Abstract
The Arf family of small GTPases regulates vesicular transport at several locations within the cell, and is in turn regulated by guanine nucleotide exchange factors (GEFs) via a conserved catalytic domain, termed the Sec7 domain. The catalytic activity of the Sec7 domain is well characterized in the context of a few GEFs acting at the periphery of the cell. This chapter describes techniques used to extend biochemical analysis of activity to the much larger GEFs acting on the Arf family in the core secretory pathway, using the activity of S. cerevisiae Sec7 on Arf1, regulating export from the trans-Golgi network (TGN), as a model. Complete methods for purification to near-homogeneity of all proteins required, including several Sec7 constructs and multiple relevant small GTPases, are detailed. These are followed by methods for quantification of the nucleotide exchange activity of Sec7 in a physiologically relevant context, including modifications required to dissect the signal integration functions of Sec7 as an effector of several other small GTPases, and methods for identifying stable Sec7-small GTPase interactions in the presence of membranes. These techniques may be extended to analysis of similar members of the Sec7 GEF subfamily in other species and acting elsewhere in the secretory pathway.
Keywords: Sec7, Arf1, vesicular transport, guanine nucleotide exchange factor (GEF), trans-Golgi network (TGN)
3 Introduction
Arf small GTPases are members of the Ras superfamily, highly conserved throughout evolution as regulators of vesicule biogenesis. Arf GTPases are regulated via bound guanine nucleotide: GTP is bound to active Arf, and Arf is inactivated by hydrolysis of GTP to GDP(Donaldson, Cassel, Kahn, & Klausner, 1992). Exchange of bound GTP for GDP, to activate Arf, is catalyzed by the Sec7 domain conserved in GEFs, including ARNO, Sec7/BIG1/2, Gea1/2/GBF1, and Yel1/EFA6, of the various Arf family proteins. The molecular mechanism of the Sec7 GEF domain is well characterized down to the structural level (Cherfils et al., 1998; Mossessova, Corpina, & Goldberg, 2003; Mossessova, Gulbis, & Goldberg, 1998; Renault, Christova, Guibert, Pasqualato, & Cherfils, 2002); regulation of its activity is achieved by regions of the protein external to the GEF domain, varying greatly between members of the Sec7 domain containing superfamily (Aizel et al., 2013; Cohen et al., 2007; Dehring, Adler, Hosseini, & Hicke, 2008; DiNitto et al., 2007; Padovani et al., 2014). Numerous functional assays and reagents have been developed for study of this regulation in model GEF proteins, notably the ARNO/Grp1/cytohesin GEFs acting at the cell periphery, including kinetic assays for Arf activation by native tryptophan fluorescence as well as more standard protein-protein interaction assays (Antonny, Huber, Paris, Chabre, & Cassel, 1997; Beraud-Dufour et al., 1998; Stalder et al., 2011).
Transport of proteins through the Golgi culminates in export from the trans-Golgi network, regulated by the Arf-GEF activity of Sec7/BIG1/2 (Morinaga, Tsai, Moss, & Vaughan, 1996; Sata, Donaldson, Moss, & Vaughan, 1998). The yeast TGN sorts not only biosynthetic traffic, but also endosomal recycling traffic, thus serving an additional role similar to that of mammalian recycling endosomes. Accordingly, in mammalian cells the Sec7 homologs BIG1/2 localize to both the TGN to recycling endosomes, with BIG2 apparently more specific for recycling endosomes (Shen, Xu, Fan, Pacheco-Rodriguez, Moss & Vaughan, 2006; Boal & Stephens, 2010; D’Souza, Semus, Billings, Meyer, Conger & Casanova, 2014). Across species, Sec7/BIG1/2 homologs range from ~1800 to ~2000 residues in size, rendering them challenging targets for biochemical purification. Thus, despite their critical role in the exocytic and recycling pathways, understanding the regulation of the Sec7/BIG1/2 Arf-GEFs has lagged behind that of the smaller peripheral Arf-GEFs.
Here, we describe in detail protocols for purifying a fully functional S. cerevisiae Sec7 construct, as well as several fragments thereof possessing partially regulated activity (McDonold & Fromme, 2014; Richardson, McDonold, & Fromme, 2012). We also describe the adjustments to previously developed GEF assays required for study of Sec7’s specific activity on Arf1. These assays form the basis for biochemical study of regulation of TGN trafficking, which may extrapolate to the study of other large proteins with similar behavior.
4 Materials
4.1 Equipment
Protocols presented here were developed on the listed brands; in most cases, alternatives with similar function are acceptable.
4.1.1 Large
New Brunswick Scientific Excella E25 refrigerated shakers
37°C incubator
- AKTA FPLC
-
○MonoQ 5/50 GL column
-
○HiTrap Phenyl HP column
-
○Superdex 200 10/300 GL column
-
○
- PTI QuantaMaster spectrofluorometer with attached water bath
-
○Hellma 105.250-QS 100 μl ultramicro quartz fluorescence cell
-
○
- Beckman Coulter Optima TLX ultracentrifuge
-
○TLA100 rotor
-
○7×20mm polycarbonate tubes (Beckman Coulter)
-
○
- Sorvall RT1 tabletop centrifuge
-
○Sorvall T41 rotor
-
○Thermo AC50.10 rotor
-
○
- Sorvall RC-3B refrigerated centrifuge
-
○H4000 rotor
-
○
Refrigerated (or cold room) microcentrifuge
- Rotovapor R II (Buchi)
-
○25 ml pear shaped flasks
-
○
Fisher Scientific Sonic Dismembrator Model 500 culture sonicator
- Avanti Mini-Extruder liposome extruder kit
-
○100 nm Nuclepore Track-Etch Membrane filters, 19 mm diameter (Whatman)
-
○10 mm PE drain disc (Whatman)
-
○
Cell culture hood
4.1.2 Consumable/semiconsumable
- Gravity flow columns
-
○Poly-prep chromatography columns (Bio-Rad)
-
○2.5 × 15 cm Econo-Column chromatography columns (Bio-Rad)
-
○
- Glass tubes and vials
-
○13×100 mm Pyrex test tubes with screw cap
-
○2 ml glass vials with screw cap
-
○
3 kD and 10 kD Amicon Ultra centrifugal filters/concentrators
SDS-PAGE gels and apparatus
Lab timer
4.2 Chemical reagents
4.2.1 Media
While the composition varies by location, tap water may often be used for synthesis of bacterial growth media to provide trace compounds helpful for growth. Naturally, all other solutions should be made with distilled water.
4.2.1.1 LB (Miller)
5 g tryptone
10 g yeast extract
5 g NaCl
Dissolve in 1 L tap water and autoclave
4.2.1.2 TB
In 900 mL tap water and autoclaved:
12 g tryptone
24 g yeast extract
8 ml 50% glycerol
Added separately, in 100 mL and filter sterilized:
12.5 g K2HPO4
2.3 g KH2PO4
4.2.2 Reagent stocks
- Lipid stocks
-
○1-25 mg/ml, as convenient, in chloroform.Use screw-top glass vial, sealed with PTFE thread sealant tape; transfer with Hamilton syringe.Store at −80°C.
-
○
Phenylmethylsulfonyl fluoride (PMSF): 100 mM in ethanol, store at −80°C.
