Abstract
Dengue has emerged to be global health problem worldwide. Hence there is an immediate need to adopt new strategies in the development of effective anti-dengue drugs. Extracts from the leaves of Azadirachta indica has been traditionally used in folk medicine for viral infections. In the present study we report the anti-viral potency of nimbin, the active compound from the neem leaf extract against the envelope protein of dengue virus. Progression of viral entry into the host cell is facilitated by the envelope protein of dengue virus, suggesting; it as an effective anti-viral target. Nimbin is found to be effective against the envelope protein of all four types of dengue virus (dengue 1–4), which is evident from our in silico analysis. Our findings suggest the clinical importance of nimbin, which can serve as effective lead compound for further analysis.
Electronic supplementary material
The online version of this article (doi:10.1007/s13337-015-0280-x) contains supplementary material, which is available to authorized users.
Keywords: Dengue, Azadirachta indica, Nimbin, Envelope, ADMET
Introduction
In accordance with World Health Organization (WHO) report, more than 50 million people are falling prey to dengue virus every year [18]. Dengue virus belongs to the family; where the mode of transmission is mainly via arthropod vectors [17]. The virus responsible for classical dengue fever, dengue shock syndrome (DSS) and dengue hemorrhagic fever (DHF) is divided into four classes based on the degree of sequence identity [20]. Classical dengue can leads to fatal conditions such as DSS and DHF when subjected to secondary infection by other serotypes. Primary infection is characterized by modest indications like fever associated with joint and muscle pain, whereas secondary infection is characterized by skin hemorrhages, damage to blood and lymph vessels leads to increase in fatal outcome rate. Dengue virus infection is considered to be fatal to the individuals with respiratory problems and other chronic diseases [13]. Envelope of dengue virus facilitates the adherence into host tissues and also responsible for the antigenic property of virus, hence considered as effective target for the development of anti-viral drugs [56]. Previous studies revealing the anti-viral properties of natural products disclose the importance of plants extracts against deadly viruses [7, 27, 52]. Azadirachta indica, a medicinal plant found to posses anti-viral property against dengue virus, small pox virus and polio virus as reported by several investigators [39, 44, 51]. At present plants are the indirect or direct sources of approximately 50 % of approved drugs [26]. Since plant derived compounds are of considerably smaller molecules and contain thousands of naturally occurring components, human body can quickly absorb and utilize these compounds. Plants have been traditional source of active substances for most therapies [3]. As per the report published by annual reports of medicinal chemistry, seven out of ten synthetic agents approved by FDA are modeled on a natural product [48]. Neem plant has been widely used as a traditional medicine for many centuries in tropical countries. Several researchers have stressed the anti-viral properties of neem against dengue virus, polio virus, HIV, coxsackie B group virus [39, 43, 44, 45, 47, 54]. In addition the virucidal activity of neem extract is proved against coxsackie B group virus in the early event of its replication cycle. Similarly experimental evidences by Parida et al. [39] proves the inhibitory potential of neem extract at the replication step of dengue virus. Recently, in vitro anti-viral activity of neem bark extract is proved against herpes simplex virus type-1 infection [53]. These evidences strongly support the significance of natural products in the development of anti-viral agents. In the present study, we report the anti-viral potential of neem leaf extracts against dengue virus envelope protein as confirmed by docking analysis and absorption, distribution, metabolism, excretion and toxicity (ADMET) studies.
Materials and methods
Active compounds
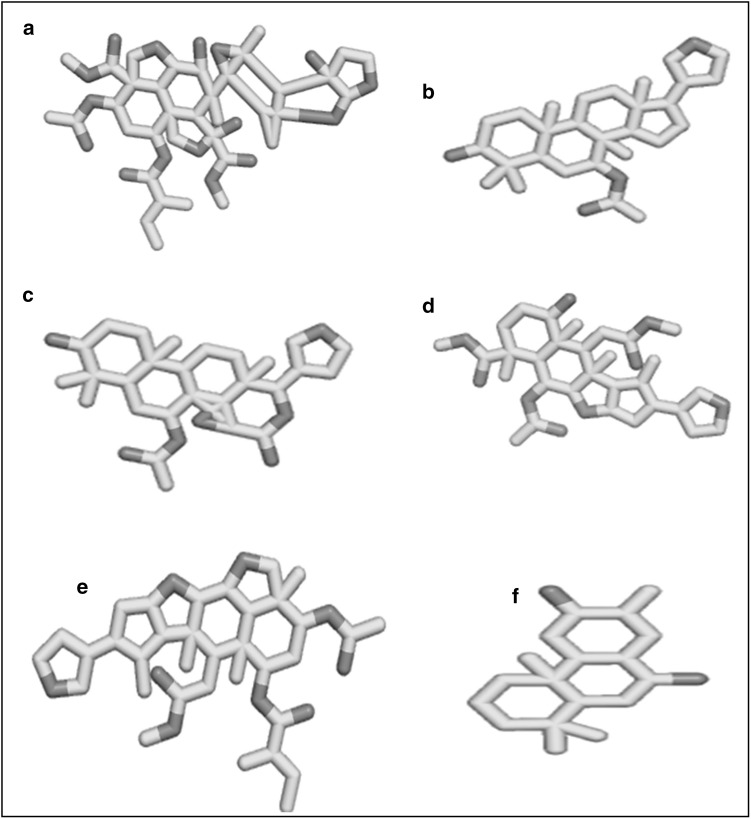
Based on literature reports [34, 42] the 3D structures of Nimbin, Ninbidol, Gedunin, Salannin, Azadirachtin and Azadirone from the leaves of Azadirachta indica, were retrieved from PubChem database and have been shown in Fig. 1.
Fig. 1.
3D structure of active compounds from the leaves of Azadirachta indica. a Azadirachtin. b Azadirone. c Gedunin. d Nimbin. e Salannin and f Ninbidol
Receptor
Dengue virus envelope plays a critical role in the dengue virus fusion with host cell membrane and thus can be considered as potential anti-dengue target. Structure of envelope protein for all the four serotypes were retrieved by Protein Data Bank (PDB) [4]. The PDB ID’s of chosen structures are shown below.
