Abstract
The inability of cells to properly fold, modify, and assemble secretory and transmembrane proteins leads to accumulation of misfolded proteins in the endoplasmic reticulum (ER). Under these conditions of “ER stress,” cell survival depends on homeostatic benefits from an intracellular signaling pathway called the unfolded protein response (UPR). When activated, the UPR induces transcriptional and translational programs that restore ER homeostasis. However, under high-level/chronic ER stress, these adaptive changes ultimately become overshadowed by alternative “Terminal UPR” signals that actively commit cells to degeneration, culminating in programmed cell death. Chronic ER stress and maladaptive UPR signaling are implicated in the etiology and pathogenesis of myriad human diseases. Naturally, this has generated widespread interest in targeting key nodal components of the UPR as therapeutic strategies. Here we summarize the state of this field with emphasis placed on two of the master UPR regulators, PERK and IRE1α, which are both capable of being drugged with small molecules.
Introduction
In eukaryotic cells, the endoplasmic reticulum (ER) comprises interconnected networks of branching tubules and sacs that are separated from the surrounding cytosol by a lipid bilayer, the ER membrane. The ER is the first organelle of the secretory pathway and therefore serves as the entryway for over a third of all cellular proteins, including those destined for secretion to the cell exterior or insertion into the plasma membrane; included in this set are proteins that ultimately will reside within the ER, the Golgi, or in lysosomes (1). Folding, post-translational modifications, structural maturation and assembly of all these proteins begins in the ER, in most cases even as they are co-translationally injected through the translocon complex into the ER. Once in the ER, these client proteins of the secretory pathway fold to their native shapes and often undergo further post-translational modifications, including glycosylation and disulfide bond formation. These folding and maturation processes are catalyzed by abundant ER-resident enzymes, which include molecular chaperones, glycosylating enzymes and oxido-reductases (2, 3). The ionic and electronic milieu of the ER is adapted to facilitate these reactions. Compared to the cytosol, the ER maintains, through energy expenditure, a much higher calcium concentration and a much more oxidizing redox potential (4, 5). Together, these enzymatic processes ensure that secretory proteins are properly folded, modified, and assembled, in some cases into multi-protein complexes, within the ER before they traffic farther downstream in the secretory pathway.
Faithful folding and maturation of proteins of the secretory pathway often fails, and because many of these proteins mediate crucial signalling roles, incompletely folded forms are not tolerated by the cell. Instead these misfolded species are disposed of by stringent quality control systems such as the ER-associated degradation (ERAD) pathway, which removes unfolded proteins from the ER to the cytosol for subsequent ubiquitylation and degradation by the 26S proteasome (6, 7). In some instances, this process may lead to a deficiency of important proteins, causing a loss of the function that they serve. On the other hand, the accumulation of unfolded proteins in the ER can also cause gain-of-function proteotoxicity.
“Professional secretory” cells, such as β-cells of the endocrine pancreas appear to work near the limits of their secretory capacity and normally secrete approximately one million molecules of insulin every minute (8). As another example, plasma cells can secrete their own weight in antibodies every day (9). Thus, such cells may routinely experience “ER stress” from secretory exhaustion (10). As a more general concept, for any type of cell, a wide range of cellular disturbances can compromise efficiency of protein-folding in the ER and lead to the accumulation of misfolded proteins within the organelle. ER stress can proceed from nutrient deprivation, hypoxia, point mutations in important secreted proteins that stabilize incomplete folding forms, and loss of calcium homeostasis, which in turn may impede proper functioning of ER-resident calcium-dependent chaperones (11–13). In the case of β-cells, ER stress can occur from the inability to fold the increased levels of proinsulin intermediates needed to maintain blood glucose during conditions of peripheral insulin resistance (14). Also, in neurons, chronic expression of folding-defective secretory proteins can saturate the protein-folding machinery and lead to ER stress (15). Under ER stress, secretory proteins start to accumulate in improperly-modified and unfolded forms within the organelle. Therefore, cells have evolved sophisticated surveillance systems to sense and respond to ER stress before cell function and survival is threatened.
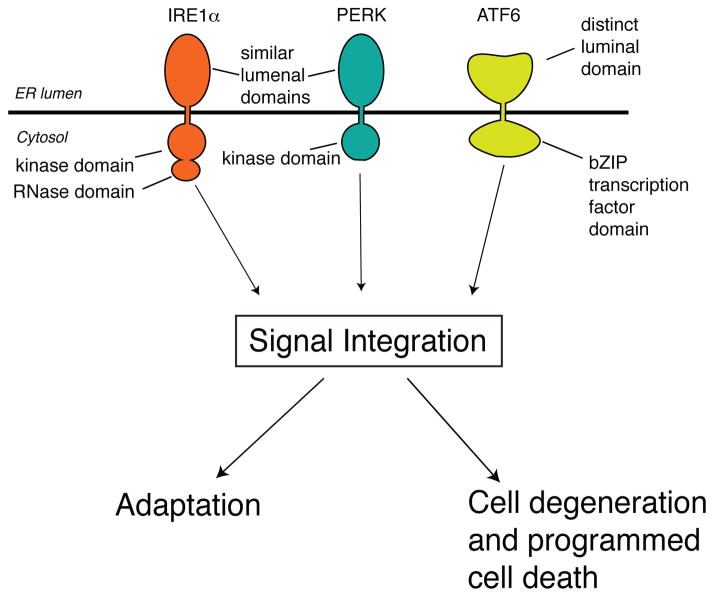
To properly match protein-folding capacity to secretory demand, cells constantly monitor the concentration of misfolded proteins in the ER lumen and initiate corrective responses. When misfolded proteins accumulate in the ER above a critical threshold, an ancient signal transduction pathway called the unfolded protein response (UPR), which is conserved in all eukaryotes, becomes activated. The UPR is triggered by the activation of three ER transmembrane proteins: IRE1α, inositol requiring enzyme 1 alpha; PERK, pancreatic endoplasmic reticulum kinase, PERK; and ATF6, activating transcription factor 6 (16). All three of these ER stress sensors contain an ER lumenal domain that directly or indirectly senses misfolded proteins (Figures 1–3). Lumenal domain sensing of misfolded proteins leads to changes in the oligomerization state of each sensor and activation of their downstream activities, hence transducing a signal from the ER lumen into the cytoplasm. For IRE1α and PERK, lumenal domain self-association, which is the initiating step, may be prevented in unstressed cells through binding of an ER chaperone called BiP (17). Furthermore, through direct binding to the lumenal domain of either IRE1α and PERK, misfolded/unfolded proteins may act as “activating ligands”, analogous to a wide range of extracellular ligands that activate various receptors on the plasma membrane (18, 19)
Figure 1. The Three Arms of the UPR.
Upon activation under ER stress, three sensors, IRE1α, PERK, and ATF6 send intracellular signals that allow the cell to either adapt or commit apoptosis.
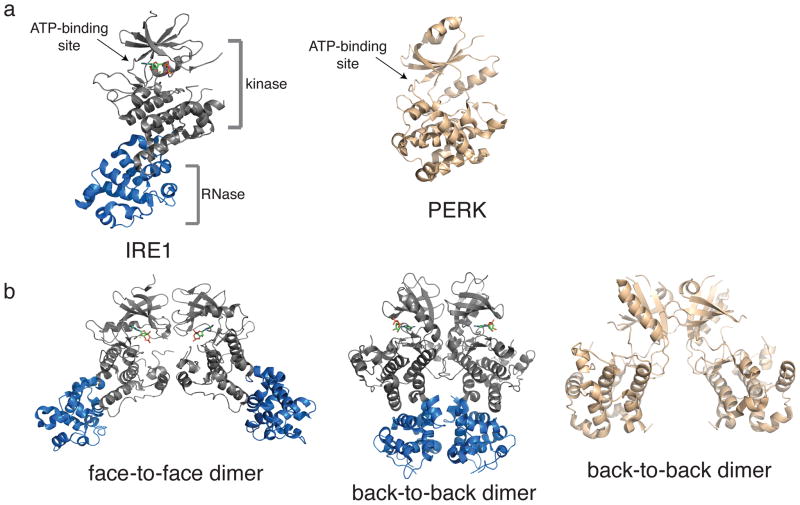
Figure 3. Domain Architecture of IRE1.
