Abstract
Gene mutations, mostly segregating with a dominant mode of inheritance, are important causes of dilated cardiomyopathy (DCM), a disease characterized by enlarged ventricular dimensions, impaired cardiac function, heart failure and high risk of death. Another myocardial abnormality often linked to gene mutations is left ventricular noncompaction (LVNC) characterized by a typical diffuse spongy appearance of the left ventricle. Here, we describe a large Bedouin family presenting with a severe recessive DCM and LVNC. Homozygosity mapping and exome sequencing identified a single gene variant that segregated as expected and was neither reported in databases nor in Bedouin population controls. The PLEKHM2 cDNA2156_2157delAG variant causes the frameshift p.Lys645AlafsTer12 and/or the skipping of exon 11 that results in deletion of 30 highly conserved amino acids. PLEKHM2 is known to interact with several Rabs and with kinesin-1, affecting endosomal trafficking. Accordingly, patients' primary fibroblasts exhibited abnormal subcellular distribution of endosomes marked by Rab5, Rab7 and Rab9, as well as the Golgi apparatus. In addition, lysosomes appeared to be concentrated in the perinuclear region, and autophagy flux was impaired. Transfection of wild-type PLEKHM2 cDNA into patient's fibroblasts corrected the subcellular distribution of the lysosomes, supporting the causal effect of PLEKHM2 mutation. PLEKHM2 joins LAMP-2 and BAG3 as a disease gene altering autophagy resulting in an isolated cardiac phenotype. The association of PLEKHM2 mutation with DCM and LVNC supports the importance of autophagy for normal cardiac function.
Introduction
Dilated cardiomyopathy (DCM) refers to a group of heterogeneous myocardial disorders that are characterized by ventricular dilation and depressed myocardial contractility in the absence of abnormal loading conditions (1). It is an important cause of heart failure (HF) and sudden cardiac death, and represents ∼30–40% of patients in multicenter HF trials (2). DCM accounts for >10 000 deaths in the USA annually and for more than half of heart transplantations in adult and children worldwide (1,3). In patients with idiopathic DCM, a common estimation is that 25–50% of cases are familial with genetic origin (4,5), although hitherto reported mutations explain only part of the familial DCM cases (6). Autosomal-dominant inheritance is the most common mode of inheritance (7), presenting usually in the second or third decade of life (8,9). Most of the known mutated genes [MIM 115200 (10)] encode structural components of the heart muscle, implying that the disease is caused by impairment of force generation, sensing and/or transmission. Autosomal-recessive mutations leading to DCM are far less common. Among the few existing cases, there are two genes encoding cardiac structural proteins (TNNI3 and TTN; [MIM 611880 and 611705, respectively]), which are also linked to dominant DCM forms of inheritance. Other described genes encode FUKUTIN (FKTN [MIM 611615]), important for properly transmitting the generated force, and TAFAZZIN (TAZ [MIM 300395]), involved in cardiolipin metabolism and ATP production (11). Similarly, we have identified a mutation in SDHA encoding the flavoprotein subunit of complex II of the mitochondrial respiratory chain (12). Mutations in SDHA, TAZ and FKTN cause additional syndromes (Leigh, Barth and muscular dystrophy dystroglycanopathy [MIM 256000; 30206; 236670]); for reviews, see (13,14).
A unique form of cardiomyopathy, that is gaining attention in regard to its genetic background, is left ventricular noncompaction (LVNC [MIM 604169]) (15,16). LVNC is a cardiomyopathy characterized by a distinctive spongy appearance of the myocardium, due to increased trabeculation and deep intertrabecular recesses in hypertrophied and hypokinetic segments of the left ventricle (LV) that communicate with the LV cavity (17). However, there is no current ‘gold standard’ for the diagnosis, which can be also challenging according to the extent of trabeculation and deep intertrabecular recesses (18,19). Clinical manifestations are highly variable, ranging from no symptoms to disabling congestive HF, arrhythmias and systemic thromboembolism (20–22). There is no specific histological finding in LVNC, although fibrosis has been described (23,24). Up to 44% of patients have a family history of cardiomyopathy (25–29). In children X-linked, autosomal-dominant and mitochondrial inheritance have been reported, whereas in adults autosomal-dominant inheritance has been described (20). Mutations in several genes that have previously been associated with hypertrophic cardiomyopathy and DCM are reported (30–36), including sarcomeric protein-encoding genes (36). These findings support the concept that LVNC is part of a diverse spectrum of cardiac pathologies triggered by sarcomeric protein gene defects, and that there is a shared molecular etiology of different cardiomyopathic phenotypes. However, genetic studies have been limited, and the genetic basis of LVNC is still unresolved in most cases.
We identified here a novel gene, in which a genetic variant leads to recessively inherited DCM and LVNC that is limited to a few segments, by using genetic mapping and exome sequencing of a highly inbred family of Bedouin ancestry.
Results
Clinical findings in affected patients
The study was initiated following several independent hospitalizations of children from a single large Bedouin family. A total of four patients at age of 7(years)–16 years who were diagnosed with DCM, focal areas of spongy LV myocardium and sustained ventricular tachycardia (VT) were examined (Fig. 1A and Table 1). Two additional family members who died prior to the study (siblings of patients II-1 and II-2, Fig. 1A) were reported to have died from sudden death.
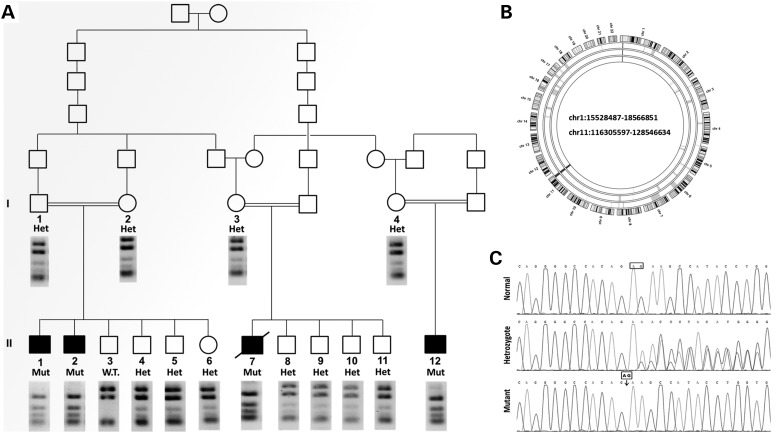
Figure 1.
Family pedigree and identification of the homozygous intervals and the mutation. (A) Pedigree: three patients (II-1, II-2 and II-7) were genotyped. The segregation of the PLEKHM 2 mutation that creates a MwoI site is presented by the restriction analysis using this enzyme, under each individual. Digestion of the 687 bp amplicon resulted in 25, 73, 75, 83, 190 and 241 bp fragments; if the mutation is present, the 239 bp fragment is cleaved into 135 and 104 bp fragments. All patients were homozygous for the mutation; all available parents and all healthy siblings were heterozygotes, except sibling II-3 who was homozygous normal. (B) AgileMultiIdeogram showing the homozygous regions of chromosomes 1 and 11 shared among patients II-1, II-2 and II-7. The total length of the chromosomal homozygous regions was 15.3 Mb. (C) Sequence chromatogram of the PCR product of genomic DNA showing the single variant compatible as a causing mutation, a deletion of two nucleotides 1:16055171_16055172delAG (GRCh37/Hg 19) in exon 12. The chromatograms present the homozygous normal sequence (individual II-3), heterozygotes (Het) and mutant (Mut).
Table 1.
