Abstract
The innate immune system is critical for the early detection of invading pathogens and for initiating cellular host defence countermeasures, which include the production of type I interferon (IFN)1–3. However, little is known about how the innate immune system is galvanized to respond to DNA-based microbes. Here we show that STING (stimulator of interferon genes) is critical for the induction of IFN by non-CpG intracellular DNA species produced by various DNA pathogens after infection4. Murine embryonic fibroblasts, as well as antigen presenting cells such as macrophages and dendritic cells (exposed to intracellular B-form DNA, the DNA virus herpes simplex virus 1 (HSV-1) or bacteria Listeria monocytogenes), were found to require STING to initiate effective IFN production. Accordingly, Sting-knockout mice were susceptible to lethal infection after exposure to HSV-1. The importance of STING in facilitating DNA-mediated innate immune responses was further evident because cytotoxic T-cell responses induced by plasmid DNA vaccination were reduced in Sting-deficient animals. In the presence of intracellular DNA, STING relocalized with TANK-binding kinase 1 (TBK1) from the endoplasmic reticulum to perinuclear vesicles containing the exocyst component Sec5 (also known as EXOC2). Collectively, our studies indicate that STING is essential for host defence against DNA pathogens such as HSV-1 and facilitates the adjuvant activity of DNA-based vaccines.
Nucleic acid species inadvertently generated by microbes after infection are potent inducers of cellular innate immune defences important for protection of the host1–3. Although considerable progress has been made into unravelling how RNA viruses induce type I IFN, required for triggering the production of anti-viral genes, little is known at the molecular level about the induction of IFN by DNA pathogens such as herpes simplex virus I (HSV-1) or by intracellular bacteria or parasites5–10. Toll-like receptor 9 (TLR9) is known to recognize CpG DNA to trigger IFN production in plasmacytoid dendritic cells (pDCs), and Z-DNA binding protein 1 (ZBP1, also known as DAI) was recently shown to be able to stimulate IFN transcription, but was found to be largely redundant in studies using DAI-deficient cells and mice11–13. Recently, a DNA receptor AIM2 was found to be important for ASC (also known as PYCARD)-dependent inflammasome mediated production of IL1β, but was not required for type I IFN production14–18. Thus, other innate signalling pathways that recognize intracellular non-CpG DNA species must exist to facilitate type I IFN production.
We previously demonstrated for the first time a role for STING (also referred to as TMEM173, MPYS and MITA), an endoplasmic reticulum (ER) resident transmembrane protein, in facilitating the production of type I IFN4,19,20. To evaluate the importance of STING in mediating DNA-induced innate immune responses, we used wild type (+/+) or Sting−/− low passage number mouse embryonic fibroblasts (MEFs) and compared the induction of type I IFN (IFNβ) in response to a variety of DNA ligands. Our results indicated that STING was essential for inducing IFNb in response to transfected viral DNA (adenovirus, Ad5; herpes simplex virus, HSV-1 and -2), purified Escherichia coli DNA, calf thymus (CT) DNA, and interferon stimulatory DNA (ISD; double-stranded 45-base-pair oligonucleotides lacking CpG sequences) (Fig. 1a). Complete abrogation of IFNβ production was also observed after transfection of synthetic double-stranded DNA (poly(dG-dC)•poly(dC-dG), hereafter referred to as poly(dGC:dGC)) in Sting−/− MEFs, and slight IFNβ production was observed using poly(dAT:dAT), probably due to STING-independent, RIG-I (also known as DDX58)-dependent signalling21,22. The loss of STING did not significantly affect poly(I:C)-mediated type I IFN production, which is largely governed by MDA5 (ref. 5). Concomitant analysis further indicated a marked reduction in IL6 production in Sting−/− MEFs compared to controls after similar DNA transfections (Fig. 1a). ISD-mediated production of Ifnb and Ifn2a messenger RNA was not detectable in Sting−/− MEFs compared to