Abstract
The microphthalmia-associated transcription factor (MITF) is a basic helix-loop-helix leucine zipper family factor that is essential for terminal osteoclast differentiation. Previous work demonstrates that phosphorylation of MITF by p38 MAPK downstream of receptor activator of NFκB ligand (RANKL) signaling is necessary for MITF activation in osteoclasts. The spontaneous Mitf cloudy eyed (ce) allele results in production of a truncated MITF protein that lacks the leucine zipper and C-terminal end. Here we show that the Mitfce allele leads to a dense bone phenotype in neonatal mice due to defective osteoclast differentiation. In response to RANKL stimulation, in vitro osteoclast differentiation was impaired in myeloid precursors derived from neonatal or adult Mitfce/ce mice. The loss of the leucine zipper domain in Mitfce/ce mice does not interfere with the recruitment of MITF/PU.1 complexes to target promoters. Further, we have mapped the p38 MAPK docking site within the region deleted in Mitfce. This interaction is necessary for the phosphorylation of MITF by p38 MAPK. Site-directed mutations in the docking site interfered with the interaction between MITF and its co-factors FUS and BRG1. MITF-ce fails to recruit FUS and BRG1 to target genes, resulting in decreased expression of target genes and impaired osteoclast function. These results highlight the crucial role of signaling dependent MITF/p38 MAPK interactions in osteoclast differentiation.
Keywords: Osteoclast, P38MAPK, Docking site, Transcription
INTRODUCTION
Normal bone modeling and remodeling is carried out by a fine tuned balance of the actions of bone-forming osteoblasts and bone-resorbing osteoclasts (Boyle et al., 2003; Ikeda and Takeshita, 2014; Teitelbaum and Ross, 2003). An imbalance in this system skewing towards the overactivity of osteoclasts is seen in debilitating diseases including osteoporosis and rheumatoid arthritis (Chen et al., 2014; Kikuta and Ishii, 2013; Novack and Teitelbaum, 2008). Osteoclasts are cells of hematopoietic origin which differentiate through the myeloid lineage and are constantly turned over in response to signals from the bone microenvironment. Osteoblasts produce two key cytokines, colony stimulating factor 1 (CSF1) and receptor activator of NF-κB ligand (RANKL) which trigger myeloid precursors to differentiate, fuse, and form multinucleate osteoclasts capable of resorbing bone (Lacey et al., 1998; Yoshida et al., 1990). The signaling cascades initiated by the binding of CSF1 and RANKL to their receptors, cFMS and RANK respectively, lead to the downstream transcription of genes necessary for osteoclast function. One downstream effector which integrates these two signaling pathways to induce the transcription of genes necessary for osteoclast function is the microphthalmia-associated transcription factor, MITF, a basic-helix-loop-helix leucine zipper (bHLHZip) family transcription factor (Hodgkinson et al., 1993).
MITF is most well known as the master regulator of the melanin producing melanocyte linage and as an oncogene in melanomas (Hemesath et al., 1994; Levy et al., 2006). However, it also plays important roles in the transcriptional regulation of genes in other diverse cell types including retinal pigmented epithelial cells of the eye, mast cells of the innate immune system, and bone resorbing osteoclasts (Moore, 1995). The distinct mechanisms by which MITF can regulate vastly different transcriptional programs in a wide variety of cell types are still being revealed. The interaction of MITF with lineage restricted co-factors partly explains its specificity in different cell types. In osteoclasts, MITF physically interacts with the Ets family factor PU.1, which is the master regulator of all myeloid lineage cells (Luchin et al., 2001). MITF/PU.1 complexes control the transcription of multiple genes necessary for osteoclast function including tartrate resistant acid phosphastase (Acp5), cathepsin K (Ctsk), the osteoclast-associated immunoglobulin receptor (Oscar), and the chloride channel encoded by Clcn7 (Luchin et al., 2001; Meadows et al., 2007; Sharma et al., 2007; So et al., 2003). In response to signaling triggered by CSF1, MITF/PU.1 complexes are recruited to the promoters of these genes, but gene transcription is not activated due to the interaction of the Ikaros family member, EOS, with MITF (Hu et al., 2007). EOS recruits co-repressors including HDAC, Sin3A, and CtBP to these sites (Hu et al., 2007). Following the downstream signaling triggered by RANKL, EOS and the repressor complex proteins dissociate and p38 MAPK phosphorylates MITF at Ser307 (Hu et al., 2007; Mansky et al., 2002). This phosphorylation allows for the recruitment of transcriptional co-activator, Fused In Sarcoma (FUS) and the SWI/SNF chromatin remodeling complex with its ATPase dependent chromatin remodeling subunit Brahma-related gene 1, BRG1 (Bronisz et al., 2014; Sharma et al., 2007). These complexes of pS307 MITF, FUS, and BRG1 subsequently recruit RNA polymerase II to initiate active gene transcription (Sharma et al., 2007).
Various spontaneous, irradiation induced or chemically induced mutations at the Mitf gene locus in mice have been identified by their coat color phenotype. Somewhat surprisingly, of the over 41 known mutant forms of MITF, only two mutations, Mitf microphthalma (mi) and Mitf oak ridge (or), have been reported to exhibit an osteopetrotic phenotype in mice (Hodgkinson et al., 1993; Sharma et al., 2009; Steingrimsson et al., 1994). Both of these mutations are in the basic domain of the MITF protein and ablate its ability to bind nascent DNA in electrophoretic mobility shift assays (Hemesath et al., 1994). The cloudy eyed (ce) mutation of MITF results in a stop codon directly after aa262, which falls between the helix-loop-helix and leucine zipper domains (Steingrimsson et al., 1994). Therefore, the protein encoded by the Mitfce mutant allele has normal basic helix-loop-helix domains, but lacks the leucine zipper and the rest of the C-terminal portion of the protein past the zipper domain as shown in Figure 1A. Thus, the Mitfce mutant model is a useful tool to study the role of co-factor interactions and signaling events which normally take place at the C-terminal end of the MITF protein. Mitfce/ce mice have a white coat due to the absence of pigment producing melanocytes and are defective in mast cell differentiation and cardiac hypertrophy response; however, a discernible bone phenotype has not been reported (Morii et al., 2001; Tshori et al., 2006; Zimring et al., 1996).
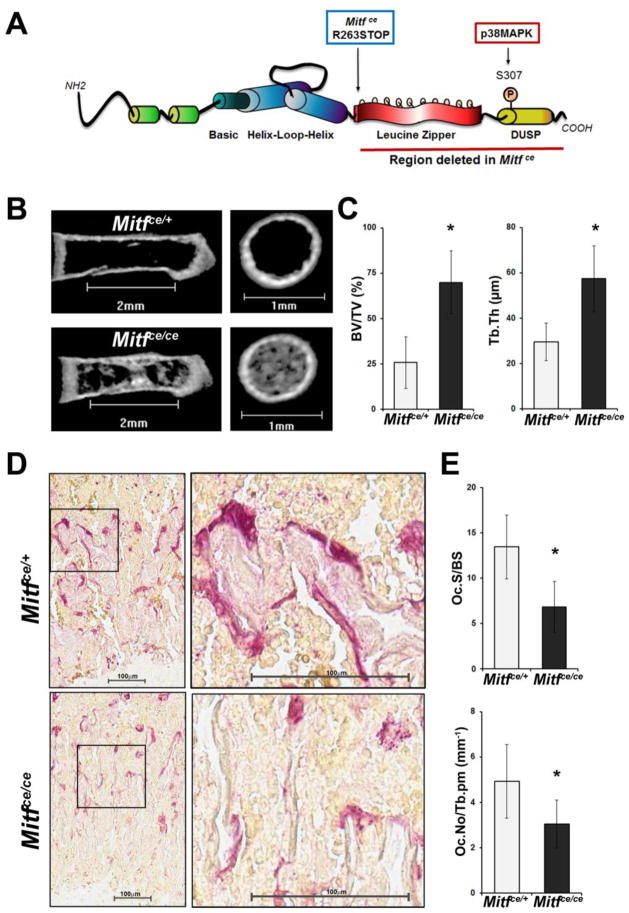
Figure 1. The cloudy eyed mutation of MITF leads to denser bone in neonatal mice due to defective osteoclastogenesis.