Isopropyl β-D-galactopyranoside (IPTG): 1 M in water, store at −20°C.
Ampicillin: 100 mg/ml in water, store at −20°C.
Kanamycin: 30 mg/ml in water, store at −20°C.
Chloramphenicol: 35 mg/ml in ethanol, store at −20°C.
Coomassie stain and destain solutions
2× and 5× SDS-PAGE loading buffer
PreScission protease
4.2.3 Commercial reagents
Rosetta 2 competent cells (EMD Millipore)
Sf9 and Tni insect cells (Expression Systems)
Cellfectin II (Life Technologies)
Protease inhibitor cocktail tablets (Roche)
ESF 921 insect cell media (Expression Systems)
Penicillin-streptomycin (MP Biomedicals)
Bradford reagent / Bio-Rad Protein Assay
Ni-NTA agarose resin (Qiagen)
Glutathione resin (G Biosciences)
DEAE-Sephacel resin (GE Healthcare)
Toyopearl Phenyl-650M resin (Tosoh Bioscience)
IRDye blue protein stain (LiCor)
4.3 Software
Microsoft Excel
GraphPad PRISM
ImageJ
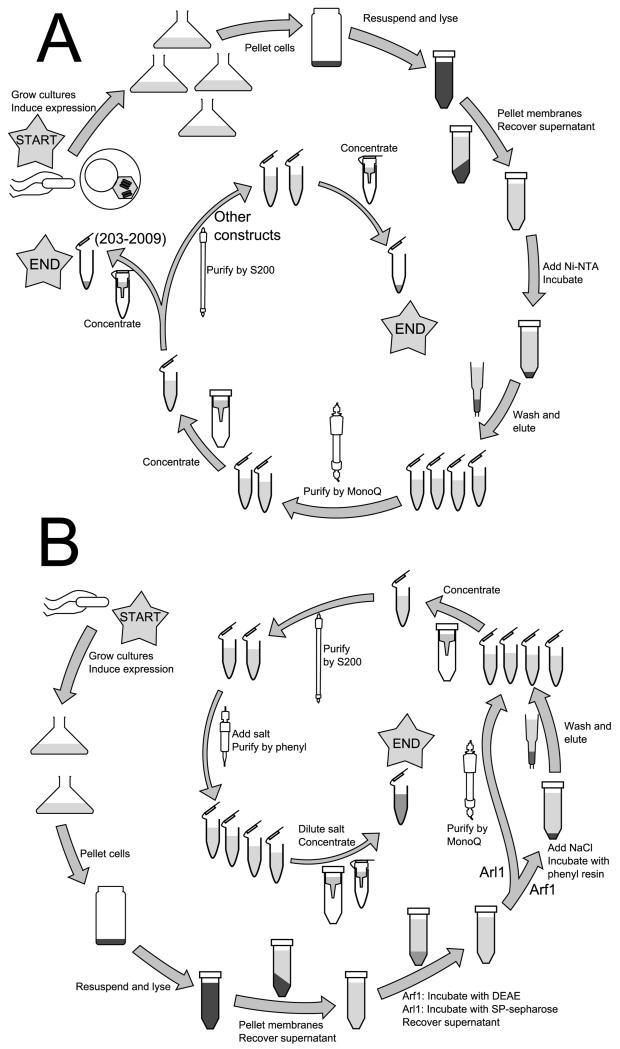
5 Purification of required proteins
Careful purification of the multiple proteins being studied is essential for obtaining quantifiable results and maintaining consistency of results over time. In most cases, affinity purification followed by one or two column purifications is sufficient to purify these constructs to homogeneity (i.e. a single band via SDS-PAGE/Coomassie); as myristoylated Arf1 and Arl1 are preferably not affinity tagged, alternative methods for purification to near-homogeneity are detailed. Excluding the time required to grow insect cells for the Sec7(203-2009) purification, all can be performed start to finish in 4-5 days, and with experience multiple constructs may be purified in parallel. Buffers used are detailed in Table 1.
Table 1.
Protein buffers
| Sec7 lysis buffer | Arf1 lysis buffer | Rab lysis buffer |
|---|---|---|
|
|
|
| Sec7 wash buffer | Low salt phenyl buffer | PreScission digest buffer |
|---|---|---|
|
|
|
| Sec7 elution buffer | High salt phenyl buffer | MonoQ buffer A |
|---|---|---|
|
|
|
| Gel filtration buffer | HK buffer | MonoQ buffer B |
|---|---|---|
|
|
|
5.1 Sec7
Full-length yeast Sec7 expresses poorly in insect cells, requiring removal of a non-essential low-complexity N-terminal region to achieve reasonable expression (Richardson et al., 2012). Following domain boundaries identified by conservation (Mouratou et al., 2005), constructs serially truncating C-terminal domains can be purified; however, with the exception of the isolated GEF domain, the N-terminal DCB and HUS domains cannot be removed without destabilizing the remainder of the construct.
5.1.1 Purification of Sec7(203-2009)
Due to its large size, Sec7(203-2009) is expressed in insect cells instead of bacteria, using the Bac-to-Bac viral expression system (Life Technologies). The protocol begins with plasmid pBCR314, Sec7(203-2009) cloned into pFastBac-HTb using BamHI and NotI to attach a TEV protease-cleavable N-terminal 6His tag.
5.1.1.1 Virus preparation
With Sec7(203-2009) cloned into pFastBac-HTb, baculovirus for infection of Tni insect cells is prepared according to the manufacturer’s directions, presented here only in overview. Plan for approximately two weeks to produce virus stock from plasmid, or 3-4 days to produce from frozen P1 viral aliquots.
5.1.1.1.1 Bacmid preparation
Transform pFastBac construct into DH10Bac and plate on LB/kanamycin/gentamycin/tetracycline.
Check colonies by colony PCR to confirm recombination; the Bac-to-Bac system supports LacZ-based screening, but this is generally not necessary.
Isolate bacmid DNA from confirmed colony culture; to assure sterility following benchtop work, stop after final ethanol/isopropanol precipitation and freeze as pellet.
5.1.1.1.2 Insect cell transfection
Transform bacmid into insect cells via Cellfectin II kit. Bacmid pellet may be suspended directly in media/Cellfectin solution. Do not include antibiotics in media!
Insect cell cultures from this point onward are infectious. Do not work side-by-side with uninfected cultures, and clean work area and tools carefully with bleach afterwards.
5.1.1.1.3 Viral amplification
Amplify to 10 ml plate culture, then 50 ml flask culture, allowing ~4 days each to produce virus.
1 ml aliquots from the 10 ml (P1) culture may be flash-frozen and stored at −80°C, allowing future P2 cultures to be produced easily.
Pellet cells from the 50 ml culture and recover virus-containing media to a clean 50 ml conical tube. This working stock of virus is typically stable for months at 4°C.
5.1.1.2 Protein expression
5.1.1.2.1 ~4-5 days beforehand
5.1.1.2.1.1 Insect cell amplification
Split high-density maintained stock of Tni cells into 100 ml ESF 921 cultures in 250 ml flask at 1*106 cells/ml.
5.1.1.2.2 1 day beforehand
5.1.1.2.2.1 Insect cell amplification
Split large stock of Tni cells into 1 L ESF 921 plus penicillin-streptomycin to a density of 0.5*106 cells/ml and shake at 27°C, 130 rpm. An autoclaved 2.6 L flask in a non-sterile shaker may be used.