α and PERK
A. Crystal structures of the cytosolic regions of IRE1 and PERK. Left: Crystal structure of the kinase/RNase module of human IRE1α (PDB ID: 3P23). The kinase domain is shown in grey and the RNase (KEN domain) is shown in marine. The ATP-binding site of IRE1α is occupied by ADP. Right: Crystal structure of the cytosolic kinase domain of PERK (PDB ID: 4G31). B. Crystal structures of IRE1 and PERK dimers. Left: The face-to-face dimer of human IRE1α (PDB ID: 3P23). Middle: The back-to-back dimer of yeast IRE1 (PDB ID: 3LJ2). Right: The back-to-back dimer of PERK (PDB ID: 4G31).
Activation and Homeostatic Signaling in the UPR
The three UPR sensors evolved outputs that initially realign protein-folding demand and protein-folding capacity, thus restoring secretory homeostasis (20). This “adaptive UPR” expands ER size and enhances the physiological functions of ER chaperones, oxidoreductases, and ER membrane biosynthetic enzymes (21). Transcriptional upregulation of ERAD components by the UPR also leads to the removal and degradation of misfolded proteins from within the ER. Furthermore, transient translational blocks occur through the UPR under ER stress, which has the effect of decreasing protein flux into the secretory pathway (22). However, through a seeming paradox, the combined outputs of the UPR can lead to various destructive outcomes including cell proliferation blocks, dedifferentiation, inflammation and programmed cell death (typically through mitochondrial apoptosis (Figure 1)).
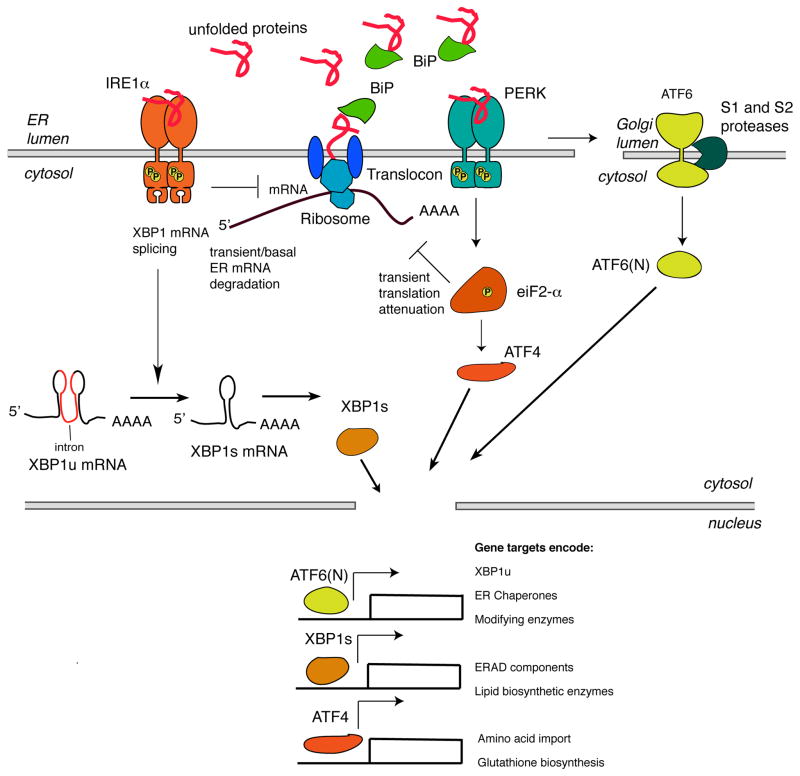
The most ancient of the UPR sensors, IRE1, exists in all eukaroytic cells. In mammalian cells, the more-widely expressed of two paralogs is IRE1α, which like all IRE1 species contains a cytoplasmic face comprising two enzymatic domains—a serine/threonine kinase domain and an endoribonuclease (RNase) domain (23, 24) (Figure 2). Upon lumenal accumulation of misfolded proteins, the lumenal domains multimerize, consequently juxtaposing IRE1α’s kinase domain which trans-autophosphorylate; this autophosphorylation event leads to conformational activation of IRE1α’s RNase domain (25). Upon activation, IRE1α’s RNase excises a 26-nucleotide intron from the mRNA encoding the XBP1 (X-box protein 1) transcription factor. Subsequent cytosolic splicing of the two resulting mRNA fragments by an unidentified RNA ligase produces the homeostatic transcription factor XBP1s that now contains a potent transactivation domain encoded in the altered reading frame (26, 27). XBP1s translocates to the nucleus and induces transcription of hundreds of genes that augment ER size and function (28).
Figure 2. Adaptive Signaling in the UPR.
Under ER stress, IRE1α’s kinase domain trans-autophosphorylates. IRE1α phosphorylation leads to allosteric activation of the adjacent RNase. The consequences of IRE1α activation vary depending on the level of ER stress. In response to low levels of ER stress, IRE1α’s RNase excises a 26-nucleotide intron from the mRNA encoding the XBP1 transcription factor to produce the homeostatic transcription factor XBP1s. XBP1s then translocates to the nucleus and induces transcription of many genes that attempt to restore ER homeostasis. In the presence of misfolded proteins, PERK dimerizes and phosphorylates eIF2α. Phosphorylation inhibits eIF2α activity and hence slows down global protein translation. In contrast, translation of the transcription factor ATF4 is selectively upregulated when the amount of active eIF2α is limiting. In the presence of misfolded proteins, ATF6 translocates to the Golgi and is cleaved by Site-1 and Site-2 proteases to release the ATF6(N) transcription factor contained within its cytoplasmic tail. Together with XBP1s, ATF6(N) increases transcription of targets that expand ER size and increase its protein-folding capacity to promote cell survival. ATF4 expression transcriptionally upregulates CHOP, which tips the ER towards homeostasis through induction of a number of corrective genes, including XBP1 and chaperones.
By analogy, the lumenal domains of PERK (evolutionarily derived from IRE1 in higher eukaryotes) multimerize upon lumenal accumulation of misfolded proteins. Similar to IRE1α, the cytosolic (Ser/Thr) kinase domains of PERK become trans-autophosphorylated, but in contrast to IRE1α, for which there is currently no evidence for a subsequent protein phosphorylation cascade, activated PERK phosphorylates the eukaryotic translation initiation factor 2α (eIF2α) on its Serine 51 residue (29, 30) (Figure 2). This event reduces cap-dependent translation, causing a global translational block, thus endowing the cell with an extended time window to fold pre-existing proteins that are already present in the ER (22). However, a subset of genes are granted translational privilege during the translational block imposed by PERK. For example, the mRNA encoding a transcription factor called ATF4 gains such translational privilege, and upon production of the ATF4 transcription factor its target genes encoding activities that increase amino acid import and glutathione biosynthesis become induced (31).
A third arm of the UPR is activated through the latent transcription factor, ATF6, which as an ER membrane protein trafficks in vesicles to the Golgi, where it is cleaved by Site-1 and Site-2 proteases to release the soluble ATF6(N) transcription factor contained on its cytoplasmic face (32) (Figure 2). Together with XBP1s, ATF6(N) increases transcription of targets that expand ER size and increase its protein-folding capacity (28). Thus, in aggregate, these transcriptional events combined with a transient translational block cause negative feedback loops to close as upstream ER stress becomes contained. If successful, these adaptive events promote cell survival and cause UPR signaling to wane.