A. Schematic representation of the MITF protein with the location of the Mitfce mutation indicated. The region deleted with the Mitfce mutation is underlined with the red bar.
B. Representative longitudinal and metaphyseal cross section images from microCT analysis of the femurs of four day old mice homozygous for the cloudy eyed mutation of MITF (Mitfce/ce) or heterozygous controls (Mitfce/+)
C. Quantification of bone volume to total volume (BV/TV) and trabecular thickness (Tb.Th) from microCT analysis of four day old Mitfce/ce mice and controls (Mitfce/+), n = 3. Data are represented as the mean +/− S.D.
D. Representative images of TRAP staining to mark mature osteoclasts in the distal femurs of four day old Mitfce/ce mice and Mitfce/+ controls. Scale bars = 100μm.
E. Histomorphometric quantification of osteoclast surface area to trabecular surface area (Oc.S/BS) and osteoclast number per trabecular perimeter (Oc.No/Tb.pm) from TRAP staining shown in C, n = 3. Data are represented as the mean +/− S.D.
The purpose of this work was to examine the effect of the cloudy eyed (ce) allele of MITF on osteoclast differentiation and function. Here we show that in Mitfce/ce mice the ability of myeloid precursors to form functional osteoclasts in vitro and in vivo is significantly decreased. MITF-ce and its osteoclast-specific co-partner, PU.1 are still recruited to the Ctsk promoter. However, MITF-ce is unable to associate with transcriptional co-activators FUS and BRG1, leading to significantly reduced expression of MITF target genes which are necessary for osteoclast function. Our results demonstrate that the loss of the p38 MAPK phosphorylation site as well as docking site within the region deleted in the MITF-ce protein is responsible for this phenotype.
MATERIALS AND METHODS
Animals
Mice harboring the Mitfce allele were maintained in the C57BL/6J background. To generate homozygous Mitfce/ce mice and Mitfce/+ control mice for experiments, Mitfce/ce males were crossed with Mitfce/+ females. All animal use and care for this study was approved by The Ohio State University Institutional Animal Care and Use Committee.
Antibodies
Antibodies developed against MITF, phospho-S307 MITF, PU.1, and BRG1 were described previously (Mansky et al., 2002; Sharma et al., 2007). Anti-GST and anti-His mouse monoclonal antibodies and RNA Polymerase II rabbit polyclonal antibody were purchased from Santa Cruz Biotechnology. Anti-FLAG M2 mouse monoclonal antibody was purchased from Sigma. Anti-V5 mouse monoclonal antibody was purchased from Invitrogen. Anti-FUS rabbit polyclonal antibody was purchased from Bethyl Laboratories, Inc. Phospho-p38 MAPK rabbit polyclonal antibody was purchased from Cell Signaling.
Plasmids and Mutagenesis
Cloning of MITF, PU.1, p38 and FUS into tagged expression vectors has been previously described (Bronisz et al., 2014; Luchin et al., 2001; Mansky et al., 2002). Single or double point mutations of MITF were generated by the QuikChange method (Stratagene).
Cell Culture and Transfection
COS-7 cells were cultured as previously described (Bronisz et al., 2014). For transient transfection assays, COS-7 cells were transfected with expression vectors using Lipofectamine (Invitrogen) according to the manufacturer’s instructions. For in vitro osteoclast differentiation, either primary BMMs or splenocytes were enriched for myeloid precursor cells by culture on non-adherent plastic plates in Dulbecco’s Modified Eagle Medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (Sigma), 50 U/ml penicillin/streptomycin (Gibco) and 50ng/ml recombinant human CSF1 (Peprotech). After three days of culture, the myeloid enriched non-adherent fraction was mechanically isolated and replated on adherent tissue culture plates at a density of 5 × 106 cells per 10cm plate in DMEM containing 10% FBS, 50 U/ml Penicillin/Streptomycin, 50 ng/ml CSF1 and 100 ng/ml recombinant human RANKL (Peprotech) and harvested at the indicated time points.
Immunoprecipitation and Western Blotting
For immunoprecipitation assays, COS-7 cell lysates were prepared as previously described (Shen et al., 2003). Lysates were immunoprecipitated with anti-FLAG antibody pre-coupled to protein A/G-agarose beads (Amersham Biosciences). Beads were washed twice with lysis buffer supplemented with 2 mM DTT, twice with lysis buffer containing 0.5 M LiCl, and finally twice with PBS. Bound proteins were then eluted directly into SDS sample buffer, subjected to SDS-PAGE gel electrophoresis, transferred onto nitrocellulose membranes and probed with the indicated antibodies.
qRT-PCR
Cells were lysed in TRIzol reagent (Invitrogen) and total RNA was purified according to manufacturer’s instructions. RNA was reversed transcribed to cDNA using SuperScript III Reverse Transcriptase (Invitrogen) and random hexamer primers. Primers for qPCR were designed with the Universal Probe Library software (Roche) and primer sequences are available upon request. qPCR was performed using Taqman Master Mix (Roche) and Universal Probe Library probes (Roche) on a StepOnePlus machine (Applied Biosystems). Relative expression of the target mRNA compared to ribosomal protein L4 internal control was calculated using a variation of the ddCt method (Livak and Schmittgen, 2001).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as described previously by Luo et al. (Luo et al., 1998). Briefly, primary murine BMMs were cultured in media supplemented with 50 ng/ml CSF1 and 100ng/ml RANKL for three days. The cells were then cross-linked with formaldehyde to a final concentration of 1% for 10 min before harvest. Nuclear extracts were prepared and sonicated with a Branson 250 digital sonifier (Branson Ultrasonics, Danbury, CT) to an average DNA fragment length of 200–600 base pairs. The sonicated soluble chromatin was pre-cleared with tRNA-blocked Protein G-agarose beads (Millipore), and ~10% of the pre-cleared chromatin was set aside as input control. 1 × 106 cells were used for each immunoprecipitation and incubated overnight at 4 °C with 5 μg of the indicated antibodies. Immune complexes were pulled down using Protein G-agarose beads, extensively washed, and eluted twice with 250 μl of elution buffer (0.1M NaHCO3, 1% SDS). Reverse cross-linking was carried out at 65 °C overnight in elution buffer supplemented with 200 mM NaCl and 20 μg of RNase A (Sigma). The decrosslinked DNA was then treated with Proteinase K (Roche) and purified with the Qiagen PCR purification kit (Invitrogen) according to manufacturer’s instructions. The purified DNA was analyzed by qPCR on a StepOnePlus machine (Applied Biosystems) using Taqman MasterMix (Roche) and Universal Probe Library probes (Roche). Primers were designed with the Universal Probe Library software (Roche) and primer sequences are available upon request. Relative enrichment compared to input control was calculated using a variation of the ddCt method (Livak and Schmittgen, 2001).