Molecular docking studies
Docking analysis was performed with Surflex-dock program incorporated in SYBYL 2.0 [23]. The surflex dock scoring function is based on Hammerhead docking system [28] and it effectively neglects the false positive results, thus helps in screening of the active compounds effectively. The Structure obtained from PDB is optimized by removing the co crystallized structures, crystallographic water molecules and other unrelated structures. Using the force field AMBER 7 FF99, the structure is subjected to brief minimization after the addition of hydrogen atoms. Surflex-Dock protomol is referred to as ‘binding pocket’, in which the active compounds are aligned. Using empirically derived scoring functions, the binding affinity of ligand and receptor is calculated. To evaluate the binding affinity of ligand-receptor interactions Consensus scoring (CScore) function were used. CScore combine the use of several scoring functions to obtain the efficient results and to avoid the disadvantages of other scoring functions. CScore is in the combination of Gold score (GScore), Dock score (DScore), Potential Mean force score (PMF score), polar score and crash score. GScore [33] and DScore [12] were used to calculate energy of hydrogen bond interaction and hydrophobic and electrostatic interactions respectively. Also the energy from lipophilic interactions, hydrogen bonding interactions was calculated using Chemscore [36]. To observe the interaction between ligand and receptor, Molecular Computer Aided Design (MOLCAD), a visualization tool is used.
Calculation of molecular properties
The tools Molinspiration and admetSAR were used to calculate the molecular properties of the active compound [9, 50]. ADMET properties include blood–brain barrier (BBB), Human intestinal absorption (HIA), Caco-2 Permeability, p-glycoprotein inhibitor, Renal organic cation transporter, CYP450 (Cytochrome P450) 2C9 Substrate, CYP450 2D6 Substrate, CYP450 3A4 Substrate, CYP450 1A2 Inhibitor, CYP450 2C9 Inhibitor, CYP450 2D6 Inhibitor, CYP450 2C19 Inhibitor, CYP450 3A4 Inhibitor and AMES Toxicity. Further, the compound nimbin is tested for their cardiotoxicity risks using the tool LabMol Pred-hERG 2.0 [5, 6].
Results
Molecular docking studies
The envelope of DENV serve as an important anti-viral target, hence docking analysis is performed on a series of active compounds against the envelope of all four DENV. In order to identify the efficiency of active compounds, our results were also compared with the reference ligand Panduratin [14]. Table 1 shows the energy of binding affinity of all the compounds against DENV-1 virus. From the results we observe that compound Nimbin shows maximum binding affinity than other compounds. Even though Azadirone and Azadirachtin form three hydrogen bonds with the active site residues, they show poor results in protein-ligand atom pair interactions. CScore of Gedunin is in the same range of Nimbin but the number of hydrogen bond interaction formed is comparatively less. Since the hydrogen bond formation makes an important contribution in protein-ligand interactions, the number of hydrogen bonds formed between the active compounds and the receptors are also listed in Table 1. Docked structure of Nimbin in the active site of all four serotypes is shown in Fig. 2. Further the hydrogen bonding interaction between the compound Nimbin and the active site residues of DENV-1 are depicted in Fig. 3. On analyzing the results, we observe that the compound Nimbin and Azadirachtin shows highest binding affinity with a maximum of four hydrogen bonds. Table 2 shows the energy of binding affinity of active compounds towards the envelope of DENV-2 virus. Pictorial representation of hydrogen bonding interaction between Nimbin and the active site residues of DENV-2 are shown in Fig. 4. Even though azadirone and azadirachtin shows equal polar interaction, the number of hydrogen bonds and CScore is comparatively low than nimbin. Since hydrogen bond act as an important contributor to free energies of biological macromolecules and macromolecular complexes, it plays an important role in determining the binding affinity of ligands. Considering the energetic contribution through van der Waals interactions the compound Gedunin shows the maximum score, but the number of hydrogen bonds formed between Gedunin and the active site residues are less comparing to Nimbin and Azadirone. Finally Salannin and Ninbidol shows the least score among the six compounds analysed indicating the poor binding affinity between the ligand and the DENV-2 virus envelope. Table 3 shows the binding affinity of active compounds and the DENV-3 virus envelope. Figure 5 represents hydrogen bonding interaction between the compound Nimbin and the active site residues of DENV-3. From the results we find that the compound Nimbin and Gedunin dominates with a maximum of three hydrogen bonds. Even though Azadirone shows maximum hydrogen bond interaction, the total score is comparatively less than Nimbin and Gedunin. Salannin and Ninbidol shows poor values in all seven parameters analysed. Results of binding pattern of DENV-4 virus are depicted in Table 4 and the hydrogen bond interactions are shown in Fig. 6. It is noteworthy to mention that Nimbin shows maximum binding affinity with DENV-4, similar to DENV-1 DENV-2 and DENV-3, whereas Salannin shows poor energy values in all the parameter analyzed. It is also worth mentioning that nimbin shows high binding affinity comparing to panduratin in all the four complexes analyzed. These results reveal the significance of nimbin as a potent anti-viral compound.
Table 1.
