGCACACCTGTAAACGGATAATGGACATG; TR30021ntc: GCACTACCAGAGCTAACTCAGATAGTACT, Origene). TL312740b TL312740d and TR30021ntc are reported in the manuscript as shGS-1 shGS-2 and shNTC respectively. Briefly, lentiviruses were produced by co-transfecting HEK293T cells with each shRNA plasmid, lentiviral plasmid psPAX2-1, and packaging plasmid pVSV-G-1, according to a standard calcium phosphate procedure. Supernatants containing virus were collected, filtered and frozen in stocks at −80°C. For infection of recipient cells the supernatant was thawed, mixed with Hexadimethrine bromide (Sigma, H9268, 8μg/ml) and incubated for 24 hours with the cells. Infected cells were selected for 2 weeks in medium containing 1-3μg/ml puromycin.
Colony forming assay
iRFP and iRFP-GS expressing cells were plated at 300cells/well in 6 wells plates in complete SMEM, in the absence or presence of 0.65mM Gln and 1mM MSO as indicated. After 3 weeks, colonies were fixed with trichloroacetic acid, stained with SRB and plates were scanned using a Licor Odyssey scanner. Images were acquired using Image Studio 2.0. SF188 and U251 cells expressing shNTC and shGS sequences were plated at 500 and 350 cells/well in 6 wells, respectively. Cells were incubated in complete SMEM and supplemented as indicated in Figure 3 legend. 12-17 days after seeding colonies were fixed, stained, and representative images obtained as described above. For the quantification of colonies surface area an ImageJ macro was designed and run. The values obtained from the macro were normalized for the exact number of days cells were incubated, and presented as % of relative control.
Reagents
U-13C5-L-Glutamine and U-13C6-D-Glucose (Cambridge Isotopes Laboratories, Inc CLM-1822, CLM-1396 respectively), 15N1 Ammonium Cloride (Sigma Aldrich 299251), sulfasalazine (Sigma Aldrich S0883), MSO (Sigma Aldrich M5379), compound 968 (Calbiochem 352010, dissolved 10mM in DMSO), BPTES (Sigma Aldrich SML0601, dissolved 10mM in DMSO) Erwinase (Eusa Pharma, an international division of Jazz Phamaceuticals), all L-amino acids, nucleosides, and nitrogenous bases utilized in the study were obtained from Sigma Aldrich.
Genome-scale constraint-based metabolic modeling
A Detailed description of metabolic modelling methods and results is provided in the Supplementary note.
Statistical Methods
No statistical method was used to predetermine sample size. For the animal studies, the experiments were not randomized, and the investigators were not blinded to allocation during experiments. The number of independent experiments performed is reported in figure legend. For experiments performed once or twice the raw data of independent repeats are provided in the statistics source data. Error bars represent standard error of the mean (S.E.M.). Two-tailed Student’s t, Pearson correlation test, and Log-rank (Mantel-Cox) test were performed with Graph Pad Prism 5.0.
Supplementary Material
ACKNOWLEDGMENTS
This study has been supported by Cancer Research UK. S.T. is recipient of an AIRC/Marie Curie International Fellowship for Cancer Research. The human and animal metabolomic studies were supported by The Norwegian Cancer Society, The Norwegian Research Council, Helse Vest, Haukeland University Hospital and the K.G-Jebsen Foundation. We acknowledge Anna Golebiewska, Virginie Baus-Talko, Niels Van Den Broek, Gillian MacKay, Colin Nixon, Elaine MacKenzie, for excellent technical assistance and Ayala King for excellent editorial work.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Supplementary information is available
REFERENCES
- 1.Moreadith RW, Lehninger AL. The pathways of glutamate and glutamine oxidation by tumor cell mitochondria. Role of mitochondrial NAD(P)+-dependent malic enzyme. The Journal of biological chemistry. 1984;259:6215–6221. [PubMed] [Google Scholar]
- 2.Yuneva MO, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell metabolism. 2012;15:157–170. doi: 10.1016/j.cmet.2011.12.015. doi:10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wise DR, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. doi:10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeBerardinis RJ, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. doi:10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tardito S, et al. L-Asparaginase and inhibitors of glutamine synthetase disclose glutamine addiction of beta-catenin-mutated human hepatocellular carcinoma cells. Current cancer drug targets. 2011;11:929–943. doi: 10.2174/156800911797264725. [DOI] [PubMed] [Google Scholar]