MicroCT
Femora were dissected from four day old mice and fixed for 24 hr in 4% paraformaldehyde, then stored in 70% ethanol. Micro-computed tomography (μCT) analysis was conducted on a Siemens Inveon Preclinical CT scanner (Siemens AG, Munich, Germany). Images were acquired in 400 projections over 360 degrees at 100 kVp, 200 MA, 1 second exposure, Bin 2, and a medium-high system magnification with a pixel width (resolution) of 19.4 μm. Image data was reconstructed using Cobra software (Exxim, Pleasanton CA) and analyzed using 3D bone morphometry analysis software (Inveon Research Workplace 3D Image Software, Siemens PreClinical).
Anatomical lengths of the femora were measured. The volume of interest (VOI) for analysis of the diaphysis was defined as the central 5% of the overall length of the bone and cropped. The distal metaphysis was defined as 12.5% of the overall length just proximal to the distal physis. The VOI for analysis of the distal metaphysis was then defined as the upper 40% of this region (5% of the overall length of the femur). Cortical bone morphometric indices were analyzed in both the diaphysis and distal metaphysis and trabecular bone morphometric indices were analyzed in the distal metaphysis. Segmentation thresholds were kept constant for all femora and bones were both cropped and analyzed in a blinded manner using predefined randomization tables.
TRAP Staining and Bone Histomorphometry
Femurs dissected from 4 day old mice and fixed in 4% paraformaldehyde were embedded in paraffin wax and cut to 4μm thick sagittal sections. Sections were stained for tartrate resistant acid phosphatase (TRAP) using a Leukocyte Acid Phosphastase kit (Sigma) to visualize osteoclasts. Stained slides were scanned on a ScanScope Slide Scanner. Osteoclast and trabecular surface area of the scanned images was quantified using ImageScope software. For TRAP staining of osteoclasts cultured from murine BMMs, BMMs were grown on adherent plates in CSF1/RANKL media for five days then fixed with 4% paraformaldehyde for 30 min at 4 °C. Fixed cells were then stained with the Leukocyte Acid Phosphatase kit (Sigma). For quantification, all cells in 5 fields per sample were counted and represented as the percentage of osteoclasts per total cells. Osteoclasts were defined as TRAP positive cells having three or more visible nuclei. At least two biological replicates were used per group.
Statistical Analysis
Unpaired student’s t-tests were performed using Microsoft Excel for all statistical analyses. All differences with p < 0.05 were considered significant.
RESULTS
The femurs of neonatal Mitfce/ce mice are denser due to defective osteoclast differentiation
We performed microCT analysis on four day old neonatal mice when rapid bone remodeling occurs due to the growth phase. This analysis demonstrated that the femurs of four day old Mitfce/ce mice are significantly denser compared to heterozygous Mitfce/+controls (Figure 1B). The distal femoral metaphyses of four day old Mitfce/ce mice have an approximately 2.5 fold higher bone volume to total volume ratio (BV/TV, p = 0.027) and contain trabeculae which are twice as thick as controls (Tb.Th, p = 0.044) (Figure 1C). Further, the bones of Mitfce/ce mice trended towards higher trabecular number (Tb.N) and decreased trabecular spacing (Tb.Sp) compared to Mitfce/+ controls in the distal femoral metaphysis (Table 1). Significant differences between the two genotypes were not observed in the diaphyseal region (Table 1).
Table 1.
Bone histomorphometric parameters of the femurs of four day old Mitfce/ce mice and corresponding controls
| Mitfce/+ | Mitfce/ce | P value | |
|---|---|---|---|
| Femoral length (mm) | 3.58 ± 0.31 | 3.53 ± 0.44 | 0.879 |
| Femoral distal metaphysis | |||
| Bone volume fraction (BV/TV, %) | 25.84 ± 14.11 | 69.93 ± 17.31 | 0.027 |
| Bone surface density (BS/TV, 1/mm) | 71.13 ± 17.35 | 36.29 ± 8.50 | 0.035 |
| Trabecular number (Tb.N, 1/mm) | 8.42 ± 2.45 | 12.21 ± 1.26 | 0.076 |
| Trabecular thickness (Tb.Th, μm) | 29.47 ±8.28 | 57.37 ± 14.46 | 0.044 |
| Trabecular separation (Tb.Sp, μm) | 97.21 ± 46.33 | 25.07 ± 15.96 | 0.063 |
| Trabecular pattern factor (Tb.Pf) | 17.90 ± 10.94 | 0.59 ± 2.01 | 0.054 |
| Average cortical thickness (Ct.Th, μm) | 79.36 ± 18.06 | 84.76 ± 10.08 | 0.675 |
| Trabecular surface area/total surface area (%) | 31.24 ± 7.08 | 42.72 ± 1.82 | 0.053 |
| Cortical surface area/total surface area (%) | 36.94 ± 4.28 | 32.30 ± 3.22 | 0.208 |
| Trabecular specific bone surface (Tb.BS/Tb.BV, 1/mm) | 71.07± 17.35 | 36.29 ± 8.49 | 0.036 |
| Cortical specific bone surface (Ct.BS/Ct.BV, 1/mm) | 39.66 ± 7.07 | 44.54 ± 6.20 | 0.420 |
| Trabecular volume/total volume (Tb.V/TV, %) | 17.23 ± 9.20 | 49.45 ± 12.43 | 0.023 |
| Marrow volume/total volume (Ma.V/TV, %) | 49.45 ± 9.77 | 21.19± 12.13 | 0.035 |
| Cortical volume/total volume (Ct.V/TV, %) | 33.33 ± 3.56 | 29.35 ± 0.37 | 0.126 |
| Bone mineral density (mg/cc) | |||
| Trabecular bone | 1318.73 ± 23.64 | 1329.19 ± 3.56 | 0.491 |
| Bone marrow | 1078.94 ± 20.14 | 1142.64 ± 25.99 | 0.028 |
| Cortical bone | 1432.96 ± 40.57 | 1419.25 ± 48.65 | 0.727 |
| Cumulative | 1239.05 ± 47.45 | 1314.26 ± 22.12 | 0.068 |
| Femoral diaphysis | |||
| Bone volume fraction (BV/TV, %) | 16.08 ± 12.45 | 38.48± 31.19 | 0.312 |
| Bone surface density (BS/TV, 1/mm) | 93.94 ± 27.33 | 64.59 ± 32.54 | 0.298 |
| Average cortical thickness (Ct.Th, μm) | 82.19 ± 13.37 | 97.75± 11.16 | 0.196 |
| Cortical surface area/total surface area (%) | 47.95 ± 7.75 | 46.25 ± 7.20 | 0.795 |
| Cortical specific bone surface (Ct.BS/Ct.BV, 1/mm) | 35.45 ± 4.91 | 36.21 ± 3.81 | 0.842 |
| Marrow volume/total volume (Ma.V/TV, %) | 44.74 ± 3.89 | 32.07 ± 13.94 | 0.204 |
| Cortical volume/total volume (Ct.V/TV, %) | 46.31 ± 3.64 | 46.12 ± 5.28 | 0.961 |
| Bone mineral density (mg/cc) | |||
| Bone marrow | 1071.97 ± 10.56 | 1091.28 ± 12.55 | 0.111 |
| Cortical bone | 1493.92 ± 52.15 | 1503.64 ± 28.45 | 0.791 |
| Cumulative | 1290.49 ± 21.89 | 1357.81 ± 70.87 | 0.191 |
To determine if defective osteoclast formation was responsible for this observed dense bone phenotype in neonatal Mitfce/ce mice, we stained femurs from these mice for tartrate resistant acid phosphatase (TRAP) to identify mature osteoclasts. The femurs of four day old Mitfce/ce mice had approximately two fold less of their trabecular surface area covered by osteoclasts (Oc.S/BS, p = 0.005) and 40% fewer osteoclasts per millimeter of trabecular perimeter (Oc.No/Tb.pm, p = 0.038) than heterozygous controls (Figure 1D, E). These results indicate that the observed denser bones in neonatal Mitfce/ce mice are due to defective osteoclastogenesis.
