Abstract
Prostate cancer (PCa) is a major health concern for men in the USA. Aberrant expression of microRNAs (miRNAs) has been associated with the pathogenesis of various cancers, including PCa. Circulatory forms of miRNAs have been detected in serum and hold promise as minimally invasive cancer biomarkers. This study aimed to identify potential circulatory miRNAs that can provide insights into new mechanisms for clinical diagnosis of PCa and can serve as potential biomarkers and/or therapeutic targets. Candidate serum miRNAs were detected by using PCR microarray in a learning set of six African American (AA) and six Caucasian American (CA) PCa patients. Discriminating performance of candidate miRNAs was validated by qRT-PCR in serum samples from 36 AA (24 PCa patients and 12 controls) and 36 CA (16 PCa patients and 20 controls). From the miRNA profiling experiments, three differentially expressed miRNAs (miR-25, miR-101, and miR-628-5p) were selected for future validation. In the validation set, there was an overall low expression of miR-25 (p<0.01), miR-101 (p<0.001), and miR-628-5p (p<0.0001) in serum of PCa patients as compared with normal individuals. Subdivision on the basis of ethnicity showed that serum expression levels of miR-628-5p were significantly downregulated in both AA and CA PCa patients when compared with their respective controls. Our results demonstrate that the three miRNAs, particularly miR-628-5p, may be further developed as a biomarker, which can serve as novel noninvasive biomarker for PCa diagnosis and prognosis.
Keywords: MicroRNA, Prostate cancer, Serum, Circulatory miRNA
Introduction
Prostate cancer (PCa) is the most common cancer in American men and the second leading cause of cancer-related deaths after lung cancer. The American Cancer Society estimates ~238,590 new cases resulting in ~29,720 deaths in 2013 [1]. African American (AA) men are 1.6 times more likely to develop PCa and 2.4 times more likely to die from it as compared with Caucasian American (CA) men. The high mortality rate can be attributed primarily to an aggravated disease and bone metastasis [2]. Current PCa prognostic modalities are mostly based on pretreatment prostate-specific antigen (PSA) levels, Gleason scores, and clinical staging, all of which are inadequate in precisely predicting the disease progression [2]. Notwithstanding the efficacy of surgery and radiation therapy for treating localized disease and earlier diagnosis through testing serum PSA levels, up to 30 % of treated PCa patients suffer relapse [3]. Therefore, the identification of biomarkers that can predict the disease at an early stage is needed for optimizing management and treatment strategies.
MicroRNAs (miRNAs) are small (18–24 nucleotides), well-conserved, non-protein-coding RNA molecules that regulate divergent cellular processes including cell cycle, differentiation, and proliferation, thus influencing tumorigenesis and disease progression [4, 5]. Bioinformatics analysis of miRNA targets suggests that over 60 % of mammalian gene transcripts are regulated by miRNAs [6]. Several miRNA expression studies and functional experiments in various cancers have shown an important role for miRNAs in cancer initiation and progression and their potential as diagnostic, prognostic, and predictive biomarkers [7]. Further, the discovery of circulating miRNAs offers interesting clinical perspectives as they can be considered representative of some pathological conditions. Moreover, their accessibility and high stability in the circulation make them perfect biomarkers, especially for surveillance of early stage, presymptomatic diseases in at-risk patients. Although the physiological significance of circulating miRNAs is not fully understood, they have attracted attention as potential diagnostic and prognostic biomarkers for various diseases, particularly cancer. In the present study, we aimed to identify circulatory miRNAs that can serve as predictors of PCa.
Material and methods
Patient samples
All the patient serum samples obtained were de-identified to protect patient confidentiality and had Georgetown University IRB approval and consent. Written informed consent was obtained from all the subjects. Serum samples from patients were obtained from the Georgetown University Hospital CyberKnife Prostate Cancer Program from 2009 to 2012. Briefly, 40 PCa patients (16 CA and 24 AA) were recruited for the study. Age-matched serum samples from 32 healthy individuals (20 CA and 12 AA) were obtained from Innovative Research (Novi, Michigan) and Georgetown University Hospital.