5.1.1.2.3 Day 1
5.1.1.2.3.1 Insect cell infection
Add 1-10 ml viral P2 stock to 1 L Tni insect cell culture, now at 1*106 cells/ml.
NOTE: The volume of virus to be added is empirically determined and varies by batch of virus. This can be tested in 100 ml cultures while larger cultures are growing, requiring only SDS-PAGE/Coomassie analysis of raw lysates 2 days post-infection.
5.1.1.2.4 Day 3
5.1.1.2.4.1 Harvesting (48 hours post-infection)
Pellet cells in 1 L centrifuge flask for 10 minutes at 600 g.
Proceed directly into purification protocol; freezing pelleted cells for storage yields poor results.
5.1.1.3 Protein purification
Due to the limited stability of Sec7(203-2009), execution in a single day is recommended. All work should be performed at 4°C or on ice, as appropriate.
5.1.1.3.1 Lysis and lysate clearing
Add one protease inhibitor tablet to 50 ml lysis buffer and rotate until dissolved (~10 min).
Resuspend pelleted cells in prepared cold lysis buffer by vortexing briefly.
Transfer to 100 ml plastic beaker on ice and sonicate in three 15 sec rounds at 20% power (insect cells are fragile and do not require extensive sonication).
Centrifuge lysate for 30 min at 12,500 g in 50 ml conical tubes and discard pellet; supernatant will remain somewhat cloudy.
Optional: The lysate may be clarified further by pelleting in reusable 30 ml tubes in an SS34 or SA600 rotor at 28,000 g; this does not, however, appear to improve significantly the results of the following batch affinity purification.
5.1.1.3.2 Batch nickel affinity purification
Add 300 μl Ni-NTA resin and rotate ~1 hr.
Spin 3 minutes at 400 g to pellet resin. Remove supernatant, resuspend in 40 ml lysis buffer, and transfer resin to gravity flow column.
Wash once more with 50 ml lysis buffer, then with 50 ml wash buffer 5-10 ml at a time.
Elute in 10 × 1 ml fractions of elution buffer into 1.5 ml Eppendorf tubes and locate protein by Bradford assay. Discard fractions without protein.
5.1.1.3.3 Anion exchange
Equilibrate MonoQ 5/5 column sequentially with 5 column volumes (CV) MonoQ buffer A, 5 CV MonoQ buffer B, and 5 mL 90% buffer A/10% buffer B.
Centrifuge nickel resin eluate 10 minutes at top speed and 4°C to pellet any solid particles.
Load onto MonoQ column and elute in a gradient of 10-50% buffer B. Sec7(203-2009) elutes late at ~33% buffer B, with little contamination.
Concentrate to ~1 mg/ml in 10 kD Amicon Ultra spin concentrator, determine concentration, and flash-freeze in aliquots.
5.1.2 Purification of Sec7(203-1220) and Sec7(203-1017)
Shorter Sec7 constructs are purified from Rosetta 2 E. coli using a pET plasmid modified to fuse a TEV protease-cleavable 6His tag to the N-terminus of the protein.
5.1.2.1 Protein expression
5.1.2.1.1 Day 1
5.1.2.1.1.1 Bacterial transformation
Transform 1 μl miniprep into 50-200 μl competent Rosetta 2 cells and plate on appropriate selective media overnight. Use of chloramphenicol in the plates for the pRARE plasmid is not necessary.
5.1.2.1.2 Day 2
5.1.2.1.2.1 Preparation for large-scale cultures
Inoculate 100 ml overnight culture of LB and appropriate antibiotics, including chloramphenicol, with streak of transformant colonies.
Prepare 8 × 1 L sterilized TB in 2.6 L culture flasks.
5.1.2.1.3 Day 3
5.1.2.1.3.1 Large-scale culture growth
Add TB salts and antibiotics to TB and inoculate 1:100 with overnight bacterial culture.
Grow to OD600 of at least ~3.5; OD is best measured at later stages at a 1:10 dilution in water. Culture growth will slow down greatly above an OD of 2.
Turn shaker temperature to 18°C and wait 45 minutes for cultures to cool.
Induce expression with 250 μM IPTG and express overnight.
5.1.2.1.4 LB variant
While TB is preferred due to the much denser final cultures, LB may be more convenient if a lower yield is acceptable. Reduce temperature of cultures at OD600 of ~.5. In lieu of growing a starter culture overnight, growth for ~5h followed by 2-3h of large-scale culture growth permits reduction of total time required by one day.
5.1.2.1.5 Culture storage
Bacterial cultures can be stored indefinitely at −80°C by pelleting, resuspending in minimal volume of lysis buffer plus 1 mM PMSF, and transferring to 50 ml conical tubes for flash-freezing in liquid nitrogen.
5.1.2.2 Protein purification
5.1.2.2.1 Day 3
5.1.2.2.1.1 Lysis and lysate clearing
All work should be performed on ice!
Following overnight expression, pellet bacteria in 1 L centrifuge bottles 15 minutes at 3000 g and 4°C. 1 L culture should yield 10-15 ml pellet, or 4 ml if LB is used.
Resuspend in 25 ml lysis buffer per L of culture and transfer to 600 ml plastic beaker.
Wash centrifuge bottles with ~25 ml lysis buffer/4 bottles to recover remaining pellet.
Add PMSF to 1 mM.
Lyse by sonication: 4 rounds of 35 pulses at 100% power, allowing it to cool for several minutes between rounds.
Pellet membranes: 20 minutes at 12,500 g; a Thermo AC50.10 rotor permits use of disposable 50 ml conical tubes for centrifugation, eliminating the need to clean dense membrane pellets from reusable centrifuge tubes; if unavailable, 30 ml reusable centrifuge tubes may be used in an SA 600 or SS 34 rotor at 28,000 g for more efficient pelleting. Pool supernatant in 50 ml conical tubes.
5.1.2.2.1.2 Batch nickel affinity purification
Add 250-500 μl Ni-NTA resin per 50 ml conical tube and rotate at 4°C ~1 h.
Spin at 400 g to pellet resin. Remove supernatant. Combine all resin in single conical tube with 45 ml lysis buffer.
Pellet and wash again with 50 ml lysis buffer.
Wash twice with 25 ml wash buffer.
Transfer to PolyPrep column using cut-off pipet tip and let drain.
Elute in 4 × 1ml elution buffer and locate protein by Bradford.
OPTIONAL: Add TEV and digest overnight to remove 6His tag.
5.1.2.2.1.3 Anion exchange
Equilibrate MonoQ 5/5 column sequentially with 5 CV MonoQ buffer A, 5 CV MonoQ buffer B, and 5 mL 90% buffer A/10% buffer B.
Centrifuge nickel resin eluate 10 minutes at top speed and 4°C to pellet any solid particles. Dilute 1:1 with MonoQ buffer A to reduce salt concentration.
Load onto MonoQ column and elute in a gradient of 10-50% buffer B. Smaller Sec7 fragments elute early in the gradient; check fractions by SDS-PAGE/Coomassie and pool fractions containing Sec7 with minimal contamination. Protein can be left overnight at 4°C.