We also examined mice homozygous for the Mitfce mutant allele at 30 days of age by microCT analysis. We found that the BV/TV ratio and other morphometric parameters in 30 day old Mitfce/ce mice trended towards more bone mass and denser bone than controls; however, these differences did not reach statistical significance (data not shown).
Defective osteoclast differentiation in Mitfce/ce mice persists into adulthood due to reduced MITF target gene expression
To further confirm if the observed dense bone phenotype in Mitfce/ce mice is a cell autonomous effect specifically in osteoclasts, we cultured enriched myeloid precursor cells from the spleens of four day old mice in vitro with CSF1/RANKL stimulation. We fixed cells after five days of culture with RANKL, performed TRAP staining and counted the number of multinucleate mature osteoclasts formed from cells derived from Mitfce/ce mice and controls. In response to RANKL stimulation, myeloid linage cells derived from the spleens of four day old Mitfce/ce mice formed 40% fewer osteoclasts than cells derived from the spleens of controls (p = 0.035) (Figure 2A). We also cultured myeloid precursor cells derived from the bone marrow of adult Mitfce/ce mice with CSF1 and RANKL and performed TRAP staining after five days in culture. Similar to the cells derived from neonatal mice, cells from the bone marrow of adult Mitfce/ce mice formed 60% fewer osteoclasts than cells derived from controls (p = 0.031) (Figure 2B). This result indicates that even though the dense bone phenotype seems to partially resolve by adulthood in Mitfce/ce mice, the defect in osteoclasts is persistent.
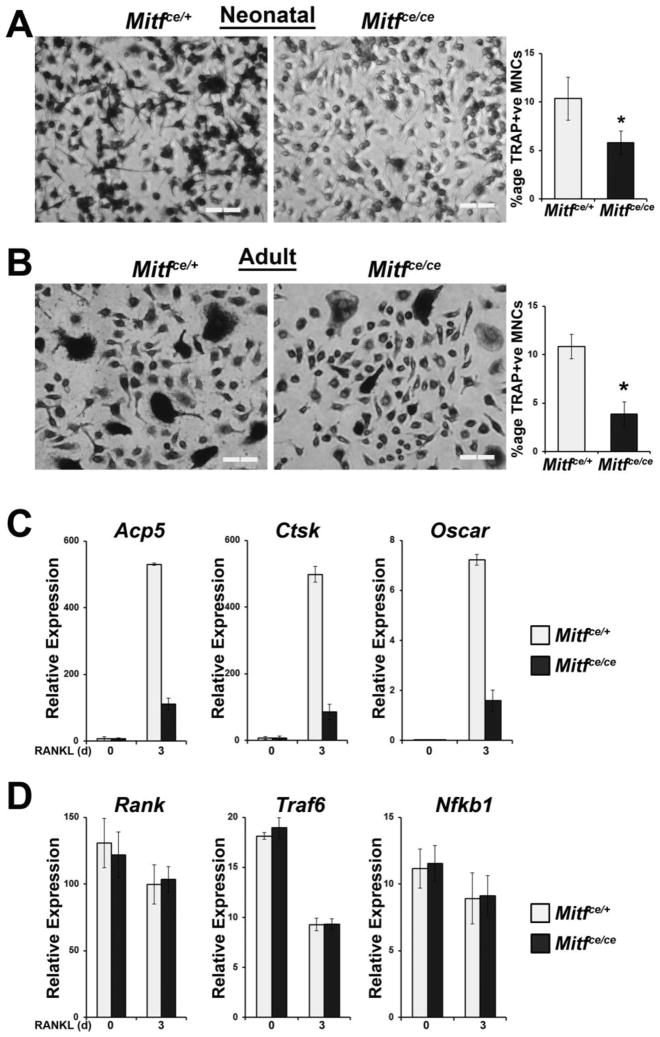
Figure 2. Defective osteoclast differentiation in Mitfce is a result of reduced expression of MITF target genes.
A. Representative images and quantification of TRAP staining of in vitro differentiated osteoclasts from splenocytes extracted from four day old Mitfce/ce mice and Mitfce/+ controls. Primary myeloid lineage cells were cultured from the spleens of four day old mice and subsequently treated in vitro with CSF1 and RANKL for 5 days before TRAP staining. Scale bars = 200μm. Graph indicates the average percentage of multinucleate (>3 nuclei) TRAP-positive osteoclasts in all treated cells. Data are represented as the mean +/− S.D.
B. Representative images and quantification of TRAP staining of in vitro differentiated osteoclasts from the bone marrow of adult Mitfce/ce mice and Mitfce/+ controls. Primary myeloid lineage cells were cultured from the bone marrow of adult mice and subsequently treated in vitro with CSF1 and RANKL for 5 days before TRAP staining. Scale bars = 200μm. Graph indicates the average percentage of multinucleate (>3 nuclei) TRAP-positive osteoclasts in all treated cells. Data are represented as the mean +/− S.D.
C. qRT-PCR gene expression analysis of MITF target genes Acp5, Ctsk, and Oscar in in vitro differentiated osteoclasts from Mitfce/ce mice and Mitfce/+ controls, n = 3. Primary bone marrow derived myeloid cells were treated in vitro with CSF1 alone or CSF1 and RANKL for 3 days. Data are represented as the mean +/− S.D.
D. qRT-PCR gene expression analysis of non-MITF target genes RANK, Traf6, and Nfkb1 in in vitro differentiated osteoclasts from Mitfce/ce mice and Mitfce/+ controls, n = 3. Primary bone marrow derived myeloid cells were treated in vitro with CSF1 alone or CSF1 and RANKL for 3 days. Data are represented as the mean +/− S.D.
To query the effects of the ce mutation on MITF target gene expression, we performed qRT-PCR analysis of the expression of osteoclast marker genes including Acp5, Ctsk, and Oscar. We have previously shown that these genes are transcriptional targets of MITF in osteoclasts in response to RANKL stimulation (Luchin et al., 2001; Sharma et al., 2007; So et al., 2003). Myeloid precursor cells derived from the bone marrow of adult Mitfce/ce mice showed approximately five fold less expression of these genes in response to RANKL stimulation compared to cells derived from control mice (Figure 2C). In contrast, the expression of genes necessary for osteoclast differentiation which are not MITF transcriptional targets was not different between osteoclasts derived from the bone marrow of Mitfce/ce mice and Mitfce/+ controls (Figure 2D). These non-MITF target genes tested include Rank (the receptor for RANKL), RANK’s signaling adapter, Traf6, and Nfkb1, a downstream effector of this pathway. This result indicates that the ce mutation significantly blunts the ability of MITF to control the transcription of the genes that are important for osteoclast function.