controls (Fig. 1b). Translocation of IRF3 or IRF7 was thus not observed in ISD-transfected Sting−/− MEFs, indicating that STING probably functions in mediating intracellular-DNA-triggered IFN production upstream of TBK1 (Fig. 1c and Supplementary Fig. 1). NF-κB signalling was also defective in Sting−/− MEFs after exposure to transfected ISD (Supplementary Fig. 1). Given this, we next examined the importance of STING in facilitating intracellular-DNA-mediated production of type I IFN in antigen presenting cells. This analysis indicated that Sting−/− macrophages transfected with ISD, or infected with the DNA pathogens HSV-1 or Listeria monocytogenes, were greatly defective in their ability to manufacture type I IFN (Fig. 1d). However, the cleavage of pro-caspase 1 and production of active IL1β, which is AIM2-dependent, was unaffected by the loss of STING (Fig. 1e and Supplementary Fig. 1). Thus, STING functions independently of the AIM2 ‘inflammasome’ pathway. Further analysis also indicated that STING was required for efficient DNA-mediated production of type I IFN in granulocyte–macrophage dendritic cells (GM-DCs), as well as pDCs (FLT3-ligand-induced dendritic cells, FLT3-DCs) (Fig. 1f, g). However, exogenous CpG DNA remained able to induce type I IFN in Sting−/− FLT3-DCs compared to controls, indicating that TLR9 functions independently of the STING pathway (Fig. 1g). The induction of IL6 in response to intracellular DNA was also reduced in Sting−/− macrophages (Supplementary Fig. 1). However, HSV-1 and CpG DNA remained able to induce IL6 in Sting−/− macrophages, probably through TLR9-dependent signalling (Supplementary Fig. 1)11. Furthermore, we noted that STING seemed to be essential for the production of type I IFN by cytomegalovirus (CMV), vaccinia virus (VVΔE3L) and baculovirus (Supplementary Fig. 1). STING therefore seems critical for intracellular-DNA-mediated production of type I IFN in fibroblasts, macrophages, conventional dendritic cells as well as pDCs.
Figure 1. STING is essential for intracellular DNA-mediated type I IFN production.
a, MEFs were transfected with 1 μg ml−1 of DNA ligands (with Lipofectamine 2000) for 16 h, and IFNβ or IL6 were measured. b, MEFs were transfected with ISD for 4 h and Ifnb or Ifna2 mRNA levels were measured. c, MEFs treated as in b were stained with an antibody for IRF3 translocation. Original magnification, ×40. d, Bone-marrow-derived macrophages were transfected with poly(dAT:dAT), poly(I:C) or ISD, or infected with HSV-1 (multiplicity of infection (m.o.i.) 10) or Listeria (m.o.i. 10) for 16 h, and IFNβ was measured. e, Macrophages were infected with HSV-1 for 16 h and IL1β was measured. f, GM-colony stimulating factor (CSF)-induced dendritic cells (GM-DCs) were treated as in d, and IFNβ or IFNα was measured after 16 h. g, FLT3-stimulated dendritic cells were treated as in f (exogenous CpG oligodeoxynucleotides (ODN) (1 μg ml−1) were also used). *P < 0.05, Student's t-test. Error bars indicate s.d. ND, not determined.
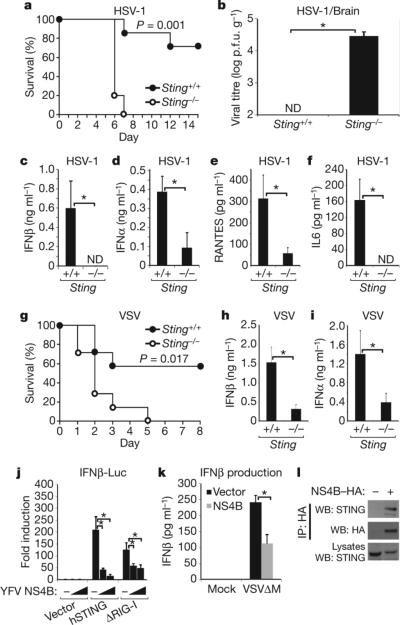
We next evaluated the in vivo importance of STING in facilitating effective host defence against select virus infection. Principally, Sting−/− or control mice were infected intravenously (i.v.) with HSV-1 and survival was monitored. The Sting-knockout mice died within 7 days of HSV-1 infection (Fig. 2a), whereas 80% of similarly infected wild-type mice survived. Significant amounts of HSV-1 were detected in the brain of infected Sting−/− mice, but not in controls at 5 days after infection (Fig. 2b). Analysis of serum from the Sting−/−-infected animals indicated a profound defect in the production of type I IFN at 6 h after infection, compared to infected control animals (Fig. 2c, d and Supplementary Fig. 2). RANTES and IL6 levels were similarly markedly reduced in Sting−/− mice at the same time point (Fig. 2e, f). Moreover, Sting−/− mice were found to be more sensitive to HSV-1 after intravaginal administration of HSV-1 (Supplementary Fig. 2). This data indicates that STING is necessary, in vivo, for the effective production of type I IFN and is essential for efficient protection against HSV-1 infection.