Domain Architecture of IRE1α and PERK
Informed by recent structural biology advances, we turn now to a delineation of the mechanisms of activation of the two components of the UPR that possess druggable enzymatic activity, IRE1α and PERK. Full length IRE1α possesses an N-terminal lumenal sensor domain and a cytosolic kinase/RNase domain that are connected by a type 1 transmembrane segment/cytosolic linker. The canonical serine/threonine protein kinase domain and unique RNase domain of IRE1α are intimately adjoined, which facilitates efficient allosteric communication (Figure 3a, left) (33). The overall architecture of IRE1α’s kinase domain resembles that of other protein kinases; consisting of a mainly β-stranded N-terminal lobe and an α-helical C-terminal lobe; PERK’s overall architecture is similar (Figure 3a, right). The ATP-binding site, which contains all of the catalytic residues necessary for phosphate transfer, is located between these two lobes. The RNase domain of IRE1α is rigidly fused to the C-terminus of the kinase domain and is composed exclusively of α helices connected by short loops. While IRE1α’s RNase domain resembles the sterile α helix motif found in ephrin receptors, its overall fold is novel and is referred to as a Kinase Extension Ribonuclease (KEN) domain.
Structural and biochemical studies with truncated IRE1 constructs that contain only the cytosolic RNase/kinase module have proven valuable in understanding the transition from monomer to higher order oligomer. Numerous studies have shown that the kinase/RNase module alone (in both yeast and mammalian orthologs) is able to form dimers and oligomers that are likely similar to the assemblies formed by full length IRE1 tethered to the ER (33–35). In the absence or ER stress, IRE1 is unphosphorylated and mainly monomeric. Indeed, analytical ultracentrifugation (AUC) and crosslinking studies have shown that unphosphorylated human and yeast IRE1 kinase/RNase domain modules form very little dimer/oligomer in solution. Upon clustering under ER stress, IRE1 undergoes multiple autophosphorylation events on its activation loop and cytosolic linker, which is believed to promote dimer/oligomer formation (36). This notion is supported by experiments showing that multiply phosphorylated kinase/RNase constructs form dimers/oligomers at lower concentrations in solution than the unphosphorylated form. Thus, IRE1 autophosphorylation serves to enhance oligomer formation, which, in turn, enhances RNase activity.
Unphosphorylated human IRE1α forms a dimer (face-to-face) with the ATP-binding site of each kinase domain directed towards its dimeric partner (Figure 3b, left) in crystal structures (37). The interface of the face-to-face dimer is over 1700 Å2 and is composed entirely of residues in the kinase domain. This overall configuration results in trans exchange of kinase activation loops and the RNase domain of each IRE1 protomer being separated in space by over 55 Å. The face-to-face dimer of IRE1 is believed to be a stable trans-autophosphorylation complex that occurs at an early stage of IRE1 activation. The reduced capacity of IRE1 face-to-face dimer interface mutants to undergo autophosphorylation is consistent with this notion.
Crystal structures of yeast IRE1 in an alternative dimeric form (back-to-back) have been reported (Figure 3b, middle), as have those of PERK (Figure 3b, right) (33, 35). In the back-to-back dimer, the kinase domain of each IRE1 protomer is oriented in the opposite direction than the face-to-face dimer. This brings the RNase domains of each protomer into close contact and prevents trans exchange of kinase activation loops. The interface of the back-to-back dimer is greater than 3800 Å2 and composed of residues in both the kinase and RNase domains. In the back-to-back dimer, all of the catalytic residues necessary for catalyzing phosphodiester hydrolysis reside in a cleft between adjoining RNase domains. Based on biochemical and structural studies a histidine and tyrosine residue that line the RNase active site are believed to serve as the general acid/base pair for catalysing RNA cleavage. In vitro kinetic analysis shows that dimers and oligomers of IRE1 are the catalytically active species for RNA cleavage, and the back-to-back dimer represents the basis for assembling an active RNase. The back-to-back dimer is also able to form a scaffold required for higher order oligomer formation.
Terminal Signaling in the UPR
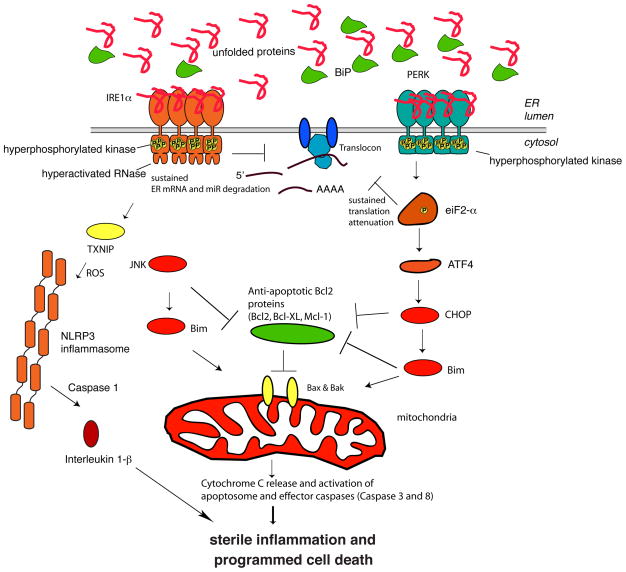
If the previously described adaptive responses fail to restore protein-folding homeostasis, UPR signaling continues to persist and eventually morphs into alternate signaling programs called the “terminal UPR” that ultimately promote programmed cell death (Figure 4) (38). Ample evidence supports that the two UPR kinases, PERK and IRE1α, have a distinct set of pro-apoptotic outputs that contribute to cell degeneration and death if ER stress cannot be resolved. For example, while a temporary pause in protein translation due to eIF2α phosphorylation can be beneficial by reducing secretory load, a protracted block in translation from sustained PERK signaling is incompatible with survival. In support of this notion, temporary forced dimerization of PERK (through an FvE-PERK construct dimerized with AP20187) affords a measure of cytoprotection against subsequent exposure to ER stress in a pre-conditioning regime, whereas tonic PERK activation through the same maneuver promotes apoptosis (39, 40). In both of these instances, eIF2α phosphorylation is activated by the FvE-PERK/AP20187 inducible system. Moreover, PERK hyperactivation can upregulate the C/EBP-homologous protein (CHOP)/GADD153 transcription factor, which inhibits expression of the gene encoding anti-apoptotic BCL-2 to hasten cell death (41, 42).
Figure 4. Terminal Signaling in the UPR.
If ER stress is irremediable, IRE1α becomes hyperactivated and undergoes homo-oligomerization. Under sustained oligomerization, IRE1α’s RNase endonucleolytically degrades hundreds of ER-localized mRNAs containing an N-terminal signal sequence, which depletes ER cargo and protein-folding components to further worsen ER stress. Moreover, when hyperactivated, IRE1α’s RNase directly cleaves select microRNAs that normally repress pro-apoptotic targets. In addition to signaling through RNA substrates, IRE1α oligomerization has been shown to induce activation or upregulation of a number of pro-inflammatory proteins. Finally, sustained IRE1α oligomerization serves as an activation platform for ASK1 and its downstream target JNK. Phosphorylation by JNK has been reported to both activate pro-apoptotic BIM and inhibit anti-apoptotic BCL-2. While a temporary pause in protein translation due to eIF2α phosphorylation can be beneficial for cells under ER stress, a protracted block in translation from sustained PERK signaling is incompatible with survival. Moreover, high levels of CHOP/GADD153 transcription factor can inhibit the expression of anti-apoptotic BCL-2 to hasten cell death and upregulate pro-apoptotic BIM to trigger activation of the mitochondrial-dependent apoptotic pathway.
Similarly, when hyperactivated by chronic ER stress, phosphorylated IRE1α transitions from homodimers into high order oligomers, allowing its RNase to acquire affinity for additional RNA substrates aside from XBP1 mRNA. Under sustained oligomerization, IRE1α’s RNase causes massive endonucleolytic decay of hundreds of ER-localized mRNAs containing an N-terminal signal sequence, which depletes ER cargo and protein-folding components to further worsen ER stress at later time points (43, 44). However, as with forced PERK activation, pre-emptive activation of IRE1α through chemical-genetic systems can partially prolong cell survival under ER stress. In support of this, we (45), and others (46), have demonstrated that pre-emptive activation of IRE1α’s homeostatic arm provides a metastable degree of cytoprotection against subsequent ER stress. Due to the highly unusual relationship between IRE1α’s kinase and RNase domains (47), the kinase domain of an IRE1α mutant can be engaged with an orthogonal kinase inhibitor called 1NM-PP1 to enforce an active ATP-binding conformation while bypassing autophosphorylation (34), which spontaneously triggers RNase activity even without ER stress. This allosteric activation forces splicing of XBP1 mRNA, resulting in the production of Xbp1. Cells subjected to these treatments pre-emptively are afforded a small, but significant, degree of cytoprotection under ER stress (45).
