Abstract
Lipid peroxidation is responsible for the generation of chemically reactive, diffusible lipid-derived electrophiles (LDE) that covalently modify cellular protein targets. These protein modifications modulate protein activity and macromolecular interactions and induce adaptive and toxic cell signaling. Protein modifications induced by LDEs can be identified and quantified by affinity enrichment and liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based techniques. Tagged LDE analog probes with different electrophilic groups can be covalently captured by Click chemistry for LC-MS/MS analyses, thereby enabling in-depth studies of proteome damage at the protein and peptide-sequence levels. Conversely, Click-reactive, thiol-directed probes can be used to evaluate LDE-thiol damage by difference. These analytical approaches permit systematic study of the dynamics of LDE-protein damage and mechanisms by which oxidative stress contributes to toxicity and diseases.
Keywords: Reactive Oxygen Species (ROS), Polyunsaturated fatty acids (PUFA), lipid derived electrophiles (LDE), affinity purification, biotin hydrazide, Click chemistry, mass spectrometry-based proteomics, stable isotope labeling
Introduction
Polyunsaturated fatty acids (PUFA), major constituents of biological membranes, are primary targets of reactive oxygen species generated by oxidative stress [1–3]. PUFA oxidation proceeds by both enzymatic and non-enzymatic pathways [4,5] to produce highly reactive electrophilic oxidation products [2]. These species are capable of interacting with and modifying various cellular targets and as a result trigger specific biochemical and cellular responses, like alterations in cell signaling and survival [6]. The endogenous formation of lipid oxidation products has been viewed as a marker of oxidative stress [7]; higher concentrations are associated with cellular damage, as reactive aldehydes are highly stable and can diffuse away from the cell membrane to attack targets far from the site of their production [8]. This cellular damage can have negative consequences for the organism, as modifications of proteins by reactive aldehyde products of lipid peroxidation contribute to neurodegenerative disorders [9], activation of kinases [10,11] and inhibition of the nuclear transcription factors [12].
Whereas the initial products of both enzymatic and non-enzymatic lipid peroxidation are fatty acid hydroperoxides, further decomposition of these products often yields reactive electrophiles, including α,β -unsaturated aldehydes, malondialdehyde, hydroxyalkenals, oxoalkenals, epoxyalkenals and γ-ketoaldehydes [13–16] (Figure 1). Here we use the term lipid-derived electrophiles (LDE) to encompass this diverse range of products. These highly reactive electrophiles can react with cellular nucleophiles through Michael addition (e.g., α, β-unsaturated carbonyls) and irreversible Schiff base formation (e.g., aldehydes). The nucleophilic amino acids cysteine, histidine and lysine are the most commonly modified by lipid-derived electrophiles, but the structure and accessibility of the amino acids in protein structures determines their vulnerability to electrophile modifications. Microenvironment-derived characteristics of the targeted nucleophile, such as hydrogen bonding with neighboring amino acids, can modulate the reactivity towards Michael adduct formation by orders of magnitude [17].
Figure 1.
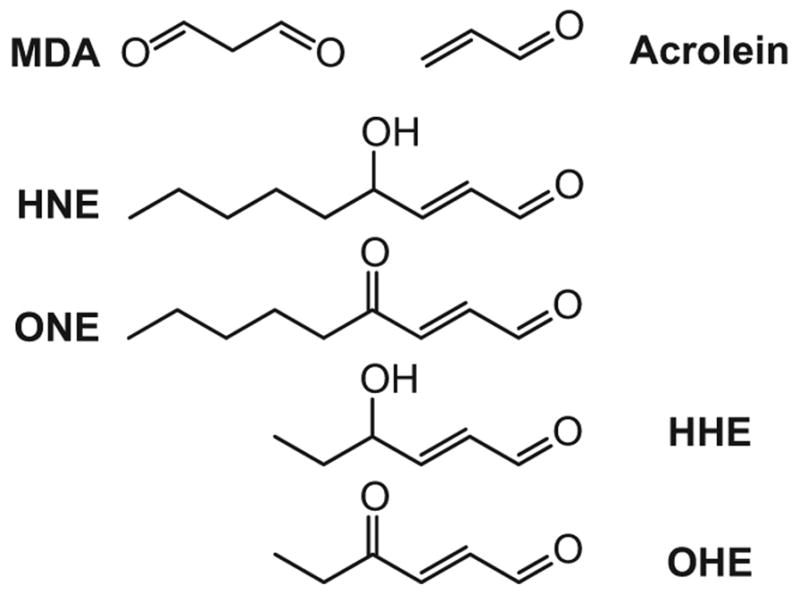
Structures of representative aldehydic products of lipid peroxidation induced by oxidative stress. These highly reactive lipid-derived electrophiles (LDE) covalently modify and as a result damage proteins. Malondialdehyde (MDA), acrolein, 4-hydroxy-nonenal (HNE), 4-oxo-nonenal (ONE), 4-hydroxy-hexanal (HHE), 4-oxo-hexanal (OHE).