Figure 2. STING is required for effective in vivo host defence.
a, Sting-deficient animals (Sting−/−) or littermate controls (Sting+/+) (n = 7; approximately 8-weeks-of-age) were infected with HSV-1 (1 × 107 i.v.) and survival was monitored. b, Sting−/− or control mice were infected with HSV-1 as in a and brains were retrieved after 5 days for HSV-1 plaque assays. p.f.u., plaque-forming units. c, d, Serum from animals (n = 3) infected with HSV-1 (1 × 107 i.v.) was analysed for IFNβ (c) or IFNα (b) production after 6 h. e, f, Serum from animals infected as in c was analysed for RANTES (e) and IL6 (f) production. g, Sting−/− or control mice (n = 6) were infected with VSV (5 × 107 i.v.) and survival was monitored. h, i, Mice (n = 3) were treated as in g and IFNβ (h) or IFNα (i) was measured after 6 h. j, Increasing amounts of YFV NS4B were co-transfected into 293T cells with human STING or the amino terminus of RIG-I (ΔRIG-I, residues 1–284) and transfected IFNβ promoter-driven luciferase (IFNβ-Luc) was measured after 36 h. k, Immortalized MEFs were transfected with YFV NS4B for 24 h, infected with VSVΔM4 (m.o.i. 1) for 16 h, and IFNβ was measured. l, 293 cells were transfected with NS4B–HA for 36 h and after immunoprecipitation (IP) with anti-haemagglutinin antibody, were analysed by western blot (WB) using anti-STING serum. *P < 0.05, Student's t-test. Error bars indicate s.d.
Because we had previously seen, in vitro, a defect in the ability of the negative-stranded virus vesicular stomatitis virus (VSV) to induce type I IFN in the absence of STING, we next examined the in vivo importance of STING in protecting against VSV-related disease4. We observed that Sting−/− animals infected with VSV was also significantly sensitive to lethal infection compared to controls (Fig. 2g). Defects in type I IFN production were seen in Sting-knockout mice at early time points (6 h), although less so at 24 h (Fig. 2h, i and Supplementary Fig. 2). Thus, STING is necessary for efficient, early induction of type I IFN production and is required for protection against infection with the negative-stranded virus VSV, possibly by regulating the RIG-I and IPS-1 (also known as MAVS, VISA and CARDIF) pathway4,6–10.
We did not observe a significant requirement for STING in facilitating poly(I:C) or EMCV (encephalomyocarditis virus, a positive-stranded flavivirus)-mediated IFN transcription, indicating that STING may not influence MDA5 function (Fig. 1a and Supplementary Fig. 2)4. However, it is known that some flaviviruses such as hepatitis C virus (HCV) can activate the RIG-I pathway, signalling which seems to be influenced by STING4,23. In this regard, databank analysis indicated that the flaviviruses yellow fever virus (YFV) and Dengue virus encode a product NS4B that exhibits strong homology with the amino terminus of STING (amino acids 125–222) (Supplementary Fig. 3). This region was found to be critical for STING function (Supplementary Fig. 3). Various flaviviral NS4B products have been shown to localize to the ER of the cell and to suppress the induction of type I IFN, although the mechanisms remain unclear24. Our analysis here indicates that that NS4B was able to inhibit STING activity, probably by direct association (Fig. 2j–l and Supplementary Fig. 3). Thus, STING may be targeted by certain viruses for suppression.