RNA extraction and miRNA expression profiling
RNA extraction from serum samples was performed using a mirVana™ miRNA Isolation Kit (Ambion, Austin, TX) following the manufacturer's instructions. RNA was quantified using Thermo Scientific NanoDrop® ND-1000 Spectrophotometer (Thermo Scientific, Waltham, MA) and Agilent 2100 Bioanalyzer (Santa Clara, CA). A comprehensive miRNA expression profiling was carried out using Applied Biosytems TaqMan® Array Human MicroRNA Cards A and B v2.0 following the manufacturer's recommendations. Briefly, total RNAs from PCa patients and healthy individuals were converted to cDNA using Megaplex™ Primer pools and Taqman miRNA Reverse Transcription Kit (Applied Biosytems, Grand Island, NY). Quantitative reverse transcriptase (qRT)-PCR was carried out to screen a total of 667 unique human miRNAs using Applied Biosystems 7900HT Fast Real-Time PCR Sequence Detection System. Data were analyzed on sequence detection system (SDS) software version 2.3 (Applied Biosytems, Grand Island, NY).
qRT-PCR
MiRNA expression levels were measured using inventoried TaqMan® miRNA assay (Applied Biosytems, Grand Island, NY) following the manufacturer's recommendations on 7300 Real-Time PCR System (Applied Biosytems, Grand Island, NY). Briefly, for reverse transcription, 10 ng of RNA was used in each reaction and mixed with the specific stem–loop primers. Subsequently, the qRT-PCR reactions were run in triplicate. No reverse transcriptase (RT) controls were used to rule out the possibility of potential genomic DNA contamination. Relative miRNA expression levels were normalized against miR-223 as endogenous control as suggested by Kroh et al., 2010. Expression of miR-223 has been observed unaltered in serum samples [8]. Expression of miR-223 was unaltered in our preliminary experiments as well, and therefore, it was used as the endogenous control in our study. MiRNAs with threshold cycle (Ct) values of ≥38 were excluded from the analysis. All samples underwent reverse transcription, and PCR was run simultaneously to minimize errors introduced by variations in reaction efficiency.
Statistical analysis
The raw profiling data from TaqMan® Array Human MicroRNA Card Set v2.0 (cards A and B) were statistically analyzed using the Integromics® RealTime StatMiner® software version 4.0 and R/Bioconductor software version 2.9.2. The 2−ΔΔCt method [9] was employed for pre-processing and fold change calculation. Differentially expressed circulatory miRNAs between AA and CA PCa serum samples were identified using limma package [10] which employs the empirical Bayesian model to deal with the small sample size compared with the relatively much larger number of miRNAs. The p values were adjusted using Benjamin–Hochberg false discovery rate (FDR) correction [11].
All qRT–PCR experiments were conducted according to the MIQE (minimum information for publication of quantitative real-time PCR experiments) guidelines [12]. Each amplification reaction was performed in triplicate, and the mean value of the three threshold cycles was used for further analysis. Data are presented as mean±SE. P value of p≤0.05 was considered statistically significant. The nonparametric Student's t test was used for comparing the two groups, and all statistics were adjusted using the Holm–Bonferonni correction for multiple comparisons.
Receiver operating characteristic (ROC) curves were constructed, and area under curve (AUC) was estimated to study the feasibility of using the particular miRNA to discriminate PCa patients from healthy controls. Logistic regression was used to construct ROC curves using miRNA expression levels. All the statistical analyses were performed using GraphPad Prism (La Jolla, CA).
Results
Expression profiling of miRNAs from serum of PCa patients
Assessing changes in miRNA expression in biofluids may offer a promising tool for identifying specific biomarkers that can aid in the diagnosis and prognosis of PCa. To identify the differentially expressed miRNA, expression profiling was performed on 12 PCa patients, six each (pooled in three groups comprising two patients each) of AA and CA. We performed miRNA profiling analysis for a large range of miRNAs (comprising 667 unique human miRNAs); however, we observed that a very limited number of miRNAs were differentially expressed between AA and CA populations. The miRNAs most differentially expressed between the two populations were miR-25, miR-101, and miR-628-5p. For validation study, we selected a total of three miRNAs (miR-25, miR-101, and miR-628-5p) based on their published role in cancer biology [13–15].