Membrane rich organelles, such as endoplasmic reticulum and mitochondria have been shown to accumulate intra-membrane lipid peroxidation derived electrophilic compounds to as high as 4.5 mM in the lipid bilayer of isolated peroxidizing microsomes [1]. The regulatory function and cytotoxicity of these aldehydes depends on abundance, reactivity and longevity. Longevity varies greatly, with compounds such as 4-oxononenal (4-ONE) and 4-hydroxynonenal (4-HNE) displaying half-lives of roughly 1 second and 2 minutes, respectively [18,19]. By persisting on a scale of minutes, some aldehydes may have a more pronounced global impact by altering a broader array of proteomic and genomic targets [3,20].
More recent publications reported a significant increase in neurotoxic by-products of lipid peroxidation, such as HNE and acrolein, when measured in healthy versus diseased states [21,22]. Using LC-MS/MS technology, Williams and colleagues analyzed levels of HNE and acrolein in three vulnerable brain regions of Alzheimer’s disease patients, such as hippocampus/parahippocampal gyrus (HPG), superior and middle temporal gyrus (SMTG) and cerebellum (CER). Data showed a statistically significant (P < 0.05) increase in the measured levels of HNE in all three studied brain regions of Alzheimer’s disease patients when compared to age-matched control subjects. Measurement of large differences in HNE levels between different diseased states allowed scientists to view HNE more as a toxin rather than as a cytoprotective electrophile. Nevertheless, the challenge remains to measure low levels of HNE and other lipid derived electrophiles different tissue compartments [23].
Lipid peroxidation has been highlighted as a key chemical mechanism driving the effects of oxidative stress on numerous disease processes, including inflammation, degenerative diseases, and tumor formation [9,24]. Covalent modification of cellular proteins by LDE is thought to be a critical mechanism by which oxidative stress contributes to these pathologies [4]. Protein adduction by LDE is thought to initiate both adaptive and toxic cellular responses that mediate resistance and susceptibility to different diseases including atherosclerosis, neurodegenerative diseases, diabetes, and cancer [9,23,25]. However, understanding the mechanistic basis for damage and its consequences requires the identification of the protein targets and sites of modification. In this review, we focus on approaches to this challenging analytical problem.
The challenge of protein damage analysis
The problem of identifying protein targets of LDE is complicated by the diversity of targets and the low levels of damage under relevant conditions, which presents a complex analytical challenge [26]. These types of post-translational modifications (PTMs), much like phosphorylation or acetylation, can alter protein binding, interacting partners or subcellular localization, even if only a small fraction of the proteome has been modified.
The low concentration of endogenously generated LDE poses multiple challenges for the identification and quantification of their cellular targets as well as the identification of exact sites of modification. Electrophiles are expected to react primarily in close proximity to sites of generation, where they would be at relatively higher concentration. Thus, for in vitro studies of cellular responses, it is not unusual to use exogenous addition of lipid electrophile in μM concentration to approximate the effects of nM endogenous electrophile concentration produced over time. In this way, susceptible proteins can be functionally altered in spite of the very low electrophile concentration detected in vivo [4,27].
Proteomics approaches to characterization of protein adducts
In the past decade, there have been remarkable advances in proteomic technologies [28]. Mass spectrometry (MS) has emerged as the preferred method for in-depth characterization of the composition, regulation and function of protein complexes in biological systems. Recent advances in MS instrumentation, protein and peptide separations and bioinformatics tools all have enabled modern proteomics approaches to characterize proteins and proteomes. Mass-spectrometry-based proteomics, including the instrumentation and the methods for data acquisition and analysis, have been discussed in several recent reviews [29–31].
A major challenge in MS-based proteomic analysis is the exceptionally wide dynamic range for protein expression; there is at least a million-fold difference in concentration between the least abundant and most abundant proteins in cells. Detection of both higher abundance and lower abundance components is thus limited by the dynamic range of the technology platform. Moreover, modified protein forms, including oxidized- or LDE-modified proteins, are typically present at low stoichiometry compared to unmodified forms. Thus, global analysis of covalentyl-modified proteins require affinity enrichment of specific adducted or modified forms and identification methods capable of resolving and detecting anywhere from dozens to thousands of different modified species [26].