While the aforementioned studies demonstrate that intermediate states are available to either PERK or IRE1α, leading to divergent cell fate outcomes, IRE1α oligomerization under irremediably high levels of ER stress has been shown to induce activation or upregulation of a number of pro-inflammatory and pro-death proteins. For example, when hyperactivated, IRE1α’s RNase also reduces the levels of select microRNAs (possibly through directly cleaving their precursors at the ER membrane) that normally repress pro-apoptotic targets such as pro-oxidant protein TXNIP (thioredoxin-interacting protein), leading to their rapid upregulation(48, 49). Increased TXNIP protein levels then activate the NLRP3 inflammasome and its Caspase-1 dependent pro-death pathway, leading to sterile inflammation and pyroptotic cell death (49). Finally, sustained IRE1α oligomerization may serve as an activation platform for apoptosis signal-regulating kinase 1 (ASK1) and its substrate c-Jun NH2-terminal kinase (JNK) (50, 51). JNK phosphorylation has been reported to result in pro-apoptotic BIM activation and anti-apoptotic BCL-2 inhibition.
Many of the pro-death signals from the UPR sensors ultimately converge on the mitochondrial apoptotic pathway, which is triggered when toxic mitochondrial proteins, such as cytochrome c and Smac/Diablo, are forcibly released into the cytoplasm, which results in activation of downstream effector caspases (e.g., Caspase-3) (52). The BCL-2 family proteins govern this “intrinsic” apoptotic pathway by regulating outer mitochondrial membrane integrity (53). The intrinsic mitochondrial apoptotic pathway is engaged when cellular damage results in the expression and/or post-translational activation of Bcl-2 homology 3 (BH3)-only proteins, which are a diverse collection of pro-death proteins that contain an α-helix called the BH3 domain that is necessary for cell death (54). In the terminal UPR, at least four different BH3-only proteins (BID, BIM, NOXA, PUMA) become activated (55–57). The mechanism of activation of each BH3-only protein under ER stress is unique. For example, PERK activity leads to transcriptional upregulation of BIM, and the resultant protein product is stabilized by ER stress-dependent JNK dephosphorylation (56). Once activated, BH3-only proteins disable mitochondrial-protective proteins (e.g., BCL-2, BCL-XL, MCL-1) and in some cases directly trigger the multidomain pro-apoptotic BAX and BAK proteins to permeabilize the outer mitochondrial membrane by a process referred to as MOMP (Mitochondrial Outer Membrane Permeabilization)
ER stress-related Diseases
Cell injury, degeneration, and programmed death due to chronic, unmitigated ER stress has been increasingly implicated as underlying the pathophysiology of a wide range of prevalent human diseases (16). ER stress and sustained UPR signalling have been well documented in affected tissues in diabetes, neurodegeneration, inflammatory disorders, cancer, pulmonary fibrosis, and heart disease. In support of the notion that ER stress and maladaptive UPR signalling can contribute to pathology, inherited mutations in the UPR pathway have been associated with rare forms of diabetes and other diseases in humans (see below). For many of the aforementioned diseases, genetic manipulation of specific UPR components has been shown to influence disease outcome in rodent models. A few such diseases most strongly associated with ER stress are discussed below although more extensive reviews focused on various diseases have been published.
Extensive studies have implicated maladaptive UPR signalling in experimental and common forms of diabetes mellitus. As professional secretory cells, pancreatic β-cells synthesize, store and secrete large amounts of the polypeptide hormone insulin; it is estimated that each human β-cell produces on average about one million molecules of insulin every minute to support normoglycemia (8). Ultimately, this glucostatic cycle is dysregulated in diabetes because the requisite amount of insulin needed to maintain normoglycemia is not produced by a depleted mass of functioning β-cells. Myriad rodent models of ER stress-induced β-cell degeneration (too numerous to list in this review) amply prove the principle that premature β-cell loss from dysregulated UPR signalling is the causative insult leading to diabetes (8, 58).
Studies have shown that the β-cells of mice are most likely operating at UPR activation levels well above those of other professional secretory cells, even in healthy states (59). Therefore, β-cells may readily cross a terminal UPR threshold that puts them at risk for de-differentiation and apoptosis without possessing a significant capacity for homeostatic adjustment. Experimental rare diseases may inform our understanding of pathophysiology in common human diabetic syndromes (i.e. types 1 and 2) and eventually lead to target identification for disease modification. Indeed, dysregulation of the PERK and IRE1α upstream UPR arms has critical consequences for β-cell survival. A striking example of UPR dysregulation in diabetes is evidenced in PERK knockout mice. Massive and rapid β-cell apoptosis, which leads to infantile diabetes, results from homozygous deletion of the PERK gene in mice (60, 61). PERK knockout mice also exhibit growth defects and develop pancreatic exocrine insufficiency at early stages of life. These effects are rationalized as being due to the malfunction and demise of several different types of professional secretory cells, but interestingly diabetes mellitus is one of the earliest and most dominant phenotypes in these knockout animals. Intriguingly, PERK gene mutations result in a rare human diabetic syndrome (called Wolcott-Rallison syndrome) that is a phenocopy of the mouse PERK gene knockout. Furthermore, while homozygous deletion of either Ire1α or Xbp1 impedes embryogenesis and secretory cell development early in embryonic life (23, 50, 62, 63), the genetic removal of Xbp1 in the β-cell compartment leads to upstream IRE1α hyperactivation, degeneration of β-cells, and hyperglycemia, supporting a role for dysregulated IRE1α signalling in ER stress-induced degenerative changes in the endocrine pancreas (64). Also clearly apparent, the original studies on the PERK gene knockout, which leads to diabetes, showed compensatory IRE1α hyperactivation in islets, which may have contributed to degenerative changes in the β-cell compartment as the basis for diabetes progression in addition to the reported mechanism of removing a check on translation (65).
For various neurodegenerative diseases, a common pathologic hallmark is the accumulation of misfolded proteins and protein aggregates within affected neurons and surrounding supporting cells (66). Accumulation of many toxic protein species can kill neurons (67), and there is growing evidence that ER stress is an important mechanism driving this neurotoxicity (68). IRE1α activation and UPR induction are present in post-mortem brain and spinal cord tissues in Alzheimer disease (AD) (69–71), Parkinson disease (PD) (72) and amyotrophic lateral sclerosis (ALS) (73). Moreover, the accumulation of protein aggregates in cellular and animal models of HD (51), PD (74) and ALS (75–77) strongly correlate with UPR activation. Importantly, UPR upregulation is observed prior to the onset of symptoms, suggesting an active role in the disease (75). Furthermore, a recent study found that oral administration of a highly selective PERK inhibitor that efficiently crosses the blood-brain barrier significantly reduced neurodegeneration and clinical disease in prion-infected mice (78).
Finally, we consider the case for UPR modulation in cancers. Tumor cells often invade or metastasize into foreign environments where unfavourable conditions, such as hypoxia, glucose deprivation, lactic acidosis, oxidative stress and inadequate amino acid supplies compromise protein folding in the ER (79–81). Indeed, many studies find evidence of sustained and high level activation of all three branches of the UPR (PERK, ATF6, IRE1α) in a wide range of primary human tumor types, including glioblastoma, multiple myeloma, and carcinomas of the breast, stomach, esophagus, and liver (18, 82–86). Also, rare somatic mutations occur in the Ire1α gene in a small percentage of human solid tumors (87). However, despite the overwhelming evidence of ongoing ER stress and UPR activation in many forms of cancer, whether the UPR ultimately inhibits or promotes tumor growth remains unresolved but is an area of active study. Recently, triple-negative breast cancer, an aggressive malignancy with few treatment options, was found to exhibit a strong XBP1-dependent gene expression signature that correlated with poor prognosis (88). Myeloma, a highly secretory tumor composed of malignant plasma cells, is another cancer for which the UPR is frequently mentioned as a potentially attractive target based on strong evidence that the UPR, through IRE1α and its homeostatic target XBP1, is essential for plasma cell development (62, 63). Interestingly, up to 50% of primary myelomas show unusually high levels of XBP1s (82). Moreover, mice expressing a transgene of Xbp1s (that is missing the 26nt intron and hence requires no further processing by IRE1α) in B lymphocytes develop a plasma cell malignancy closely resembling myeloma (82). There is also evidence to suggest that proteasome inhibition with bortezomib (Velcade), which is FDA-approved as first line therapy for myeloma, leads to myeloma cell death in part by preventing disposal of misfolded proteins through the ERAD pathway and thus triggering ER stress-induced apoptosis (89). On the basis of these findings, several pharmacologic inhibitors of the IRE1α RNase activity have recently been tested on human myeloma xenografts and found to have antimyeloma activity (90, 91); however, the specificity and off-target effects of these pharmacological agents are not yet well understood.
While the above findings suggest an oncogenic role for XBP1s in the development of myeloma, recent data have emerged that challenge this notion. First, downregulation of XBP1s expression in myeloma correlates with resistance to bortezomib (92, 93). Second, using a combination of whole-genome and whole-exome sequencing of primary tumors from 38 myeloma patients, researchers discovered XBP1 mutations in two of these patients (94). On further analysis, these mutations were shown to inactivate XBP1s, arguing against an obligate role for this transcription factor in myeloma. Also, arguing against a direct cytoprotective role for IRE1α in various human cancers, many rare somatic mutations in the Ire1α gene found in glioblastoma, ovarian cancer, lung cancer and gastric cancer encode hypomorphic IRE1α variants that preserve XBP1 mRNA splicing but prevent the higher-order homo-oligomerization needed to activate extra-XBP1 RNase activity that promotes apoptosis (87, 95). Thus, hypomorphic IRE1α variants may provide a survival advantage by disabling or co-opting the terminal UPR. Furthermore, proliferative blocks through IRE1α that normally result from ER stress are overcome in the cancer mutants (95). Finally, it was recently reported that genetic knockdown of IRE1α or XBP1 in human myeloma cell lines is well tolerated and leads to bortezomib resistance (96), challenging the rationale for using IRE1α inhibitors in this disease.
Overall, the lessons from myeloma to date suggest that the effects of the UPR (or at least its IRE1α/XBP1 branch) on tumor development and maintenance are more complicated and nuanced than originally anticipated and that the role of the UPR is perhaps less well understood in cancers than in cell degenerative diseases.
Pharmacological Modulators of IRE1α and PERK
Does the UPR present tractable drug targets? Given the strong evidence of UPR deregulation across a range of human disease, there is great interest in the possibility of pharmacologically modulating its outputs to control cell fate under ER stress. Pushing the UPR’s homeostatic-apoptotic switch towards cell survival could potentially be therapeutically beneficial in cell degenerative diseases such as type 2 diabetes and neurodegeneration. However, the parallel and cross-wired networking of the UPR may require the simultaneous targeting of multiple nodes in order to obtain desired benefits. One approach may be to lengthen the adaptive phases of the UPR in order to increase chances of recovery (the transcription factors XBP1 and ATF6 are targets of this kind); such a regime may involve pre-conditioning the secretory pathway by pre-emptive UPR activation in order to make it more robust. A different approach is to inhibit key mediators of apoptosis (CHOP, thioredoxin-interacting protein (TXNIP)). Perhaps, potential timers of the UPR such as GADD34 and p58IPK may also be appealing targets for intervention. In this vein, a pharmacological agent called salubrinal has been demonstrated to inhibit eIF2α dephosphorylation and hence result in enhanced cell survival under ER stress (97).
As previously mentioned, pre-emptive preconditioning was demonstrated to be partially cytoprotective in cell culture models of ER and oxidative stress utilizing dimerizable versions of PERK in combination with small molecule dimerizers (39). However, it is unclear whether the translational inhibition that results from prolonged PERK activation would be efficacious in vivo. Related to this, we (45), and others (46), have demonstrated that pre-emptive activation of IRE1α’s homeostatic mode using chemical-genetics can partially prolong cell survival under ER stress. However, at best, pre-emptive activation through IRE1α provides only a small and temporary measure of cytoprotection, and furthermore has not been demonstrated to be efficacious in an animal model. Irremediable ER stress hyperactivates both PERK and IRE1α, leading to entry into apoptosis. Thus a diametrically opposite strategy is to inhibit the hyperactivated state using inhibitors of PERK (98), RNase inhibitors of IRE1α (90), or allosteric RNase-inhibitory type II kinase inhibitors of IRE1α (34).
The enzyme active sites of IRE1α and PERK represent attractive targets for the development of small molecule modulators of the UPR. Ligands that interact with PERK’s kinase domain and IREα’s kinase and RNase domains have been identified (Figure 5). These small molecule modulators not only represent promising starting points for the development of therapeutics that target the UPR, but are also useful regents for gaining insight into the functional roles that these multi-domain proteins play in responding to ER stress.
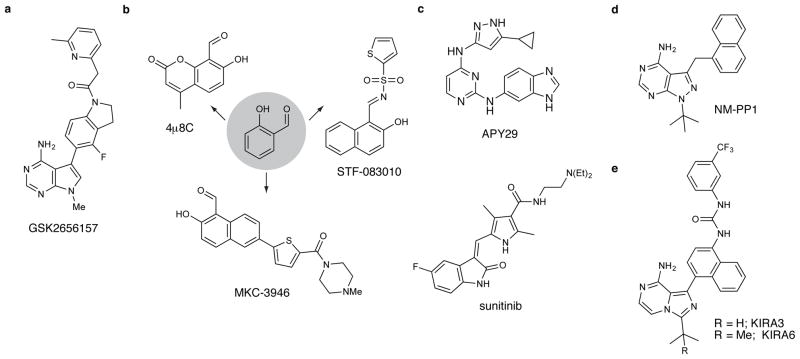
Figure 5. Small Molecule Modulators of PERK and IRE1α.
a. Chemical structure of the ATP-competitive PERK inhibitor GSK2656157. b. Salicylaldehyde-based inhibitors of the RNase domain of IRE1α. c. Orthogonal type I ATP-competitive inhibitor 1NM-PP1. d. Type I kinase inhibitors that increase the RNase activity of IRE1α. e. Type II kinase inhibitors that inhibit the RNase activity of IRE1α.
ATP-competitive PERK inhibitors
Efforts have been made to identify compounds that inhibit the ability of PERK to phosphorylate eIF2α. Towards this goal, GlaxoSmithKline used structure-based design to develop highly potent and selective inhibitors of PERK’s kinase domain from an initial screening hit (98). Optimized ATP-competitive inhibitors, like GSK2656157, are based on a pyrrolopyrimidine scaffold that displays an indoline-arylactetamide substituent from its C-3 position (Figure 5a). The pyrrolopyrimidine scaffold of GSK2656157 sits in the adenine-binding pocket and the indoline-arylactetamide moiety projects towards PERK’s helix αC, stabilizing an inactive conformation of this structural element. Helix αC forms part of the back-to-back dimer interface that the kinase domain of PERK forms, but it is unclear whether the displacement stabilized by GSK2656157 and its analogs disfavour dimerization. In cells, GSK2656157 potently inhibits ER stress-induced PERK autophosphorylation and the phosphorylation of eIF2α. This inhibitory activity corresponds to an observed decrease in ATF4 and CHOP. While early reports suggest that a PERK inhibitor may protect against pre-clinical models of neurodegeneration (78), much more work needs to be done to understand the potential benefits and risks of inhibiting ER stress-induced cell degeneration in vivo, especially because GSK PERK inhibitors mimic the pancreatic cell loss evident in the murine Wolcott-Rallison syndrome model (deletion of Perk, and consequent compensatory activation of IRE1α in islets).