Clinical evaluation of patients
| Patient | Age at onset (years) | Presenting symptoms at onset | Echo at onset | Age at follow-up and present situation | Echo at follow-up | CMRI cine-SSFP LVEDV (ml/sqs) LVESV (ml/sqs) EF (%) |
CMRI description Cine-SSFP four chambers short axis |
Late gadolinium enhancement (LGE) |
|---|---|---|---|---|---|---|---|---|
| II-1 | 16 | Ventricular tachycardia | LVESD 59 mm LVEDD 68 mm EF—35% |
18 ICD followed by heart transplantation, presently 22 years |
LVESD 60 mm LVEDD 73 EF—35% |
Severe dilatation LV Severe global LV dysfunction LVEDV—181 LVESV—122 EF—30 |
Noncompaction on mid-lateral anterior and posterior segments Compact—3.4 mm Noncompact—10 mm |
Large patches of diffuse/transmural and longitudinal striae of mid-wall enhancement |
| II-2 | 13 | Asymptomatic familial cardiomyopathy checkout | LVESD 34 mm LVEDD 51 mm EF—58% |
18 ICD at 17.5 years due to VPBS Asymptomatic |
LVESD 57 mm LVEDD 70 mm EF—35% |
Severe dilatation LV Severe global LV dysfunction LVEDV—176 LVESV—114 EF—35 |
Noncompaction on mid-lateral anterior and posterior segments and apex Compact—6.9 mm Noncompact—14 mm |
Large transmural patches in apical and mid-anterior lateral segments. Longitudinal striae in the mid-wall of the septum |
| II-7 | 16 | Exercise induced ventricular tachycardia | LVESD 38 mm LVEDD 54 mm EF 56% Mid-lateral trabeculations |
21 ICD at 17.5 years Died at 22 while waiting for heart transplantation |
LVESD 59 mm LVEDD 65 mm EF < 11% |
Not done | ||
| II-12 | 7 | Asymptomatic familial cardiomyopathy checkout | LVESD 24 mm LVEDD 43 mm EF—70% |
14 asymptomatic, fully active |
LVESD 32 mm LVEDD 59 mm EF—70% |
Moderate dilatation LV Normal global LV function LVEDV 91 mm LVESV 39 mm EF—65 |
Noncompaction on mid-lateral anterior and posterior segments Compact—6.4 mm Noncompact—16 mm |
No enhancement |
The annotation of the patients is according to Figure 1A.
LVESD: left ventricular end-systolic diameter; LVEDD: left ventricular end-diastolic diameter; EF: ejection fraction; ICD: implantable cardioverter-defibrillator; VPBS: ventricular prematur beats.
In terms of cardiac morphology, two-dimensional (2D) and Doppler echocardiography (TTE) in the apical four-chamber and parasternal short-axis images at the level of the ventricles showed dilatation of both ventricles with global severe depression of LV function (Table 1). In addition, 4–5 trabeculae and intertrabecular recesses in inferior and lateral walls of the LV were noted with normal origins of the coronary arteries. In these lacunar regions, the ratio of compacted versus noncompacted myocardium was 1 : 2. Cardiac magnetic resonance imaging (CMRI) revealed, in short-axis axial and four-chamber images, severe dilatation of the LV with severely depressed function. Clinically, the two older patients (II1 and II7, Table 1) were frequently suffering from VT and, on multiple occasions, underwent recurrent electrical cardioversion and intravenous administration of amiodarone. They have undergone implantable cardioverter-defibrillator (ICD) implantation and one patient had a heart transplant (Table 1).
Homozygosity mapping and identification of the mutation in PLEKHM2
To pursue a molecular diagnosis in this family, genomic DNA was isolated from blood of all numbered individuals (Fig. 1A), lymphoblastoid cells were established for all patients and fibroblast cultures were established from patients II1 and II2. Assuming homozygosity by descent of a recessive mutation as the likely cause of the disorder, we have genotyped three patients (II-1, II-2 and II-7) and identified the autozygosity regions shared by the three patients as: chr1:15528487–18566851 and chr11:116305597–128546634 (GRCh37/Hg 19, total of 15.3 Mb; Fig. 1B). Exome sequencing was performed on patient II-1. Analysis of the variations in the homozygous regions revealed 14 homozygous variants. Six of them were reported in public databases (ExAc browser, 1000 Genomes, dbSNP, Exome Variant Server) with a high frequency, and seven were present in homozygosity in our collection of Bedouin exomes and were not associated with any type of cardiac abnormality. Only one variant was novel, a deletion of two nucleotides in Chr 1:16055171_16055172delAG (GRCh37/Hg 19) in exon 12 and in the coding region of PLEKHM2 (pleckstrin homology domain containing, family M member 2) cDNA2156_2157delAG, causing the frameshift p.Lys645AlafsTer12. This novel variant segregated as expected in the large family, namely homozygous in the patients but not in the parents or in any of the eight healthy siblings (Fig. 1A and C). The variant was excluded as a population-specific variation by not being present in 102 control Bedouins of the Negev, composed of our Bedouin exome collection and additional samples that were analyzed by restriction analysis with MwoI, a site created by the variation (Fig. 1A). Using RT-qPCR analysis of several tissues, we confirmed that PLEKHM2 is transcribed in the human heart at intermediate levels in the myocardium, although transcription is most prominent in brain and testis (Supplementary Material, Fig. S1).
PLEKHM2, also known as SKIP (SifA and kinesin-interacting protein), was originally identified as a target of the Salmonella effector protein SifA and found to bind the light chain of kinesin-1 to activate the motor on the bacteria's replicative vacuole (37). In infected cells, SifA contributed to the fission of vesicles from the bacterial vacuole and the SifA/PLEKHM2 complex was required for the formation and/or the anterograde transport of kinesin-1-enriched vesicles (38). PLEKHM2 has an N-terminal RUN domain, two WD domains (39) and a carboxy-terminal PH domain. The position of the frameshift deletion is in the middle of the protein, and thus, truncation of the protein would eliminate the PH domain (Fig. 2C).
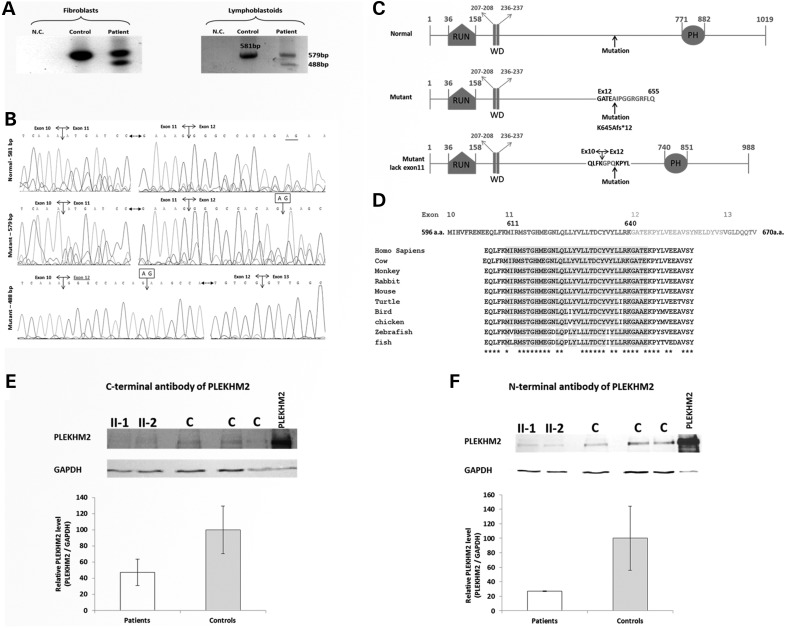
Figure 2.
Expression of PLEKHM2 gene in patients' cells. (A) RT-PCR products. RNA was extracted from lymphoblastoid cells and fibroblasts of patient II-1 and from comparable cells of control individuals. PCR was performed on the cDNA using primers in exons 10 and 15, flanking exon 12 that contains the mutation and the PCR products were separated on 2% agarose gel. The patient cells exhibited two fragments in contrast to control cells that had only one. The large PCR product of the patient (579 bp) contains the deletion of AG in exon 12 and the smaller fragment of 488 bp that appears only in patients' cells results from skipping of exon 11. (B) Sequence chromatogram of the 581 bp PCR product of the control cells (upper), the 579 bp PCR product of the patient (middle) and the 448 bp PCR product showing the skipping of exon 11 (exon 12 joining directly to exon 10 is marked by darker letters). (C) Diagram of the domains of the protein and the location of the mutation. RUN-PH-WD detail of names of domains is according to the predictions by UniProt and literature (39). The mutant diagram presents the result of the frameshift mutation that will be created by the PLEKHM2 mRNA that produces the 579 bp RT-PCR product. The 12 amino acids marked in darker are created by the frameshift translation. The diagram of the mutant lacking exon 11 presents the amino acids flanking the deletion and in darker are the amino acids that are produced by the frameshift that is created by splicing exon 10–12 until the deletion of the AG nucleotide rescues the frameshift. (D) Sequence conservation of exon 11 and the first amino acids of exon 12 that are missing in the patients. The amino acids that are deleted by the mutation are shaded by gray. (E) PLEKHM2 protein is expressed in fibroblasts cells of patients II-1 and II-2 as demonstrated by western blot using an antibody to the C-terminal region. Control cells (C) are from fibroblasts from two control individuals as well as commercial fibroblasts. PLEKHM2 protein sample produced by expression of plasmid Pcmv-Myc-PLEKHM2 was loaded in parallel for use as size migration control (PLEKHM2). (F) Same as (E) with an antibody to the N-terminal region.