- 6.Son J, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. doi:10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gameiro PA, et al. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell metabolism. 2013;17:372–385. doi: 10.1016/j.cmet.2013.02.002. doi:10.1016/j.cmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willems L, et al. Inhibiting glutamine uptake represents an attractive new strategy for treating acute myeloid leukemia. Blood. 2013 doi: 10.1182/blood-2013-03-493163. doi:10.1182/blood-2013-03-493163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu M, et al. Glutamine depletion by crisantaspase hinders the growth of human hepatocellular carcinoma xenografts. British journal of cancer. 2014;111:1159–1167. doi: 10.1038/bjc.2014.425. doi:10.1038/bjc.2014.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang JB, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. doi:10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki S, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A. 2010;107:7461–7466. doi: 10.1073/pnas.1002459107. doi:10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dal Bello B, et al. Glutamine synthetase immunostaining correlates with pathologic features of hepatocellular carcinoma and better survival after radiofrequency thermal ablation. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16:2157–2166. doi: 10.1158/1078-0432.CCR-09-1978. doi:10.1158/1078-0432.CCR-09-1978. [DOI] [PubMed] [Google Scholar]
- 13.Krebs HA. Metabolism of amino-acids: The synthesis of glutamine from glutamic acid and ammonia, and the enzymic hydrolysis of glutamine in animal tissues. The Biochemical journal. 1935;29:1951–1969. doi: 10.1042/bj0291951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van den Berg CJ, Garfinkel D. A stimulation study of brain compartments. Metabolism of glutamate and related substances in mouse brain. The Biochemical journal. 1971;123:211–218. doi: 10.1042/bj1230211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takano T, et al. Glutamate release promotes growth of malignant gliomas. Nature medicine. 2001;7:1010–1015. doi: 10.1038/nm0901-1010. doi:10.1038/nm0901-1010. [DOI] [PubMed] [Google Scholar]
- 16.Scott DA, et al. Comparative metabolic flux profiling of melanoma cell lines: beyond the Warburg effect. The Journal of biological chemistry. 2011;286:42626–42634. doi: 10.1074/jbc.M111.282046. doi:10.1074/jbc.M111.282046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. doi:10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamphorst JJ, et al. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci U S A. 2013;110:8882–8887. doi: 10.1073/pnas.1307237110. doi:10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan J, Kamphorst JJ, Rabinowitz JD, Shlomi T. Fatty acid labeling from glutamine in hypoxia can be explained by isotope exchange without net reductive isocitrate dehydrogenase (IDH) flux. The Journal of biological chemistry. 2013;288:31363–31369. doi: 10.1074/jbc.M113.502740. doi:10.1074/jbc.M113.502740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mullen AR, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. doi:10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewerenz J, et al. The Cystine/Glutamate Antiporter System x(c)(−) in Health and Disease: From Molecular Mechanisms to Novel Therapeutic Opportunities. Antioxidants & Redox Signaling. 2013;18:522–555. doi: 10.1089/ars.2011.4391. doi:DOI 10.1089/ars.2011.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duarte NC, et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci U S A. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. doi:10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–U100. doi: 10.1038/nature07823. doi:Doi 10.1038/Nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. Journal of Cell Biology. 2007;178:93–105. doi: 10.1083/jcb.200703099. doi:DOI 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bax DA, et al. Molecular and phenotypic characterisation of paediatric glioma cell lines as models for preclinical drug development. PloS one. 2009;4:e5209. doi: 10.1371/journal.pone.0005209. doi:10.1371/journal.pone.0005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filonov GS, et al. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nature biotechnology. 2011;29:757–761. doi: 10.1038/nbt.1918. doi:10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hock AK, et al. iRFP is a sensitive marker for cell number and tumor growth in high-throughput systems. Cell cycle. 2013;13 doi: 10.4161/cc.26985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin Y, et al. Glutamine synthetase functions as a negative growth regulator in glioma. Journal of Neuro-Oncology. 2013;114:59–69. doi: 10.1007/s11060-013-1168-5. doi:DOI 10.1007/s11060-013-1168-5. [DOI] [PubMed] [Google Scholar]