Ligands that interact with the RNase domain of IRE1α
A number of efforts have been made to identify small molecules that directly inhibit the RNase activity of IRE1α through its RNase domain (90, 91, 99, 100). To date, all of the inhibitors identified contain a reactive electrophile that most likely covalently modifies IRE1α’s RNase active site. The fact that all of these inhibitors contain a reactive moiety is not surprising as the active site of IRE1α’s RNase is relatively shallow and polar, which presents minimal opportunities for high affinity interactions with small molecules. The most effective pharmacophore for targeting the RNase domain of IRE1α has proven to be a salicylaldehyde (Figure 5b). Inhibitors of this class are exemplified by STF-083010, MKC-3946, and 4μ8c. MKC-3946 and 4μ8c were identified for their abilities to prevent a purified, recombinant RNase/kinase IRE1α construct from cleaving a fluorescently-labelled XBP1 RNA mini-substrate in vitro, while STF-083010 was a hit in a high-throughput, cell-based reporter gene assay (90, 91, 99, 100). In cells, inhibitors based on the salicylaldehyde pharmacophore inhibit the RNase activity of IRE1α without affecting its ability to autophosphorylate. Presumably, these inhibitors also do not prevent IRE1α dimerization and oligomerization under ER stress.
The similar potencies of structurally diverse salicylaldehyde-based inhibitors suggests that the efficacy of these compounds is likely driven by reactivity more than binding affinity. Indeed, the IC50s of STF-083010, MKC-3946, and 4μ8c are within 10–20-fold in in vitro RNase activity assays. Several studies have shown that salicylaldehyde-based inhibitors form a Schiff base with a lysine in the RNase active site (100, 101). As other salicylaldehyde-based inhibitors compete with 4μ8c for Schiff base formation, it is likely that lysine 907 is generally targeted by this inhibitor class. Several recently reported crystal structures of murine IRE1α bound to salicylaldehyde-based inhibitors show that lysine 907 is indeed involved in Schiff base formation (101). Furthermore, these structural studies provide insight into how covalent bond formation interferes with RNase activity.
Ligands that interact with the kinase domain of IRE1α
Several ATP-competitive inhibitors that target the kinase domain of IRE1α have been identified. While all of these ligands inhibit the kinase activity of IRE1α, occupation of the ATP-binding site can have profoundly divergent effects on the RNase domain. Allosteric communication between the ATP-binding site and RNase domain was first discovered in a yeast IRE1 mutant that contains an enlarged (“holed”) ATP-binding pocket, which can be selectively complemented with the orthogonal type I ATP-competitive inhibitor 1NM-PP1 (Figure 5c) (47). Holed ATP-binding site mutants of IRE1 possess crippled kinase and RNase activities, but binding of 1NM-PP1 to IRE1’s ATP-binding site restores RNase function. Thus, an ATP-competitive ligand is able to allosterically activate IRE1’s RNase domain in the absence of kinase autophosphorylation. This allosteric relationship is maintained in human IRE1α, as 1NM-PP1 is able to restore “holed” human IRE1α’s ability to cleave RNA in vitro and in cells. ATP-competitive inhibitors of wild-type IRE1α are also able to activate RNase activity while inhibiting kinase activity. For example, the promiscuous ATP-competitive inhibitor APY29 and the clinically-approved drug sunitinib activate IRE1α’s RNase domain (Figure 5c) (34, 35, 43). In addition, these ATP-competitive inhibitors increase the dimerization/oligomerization state of IRE1α, which most likely contributes to the observed enhancement of RNase activity.
Recently, ATP-competitive ligands that inhibit IRE1α’s RNase activity through the kinase domain have been identified (Figure 5d,e) (34). These kinase inhibiting RNase attenuators (KIRAs) were discovered by screening type II ATP-competitive ligands, which stabilize inactive ATP-binding site conformations, for their abilities to block the RNase activity of a recombinant IRE1α kinase/RNase construct. Optimization of an initial hit based on a pyrazolopyrimidine scaffold resulted in KIRA3 (Figure 5d), which inhibits the kinase and RNase activities of IRE1α in vitro and in cells. Consistent with its divergent behaviour relative to type I ATP-competitive inhibitors, KIRA3 directly opposes the ability of APY29 to activate the RNase domain of IRE1α. Furthermore, and in contrast to sunitinib and APY29, KIRA3 suppresses the dimerization/oligomerization of IRE1α by stabilizing the monomeric form of this protein. While a structure of KIRA3 bound to IRE1α has not yet been determined, stabilization of an inactive, ATP-binding site conformation most likely leads to stabilization of monomeric IRE1α and suppression of RNase inhibition.
A more advanced KIRA, KIRA6 (Figure 5e), with suitable pharmacokinetic properties for in vivo studies showed greater potency and dose-dependently reduced IRE1α phosphorylation, oligomerization, and RNase activation in vitro and in vivo to preserve both cell viability and function. Importantly, KIRA6 showed efficacy in two different animal models of ER stress-induced cell degeneration: Specifically, KIRA6 preserved photoreceptor functional viability in rat models of ER stress-induced retinal degeneration when injected into the vitreous. Moreover, KIRA6 preserved pancreatic β-cells, increased insulin, and reduced hyperglycemia in Akita diabetic mice, which spontaneously develop diabetes in neonatal life due to their harbouring a mutant proinsulin that is unable to complete oxidative folding, resulting in chronic ER stress and β-cell apoptosis (49, 95).
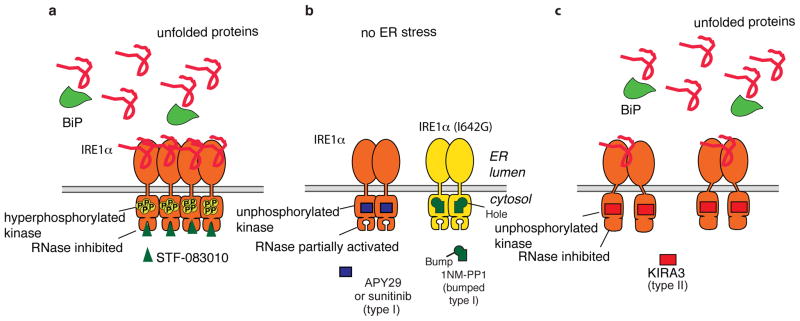
The availability of diverse ligands that target two distinct active sites, presents unique opportunities to pharmacologically tune the intra-cellular behaviour of IRE1α, perhaps even combinatorially (95). Under ER stress, salicylaldehyde-based inhibitors will directly inhibit the RNase activity of IRE1α but not prevent autophosphorylation or the phosphorylation of any potential non-autonomous protein substrates (Figure 6a). Furthermore, direct inhibition of the RNase domain most likely does not prevent dimerization/oligomerization. Type I inhibitors, like APY29 and sunitinib, will prevent IRE1α autophosphorylation, but activate the RNase domain, even in the absence of ER stress (Figure 6b). Furthermore, inhibitors of this class should enhance IRE1α dimerization/oligomerization. In contrast, type II inhibitors, like KIRA3 and KIRA6, inhibit both the kinase and RNase activities of IRE1α (Figure 6c). Additionally, type II IRE1α inhibitors will stabilize the monomeric form and oppose dimerization/oligomerization induced by ER stress.
Figure 6. Modes of Pharmacological Modulation of IRE1α.
a. Model of how salicylaldehyde-based RNase inhibitors affect intra-cellular IRE1α. b. Model of how type I kinase inhibitors affect intra-cellular IRE1α. c. Model of how type II kinase inhibitors affect intra-cellular IRE1α.