Validation of miRNAs by qRT-PCR
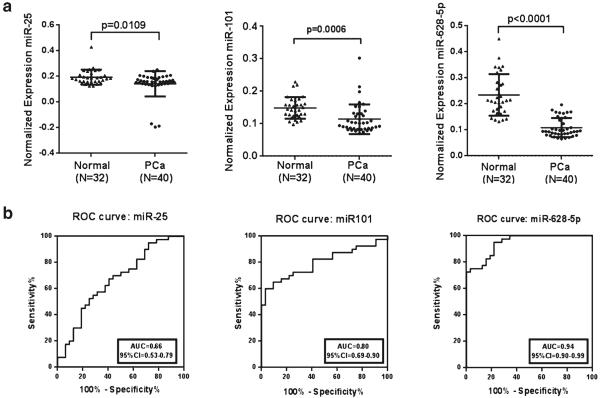
In order to compare the expression level of these circulatory miRNAs in serum of PCa patients to that of normal individuals of their respective population, healthy individuals were recruited. The selected three miRNAs (miR-25, miR-101, and miR-628-5p) were validated in 40 PCa patients and 32 healthy individuals. Table 1 shows the clinical pathological characteristics of the patients and healthy individuals. The qRT-PCR results showed that the expression levels of miR-25 (p<0.01), miR-101 (p<0.001), and miR-628-5p (p<0.0001) are significantly low in the PCa patients when compared with the healthy individuals (Fig. 1a).
Table 1.
Clinicopathological characteristics of the participants for serum sample
| Characteristics | PCa patients n (%) | Healthy control n (%) |
|---|---|---|
| No. of patients | 40 | 32 |
| Ethnicity | ||
| Caucasian American | 16 (40) | 20 (62.5) |
| African American | 24 (60) | 12 (37.5) |
| Age (yr) | ||
| Mean±SD | 70±7 | 49.3±9.6 |
| Median (range) | 69 (58–86) | 48(33–70) |
| Primary tumor | ||
| pT1–pT2 | 40 (100) | – |
| pT3–pT4 | 0 (0) | – |
| Hormone therapy | ||
| Yes | 12 (30) | – |
| No | 28 (70) | – |
| Gleason sum | ||
| ≤6 | 13 (32.5) | – |
| =7 | 16 (40) | – |
| ≥8 | 11 (27.5) | – |
| Lymph node metastasis | ||
| Nx | 27 (67.5) | – |
| N0 | 13 (32.5) | – |
| N1–N4 | 0 (0) | – |
| Distant metastasis | ||
| Mx | 24 (60) | – |
| M0 | 16 (40) | – |
| M1 | 0 (0) | – |
Fig. 1.
Validation of selected miRNAs in serum. a Scatter plots representing the serum expression level of three miRNAs (miR-25, miR-101, and miR-628-5p) from 40 PCa patients and 32 healthy individuals as assessed by qRT-PCR. Expression levels of the miRNAs are normalized to miR-223 as the endogenous control. Statistically significant differences were determined using Student's t test. b Receiver operating characteristic (ROC) curve analysis of three miRNAs was used to differentiate the PCa patients from healthy individuals. The area under the ROC curve (AUC) for each miRNA conveys its accuracy for differentiation of PCa patients and healthy subjects in terms of sensitivity and specificity
ROC curves were constructed to explore the sensitivity and specificity of miR-25, miR-101, and miR-628-5p as potential biomarkers in discriminating between individuals with cancer and disease-free individuals (Fig. 1b). The results suggest that the three miRNAs can discriminate between the two groups with high precision: miR-25 AUC=0.66 (95 % CI 0.53–0.79), miR-101 AUC=0.80 (95 % CI 0.69–0.90), and miR-628-5p AUC=0.94 (95 % CI 0.90–0.99) (Fig. 1b).
Ethnic variation in miRNA expression
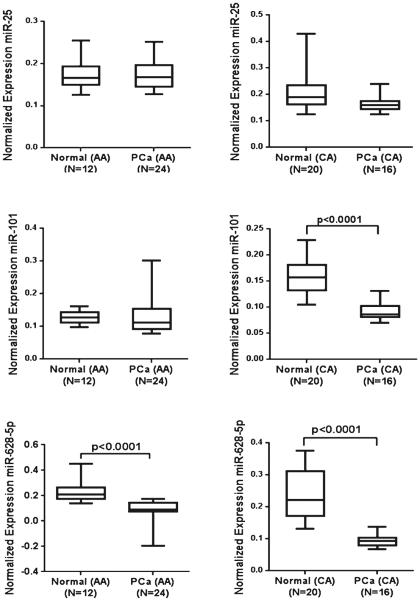
AA men have a greater incidence of PCa as compared with CA men. To determine if the expression levels of miR-25, miR-101, and miR-628-5p are different between the two populations, we subdivided the PCa patients and healthy individuals on the basis of ethnicity and compared the expression level of each miRNA. As shown in Fig. 2, there was no significant difference in expression pattern of miR-25 between CA and AA population. However, a significantly decreased expression (p<0.0001) of miR-101 was observed in the serum of CA PCa patients as compared with that of CA healthy individuals. However, no difference in expression of miR-101 was observed in AA PCa patients when compared with AA healthy individuals. Expression of miR-628-5p was low in both CA (p<0.0001) and AA (p<0.0001) populations as compared with their respective healthy controls (Fig. 2).