Application of biotin hydrazide affinity capture to identify protein targets of LDE
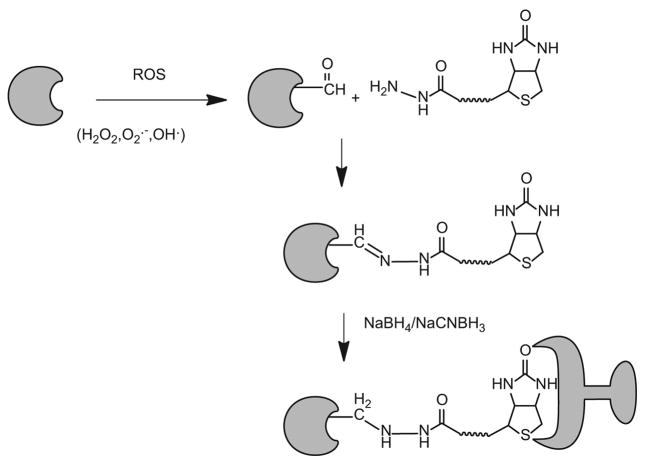
One of the unique features of the wide variety of proteins oxidized or modified by LDE is the presence of carbonyl groups. Protein carbonyl groups can react with hydrazides to form hydrazones, which can be readily reduced by borohydride to stable secondary amines [32]. Soreghan and co-workers used a functional proteomics approach combining biotin hydrazide and streptavidin capture methodology with LC-MS/MS analysis to identify oxidized proteins in aged mice [32]. They identified at least 100 carbonylated proteins in a single LC-MS/MS experiment. Target proteins ranged from high abundance cytoplasmic proteins to several low-abundant receptor proteins, mitochondrial proteins involved in glucose and energy metabolism, as well as receptors and tyrosine phosphatases known to be associated with cell-signaling pathways [32]. As in all studies employing this methodology, identifications were made at the protein level, so it is not clear whether the labeled sites were carbonyls generated by oxidation of the proteins or by covalent protein modification by LDE.
An important means of introduction of carbonyl groups on proteins is through covalent addition of LDE such as acrolein, malonaldehyde, 4-hydroxy-2-nonenal (HNE) and other hydroxyalkenals. The most extensively studied LDE, HNE is formed by oxidation of arachidonic and linoleic acid and it is one of the most reactive [33]. HNE is a bifunctional electrophile that modifies proteins either by Michael addition and Schiff base mechanisms. Although Michael reaction-derived mono adducts are the major HNE protein modifications, one molecule of HNE can react with two residues belonging to the same protein or two different proteins and cause intra or intermolecular crosslinking [10]. The one described crosslink modification known to occur from lipid peroxidation is formed between two lysine residues joined by HNE [34] to form a fluorescent compound similar to lipofuscin. The bifunctional aspect of HNE allows it to crosslink proteins by conjugate addition of Cys, His, or Lys at C3 and Schiff base condensation with Lys at the C1 carbonyls [35]. The newly formed fluorophore (with excitation and emission at 360 nm and 430 nm, respectively) contributes to the fluorescence and crosslinking that arises when proteins are exposed to HNE or ONE [36]. Among the HNE modifications, protein crosslinking, intra-and inter-peptide links can greatly increase insolubility and resistance to degradation [37].
Because lipid peroxidation may contribute to the pathology associated with Alzheimer’s disease (AD) and aging due to the high content of highly unsaturated fatty acids, brain tissue from AD and control patients was examined by immunocytochemistry and immunoelectron microscopy for evidence of HNE-crosslinking modifications [37]. The results indicated that lipid peroxidation and subsequent HNE modification of lysine residues occurs in target proteins for degradation in AD and that these accumulate in the lysosomal/proteasomal degradation pathways. Although mass spectrometry has been used to identify lipid derived electrophile adducts and crosslinks generated with isolated, purified macromolecules, the analysis of adducts in complex systems still poses major challenges [36].
Goodlett and coworkers described a new open-modification search based pipeline for characterizing both chemical as well as native cross-links (CXMS) in proteins by mass spectrometry [38]. The new CXMS method offers several advantages over previously reported techniques, being able to unequivocally identify the chemical, as well as native cross-links, inter and intra protein formed, thus allowing potential application to analysis of multiprotein complexes. A recent report on the mass spectrometric analysis of cross-linked proteins described a suite of tools named Hekate[39], which address challenges involved in analyzing protein cross-linking. The software provides an integrated pipeline for automation of data analysis workflow and a novel scoring system based on principles of linear peptide analysis [39].