Conclusions
The UPR is a highly conserved signaling pathway that is activated when cells are not able to keep pace with the protein folding demands of the ER—a form of cell injury called ER stress. Under ER stress, the UPR initially sends out adaptive outputs that reduce the protein-folding load and expand the capacity of the ER secretory pathway. However, under irremediable ER stress, the UPR assembles into a platform that sends out pro-inflammatory and pro-death signals to cause cell demise. Cell injury from chronic ER stress is emerging as central to the pathophysiology of a wide range of prevalent human diseases, including diabetes, neurodegeneration, stroke, and cancer. Recent advances in our understanding of how the UPR switches from life to death signaling may lead to new strategies to combat these ER stress-associated diseases. In this regard, the recent findings from many research groups demonstrating (A) that two master regulators of the UPR, PERK and IRE1α, akin to cell surface death receptors, have binary outputs promoting either life or death decisions for the ER stressed cell, combined with (B) the identification of novel small molecule modulators of these proteins offers rich opportunities to dissect the contribution of these signalling proteins to pathogenesis in myriad models of human ER stress-related diseases. As such, future studies will be enabled by optimizing compounds directed towards these master UPR regulators to (A) determine whether PERK and IRE1α are viable targets for disease-modification, and (B) may serve as starting points for first-in-class series of drugs. Furthermore, the combined application of PERK and IRE1α inhibitors may prevent compensatory activation resulting from inhibition of either kinase in isolation. Thus, combination therapy against the two druggable kinases of the UPR may emerge someday as a useful means to ameliorate the critical downstream terminal signalling events that are linked to cell demise in myriad diseases proceeding from unchecked ER stress.
Acknowledgments
Work was supported by NIH: DP2OD001925 (F.R.P), RO1DK080955 (F.R.P), P30DK063720 (F.R.P.), UO1DK089541 (F.R.P.), R01DK100623 (F.R.P and D.J.M.), R01GM086858 (D.J.M.); Burroughs Wellcome Foundation (F.R.P.); Juvenile Diabetes Research Foundation (F.R.P.); Harrington Discovery Institute Scholar-Innovator Award (F.R.P.); Alfred P. Sloan Foundation (D.J.M.); Camille and Henry Dreyfus Foundation (D.J.M.).
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Anelli T, Sitia R. Protein quality control in the early secretory pathway. EMBO J. 2008;27:315–327. doi: 10.1038/sj.emboj.7601974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tu BP, Weissman JS. Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sevier CS, Kaiser CA. Formation and transfer of disulphide bonds in living cells. Nature Rev Mol Cell Biol. 2002;3:836–847. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- 4.van Anken E, Braakman I. Versatility of the endoplasmic reticulum protein folding factory. Crit Rev Biochem Mol Biol. 2005;40:191–228. doi: 10.1080/10409230591008161. [DOI] [PubMed] [Google Scholar]
- 5.Merksamer PI, Trusina A, Papa FR. Real-Time Redox Measurements during Endoplasmic Reticulum Stress Reveal Interlinked Protein Folding Functions. Cell. 2008;135:933–947. doi: 10.1016/j.cell.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 8.Scheuner D, Kaufman RJ. The unfolded protein response: a pathway that links insulin demand with beta-cell failure and diabetes. Endocr Rev. 2008;29:317–333. doi: 10.1210/er.2007-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Anken E, et al. Efficient IgM assembly and secretion require the plasma cell induced endoplasmic reticulum protein pERp1. Proc Nat Acad Sci USA. 2009;106:17019–17024. doi: 10.1073/pnas.0903036106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13:184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oyadomari S, et al. Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. J Clin Invest. 2002;109:525–532. doi: 10.1172/JCI14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang HM, et al. Coxsackievirus B3 infection activates the unfolded protein response and induces apoptosis through downregulation of p58IPK and activation of CHOP and SREBP1. J Virol. 2010;84:8446–8459. doi: 10.1128/JVI.01416-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Y, Hendershot LM. ER chaperone functions during normal and stress conditions. J Chem Neuroanat. 2004;28:51–65. doi: 10.1016/j.jchemneu.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Flamment M, Hajduch E, Ferre P, Foufelle F. New insights into ER stress-induced insulin resistance. Trends Endocrinol Metab. 2012;23:381–390. doi: 10.1016/j.tem.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Gestwicki JE, Garza D. Protein quality control in neurodegenerative disease. Prog Mol Biol Transl Sci. 2012;107:327–353. doi: 10.1016/B978-0-12-385883-2.00003-5. [DOI] [PubMed] [Google Scholar]
- 16.Wang S, Kaufman RJ. The impact of the unfolded protein response on human disease. J Cell Biol. 2012;197:857–867. doi: 10.1083/jcb.201110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pincus D, et al. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol. 2010;8:e1000415. doi: 10.1371/journal.pbio.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardner BM, Walter P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science. 2011;333:1891–1894. doi: 10.1126/science.1209126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci USA. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 21.Travers KJ, et al. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 22.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 23.Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang XZ, et al. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korennykh A, Walter P. Structural basis of the unfolded protein response. Annu Rev Cell Develop Biol. 2012;28:251–277. doi: 10.1146/annurev-cellbio-101011-155826. [DOI] [PubMed] [Google Scholar]
- 26.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto K, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 29.Liu CY, Schroder M, Kaufman RJ. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J Biol Chem. 2000;275:24881–24885. doi: 10.1074/jbc.M004454200. [DOI] [PubMed] [Google Scholar]
- 30.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 31.Harding HP, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 32.Ye J, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 33.Lee KP, et al. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell. 2008;132:89–100. doi: 10.1016/j.cell.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, et al. Divergent allosteric control of the IRE1alpha endoribonuclease using kinase inhibitors. Nat Chem Biol. 2012;8:982–989. doi: 10.1038/nchembio.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korennykh AV, et al. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2009;457:687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korennykh AV, et al. Cofactor-mediated conformational control in the bifunctional kinase/RNase Ire1. BMC Biol. 2011;9:48. doi: 10.1186/1741-7007-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ali MM, et al. Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. EMBO J. 2011;30:894–905. doi: 10.1038/emboj.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shore GC, Papa FR, Oakes SA. Signaling cell death from the endoplasmic reticulum stress response. Curr Opin Cell Biol. 2011;23:143–149. doi: 10.1016/j.ceb.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu PD, et al. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 2004;23:169–179. doi: 10.1038/sj.emboj.7600030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin JH, Li H, Zhang Y, Ron D, Walter P. Divergent effects of PERK and IRE1 signaling on cell viability. PloS One. 2009;4:e4170. doi: 10.1371/journal.pone.0004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Molec Cellular Biol. 2001;21:1249–1259. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marciniak SJ, et al. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han D, et al. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 45.Han D, et al. A kinase inhibitor activates the IRE1alpha RNase to confer cytoprotection against ER stress. Biochem Biophys Res Comm. 2008;365:777–783. doi: 10.1016/j.bbrc.2007.11.040. [DOI] [PubMed] [Google Scholar]
- 46.Lin J, et al. IRE1 Signaling Affects Cell Fate During the Unfolded Protein Response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papa FR, Zhang C, Shokat K, Walter P. Bypassing a kinase activity with an ATP-competitive drug. Science. 2003;302:1533–1537. doi: 10.1126/science.1090031. [DOI] [PubMed] [Google Scholar]
- 48.Upton JP, et al. IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 2012;338:818–822. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lerner AG, et al. IRE1alpha induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab. 2012;16:250–264. doi: 10.1016/j.cmet.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urano F, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 51.Nishitoh H, et al. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002;16:1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang C, Youle RJ. The role of mitochondria in apoptosis. Annu Rev Genet. 2009;43:95–118. doi: 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giam M, Huang DC, Bouillet P. BH3-only proteins and their roles in programmed cell death. Oncogene. 2008;27(Suppl 1):S128–S135. doi: 10.1038/onc.2009.50. [DOI] [PubMed] [Google Scholar]