TBK1 has been shown to have an important role in mediating the adjuvant activity of DNA vaccines in vivo12. TBK1 activation in response to plasmid DNA was found to occur in the absence of the DNA sensors TLR9 or DAI, indicating that other pathways exist to facilitate DNA-mediated immunization12,25. To evaluate whether STING was involved in this signalling pathway, Sting−/− or control mice were immunized with plasmid DNA encoding the ovalbumin gene. Although we noted normal B- and T-cell subsets in unstimulated Sting−/− animals, after immunization Sting−/− mice showed significantly less serum ovalbumin (OVA)-specific IgG compared to controls (Fig. 3a and Supplementary Fig. 4). Furthermore, spleen CD8+ T-cell frequency and IFNγ secretion was markedly reduced in Sting−/− mice after immunization, compared to wild-type mice (Fig. 3b, c). Because immunoglobulin responses to OVA peptide were normal, these data emphasize that the STING-governed DNA sensor pathway is essential for efficient DNA-vaccine-induced T-cell responses to antigen (Fig. 3 and Supplementary Fig. 4). Similar studies also indicated that STING had a key role in facilitating T-cell responses to the DNA virus vaccinia expressing ovalbumin (VV-OVA). Our data emphasizes the importance of STING in innate immune signalling processes required for DNA adjuvant activity (Fig. 3d).
Figure 3. STING is required for effective DNA-mediated adaptive immune responses.

a, Sting−/− or control (Sting+/+) mice (n = 5; approximately 8-weeks-of-age) were immunized twice (100 μg i.m.) by electroporation with a DNA vaccine encoding ovalbumin. Serum was measured for anti-OVA IgG. b, c, Mice were treated as in a and spleen CD8+ IFNγ+ cells were measured by fluorescence-activated cell sorting (FACS; b), and anti-OVA-specific IFNγ production was measured by ELISA after stimulation of splenocytes using SIINFEKL peptide (c). d, Sting−/− mice or controls (n = 4; approximately 8-weeks-of-age) were infected with vaccinia expressing ovalbumin (VV-OVA; 5 × 106 i.v.) and spleen anti-OVA-specific IFNγ production was measured by ELISA. *P < 0.05, Student's t-test. Error bars indicate s.d. All experiments were repeated twice.
We previously demonstrated that STING is an ER resident protein and member of the TRAP(translocon associated protein) complex that can associate with RIG-I and the mitochondrial innate immune signalling adaptor IPS-1 (refs 4, 26). Physical association of mitochondria and the ER, referred to as mitochondria-associated ER membrane (MAM), is important for transmission of Ca2+ to the mitochondria and for oxidative metabolism27. We thus examined whether STING could associate with MAMs. First, we reconstituted haemagglutinin (HA)-tagged STING into Sting−/− MEFs to follow endogenous STING localization using a haemagglutinin antibody. This analysis confirmed that STING is predominantly associated with the ER as determined by calreticulin marker co-staining (Fig. 4a). Mitotracker co-staining also indicated that STING may co-localize with mitochondria associated with the ER (Fig. 1b). The association of endogenous STING with the ER was also confirmed using anti-STING serum (Supplementary Fig. 5). Fractionation analysis subsequently demonstrated that STING is associated with microsomes, a complex of continuous membranes that comprise the ER, Golgi and transport vesicles (Fig. 4c). Endogenous STING was found to fractionate with MAMs and mitochondria fractions under non-stimulated conditions in MEFs (Fig. 4c). Calreticulin, known to be a chaperone involved in regulating the association of the ER and mitochondria, was observed to fractionate similarly27. This data may indicate that STING could associate with IPS-1 by MAM interaction4. Interestingly, after HSV-1 infection, STING was shown to become predominantly associated only with microsome fractions (Fig. 4c). To clarify these observations, we infected STING–HA MEFs with HSV-1, or transfected these cells with stimulatory ISD or negative-control single-stranded DNA (ssDNA). These results indicated that in response to HSV-1 infection or ISD transfection, STING translocated from the ER and predominantly congregated to perinuclear, non-ER microsome compartments in the cell (Fig. 4d and Supplementary Figs 5 and 6). Brefeldin A, but not chloroquine, blocked STING trafficking, indicating that STING locates from the ER via the Golgi to vesicles in the perinuclear region (Supplementary Fig. 5). This trafficking, in response to intracellular DNA, was similarly observed for TBK1, which we have previously shown to associate with STING4 (Fig. 4e). Notably, in the absence of STING, TBK1 failed to relocate to perinuclear regions in response to ISD transfection (Supplementary Fig. 7).