Binding affinity of active compounds towards the envelope of DENV-1 virus
| S. No | Compounds | CScorea | Crash scoreb | Polar scorec | G scored | PMF scoree | D scoref | Chem scoreg | No. of hydrogen bonds |
|---|---|---|---|---|---|---|---|---|---|
| 1. | Nimbin | 3.68 | −1.14 | 2.33 | −100.07 | −72.670 | −149.12 | −17.988 | 3 |
| 2. | Gedunin | 3.46 | −1.63 | 1.19 | −110.53 | −41.870 | −195.83 | −13.662 | 2 |
| 3. | Azadirone | 2.90 | −0.95 | 0.21 | −78.466 | −38.585 | −163.55 | −12.366 | 3 |
| 4. | Salannin | 1.95 | −1.11 | 1.22 | −133.36 | −69.767 | −177.73 | −21.510 | 2 |
| 5. | Ninbidol | 1.72 | −1.09 | 1.78 | −344.08 | −97.851 | −150.06 | −22.381 | 1 |
| 6. | Azadirachtin | 3.33 | −1.75 | 2.19 | −115.23 | −40.890 | −156.32 | −14.335 | 3 |
| 7. | Panduratin* | 2.33 | −1.15 | 1.35 | −65.92 | −27.512 | −158.31 | −11.266 | 1 |
* Reference ligand
aCScore is a consensus scoring which uses multiple types of scoring functions to rank the affinity of ligands
bCrash-score revealing the inappropriate penetration into the binding site
CPolar region of the ligand
dG-score showing hydrogen bonding, complex (ligand–protein), and internal (ligand–ligand) energies
ePMF-score indicating the Helmholtz free energies of interactions for protein–ligand atom pairs (Potential of Mean Force, PMF)
fD-score for charge and van der Waals interactions between the protein and the ligand
gChem-score points for hydrogen bonding, lipophilic contact, and rotational entropy, along with an intercept term
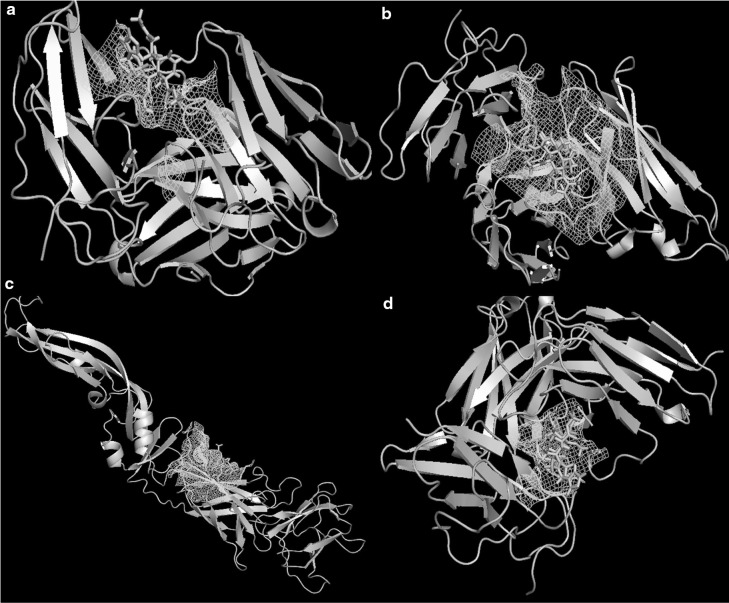
Fig. 2.
Docked structure of the compound Nimbin in the active site of a DENV-1. b DENV-2. c DENV-3 and d DENV-4. Active site of the receptor is shown in mesh representations
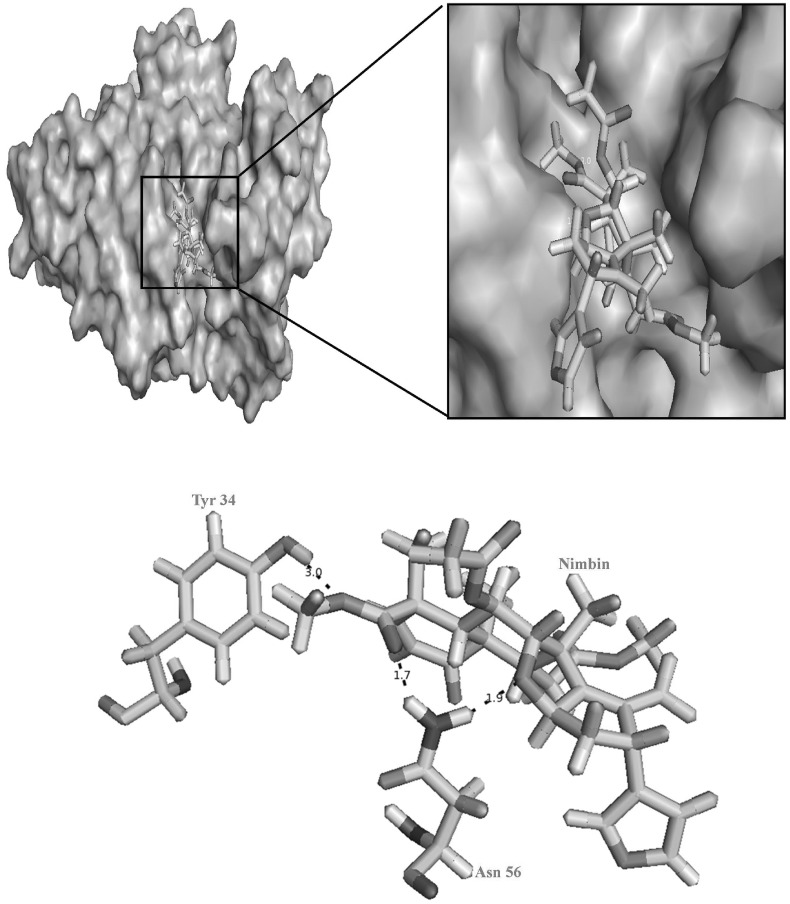
Fig. 3.
Pictorial representation hydrogen bond interactions between nimbin and the active site residues of DENV-1. Dotted lines represents the hydrogen bonds interactions between H atom of Tyr 34 with O atom of nimbin, HD2 atom of Asn 56 with O atom of nimbin and H atom of Asn 56 with O atom of nimbin
Table 2.
Binding affinity of active compounds towards the envelope of DENV-2 virus
| S. No | Compounds | CScorea | Crash scoreb | Polar scorec | G scored | PMF scoree | D scoref | Chem scoreg | No. of hydrogen bonds |
|---|---|---|---|---|---|---|---|---|---|
| 1. | Nimbin | 3.26 | −1.11 | 0.01 | −90.30 | −10.14 | −191.47 | −15.507 | 4 |
| 2. | Azadirone | 3.15 | −1.11 | 0.01 | −93.30 | −15.143 | 175.23 | −21.252 | 2 |
| 3. | Gedunin | 3.11 | −2.52 | 2.03 | −104.87 | −19.18 | −206.00 | −20.252 | 2 |
| 4. | Salannin | 1.49 | −1.34 | 1.47 | −272.24 | −28.690 | −155.09 | −22.227 | 1 |
| 5. | Ninbidol | 1.44 | −0.86 | 1.85 | −206.94 | 8.424 | −149.46 | −20.544 | 1 |
| 6. | Azadirachtin | 3.23 | −1.10 | 0.01 | −95.8 | −13.52 | −190.47 | −16.206 | 4 |
| 7. | Panduratin* | 1.25 | −0.95 | 1.15 | −205.91 | −17.512 | −138.32 | −20.266 | 1 |
* Reference ligand
aCScore is a consensus scoring which uses multiple types of scoring functions to rank the affinity of ligands
bCrash-score revealing the inappropriate penetration into the binding site
CPolar region of the ligand
dG-score showing hydrogen bonding, complex (ligand–protein), and internal (ligand–ligand) energies
ePMF-score indicating the Helmholtz free energies of interactions for protein–ligand atom pairs (Potential of Mean Force, PMF)
fD-score for charge and van der Waals interactions between the protein and the ligand
gChem-score points for hydrogen bonding, lipophilic contact, and rotational entropy, along with an intercept term
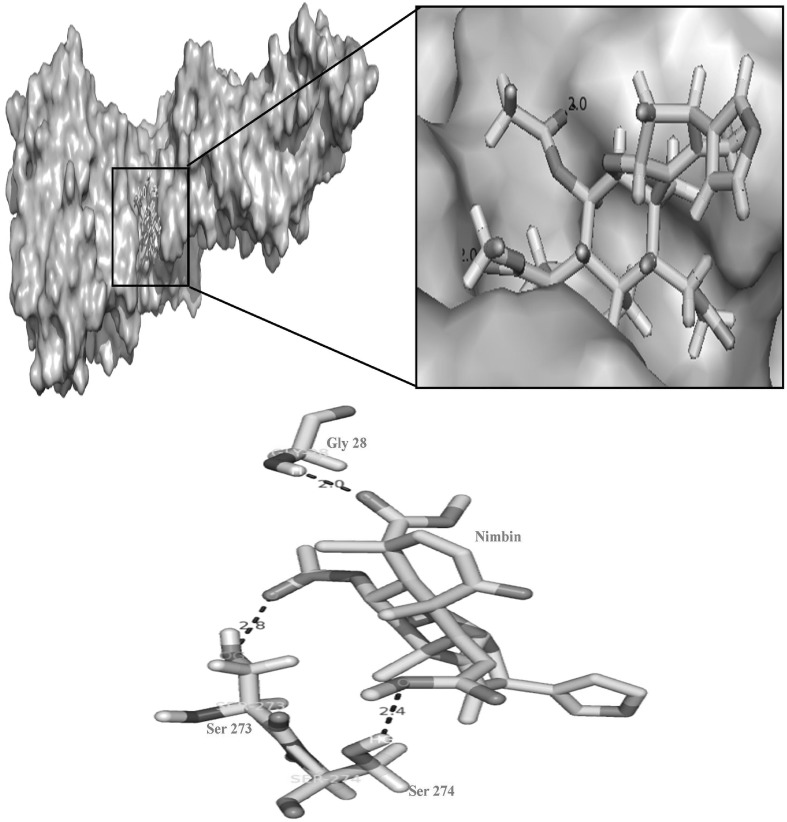
Fig. 4.
Pictorial representation hydrogen bond interactions between nimbin and the active site residues of DENV-2. Dotted lines represents the hydrogen bonds interactions between H atom of Gly 28 with O atom of nimbin, OG atom of Ser 273 with O atom of nimbin and HG atom of Ser 274 with O atom of nimbin
Table 3.
Binding affinity of active compounds towards the envelope of DENV-3 virus
| S. No | Compounds | CScorea | Crash scoreb | Polar scorec | G scored | PMF scoree | D scoref | Chem scoreg | No. of hydrogen bonds |
|---|---|---|---|---|---|---|---|---|---|
| 1. | Nimbin | 3.79 | −0.76 | 2.17 | −76.666 | −15.983 | −165.24 | −16.325 | 3 |
| 2. | Gedunin | 2.69 | −1.00 | 0.99 | −70.787 | −30.324 | −173.05 | −12.388 | 3 |
| 3. | Azadirone | 2.50 | −1.32 | 0.96 | −94.813 | −25.101 | −178.37 | −17.401 | 3 |
| 4. | Ninbidol | 1.20 | −0.92 | 1.11 | −210.08 | −7.003 | −155.32 | −22.227 | 2 |
| 5. | Salannin | 1.07 | −1.21 | 0.44 | −235.45 | −34.09 | −176.33 | −19.688 | 1 |
| 6. | Azadirachtin | 2.65 | −1.05 | 0.95 | −80.236 | −28.411 | −160.58 | −15.603 | 2 |
| 7. | Panduratin* | 2.15 | −1.05 | 1.15 | −85.91 | −27.422 | −168.51 | −12.254 | 2 |
* Reference ligand
aCScore is a consensus scoring which uses multiple types of scoring functions to rank the affinity of ligands
bCrash-score revealing the inappropriate penetration into the binding site
CPolar region of the ligand
dG-score showing hydrogen bonding, complex (ligand–protein), and internal (ligand–ligand) energies
ePMF-score indicating the Helmholtz free energies of interactions for protein–ligand atom pairs (Potential of Mean Force, PMF)
fD-score for charge and van der Waals interactions between the protein and the ligand
gChem-score points for hydrogen bonding, lipophilic contact, and rotational entropy, along with an intercept term
Fig. 5.
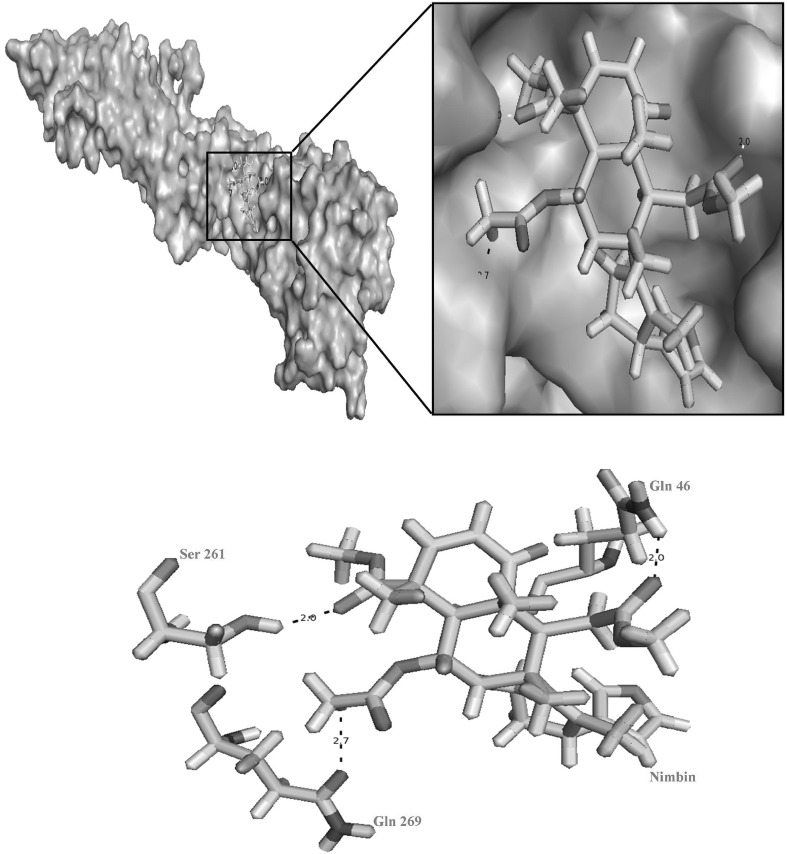
Pictorial representation hydrogen bond interactions between nimbin and the active site residues of DENV-3. Dotted lines represents the hydrogen bonds interactions between HG atom of Ser 261 with O atom of nimbin, OE1 atom of Gln 269 with H atom of nimbin and HE1 atom of Gln 46 with O atom of nimbin
Table 4.
Binding affinity of active compounds towards the envelope of DENV-4 virus
| S. No | Compounds | CScorea | Crash scoreb | Polar scorec | G scored | PMF scoree | D scoref | Chem scoreg | No. of hydrogen bonds |
|---|---|---|---|---|---|---|---|---|---|
| 1. | Nimbin | 3.37 | −0.77 | 1.10 | −85.944 | 13.383 | −174.23 | −18.83 | 4 |
| 2. | Azadirone | 3.11 | −0.99 | 1.28 | −92.730 | 1.031 | −162.25 | −11.90 | 2 |
| 3. | Gedunin | 3.06 | −0.76 | 2.72 | −83.563 | 0.991 | −144.51 | −18.825 | 3 |
| 4. | Ninbidol | 1.04 | −1.20 | 2.62 | −347.54 | 8.945 | −128.58 | −21.90 | 2 |
| 5. | Salannin | 0.71 | −1.62 | 1.25 | −172.65 | 10.358 | −163.34 | −23.687 | 1 |
| 6. | Azadirachtin | 3.05 | −0.82 | 1.35 | −87.378 | 2.274 | −158.75 | −16.423 | 4 |
| 7. | Panduratin* | 1.75 | −1.27 | 2.17 | −285.31 | 7.425 | −159.27 | −22.584 | 1 |
* Reference ligand
a CScore is a consensus scoring which uses multiple types of scoring functions to rank the affinity of ligands
b Crash-score revealing the inappropriate penetration into the binding site
C Polar region of the ligand
d G-score showing hydrogen bonding, complex (ligand–protein), and internal (ligand–ligand) energies
e PMF-score indicating the Helmholtz free energies of interactions for protein–ligand atom pairs (Potential of Mean Force, PMF)
f D-score for charge and van der Waals interactions between the protein and the ligand
g Chem-score points for hydrogen bonding, lipophilic contact, and rotational entropy, along with an intercept term
Fig. 6.
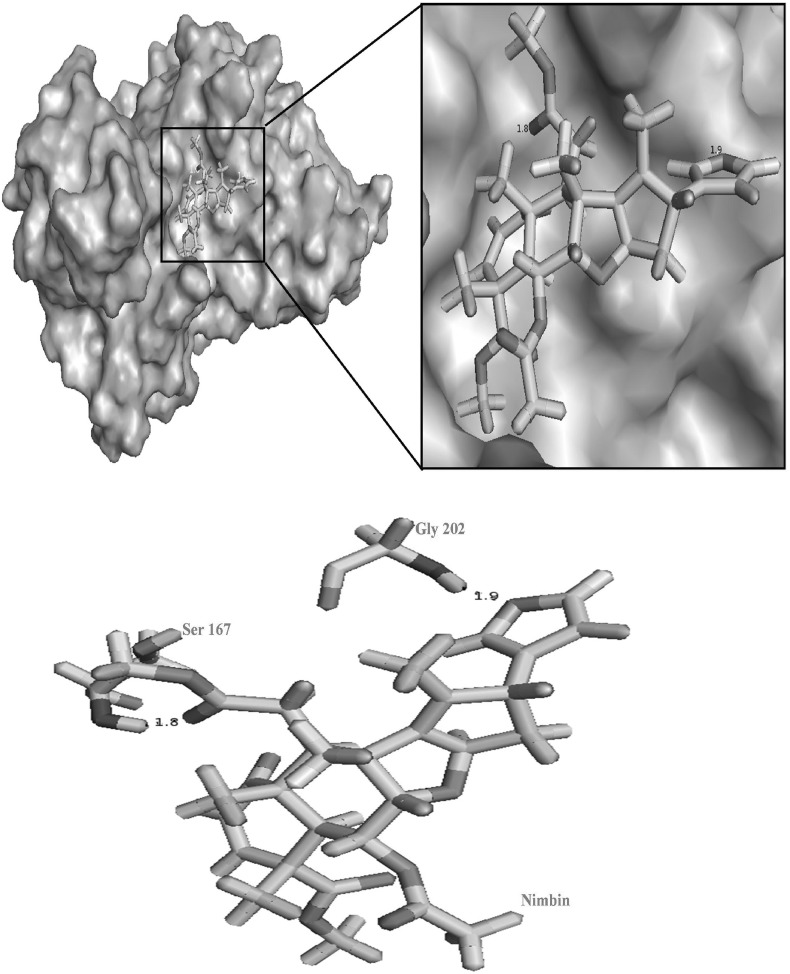
Pictorial representation hydrogen bond interactions between nimbin and the active site residues of DENV-4. Dotted lines represents the hydrogen bonds interactions between H atom of Ser 167 with O atom of nimbin and H atom of Gly 202 with O atom of nimbin
ADMET profiling
The physiochemical property and biological activity of the active compound Nimbin is analyzed based on Lipinski’s rule of 5. Our analysis shows that the compound Nimbin successfully passed through the Lipinski’s filter with minor violation. The results of the molecular properties and drug likeness of Nimbin is presented in Supplementary Table 5. Further ADMET profiling of Nimbin is performed and the results are tabulated in Supplementary Table 6. ADMET profiling is carried out for the parameters including blood–brain barrier penetration, Caco2 permeability, p-glycoprotein inhibitor, renal organic cation transporters and toxicity studies. Further predicted hERG risk for active compound shows favorable results which reveal the efficiency of nimbin to act as drug candidate.