We applied a biotin hydrazide affinity labelling and capture approach to enable the selective enrichment and analysis of proteins adducted by HNE [40] (Figure 2). HNE Michael adducts from human colorectal carcinoma (RKO) cells exposed to increasing concentrations of electrophile (0, 50 and 100 μM) for 1 h at 37 °C, were biotinylated by reaction with biotinamidohexanoic acid hydrazide and captured with streptavidin. Captured proteins were resolved by one dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis, digested with trypsin, and identified by LC-MS/MS. Affinity capture methods in complex proteomes entail the potential for many false-positive identifications due to nonspecific binding to the affinity support. We thus used a label-free approach to quantify captured proteins as a function of electrophile concentration and then identified protein targets demonstrating concentration-dependent adduction. The levels of adduction were estimated from spectral counts, which increased for detected peptides from true protein targets with increasing HNE treatment concentration [41]. Of the 1500+ proteins identified, 417 displayed a statistically significant increase in spectral counts with increasing HNE exposure concentration [40]. This relationship distinguishes true adducts from proteins nonspecifically captured with streptavidin or proteins containing carbonyls not derived from HNE treatment. By mapping the identified HNE adducted proteins to a human protein interaction network, we were able to provide systems-level insight and characterized protein networks targeted by HNE and suggested cellular processes that may be affected by LDE.
Figure 2.
Biotinylation of protein adducted by reactive carbonyls with biotin hydrazide [32].
Application of click chemistry to identify protein targets of LDE
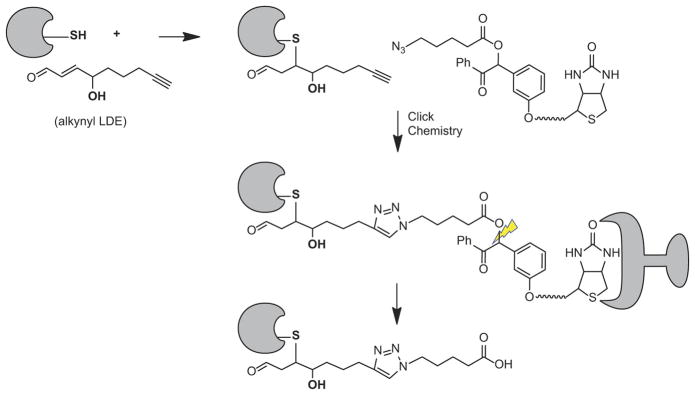
Although the biotin hydrazide capture approach provided new information about the cellular scope of HNE targets, the approach detects other carbonyl modifications and is subject to a high background. We therefore explored alkynyl analogs of LDE, which form adducts that can be post labeled with azido-biotin tags by Cu+-catalyzed cycloaddition (Click chemistry) [42]. In addition, an enhanced selectivity of adduct capture can be achieved by using an azido-biotin reagent with a photocleavable linker, which allows efficient recovery of adducted proteins and peptides under mild conditions [43,44]. This approach provides both the identification of protein targets of LDE and sequence mapping of adducts from cellular lysates and human plasma [45,46]. The residual tag after photocleavage is small (311 mass units for alkynyl HNE adducts), which minimizes interference with adduct analysis by LC-MS/MS. This approach was successfully applied to human plasma exposed to alkynyl HNE and then post labeled by Click chemistry with the photocleavable azido-biotin-linker to profile both proteins and peptides adducted [44]. These analyses identified with high confidence 14 plasma proteins as LDE targets and mapped 50 specific adduction sites. The combination of Click chemistry with LDE probes has proven useful for inventory of large numbers of proteins targets of LDEs in cells or plasma [40,42,44,47–49].
With a powerful global LDE adduct profiling platform, a major challenge is to distinguish toxic from nontoxic protein alkylation damage, given the hypothesis that only a subset of alkylation events contribute to injury. We employed alkynyl analogs of the prototypical lipid electrophiles HNE and 4-oxo-2-nonenal (ONE) to evaluate the systems-level impact of LDE protein alkylation damage [20] (Figure 3). Data obtained from two different cell lines with a large fold difference in GSH content showed that GSH levels dramatically affected profiles of covalent protein alkylation, but did not produce a difference in toxicity. Analysis of spectral count data for the adducted proteins revealed distinct patterns of protein adduction with increasing electrophile concentration. Protein targets of alkynyl analogs of HNE and ONE thus were grouped into three major classes reflecting the concentration dependence of adduct accumulation. The highest reactivity class was substantially modified at sub-toxic alkynyl HNE and ONE concentrations, whereas the lowest reactivity class was modified at toxic concentrations. Comparisons of the proteins in these distinct classes revealed that covalent adduct formation at a sub toxic level may largely be survivable damage to cytoskeletal networks. In contrast, covalent binding at toxic LDE concentrations produces lethal injury by targeting protein synthesis and catabolism [20]. This study defined a functional systems hierarchy for accumulation of electrophile-mediated protein damage. A hierarchal protein damage susceptibility may represent the evolution of functional cellular systems to separate survivable from lethal injury, particularly in the context of oxidative stress.