Fig. 2.
Comparison of miRNA expression on the basis of ethnicity. Box plots represent the differences in expression levels of three miRNAs in the serum of PCa patients as compared with their normal adjacent counterpart in African American (AA) and Caucasian American (CA) populations. Expression levels of the miRNAs were normalized to miR-223 as the endogenous control. Statistically significant differences were determined using unpaired Student's t test
Discussion
MicroRNAs emerged as novel biological entity with prospective use as tumor biomarkers, which can improve diagnosis, prognosis, and monitoring of treatment response for human cancers. Circulating miRNAs are abundantly present in many body fluids and represent reliable markers for several physio-pathological disorders, including cancer. In many recent studies, individual miRNA proved to provide diagnostic and prognostic serum/plasma markers for various cancers. Being easily accessible and collected routinely as part of medical assessments, plasma and serum represent the most promising and best studied source of cell-free miRNAs.
In this study, we aimed to study the differential expression of circulatory miRNAs between AA and CA PCa patients. We also compared the expression levels of PCa patients with those of normal individuals of the same ethnicity. Serum expression levels of miR-25 were significantly downregulated in PCa patients. In previous studies, miR-106b~25 clusters have been associated with PCa pathogenesis and shown to be aberrantly overexpressed in PCa. The miR-106b~25 locus on chromosome 7 is entirely composed of PTEN-targeting miRNAs (miR-106b, miR-93, and miR-25) and is markedly overexpressed and genetically amplified in PCa [16]. Serum miR-25 levels have been suggested to serve as biomarker for HCC diagnosis [17], while the downregulation of miR-25 has been shown to contribute to the process of thyroid cancer progression, leading to the development of anaplastic carcinomas [13]. Plasma levels of miR-25 were not significantly different between gastric cancer patients and healthy controls [18]. MiR-25 has been observed to be upregulated in breast cancer [19, 20], advanced gastric carcinoma [21, 22], esophageal squamous cell carcinoma [23, 24], hepatocellular carcinoma [25], lung carcinoma [26], cholangiocarcinoma [27], and in ovarian cancer tissues [28]. Our observation that miR-25 is downregulated in serum from PCa patients is intriguing and needs further validation in larger set of samples.
Another significantly downregulated miRNA identified by miRNA profiling in the serum of PCa patients was miR-101. The levels of miR-101 were even lower in CA patients as compared with AA patients. MiR-101 has been shown to target EZH2 and to decrease the invasiveness of PCa cells; also, loss of miR-101 is concomitant with the overexpression of EZH2. Genomic loci encoding miR-101 were found to be lost both in clinically localized PCa (37.5 %) and in metastatic cancer (66.7 %) [29–31]. Exogenous miR-101 is able to effectively suppress the growth of cultured PCa cells and prostate tumor xenografts [32]. MiR-101 has also been reported to be downregulated in various other cancers including colon cancer, nasopharyngeal carcinoma, neuroblastoma, gastric, and prostate cancer and reported to exert its effect by altering cell proliferation, invasion migration, metastasis, and angiogenesis [14, 33–41]. Mir-101 has also been reported to affect various cellular processes including inhibition of autophagy, promoting apoptosis, suppression of tumorigenicity, activation of p23 pathway, Wnt/β-catenin signaling, etc. [42–45]. Although miR-101 expression has been associated with various cancers, this is the first time that we show the differential expression of miR-101 in the serum of PCa patients.