- 55.Upton JP, et al. Caspase-2 cleavage of BID is a critical apoptotic signal downstream of endoplasmic reticulum stress. Mol Cell Biol. 2008;28:3943–3951. doi: 10.1128/MCB.00013-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Puthalakath H, et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129:1337–1349. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 57.Li J, Lee B, Lee AS. Endoplasmic reticulum stress-induced apoptosis: multiple pathways and activation of p53-up-regulated modulator of apoptosis (PUMA) and NOXA by p53. J Biol Chem. 2006;281:7260–7270. doi: 10.1074/jbc.M509868200. [DOI] [PubMed] [Google Scholar]
- 58.Papa FR. Endoplasmic reticulum stress, pancreatic beta-cell degeneration, and diabetes. Cold Spring Harbor Perspect Med. 2012;2:a007666. doi: 10.1101/cshperspect.a007666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iwawaki T, Akai R, Kohno K, Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med. 2004;10:98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- 60.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 61.Delepine M, et al. EIF2AK3, encoding translation initiation factor 2-alpha kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat Genet. 2000;25:406–409. doi: 10.1038/78085. [DOI] [PubMed] [Google Scholar]
- 62.Reimold AM, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 63.Zhang K, et al. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J Clin invest. 2005;115:268–281. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee AH, Heidtman K, Hotamisligil GS, Glimcher LH. Dual and opposing roles of the unfolded protein response regulated by IRE1alpha and XBP1 in proinsulin processing and insulin secretion. Proc Natl Acad Sci USA. 2011;108:8885. doi: 10.1073/pnas.1105564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harding HP, et al. Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 66.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10:S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 67.Taylor JP, Hardy J, Fischbeck KH. Toxic proteins in neurodegenerative disease. Science. 1991;296 doi: 10.1126/science.1067122. need pages. [DOI] [PubMed] [Google Scholar]
- 68.Roussel BD, et al. Endoplasmic reticulum dysfunction in neurological disease. Lancet Neurol. 2013;12:105–118. doi: 10.1016/S1474-4422(12)70238-7. [DOI] [PubMed] [Google Scholar]
- 69.Hamos JE, et al. Expression of heat shock proteins in Alzheimer’s disease. Neurology. 1991;41:345–350. doi: 10.1212/wnl.41.3.345. [DOI] [PubMed] [Google Scholar]
- 70.Hoozemans JJ, et al. The unfolded protein response is activated in pretangle neurons in Alzheimer’s disease hippocampus. Am J Pathol. 2009;174:1241–1251. doi: 10.2353/ajpath.2009.080814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Unterberger U, et al. Endoplasmic reticulum stress features are prominent in Alzheimer disease but not in prion diseases in vivo. J Neuropathol Exp Neurol. 2006;65:348–357. doi: 10.1097/01.jnen.0000218445.30535.6f. [DOI] [PubMed] [Google Scholar]
- 72.Wang HQ, Takahashi R. Expanding insights on the involvement of endoplasmic reticulum stress in Parkinson’s disease. Antioxid Redox Signal. 2007;9:553–561. doi: 10.1089/ars.2006.1524. [DOI] [PubMed] [Google Scholar]
- 73.Atkin JD, et al. Endoplasmic reticulum stress and induction of the unfolded protein response in human sporadic amyotrophic lateral sclerosis. Neurobiol Dis. 2008;30:400–407. doi: 10.1016/j.nbd.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 74.Holtz WA, O’Malley KL. Parkinsonian mimetics induce aspects of unfolded protein response in death of dopaminergic neurons. J Biol Chem. 2003;278:19367–19377. doi: 10.1074/jbc.M211821200. [DOI] [PubMed] [Google Scholar]
- 75.Atkin JD, et al. Induction of the unfolded protein response in familial amyotrophic lateral sclerosis and association of protein-disulfide isomerase with superoxide dismutase 1. J Biol Chem. 2006;281:30152–30165. doi: 10.1074/jbc.M603393200. [DOI] [PubMed] [Google Scholar]
- 76.Kikuchi H, et al. Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. Proc Natl Acad Sci USA. 2006;103:6025–6030. doi: 10.1073/pnas.0509227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nishitoh H, et al. ALS-linked mutant SOD1 induces ER stress- and ASK1-dependent motor neuron death by targeting Derlin-1. Genes Dev. 2008;22:1451–1464. doi: 10.1101/gad.1640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moreno JA, et al. Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci Transl Med. 2013;5:206ra138. doi: 10.1126/scitranslmed.3006767. [DOI] [PubMed] [Google Scholar]
- 79.Lee AS, Hendershot LM. ER stress and cancer. Cancer Biol Ther. 2006;5:721–722. doi: 10.4161/cbt.5.7.3120. [DOI] [PubMed] [Google Scholar]
- 80.Koumenis C. ER stress, hypoxia tolerance and tumor progression. Curr Mol Med. 2006;6:55–69. doi: 10.2174/156652406775574604. [DOI] [PubMed] [Google Scholar]
- 81.Moenner M, Pluquet O, Bouchecareilh M, Chevet E. Integrated endoplasmic reticulum stress responses in cancer. Cancer Res. 2007;67:10631–10634. doi: 10.1158/0008-5472.CAN-07-1705. [DOI] [PubMed] [Google Scholar]
- 82.Carrasco DR, et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11:349–360. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fernandez PM, et al. Overexpression of the glucose-regulated stress gene GRP78 in malignant but not benign human breast lesions. Breast Cancer Res Treat. 2000;59:15–26. doi: 10.1023/a:1006332011207. [DOI] [PubMed] [Google Scholar]
- 84.Shuda M, et al. Activation of the ATF6, XBP1 and grp78 genes in human hepatocellular carcinoma: a possible involvement of the ER stress pathway in hepatocarcinogenesis. J Hepatol. 2003;38:605–614. doi: 10.1016/s0168-8278(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 85.Song MS, Park YK, Lee JH, Park K. Induction of glucose-regulated protein 78 by chronic hypoxia in human gastric tumor cells through a protein kinase C-epsilon/ERK/AP-1 signaling cascade. Cancer Res. 2001;61:8322–8330. [PubMed] [Google Scholar]
- 86.Chen X, Ding Y, Liu CG, Mikhail S, Yang CS. Overexpression of glucose-regulated protein 94 (Grp94) in esophageal adenocarcinomas of a rat surgical model and humans. Carcinogenesis. 2002;23:123–130. doi: 10.1093/carcin/23.1.123. [DOI] [PubMed] [Google Scholar]
- 87.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen X, et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature. 2014;508:103–107. doi: 10.1038/nature13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Voorhees PM, Orlowski RZ. The proteasome and proteasome inhibitors in cancer therapy. Annu Rev Pharmacol Toxicol. 2006;46:189–213. doi: 10.1146/annurev.pharmtox.46.120604.141300. [DOI] [PubMed] [Google Scholar]
- 90.Papandreou I, et al. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117:1311–1314. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mimura N, et al. Blockade of XBP1 splicing by inhibition of IRE1alpha is a promising therapeutic option in multiple myeloma. Blood. 2012;119:5772–5781. doi: 10.1182/blood-2011-07-366633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gu JL, et al. Differentiation induction enhances bortezomib efficacy and overcomes drug resistance in multiple myeloma. Biochem Biophys Res Commun. 2012;420:644–650. doi: 10.1016/j.bbrc.2012.03.056. [DOI] [PubMed] [Google Scholar]
- 93.Ling SC, et al. Response of myeloma to the proteasome inhibitor bortezomib is correlated with the unfolded protein response regulator XBP-1. Haematologica. 2012;97:64–72. doi: 10.3324/haematol.2011.043331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ghosh R, et al. Allosteric Inhibition of the IRE1alpha RNase Preserves Cell Viability and Function during Endoplasmic Reticulum Stress. Cell. 2014;158:534–548. doi: 10.1016/j.cell.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leung-Hagesteijn C, et al. Xbp1s-negative tumor B cells and pre-plasmablasts mediate therapeutic proteasome inhibitor resistance in multiple myeloma. Cancer Cell. 2013;24:289–304. doi: 10.1016/j.ccr.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Boyce M, et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 98.Atkins C, et al. Characterization of a novel PERK kinase inhibitor with antitumor and antiangiogenic activity. Cancer Res. 2013;73:1993–2002. doi: 10.1158/0008-5472.CAN-12-3109. [DOI] [PubMed] [Google Scholar]
- 99.Volkmann K, et al. Potent and selective inhibitors of the inositol-requiring enzyme 1 endoribonuclease. J Biol Chem. 2011;286:12743–12755. doi: 10.1074/jbc.M110.199737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cross BC, et al. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc Natl Acad Sci USA. 2012;109:E869–E878. doi: 10.1073/pnas.1115623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sanches M, et al. Structure and mechanism of action of the hydroxy-aryl-aldehyde class of IRE1 endoribonuclease inhibitors. Nat Commun. 2014;5:4202. doi: 10.1038/ncomms5202. [DOI] [PMC free article] [PubMed] [Google Scholar]