Figure 3.
Biotinylation of protein adducted by LDE using Click chemistry and a photocleavable linker [20].
Application of click chemistry to quantify LDE-cysteine reactions
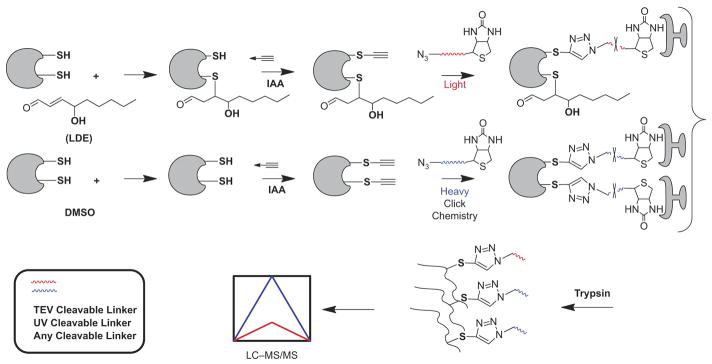
Another important dimension of the LDE-protein reactivity problem is the functional annotation of proteins with alkylation-susceptible thiols [50–52]. Cravatt and colleagues have applied activity based proteome profiling strategies to study thiol reactivity across proteomes. Weerapana and coworkers [51] described an approach termed isoTOP-ABPP (isotopic tandem orthogonal proteolysis–activity-based protein profiling), which allowed them to globally characterize cysteine functionality in native proteomes based on quantitative reactivity profiling with isotopically labelled, small-molecule electrophiles. Their approach employed an electrophilic iodoacetamide (IA) probe to label cysteine residues in different proteomes. This generic cysteine-alkylating reagent can stably modify cysteine residues in proteins [53] and has been used to assess nucleophilicity of cysteine residues [54] in purified proteins. A modified IA probe with an alkynyl tag allowed for Click chemistry conjugation of probe-labelled proteins [55] to an azido-biotin-linker functionalized with a tobacco etch virus (TEV)-cleavable site for further streptavidin enrichment of modified proteins [50]. After TEV cleavage, the released, modified peptides contain an isotopically (light/heavy) labelled valine residue, which enables quantitative mass spectrometry comparisons of IA-labelled peptides across multiple proteomes. Using two different concentrations of the IA probe to label the proteomes and measured ratios between the light and heavy modified probe for each identified cysteine, they were able to estimate the relative reactivity of cysteine residues on a global scale. The same strategy can be generalized to profile cysteine reactivity towards LDEs as the chemical modifiers, alone or in combinations with other techniques appropriate for a more complex biological system.
Wang and coworkers extended this approach by combining IA probes and Click chemistry to label protein thiols and then quantify thiol reactions with LDE via their impact on IA probe labeling [56]. This competitive activity-based protein profiling assay (ABPP) quantified the relative reactivity of more than 1000 cysteines from the whole proteome in parallel, against specific LDEs. This method not only identifies those cysteine residues targeted by different LDEs, but circumvents the requirement for of a series of Clickable LDE probes. The same isotopic labeling strategy described above then provides estimates of relative reactivity of different cysteine residues for each of a series of LDE. A comparison of the measurements between treatments with three different LDE (HNE, 15d-PGJ2, and 2-HD), all of which have α, β-unsaturated carbonyls, revealed significant reactivity of just a few cysteine residues towards LDE.
This type of competition-based platform allows for a global approach to identify and quantify LDE-reactive hot spots in the human proteome (Figure 4). A general approach to quantify the sensitivity of unique cysteine residues towards LDE could have broad applicability to profiling LDE-protein reactions in primary cells and tissues [51].
Figure 4.
Competitive isoTOP-ABPP for quantitative mapping of cysteine-lipid-derived electrophile (LDE) reactions in proteomes. Proteomes are treated with DMSO (vehicle control) or LDE, labeled with an IA-alkynyl (IA) probe, biotinylated using Click chemistry with isotopically-labeled cleavable biotin tags, enriched with streptavidin, released from the beads, and digested with trypsin to obtain probe-labeled peptides for LC-MS/MS analysis.
Footnotes
Declaration of Interest
This work was supported by National Institutes of Health Grants ES013125, ES000267 and ES007028.
References
- 1.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11(1):81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 2.Fritz KS, Petersen DR. An overview of the chemistry and biology of reactive aldehydes. Free Radic Biol Med. 2013;59:85–91. doi: 10.1016/j.freeradbiomed.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fritz KS, Petersen DR. Exploring the biology of lipid peroxidation-derived protein carbonylation. Chem Res Toxicol. 2011;24(9):1411–9. doi: 10.1021/tx200169n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schopfer FJ, Cipollina C, Freeman BA. Formation and signaling actions of electrophilic lipids. Chem Rev. 2011;111(10):5997–6021. doi: 10.1021/cr200131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown HA, Marnett LJ. Introduction to lipid biochemistry, metabolism, and signaling. Chem Rev. 2011;111(10):5817–20. doi: 10.1021/cr200363s. [DOI] [PubMed] [Google Scholar]
- 6.West JD, Marnett LJ. Endogenous reactive intermediates as modulators of cell signaling and cell death. Chem Res Toxicol. 2006;19(2):173–94. doi: 10.1021/tx050321u. [DOI] [PubMed] [Google Scholar]
- 7.Boveris A, Navarro A. Brain mitochondrial dysfunction in aging. IUBMB Life. 2008;60(5):308–14. doi: 10.1002/iub.46. [DOI] [PubMed] [Google Scholar]
- 8.Esterbauer H. Estimation of peroxidative damage. A critical review. Pathol Biol (Paris) 1996;44(1):25–8. [PubMed] [Google Scholar]
- 9.Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39(1):44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Uchida K. 4-Hydroxy-2-nonenal: a product and mediator of oxidative stress. Prog Lipid Res. 2003;42(4):318–43. doi: 10.1016/s0163-7827(03)00014-6. [DOI] [PubMed] [Google Scholar]
- 11.Uchida K, Shiraishi M, Naito Y, Torii Y, Nakamura Y, Osawa T. Activation of stress signaling pathways by the end product of lipid peroxidation. 4-hydroxy-2-nonenal is a potential inducer of intracellular peroxide production. J Biol Chem. 1999;274(4):2234–42. doi: 10.1074/jbc.274.4.2234. [DOI] [PubMed] [Google Scholar]
- 12.Camandola S, Poli G, Mattson MP. The lipid peroxidation product 4-hydroxy-2,3-nonenal inhibits constitutive and inducible activity of nuclear factor kappa B in neurons. Brain Res Mol Brain Res. 2000;85(1–2):53–60. doi: 10.1016/s0169-328x(00)00234-5. [DOI] [PubMed] [Google Scholar]
- 13.Gueraud F, Atalay M, Bresgen N, Cipak A, Eckl PM, Huc L, Jouanin I, Siems W, Uchida K. Chemistry and biochemistry of lipid peroxidation products. Free Radic Res. 2010;44(10):1098–124. doi: 10.3109/10715762.2010.498477. [DOI] [PubMed] [Google Scholar]
- 14.Uchida K, Kanematsu M, Sakai K, Matsuda T, Hattori N, Mizuno Y, Suzuki D, Miyata T, Noguchi N, Niki E, et al. Protein-bound acrolein: potential markers for oxidative stress. Proc Natl Acad Sci U S A. 1998;95(9):4882–7. doi: 10.1073/pnas.95.9.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niki E, Yoshida Y, Saito Y, Noguchi N. Lipid peroxidation: mechanisms, inhibition, and biological effects. Biochem Biophys Res Commun. 2005;338(1):668–76. doi: 10.1016/j.bbrc.2005.08.072. [DOI] [PubMed] [Google Scholar]
- 16.Porter NA, Caldwell SE, Mills KA. Mechanisms of free radical oxidation of unsaturated lipids. Lipids. 1995;30(4):277–90. doi: 10.1007/BF02536034. [DOI] [PubMed] [Google Scholar]
- 17.Nagahara N, Matsumura T, Okamoto R, Kajihara Y. Protein cysteine modifications: (1) medical chemistry for proteomics. Curr Med Chem. 2009;16(33):4419–44. doi: 10.2174/092986709789712880. [DOI] [PubMed] [Google Scholar]
- 18.Benedetti A, Comporti M, Fulceri R, Esterbauer H. Cytotoxic aldehydes originating from the peroxidation of liver microsomal lipids. Identification of 4,5-dihydroxydecenal. Biochim Biophys Acta. 1984;792(2):172–81. doi: 10.1016/0005-2760(84)90219-4. [DOI] [PubMed] [Google Scholar]
- 19.Doorn JA, Petersen DR. Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chem Res Toxicol. 2002;15(11):1445–50. doi: 10.1021/tx025590o. [DOI] [PubMed] [Google Scholar]
- 20.Codreanu SG, Ullery JC, Zhu J, Tallman KA, Beavers WN, Porter NA, Marnett LJ, Zhang B, Liebler DC. Alkylation damage by lipid electrophiles targets functional protein systems. Mol Cell Proteomics. 2014;13(3):849–59. doi: 10.1074/mcp.M113.032953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams TI, Lynn BC, Markesbery WR, Lovell MA. Increased levels of 4-hydroxynonenal and acrolein, neurotoxic markers of lipid peroxidation, in the brain in Mild Cognitive Impairment and early Alzheimer’s disease. Neurobiol Aging. 2006;27(8):1094–9. doi: 10.1016/j.neurobiolaging.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Markesbery WR, Lovell MA. Four-hydroxynonenal, a product of lipid peroxidation, is increased in the brain in Alzheimer’s disease. Neurobiol Aging. 1998;19(1):33–6. doi: 10.1016/s0197-4580(98)00009-8. [DOI] [PubMed] [Google Scholar]
- 23.Butterfield DA, Bader Lange ML, Sultana R. Involvements of the lipid peroxidation product, HNE, in the pathogenesis and progression of Alzheimer’s disease. Biochim Biophys Acta. 2010;1801(8):924–9. doi: 10.1016/j.bbalip.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stadtman ER, Berlett BS. Reactive oxygen-mediated protein oxidation in aging and disease. Chem Res Toxicol. 1997;10(5):485–94. doi: 10.1021/tx960133r. [DOI] [PubMed] [Google Scholar]
- 25.Grimsrud PA, Picklo MJ, Sr, Griffin TJ, Bernlohr DA. Carbonylation of adipose proteins in obesity and insulin resistance: identification of adipocyte fatty acid-binding protein as a cellular target of 4-hydroxynonenal. Mol Cell Proteomics. 2007;6(4):624–37. doi: 10.1074/mcp.M600120-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Liebler DC. Protein damage by reactive electrophiles: targets and consequences. Chem Res Toxicol. 2008;21(1):117–28. doi: 10.1021/tx700235t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh JY, Giles N, Landar A, Darley-Usmar V. Accumulation of 15-deoxy-delta(12,14)-prostaglandin J2 adduct formation with Keap1 over time: effects on potency for intracellular antioxidant defence induction. Biochem J. 2008;411(2):297–306. doi: 10.1042/bj20071189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cravatt BF, Simon GM, Yates JR., 3rd The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450(7172):991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 29.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 30.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312(5771):212–7. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 31.Yates JR., 3rd Mass spectral analysis in proteomics. Annu Rev Biophys Biomol Struct. 2004;33:297–316. doi: 10.1146/annurev.biophys.33.111502.082538. [DOI] [PubMed] [Google Scholar]
- 32.Soreghan BA, Yang F, Thomas SN, Hsu J, Yang AJ. High-throughput proteomic-based identification of oxidatively induced protein carbonylation in mouse brain. Pharm Res. 2003;20(11):1713–20. doi: 10.1023/b:pham.0000003366.25263.78. [DOI] [PubMed] [Google Scholar]
- 33.Mali VR, Palaniyandi SS. Regulation and therapeutic strategies of 4-hydroxy-2-nonenal metabolism in heart disease. Free Radic Res. 2014;48(3):251–63. doi: 10.3109/10715762.2013.864761. [DOI] [PubMed] [Google Scholar]
- 34.Tsai L, Szweda PA, Vinogradova O, Szweda LI. Structural characterization and immunochemical detection of a fluorophore derived from 4-hydroxy-2-nonenal and lysine. Proc Natl Acad Sci U S A. 1998;95(14):7975–80. doi: 10.1073/pnas.95.14.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu G, Sayre LM. Structural characterization of a 4-hydroxy-2-alkenal-derived fluorophore that contributes to lipoperoxidation-dependent protein cross-linking in aging and degenerative disease. Chem Res Toxicol. 1998;11(4):247–51. doi: 10.1021/tx980003d. [DOI] [PubMed] [Google Scholar]
- 36.Sayre LM, Lin D, Yuan Q, Zhu X, Tang X. Protein adducts generated from products of lipid oxidation: focus on HNE and one. Drug Metab Rev. 2006;38(4):651–75. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- 37.Zhu X, Castellani RJ, Moreira PI, Aliev G, Shenk JC, Siedlak SL, Harris PL, Fujioka H, Sayre LM, Szweda PA, et al. Hydroxynonenal-generated crosslinking fluorophore accumulation in Alzheimer disease reveals a dichotomy of protein turnover. Free Radic Biol Med. 2012;52(3):699–704. doi: 10.1016/j.freeradbiomed.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh P, Shaffer SA, Scherl A, Holman C, Pfuetzner RA, Larson Freeman TJ, Miller SI, Hernandez P, Appel RD, Goodlett DR. Characterization of protein cross-links via mass spectrometry and an open-modification search strategy. Anal Chem. 2008;80(22):8799–806. doi: 10.1021/ac801646f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holding AN, Lamers MH, Stephens E, Skehel JM. Hekate: software suite for the mass spectrometric analysis and three-dimensional visualization of cross-linked protein samples. J Proteome Res. 2013;12(12):5923–33. doi: 10.1021/pr4003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Codreanu SG, Zhang B, Sobecki SM, Billheimer DD, Liebler DC. Global analysis of protein damage by the lipid electrophile 4-hydroxy-2-nonenal. Mol Cell Proteomics. 2009;8(4):670–80. doi: 10.1074/mcp.M800070-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76(14):4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 42.Vila A, Tallman KA, Jacobs AT, Liebler DC, Porter NA, Marnett LJ. Identification of protein targets of 4-hydroxynonenal using click chemistry for ex vivo biotinylation of azido and alkynyl derivatives. Chem Res Toxicol. 2008;21(2):432–44. doi: 10.1021/tx700347w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Codreanu SG, Kim HY, Porter NA, Liebler DC. Biotinylated probes for the analysis of protein modification by electrophiles. Methods Mol Biol. 2012;803:77–95. doi: 10.1007/978-1-61779-364-6_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim HY, Tallman KA, Liebler DC, Porter NA. An azido-biotin reagent for use in the isolation of protein adducts of lipid-derived electrophiles by streptavidin catch and photorelease. Mol Cell Proteomics. 2009;8(9):2080–9. doi: 10.1074/mcp.M900121-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szapacs ME, Kim HY, Porter NA, Liebler DC. Identification of proteins adducted by lipid peroxidation products in plasma and modifications of apolipoprotein A1 with a novel biotinylated phospholipid probe. J Proteome Res. 2008;7(10):4237–46. doi: 10.1021/pr8001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tallman KA, Kim HY, Ji JX, Szapacs ME, Yin H, McIntosh TJ, Liebler DC, Porter NA. Phospholipid-protein adducts of lipid peroxidation: synthesis and study of new biotinylated phosphatidylcholines. Chem Res Toxicol. 2007;20(2):227–34. doi: 10.1021/tx600331s. [DOI] [PubMed] [Google Scholar]
- 47.Connor RE, Codreanu SG, Marnett LJ, Liebler DC. Targeted protein capture for analysis of electrophile-protein adducts. Methods Mol Biol. 2013;987:163–76. doi: 10.1007/978-1-62703-321-3_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han B, Hare M, Wickramasekara S, Fang Y, Maier CS. A comparative ‘bottom up’ proteomics strategy for the site-specific identification and quantification of protein modifications by electrophilic lipids. J Proteomics. 2012;75(18):5724–33. doi: 10.1016/j.jprot.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roe MR, Xie H, Bandhakavi S, Griffin TJ. Proteomic mapping of 4-hydroxynonenal protein modification sites by solid-phase hydrazide chemistry and mass spectrometry. Anal Chem. 2007;79(10):3747–56. doi: 10.1021/ac0617971. [DOI] [PubMed] [Google Scholar]
- 50.Weerapana E, Speers AE, Cravatt BF. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)--a general method for mapping sites of probe modification in proteomes. Nat Protoc. 2007;2(6):1414–25. doi: 10.1038/nprot.2007.194. [DOI] [PubMed] [Google Scholar]
- 51.Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468(7325):790–5. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eisenberg D, Marcotte EM, Xenarios I, Yeates TO. Protein function in the post-genomic era. Nature. 2000;405(6788):823–6. doi: 10.1038/35015694. [DOI] [PubMed] [Google Scholar]
- 53.Shiio Y, Aebersold R. Quantitative proteome analysis using isotope-coded affinity tags and mass spectrometry. Nat Protoc. 2006;1(1):139–45. doi: 10.1038/nprot.2006.22. [DOI] [PubMed] [Google Scholar]
- 54.Voss AA, Lango J, Ernst-Russell M, Morin D, Pessah IN. Identification of hyperreactive cysteines within ryanodine receptor type 1 by mass spectrometry. J Biol Chem. 2004;279(33):34514–20. doi: 10.1074/jbc.M404290200. [DOI] [PubMed] [Google Scholar]
- 55.Speers AE, Adam GC, Cravatt BF. Activity-based protein profiling in vivo using a copper(i)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Am Chem Soc. 2003;125(16):4686–7. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 56.Wang C. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat Methods. 2014;11:78–84. doi: 10.1038/nmeth.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]