Figure 4. STING translocates from the ER to Sec5-containing vesicles.
a, Sting−/− MEFs, stably reconstituted with haemagglutinin-tagged mouse STING (mSTING–HA) were stained using haemagglutinin (green) and a calreticulin (red) antibody. b, STING–HA MEFs were stained for STING–HA (green), calreticulin (blue) or Mitotracker (red) and three-dimensional reconstruction images were taken. c, Immunoblot analysis of fractionation experiments of uninfected or HSV-1-infected (m.o.i. 10; 4 h) MEFs. Endogenous STING was detected using an anti-STING antibody. Calreticulin detects ER, Sigma1R detects MAM, and COXIV detects mitochondria. d, Haemagglutinin (green) or calreticulin (red) staining of mSTING–HA MEFs after treatment with transfected ISD (1 μg ml−1), transfected ssDNA (1 μg ml−1) or HSV-1 infection as in c. e, mSTING–HA MEFs were transfected with or without ISD and cells were stained with haemagglutinin (green), calreticulin (blue) and a TBK1 (red) antibody. f, mSTING–HA MEFs were transfected as in e and stained with haemagglutinin (green) and a TFR (red) antibody. g, mSTING–HA MEFs were transfected as in e and stained with haemagglutinin (green) and a Sec5 antibody (red). h, i, MEFs were treated with RNAi to Trapb, Sting or Sec5 for 72 h and transfected with ISD. IFNβ mRNA and protein were measured at 4 and 16 h, respectively. *P < 0.05, Student's t-test. Error bars indicate s.d. Scale bars, 10 μm.
We further observed that in the presence of DNA, STING mostly localized with the early endosome marker protein EEA1 and recycling endosome marker transferrin receptor (TFR; Fig. 4f and Supplementary Fig. 6). TBK1 has also been demonstrated to associate with Sec5, a component of the excocyst 8 subunit complex that facilitates vesicular transport processes28. After intracellular DNA stimulation, STING was found to strongly colocalize with Sec5, which has also been demonstrated to associate in perinuclear endosome compartments (Fig. 4g)29. The RALB and Sec5 pathway has been previously shown to be required for efficient Sendai-virus-mediated type I IFN production28. However, our data here indicates that STING and TBK1 complexes may traffic to endosome compartments to associate with Sec5/exocyst components and facilitate the production of type I IFN in response to intracellular DNA. To evaluate whether Sec5 also modulates the production of IFNβ in response to ISD, we suppressed Sec5 production in normal MEFs using RNA interference (RNAi). This study indicated that in the absence of Sec5, ISD-mediated IFN production was significantly impaired (Fig. 4h, i). A similar effect was observed after knockdown of Trapb (also known as Ssr2) and Sec61b, components of the TRAP complex (Fig. 4h, i and Supplementary Fig. 8). Our data thus indicates that intracellular DNA may induce STING to complex with TBK1 and traffic to Sec5-containing endosome compartments—events that facilitate the production of type I IFN.
In conclusion, we demonstrate that STING is essential for the recognition of intracellular DNA and efficient production of type I IFN in all cell types examined. Loss of STING renders mice susceptible to lethal DNA virus infection (HSV-1). However STING also facilitates host defence responses to negative-stranded viruses such as VSV, plausibly through RIG-I and IPS-1–MAM translocon interactions. Although STING-independent, VSV-mediated type I IFN-induction pathways clearly exist, they do not seem to be sufficient on their own to protect mice against lethal VSV infection. We conclude that in response to intracellular DNA, STING and TBK1 complexes traffic to endosomal compartments to associate with exocyst components including Sec5, resulting in the induction of type I IFN.
METHODS
Mice, cells, viruses and reagents
Sting-knockout mice on a 129SvEv3 × C57BL/6J background have been described previously4. MEFs, bone-marrow-derived macrophages and GM-DCs were prepared as described previously4. To prepare FLT3-DCs, bone marrow cells were cultured in RPMI 1640 medium supplemented with 10% FBS, 50 μM 2-mercaptoethanol, 10 mM HEPES, pH 7.4, and 100 ng ml−1 human FLT3 ligand (Peprotech) for 8 days. 293T cells were obtained from the American Type Culture Collection (ATCC) and were maintained in DMEM medium supplemented with 10% FBS. VSV (Indiana strain), VSVΔM and EMCV were described previously4. HSV-1 (KOS strain) and Listeria monocytogenes (10403 serotype) were obtained from ATCC. Vaccinia virus encoding chicken ovalbumin (VV-OVA), vaccinia virus E3L deletion mutant (VVΔE3L), human cytomegalovirus (AD169 strain), and baculovirus (Autographa californica M nucleopolyhedrovirus)were gifts from J. Yewdell, B. Jacobs, K. Frueh and M. Kobayashi, respectively. Genomic DNA was obtained from following sources: HSV-1, HSV-2, adenovirus type 5 (ATCC); E. coli, and calf thymus (Sigma). Poly(dAT:dAT) and poly(I:C) were obtained from Amersham Biosciences. Poly(dGC:dGC) and poly(dA) were obtained from Sigma. CpG ODN (ODN 1585) was obtained from Invivogen. For stimulation of cells, genomic DNA, polydeoxynucleotides or poly(I:C) were mixed with Lipofectamine 2000 (Invitrogen) at a ratio of 1:1 (v/w), and then added to cells at a final concentration of 1 μg ml−1. LPS was obtained from Invivogen. Brefeldin Aand chloroquine were obtained from Sigma.