The frameshift mutation in exon 12, which is an inner exon of the gene, may lead to either nonsense-mediated decay of the mRNA (40) or to rescue of translation by exon skipping. Thus, we verified the effect of the gene variant on mRNA level. RNA was extracted from lymphoblastoid cells and fibroblasts of patient II-1 and from comparable cells of control individuals, and cDNA was prepared. PCR was performed on the cDNA using primers in exons 10 and 15, flanking exon 12 that contains the mutation. The RT-PCR products of the patient-derived cells demonstrate two fragments in contrast to the one expected fragment of normal splicing shown in control cells (Fig. 2A). Sanger sequencing of the PCR products demonstrated that the large fragment of the patient (579 bp) contains exon 12 with the deleted AG and the smaller fragment of 488 bp that appears only in patient cells reflecting skipping of exon 11 (Fig. 2B). The expected proteins are presented in Figure 2C. Conventional splicing leading to the large fragment would produce the truncated protein p.Lys645AlafsTer12, whereas the mutation-induced splicing with the skipping of exon 11 would result in a smaller fragment rescuing the reading frame of translation. However, the later protein product is missing 30 highly conserved amino acids encoded by exon 11 and 4 amino acids of exon 12 that are replaced by three novel amino acids. Figure 2D represents the evolutionary conservation and Supplementary Material, Figure S2 represents the 3D structure of PLEKHM2 using the modeller v9.14 program showing the location of the deleted amino acids. To further ascertain that the PLEKHM2 protein missing exon 11 is expressed in patients II-1 and II-2, we performed western blot on protein lysates of fibroblasts of these patients in comparison with control fibroblasts of two control individuals as well as commercially available fibroblasts (ScienCell-HDF-a cat. no. 2320). Using antibodies directed against the C-terminal region (Fig. 2E) and the N-terminal region (Fig. 2F) of the protein, PLEKHM2 was detected in patients' cells. The 30 amino acids difference (encoded by exon 11) could not be distinguished from the 1019 amino acids protein while separated on 12% polyacrylamide gel. The reduction in the total protein quantity is, however, in agreement with the RT-PCR result, showing that at least half the transcripts would produce the truncated protein (Fig. 2E and F).
Effect of the mutation on patients' cells
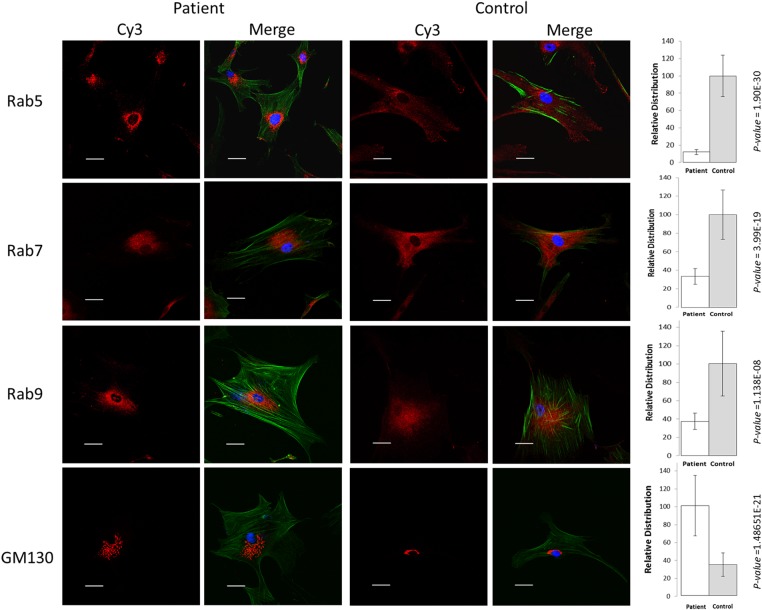
Various lines of evidence may support the notion that PLEKHM2 is vital for endocytotic trafficking. A specific and direct interaction between the late endosomal GTPase Rab9 and the PH domain of PLEKHM2 was demonstrated in a GTP-dependent manner. This interaction was required for the peripheral distribution of lysosomes (41). Rab7 also seems to interact with PLEKHM2, being pulled down by SifA in cellular lysates, though no direct interaction with the PH domain was demonstrated (41,42). In addition, PLEKHM2 was demonstrated to directly interact with Rab1a by its RUN domain (43). In regard to endocytotic trafficking, the Rab family members of proteins have been implicated as critical regulators of vesicle formation, intracellular transport (including binding to motor proteins or motor adaptors), vesicle tethering and vesicle fusion (44). They act through the GTP-dependent recruitment of protein ligands at the appropriate time and place. In addition to its direct interaction with the Rab proteins, PLEKHM2 was suggested to be involved in the movement of endosomes through its interaction with kinesin since its overexpression induced a microtubule- and kinesin-1-dependent anterograde movement of late endosomal/lysosomal compartments, whereas the opposite was found for siRNA-induced knockdown of PLEKHM2 levels (37,38). Thus, we evaluated whether the mutation affects the subcellular distribution of the Rabs reported to interact with PLEKHM2. Compared distribution of Rabs in fibroblasts of patients relative to control fibroblasts of comparable passages (15) by immunofluorescent labeling demonstrated that this is indeed the case. The qualitative and quantitative results presenting the subcellular distribution of Rab5, Rab7 and Rab9 for patient II-1 relative to commercial fibroblast are presented in Figure 3. The distribution of the endosomes labeled with different Rab antibodies presents a perinuclear localization in patients' fibroblasts relative to controls. Similar results were observed in cells from patient II-2 in comparison with a healthy control (Supplementary Material, Fig. S3). In contrast, we could not identify differences in the subcellular distribution of Rab1, Rab4a and Rab11 (not shown). The Golgi apparatus, detected using an antibody against GM130, exhibited a significant dispersion in patients when compared with controls (Fig. 3, for patient II-1 and Supplementary Material, Fig. S3 for patient II-2). This is in agreement with the findings in the literature using siRNA-induced knockdown of PLEKHM2 levels (37).
Figure 3.
Subcellular distribution of organelles stained with Rabs and Golgi. Representative photographs of the distribution of the endosomes in primary fibroblasts of patient II-1 relative to control fibroblasts labeled with Rab 5, Rab7 and Rab9. Note perinuclear localization of the various rab in the patient' cells relative to control cells and fragmentation and spreading of the Golgi apparatus in patient' cells relative to control cells (comparable results were obtained for patient II-2, Supplementary Material, Fig. S3). Blue is DAPI staining for the nuclei, and green is AlexaFluor 488 Phalloidin used to mark the cells' borders. Dimension bar is 40 μm long. Relative distribution around nuclei, presented at the right side of the respective images, was calculated for a set intensity around the nucleus, and the area giving this intensity was calculated for each cell. The intensity was measured using ImageJ software (NIH) with a constant threshold. At least 20 cells of each patient and control with the specific staining were measured. Data are expressed as mean ± SD, and P-values are indicated at the right of the bars (Student's t-test).
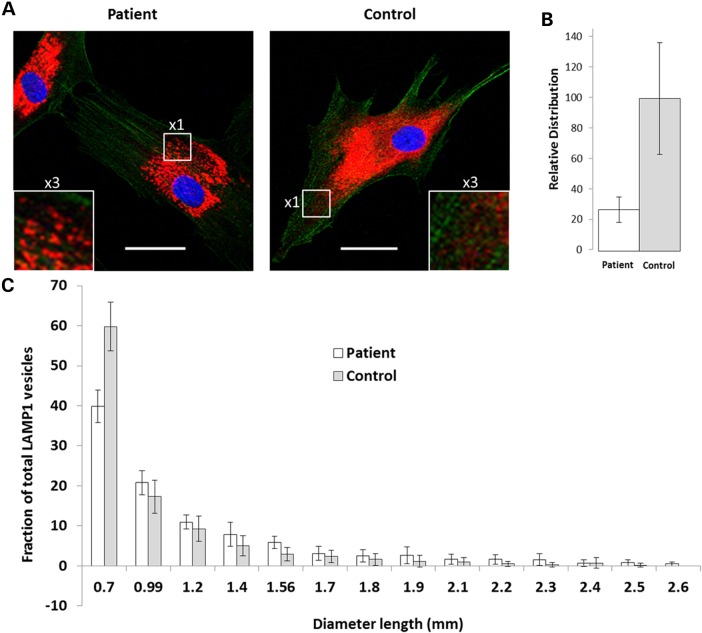
The interaction of PLEKHM2 with lysosomes is mediated through the binding of the constitutive lysosomal membrane protein Arl8. Both PLEKHM2 and Arl8 are required for lysosomes to distribute away from the microtubule-organizing center (39). Thus, we further evaluated the effect of the mutation on the distribution of lysosomes using LAMP1 antibodies and found that lysosomes of patient cells are more concentrated near the nucleus compared with controls (Fig. 4A and Supplementary Material, Fig. S4). This finding was supported by quantitative analysis as shown in Figure 4B and Supplementary Material, Figure S4, and is in agreement with the findings in the literature using siRNA-induced knockdown of PLEKHM2 levels (37,38). In addition, the size of the lysosomes in patient cells was larger (Fig. 4A and C), suggesting a possible disruption of function.
Figure 4.
Subcellular and size distribution of lysosomes. Lysosomes were visualized using antibodies to LAMP1. Acquirement of the images is as detailed in Figure 3. (A) A representative photograph of patient's cells shows a perinuclear localization of lysosomes, and the relative distribution around the nuclei is presented at the right. Dimension bar shows 40 μm. (B) Relative distribution around nuclei was analyzed as in Figure 3. (C) Analysis of lysosomal size indicates a higher proportion of large lysosomes in patient compared with control fibroblasts. Analysis was done using the ‘analyze particles’ plugin of ImageJ with size settings from 0–infinity and circularity 0–1.
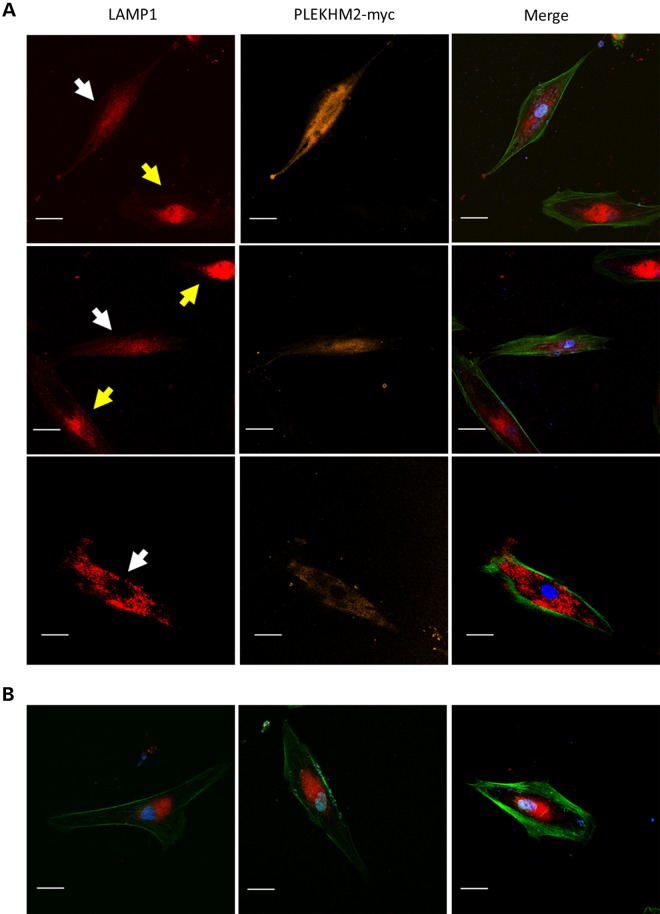
To further evaluate the cause and effect relationship between the PLEKHM2 mutation and lysosomal distribution, we tested whether exogenous expression of wild-type PLEKHM2 in the patient fibroblasts may correct the abnormality of lysosome distribution. For that purpose, the fibroblasts of patient II-1 were transfected with the PLEKHM2-myc plasmid and lysosomes were visualized by LAMP1-specific antibodies. Indeed, the results indicated that transfected cells which expressed the normal PLEKHM2 converted their distribution of LAMP1 toward the dispersed distribution normally seen in healthy fibroblasts. In contrast, cells which were not transfected demonstrated abnormal distribution of LAMP1 (Fig. 5). Cells transfected with a control vector expressing GFP only also demonstrated abnormal distribution of LAMP1 (not shown).
Figure 5.
Correction of the distribution of lysosomes in patient fibroblasts by overexpression of normal PLEKHM2 protein. (A) The fibroblasts of patient II-1 were transfected with PLEKHM2-myc plasmid and lysosomes visualized by LAMP1-specific antibodies as detailed in Figure 4. Myc was visualized by monoclonal antibody and secondary antibody was Cy5. The white arrows point cells transfected with the plasmid and the yellow arrow points untransfected cells. (B) Patient's cells in the same experiment that were not transfected. Dimension bar is 40 μm long.
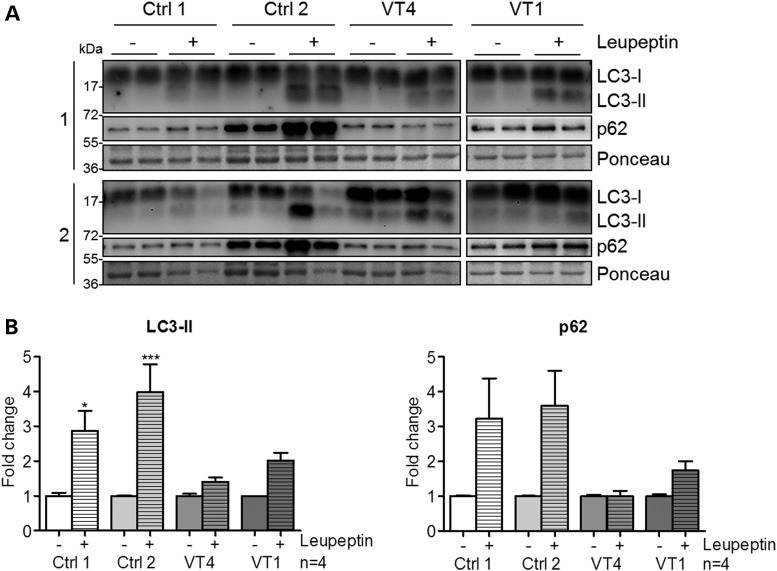
Autophagy is highly dependent on Rab proteins at various stages. Rab1, Rab5, Rab7, Rab9A, Rab11, Rab23, Rab32 and Rab33B participate in autophagosome formation (45), whereas Rab9 is required in noncanonical autophagy. Rab7, in addition to Rab8B and Rab24, has a key role in autophagosome maturation (45). Additionally, autophagy depends on lysosomal function (46,47). Thus, we evaluated the autophagic flux in fibroblasts with mutated PLEKHM2 derived from two of the patients in comparison with those from two controls. The accumulation of two proteins widely used for monitoring autophagy flux LC3-II and p62 (48) upon treatment of the cells with leupeptin was measured by Western blotting using specific antibodies. The result showed a marked decrease in the ability of fibroblasts from the patients to accumulate these proteins (Fig. 6), suggesting marked impairment in autophagic flux.
Figure 6.
Autophagic flux was impaired in patient fibroblasts. (A) Representative immunoblot performed on crude proteins from fibroblasts of two patients [VT1 (II-1) and VT4 (II-2)] and two controls. Cells were treated or not with 100 µM leupeptin for 24 h. Immunoblots were stained with antibodies against LC3 or p62. (B) Quantification of LC3-II and p62 normalized to Ponceau as a loading control. Treated samples were related to untreated controls for each cell line to compare the autophagic flux independent of varying basal expression levels among cell lines. Bars represent mean ± SEM with *P < 0.05 and ***P < 0.001 versus untreated control, one-way ANOVA with Bonferroni's post-test (n = 4 per group).
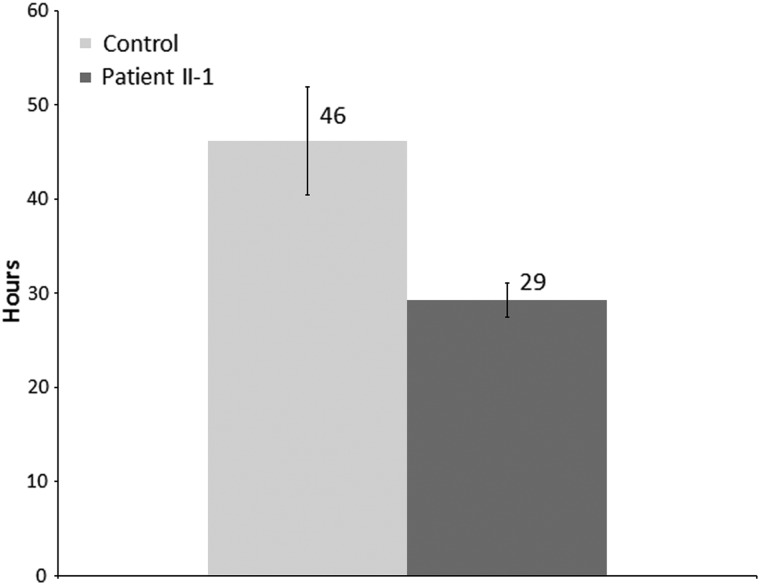
Inhibition of autophagy was reported by several studies to enhance migration of both normal (49–51) and tumor cells (50–53). Thus, we aimed to determine whether the fibroblasts with mutated PLEKHM2 demonstrate changes in cell migration. To this end, fibroblasts of identical passages (16–18) from patient II-1 and control fibroblasts were plated with culture inserts of 500 μm. When the cell density reached ∼80%, the inserts were removed, thus creating a ∼500 μm-wide gap within the culture. Fibroblast movement toward closing the gap was traced by time-lapse live cell microscopy. Indeed, the patient's fibroblasts bridged the gap within 29 ± 2 h in comparison with 46 ± 6 h for control fibroblasts (Fig. 7, P = 0.002), indicating faster movement of the fibroblasts with mutated PLEKHM2 compared with controls.
Figure 7.
PLEKHM2 patient's fibroblasts migrate faster than control fibroblasts. Fibroblasts of patient II-1 and control fibroblasts were plated with 500 μm culture inserts. When the density reached ∼80%, the inserts were removed and fibroblasts movement was monitored using time-lapse live cell microscopy for 65–70 h. The DIC images were acquired every 15 min. The bridging time was defined as the time passed from the insert removal till formation of a continuous string of cells across the gap. The results represent four independent experiments each in quadruplicates. Data are expressed as mean ± SEM, P = 0.002 (Student's t-test).
Discussion
Our results demonstrate that a mutation in PLEKHM2 causes recessive DCM with LVNC that is limited to a few segments in our patients, and indicates that a likely mechanistic explanation for this phenotype involves perturbation of lysosomal function and autophagy. This is the first report implicating a mutation in PLEKHM2 in any human disease or mouse phenotype. Our data indicate the importance of PLEKHM2 for normal cardiac function. Moreover, the link between PLEKHM2 and DCM contributes to the notion that tightly controlled regulation of autophagy is important for maintaining cardiac homeostasis, and that dysregulation of this pathway may have deleterious effects (54). Autophagy is a dynamic process that ensures cellular homeostasis by eliminating damaged organelles, long-lived proteins and toxic protein aggregates, as well as recycling nutrients during starvation and stress (47). Tightly controlled regulation of autophagy is important for maintaining cardiac homeostasis, and dysregulation of this pathway resulting in either excessive or insufficient autophagy is suggested to have deleterious effects (54). Altered autophagy has been observed in many cardiovascular diseases in response to pathological stimuli, including cardiac hypertrophy and HF (55). Animal models suggest that impaired autophagy may lead to cardiac dysfunction over time (56). PLEKHM2 joins two other genes (LAMP2 and BAG3) with a role in autophagy as causes of cardiomyopathy. LAMP2 is an important constituent of the lysosomal membrane; mutations in LAMP2 were demonstrated to cause Danon disease, a condition of severe and progressive myopathy and cardiomyopathy with multisystem glycogen-storage disease (OMIM 300257) associated with accumulation of autophagic material in striated myocytes (57), hypertrophic cardiomyopathy with skeletal muscle weakness (58) and prominent cardiac hypertrophy and electrophysiological abnormalities (52). Mutations in BAG3 can cause childhood onset of rapidly progressive myofibrillar myopathy (MFN) (59,60) or autosomal-dominant DCM (61–63). However, human BAG3 mutations have not been characterized sufficiently, and two DCM-associated mutations rather fall into the category of stress-induced apoptosis (61). After gene transfer of BAG3 mutations in neonatal rat cardiomyocytes or H9c9 cells, the assembly and integrity of Z-discs and nuclear localization of BAG3 were disturbed and cells exhibited a higher susceptibility to stress-induced apoptosis (61). On the contrary, myotube formation was disturbed after gene transfer of MFN-associated, but not DCM-associated BAG3, mutations in C2C12 myoblasts (61).
LVNC has been considered to be a developmental failure of the heart to fully form the compact myocardium during the later stages of cardiac development resulting in persistence of multiple prominent ventricular trabeculations and deep intertrabecular recesses (64). Several genetically engineered mouse models that have defects in cardiac trabeculation and compaction indicate that cellular growth and differentiation signaling pathways are keys in these ventricular morphogenetic events and point to several important genes participating in the NOTCH pathway (65). One study reported that inactivating mutations in the NOTCH pathway regulator MIB1 cause LVNC in humans (66). However, the etiology and pathogenesis causing LVNC are not known (64). Although LVNC is classified as a primary genetic cardiomyopathy by the American Heart Association (1), it is still considered unclassified in the European Society of Cardiology classification, as it remains unclear whether it represents a distinct disease process or a morphological trait shared by many phenotypically different cardiomyopathies (2). Mutations in 24 nuclear genes, in addition to mitochondrial disorders and chromosome abnormalities, present with LVNC together with multiple cardiac disorders. The extensive genetic heterogeneity of LVNC suggests that the uniform morphology of LVNC is attributable to embryonic noncompaction, as well as from induction of hypertrabeculation as a compensatory reaction of an impaired myocardium (67). This suggestion is supported by the observations that in some patients, LVNC develops later in life, can be acquired by intense athletic training or as an adaptive mechanism of an impaired myocardium, such as ischemic heart disease or trauma, and may even disappear (24,67,68). Cardiac fibroblasts appear to be of critical importance in heart repair and cell migration is essential in wound healing (69). In this regard, inhibition of autophagy was reported by several studies to enhance migration of both normal (49–51) and tumor cells (50–53). This mechanism is suggested as ‘signalphagy’, where active signaling proteins are not degraded by autophagy, for example RHOA-GTP (70) or the oncogenic transcription factor, Twist1 (49). Since derangements in cell movement could contribute to the noncompaction observed in patients, we tested the migration of fibroblasts of a patient compared with fibroblasts of a control. Our data clearly indicate enhanced movement of the patient's fibroblast relative to controls. Thus, although noncompaction was observed at age 7 in our youngest patient (II-12, Table 1) and thus could be caused by a defect in development, it is plausible that this abnormally facilitated migration may contribute to the noncompaction morphology.
The current study associates a mutation disrupting lysosomes and autophagy with pathology selectively affecting the human heart. Thus, these results may pave the way for better understanding of the apparently critical link between autophagy and normal cardiac function.
Materials and Methods
Patients
The study was approved by the Soroka Medical Center institutional review board, and all participants gave written informed consent prior to participation. Genomic DNA was extracted from white blood cells by using standard procedures. Medical records were carefully reviewed, and details of somatic growth, psychomotor development, clinical course, hospitalizations and laboratory results were obtained. Parents and siblings were interviewed and underwent a complete physical examination, which particularly focused on the cardiac and neuromuscular findings. Cardiac evaluation included standard transthoracic 2D and Doppler echocardiography (TTE) using a Vivid 7 system (GE Medical Systems, Saskatchewan, Canada) and CMRI using a 1.5 T scanner (Achieva, Philips Medical Systems, Best, The Netherlands), which included phase contrast images. TTE measurements of LV end-diastolic dimension (LVED) and LV end-systolic dimension (LVES) were obtained in accordance with the recommendations of the American Society of Echocardiography (71). Dimensions were corrected for age and bovine serum albumin (BSA) according to the formula of Henry et al. (72): LVED (percent predicted) = (measured LVED/predicted LVED) × 100; predicted LVED = (45.3 × BSA0.3) − (0.03 × Age) − 7.2. LV abnormalities were classified as follows: DCM, LVED ≥117% predicted and fractional shortening <25% in the absence of known causes of ventricular dilatation (72,73). LV noncompaction evaluation was performed according to the echocardiographic criteria described by Jenni et al. (17,74).
Genomic DNA was isolated from blood by standard procedures. Lymphoblastoid cells and fibroblast cultures were established by standard procedures.
Genotyping and exome sequencing
Genotyping was performed using Affymetrix (CA, USA) Gene-Chip Human SNP5 array. We determined the genotype calls by using Affymetrix GeneChip Genotyping Console Analysis Software (GTYPE) and identified the autozygosity using AgileMultiIdeogram (http://dna.leeds.ac.uk/agile/AgileMultiIdeogram/). Whole exome sequencing was performed using NimbleGen v2 exome capture followed by 50 bp paired-end sequencing using an Illumina HiSeq Sequencing System. The resulting sequence data were analyzed as described (75).
Verification of a mutation
PCR amplification of exon 12 was performed using primers forward: TTGTAGACGAGGCTGACTCTCA and reverse: CAGAAACCACACCGTGACAT (annealing temperature 61°C). Direct sequencing the PCR products was performed on an ABI PRISM 3100 DNA Analyzer with the BigDye Terminator v.1.1 Cycle Sequencing Kit (Applied Biosystems, CA, USA) according to the manufacturer's protocol. MwoI (NEB) digestion was done according to the manufacturer's instruction. Restriction digest of the 687 bp amplicon resulted in 25, 73, 75, 83, 190 and 241 bp fragments; if the mutation is present, the 239 bp fragment is cleaved into 135 and 104 bp fragments. Digested products were separated by electrophoresis on 2% agarose gel.
RT-qPCR analysis
About 500 ng of total RNA from different human tissues obtained from Clontech (Takara Bio Europe/Clontech) was primed with random hexamers and reverse-transcribed with the SuperScript Reverse Transcription Kit from Invitrogen, following the manufacturer's instructions. cDNAs were PCR-amplified in the ABI 7500 Software V2.0.3 (Applied Biosystems) with the SYBR Green (Applied Biosystems). The ubiquitously expressed GAPDH gene was used as an internal control to normalize the data. The results represent one experiment done in duplicates, means ± sem (not visible). The sequences of the primers used in the experiment are the following: PLEKHM2 (Ex11–Ex12 + 13): forward: 5′ TGCTGCTCACAGACTGCTAT 3′; reverse: 5′ AAGGCCAACCGACACATA 3′. GAPDH forward: 5′ AGAAGGCTGGGGCTCATTTG 3′; reverse: 5′ GGGGCCATCCACAGTCTTC 3′.The PLEKHM2 primers yielded a linear standard curve with an R2 = 0.99.
Analysis of splicing
RNA was extracted from lymphoblastoid cells and fibroblasts of patient II-1 and from comparable cells of control individuals using the EZ-RNA II Kit (Biological Industries, Israel). cDNA was prepared using the SuperScript Kit (Invitrogen), PCR was performed on the cDNA using primers in exons 10 and 15, forward: TCGGAGTTCAGAGTAGACAACAA and reverse: ATGCCTTCTTTGGTGATGGT flanking exon 12 that contains the mutation and the PCR products were separated on 2% agarose gel.
Protein structure
The predictions of the domains of PLEKHM2 were retrieved from UniProt (http://www.uniprot.org/uniprot/). The 3D structure modeling was created by using the modeller v9.14 program (http://toolkit.tuebingen.mpg.de/modeller) as described in the manual of the manufacturer. The amino acid sequence of human PLEKHM2 was retrieved from the NCBI protein database (NP_055979.2). A sequence similarity search was performed using the NCBI BLASTP server (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&BLAST_SPEC=&LINK_LOC=blasttab&LAST_PAGE=blastn). Based on the sequence identity, 3CXB and 3HW2 were chosen as the most suitable templates for homology modeling. Color modifications on the protein was changed using RasMol v2.7.5.2 (http://rasmol.org/).
Western blot analyses
Cells were lysed in RIPA lysis buffer (20 mm Tris pH 7.5, 150 mm NaCl, 1% Triton, 1 mm EDTA, 1 mm EGTA, 2.5 mm sodium pyrophosphate and 1 mm Na3VO4) with 1 : 100 PMSF and a protease inhibitor (Sigma cat. No. S8830). Prior to separation, Laemmli sample buffer (62.5 mm Tris–HCl, pH 6.8; 25% glycerol; 2% SDS; 0.01% bromophenol blue and β-mercaptoethanol for a final concentration of 5% was added) was added in a 1 : 1 ratio. Subsequently, the samples were heated for 5 min at 95°C. About 35 µg of protein tissue extract was loaded on 12% SDS–PAGE. Blotting was done into the nitrocellulose membrane, and the blots were blocked with 5% nonfat dry milk in TBST (Tris-buffered saline, 0.05% Tween) and incubated with the primary antibody (at 4°C, for overnight). For PLEKHM2 C-terminal region (Thermo PA5-20850) and for the N-terminal region (Santa Cruz sc-136808) and respective HRP-conjugated secondary antibody anti-rabbit IgG (Jackson, USA). Signal intensities of PLEKHM2 bands were normalized against the internal control GAPDH (Millipore MAB374). Commercial fibroblasts were purchased from ScienCell-HDF-a (cat. no. 2320).
Immunofluorescence
Fibroblast cells were grown in an eight-well slide (ibidi 80826) in 10% DMEM. Cells were fixed with 4% formaldehyde solution, incubated with 0.1% Triton X-100 for 5 min and blocked with 1% (BSA; Sigma-Aldrich) in PBS for 30 min. Samples were incubated at room temperature for 1 h with the primary antibody. All primary antibodies except Rab4a (Santa Cruz biotechnology Ab sc-312) were from Cell Signaling technology: Rab1 Ab #13075, Rab5 #3547, Rab7 #9367, Rab9 #5118, Rab11 #5589, LAMP1 #9091 and GM130 #12480 secondary antibody Cy3 (Thermo SA5-10169). Blue is DAPI (Vectorlabs H-1200) staining for the nuclei, and green is AlexaFluor 488 Phalloidin (Molecular probes A12379) to mark the cells' borders. Images were acquired with the confocal FluoView 1000 fluorescence microscope (OLYMPUS), with a 40× objective and with the manufacturer's software FV1000.
Overexpression of normal PLEKHM2 protein
Fibroblast cells of patient II-1 were transfected using Turbofect (Thermo #R0531) with PLEKHM2-myc plasmid (pcDNA3.1, gift of Dr Stéphane Méresse). Myc was visualized by primary anti-myc antibody (monoclonal, gift of Dr Noah Isakov) and secondary cy5 anti-mouse antibody (Thermo SA5-10169), and lysosomes visualized by LAMP1 antibody and cy3 anti-rabbit antibody.
Autophagic flux
Patients' fibroblasts were plated at a density of 30 000 cells per 24-well plate well. The next day, cells were treated or not with 100 µM leupeptin (Sigma, L-8511) and harvested 24 h after treatment in lysis buffer (3% SDS, 30 mm Tris-base, pH 8.8, 5 mm EDTA, 30 mm NaF, 10% glycerol and 1 mm DTT). About 10 µg of protein lysate was used for immunoblots, which were stained with antibodies against LC3 (Novus Biologicals, NB100-2331) or p62 (Sigma, P0067).
Cell migration assay
Fibroblasts of identical passages (16–18) of patient II-1 and control fibroblasts were plated in eight-wells μ-slides (ibidi GmBh Germany, Cat #80826) with culture inserts of 500 μm (ibidi GmBh Germany, cat. #80209). When the density reached ∼80%, the inserts were removed and fibroblast movement was monitored using time-lapse live cell microscopy for 65–70 h. The differential interference contrast (DIC) images were acquired every 15 min. The bridging time was defined as the time passed from the insert removal until formation of a continuous string of cells across the gap.
Supplementary Material
Funding
This work was partly supported by an internal grant from Ben- Gurion University of the Negev, Faculty of Health Sciences. This work was partly supported by the Leducq Foundation (research grant no. 11, CVD O4 to L.C.). V.C.S. was supported by the Howard Hughes Medical Institute.
Supplementary Material
Acknowledgements
We are grateful to the affected persons and their families, whose cooperation made this study possible. We are very grateful to Dr Stéphane Méresse from Centre d'Immunologie de Marseille-Luminy for his gift of the PLEKHM2-myc plasmid.
Conflict of Interest statement. None declared.
References
- 1.Yancy C.W., Jessup M., Bozkurt B., Butler J., Casey D.E. Jr, Drazner M.H., Fonarow G.C., Geraci S.A., Horwich T., Januzzi J.L. et al. (2013) 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology Foundation/American Heart Association Task Force on practice guidelines. Circulation, 128, e240–e327. [DOI] [PubMed] [Google Scholar]
- 2.McMurray J.J., Adamopoulos S., Anker S.D., Auricchio A., Bohm M., Dickstein K., Falk V., Filippatos G., Fonseca C., Gomez-Sanchez M.A. et al. (2012) ESC guidelines for the diagnosis and treatment of acute and chronic heart failure 2012: the task force for the diagnosis and treatment of Acute and Chronic Heart Failure 2012 of the European Society of Cardiology. Developed in collaboration with the Heart Failure Association (HFA) of the ESC. Eur. J. Heart Fail., 14, 803–869. [DOI] [PubMed] [Google Scholar]
- 3.Piran S., Liu P., Morales A., Hershberger R.E. (2012) Where genome meets phenome: rationale for integrating genetic and protein biomarkers in the diagnosis and management of dilated cardiomyopathy and heart failure. J. Am. Coll. Cardiol., 60, 283–289. [DOI] [PubMed] [Google Scholar]
- 4.Gregori D., Rocco C., Miocic S., Mestroni L. (2001) Estimating the frequency of familial dilated cardiomyopathy in the presence of misclassification errors. J. Appl. Stat., 28, 53–62. [Google Scholar]
- 5.Petretta M., Pirozzi F., Sasso L., Paglia A., Bonaduce D. (2011) Review and metaanalysis of the frequency of familial dilated cardiomyopathy. Am. J. Cardiol., 108, 1171–1176. [DOI] [PubMed] [Google Scholar]
- 6.Karkkainen S., Peuhkurinen K. (2007) Genetics of dilated cardiomyopathy. Ann. Med., 39, 91–107. [DOI] [PubMed] [Google Scholar]
- 7.McNally E.M., Golbus J.R., Puckelwartz M.J. (2013) Genetic mutations and mechanisms in dilated cardiomyopathy. J. Clin. Invest., 123, 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grunig E., Tasman J.A., Kucherer H., Franz W., Kubler W., Katus H.A. (1998) Frequency and phenotypes of familial dilated cardiomyopathy. J. Am. Coll. Cardiol., 31, 186–194. [DOI] [PubMed] [Google Scholar]
- 9.Mestroni L., Rocco C., Gregori D., Sinagra G., Di Lenarda A., Miocic S., Vatta M., Pinamonti B., Muntoni F., Caforio A.L. et al. (1999) Familial dilated cardiomyopathy: evidence for genetic and phenotypic heterogeneity. Heart Muscle Disease Study Group. J. Am. Coll. Cardiol., 34, 181–190. [DOI] [PubMed] [Google Scholar]
- 10.OMIM. Online Mendelian Inheritance in Man. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University, Baltimore, MD: and National Center for Biotechnology Information, National Library of Medicine, Bethesda, MD; http://www.ncbi.nlm.nih.gov/OMIM, in press. [Google Scholar]
- 11.He Q. (2010) Tafazzin knockdown causes hypertrophy of neonatal ventricular myocytes. Am. J. Physiol.. Heart Circ. Physiol., 299, H210–H216. [DOI] [PubMed] [Google Scholar]
- 12.Levitas A., Muhammad E., Harel G., Saada A., Caspi V.C., Manor E., Beck J.C., Sheffield V., Parvari R. (2010) Familial neonatal isolated cardiomyopathy caused by a mutation in the flavoprotein subunit of succinate dehydrogenase. Eur. J. Hum. Genet., 18, 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parvari R., Levitas A. (2012) The mutations associated with dilated cardiomyopathy. Biochem. Res. Int., 2012, 639250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedrich F.W., Carrier L. (2012) Genetics of hypertrophic and dilated cardiomyopathy. Curr. Pharmaceut. Biotechnol., 13, 2467–2476. [DOI] [PubMed] [Google Scholar]
- 15.Moric-Janiszewska E., Markiewicz-Loskot G. (2008) Genetic heterogeneity of left-ventricular noncompaction cardiomyopathy. Clin. Cardiol., 31, 201–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W., Chen H., Qu X., Chang C.P., Shou W. (2013) Molecular mechanism of ventricular trabeculation/compaction and the pathogenesis of the left ventricular noncompaction cardiomyopathy (LVNC). Am. J. Med. Genet. C Semin. Med. Genet., 163c, 144–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenni R., Oechslin E.N., van der Loo B. (2007) Isolated ventricular non-compaction of the myocardium in adults. Heart, 93, 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paterick T.E., Tajik A.J. (2012) Left ventricular noncompaction: a diagnostically challenging cardiomyopathy. Circ. J., 76, 1556–1562. [DOI] [PubMed] [Google Scholar]
- 19.Arbustini E., Weidemann F., Hall J.L. (2014) Left ventricular noncompaction: a distinct cardiomyopathy or a trait shared by different cardiac diseases? J. Am. Coll. Cardiol., 64, 1840–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maron B.J., Towbin J.A., Thiene G., Antzelevitch C., Corrado D., Arnett D., Moss A.J., Seidman C.E., Young J.B. (2006) Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation, 113, 1807–1816. [DOI] [PubMed] [Google Scholar]
- 21.Fazio G., Corrado G., Pizzuto C., Zachara E., Rapezzi C., Sulafa A.K., Sutera L., Stollberger C., Sormani L., Finsterer J. et al. (2008) Supraventricular arrhythmias in noncompaction of left ventricle: is this a frequent complication? Int. J. Cardiol., 127, 255–256. [DOI] [PubMed] [Google Scholar]
- 22.Stollberger C., Winkler-Dworak M., Blazek G., Finsterer J. (2007) Prognosis of left ventricular hypertrabeculation/noncompaction is dependent on cardiac and neuromuscular comorbidity. Int. J. Cardiol., 121, 189–193. [DOI] [PubMed] [Google Scholar]
- 23.Burke A., Mont E., Kutys R., Virmani R. (2005) Left ventricular noncompaction: a pathological study of 14 cases. Hum. Pathol., 36, 403–411. [DOI] [PubMed] [Google Scholar]
- 24.Finsterer J., Stollberger C., Feichtinger H. (2002) Histological appearance of left ventricular hypertrabeculation/noncompaction. Cardiology, 98, 162–164. [DOI] [PubMed] [Google Scholar]
- 25.Chin T.K., Perloff J.K., Williams R.G., Jue K., Mohrmann R. (1990) Isolated noncompaction of left ventricular myocardium. A study of eight cases. Circulation, 82, 507–513. [DOI] [PubMed] [Google Scholar]
- 26.Ritter M., Oechslin E., Sutsch G., Attenhofer C., Schneider J., Jenni R. (1997) Isolated noncompaction of the myocardium in adults. Mayo Clin. Proceed., 72, 26–31. [DOI] [PubMed] [Google Scholar]
- 27.Oechslin E.N., Attenhofer Jost C.H., Rojas J.R., Kaufmann P.A., Jenni R. (2000) Long-term follow-up of 34 adults with isolated left ventricular noncompaction: a distinct cardiomyopathy with poor prognosis. J. Am. Coll. Cardiol., 36, 493–500. [DOI] [PubMed] [Google Scholar]
- 28.Ichida F., Hamamichi Y., Miyawaki T., Ono Y., Kamiya T., Akagi T., Hamada H., Hirose O., Isobe T., Yamada K. et al. (1999) Clinical features of isolated noncompaction of the ventricular myocardium: long-term clinical course, hemodynamic properties, and genetic background. J. Am. Coll. Cardiol., 34, 233–240. [DOI] [PubMed] [Google Scholar]
- 29.Sasse-Klaassen S., Gerull B., Oechslin E., Jenni R., Thierfelder L. (2003) Isolated noncompaction of the left ventricular myocardium in the adult is an autosomal dominant disorder in the majority of patients. Am. J. Med. Genet. A, 119A, 162–167. [DOI] [PubMed] [Google Scholar]
- 30.Ichida F., Tsubata S., Bowles K.R., Haneda N., Uese K., Miyawaki T., Dreyer W.J., Messina J., Li H., Bowles N.E. et al. (2001) Novel gene mutations in patients with left ventricular noncompaction or Barth syndrome. Circulation, 103, 1256–1263. [DOI] [PubMed] [Google Scholar]
- 31.Vatta M., Mohapatra B., Jimenez S., Sanchez X., Faulkner G., Perles Z., Sinagra G., Lin J.H., Vu T.M., Zhou Q. et al. (2003) Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J. Am. Coll. Cardiol., 42, 2014–2027. [DOI] [PubMed] [Google Scholar]
- 32.Kenton A.B., Sanchez X., Coveler K.J., Makar K.A., Jimenez S., Ichida F., Murphy R.T., Elliott P.M., McKenna W., Bowles N.E. et al. (2004) Isolated left ventricular noncompaction is rarely caused by mutations in G4.5, alpha-dystrobrevin and FK binding protein-12. Mol. Genet. Metab., 82, 162–166. [DOI] [PubMed] [Google Scholar]
- 33.Chen R., Tsuji T., Ichida F., Bowles K.R., Yu X., Watanabe S., Hirono K., Tsubata S., Hamamichi Y., Ohta J. et al. (2002) Mutation analysis of the G4.5 gene in patients with isolated left ventricular noncompaction. Mol. Genet. Metab., 77, 319–325. [DOI] [PubMed] [Google Scholar]
- 34.Xing Y., Ichida F., Matsuoka T., Isobe T., Ikemoto Y., Higaki T., Tsuji T., Haneda N., Kuwabara A., Chen R. et al. (2006) Genetic analysis in patients with left ventricular noncompaction and evidence for genetic heterogeneity. Mol. Genet. Metab., 88, 71–77. [DOI] [PubMed] [Google Scholar]
- 35.Hermida-Prieto M., Monserrat L., Castro-Beiras A., Laredo R., Soler R., Peteiro J., Rodriguez E., Bouzas B., Alvarez N., Muniz J. et al. (2004) Familial dilated cardiomyopathy and isolated left ventricular noncompaction associated with lamin A/C gene mutations. Am. J. Cardiol., 94, 50–54. [DOI] [PubMed] [Google Scholar]
- 36.Klaassen S., Probst S., Oechslin E., Gerull B., Krings G., Schuler P., Greutmann M., Hurlimann D., Yegitbasi M., Pons L. et al. (2008) Mutations in sarcomere protein genes in left ventricular noncompaction. Circulation, 117, 2893–2901. [DOI] [PubMed] [Google Scholar]
- 37.Boucrot E., Henry T., Borg J.P., Gorvel J.P., Meresse S. (2005) The intracellular fate of Salmonella depends on the recruitment of kinesin. Science, 308, 1174–1178. [DOI] [PubMed] [Google Scholar]
- 38.Dumont A., Boucrot E., Drevensek S., Daire V., Gorvel J.P., Pous C., Holden D.W., Meresse S. (2010) SKIP, the host target of the Salmonella virulence factor SifA, promotes kinesin-1-dependent vacuolar membrane exchanges. Traffic, 11, 899–911. [DOI] [PubMed] [Google Scholar]
- 39.Rosa-Ferreira C., Munro S. (2011) Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev. Cell, 21, 1171–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maquat L.E. (2004) Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol., 5, 89–99. [DOI] [PubMed] [Google Scholar]
- 41.Jackson L.K., Nawabi P., Hentea C., Roark E.A., Haldar K. (2008) The Salmonella virulence protein SifA is a G protein antagonist. Proc. Natl. Acad. Sci. USA, 105, 14141–14146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harrison R.E., Brumell J.H., Khandani A., Bucci C., Scott C.C., Jiang X., Finlay B.B., Grinstein S. (2004) Salmonella impairs RILP recruitment to Rab7 during maturation of invasion vacuoles. Mol. Biol. Cell, 15, 3146–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fukuda M., Kobayashi H., Ishibashi K., Ohbayashi N. (2011) Genome-wide investigation of the Rab binding activity of RUN domains: development of a novel tool that specifically traps GTP-Rab35. Cell Struct. Funct., 36, 155–170. [DOI] [PubMed] [Google Scholar]
- 44.Grosshans B.L., Ortiz D., Novick P. (2006) Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. USA, 103, 11821–11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bento C.F., Puri C., Moreau K., Rubinsztein D.C. (2013) The role of membrane-trafficking small GTPases in the regulation of autophagy. J. Cell Sci., 126, 1059–1069. [DOI] [PubMed] [Google Scholar]
- 46.Levine B., Kroemer G. (2008) Autophagy in the pathogenesis of disease. Cell, 132, 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie M., Morales C.R., Lavandero S., Hill J.A. (2011) Tuning flux: autophagy as a target of heart disease therapy. Curr. Opin. Cardiol., 26, 216–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klionsky D.J., Abdalla F.C., Abeliovich H., Abraham R.T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J.A. et al. (2012) Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy, 8, 445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiang L., Zhao B., Ming M., Wang N., He T.C., Hwang S., Thorburn A., He Y.Y. (2014) Regulation of cell proliferation and migration by p62 through stabilization of Twist1. Proc. Natl. Acad. Sci. USA, 111, 9241–9246. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Belaid A., Cerezo M., Chargui A., Corcelle-Termeau E., Pedeutour F., Giuliano S., Ilie M., Rubera I., Tauc M., Barale S. et al. (2013) Autophagy plays a critical role in the degradation of active RHOA, the control of cell cytokinesis, and genomic stability. Cancer Res., 73, 4311–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Belaid A., Ndiaye P.D., Cerezo M., Cailleteau L., Brest P., Klionsky D.J., Carle G.F., Hofman P., Mograbi B. (2014) Autophagy and SQSTM1 on the RHOA(d) again: emerging roles of autophagy in the degradation of signaling proteins. Autophagy, 10, 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arad M., Maron B.J., Gorham J.M., Johnson W.H. Jr, Saul J.P., Perez-Atayde A.R., Spirito P., Wright G.B., Kanter R.J., Seidman C.E. et al. (2005) Glycogen storage diseases presenting as hypertrophic cardiomyopathy. N Engl. J. Med., 352, 362–372. [DOI] [PubMed] [Google Scholar]
- 53.Bai H., Li H., Li W., Gui T., Yang J., Cao D., Shen K. (2015) The PI3 K/AKT/mTOR pathway is a potential predictor of distinct invasive and migratory capacities in human ovarian cancer cell lines. Oncotarget, 22, 25520–25532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gustafsson A.B., Gottlieb R.A. (2009) Autophagy in ischemic heart disease. Circ. Res., 104, 150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma S., Wang Y., Chen Y., Cao F. (2014) The role of the autophagy in myocardial ischemia/reperfusion injury. Biochim. Biophys. Acta, 1852, 271–276. [DOI] [PubMed] [Google Scholar]
- 56.Taneike M., Yamaguchi O., Nakai A., Hikoso S., Takeda T., Mizote I., Oka T., Tamai T., Oyabu J., Murakawa T. et al. (2010) Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy, 6, 600–606. [DOI] [PubMed] [Google Scholar]
- 57.Nishino I., Fu J., