Abstract
Background
The RNA-guided Cas9 system represents a flexible approach for genome editing in plants. This method can create specific mutations that knock-out or alter target gene function. It provides a valuable tool for plant research and offers opportunities for crop improvement.
Results
We investigate the use and target specificity requirements of RNA-guided Cas9 genome editing in barley (Hordeum vulgare) and Brassica oleracea by targeting multicopy genes. In barley, we target two copies of HvPM19 and observe Cas9-induced mutations in the first generation of 23 % and 10 % of the lines, respectively. In B. oleracea, targeting of BolC.GA4.a leads to Cas9-induced mutations in 10 % of first generation plants screened. In addition, a phenotypic screen identifies T0 plants with the expected dwarf phenotype associated with knock-out of the target gene. In both barley and B. oleracea stable Cas9-induced mutations are transmitted to T2 plants independently of the T-DNA construct. We observe off-target activity in both species, despite the presence of at least one mismatch between the single guide RNA and the non-target gene sequences. In barley, a transgene-free plant has concurrent mutations in the target and non-target copies of HvPM19.
Conclusions
We demonstrate the use of RNA-guided Cas9 to generate mutations in target genes of both barley and B. oleracea and show stable transmission of these mutations thus establishing the potential for rapid characterisation of gene function in these species. In addition, the off-target effects reported offer both potential difficulties and specific opportunities to target members of multigene families in crops.
Electronic supplementary material
The online version of this article (doi:10.1186/s13059-015-0826-7) contains supplementary material, which is available to authorized users.
Keywords: Genome editing, CRISPR/Cas9, Barley, Brassica, PM19, GA4, Crops, Mutations, Breeding, Off-target
Background
Genetic modification is a key research tool for advancing knowledge of gene function as well as allowing the development of crops with valuable traits. Genetic modification enables the introduction of genes of interest or the reduction in expression of endogenous genes (RNAi approaches) through the insertion of transgenic sequences at random locations within the plant genome. Genetic modification technologies have advanced substantially over the past 30 years, but more recently, a series of exciting developments offer significant opportunities for the analyses of plant genomes, as well as having applications in crop improvement [1]. These approaches, collectively called genome editing, provide the opportunity to make precise changes at specific genomic locations. Genome editing may be used to induce gene insertions, gene replacements, or insertions or deletions that disrupt the function of a specific gene [2]. This latter application, leading to knock-out of target genes, has enormous benefits for research in plants, especially in crops that lack genetic resources such as knock-out libraries.
Genome editing requires a site-directed nuclease to introduce one or more breaks in the DNA at the target locus. The cell’s endogenous DNA repair mechanisms repair these breaks; imperfect repair can produce mutations or deletions in the genes of interest. To generate site-specific breaks, different approaches have employed different combinations of nucleases fused to programmable DNA binding domains including Zinc Finger Nucleases (ZFNs) and Transcription-Activator Like Effector Nucleases (TALENs). More recently, the Cas9 protein associated with Type II Clustered Regulatory Interspaced Short Palindromic Repeats (CRISPR) found in bacteria has been repurposed for genome editing in eukaryotes [3, 4]. The RNA-guided Cas9 system uses a small non-coding RNA, known as the single guide RNA (sgRNA), to direct the Cas9 nuclease to the DNA target of interest. Being small and easy to reprogram, this offers a flexible, easy-to-implement and relatively cheap method for genome editing [5]. The first applications of RNA-guided Cas9 in plants were described in 2013 [6–8] using transient systems. Inheritance of induced mutations in progeny plants was demonstrated for the first time in Arabidopsis by Feng et al. [9] and heritable changes have also been shown in rice [10, 11]. In wheat (Triticum aestivum), RNA-guided Cas9 has been used to mutate a single homoeologue of the mildew resistance locus MLO in stable T0 transgenic plants although no information was provided regarding the inheritance of the mutant alleles [12].
Very few studies have described the inheritance of RNA-guided Cas9-induced mutations and questions remain regarding its efficiency, especially in crop plants. In addition, the frequency with which the nuclease induces mutations in unintended targets (known as off-targets) has yet to be extensively investigated across plant species. The aim of this study was therefore to use RNA-guided Cas9 for targeted mutagenesis in both monocotyledonous and dicotyledonous crop species, demonstrating for the first time its application in both barley (Hordeum vulgare) and Brassica oleracea. In addition, we aimed to assess the efficiency of mutagenesis and test whether off-target effects occurred.
Arabidopsis GA4 is involved in the gibberellin biosynthesis pathway and GA4 loss-of-function mutants have dwarf stature and reduced fruit dehiscence [13, 14]. Since plant architecture and seed dispersal are important targets for crop improvement in Brassicas, we tested the effect of mutating GA4 orthologues in B. oleracea. In barley, we chose HvPM19 as our target. HvPM19 encodes an ABA-inducible plasma membrane protein [15], which in wheat acts as a positive regulator of grain dormancy [16], an important agronomic trait in cereals.
Here we demonstrate the successful use of RNA-guided Cas9 genome editing to knockout the function of target genes in both barley and B. oleracea. We show transmission of the mutation to progeny plants in both species and we demonstrate the segregation of the transgenic locus (encoding the nuclease and sgRNA) from the mutation, resulting in transgene-free plants that contain the desired mutation.
Results and discussion
RNA-guided Cas9-induced genome editing in barley
We investigated the use and target specificity requirements of RNA-guided Cas9 genome editing in barley by focusing on a multi-copy gene. We selected HvPM19, which is present as four copies within a single barley BAC clone from the cultivar ‘Morex’ (HvPM19-1 to HvPM19-4; Fig. 1a). Relative to HvPM19-1, the HvPM19-2, HvPM19-3 and HvPM19-4 loci have sequence identities of 89.8 %, 89.5 % and 88.6 %, respectively, whereas HvPM19-3 and HvPM19-4 have greater sequence identity to HvPM19-2 (98.4 % and 99.6 %). This suggests that HvPM19-1 was involved in the more ancestral duplication event and that there was a series of very recent duplication events between HvPM19-2, HvPM19-3 and HvPM19-4.
Fig. 1.
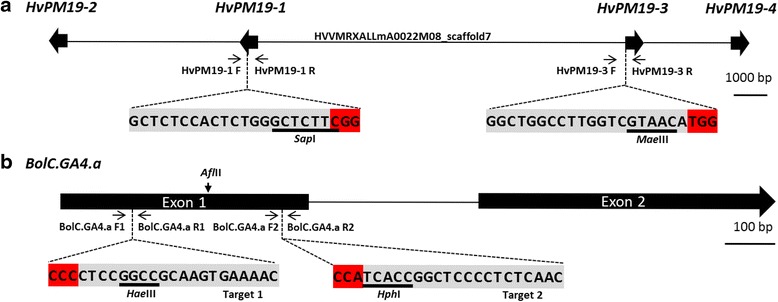
Barley HvPM19 and B. oleracea BolC.GA4.a gene models and target sequences. a Morex HVVMRXALLmA0022M08_scaffold7 sequence contains the four barley HvPM19 gene copies (filled arrows). The target sequences for sgRNAHvPM19-1 and sgRNAHvPM19-3 (grey highlight) are shown below their respective gene models, with the protospacer-adjacent motif (PAM) highlighted in red. Recognition sequences for the restriction endonucleases SapI and MaeIII are underlined. b The B. oleracea BolC.GA4.a gene model includes two exons (filled boxes) separated by an intron (represented by a solid line). The B. oleracea BolC.GA4.a sequences for sgRNA1BolC.GA4.a (Target 1) and sgRNA2BolC.GA4.a (Target 2) are shown below the target regions in grey highlight with the PAM highlighted in red. Recognition sequences for the restriction endonucleases AflII, HaeIII and HphI are underlined. Primers for mutant detection are shown in both panels and detailed in Additional file 3
We independently targeted two ancestral HvPM19 gene copies (HvPM19-1 and HvPM19-3) in the spring barley cultivar ‘Golden Promise’ which is amenable to Agrobacterium-mediated transformation. We were able to amplify HvPM19-4 from Golden Promise, but unable to amplify HvPM19-2 suggesting that this cultivar lacks this copy of HvPM19. Two binary constructs, sgRNAHvPM19-1, referred to as pPM19-1 and sgRNAHvPM19-3, referred to as pPM19-3 (Fig. 2a), were designed to independently target HvPM19-1 and HvPM19-3, respectively. The 20 base-pair target sequence in pPM19-1 has a single nucleotide mismatch with each of the corresponding sequences in HvPM19-3 and HvPM19-4, while the target sequence in pPM19-3 has three mismatches with HvPM19-1 and one mismatch with HvPM19-4 (Fig. 3a).
Fig. 2.
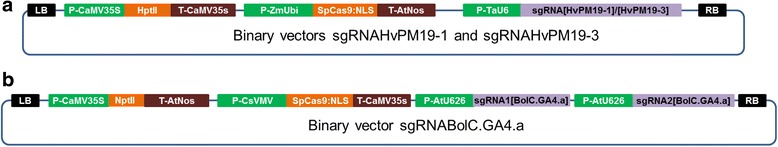
Schematic of binary plasmid vectors delivered to barley and B. oleracea. Transcription units were assembled into the binary plasmid backbone pAGM4723 or pAGM8031 using Golden Gate Modular Cloning. a The barley constructs, sgRNAHvPM19-1 and sgRNAHvPM19-3 house a hygromycin resistance cassette consisting of the hygromycin phosphotransferase coding sequence (hptII) driven and terminated by the 35 s promoter (P-CaMV35s) and terminator (T-CaMV35s) from Cauliflower mosaic virus; a Cas9 expression cassette consisting of sequence encoding Cas9 from Streptococcus pyogenies with a carboxy-terminal nuclear-localization signal from Simian vacuolating virus 40 (SpCas9:NLS) driven by a ubiquitin promoter from Zea mays (P-ZmUbi) and terminated by a nopaline synthase terminator from Agrobacterium tumefaciens (T-AtNos); and single guide RNA (sgRNAHvPM19-1 or sgRNAHvPM19-3) driven by a Triticum aestivum U6 promoter (P-TaU6). b The Brassica construct, sgRNABolC.GA4.a, houses a kanamycin resistance cassette consisting of the neomycin phosphotransferase coding sequence (nptII) driven and terminated by P-CaMV35S and T-AtNos; SpCas9:NLS driven by a constitutive promoter from Cassava Vein Mosaic Virus (P-CsVMV) and a tandem pair of single guide RNAs (sgRNA1BolC.GA4.a and sgRNA2BolC.GA4.a) driven by the U626 promoter from Arabidopsis (P-AtU626)
Fig. 3.
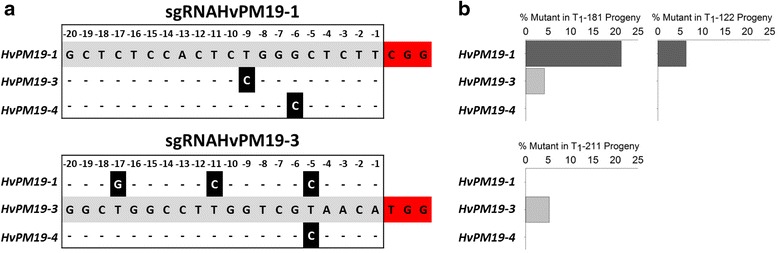
Frequency of on-target and off-target Cas9 activity on the HvPM19 gene copies at T1. a Alignment of sgRNAHvPM19-1 and sgRNAHvPM19-3 target sequences (grey highlight) with the corresponding sequences of the other copies of HvPM19. Hyphens represent alignment matches while mismatches are shown in black highlight and white font. The PAM is highlighted in red and the numbering of nucleotides is relative to the PAM. b Percentage of T1 plants with mutations in the corresponding copies of HvPM19 for sgRNAHvPM19-1 (T0-181 and T0-122) and sgRNAHvPM19-3 (T0-211). Dark and light grey bars represent the percentages for HvPM19-1 and HvPM19-3 editing, respectively
The two constructs were independently transformed into immature barley embryos to generate 28 and 20 independent transgenic lines for pPM19-1 and pPM19-3, respectively. T0 regenerated plantlets were screened for mutations using a restriction digest/PCR assay. We detected deletions in HvPM19-1 in three out of 13 pPM19-1 T0 lines screened (T0-181, T0-122 and T0-191). Similarly, out of the 10 pPM19-3 T0 plantlets screened, one line (T0-211) showed an insertion in HvPM19-3. Therefore, the frequency of Cas9-induced mutations in the first generation was 23 % for pPM19-1 and 10 % for pPM19-3. These mutation frequencies are comparable to those reported in stable T0 transformants from other monocotyledonous species such as wheat [12], rice (Oryza sativa; reviewed in [17]) and sorghum (Sorghum bicolor; [18]).
As is characteristic of Cas9-induced mutations [3, 4, 19], all the insertions or deletions (in-dels) detected were at the 3' end of the target region, 3 or 4 bp upstream of the Protospacer Adjacent Motif (PAM; Additional file 1). In the T0 plants, we detected in-dels only after enriching for the mutation by the restriction digest/PCR assay presumably because only a small proportion of the cells had been mutated at the time of sampling. To further characterise the events, we measured T-DNA copy number in the T0 lines and found that T0-181, T0-191 and T0-211 each contained a single copy of the T-DNA whereas T0-122 contained two copies.
On-target and off-target activity of RNA-guided Cas9 in T1 transgenic barley plants
Next, we examined the T1 progenies of T0-181, T0-122, T0-191 and T0-211. Twenty out of 93 T1 progenies of T0-181 contained in-dels in the target HvPM19-1 sequence as determined by Sanger sequencing. Of these, two were homozygous and 18 were heterozygous mutants determined by the presence of double peaks in the sequencing chromatogram beginning from the site of the in-del. For T0-122, only six out of 95 T1 progenies had in-dels in the target HvPM19-1 sequence, with all being heterozygous mutations. This represents mutation frequencies of 22 % in T0-181 and 6 % in T0-122 progenies. For line T0-211, which showed Cas9 activity for HvPM19-3, we detected four mutant plants out of 76 T1 progenies tested, all of which had heterozygous mutations (mutation frequency of 5 %). Line T0-191 showed mutations in seven out of 90 T1 plants, but was not analysed further. As in the T0 generation, all the in-dels were observed to occur in the 3–4 bp adjacent to the PAM. All the T1 plants with Cas9-induced mutations retained their corresponding T-DNA construct, while there was segregation in the non-mutated T1 plants. This indicated that the mutations could still be the product of sgRNA/Cas9 expression in somatic cells rather than due to germline inheritance.
To assess the specificity of the T-DNA constructs, we sequenced HvPM19-3 and HvPM19-4 in the progenies of T0-181 and T0-122 (designed to target HvPM19-1). We found no off-target activity in the T1 progenies of T0-122; whereas three T0-181 progeny from 72 tested (4.2 %) had off-target activity on HvPM19-3 (Fig. 3b). By contrast, we observed no off-target activity on HvPM19-1 and HvPM19-4 in the 73 T1 progenies of T0-211 that contained the T-DNA designed to target HvPM19-3.
Cas9-induced mutations are stably transmitted to T2 barley plants independently of the T-DNA construct
The mutation in the target gene theoretically should segregate independently of the T-DNA that encodes the nuclease and sgRNA. We observed complete co-segregation of the Cas9-induced mutations with the T-DNA construct in the T1 transgenic lines. We therefore tested the T2 progenies of several T1 lines to determine if the mutations could be stably inherited and segregate independently from the T-DNA construct. We screened for the presence of the T-DNA through PCR and qPCR assays and determined the mutation status in the T2 progenies of T1-181, T1-122 and T1-211 lines (T1 mutant lines originating from the corresponding T0 lines; Table 1). The T-DNA segregated in the progeny of some, but not all, of these T1 lines. Segregation of pPM19-1 was detected in 11 out of 19 T1-181 lines, whereas pPM19-1 segregated in the progeny of four out of six T1-122 lines. However, only one out of three T1-211 lines tested showed segregation of the pPM19-3 construct. A 3:1 presence:absence ratio was confirmed using a χ2 test in all progenies in which the T-DNA segregated (P >0.44 or higher).
Table 1.
Summary of transgenerational RNA-guided Cas9 activity and segregation in barley
| T-DNA Construct | T0 line | T1 mutation type | T1 line | Number of T2 plants screened for T-DNA | Plants without T-DNA | Plants without T-DNA and with (homozygous/heterozygous) mutationsa |
|---|---|---|---|---|---|---|
| pPM19-1 | T0-181 | Homozygous | T1-181_B5 | 4 | 0 | - |
| T1-181_E1 | 12 | 5 | 5/0 | |||
| Heterozygous | T1-181_A11 | 12 | 3 | 1/1 | ||
| T1-181_B1 | 8 | 2 | 0/0 | |||
| T1-181_B8 | 11 | 0 | - | |||
| T1-181_C1 | 9 | 0 | - | |||
| T1-181_C12 | 1 | 0 | - | |||
| T1-181_C3 | 12 | 0 | - | |||
| T1-181_C4 | 12 | 1 | 0/0 | |||
| T1-181_C9 | 11 | 1 | 0/1 | |||
| T1-181_D11 | 12 | 4 | 1/1 | |||
| T1-181_D2 | 12 | 3 | 2/0 | |||
| T1-181_D9 | 2 | 0 | - | |||
| T1-181_E4 | 12 | 0 | - | |||
| T1-181_G4 | 11 | 3 | 0/1 | |||
| T1-181_G5 | 12 | 1 | 0/0 | |||
| T1-181_H2 | 12 | 3 | 1/1 | |||
| T1-181_H5 | 12 | 0 | - | |||
| T1-181_H9 | 12 | 4 | 0/0 | |||
| T0-122 | Heterozygous | T1-122_B11 | 12 | 4 | 2/1 | |
| T1-122_C6 | 12 | 1 | 0/1 | |||
| T1-122_F12 | 12 | 4 | 0/3 | |||
| T1-122_H2 | 12 | 0 | - | |||
| T1-122_H4 | 12 | 3 | 0/0 | |||
| T1-122_H9 | 12 | 0 | - | |||
| pPM19-3 | T0-211 | Heterozygous | T1-211_B11 | 12 | 3 | 0/0 |
| T1-211_D10 | 12 | 0 | - | |||
| T1-211_G4 | 7 | 0 | - |
aHyphens (-) indicate that all plants had presence of the T-DNA construct, and thus were not tested
We next sequenced HvPM19-1 and HvPM19-3 from all 45 T2 plants that had not inherited the T-DNA. In these plants, we detected mutations in HvPM19-1 in 15 T2 progenies originating from seven independent T1-181 lines and seven T2 progenies originating from three independent T1-122 lines (Table 1). A single progeny of T1-181_H2 showed an off-target mutation in HvPM19-3 in addition to the on-target HvPM19-1 mutation. Interestingly, T1-181_H2 is one of the three T0-181 lines that showed off-target activity of pPM19-1 in the T1 generation. For pPM19-3, we did not detect mutations in HvPM19-3 (and HvPM19-1) in the absence of the T-DNA in the progenies of any of the T1-211 lines.
We found that the mutations detected in the T2 progenies matched those observed in the corresponding T1 parent in all cases examined. For instance, the homozygous 1-bp deletion observed in line T1-181_E1 was also present in all its T2 progenies that segregated away from the T-DNA construct (Table 1; Fig. 4a). Likewise, a range of mutations found in heterozygous T1 plants were also identified in homozygous T2 individuals in the absence of the T-DNA construct (Fig. 4b). Taken together, the T1 and T2 sequence data from six lines, originating from two independent T0 events (T0-181 and T0-122), provide strong evidence of stable germline transmission of Cas9-induced mutations in barley in the absence of the T-DNA.
Fig. 4.

Germline transmission of Cas9 induced mutations from T1 to T2 plants in barley and B. oleracea in the absence of the T-DNA construct. a Sequence alignment of T1-181_E1 and five homozygous T2 progeny with homozygous 1-bp deletion in HvPM19_1. b Sequence alignment from representative clones of T1 heterozygote mutants and direct Sanger sequencing of their T2 progeny with homozygous mutations in the absence of the T-DNA. The numbers of clones supporting T1 mutant alleles are indicated on the right. c Sequence alignments of BolC.GA4.a Target 2 in homozygous T1 and T-DNA free T2 plants. Across panels the target sequences for sgRNAHvPM19-1 and sgRNABolC.GA4.a (grey) and PAM (red) are highlighted and Cas9 induced insertions and deletions are indicated by red font or red hyphens, respectively. Names of homozygous T2 plants that lack the presence of the T-DNA construct are indicated in blue; individual homozygous plants with the same allele are shown on the same row and are labelled with a ‘p’ prefix
The ability to develop transgene-free and stable germline mutations is of considerable interest in crop species given the current regulatory framework for deployment of transgenic crops in the field. Although regulation of edited crops is still being debated [20], crops free of transgenes may not be subject to existing regulations on genetic modification. Here, we demonstrate that in several instances there was stable germline-transmitted inheritance of Cas9-induced mutations in barley from the T1 to the T2 generation in the absence of the T-DNA construct. This supports previous studies in plants (Arabidopsis, tomato, tobacco and rice) that have documented transgene-free inheritance of Cas9-induced mutations in the T1 and T2 generations. No description of germline inheritance has been previously reported for Triticeae [12]. We also identified a single plant with an off-target mutation in the T2 generation in the absence of the T-DNA construct. This plant had mutations in both HvPM19-1 and HvPM19-3, suggesting that tandemly duplicated genes can be knocked-out with a single sgRNA, although we have yet to establish if these mutations are in cis or on homologous chromosomes. Previous work in rice had identified off-target mutations only in the T1 generation and in the presence of the sgRNA/Cas9 construct [21].
RNA-guided Cas9 induced genome editing in Brassica oleracea
RNA-guided Cas9-induced genome editing was performed in B. oleracea DH1012 [22] by targeting BolC.GA4.a (Bol038154) located on chromosome 5. This gene is an orthologue of Arabidopsis GA4 which encodes AtGA3OX1, the last enzyme in the biosynthesis of bioactive gibberellins. In Arabidopsis, ga4 loss-of-function mutants show a semi-dwarf phenotype [23] and this gene is required for efficient seed dispersal as it promotes specification of the dehiscence zone in siliques [13]. BolC.GA4.a has a paralog on chromosome 8, designated BolC.GA4.b (Bol031570), which shares 90 % DNA sequence identity. To generate Cas9 induced mutations in BolC.GA4.a, we designed a binary construct containing two sgRNAs (sgRNA1BolC.GA4.a and sgRNA2BolC.GA4.a) that target separate regions (Target 1 and Target 2, respectively) in the first exon of BolC.GA4.a (Figs. 1b and 2b).
Eighty independent transgenic lines were generated by Agrobacterium-mediated transformation, and 20 of these T0 plantlets were screened by the restriction digest/PCR assay to detect mutations in the target sequences. We identified in-dels at the target sites in BolC.GA4.a in two out of 20 T0 lines (L2F1_8.2 and L2E1_17.1). Mutations in L2E1_17.1 were confirmed by TA cloning and Sanger sequencing of the PCR products (Fig. 5a). Line L2F1_8.2 showed a 282-bp deletion that corresponds to re-joining of the DNA at exactly 3 bp from the PAM in both target regions. As in barley, the detection of the mutations required an enrichment of the target by restriction digest prior to PCR.
Fig. 5.
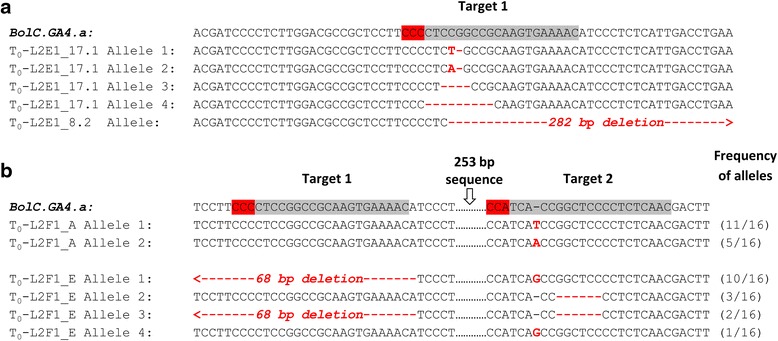
Mutant alleles detected in T0 B. oleracea. Alignment of wild-type and mutant sequences surrounding the target sequences (grey highlight) and PAM (red highlight) in mutants identified by restriction digest/PCR screen (a) and by phenotypic screen (b). Insertions and deletions are indicated by red font or red hyphens, respectively. For large deletions, red arrows indicate the direction of the deletions. For each line in panel b (L2F1_A and L2F1_E), 16 clones were examined and the frequencies of each mutant allele (represented as clones with mutant allele/total number of clones examined) are indicated at the right side of the panel
We also hypothesised that plants with homozygous mutations in BolC.GA4.a would show a dwarf phenotype similar to that observed in Arabidopsis ga4 mutants. Therefore, we performed a phenotypic screen of the 80 T0B. oleracea plants. All 80 T0 lines were grown to maturity, and at flowering two lines not previously characterised by the restriction digest/PCR assay were observed to be dwarf in stature (L2F1_A and L2F1_E; Fig. 6a). The BolC.GA4.a sequences from both dwarf plants were found to contain a series of mutant alleles in Target 1 and Target 2, in two independent leaf samples from each plant (Fig. 5b, Additional file 2). In addition, the mutation was restricted to BolC.GA4.a, as we were unable to detect any mutation in BolC.GA4.b. The identification of T0 plants with a visible knockout phenotype has also been reported in rice, tomato, and Arabidopsis [8, 10, 24].
Fig. 6.
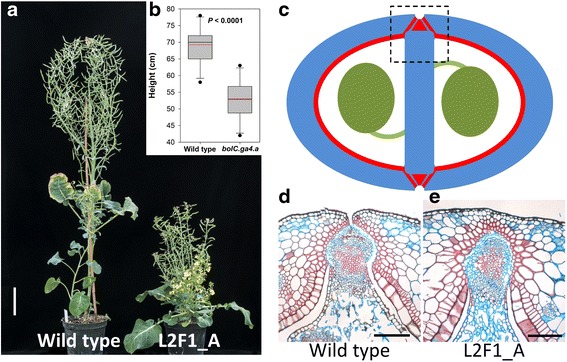
Mutations in BolC.GA4.a result in dwarf stature and affect the pod valve margin. a Wild-type B. oleracea DH1012 (left) and L2F1_A with a mutation in BolC.GA4.a showing a severe dwarf phenotype. Scale bar 10 cm. b Height of homozygous T1 plants with wild type (n = 11) or bolC.ga4.a mutant (n = 16) alleles. c Schematic cross section of B. oleracea pod with replum/valve margin region indicated by dashed square. Lignified tissue is indicated in red, unlignified cells are indicated in blue, and developing seeds are in green. d, e Cross-section of replum valve margin region of B. oleracea wild-type pod (d) and L2F1_A mutant pod (e); scale bars 200 μm
The 80 T0B. oleracea plants described above originated from the same transformation experiment, but differed in their culture period. A first batch of 41 T0 shoots was isolated four weeks after Agrobacterium inoculation, whereas a second batch of 39 T0 shoots was isolated 7 weeks after inoculation. Both dwarf lines were derived from the 7-week batch, supporting a recent report in rice [25] that obtained an increased proportion and variety of mutated cells by extending the culture period of rice calli by 4 weeks. Across different target genes, Mikami et al. [25] found a 3.7-fold increase in mutation frequencies between rice calli cultured for 1 month compared to 2 months. They hypothesize that this is due to a greater chance of inducing novel mutations in non-mutated cells [25]. Our results are consistent with this hypothesis which also implies that shorter selection periods during culture of calli could reduce the number of off-target mutations.
Cas9-induced mutations are stably transmitted to T2B. oleracea plants independently of the T-DNA construct
To examine the mutation frequency of the target locus BolC.GA4.a, the T1 progenies of lines L2F1_8.2 and L2E1_17.1 were screened for Cas9-induced mutations in Target 1 and 2 by PCR amplification of BolC.GA4.a followed by direct sequencing. Using the sequencing chromatograms it was possible to identify homozygous and heterozygous mutations. We detected mutations in the T1 progenies of L2F1_8.2, but not in L2E1_17.1. Heterozygous in-dels were observed in 68 of 90 L2F1_8.2 T1 progenies; however, no homozygous mutations were identified. Of these 68 T1 plants, 35 had mutations in Target 1, whereas Target 2 was mutated in 67 lines, suggesting a higher efficiency of the Target 2 sgRNA (Fig. 7b). None of the 90 T1 progenies inherited the complete 282-bp deletion between the two BolC.GA4.a target regions that was observed in the T0 generation.
Fig. 7.
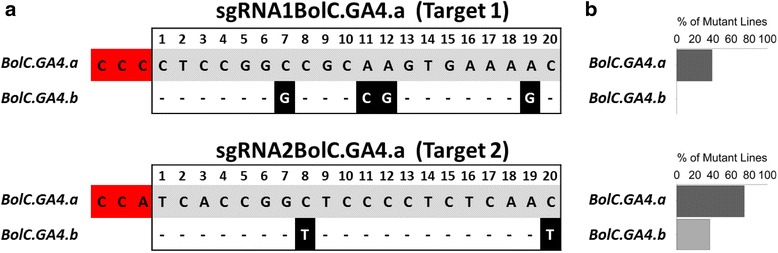
Frequency of on- and off-target Cas9 activity in L2F1_8.2 T1 Brassica plants. a The alignment of sgRNA1BolC.GA4.a and sgRNA2BolC.GA4.a target sequences in BolC.GA4.a with their corresponding sequences in BolC.GA4.b. Hyphens represent alignment matches while mismatches are shown in black highlight and white font. The PAM is highlighted in red and numbering of nucleotides is relative to the PAM. b Percentage of the T1 plants with mutations in BolC.GA4.a and BolC.GA4.b. Dark and light grey bars represent the percentages of BolC.GA4.a and BolC.GA4.b editing, respectively. N = 90 plants
The sgRNA targets were also sequenced in the T1 progenies of lines L2F1_A and L2F1_E that showed a dwarf phenotype. T1 plants from each of L2F1_A and L2F1_E were screened and found to carry a range of either homozygous or heterozygous mutations across the target regions in BolC.GA4.a (Additional file 2). Of the 39 plants screened, 20 had either homozygous mutations or a combination of two mutant alleles previously identified in the T0 plants; all of these plants displayed the dwarf phenotype at maturity (Fig. 6b). In the remaining 19 heterozygous lines we also identified the same mutations as in the T0 plants, including the large 68-bp deletion across Target 1 (Fig. 5b; Additional file 2). These results are consistent with stable transmission of Cas9-induced mutant alleles in B. oleracea.
To determine germline inheritance, we screened for the presence of the T-DNA construct in 12 individual T2 plants derived from eight homozygous mutant T1 lines (96 plants in total). Nine T2 plants which lacked the T-DNA construct were recovered (Fig. 4c). These plants all carried the same 6-bp deletion in Target 2, and wild-type allele in Target 1, as in the parental T1 plants (L2F1_E_B6, L2F1_E_C7, and L2F1_E_D8). The fact that the mutations in T1 plants were stably transmitted to the T2 generation in the absence of the T-DNA construct supports the germline inheritance of the Cas9- induced mutations in B. oleracea.
Off-target activity of RNA-guided Cas9 in T1 transgenic B. oleracea plants
Sequencing of BolC.GA4.b in the T1 progenies of T0-L2F1_8.2 revealed off-target activity in 32 out of 88 plants (36.4 %; Fig. 7b). This was restricted to Target 2 where the sgRNA contained two mismatches with BolC.GA4.b (Fig. 7a). No mutations were observed across Target 1 where the sgRNA contained four mismatches in the target region compared to BolC.GA4.b. In T0 lines with the dwarf phenotype (T0 lines L2F1_A and L2F1_E), we detected no off-target activity in BolC.GA4.b, indicating that the dwarf phenotype was due to mutations in BolC.GA4.a only.
In this study, we show that a single sgRNA (sgRNA2BolC.GA4.a) can simultaneously target two copies of GA4 despite the presence of a mismatch between the sgRNA and the BolC.GA4.b off-target sequence 8 bp from the PAM (Fig. 7a). This observation mirrors our results in barley, in which off-target activity was detected in HvPM19-3 due to the sgRNA in pPM19-1, designed to target HvPM19-1 and which has a mismatch 9 bp from the PAM (Fig. 3). This off-target activity was detected only in the progeny of the B. oleracea and barley lines with high on-target mutation frequencies. Given these results and the idea that on-target mutations may precede off-target mutations [26], it is tempting to speculate that higher on-target Cas9 activity positively correlates with higher off-target mutation frequencies.
These results differ from reports in wheat, where a single mismatch 3 bp from the PAM between MLO homoeologues limited off-target activity, although on-target mutation frequencies were relatively low (5.6 %; [12]). Previous studies found that a single mismatch within the 12 bp adjacent to the PAM could confer specificity in humans and other systems [3, 27]. However, others [28, 29] have shown that up to two mismatches, as well as small insertions and deletions, are tolerated within this sequence. Taken together, these results suggest that additional work is needed to decipher the key design rules and experimental parameters relating to on- and off-target mutations using the Cas9 system.
The presence of off-target activity can be considered a negative feature of the Cas9 system when specificity is sought. Several approaches have been suggested for the reduction of off-target activity. These include using truncated sgRNAs [30], a pair of Cas9 nickase mutants directed to opposing strands that require a pair of correctly positioned 20 bp DNA targets to produce a DSB [31, 32], and also the fusion of catalytically dead Cas9 (dCas9) to homodomains of a FokI nuclease dimer that will also only produce a DSB when both targets are in correct proximity [33, 34]. However, off-target activity can also be beneficial for targeting gene families [26] or closely related sequences. Our results suggest that a single sgRNA can simultaneously target multiple gene copies facilitating gene functional analysis by overcoming possible redundancy between the closely related sequences [35]. Importantly, we identified an individual transgene-free barley plant that had concurrent heterozygous mutations in the target (HvPM19-1) and off-target (HvPM19-3) genes. Many crop species are polyploid (for example, wheat, potato), have undergone recent whole-genome duplication events (for example, Brassica, maize; [36]), or have a high number of tandemly duplicated genes [37], such as the HvPM19 locus investigated in this study. Therefore, the potential to generate progeny with mutations limited to on-target sites, as well as progeny with both on- and off-target mutations, makes the RNA-guided Cas9 system especially relevant for functional analyses in crops.
Mutations in BolC.GA4.a affect the pod valve margin
Tissue patterning in the fruits of Arabidopsis and members of the Brassica genus is highly similar reflecting their close evolutionary relationship [38]. Seed dispersal in these species depends on formation of valve margin cells along the valve and replum borders that mediate fruit opening [39]. Since valve margins from Arabidopsis ga4 mutants fail to mediate efficient seed dispersal [13], we tested if the B. oleracea Cas9 lines presented here suffered from similar defects. Cross-sections stained with a combination of Safranin O and Alcian Blue revealed that in comparison to wild-type fruits, fruits from L2F1_A failed to pattern the valve margin region properly, such that valve cells replaced the valve margin cells in this line (Fig. 6c-e). As a result, these fruits would disperse their seeds less efficiently than wild type. Although less severe, this phenotype resembled the phenotype observed when another regulator of valve margin formation, BolC.IND.a, was downregulated by RNAi [38]. These data therefore demonstrate that the BolC.GA4.a function is conserved between B. oleracea and Arabidopsis and likely regulated in a similar fashion. They also demonstrate the potential for the use of RNA-guided Cas9 to target important traits in Brassica crops based on knowledge of gene function from model plants.
Conclusions
In this study, we demonstrate the use of RNA-guided Cas9 to induce targeted mutations in two crop species, B. oleracea and barley, and report stable transmission of the mutations across generations. We show that knock-out phenotypes can be recovered as early as the primary T0 generation, exemplifying the use of this technology for rapid analyses of gene function. We produced transgene-free barley and B. oleracea plants with stably-inherited mutations in the target gene, supporting the potential for downstream biotechnological applications. Both species showed off-target activity, despite the presence of at least one mismatch between the sgRNA and the paralogous gene. This led to the identification of a single barley plant with concurrent mutations in the target and off-target gene in the absence of the T-DNA construct. Our results suggest that experimental parameters relating to on- and off-target mutations need to be carefully considered and monitored, and that a single sgRNA has the potential to generate progeny with simultaneous knock-out mutations in paralogous genes. Given that crop genomes commonly contain multiple closely related sequences, the features described herein make RNA-guided Cas9 especially relevant for functional analyses in these species.
Materials and methods
Target locus selection and sgRNA design
Gene sequences for B. oleracea BolC.GA4.a (Bol038154) and barley HvPm19 (AF218627.1; [15]) were obtained from The Brassica Database [40] and the International Barley Sequencing Consortium [41] databases. For barley, sequence of the BAC clone HVVMRXALLmA0022M08 from the cultivar ‘Morex’ was kindly provided pre-publication by Dr Nils Stein (IPK). This BAC was annotated and four copies of HvPm19 were identified (HvPM19-1 to HvPM19-4). Target sequences that conformed to G(N)20GG were identified on sense and antisense strands in the coding sequence for BolC.GA4.a and for HvPM19-1 and HvPM19-3 and potential off-target sequences were detected via BLAST searches [40, 41]. Potential targets were also evaluated for the presence of non-CpG sensitive restriction site sequences predicted to be disrupted by Cas9 induced in-dels, which also had to be unique within a PCR amplicon. Final target sequences were chosen to be as specific as possible to the intended target sequence (that is, keeping the number of mismatches to off-target sequences high), close to the start codon, and to include an appropriate restriction site (Fig. 1). These targets were checked by PCR and Sanger sequencing (Additional file 3) in the varieties to be transformed (spring barley cultivar ‘Golden Promise’ and Brassica oleracea DH1012) to ensure that no polymorphisms existed between the sgRNA and the target G(N)20GG sequences. Single sgRNAs were used for barley HvPM19-1 and HvPM19-3, whereas two independent sgRNAs were targeted to the first exon of Brassica BolC.GA4.a. Barley ‘Golden Promise’ sequences for the three HvPM19 genes were deposited in GenBank (accession numbers KT336449-KT336451).
Construct assembly
The binary plasmid vector constructs were assembled using Golden Gate Modular Cloning (MoClo) [42]. We used Level 0 parts from the Golden Gate MoClo Plant Parts Kit (Addgene kit # 1000000047) and plasmids from Golden Gate MoClo Plant Toolkit (Addgene kit # 1000000044) described in Engler et al. [43]. Level 1 transcriptional units were assembled from Level 0 parts and these were subsequently assembled to make the plasmids vectors shown in Fig. 2. A detailed protocol for the assembly of binary vectors with multiple sgRNAs using the Golden Gate MoClo ToolKit and the identity of all plasmids used are given in Additional file 4. Annotated sequences of the plasmids made in this study are provided in Additional file 5 and are available at the non-profit plasmid depository AddGene (https://www.addgene.org/browse/article/14759/).
Plant transformation and screening of transgenic material
Barley (cv. ‘Golden Promise’) was transformed by Agrobacterium tumefaciens-mediated transformation of immature embryos as described by Harwood [44]. Brassica oleracea (DH1012) was transformed by Agrobacterium tumefaciens infection of 4-day-old cotyledonary petioles according to Hundleby and Irwin [45].
Primary transgenic T0 materials were screened using a modified restriction enzyme site loss method [46]. Briefly, for single sgRNA targets, genomic DNA was digested prior to PCR with a CpG-insensitive enzyme to remove wild-type template and thus favour the PCR amplification of mutant DNA where the restriction site had been lost. For Brassica, where a pair of sgRNAs was used, an additional screen was implemented; a CpG-insensitive restriction enzyme (AflII) was used prior to PCR to enrich for mutant DNA where the fragment between the two guides had been removed. PCR amplification across the region thus led to shorter PCR products than expected from a wild-type individual.
DNA was extracted according to Edwards et al. [47] from rooted shoots of less than 10 cm in height and quantified using a Nanodrop 8000 (Thermo Scientific). Genomic DNA (100 ng) was digested overnight with 20 units of the appropriate restriction enzyme shown in Fig. 1 (SapI, HaeIII, HphI, AflII (NEB); MaeIII (Roche)) and then purified using a Qiagen QIAquick Gel Extraction Kit (final elution with 25 uL of water). Purified digested DNA (5 μL) was used as PCR template to amplify across the target regions using gene-specific primers (Additional file 3). PCR products were confirmed by agarose gel electrophoresis, purified using the QIAquick Gel Extraction Kit, and Sanger sequenced (Eurofins MWG) to confirm the presence of in-dels. Where the amplicon was too short for direct sequencing, the PCR product was first cloned using the pGEMT-Easy kit (Promega) according to the manufacturer’s instructions and then sequenced with M13 universal primers.
The detection of mutations in T1 and T2 transgenic lines was performed though Sanger sequencing of PCR amplicons produced using genomic DNA template that was not digested prior to PCR (Additional file 3). Sequences were compared to wild type to detect the presence of homozygous in-dels. Chromatograms were also examined to identify overlapping traces in the region surrounding the PAM, indicative of the presence of mutations. The presence of the T-DNA construct was assessed in progenies of active lines by PCR amplification of the nptII CDS in Brassica and hptII CDS in barley (Additional file 3).
Phenotyping of B. oleracea transgenic lines
The 80 primary T0 transgenic lines and corresponding controls were grown in a controlled environment room with 16 h light (high-pressure sodium lamps with an average bench reading of 200 μmol/m2/s) at 12 °C and 8 h dark at 12 °C and constant 65–75 % humidity. Plant height was measured at final maturity. Seed pods at developmental stage 17 [48] were collected from dwarf line L2F1_A and the wild-type DH1012 control. Pods were fixed for 16 h in FAA solution (3.7 % formaldehyde, 5 % acetic acid, 50 % ethanol) and subsequently dehydrated through an ethanol series consisting of 50 %, 60 %, 70 %, 80 %, 90 %, 95 %, and 100 % ethanol for 30 min each at room temperature. The tissues were cleared with Histoclear (National Diagnostics,) and embedded in paraffin wax. Transverse sections 8 μm thick were cut using an RM 2055 rotary microtome (Leica) and mounted on Polysine™ slides (VWR International).The wax was removed using Histoclear and sections stained using an Alcian Blue/Safranin-O solution (0.05 % Alcian Blue and 0.01 % Safranin-O in 0.1 M acetate buffer (pH5.0)) as described by Østergaard et al. [49]. Sections were examined by light microscopy using a Zeiss Axioplan microscope and images captured using a Leica DFC 320 camera with Leica Application Suite software.
T-DNA copy number and presence/absence determination in transgenic barley
Quantitative real-time PCR was used to determine copy number (T0) and presence/absence (T2) of the T-DNA in transgenic barley and B. oleracea lines. The reaction compared the Cq values of an HptII (Fig. 2a) amplicon to a single-copy barley gene CO2 (Constans-like, AF490469) amplicon and the Cq values of an NptII amplicon to a single-copy B. oleracea gene GL2-like (Bol021421) within a single multiplexed assay (Additional file 3). The reactions used Thermo ABGene Absolute QPCR Rox Mix (Cat number AB1139) with the probes and primers at a final concentration of 200 nM (HptII and NptII) and 100nM (CO2 and GL2). The assay contained 5 μL DNA solution, and was optimised for final DNA concentrations of 1 to 10 ng/μL (5 to 50 ng DNA in the assay). PCRs were carried out in a Bio-Rad CFX96 machine (C1000 Touch). The detectors used were FAM-TAMRA and VIC-TAMRA for barley and HEX-BHQ1 and FAM-BHQ1 for B. oleracea. The PCR cycling conditions were 95 °C for 15 min (enzyme activation), 40 cycles of 95 °C for 15 s, and 60 °C for 60 s. Each sample was analysed twice and for presence/absence determinations, two independent DNA extractions of the T2 transgenic plants were used. Cq values were determined using the accompanying CFX96 software (version 3.1), with Cq determination set to regression mode. Values obtained were used to calculate T-DNA copy number according to published methods [50].
Acknowledgments
The authors would like to thank Dr Nils Stein (IPK), Xiao-Qi Zhang (Murdoch University), Guoping Zhang (Zhejiang University) and Mr Simon Lee (BGI-Shenzhen) for providing pre-publication access to barley Morex BAC sequences.
Funding
This work was supported by the Institute Strategic Programme grant BB/J004553/1 from the UK Biotechnology and Biological Sciences Research Council (BBSRC), the BBSRC and Engineering and Physics Research Council (EPSRC) Synthetic Biology Research Centre award (BB/L014130/1), by the John Innes Centre Innovation fund, the John Innes Foundation and the Gatsby Charitable Foundation.
Additional files
Chromatogram traces of T 0 -181 and T 0 -191 plants. (JPG 439 kb)
Analysis of BolC.GA4.a sequences in L2F1_A and L2F1_E T 0 and T 1 plants. (DOCX 25 kb)
Primers and probes used in this study. (XLSX 12 kb)
Detailed methods for assembly of binary vectors with multiple sgRNAs using the Golden Gate MoCloToolKit. (DOCX 28 kb)
Genbank files (.gb) of the 14 plasmids submitted to AddGene. (ZIP 41 kb)
Footnotes
Tom Lawrenson and Oluwaseyi Shorinola contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TL, OS and NP designed and generated the constructs used in this study; TL generated the transgenic lines; TL and OS performed the molecular characterisation of the transgenic lines; NS performed the phenotypic characterisation of B. oleracea lines; CL provided pre-publication access to barley BAC sequence; WH and CU designed the experiments. TL, OS, NS, LO, NP, CU and WH contributed to the writing of the manuscript and all authors reviewed the final version. All authors read and approved the final manuscript.
Contributor Information
Tom Lawrenson, Email: tom.lawrenson@jic.ac.uk.
Oluwaseyi Shorinola, Email: oluwaseyi.shorinola@jic.ac.uk.
Nicola Stacey, Email: nicola.stacey@jic.ac.uk.
Chengdao Li, Email: C.Li@murdoch.edu.au.
Lars Østergaard, Email: lars.ostergaard@jic.ac.uk.
Nicola Patron, Email: nicola.patron@sainsbury-laboratory.ac.uk.
Cristobal Uauy, Email: cristobal.uauy@jic.ac.uk.
Wendy Harwood, Email: wendy.harwood@jic.ac.uk.
References
- 1.Voytas DF, Gao C. Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol. 2014;12:e1001877. doi: 10.1371/journal.pbio.1001877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu Patrick D, Lander Eric S, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–78. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belhaj K, Chaparro-Garcia A, Kamoun S, Patron NJ, Nekrasov V. Editing plant genomes with CRISPR/Cas9. Curr Opin Biotechnol. 2015;32:76–84. doi: 10.1016/j.copbio.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Li J-F, Norville JE, Aach J, McCormack M, Zhang D, Bush J, et al. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotech. 2013;31:688–91. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotech. 2013;31:691–3. doi: 10.1038/nbt.2655. [DOI] [PubMed] [Google Scholar]
- 8.Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotech. 2013;31:686–8. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- 9.Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang D-L, et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci. 2014;111:4632–7. doi: 10.1073/pnas.1400822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J. 2014;12:797–807. doi: 10.1111/pbi.12200. [DOI] [PubMed] [Google Scholar]
- 11.Zhou H, Liu B, Weeks DP, Spalding MH, Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014;42:10903–14. doi: 10.1093/nar/gku806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotech. 2014;32:947–51. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- 13.Arnaud N, Girin T, Sorefan K, Fuentes S, Wood TA, Lawrenson T, et al. Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 2010;24:2127–32. doi: 10.1101/gad.593410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiang H-H, Hwang I, Goodman HM. Isolation of the Arabidopsis GA4 locus. Plant Cell. 1995;7:195–201. doi: 10.1105/tpc.7.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ranford JC, Bryce JH, Morris PC. PM19, a barley (Hordeum vulgare L.) gene encoding a putative plasma membrane protein, is expressed during embryo development and dormancy. J Exp Bot. 2002;53:147–8. doi: 10.1093/jexbot/53.366.147. [DOI] [PubMed] [Google Scholar]
- 16.Barrero JM, Cavanagh C, Verbyla KL, Tibbits JFG, Verbyla AP, Huang BE, et al. Transcriptomic analysis of wheat near-isogenic lines identifies PM19-A1 and A2 as candidates for a major dormancy QTL. Genome Biol. 2015;16:93. doi: 10.1186/s13059-015-0665-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bortesi L, Fischer R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv. 2015;33:41–52. doi: 10.1016/j.biotechadv.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41(20):e188. doi: 10.1093/nar/gkt780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA Endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartung F, Schiemann J. Precise plant breeding using new genome editing techniques: opportunities, safety and regulation in the EU. Plant J. 2014;78:742–52. doi: 10.1111/tpj.12413. [DOI] [PubMed] [Google Scholar]
- 21.Xu R-F, Li H, Qin R-Y, Li J, Qiu C-H, Yang Y-C, et al. Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci Rep. 2015;5:11491. doi: 10.1038/srep11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sparrow P, Dale P, Irwin J. Brassica oleracea. In: Wang K, editor. Agrobacterium Protocols. New York: Humana Press; 2006. pp. 417–26. [Google Scholar]
- 23.Mitchum MG, Yamaguchi S, Hanada A, Kuwahara A, Yoshioka Y, Kato T, et al. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 2006;45:804–18. doi: 10.1111/j.1365-313X.2005.02642.x. [DOI] [PubMed] [Google Scholar]
- 24.Brooks C, Nekrasov V, Lippman ZB, Van Eck J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014;166:1292–7. doi: 10.1104/pp.114.247577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikami M, Toki S, Endo M. Parameters affecting frequency of CRISPR/Cas9 mediated targeted mutagenesis in rice. Plant Cell Rep. 2015;34:1807–15. doi: 10.1007/s00299-015-1826-5. [DOI] [PubMed] [Google Scholar]
- 26.Endo M, Mikami M, Toki S. Multi-gene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol. 2015;56:41–7. doi: 10.1093/pcp/pcu154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotech. 2013;31:233–9. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584–92. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y, Cradick TJ, Brown MT, Deshmukh H, Ranjan P, Sarode N, et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473–85. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–84. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ran FA, Hsu Patrick D, Lin C-Y, Gootenberg Jonathan S, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–9. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen B, Zhang W, Zhang J, Zhou J, Wang J, Chen L, et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Meth. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 33.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotech. 2014;32:577–82. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotech. 2014;32:569–76. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borrill P, Adamski N, Uauy C. Genomics as the key to unlocking the polyploid potential of wheat. New Phytol. 2015;208:1008–22. doi: 10.1111/nph.13533. [DOI] [PubMed] [Google Scholar]
- 36.Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8:135–41. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 37.The International Wheat Genome Sequencing Consortium A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345:1251788. doi: 10.1126/science.1251788. [DOI] [PubMed] [Google Scholar]
- 38.Girin T, Stephenson P, Goldsack CMP, Kempin SA, Perez A, Pires N, et al. Brassicaceae INDEHISCENT genes specify valve margin cell fate and repress replum formation. Plant J. 2010;63:329–38. doi: 10.1111/j.1365-313X.2010.04244.x. [DOI] [PubMed] [Google Scholar]
- 39.Spence J, Vercher Y, Gates P, Harris N. ‘Pod shatter’ in Arabidopsis thaliana, Brassica napus and B. juncea. J Microsc. 1996;181:195–203. doi: 10.1046/j.1365-2818.1996.111391.x. [DOI] [Google Scholar]
- 40.The Brassica Database. [http://brassicadb.org/brad/blastPage.php].
- 41.International Barley Sequencing Consortium Database [http://webblast.ipk-gatersleben.de/barley/]
- 42.Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S. A modular cloning system for standardized assembly of multigene constructs. PLoS One. 2011;6:e16765. doi: 10.1371/journal.pone.0016765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engler C, Youles M, Gruetzner R, Ehnert T-M, Werner S, Jones JDG, et al. A golden gate modular cloning toolbox for plants. ACS Synthetic Biol. 2014;3:839–43. doi: 10.1021/sb4001504. [DOI] [PubMed] [Google Scholar]
- 44.Harwood W. A Protocol for High-Throughput Agrobacterium-Mediated Barley Transformation. In: Henry RJ, Furtado A, editors. Cereal Genomics. New York: Humana Press; 2014. pp. 251–60. [DOI] [PubMed] [Google Scholar]
- 45.Hundleby PAC, Irwin J. Brassica oleracea and B. napus. In: Wang K, editor. Agrobacterium Protocols. New York: Springer; 2015. pp. 287–97. [Google Scholar]
- 46.Voytas DF. Plant genome engineering with sequence-specific nucleases. Annu Rev Plant Biol. 2013;64:327–50. doi: 10.1146/annurev-arplant-042811-105552. [DOI] [PubMed] [Google Scholar]
- 47.Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–67. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Østergaard L, Kempin SA, Bies D, Klee HJ, Yanofsky MF. Pod shatter-resistant Brassica fruit produced by ectopic expression of the FRUITFULL gene. Plant Biotechnol J. 2006;4:45–51. doi: 10.1111/j.1467-7652.2005.00156.x. [DOI] [PubMed] [Google Scholar]
- 50.Weng H, Pan A, Yang L, Zhang C, Liu Z, Zhang D. Estimating number of transgene copies in transgenic rapeseed by real-time PCR assay with HMG I/Y as an endogenous reference gene. Plant Mol Biol Rep. 2004;22:289–300. doi: 10.1007/BF02773139. [DOI] [Google Scholar]