Plasmids
YFV NS4B sequence was amplified by PCR using pYFM5.2 encoding the complete YFV-17D sequence as a template, and was cloned into a pcDNA3 (Invitrogen) plasmid to generate carboxy-terminally haemagglutinin-tagged expression construct. C-terminally haemagglutinin-tagged STINGΔSP (Δ1–36 amino acids) and STINGΔTM5 (Δ153–173 amino acids) were amplified by PCR and cloned into a pcDNA3 plasmid. The expression plasmid containing chicken ovalbumin (OVA) complementary DNA was constructed by cloning of PCR-amplified OVA cDNA into pCDNA3. Expression plasmids encoding haemagglutinin-tagged murine STING (mSTING–HA), Flag-tagged ΔRIG-I (amino acids 1–284), ΔMDA5 (amino acids 1–349) and IRF-7 were described previously4. p110-Luc (IFNβ-Luc) was obtained from T. Maniatis. pUNO-hsaIRF3 (IRF3SA) and pUNO-hsaIRF7Δ (IRF7SA) were obtained from Invivogen. pCMV-SPORT6 containing murine DAI was obtained from Open Biosystems.
Primers
The following primers were used for cloning: YFV NS4B forward, 59′-GGGGTACCATGAACGAGCTAGGCATGCTGGAG-3′; YFV NS4B reverse, 5′-CCGCTCGAGCCGGCGTCCAGTTTTCATCTTC-3′; STINGΔSP forward, 5′-CCCAAGCTTGCCGCCACCATGCTAGGAGAGCCACCAGAGCAC-3′;STINGΔSP reverse, 5′-CCGCTCGAGAGAGAAATCCGTGCGGAGAG-3′; OVA forward, 5′-ATGGGCTCCATCGGCGCAGCAA-3′; OVA reverse, 5′-TTAAGGGGAAACACATCTGCC-3′.
Antibodies and ELISA
Rabbit polyclonal antibody against STING was described previously4. The antibody against STING-C was generated by immunizing rabbit with recombinant glutathione S-transferase (GST)–hSTING-C (amino acids 173–379) produced in E. coli. Rabbit polyclonal antibody against Sec5 was a gift from H. Horiuchi. Other antibodies were obtained from following sources: caspase-1 p10 (Santa Cruz Biotechnology), calreticulin (ab14234; Abcam), Sigma1 receptor (ab53852; Abcam), TBK1 (EP611Y, Abcam), COXIV (ab16056, Abcam), rabbit polyclonal HA (ab9110; Abcam), transferrin receptor (H68.4; Invitrogen), mouse monoclonal haemagglutinin (Sigma), Flag (M2; Sigma), IRF3 (ZM3; Zymed), TGN46 (ab16059; Abcam), giantin (ab24586; Abcam), EEA1 (no.2441; Cell Signaling), LAMP1 (NB120; Novus Biologicals) and Sec61β (Upstate). ELISA kits were obtained from following sources: murine IFNβ and IFNα (PBL), murine IL6 (R&D systems or Quansys Biosciences), murine IL1β and IFNγ (R&D systems), active NF-κB p65 (Active Motif), and murine RANTES (Quansys Biosciences).
Real-time PCR
Fluorescence real-time PCR analysis was performed using a LightCycler 2.0 instrument (Roche Molecular Biochemicals) and the following TaqMan Gene Expression Assays (Applied Biosystems): IFNβ (Mm00439546_s1), IFNα2 (Mm00833961_s1) and TRAPβ (Mm00481383_m1). Relative amounts of mRNA were normalized to the 18S ribosomal RNA levels in each sample.