The reduced MITF target gene expression in Mitfce/ce mice is not an effect of MITF/PU.1 recruitment to target promoters
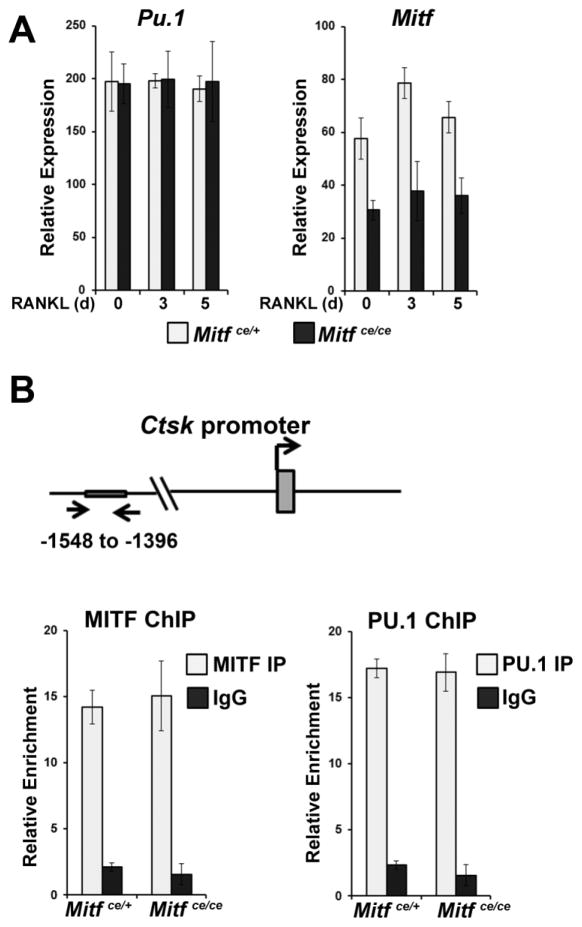
We have previously shown that MITF and PU.1 are co-recruited to target gene enhancers during osteoclast differentiation (Luchin et al., 2001; Sharma et al., 2007). Additionally, Mitf mRNA transcribed from the ce mutant allele may be less stable than wildtype Mitf mRNA (Steingrimsson et al., 1994). Thus, we evaluated whether the dense bone phenotype observed in Mitfce/ce mice was due to decreased expression of Mitf and consequent lower recruitment of the MITF and PU.1 complexes to target genes in Mitfce/ce osteoclasts. To examine this, we tested the expression of Mitf and Pu.1 mRNA in differentiating osteoclasts derived from the bone marrow of adult Mitfce/ce mice using qRT-PCR. We found that cells derived from Mitfce/ce mice expressed the same amount of Pu.1 mRNA throughout osteoclast differentiation in vitro as cells derived from controls (Figure 3A). In contrast, cells derived from Mitfce/ce mice expressed approximately 50% less Mitf mRNA compared to cells derived from controls (Figure 3A).
Figure 3. The MITF-ce mutant protein and its osteoclast-specific co-partner, PU.1, are recruited to target gene promoters.
A. qRT-PCR gene expression analysis of PU.1 and MITF mRNA in in vitro differentiated osteoclasts from Mitfce/ce mice and Mitfce/+ controls, n = 3. Primary bone marrow derived myeloid cells were treated in vitro with CSF1 alone for 1 day or CSF1 and RANKL for 3 or 5 days as indicated. Data are represented as the mean +/− S.D.
B. ChIP-qPCR analysis of PU.1 and MITF binding to the Ctsk promoter in in vitro differentiated osteoclasts from Mitfce/ce mice and Mitfce/+ controls, n = 2. Primary bone marrow derived myeloid cells were treated in vitro with CSF1 and RANKL for 3 days. Data are represented as the mean +/− S.D.
To evaluate whether the lower expression of Mitf mRNA impacted recruitment of MITF/PU.1 complexes to target gene enhancers, we utilized chromatin immunoprecipitation. These experiments demonstrated that MITF and PU.1 bound to the Ctsk promoter at similar levels in osteoclasts derived from both Mitfce/ce mice and controls (Figure 3B), indicating that the ce mutation does not affect the recruitment of these factors to osteoclast specific target genes.
The truncated portion of the Mitfce mutant protein harbors the docking site for p38 MAPK
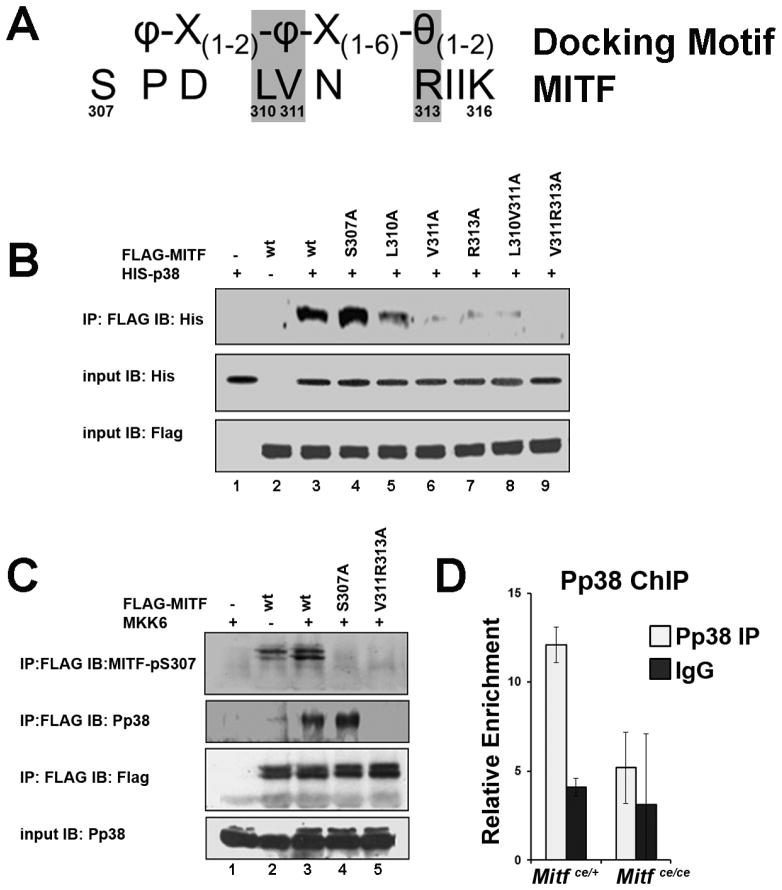
P38 MAPK phosphorylates MITF at Ser307 and this phosphorylation is necessary to initiate target gene transcription (Sharma et al., 2007). Since the binding of MITF and PU.1 to target gene promoters was not affected by the ce mutation of MITF, we next examined whether activation of MITF-ce by p38 MAPK was affected. While MITF Ser307 is absent from the MITF-ce protein, there are potentially other MAP kinase phosphorylation sites present in MITF. Therefore, we set out to map the p38 MAPK docking site necessary for p38 MAPK phosphorylation of MITF and determine whether the Mitfce mutation affected it. Like other MAPKs including ERK and JNK, p38 MAPK uses a D type docking motif (Garai et al., 2012). The D type docking motif takes the form, θ(1–2)-X(1–6)-φ-X(1–2)- φ, where θ is a positively charged residue (R or K), φ is a hydrophobic residue and X can be any amino acid. However, non-canonical docking motifs can be in reverse orientation (see Figure 4A) (Garai et al., 2012). We found MITF potentially contained a non-canonical reversed D type docking motif proximal to the S307 residue that is phosphorylated by p38 MAPK (Figure 4A). To query if these residues form a docking motif for p38 MAPK on MITF, we mutated key residues in the motif, L310, V311, or R313 to alanine. We overexpressed wildtype FLAG-tagged MITF or FLAG-MITF containing these point mutations in COS-7 cells with His-tagged p38 and used co-immunoprecipitation assays to query the effect of these mutations on MITF/p38 interaction. As shown in Figure 4B, p38 MAPK was robustly co-precipitated with wildtype FLAG-MITF and with FLAG-MITF S307A, a mutated protein lacking the p38 MAPK phosphorylation site (Figure 4B, see lanes 3 and 4). In contrast, the V311A and R313A single point mutations significantly interfered with the MITF/p38 interaction (Figure 4B compare lane 3 to lanes 6 and 7). In combination, the V311A and R313A mutations completely abolished the interaction (Figure 4B, compare lanes 3 and 9). The L310A single point mutation, alone or in combination with V311A, only slightly affected the ability of MITF to interact with p38 (Figure 4B, compare lanes 3 and 5). These results indicate that V311 and R313 are necessary for the docking of p38 MAPK to MITF.