5.1.2.2.2 Day 4
5.1.2.2.2.1 Gel filtration
Equilibrate Superdex 200 10/300 column with 1.2-2 CV 15% MonoQ buffer B/85% MonoQ buffer A.
Meanwhile, concentrate protein in 10 kD Amicon Ultra concentrator to .5-1.5 ml.
Run protein over column 0.5 ml at a time, collecting 0.5 ml fractions; multiple runs may be collected into the same tubes.
Locate protein by SDS-PAGE/Coomassie, concentrate to 1 mg/ml or higher (Table 2), and flash-freeze in aliquots.
Table 2.
TGN liposome composition
| Lipid | Molar % |
|---|---|
| DOPC | 24 |
| POPC | 6 |
| DOPE | 7 |
| POPE | 3 |
| DOPS | 1 |
| POPS | 2 |
| DOPA | 1 |
| POPA | 2 |
| PI | 30 |
| PI(4)P | 1 |
| CDP+DAG | 2 |
| PO-DAG | 4 |
| DO-DAG | 2 |
| Ceramide (C18) | 5 |
| Cholesterol | 10 |
5.2 Small GTPases
5.2.1 Purification of soluble constructs
S. cerevisiae ΔN17-Arf1 and ΔN18-Arl1, lacking their membrane anchor, may be purified as 6His-tagged fusions following the protocol for bacterial expression of Sec7 fragments. Add 1 mM MgCl2 to all buffers, and use 3 kD concentrators in place of 10 kD to avoid protein loss.
5.2.2 Purification of myristoylated Arf1
Full-length myristoylated S. cerevisiae Arf1 is purified from E. coli by a modification of existing procedures for untagged protein purification (Ha, Thomas, Stauffer, & Randazzo, 2005), using a strain co-expressing Arf1 and NMT1. The protocol scales well to volume of culture, but is limited by the amount of DEAE-sephacel on hand for later steps. Freezing of cultures following expression reduces yield and purity and is not recommended.
5.2.2.1 Protein expression
5.2.2.1.1 Day 1
5.2.2.1.1.1 Thaw bacteria
Streak out bacteria from frozen glycerol stock on LB/ampicillin/kanamycin and grow overnight.
Alternatively, freshly cotransform competent BL21(DE3) (not Rosetta!) strain with GTPase expression plasmid and pNMT1.
5.2.2.1.2 Day 2
5.2.2.1.2.1 Grow cultures
Inoculate 100 ml LB/ampicillin/kanamycin and grow ~5 hours.
Meanwhile, autoclave 2 × 1L LB in 2.6 L shaker flasks.
Once starter culture density reaches or exceeds an OD600 of ~0.5, add ampicillin and kanamycin to 1 L cultures, inoculate 1:100 with starter culture, and shake at 37° until OD600 reaches ~.6 (3-5 hours).
Add small spatula tip of solid myristate (~ 10 mg for ~50 μM final concentration).
Turn temperature to 18°C, wait 45 minutes for cultures to cool, and induce expression with 1 mM IPTG. Express overnight.
5.2.2.2 Protein purification
5.2.2.2.1 Day 3
5.2.2.2.1.1 Cell lysis
All work should be performed on ice.
Following overnight expression, pellet bacteria in 1 L centrifuge bottles 15 minutes at 3000 g and 4°C.
Resuspend in 20 mL lysis buffer per liter of culture and pool; add PMSF to 1 mM.
Sonicate to lyse: 3 rounds of 20 1-second pulses at 100% power, allowing to cool between rounds.
Pellet membranes 20 minutes at 12,500 g.
5.2.2.2.1.2 DEAE purification in batch
Note: Arf1 does not bind DEAE in lysis buffer; this step removes much of the primary contaminant, EF-Tu.
Note: DEAE, once used, may be saved and regenerated for reuse.
Equilibrate 12.5 ml DEAE per liter of culture in lysis buffer in batch.
Add DEAE to cleared lysate and rotate ~1 h at 4°C.
Pellet DEAE 5 minutes at 400 g; remove and save supernatant.
Wash DEAE with equal volume lysis buffer, spin, and combine supernatant with previous.
Centrifuge pooled supernatant at 2700 g 10 minutes to remove residual DEAE.
5.2.2.2.1.3 Phenyl purification in batch
Bring salt concentration in pooled supernatant to 3 M by addition of solid NaCl.
Quantify protein concentration by Bradford.
Equilibrate enough phenyl resin to bind protein in high salt phenyl buffer.
Add phenyl resin to protein solution and rotate ~1 h at 4°C.
Transfer to gravity flow column and let drain.
Wash with 50 ml high salt phenyl buffer.
Elute with phenyl elution buffer in 6-10 × 1 CV fractions; locate protein by small-scale Bradford.
5.2.2.2.2 Day 4
5.2.2.2.2.1 Gel filtration
NOTE: As time permits, gel filtration may be performed on day 3, or started and run overnight.
During the phenyl purification, equilibrate Superdex 200 10/300 column (or, optionally, Superdex 75) with 1.2-2 CV gel filtration buffer.
Concentrate phenyl eluate to 1 ml in 3 kD Amicon Ultra concentrator; or, if the starting volume is large, a pressure cell concentrator may be more convenient.
Run in 2 × 0.5 ml loads over S200 column, collecting .5 ml fractions in 1.5 ml Eppendorf tubes; both runs may be collected into the same tubes.
Check fractions by SDS-PAGE/Coomassie and combine appropriate fractions.
5.2.2.2.2.2 Gradient phenyl purification
Equilibrate 1 ml HiTrap phenyl column with 5 CV high salt phenyl buffer, 5 CV low salt phenyl buffer, and 5 CV high salt phenyl buffer again.
Bring [NaCl] of pooled gel filtration fractions to 3M with solid NaCl.
Run protein over phenyl column in gradient from high salt buffer to low salt buffer.
Check fractions by SDS-PAGE/Coomassie and combine appropriate fractions; peak is likely to be near the end of the gradient and fairly wide.
Dilute protein in low salt buffer to reduce [NaCl] to ~150 mM. This may be performed over multiple concentration/dilution repeats.
5.2.2.2.2.3 Final cleanup
Concentrate protein to desired final concentration.
Aliquot and flash-freeze; protein is quite stable and can withstand multiple freeze/thaw cycles or at least a week of storage at 4°C.
5.2.3 Purification of myristoylated Arl1
Myristoylated full-length S. cerevisiae Arl1 may be purified using the myrArf1 protocol with the following modifications:
Use 1 ml SP-sepharose resin per 7 mg protein in lysate in place of DEAE resin.
Omit batch phenyl resin step.
Optional: after SP-sepharose, run over 1 ml MonoQ FPLC column; will elute early and possibly flow through.
Concentrate to the extent practical and gel filter via S200 (will require several runs!).
Optional: reconcentrate gel filtered fractions and gel filter a second time.
5.2.4 Rab GTPases
Rab small GTPases, which constitutively localize to membranes via a C-terminal prenylation site, are purified via an N-terminally fused cleavable GST tag, and with a C-terminal 7His tag replacing the prenylation site to facilitate membrane recruitment in vitro. pGEX-6P-1 with a PreScission-cleavable GST tag is the preferred host vector; use of vectors with alternative site-specific cleavage sites is effective, but requires an additional step to remove the protease following cleavage.
5.2.4.1 Days 1-3
Follow protocol for growth and expression of Sec7 fragments in TB with the following modifications:
Substitute pGEX GTPase plasmid for Sec7 plasmid.
Use ampicillin and chloramphenicol for selection.
Reduce culture volume to 1-2 L.
Increase IPTG to 500 μM in induction.
5.2.4.2 Day 4
5.2.4.2.1 Cell lysis
As per Sec7 fragments, substituting Rab lysis buffer.
5.2.4.2.2 GSH-agarose affinity purification
Equilibrate 500 μl GSH-agarose via 3 × 1 ml washes in Rab lysis buffer and add to lysate.
Rotate 3 hr or more at 4°C.
Pellet resin, remove supernatant, and transfer resin to 15 ml conical tube.
Wash 3 × 5 ml with lysis buffer.
Wash with 5 ml PreScission digest buffer.
Resuspend resin in 500 μl PreScission digest buffer, add 25 μg PreScission protease, and rotate at 4°C overnight.
Elute supernatant, wash with additional 500 μl digest buffer and pool.
Optional: exchange into HKM or other assay buffer.
Concentrate to desired concentration using 3 kD filter, aliquot, and flash-freeze for storage at −80°C.
6 Liposomes
6.1 Composition
Membrane lipid composition varies considerably by organelle (Klemm et al., 2009); while simple PC/PS or Folch liposomes are commonly used for convenience, for best results lipids should be mixed to match the organelle being studied. An appropriate mix for study of TGN-localized proteins is given in Table 2.
6.1.1 Additives
Lipid components can be adjusted as desired to add or remove components; adjust molar fraction of PI and/or PC to compensate.
6.1.1.1 Nickel
As an alternative to native recruitment of small GTPases to membranes, Ni2+-DOGS may be added to liposomes at 5% total lipid to recruit proteins fused to a 6His tag. This is convenient for studying nucleotide-independent protein binding or Rabs in the absence of their GDIs, but requires extra care to be taken to completely cleave the frequently used 6His tags from all other proteins!
6.1.1.2 DiR
For quantification of lipids in solution, the lipophilic dye DiR (Life Technologies) or other lipid-segregating fluorophore may be added to liposomes at 1% total lipid. DiR does not interfere with any protocol presented here.
6.2 Size
Liposome preparations are notoriously heterogenous and difficult to compare batch-to-batch. One significant confounding factor is the generation of multilamellar vesicles, decreasing the effective liposome concentration by sequestering a significant fraction of the surface area. This can be minimized by extruding liposomes through a 100 nm filter in place of the more commonly used 400 nm filter.
6.3 Liposome production
6.3.1 Lipid mixes
The chloroform-lipid solutions must not contact plastic!
Set up a simple wash station for Hamilton syringes with two bottles of chloroform labeled Wash A and Wash B and an empty bottle for disposal.
One component at a time, transfer lipid solutions from storage tube to a glass test tube, and wash the Hamilton syringe twice in Wash A and twice in Wash B, discarding into the disposal bottle each time.
.5 ml of 2 mM lipid solution generates 1 ml of 1 mM liposome working stock. Using large Hamilton syringe, transfer into 25 ml pear flask for immediate liposome generation, or 1 ml screwtop glass vial for long-term storage at −80°C.
Evaporate chloroform via rotovap. When storing aliquots in glass vials, monitor to ensure that the boiling solution does not splash out of the vial.
6.3.2 Solubilization and extrusion
Add 1 ml appropriate buffer to dried lipid in pear shaped flask and incubate 1 h to overnight at 37°C. Once ready, lipid should resuspend with gentle swirling of the flask.
Assemble Avanti lipid extruder as per instructions, including filters and filter supports.
Wash twice with water, and twice with buffer.
Extrude lipid suspension back and forth for 25 passes through the filter; an odd number of passes is essential to ensure that any unextrudable material is separated from the final liposome stock.
Store liposome stock in 1.5 ml Eppendorf at 4°. Liposomes are typically stable for at least a month without obvious aggregation or changes in assay behavior.
Clean extruder and syringes twice with water and twice with 10% ethanol. Dry and store.
7 Measuring GEF activity
7.1 Native tryptophan fluorescence of Arf1
Conveniently, Arf1 exhibits a change in tryptophan fluorescence when transitioning between GDP- and GTP-bound states (Antonny et al., 1997), presumably largely due to the disruption of mutual quenching of tryptophans 66 and 78. This permits study of the nucleotide bound state of the unmodified protein in real time.
7.1.1 Working stocks
For convenience, working stocks of all components should be made at 15× final concentration to eliminate the need to adjust pipettor volume during the protocol. Make enough to do all trials in triplicate, plus overhead.
Results are not necessarily directly comparable between liposome batches; if necessary, prepare and pool multiple 1 ml liposome batches before starting!
Optimal concentration of each component varies by experiment, but sensible starting points are as follows:
Sec7 constructs: 1.5 μM stock for 100 nM final concentration.
Arf1: 10 μM stock for 667 nM final concentration.
GTP: 3 mM stock for 100 μM final concentration.
7.1.2 Data collection
7.1.2.1 Notes
Using a 150 μl reaction volume in a 100 μl cuvette reduces the risk of the meniscus confounding data collection.
Exact timing of reagent addition to the reaction, followed by quick and consistent pipetting to mix, is enormously helpful in data analysis later to identify time of addition within the ~10s gap in data collection. Open the fluorometer with a few seconds to spare, and keep a lab timer running during the experiment to assist in timing. While stirred cuvettes and injection systems are available that eliminate this gap, they also require 5-6× the reaction volume (Antonny et al., 1997).
2 minutes between reagent additions is typically sufficient for the trace to stabilize; adjust as necessary.
Each separate condition should be collected in at least triplicate. Collect each condition once before moving on to the next set of duplicates, and vary order of collection, to minimize systematic error.
A downward drift of the exchange reaction may be observed in data collection, potentially due to photobleaching or settling of liposomes. This is typically linear in nature (which can be confirmed by continued monitoring of a completed reaction), and can easily be corrected for in data analysis.
7.1.2.2 Before starting
Turn on fluorometer and light source according to manufacturer’s directions.
Turn on water bath to pre-equilibrate system to 30°C.
Thaw all reagents, spin 10 minutes at top speed in refrigerated microfuge, and prepare working stocks.
7.1.2.3 Protocol
Collect native tryptophan fluorescence at 297.5 nm excitation, 340 nm emission according to fluorometer instructions.
Add 90 μl HKM and 30 μl liposomes to 100 μl fluorometer cuvette. Set P200 volume to 80 μl and pipet to mix. Insert cuvette into fluorometer.
Simultaneously start fluorometer trace and lab timer, counting upwards from 0.
30 seconds: add 10 μl GEF construct and pipet to mix.
2:30: add 10 μl Arf1 and pipet to mix.
4:30: add 10 μl GTP and pipet to mix.
Collect trace until reaction is complete.
Empty cuvette and wash with 3 × 150 μl HKM buffer or vacuum wash apparatus.
7.1.2.4 Optional: protein fluorescence calibration
To compensate for pipetting error, exact GEF concentration may be calculated by its native tryptophan fluorescence.
Prepare 3-4 concentrations of GEF solution bracketing the working concentration.
Prepare exchange reaction in fluorometer as usual up to the point of GEF addition.
Collect baseline in fluorometer, add GEF dilution, and collect trace until ~30s after it stabilizes. Wash cuvette and repeat.
Plot protein concentration vs. background-subtracted protein fluorescence to obtain a standard curve usable to calibrate further assays.
7.1.3 Data analysis
With the exception of the curve fitting, analysis may be performed in the spreadsheet software of choice; here, Excel is assumed due to general familiarity and ubiquity.
Curve fitting and statistical analysis are performed in GraphPad PRISM, also capable of plotting the data if desired.
7.1.3.1 Curve extraction
Extract the trace from addition of GTP to the end to a new set of columns; delete data points preceding closing the fluorometer.
Set addition of GTP as t=0 (here, subtracting 270 sec from all timepoints).
Subtract native fluorescence preceding GTP addition to zero baseline, accounting for protein dilution (here, average fluorescence from 236 sec to 265 sec and multiply by 140/150 to obtain baseline; subtract this value from fluorescence).
If protein concentration has been calibrated to fluorescence, average its stabilized baseline, subtract background, and compare to the standard curve to obtain concentration.
7.1.3.2 Curve fitting
Copy zeroed curves to PRISM.
Perform a nonlinear regression to fit the curves to Fluorescence = Baseline + Plateau(1 – e(kreact*[GEF])*t) – Drift * t, in which all values but fluorescence and t are constants. If no downward drift is seen in the data, the final term should be eliminated. [GEF] should be set to the expected final concentration, or if available to the value calculated from the early fluorescence trace. Remaining values should be fit to the data. If fits are good, the baseline should be below 0 (due to the delay in beginning data collection) and plateaus should be reasonably consistent within a given condition (but will vary significantly between different Sec7 constructs).
Visually check the fit of each calculated curve to the raw data. Divergence may occur at short time points, an effect either of reduced initial rate due to positive feedback or of misfitting of the linear drift term. In the former case, redo the curve fit using only later time points; in the latter case, extending the data collection time well past completion will make the linear drift component of the curve more obvious and easier to fit.
7.1.3.3 Curve comparison
To compare triplicate experiments, curves must be individually fit and the calculated kreact values subsequently subjected to statistical analysis. As baseline and plateau values vary by trace, plotting raw traces together on a chart is often not helpful. However, taking baseline fluorescence as fully GDP-bound Arf1, and (baseline+plateau) fluorescence as fully GTP-bound Arf1, traces may be scaled together by plotting vs. time; if desired, triplicate curves may be averaged point-by-point here for presentation.
7.2 Variation: Small GTPase crosstalk
Measurement of the effect of another protein on Sec7-mediated nucleotide exchange is easily accomplished by addition of another step preceding Arf1 addition in which the additional protein is added and the trace is allowed to stabilize; initial HKM volume must of course be reduced to compensate.
This process becomes more complicated when a small GTPase itself requiring activation by GTP exchange in the presence of membranes is used to stimulate Sec7. Here, the protocol is altered to the following:
Start: 60 μl HK, 30 μl liposomes, 10 μl .05 M EDTA
00:30 – Add 10 μl GTP
01:30 – Add 10 μl GEF construct
03:30 – Add 10 μl small GTPase
Allow exchange to complete and add 10 μl .2 M MgCl2.
+02:00 – Add 10 μl myrArf1
Collect trace until completion of exchange.
Several variations on this protocol are possible:
If the GEF construct is particularly unstable at reaction temperature, adding it after addition of MgCl2 may improve results.
If the nucleotide-bound state of the first GTPase cannot be determined by tryptophan fluorescence, typically a 15 minute EDTA-mediated exchange is sufficient.
Addition of EDTA after the GEF-mediated exchange and monitoring subsequent exchange activity for a third time may be used to confirm that the reaction has proceeded to completion.
When Arf1 is itself used for monitoring of its positive feedback behavior, HKM may be used in place of HK and EDTA/MgCl2 may be omitted due to the presence of Sec7 to mediate both exchange reactions.
8 Identifying protein-Arf1 interactions
8.1 Notes
For assessment of protein-mediated recruitment to liposomes, two methods of vesicle-specific recovery are available, each with its own advantages and disadvantages. Flotation of liposomes on a sucrose gradient yields exceedingly clean results, but demands some degree of manual dexterity and protein input must be balanced between using enough protein to detect on a gel and not using so much that the liposomes no longer float. Pelleting of liposomes is a more straightforward and frequently more quantitative assay, but is easily confounded by high background levels of protein pelleting even in the absence of liposomes. In the case of Sec7, this background pelleting is such that liposome floatation is strongly recommended, and is solely presented here.
8.2 Liposome flotation
Typically, liposome flotation will compare binding of several constructs with GDP and GMP-PNP bound small GTPase.
Liposomes must include DiR dye (or alternative) for quantification of recovery.
8.2.1 Nucleotide exchange
Set up exchange reactions and incubate 1 h on benchtop or 20 min at 30°C, covered with foil:
20 μl 1 mM liposomes in HK buffer
6 μg small GTPase
1 μl .05M EDTA
1 μl 10 mM nucleotide
HK buffer to 80 μl minus volume to be added in following step
8.2.2 Liposome binding
Add remaining components to reactions and incubate 10 min to 1 h on benchtop, covered with foil:
1 μl .2M MgCl2
6 μg protein to be assayed for interaction (adjust as necessary)
8.2.3 Discontinuous sucrose gradient flotation
Using cutoff pipet tip due to viscosity, add 50 μl 2.5 M sucrose in HKM to each sample, mixing thoroughly.
Transfer 100 μl of mixture to Beckman 7 × 20 mm PC ultracentrifuge tube, saving remainder as input sample; ultracentrifuge tubes can be conveniently held in empty 1.5 ml Eppendorf tubes.
Carefully overlay with 100 μl .75M sucrose in HKM.
Carefully overlay with 20 μl HKM.
Load into TLA100 rotor and ultracentrifuge 100 krpm at 20°C for 20 min; set acceleration and deceleration speed to as slow as permitted by the ultracentrifuge.
Recover 30 μl from top of gradient. If possible, visually locate liposomes, which may clump and/or adhere to the side of the tube, and recover appropriately; otherwise, liposomes are dispersed in the top fraction of the gradient.
8.2.4 Quantification of recovery
Transfer 2 μl each input and float fraction to 48 μl .1% Triton X-100 in 96-well plate.
Quantify fluorescence at 800 nm in LiCor Odyssey (or other plate imager appropriate to the dye used) to quantify relative lipid recovery of each flotation reaction.
8.2.5 Analysis
Add 5× SDS-PAGE sample buffer to float samples, and 2× SDS-PAGE sample buffer to input samples.
Load float samples on SDS-PAGE such that all lanes contain the same amount of recovered lipid, and as much as possible of the sample with lowest recovery is loaded (typically 15 μl, but dependent on apparatus). Load 5 μl each input.
Stain with Coomassie, or if necessary analyze by Western. Silver stain typically works poorly with the float fractions. A representative result is shown in Figure 3.
Figure 3.
Demonstration of interaction of Sec7 and Arf1 by liposome flotation
Liposomes of TGN composition are incubated with Arf1 possessing or lacking its membrane interaction domain and binding either GDP (inactive) or GMPPNP (GTP*, active). Sec7(203-2009) is added and flotation performed, followed by SDS-PAGE and Coomassie staining.
Optional: band quantification can be improved by use of a fluorescent protein stain such as IRDye in place of Coomassie, followed by fluorescent imaging.
9 Supporting protocol
9.1 Protein concentration quantification
9.1.1 Quick analysis of elution fractions
Per fraction, combine 7 μl water with 2 μl Bradford reagent.
Pipet onto parafilm as single drop
Add 1 μl eluate and briefly mix; incubate ~30 seconds.
Blue color indicates presence of protein; clear color indicates minimal protein.
9.1.2 Bradford semiquantitative analysis
In a 1 ml disposable spectroscopy cuvette, combine 800 μl water with 200 μl Bradford reagent.
Add protein sample until bluish-grey, mixing well; depending on sample, may require anywhere from 1 μl to 100 μl.
Incubate 5-10 minutes.
Measure A595. Protein concentration in mg/ml roughly equals 20 * A595 / μl added.
OPTIONAL: compare to standard curve of lysozyme or other protein (not BSA!). In practice, however, protein-to-protein variability is such that if greater accuracy than the rough calculation above is desired, A280 is preferable.
9.1.3 A280
Calculating protein concentration by A280 is the method of choice for highly purified protein solutions.
Blank a 100 μl UV-transparent cuvette with 150 μl matched protein buffer; ideally, quantify immediately after concentration and use the concentrator buffer to blank.
Add 3-10 μl protein and measure A280.
Use ProtParam (Gasteiger et al., 2005) and the primary sequence of the protein being measured, including tags if present, to calculate the expected A280 at 1 mg/ml; values for constructs used here are presented in Table 3.
Table 3.
Physical properties of protein constructs
| Construct | MW (kDa) | A280 at 1 mg/ml * |
|---|---|---|
| 6His-TEV-Sec7 203-2009 | 208.8 | .731 |
| 6His-TEV-Sec7 203-1220 | 116.6 | .530 |
| 6His-TEV-Sec7 203-1017 | 93.8 | .462 |
| myrArf1 | 20.5 | 2.14 |
| 6His-TEV-ΔN17Arf1 | 21.1 | 2.16 |
| ΔN17Arf1 | 18.8 | 2.34 |
| myrArl1 | 20.4 | 2.15 |
| 6His-TEV-ΔN18Arl1 | 20.8 | 1.92 |
| ΔN18Arl1 | 18.5 | 2.08 |
A280 is estimated for the primary sequence using ProtParam (Gasteiger et al., 2005), with absorbance of a single guanine nucleotide (9000 M−1 cm−1) (Sontag, Sage, & Erickson, 2009) added for the small GTPases.
9.1.4 Coomassie gel analysis
If necessary, protein samples with significant impurities may be quantified by Coomassie staining of an SDS-PAGE gel if a very similar pure construct is available, such as when comparing activity of partially destabilized mutants with bound bacterial chaperones to activity of the fully purified wild-type construct. Comparison of band density of the impure construct to that of the pure construct with known concentration permits quantification of only the desired component of the impure mixture of proteins.
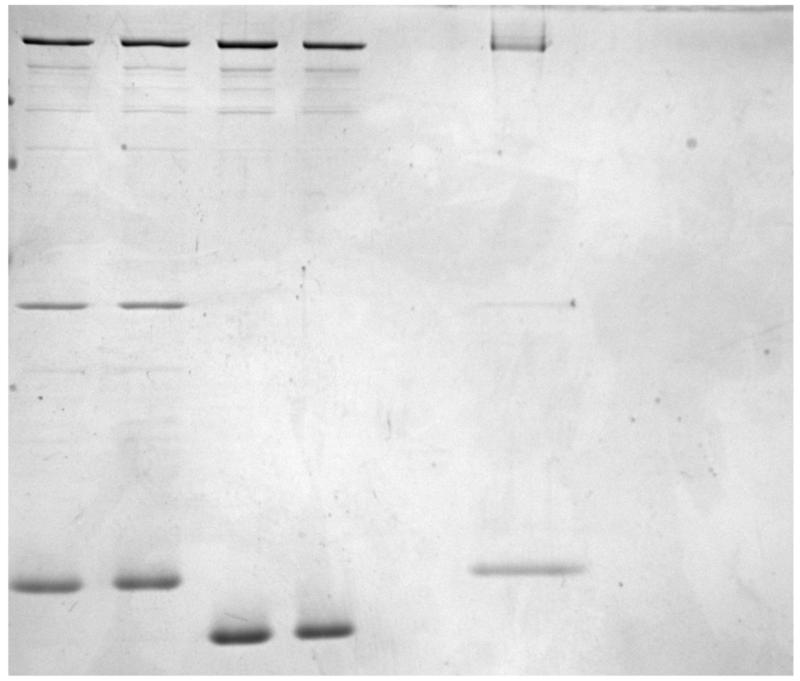
Figure 1.
- Purification of Sec7 constructs.
- Purification of Arf1 and Arl1 small GTPases.
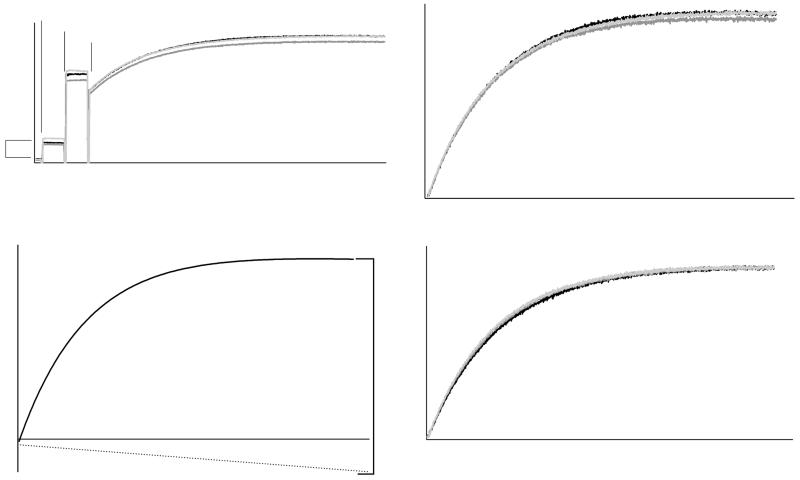
Figure 2.
- Unprocessed tryptophan fluorescence of Sec7(203-1220)-mediated exchange of Arf1 nucleotide in triplicate. Components are sequentially added as indicated; Sec7 concentration is proportional to the indicated difference in fluorescence.
- Arf1 nucleotide exchanges from (a) extracted and zeroed to baseline.
- One exchange reaction from (b) fit to a single exponential with linear drift component.
- Exchange reactions from (b) normalized for publication via curve fitting as per (c).
Acknowledgments
We thank Caitlin M. McDonold and Jon E. Paczkowski for development of parts of the small GTPase purification protocols presented here, and Maggie A. Gustafson for proofreading of the manuscipt. The authors are supported by NIH/NIGMS grant R01GM098621.
References
- Aizel K, Biou V, Navaza J, Duarte LV, Campanacci V, Cherfils J, Zeghouf M. Integrated Conformational and Lipid-Sensing Regulation of Endosomal ArfGEF BRAG2. PLoS Biol. 2013;11(9):e1001652. doi: 10.1371/journal.pbio.1001652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonny B, Huber I, Paris S, Chabre M, Cassel D. Activation of ADP-ribosylation Factor 1 GTPase-Activating Protein by Phosphatidylcholine-derived Diacylglycerols. Journal of Biological Chemistry. 1997;272(49):30848–30851. doi: 10.1074/jbc.272.49.30848. [DOI] [PubMed] [Google Scholar]
- Beraud-Dufour S, Robineau S, Chardin P, Paris S, Chabre M, Cherfils J, Antonny B. A glutamic finger in the guanine nucleotide exchange factor ARNO displaces Mg2+ and the [beta]-phosphate to destabilize GDP on ARF1. EMBO J. 1998;17(13):3651–3659. doi: 10.1093/emboj/17.13.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boal F, Stephens DJ. Specific functions of BIG1 and BIG2 in endomembrane organization. PLoS One. 2010;5:e9898. doi: 10.1371/journal.pone.0009898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J, Menetrey J, Mathieu M, Le Bras G, Robineau S, Beraud-Dufour S, Chardin P. Structure of the Sec7 domain of the Arf exchange factor ARNO. Nature. 1998;392(6671):101–105. doi: 10.1038/32210. [DOI] [PubMed] [Google Scholar]
- Cohen LA, Honda A, Varnai P, Brown FD, Balla T, Donaldson JG. Active Arf6 Recruits ARNO/Cytohesin GEFs to the PM by Binding Their PH Domains. Molecular Biology of the Cell. 2007;18(6):2244–2253. doi: 10.1091/mbc.E06-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehring DAK, Adler AS, Hosseini A, Hicke L. A C-terminal Sequence in the Guanine Nucleotide Exchange Factor Sec7 Mediates Golgi Association and Interaction with the Rsp5 Ubiquitin Ligase. Journal of Biological Chemistry. 2008;283(49):34188–34196. doi: 10.1074/jbc.M806023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNitto JP, Delprato A, Gabe Lee M-T, Cronin TC, Huang S, Guilherme A, Lambright DG. Structural Basis and Mechanism of Autoregulation in 3-Phosphoinositide-Dependent Grp1 Family Arf GTPase Exchange Factors. Molecular Cell. 2007;28(4):569–583. doi: 10.1016/j.molcel.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Cassel D, Kahn RA, Klausner RD. ADP-ribosylation factor, a small GTP-binding protein, is required for binding of the coatomer protein beta-COP to Golgi membranes. Proceedings of the National Academy of Sciences. 1992;89(14):6408–6412. doi: 10.1073/pnas.89.14.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza RS, Semus R, Billings EA, Meyer CB, Conger K, Casanova JE. Rab4 Orchestrates a Small GTPase Cascade for Recruitment of Adaptor Proteins to Early Endosomes. Curr. Biol. 2014 doi: 10.1016/j.cub.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. The Proteomics Protocols Handbook. Humana Press; 2005. Protein Identification and Analysis Tools on the ExPASy Server; pp. 571–607. [Google Scholar]
- Ha VL, Thomas GMH, Stauffer S, Randazzo PA. Preparation of myristoylated Arf1 and Arf6. Methods in Enzymology. 2005;404:164–174. doi: 10.1016/S0076-6879(05)04016-4. [DOI] [PubMed] [Google Scholar]
- Klemm RW, Ejsing CS, Surma MA, Kaiser H-J, Gerl MJ, Sampaio JL, Simons K. Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J. Cell Biol. 2009;185(4):601–612. doi: 10.1083/jcb.200901145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonold CM, Fromme JC. Four GTPases Differentially Regulate the Sec7 Arf-GEF to Direct Traffic at the trans-Golgi Network. Developmental Cell. 2014;30(6):759–767. doi: 10.1016/j.devcel.2014.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaga N, Tsai S-C, Moss J, Vaughan M. Isolation of a Brefeldin A-Inhibited Guanine Nucleotide-Exchange Protein for ADP Ribosylation Factor (ARF) 1 and ARF3 That Contains a Sec7-Like Domain. Proceedings of the National Academy of Sciences. 1996;93(23):12856–12860. doi: 10.1073/pnas.93.23.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E, Corpina RA, Goldberg J. Crystal Structure of ARF1-Sec7 Complexed with Brefeldin A and Its Implications for the Guanine Nucleotide Exchange Mechanism. Molecular Cell. 2003;12(6):1403–1411. doi: 10.1016/s1097-2765(03)00475-1. [DOI] [PubMed] [Google Scholar]
- Mossessova E, Gulbis JM, Goldberg J. Structure of the Guanine Nucleotide Exchange Factor Sec7 Domain of Human Arno and Analysis of the Interaction with ARF GTPase. Cell. 1998;92(3):415–423. doi: 10.1016/s0092-8674(00)80933-2. [DOI] [PubMed] [Google Scholar]
- Mouratou B, Biou V, Joubert A, Cohen J, Shields D, Geldner N, Cherfils J. The domain architecture of large guanine nucleotide exchange factors for the small GTP-binding protein Arf. BMC Genomics. 2005;6(1):20. doi: 10.1186/1471-2164-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padovani D, Folly-Klan M, Labarde A, Boulakirba S, Campanacci V, Franco M, Cherfils J. EFA6 controls Arf1 and Arf6 activation through a negative feedback loop. Proceedings of the National Academy of Sciences. 2014;111(34):12378–12383. doi: 10.1073/pnas.1409832111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault L, Christova P, Guibert B, Pasqualato S, Cherfils J. Mechanism of Domain Closure of Sec7 Domains and Role in BFA Sensitivity†. Biochemistry. 2002;41(11):3605–3612. doi: 10.1021/bi012123h. [DOI] [PubMed] [Google Scholar]
- Richardson BC, McDonold CM, Fromme JC. The Sec7 Arf-GEF Is Recruited to the trans-Golgi Network by Positive Feedback. Developmental Cell. 2012;22(4):799–810. doi: 10.1016/j.devcel.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sata M, Donaldson JG, Moss J, Vaughan M. Brefeldin A-inhibited guanine nucleotide-exchange activity of Sec7 domain from yeast Sec7 with yeast and mammalian ADP ribosylation factors. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(8):4204–4208. doi: 10.1073/pnas.95.8.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Xu KF, Fan Q, Pacheco-Rodriguez G, Moss J, Vaughan M. Association of brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BIG2) with recycling endosomes during transferrin uptake. Proc. Natl. Acad. Sci. USA. 2006;103:2635–2640. doi: 10.1073/pnas.0510599103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag CA, Sage H, Erickson HP. BtubA-BtubB Heterodimer Is an Essential Intermediate in Protofilament Assembly. PLoS ONE. 2009;4(9):e7253. doi: 10.1371/journal.pone.0007253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder D, Barelli H, Gautier R, Macia E, Jackson CL, Antonny B. Kinetic Studies of the Arf Activator Arno on Model Membranes in the Presence of Arf Effectors Suggest Control by a Positive Feedback Loop. Journal of Biological Chemistry. 2011;286(5):3873–3883. doi: 10.1074/jbc.M110.145532. [DOI] [PMC free article] [PubMed] [Google Scholar]