Reporter analysis
293T cells seeded on 24-well plates were transiently transfected with 50 ng of the luciferase reporter plasmid together with a total of 600 ng of various expression plasmids or empty control plasmids. As an internal control, 10 ng pRL-TK was transfected simultaneously. Then, 24 or 36 h later, the luciferase activity in the total cell lysate was measured.
DNA vaccine
Mice were immunized with a plasmid encoding OVA by i.m. electroporation (100 μg per mouse). The booster immunization was given within 4 weeks of the primary immunization.
Measurement of OVA-specific immune response
Spleens were extracted 2 weeks after the second immunization and 5 × 105 spleen cells were seeded on 96-well plates and then stimulated with synthetic peptide for OVA (H-2Kb SIINFEKL, Proimmune) at 10 μg ml−1. After 3 days, the cell culture supernatants were collected and analysed for the IFNγ titre by ELISA (R&D systems). For intracellular IFNγ staining, stimulated splenocytes were stained using FITC-labelled anti-CD8 antibody (BD). After washing, cells were fixed and permeabilized. Then cells were stained using phycoerythrin (PE)-labelled anti-IFNγ antibody (BD). Flow cytometric analysis was performed on a FACScaliber instrument (BD). The serum anti-OVA antibody titre was measured by ELISA. In brief, 96-well plates were coated with an OVA protein at 1 μg ml−1 and then blocked with PBS containing 5% skimmed milk. Plates were washed and overlaid with serially diluted serum for 1 h at room temperature. After washing, antibodies were detected using goat anti-mouse IgG conjugated to horseradish peroxidase (Jackson Immuno Research). After further washing, the plates were stained using 3,3′,5,5′-tetramethylbenzidine (TMB, Sigma) as a substrate. The reaction was stopped with 1 M H2SO4 and the absorbance was measured. Antibody titres were expressed as the reciprocal of the endpoint dilution after background subtraction.
Fractionation
MAM, mitochondria and microsomes were isolated from Sting−/− MEFs stably transfected with mSTING–HA plasmid as previously described30. In brief, cells were washed in PBS and pelleted by centrifugation at 1,000g for 10 min. The pellet was resuspended in sucrose homogenization buffer (0.25 M sucrose, 10 mM HEPES, pH 7.4), and cells were lysed by using a dounce homogenizer. Lysed cells were centrifuged at 500g for 10 min, and the supernatant was collected. The supernatant was then centrifuged at 10,300g for 10 min to separate the crude microsomal (microsome and cytosol) from the crude mitochondrial (MAM and mitochondria) fraction, and the crude microsomal fraction (supernatant) was subjected to ultracentrifugation at 100,000g for 60 min. The crude mitochondrial fraction (pellet) was resuspended in ice-cold mannitol buffer A (0.25 M mannitol, 5 mM HEPES, 0.5 mM EDTA) and layered on top of a 30% Percoll in mannitol buffer B (0.225 M mannitol, 25 mM HEPES, 1 mM EDTA). Mitochondria and MAM fractions were separated by ultracentrifugation at 95,000g for 65 min. Both isolated fractions were diluted with mannitol buffer B and centrifuged at 6,300g for 10 min. The supernatant of MAM centrifugation was further separated by centrifugation at 100,000g for 60 min and the pellet was used for the MAM fraction, whereas the pellet of the mitochondria centrifugation was used as the mitochondria fraction. All of the fractions were resuspended in mannitol buffer B.
Confocal microscopy
For localization of Sec5 and LAMP1, cells grown on coverslips were fixed in 80%/20% methanol/acetone at −20 °C for 5 min. For EEA1 staining, cells were fixed with 4% paraformaldehyde in PBS for 15 min at 37 °C, and were permeabilized in 0.2% Triton X-100. For staining of other proteins, cells were fixed by 4% formaldehyde in DMEM for 15 min at 37 °C, and were permeabilized by 0.2% Triton X-100. For mitochondria staining, living cells were incubated with 300 nM of Mito Tracker Red (Invitrogen) for 45 min at 37 °C. Fixed and permeabilized cells were pre-incubated with 0.1% BSA in PBS and were incubated with primary antibodies. Cells were then incubated with secondary antibodies conjugated with FITC, Cy3 or Cy5 (Sigma).