In our study, the most significantly downregulated miRNA in PCa patients, overall, was miR-628-5p. Also, decreased expression of miR-628-5p was observed in both populations (AA and CA) which emphasize its importance in PCa. MiR-628-5p is a relatively unexplored, miRNA and to the best of our knowledge, this is the first time that miR-628-5p has been reported as differentially expressed in PCa. There are very few reports exploring the role of miR-628-5p. One recent report suggests that decreased levels of miR-628-5p caused by IL-3 signaling in leukemic progenitors might be responsible for promoting increased FOXO3a expressions in acute myeloid leukemia [46]. Recently, in an attempt to generate a miRNA profile of chemo-resistant blastemal cells in high-risk Wilms' tumors (nephroblastoma), Watson et al. [15] observed upregulation of miR-628-5p. Our results also emphasize the potential role of miR-628-5p in PCa pathogenesis and diagnosis.
We acknowledge the limitation of our study that our small sample size is not large and which may be the reason for the lack of clear depiction of differential expression of miRNAs in subpopulations. Nevertheless, our study provides proof of concept that miR-25, miR-101, and miR-628-5p are down-regulated in PCa which can be used to differentiate PCa patients from healthy controls. To the best of our knowledge, this is the first time that the change in expression levels of miR-628-5p has been associated with PCa indicating that the levels of circulating miR-628-5p in serum hold promise to be used as biomarker to differentiate between healthy individuals and PCa patients. Expression of miR-628-5p needs to be studied in a larger number of samples to determine its prognostic, predictive, or therapeutic value. Association of miR-628-5p with PCa is highly significant, and in view of its differential expression in both ethnic groups, further studies in our lab are directed toward characterizing the role of miR-628-5p in PCa.
In conclusion, the observed differentially expressed miRNAs in our study, miR-25 and miR-101, are relatively well characterized and have a distinct role in cancer. However, their role as circulatory miRNAs is relatively new in PCa. The most noticeable outcome of this study was miR-628-5p, which emerged as the most important differentially expressed miRNA in our study due to its more pronounced and consistent low expression in CA as well as in AA population. The differentially expressed miRNAs appear to significantly affect PCa pathogenesis and hold promise to enclose clinical utility as a supplement to PSA testing and to serve as biomarkers for screening PCa patients.
Acknowledgments
We acknowledge the support of the Genomic and Epigenetic Shared Resource (GESR) and Biostatistics and Bioinformatics Shared Resource (BBSR) at the LCCC. Assistance by Ms. Sunghae Uhm is gratefully acknowledged. Funding support: NIH: CA162264 and CA141935; PCRP: PC111314.
Footnotes
Conflicts of interest None
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Yaman Agaoglu F, Kovancilar M, Dizdar Y, Darendeliler E, Holdenrieder S, Dalay N, et al. Investigation of mir-21, mir-141, and mir-221 in blood circulation of patients with prostate cancer. Tumor Biol. 2011;32:583–8. doi: 10.1007/s13277-011-0154-9. [DOI] [PubMed] [Google Scholar]
- 3.Roberts WW, Bergstralh EJ, Blute ML, Slezak JM, Carducci M, Han M, et al. Contemporary identification of patients at high risk of early prostate cancer recurrence after radical retropubic prostatectomy. Urology. 2001;57:1033–7. doi: 10.1016/s0090-4295(01)00978-5. [DOI] [PubMed] [Google Scholar]
- 4.Shariat SF, Semjonow A, Lilja H, Savage C, Vickers AJ, Bjartell A. Tumor markers in prostate cancer I: blood-based markers. Acta Oncol (Formerly: Acta Radiol Oncol) 2011;50:61. doi: 10.3109/0284186X.2010.542174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferracin M, Veronese A, Negrini M. Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert Rev Mol Diagn. 2010;10:297–308. doi: 10.1586/erm.10.11. [DOI] [PubMed] [Google Scholar]
- 8.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 10.Wettenhall JM, Smyth GK. Limmagui: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–6. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- 11.Verhoeven KJF, Simonsen KL, McIntyre LM. Implementing false discovery rate control: increasing your power. Oikos. 2005;108:643–7. [Google Scholar]
- 12.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 13.Esposito F, Tornincasa M, Pallante P, Federico A, Borbone E, Pierantoni GM, et al. Down-regulation of the miR-25 and miR-30d contributes to the development of anaplastic thyroid carcinoma targeting the polycomb protein EZH2. J Clin Endocrinol Metab. 2012;97:E710–8. doi: 10.1210/jc.2011-3068. [DOI] [PubMed] [Google Scholar]
- 14.Wang HJ, Ruan HJ, He XJ, Ma YY, Jiang XT, Xia YJ, et al. MicroRNA-101 is down-regulated in gastric cancer and involved in cell migration and invasion. Eur J Cancer. 2010;46:2295–303. doi: 10.1016/j.ejca.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 15.Watson JA, Bryan K, Williams R, Popov S, Vujanic G, Coulomb A, et al. miRNA profiles as a predictor of chemoresponsiveness in Wilms' tumor blastema. PLoS One. 2013;8:e53417. doi: 10.1371/journal.pone.0053417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3:ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, et al. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70:9798–807. doi: 10.1158/0008-5472.CAN-10-1001. [DOI] [PubMed] [Google Scholar]
- 18.Li BS, Zhao YL, Guo G, Li W, Zhu ED, Luo X, et al. Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PLoS One. 2012;7:e41629. doi: 10.1371/journal.pone.0041629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Q, Wang C, Lu Z, Guo L, Ge Q. Analysis of serum genome-wide microRNAs for breast cancer detection. Clin Chim Acta. 2012;413:1058–65. doi: 10.1016/j.cca.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 20.Hu Z, Dong J, Wang LE, Ma H, Liu J, Zhao Y, et al. Serum microRNA profiling and breast cancer risk: the use of mir-484/191 as endogenous controls. Carcinogenesis. 2012;33:828–34. doi: 10.1093/carcin/bgs030. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Zhang Y, Zhang H, Liu X, Gong T, Li M, et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9:824–33. doi: 10.1158/1541-7786.MCR-10-0529. [DOI] [PubMed] [Google Scholar]
- 22.Kim BH, Hong SW, Kim A, Choi SH, Yoon SO. Prognostic implications for high expression of oncogenic microRNAs in advanced gastric carcinoma. J Surg Oncol. 2013;107:505–10. doi: 10.1002/jso.23271. [DOI] [PubMed] [Google Scholar]
- 23.Zhu L, Yan W, Rodriguez-Canales J, Rosenberg AM, Hu N, Goldstein AM, et al. MicroRNA analysis of microdissected normal squamous esophageal epithelium and tumor cells. Am J Cancer Res. 2011;1:574–84. [PMC free article] [PubMed] [Google Scholar]
- 24.Fang W-K, Liao L-D, Li L-Y, Xie Y-M, Xu X-E, Zhao W-J, et al. Down-regulated desmocollin-2 promotes cell aggressiveness through redistributing adherens junctions and activating beta-catenin signalling in oesophageal squamous cell carcinoma. J Pathol. 2013;231:257–70. doi: 10.1002/path.4236. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Tan W, Neo TW, Aung MO, Wasser S, Lim SG, et al. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci. 2009;100:1234–42. doi: 10.1111/j.1349-7006.2009.01164.x. [DOI] [PubMed] [Google Scholar]
- 26.Dacic S, Kelly L, Shuai Y, Nikiforova MN. miRNA expression profiling of lung adenocarcinomas: correlation with mutational status. Mod Pathol. 2010;23:1577–82. doi: 10.1038/modpathol.2010.152. [DOI] [PubMed] [Google Scholar]
- 27.Razumilava N, Bronk SF, Smoot RL, Fingas CD, Werneburg NW, Roberts LR, et al. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology. 2012;55:465–75. doi: 10.1002/hep.24698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Zuo Z, Lu X, Wang L, Wang H, Zhu Z. MiR-25 regulates apoptosis by targeting Bim in human ovarian cancer. Oncol Rep. 2012;27:594–8. doi: 10.3892/or.2011.1530. [DOI] [PubMed] [Google Scholar]
- 29.Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–9. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pang Y, Young CY, Yuan H. MicroRNAs and prostate cancer. Acta Biochim Biophys Sin (Shanghai) 2010;42:363–9. doi: 10.1093/abbs/gmq038. [DOI] [PubMed] [Google Scholar]