Discussion
Currently no effectual treatment is available to conflict the dengue viral infection therefore development of effective medicine without any adverse side effects is necessary [19]. Hence in the present study we have explored anti-viral property of active compounds from Azadirachta indica through docking studies. Studies on natural compounds reveal the anti-viral property of plant derived products [29]. It is also interestingly reported that higher plants exhibits greater inhibitory property against viruses [2]. Anti-viral properties of Azadirachta indica is reported by several investigators [15, 39]. Presence of polyphenolic substances in the leaves of Azadirachta indica is responsible for the pharmacological properties of neem trees [49]. Our analysis reveals that active compound Nimbin shows high binding affinity against the DENV envelope. Nimbi is the major compound found in Azadirachta indica is reported to elucidate the pesticida properties of neem leaf extracts [55].
As proved in earlier studies “Rule of Three” could be useful when constructing fragment libraries for efficient lead discovery [46]. Our study involves the evaluation of the conformations of the whole ligand without ligand fragmentation. Docking of the ligands to the target was performed using a whole molecule approach, as described previously [10, 25]. Hence physiochemical property and biological activity of the active compound Nimbin is analyzed based on Lipinski’s rule of 5. As a general rule, molecular weight of the compound should be not more than 500 daltons. Even though molecular weight of the compound Nimbin isgreater than 500 daltons it can exempted from violation as per the rules stated by GlaxoSmithKline (GSK) groups [30]. This rule states that the compound with molecular weight >500 but with constrained polar surface (PSA) may also show good oral bioavailability [41]. According to the earlier reports [8] compounds with polar surface area of >140 Å leads to poor oral bioavailability, since the PSA of Nimbin is below the cut off value, the compound should be considered to have good oral bioavailability. Pharmacokinetic analysis on drug discovery states that molecular weight of the drug within the range of 150 and 550 Da exhibits good bioavailability [1, 8]. Physiochemical properties of compound determine the capability of drug to pass the blood brain barrier [16]. Since dengue infection is also associated with central nervous system [38], the drug should possess the capability to cross blood brain barrier and exhibit CNS activity. Compounds within the molecular range of 400–600 Da are efficiently transported through the blood–brain barrier [37]. The BBB efficiency of the compound Nimbin is analysed and our reports shows the positive results. Polar surface area of the compound efficiently determines the intestinal absorption of drugs [21]. As stated earlier, PSA value of Nimbin is within the threshold indicates the greater absorption of the compound. In earlier studies it has been proved that esters or amides are introduced to the drug molecule and hence enhance the aqueous solubility [31, 32]. An ester functional group has the potential to interact with binding site as a hydrogen bond acceptor. Esters are susceptible to hydrolysis in vivo by metabolic enzymes called esterases, which may decrease the life span of the drug in vivo. Even though esterase plays an important role in drug metabolism, there is a strong support from experimental studies which proves that those ester groups in drugs are stabilized by electrostatic and steric stabilization [40]. Intestinal esterase acts on ester type drugs administered orally at the beginning of ester type drug metabolism. It is also proved that pharmacological effects were influenced by the hydrolyzing activity of intestinal esterase [22, 57]. It is interestingly observed that the active compound Nimbin consists of three ester moieties, which can leads to increased absorption and oral bioavailability of compound. As proved in earlier studies presence of ester groups in drugs increases the absorption and oral bioavailability of compound [31, 32]. Human intestinal absorption of drugs can be effectively predicted using Caco2 cells. In silico prediction of Caco2 permeability of Nimbin shows good correlation with the human intestinal absorption. Metabolism of drugs and other foreign particles is carried out by the enzyme cytochrome P450. It is also reported that majority of the drugs are metabolized by the following isoforms, CYP1A2, CYP2C9, CYP2C18, CYP2C19, CYP2D6, CYP2E1 and CYP3A4. ADMET properties of Nimbin on different models such as BBB penetration, P-glycoprotein substrate, renal organic cation transporter, human intestinal absorption and Caco2 permeability shows positive results suggesting Nimbin as an effective anti-dengue compound. On the whole we conclude that further optimization of Nimbin can contribute significantly in reducing the morbidity and mortality of dengue infection.
Electronic supplementary material
Acknowledgments
S.R and A.A gratefully acknowledges the Indian Council of Medical Research (ICMR), Government of India Agency for the research Grant [IRIS ID: 2014-0099] to carry out this research. P. Lavanya thanks ICMR for the research fellowship. We would like to thank the management of VIT University for providing us the necessary facilities to carry out this research project.