- 29.Reznik E, Mehta P, Segre D. Flux imbalance analysis and the sensitivity of cellular growth to changes in metabolite pools. PLoS computational biology. 2013;9:e1003195. doi: 10.1371/journal.pcbi.1003195. doi:10.1371/journal.pcbi.1003195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calise SJ, et al. Glutamine deprivation initiates reversible assembly of mammalian rods and rings. Cellular and molecular life sciences: CMLS. 2014;71:2963–2973. doi: 10.1007/s00018-014-1567-6. doi:10.1007/s00018-014-1567-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carruthers R, et al. Abrogation of radioresistance in glioblastoma stem-like cells by inhibition of ATM kinase. Molecular oncology. 2015;9:192–203. doi: 10.1016/j.molonc.2014.08.003. doi:10.1016/j.molonc.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyerand ME, Pipas JM, Mamourian A, Tosteson TD, Dunn JF. Classification of biopsy-confirmed brain tumors using single-voxel MR spectroscopy. AJNR. American journal of neuroradiology. 1999;20:117–123. [PubMed] [Google Scholar]
- 33.Parmentier JH, et al. Glutaminase activity determines cytotoxicity of L-asparaginases on most leukemia cell lines. Leukemia Research. 2015;39:757–762. doi: 10.1016/j.leukres.2015.04.008. doi:DOI 10.1016/j.leukres.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Had-Aissouni L. Toward a new role for plasma membrane sodium-dependent glutamate transporters of astrocytes: maintenance of antioxidant defenses beyond extracellular glutamate clearance. Amino acids. 2012;42:181–197. doi: 10.1007/s00726-011-0863-9. doi:10.1007/s00726-011-0863-9. [DOI] [PubMed] [Google Scholar]
- 35.Rosati A, et al. Glutamine synthetase expression as a valuable marker of epilepsy and longer survival in newly diagnosed glioblastoma multiforme. Neurooncology. 2013;15:618–625. doi: 10.1093/neuonc/nos338. doi:10.1093/neuonc/nos338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marin-Valencia I, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell metabolism. 2012;15:827–837. doi: 10.1016/j.cmet.2012.05.001. doi:10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maher EA, et al. Metabolism of [U-13 C]glucose in human brain tumors in vivo. NMR in biomedicine. 2012;25:1234–1244. doi: 10.1002/nbm.2794. doi:10.1002/nbm.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bagga P, et al. Characterization of cerebral glutamine uptake from blood in the mouse brain: implications for metabolic modeling of 13C NMR data. Journal of cerebral blood flow and metabolism: official journal of the International Society of Cerebral Blood Flow and Metabolism. 2014;34:1666–1672. doi: 10.1038/jcbfm.2014.129. doi:10.1038/jcbfm.2014.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosati A, et al. Epilepsy in glioblastoma multiforme: correlation with glutamine synthetase levels. Journal of Neuro-Oncology. 2009;93:319–324. doi: 10.1007/s11060-008-9794-z. doi:10.1007/s11060-008-9794-z. [DOI] [PubMed] [Google Scholar]
- 40.Bannai S, Ishii T. A novel function of glutamine in cell culture: utilization of glutamine for the uptake of cystine in human fibroblasts. Journal of cellular physiology. 1988;137:360–366. doi: 10.1002/jcp.1041370221. doi:10.1002/jcp.1041370221. [DOI] [PubMed] [Google Scholar]
- 41.Noble M, Murray K. Purified astrocytes promote the in vitro division of a bipotential glial progenitor cell. Embo J. 1984;3:2243–2247. doi: 10.1002/j.1460-2075.1984.tb02122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keunen O, et al. Anti-VEGF treatment reduces blood supply and increases tumor cell invasion in glioblastoma. Proc Natl Acad Sci U S A. 2011;108:3749–3754. doi: 10.1073/pnas.1014480108. doi:10.1073/pnas.1014480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanzey M, et al. Comprehensive analysis of glycolytic enzymes as therapeutic targets in the treatment of glioblastoma. PloS one. 2015;10:e0123544. doi: 10.1371/journal.pone.0123544. doi:10.1371/journal.pone.0123544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.