Figure 4. A non-canonical reversed docking motif in MITF is essential for MITF-p38MAPK interactions.
A. MITF amino acid sequence near the S307 p38 phosphorylation site, aligned with the D type docking motif. The residues where point mutations were introduced for the experiments described in B are shaded grey. Amino acids indicated as φ are hydrophobic, θ indicates a charged residue, and X can be any amino acid.
B. Lysates from COS-7 cells overexpressing His-p38 and FLAG-MITF or FLAG-MITF with various single or double point mutations in the putative p38 docking site as indicated were immunoprecipitated (IP) with anti-FLAG antibody and immunoblotted (IB) with anti-His antibody (top panel). Anti-His and anti-FLAG input controls are shown in the bottom two panels.
C. Lysates from COS-7 cells overexpressing MKK6 and wildtype (wt) FLAG-MITF, FLAG-MITF with the S307A point mutation or FLAG-MITF with the p38 docking site ablating double mutation V311A;R313A as indicated were immunoprecipitated (IP) with anti-FLAG antibody and immunoblotted (IB) with anti-phospho-S307 MITF antibody (top panel), anti-Pp38 antibody (second panel), or anti-FLAG antibody for pulldown control (third panel from top). Anti-Pp38 input control is shown in the bottom panel.
D. ChIP-qPCR analysis of Pp38 binding to the Ctsk promoter in in vitro differentiated osteoclasts from Mitfce/ce mice and Mitfce/+ controls, n = 2. Primary bone marrow derived myeloid cells were treated in vitro with CSF1 and RANKL for 3 days. Data are represented as the mean +/− S.D.
We next asked if p38 MAPK docking was necessary for the phosphorylation of MITF at Ser307. To answer this question, we over expressed MKK6 to activate p38 in COS-7 cells with wildtype FLAG-MITF, FLAG-MITF S307A, or FLAG-MITF V311A;R313A combination mutation. Immunoprecipitates pulled down with anti-FLAG antibody were probed with an antibody specific for the phosphorylation of S307 on MITF (Mansky et al., 2002). As expected, wildtype FLAG-MITF was phosphorylated by p38 at Ser307 (Figure 4C, see lane 2 in the top panel), and MKK6 overexpression increased this phosphorylation (Figure 4C, compare lanes 2 and 3 in the top panel). FLAG-MITF S307A was not phosphorylated in this assay, even though it still interacted with activated p38 MAPK (Figure 3C, compare lanes 3 and 4). Further, we found that FLAG-MITF V311A;R313A with the mutated p38 docking site was also unable to be phosphorylated by p38 (Figure 4C, compare lanes 3 and 5 in the top panel) indicating that physical interaction between p38 and MITF is necessary for phosphorylation at Ser307 on MITF.
We have previously shown that phosphorylated p38 MAPK is recruited to target gene promoters including Acp5 and Ctsk during osteoclast differentiation in response to RANKL signaling (Sharma et al., 2007). Since the region containing the p38 MAPK docking site is lost in the ce mutant form of MITF, we examined if phospho-p38 MAPK could be recruited to target gene promoters in response to RANKL signaling in BMMs derived from Mitfce/ce mice. ChIP-qPCR assays demonstrated that phospho-p38 MAPK was not recruited to the Ctsk promoter in osteoclasts derived from Mitfce/ce mice in contrast to controls (Figure 4D).
P38 MAPK docking to MITF is necessary for the recruitment of transcriptional co-activators FUS and BRG1
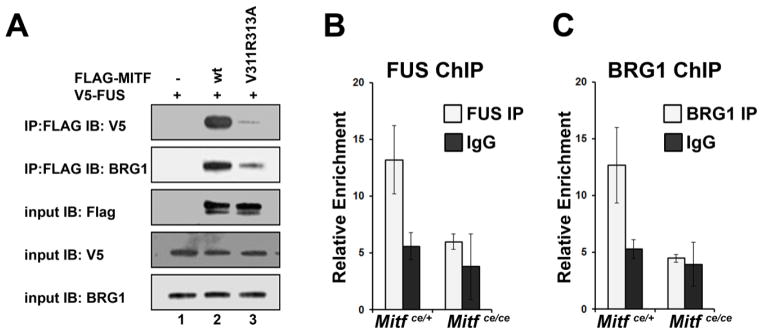
Phosphorylation of MITF at Ser307 by p38 MAPK is necessary for recruitment of transcriptional co-activators FUS and BRG1 (Bronisz et al., 2014). Since p38 MAPK docking to MITF is necessary for this phosphorylation to occur, we next asked if FUS and BRG1 could still interact with MITF lacking the p38 MAPK docking site. To address this, we overexpressed V5-FUS and wildtype FLAG-MITF or FLAG-MITF V311A;R313A p38 docking site mutation in COS-7 cells and determined the ability of MITF to interact with FUS and BRG1 using co-immunoprecipitation assays. As expected, V5-FUS and BRG1 co-precipitated with wildtype MITF (Figure 5A, lane 2 on the top two panels). However, the V311R313A p38 docking site mutation interfered with the ability of MITF to interact with BRG1 (Figure 5A, compare lanes 2 and 3 on the second panel from the top), and nearly completely ablated the interaction of MITF with FUS (Figure 5A, compare lanes 2 and 3 in the top panel).
Figure 5. MITF-ce cannot recruit transcriptional co-activators, FUS and BRG1.
A. Lysates from COS-7 cells overexpressing V5-FUS and wildtype (wt) FLAG-MITF or FLAG-MITF with the p38 docking site ablating double mutation V311R313A as indicated were immunoprecipitated (IP) with anti-FLAG antibody and immunoblotted (IB) with anti-V5 antibody (top panel) or anti-BRG1antibody (second panel). Input controls for FLAG, V5, and BRG1 are shown in the bottom three panels.
B., C. ChIP-qPCR analysis of BRG1 and FUS binding to the Ctsk promoter in in vitro differentiated osteoclasts from Mitfce/ce mice and Mitfce/+ controls, n = 2. Primary bone marrow derived myeloid cells were treated in vitro with CSF1 and RANKL for 3 days. Data are represented as the mean +/− S.D.
We used ChIP-qPCR assays to determine if the MITF-ce protein could recruit transcriptional co-activators FUS and BRG1 to target gene promoters. As predicted, these assays demonstrated that FUS and BRG1 were not effectively recruited to the Ctsk promoter in osteoclasts derived from Mitfce/ce mice, while these important co-activators were recruited in osteoclasts derived from controls (Figure 5B, C).
DISCUSSION
The initial characterization of the Mitfce allele reported that adult mice homozygous for the cloudy eyed (ce) mutation of MITF do not have a discernible bone phenotype (Zimring et al., 1996). Here, using microCT and bone histomorphometry, we have demonstrated that neonatal Mitfce/ce mice display abnormal bone remodeling which partially resolves with age. Similarly, age-resolving osteopetrosis is observed in mouse and rat models with other mutations at the Mitf locus (Cielinski and Marks, 1994; Sharma et al., 2009; Weilbaecher et al., 1998). This effect is most likely caused by the faster rate of bone remodeling in neonatal mice due to their rapid growth at this stage. Additionally, we observed defective osteoclast differentiation in vitro in myeloid precursors derived from both neonatal and adult Mitfce/ce mice. Therefore, although the severity of the dense bone phenotype decreases with age, osteoclasts homozygous for the Mitfce allele continue to exhibit defective differentiation into adulthood.
Previous reports have shown that the MITF-ce protein, as well other MITF mutant proteins, including MITF-mi and MITF-or, fail to bind to nascent DNA in EMSA assays (Hemesath et al., 1994). However, recent results indicate MITF binding at target promoters in the full chromatin context is more complex than previously understood. We have previously shown that MITF-mi and MITF-or mutated proteins are recruited to target gene promoters in osteoclasts, albeit with decreased efficiency than wildtype MITF (Sharma et al., 2009). However, these MITF mutant proteins cannot effectively recruit MITF’s osteoclast specific co-partner, PU.1 to target promoters. The MITF-ce mutant protein, along with its osteoclast specific co-partner, PU.1 is recruited to chromatin at target promoter sites with equal efficiency as controls. Therefore, the loss of the leucine zipper domain in MITF-ce protein does not interfere with the ability of MITF/PU.1 complexes to get recruited to target promoters.
We have presented evidence that the mutant MITF-ce protein leads to defective osteoclast differentiation because it fails to integrate the signaling from RANKL to MITF target gene transcription in osteoclasts. Thus, we have utilized mice with the Mitfce mutation as a valuable tool to examine how the RANKL/p38 MAPK signaling axis affects MITF function in vitro and in vivo. We have previously shown that phosphorylation of MITF by p38 MAPK is necessary for the recruitment of MITF transcriptional co-activators FUS and BRG1 (Bronisz et al., 2014; Sharma et al., 2007). However, a direct interaction of MITF and p38 MAPK had not been reported. Physical interaction of MAP kinases and their substrates can be fostered by docking motifs present on the substrate. Typically found 50–100 residues from the phosphorylation site (Ubersax and Ferrell, 2007), these docking sites effectively increase the concentration of the substrate near the kinase (Deshaies and Ferrell, 2001) and might play a role in substrate specificity (Bardwell et al., 2009). Here, we demonstrate that MITF possesses an atypical reverse orientation docking site next to the target serine of p38 MAPK. Mutating two amino acids (V311 and R313) in the potential docking site completely abrogates MITF-p38 MAPK interactions. MITF is also phosphorylated at Ser73 by ERK signaling in melanocytes and osteoclasts (Wu et al., 2000). Unlike the ERK binding motif which spans 27 amino acids (Molina et al., 2005), the mutation of only two amino acids in the p38 MAPK docking site completely abolishes the binding of p38 MAPK to MITF.
The search for docking motifs and kinase-substrate interactions has been an area of interest because of their potential as to serve as therapeutic targets in various diseases. To date, direct clinical inhibition of p38 MAPK has been largely unsuccessful due to off target effects including toxicities in liver and brain and the upregulation of other compensatory signaling pathways (Sweeney and Firestein, 2006). In this respect, targeting a specific docking site on a substrate of p38 MAPK is a more attractive alternative for disease-specific therapy. The work presented here sheds light on the key residues involved in MITF/p38 MAPK interactions. Since this interaction has only been reported in osteoclasts, the mild bone phenotype seen in adult Mitfce/ce mice makes the p38 MAPK docking site a suitable target for therapeutic applications by modulating excessive osteoclast differentiation and function.
Additionally, we have shown that site-directed mutations which ablate the p38 MAPK docking site on MITF strongly impair the interaction of MITF with its co-activators, FUS and BRG1. Further, the MITF-ce protein, which lacks the docking site, fails to recruit p38 MAPK, FUS and BRG1 to target promoters in differentiating osteoclasts. We have previously shown that recruitment of these co-activators is necessary for the induction of MITF target gene expression in response to RANKL signaling (Bronisz et al., 2014). The inability of MITF-ce to recruit these co-activators to target gene promoters is most likely causing the reduced induction of MITF target genes in myeloid precursors derived from Mitfce/ce mice upon RANKL stimulation. We have previously shown that MITF interacts with FUS through its C-terminal domain (Bronisz et al., 2014). Therefore, the lack of FUS recruitment to MITF target promoters in MITF-ce osteoclasts could be attributed to abrogation of the direct interaction between MITF-ce and FUS since MITF-ce lacks the C-terminal end. However, the mutation of only the p38 MAPK docking site on MITF severely hampers MITF/FUS interactions (Figure 5A), indicating that p38 MAPK docking and phosphorylation of MITF are key events responsible for the recruitment of MITF co-factors, FUS and BRG1. Furthermore, it is plausible that in addition to MITF, p38 MAPK may also function to directly activate its co-partners, FUS, BRG1, and PU.1. Future experiments can be directed at answering these questions.
In conclusion, we have presented mechanistic evidence explaining the defective osteoclast function caused by the Mitfce mutation and the dense bone phenotype seen in neonatal Mitfce/ce mice. MITF is essential for the differentiation of a wide variety of cell linages and attains its specificity partly through interaction with lineage specific co-factors and additionally by its response to specific signaling cascades. This work contributes important information to our knowledge of how MITF directs an osteoclast-specific gene program in response to signaling unique to the bone microenvironment. Understanding the specificity of MITF’s interaction with effector kinases in different cell types is vital to comprehending the biology of differentiation programs in higher eukaryotes.
Acknowledgments
Contract grant sponsor: NIAMS
Contract grant number: 2R01AR044719-15A
The authors are grateful to Raksha Adhikari and Jiang Yue for technical assistance, Dr. Said Sif for providing the BRG1 antibody and Subhasree Balakrishnan for graphical representation. This work was supported by NIAMS, National Institutes of Health Grant 2R01AR044719-15A to MCO and SSM.
References
- Bardwell AJ, Frankson E, Bardwell L. Selectivity of docking sites in MAPK kinases. The Journal of biological chemistry. 2009;284(19):13165–13173. doi: 10.1074/jbc.M900080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423(6937):337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- Bronisz A, Carey HA, Godlewski J, Sif S, Ostrowski MC, Sharma SM. The multifunctional protein fused in sarcoma (FUS) is a coactivator of microphthalmia-associated transcription factor (MITF) The Journal of biological chemistry. 2014;289(1):326–334. doi: 10.1074/jbc.M113.493874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Qiao H, Su Z, Li H, Ping Q, Zong L. Emerging therapeutic targets for osteoporosis treatment. Expert opinion on therapeutic targets. 2014;18(7):817–831. doi: 10.1517/14728222.2014.912632. [DOI] [PubMed] [Google Scholar]
- Cielinski MJ, Marks SC., Jr Neonatal reductions in osteoclast number and function account for the transient nature of osteopetrosis in the rat mutation microphthalmia blanc (mib) Bone. 1994;15(6):707–715. doi: 10.1016/8756-3282(94)90321-2. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Ferrell JE., Jr Multisite phosphorylation and the countdown to S phase. Cell. 2001;107(7):819–822. doi: 10.1016/s0092-8674(01)00620-1. [DOI] [PubMed] [Google Scholar]
- Garai A, Zeke A, Gogl G, Toro I, Fordos F, Blankenburg H, Barkai T, Varga J, Alexa A, Emig D, Albrecht M, Remenyi A. Specificity of linear motifs that bind to a common mitogen-activated protein kinase docking groove. Science signaling. 2012;5(245):ra74. doi: 10.1126/scisignal.2003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE. microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes & development. 1994;8(22):2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74(2):395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- Hu R, Sharma SM, Bronisz A, Srinivasan R, Sankar U, Ostrowski MC. Eos, MITF, and PU.1 recruit corepressors to osteoclast-specific genes in committed myeloid progenitors. Molecular and cellular biology. 2007;27(11):4018–4027. doi: 10.1128/MCB.01839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Takeshita S. Factors and mechanisms involved in the coupling from bone resorption to formation: how osteoclasts talk to osteoblasts. Journal of bone metabolism. 2014;21(3):163–167. doi: 10.11005/jbm.2014.21.3.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuta J, Ishii M. Osteoclast migration, differentiation and function: novel therapeutic targets for rheumatic diseases. Rheumatology. 2013;52(2):226–234. doi: 10.1093/rheumatology/kes259. [DOI] [PubMed] [Google Scholar]
- Lacey DL, Timms E, Tan HL, Kelley MJ, Dunstan CR, Burgess T, Elliott R, Colombero A, Elliott G, Scully S, Hsu H, Sullivan J, Hawkins N, Davy E, Capparelli C, Eli A, Qian YX, Kaufman S, Sarosi I, Shalhoub V, Senaldi G, Guo J, Delaney J, Boyle WJ. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93(2):165–176. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends in molecular medicine. 2006;12(9):406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luchin A, Suchting S, Merson T, Rosol TJ, Hume DA, Cassady AI, Ostrowski MC. Genetic and physical interactions between Microphthalmia transcription factor and PU.1 are necessary for osteoclast gene expression and differentiation. The Journal of biological chemistry. 2001;276(39):36703–36710. doi: 10.1074/jbc.M106418200. [DOI] [PubMed] [Google Scholar]
- Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92(4):463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- Mansky KC, Sankar U, Han J, Ostrowski MC. Microphthalmia transcription factor is a target of the p38 MAPK pathway in response to receptor activator of NF-kappa B ligand signaling. The Journal of biological chemistry. 2002;277(13):11077–11083. doi: 10.1074/jbc.M111696200. [DOI] [PubMed] [Google Scholar]
- Meadows NA, Sharma SM, Faulkner GJ, Ostrowski MC, Hume DA, Cassady AI. The expression of Clcn7 and Ostm1 in osteoclasts is coregulated by microphthalmia transcription factor. The Journal of biological chemistry. 2007;282(3):1891–1904. doi: 10.1074/jbc.M608572200. [DOI] [PubMed] [Google Scholar]
- Molina DM, Grewal S, Bardwell L. Characterization of an ERK-binding domain in microphthalmia-associated transcription factor and differential inhibition of ERK2-mediated substrate phosphorylation. The Journal of biological chemistry. 2005;280(51):42051–42060. doi: 10.1074/jbc.M510590200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KJ. Insight into the microphthalmia gene. Trends in genetics : TIG. 1995;11(11):442–448. doi: 10.1016/s0168-9525(00)89143-x. [DOI] [PubMed] [Google Scholar]
- Morii E, Ogihara H, Kim DK, Ito A, Oboki K, Lee YM, Jippo T, Nomura S, Maeyama K, Lamoreux ML, Kitamura Y. Importance of leucine zipper domain of mi transcription factor (MITF) for differentiation of mast cells demonstrated using mi(ce)/mi(ce) mutant mice of which MITF lacks the zipper domain. Blood. 2001;97(7):2038–2044. doi: 10.1182/blood.v97.7.2038. [DOI] [PubMed] [Google Scholar]
- Novack DV, Teitelbaum SL. The osteoclast: friend or foe? Annual review of pathology. 2008;3:457–484. doi: 10.1146/annurev.pathmechdis.3.121806.151431. [DOI] [PubMed] [Google Scholar]
- Sharma SM, Bronisz A, Hu R, Patel K, Mansky KC, Sif S, Ostrowski MC. MITF and PU.1 recruit p38 MAPK and NFATc1 to target genes during osteoclast differentiation. The Journal of biological chemistry. 2007;282(21):15921–15929. doi: 10.1074/jbc.M609723200. [DOI] [PubMed] [Google Scholar]
- Sharma SM, Sif S, Ostrowski MC, Sankar U. Defective co-activator recruitment in osteoclasts from microphthalmia-oak ridge mutant mice. Journal of cellular physiology. 2009;220(1):230–237. doi: 10.1002/jcp.21755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YH, Godlewski J, Bronisz A, Zhu J, Comb MJ, Avruch J, Tzivion G. Significance of 14-3-3 self-dimerization for phosphorylation-dependent target binding. Molecular biology of the cell. 2003;14(11):4721–4733. doi: 10.1091/mbc.E02-12-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So H, Rho J, Jeong D, Park R, Fisher DE, Ostrowski MC, Choi Y, Kim N. Microphthalmia transcription factor and PU.1 synergistically induce the leukocyte receptor osteoclast-associated receptor gene expression. The Journal of biological chemistry. 2003;278(26):24209–24216. doi: 10.1074/jbc.M302940200. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Moore KJ, Lamoreux ML, Ferre-D’Amare AR, Burley SK, Zimring DC, Skow LC, Hodgkinson CA, Arnheiter H, Copeland NG, et al. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nature genetics. 1994;8(3):256–263. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- Sweeney SE, Firestein GS. Mitogen activated protein kinase inhibitors: where are we now and where are we going? Annals of the rheumatic diseases. 2006;65(Suppl 3):iii83–88. doi: 10.1136/ard.2006.058388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitelbaum SL, Ross FP. Genetic regulation of osteoclast development and function. Nature reviews Genetics. 2003;4(8):638–649. doi: 10.1038/nrg1122. [DOI] [PubMed] [Google Scholar]
- Tshori S, Gilon D, Beeri R, Nechushtan H, Kaluzhny D, Pikarsky E, Razin E. Transcription factor MITF regulates cardiac growth and hypertrophy. The Journal of clinical investigation. 2006;116(10):2673–2681. doi: 10.1172/JCI27643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax JA, Ferrell JE., Jr Mechanisms of specificity in protein phosphorylation. Nature reviews Molecular cell biology. 2007;8(7):530–541. doi: 10.1038/nrm2203. [DOI] [PubMed] [Google Scholar]
- Weilbaecher KN, Hershey CL, Takemoto CM, Horstmann MA, Hemesath TJ, Tashjian AH, Fisher DE. Age-resolving osteopetrosis: a rat model implicating microphthalmia and the related transcription factor TFE3. The Journal of experimental medicine. 1998;187(5):775–785. doi: 10.1084/jem.187.5.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Hemesath TJ, Takemoto CM, Horstmann MA, Wells AG, Price ER, Fisher DZ, Fisher DE. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes & development. 2000;14(3):301–312. [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Hayashi S, Kunisada T, Ogawa M, Nishikawa S, Okamura H, Sudo T, Shultz LD, Nishikawa S. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature. 1990;345(6274):442–444. doi: 10.1038/345442a0. [DOI] [PubMed] [Google Scholar]
- Zimring DC, Lamoreux ML, Millichamp NJ, Skow LC. Microphthalmia cloudy-eye (mi(ce)): a new murine allele. The Journal of heredity. 1996;87(4):334–338. doi: 10.1093/oxfordjournals.jhered.a023009. [DOI] [PubMed] [Google Scholar]