References
- 1.Alavijeh MS, Chishty M, Palmer AM. Drug metabolism and pharmacokinetics, the blood–brain barrier, and central nervous system drug discovery. NeuroRx. 2005;2:554–571. doi: 10.1602/neurorx.2.4.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badam L, Joshi SP, Bedekar SS. In vitro antiviral activity of neem (Azadirachta indica, A. Juss) leaf extract against group BCoxsackie viruses. J Commun Dis. 1999;31:79–90. [PubMed] [Google Scholar]
- 3.Balunas MJ, Kinghorn AD. Drug discovery from medicinal plants. Life Sci. 2005;78:431–441. doi: 10.1016/j.lfs.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 4.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyallow IN, Bourne PE. The Protein data bank. Nucleic Acid Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braga RC, Alves VM, Silva MFB, Muratov E, Fourches D, Tropsha A, Andrade CH. Tuning hERG out: antitarget QSAR models for drug development. Curr Top Med Chem. 2014;14:1399–1415. doi: 10.2174/1568026614666140506124442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braga RC, Alves VM, Silva MFB, Muratov E, Fourches D, Liao LM, Tropsha A, Andrade CH. Pred-hERG: a novel web-accessible computational tool for predicting cardiac toxicity. Mol Inf. 2015 doi: 10.1002/minf.201500040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chattopadhyay D, Sarkar MC, Chatterjee T, Dey SR, Bag P, Chakraborti S, Khan MT. Recent advancements for the evaluation of anti-viral activities of natural products. N Biotechnol. 2009;25:347–368. doi: 10.1016/j.nbt.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaturvedi PR, Decker CJ, Odinecs A. Prediction of pharmacokinetic properties using experimental approaches during early drug discovery. Curr Opin Chem Biol. 2001;5:452–463. doi: 10.1016/S1367-5931(00)00228-3. [DOI] [PubMed] [Google Scholar]
- 9.Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, Lee PW, Tang Y. admetSAR: a comprehensive source and free tool for evaluating chemical ADMET properties. J Chem Inf Model. 2012;52:3099–3105. doi: 10.1021/ci300367a. [DOI] [PubMed] [Google Scholar]
- 10.Cleves AE, Jain AN. Effects of inductive bias on computational evaluations of ligand-based modeling and on drug discovery. J Comput Aided Mol Des. 2008;22:147–159. doi: 10.1007/s10822-007-9150-y. [DOI] [PubMed] [Google Scholar]
- 11.Cockburn JJ, Sanchez NME, Fretes N, Urvoas A, Staropoli I, Kikuti CM, Coffey LL, Seisdedos FA, Bedouelle H, Rey FA. Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody. Structure. 2012;20(2):303–314. doi: 10.1016/j.str.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J Comput Aided Mol Des. 1997;11:425–445. doi: 10.1023/A:1007996124545. [DOI] [PubMed] [Google Scholar]
- 13.Figueiredo MAA, Rodrigues LC, Barreto ML, Wellington JO, Costa MCN, Morata V, Vasconcelos PFC, Nunes MRT, Teoxeora MG. Allergies and diabetes as risk factors for dengue hemorrhagic fever: results of a case control study. PLoS Negl Trop Dis. 2010;4:e699. doi: 10.1371/journal.pntd.0000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frimayanthi N, Chee CF, Zain SM, Rahman NA. Design of new competitive dengue Ns2b/Ns3 protease inhibitors—a computational approach. Int J Mol Sci. 2011;12:1089–1100. doi: 10.3390/ijms12021089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghimeray AK, Jin C, Ghimire BK, Cho DH. Antioxidant activity and quantitative estimation of azadirachtin and nimbin in Azadirachta IndicaA. Juss grown in foothills of Nepal. Afri. J Biol. 2009;8:3084–3091. [Google Scholar]
- 16.Goswami RP, Mukherjee A, Biswas T, Karmakar PS, Ghosh A. Two cases of dengue meningitis: a rare first presentation. J Infect Dev Ctries. 2012;6:208–211. doi: 10.3855/jidc.2241. [DOI] [PubMed] [Google Scholar]
- 17.Gupta N, Srivastava S, Jain A, Chaturvedi UC. Dengue in India. Indian J Med Res. 2012;136:373–390. [PMC free article] [PubMed] [Google Scholar]
- 18.Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, Hunsperger E, Kroeger A, Margolis HS, Martínez ES, Nathan MB, Pelegrino JL, Simmons C, Yoksan S, Peeling RW. Dengue: a continuing global threat. Nat Rev Microbiol. 2010;8:S7–S16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermann EC, Muccra J, Kucera LS. Antiviral substances in plants of the mint family (Labiatae) Proc Soc Exp Biol Med. 1967;124:865–874. doi: 10.3181/00379727-124-31872. [DOI] [PubMed] [Google Scholar]
- 20.Hesse R. Molecular evolution and distribution of dengue viruses type 1 and 2 in nature. Virology. 1990;174:479. doi: 10.1016/0042-6822(90)90102-W. [DOI] [PubMed] [Google Scholar]
- 21.Huttunen KM, Raunio H, Rautio J. Prodrugs—from serendipity to rational design. Pharamacol Rev. 2011;63:750–771. doi: 10.1124/pr.110.003459. [DOI] [PubMed] [Google Scholar]
- 22.Inoue M, Morikawa M, Tsuboi M, Yamada T, Sugiura M. Hydrolysis of ester-type drugs by the purified Esterase from human intestinal mucosa. Jpn J Pharmacol. 1979;29:17–25. doi: 10.1254/jjp.29.17. [DOI] [PubMed] [Google Scholar]
- 23.Jain AN. Scoring noncovalent protein-ligand interactions: a continuous differentiable function tuned to compute binding affinities. J Comput Aided Mol Des. 1996;10:427–440. doi: 10.1007/BF00124474. [DOI] [PubMed] [Google Scholar]
- 24.Jain AN. Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem. 2003;46:499–511. doi: 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]
- 25.Jain AN. Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem. 2003;46:499–511. doi: 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]
- 26.Ji HF, Li XJ, Zhang HY. Natural products and drug discovery. Can thousands of years of ancient medical knowledge lead us to new and powerful drug combinations in the fight against cancer and dementia? EMBO Rep. 2009;10:194–200. doi: 10.1038/embor.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johari J, Kianmehr A, Mustafa MR, Abubakar S, Zandi K. Antiviral activity of baicalein and quercetin against the Japanese encephalitis virus. Int J Mol Sci. 2012;13:16785–16795. doi: 10.3390/ijms131216785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones G, Willett P, Glen RC. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J Mol Biol. 1995;245:43–53. doi: 10.1016/S0022-2836(95)80037-9. [DOI] [PubMed] [Google Scholar]
- 29.Kamb KM, Amoral M, Girre L. Search for non-antiviral agents of plant origin. Pharmacol Acta Helv. 1992;67:130–147. [PubMed] [Google Scholar]
- 30.Keller TH, Pichota A, Yin Z. A practical view of ‘druggability’. Curr Opin Chem Biol. 2006;10:357–361. doi: 10.1016/j.cbpa.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 31.Kerns EH, Di L. Drug-like properties: concepts, structure design and methods—from ADMET to toxicity optimization. 1. London: Academic Press; 2010. pp. 426–438. [Google Scholar]
- 32.Lin JH. Role of P-glycoprotein in pharmacokinetics clinical implications. Clin Pharmacokinet. 2003;42:59–98. doi: 10.2165/00003088-200342010-00003. [DOI] [PubMed] [Google Scholar]
- 33.Meng EC, Shoichet BK, Kuntz ID. Automated docking with grid-based energy evaluation. J Comput Chem. 1992;13:505–524. doi: 10.1002/jcc.540130412. [DOI] [Google Scholar]
- 34.Mitchell MJ, Smith SL, Johnson S, Morgan ED. Effects of the neem tree compounds azadirachtin, salannin, nimbin, and 6-desacetylnimbin on ecdysone 20-monooxygenase activity. Arch Insect Biochem Physiol. 1997;35:199–209. doi: 10.1002/(SICI)1520-6327(1997)35:1/2<199::AID-ARCH18>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 35.Modis Y, Ogata S, Clements D, Harrison SC. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J Virol. 2005;79(2):1223–12231. doi: 10.1128/JVI.79.2.1223-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molinspiration Cheminformatics, Bratislava, Slovak Republic. www.molinspiration.com/services/properties.html. Accessed Sept 2013.