RNA interference
Chemically synthesized 21-nucleotide siRNA duplexes were obtained from Dharmacon, Inc. The sequences of each siRNA oligonucleotide used in this study are follows: murine Trapb siRNA, 5′-UGAAAGAGAGGACGGGUUAUU-3′; murine Sec5 siRNA, 5′-AGAAGUAUUAGGUCGGAAA-3′, 5′-UCAACGUACUUCAGCGAUU-3′, 5′-CAGCAGAGAUUACACGUCA-3′, 5′-GUGAGUGGCUUGCGCAGUA-3′; murine Sting siRNA, 5′-CCAACAGCGUCUACGAGA-3′; murine Sec61b siRNA, 5′-GCAAGUACACGCGAUCAUA-3′, 5′-CAUCGCUGCUGUAUUUAUG-3′, 5′-CCACUGUUCGGCAGAGAAA-3′, 5′-GGCGAUUCUACACGGAAGA-3′. Control siRNA was obtained from Dharmacon (D-001206–01-80). MEFs were transfected by using an Amaxa nucleofector apparatus (program A-023) and Amaxa MEF nucleofector kit 1 according to the manufacturer's instructions. L929 cells were transfected using Lipofectamine RNAiMAX (Invitrogen). At 72 h after transfection, cells were used for further experiments.
Statistics
Student's t-test was used to analyse data.
Supplementary Material
Acknowledgements
We thank J. Yewdell for VV-OVA, B. Jacobs for VVΔE3L, K. Frueh for HCMV, M. Kobayashi for baculovirus, H. Horiuchi for the Sec5 antibody, Y. C. Weh for Tbk1-knockout MEFs, and S. Nagata, T. Maniatis, J. Hiscott and N. Reich for plasmid constructs. This work was supported by NIH grant AI079336.
Footnotes
Author Contributions H.I. and G.N.B. designed the research and analysed the data. H.I. performed most experiments. Z.M. performed experiments related to YFV NS4B, carried out exocyst RNAi studies and helped with experiments. G.N.B. wrote the paper.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol. Rev. 2009;227:221–233. doi: 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi O, Akira S. Innate immunity to virus infection. Immunol. Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beutler BA. TLRs and innate immunity. Blood. 2009;113:1399–1407. doi: 10.1182/blood-2008-07-019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 6.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nature Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 7.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nature Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 8.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 10.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-b signaling. Mol. Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 11.Bauer S, Pigisch S, Hangel D, Kaufmann A, Hamm S. Recognition of nucleic acid and nucleic acid analogs by Toll-like receptors 7, 8 and 9. Immunobiology. 2008;213:315–328. doi: 10.1016/j.imbio.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 12.Ishii KJ, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 13.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 14.Muruve DA, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 15.Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 16.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bürckstüümmer T, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nature Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 19.Jin L, et al. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol. Cell. Biol. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nature Immunol. doi: 10.1038/ni.1779. doi:10.1038/ni.1779 (16 July 2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic dna and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muñoz-Jordan JL, et al. Inhibition of a/b interferon signaling by the NS4B protein of flaviviruses. J. Virol. 2005;79:8004–8013. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spies B, et al. Vaccination with plasmid DNA activates dendritic cells via Toll-like receptor 9 (TLR9) but functions in TLR9-deficient mice. J. Immunol. 2003;171:5908–5912. doi: 10.4049/jimmunol.171.11.5908. [DOI] [PubMed] [Google Scholar]
- 26.Ménétret JF, et al. Single copies of Sec61 and TRAP associate with a nontranslating mammalian ribosome. Structure. 2008;16:1126–1137. doi: 10.1016/j.str.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayashi T, Rizzuto R, Hajnoczky G, Su TP. MAM: more than just a housekeeper. Trends Cell Biol. 2009;19:81–88. doi: 10.1016/j.tcb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chien Y, et al. RalB GTPase-mediated activation of the IkB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–170. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 29.Spiczka KS, Yeaman C. Ral-regulated interaction between Sec5 and paxillin targets Exocyst to focal complexes during cell migration. J. Cell Sci. 2008;121:2880–2891. doi: 10.1242/jcs.031641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mavinakere MS, Williamson CD, Goldmacher VS, Colberg-Poley AM. Processing of human cytomegalovirus UL37 mutant glycoproteins in the endoplasmic reticulum lumen prior to mitochondrial importation. J. Virol. 2006;80:6771–6783. doi: 10.1128/JVI.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.