- 31.Ren G, Baritaki S, Marathe H, Feng J, Park S, Beach S, et al. Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer Res. 2012;72:3091–104. doi: 10.1158/0008-5472.CAN-11-3546. [DOI] [PubMed] [Google Scholar]
- 32.Hao Y, Gu X, Zhao Y, Greene S, Sha W, Smoot DT, et al. Enforced expression of miR-101 inhibits prostate cancer cell growth by modulating the COX-2 pathway in vivo. Cancer Prev Res (Phila) 2011;4:1073–83. doi: 10.1158/1940-6207.CAPR-10-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alajez NM, Lenarduzzi M, Ito E, Hui AB, Shi W, Bruce J, et al. MiR-218 suppresses nasopharyngeal cancer progression through down-regulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res. 2011;71:2381–91. doi: 10.1158/0008-5472.CAN-10-2754. [DOI] [PubMed] [Google Scholar]
- 34.Cao P, Deng Z, Wan M, Huang W, Cramer SD, Xu J, et al. MicroRNA-101 negatively regulates EZH2 and its expression is modulated by androgen receptor and HIF-1alpha/HIF-1beta. Mol Cancer. 2010;9:108. doi: 10.1186/1476-4598-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strillacci A, Griffoni C, Sansone P, Paterini P, Piazzi G, Lazzarini G, et al. MiR-101 downregulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp Cell Res. 2009;315:1439–47. doi: 10.1016/j.yexcr.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 36.Buechner J, Tomte E, Haug BH, Henriksen JR, Lokke C, Flaegstad T, et al. Tumour-suppressor microRNAs let-7 and miR-101 target the proto-oncogene MYCN and inhibit cell proliferation in MYCN-amplified neuroblastoma. Br J Cancer. 2011;105:296–303. doi: 10.1038/bjc.2011.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carvalho J, van Grieken NC, Pereira PM, Sousa S, Tijssen M, Buffart TE, et al. Lack of microRNA-101 causes E-cadherin functional deregulation through EZH2 up-regulation in intestinal gastric cancer. J Pathol. 2012;228:31–44. doi: 10.1002/path.4032. [DOI] [PubMed] [Google Scholar]
- 38.Smits M, Mir SE, Nilsson RJ, van der Stoop PM, Niers JM, Marquez VE, et al. Down-regulation of miR-101 in endothelial cells promotes blood vessel formation through reduced repression of EZH2. PLoS One. 2011;6:e16282. doi: 10.1371/journal.pone.0016282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang JG, Guo JF, Liu DL, Liu Q, Wang JJ. MicroRNA-101 exerts tumor-suppressive functions in non-small cell lung cancer through directly targeting enhancer of zeste homolog 2. J Thorac Oncol. 2011;6:671–8. doi: 10.1097/JTO.0b013e318208eb35. [DOI] [PubMed] [Google Scholar]
- 40.Sakurai T, Bilim VN, Ugolkov AV, Yuuki K, Tsukigi M, Motoyama T, et al. The enhancer of zeste homolog 2 (EZH2), a potential therapeutic target, is regulated by miR-101 in renal cancer cells. Biochem Biophys Res Commun. 2012;422:607–14. doi: 10.1016/j.bbrc.2012.05.035. [DOI] [PubMed] [Google Scholar]
- 41.Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, et al. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–42. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 42.Frankel LB, Wen J, Lees M, Hoyer-Hansen M, Farkas T, Krogh A, et al. microRNA-101 is a potent inhibitor of autophagy. EMBO J. 2011;30:4628–41. doi: 10.1038/emboj.2011.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y, An Y, Wang Y, Zhang C, Zhang H, Huang C, et al. miR-101 inhibits autophagy and enhances cisplatin-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep. 2013;29:2019–24. doi: 10.3892/or.2013.2338. [DOI] [PubMed] [Google Scholar]
- 44.Liu X, Zou L, Zhu L, Zhang H, Du C, Li Z, et al. miRNA mediated up-regulation of cochaperone p23 acts as an anti-apoptotic factor in childhood acute lymphoblastic leukemia. Leuk Res. 2012;36:1098–104. doi: 10.1016/j.leukres.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 45.Strillacci A, Valerii MC, Sansone P, Caggiano C, Sgromo A, Vittori L, et al. Loss of miR-101 expression promotes Wnt/β-catenin signalling pathway activation and malignancy in colon cancer cells. J Pathol. 2013;229:379–89. doi: 10.1002/path.4097. [DOI] [PubMed] [Google Scholar]
- 46.Favreau AJ, Sathyanarayana P. miR-590-5p, miR-219-5p, miR-15b and miR-628-5p are commonly regulated by IL-3, GM-CSF and G-CSF in acute myeloid leukemia. Leuk Res. 2012;36:334–41. doi: 10.1016/j.leukres.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]