- 37.Palm K, Stenberg P, Luthman K, Artursson P. Polar molecular surface properties predict the intestinal absorption of drugs in humans. Pharm Res. 1997;14:568–571. doi: 10.1023/A:1012188625088. [DOI] [PubMed] [Google Scholar]
- 38.Pardridge WM. CNS drug design based on principles of blood–brain barrier transport. J Neurochem. 1998;70:1781–1792. doi: 10.1046/j.1471-4159.1998.70051781.x. [DOI] [PubMed] [Google Scholar]
- 39.Parida MM, Upadhyay C, Pandya G, Jana AM. Inhibitory potential of neem (Azadirachta indica Juss) leaves on dengue virus type-2 replication. J Ethnopharmacol. 2002;79:273–278. doi: 10.1016/S0378-8741(01)00395-6. [DOI] [PubMed] [Google Scholar]
- 40.Patrick GL. An introduction to medicinal chemistry. New York: Oxford University Press; 2001. [Google Scholar]
- 41.Plasencia C, Dayam R, Wang Q, Pinski J, Burke TR, Quinn DI, Neamati N. Discovery and preclinical evaluation of a novel class of small-molecule compounds in hormone-dependent and -independent cancer cell lines. Cancer Ther. 2005;4:110. doi: 10.1158/1535-7163.MCT-04-0288. [DOI] [PubMed] [Google Scholar]
- 42.Pramely R, Raj TLS. Prediction of biological activity spectra of a few phytoconstituents of Azadirachta indicia Juss. J Biol Technol. 2012;3:375–379. [Google Scholar]
- 43.Rai AR, Sethi M. Screening of some plants for their activity against vaccinia and fowl pox viruses. Indian J Anim Sci. 1972;42:1066–1070. [Google Scholar]
- 44.Rao AR, Kumar SSV, Paramsivam TR, Kamalakshi S, Parashuraman AR, Shanta B. Study of antiviral activity of leaves of margosa tress on vaccinia and variola viruses—a preliminary report. Indian J Med Res. 1969;57:495–502. [PubMed] [Google Scholar]
- 45.Reddy AB, Sethi MS. Antiviral effects of some indigenious plant extracts on vaccinia and fowl pox viruses on chick embryo fibroblasts. Indian J Exp Biol. 1974;12:572–579. [PubMed] [Google Scholar]
- 46.Rees DC, Congreve M, Murray CW, Carr R. Fragment-based lead discovery. Nat Rev Drug Discov. 2004;3:660–672. doi: 10.1038/nrd1467. [DOI] [PubMed] [Google Scholar]
- 47.Sai Ram M, Ilavazhagan G, Sharma SK, Dhanraj SA, Saresh B, Parida MM, Jan AM, Davendra K, Selvanmurthy W. Antimicrobial activity of a new vaginal contraceptive NIM. 76 from neem oil (Azadirachta indica) J Ethnopharmacol. 2000;1:377–382. doi: 10.1016/S0378-8741(99)00211-1. [DOI] [PubMed] [Google Scholar]
- 48.Sayet KA. Natural products as antiviral agents. Stud Nat Prod Chem. 2000;24:473–572. doi: 10.1016/S1572-5995(00)80051-4. [DOI] [Google Scholar]
- 49.Sidhu O, Kumar VP, Hari MB. Variability in triterpenoids (Nimbin and Salanin) composition of neem among different provenances of India. Ind Crops Prod. 2004;19:69–75. doi: 10.1016/j.indcrop.2003.07.002. [DOI] [Google Scholar]
- 50.Sim S, Ramirez JL, Dimopoulos G. Dengue virus infection of the Aedes aegypti salivary gland and chemosensory apparatus induces genes that modulate infection and blood-feeding behavior. PLoS Pathog. 2012;8:e1002631. doi: 10.1371/journal.ppat.1002631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subapriya R, Nagini S. Medicinal properties of neem leaves: a review. Curr Med Chem Anticancer Agents. 2005;5(2):149–156. doi: 10.2174/1568011053174828. [DOI] [PubMed] [Google Scholar]
- 52.Tang LI, Ling AP, Koh RY, Chye SM, Voon KG. Screening of anti-dengue activity in methanolic extracts of medicinal plants. Bmc Complem Altern Med. 2012;12:3. doi: 10.1186/1472-6882-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tiwari V, Darmani NA, Yue BYUT, Shukla D. In vitro antiviral activity of neem (Azardirachta indica L.) bark extract against herpes simplex virus type-1 infection. Phytother Res. 2010;24:1132–1140. doi: 10.1002/ptr.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Upadhyay SN, Dhawan S, Garg S, Wali N, Tusker L, Anderson DJ. Immunomodulatory properties of neem. In: World Neem Conference: Bangalore, India. 1993 (p. abstract no. 80).
- 55.Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability or drugs. J Med Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 56.Wang QY, Patel SJ, Vangrevelinghe E, Xu HY, Rao R, Jaber D, Schul W, Gu F, Heudi O, Ma NL, Poh MK, Phong WY, Keller TH, Jacoby E, Vasudevan SG. A small-molecule dengue virus entry inhibitor. Antimicrob Agents Chemother. 2009;53:1823–1831. doi: 10.1128/AAC.01148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams FM. Clinical significance of esterases in man. Clin Pharmacokinet. 1985;10(5):392–403. doi: 10.2165/00003088-198510050-00002. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, Kuhn RJ, Rossman MJ. Conformational changes of the Flavivirus E glycoprotein. Structure. 2004;12(9):1607–1608. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.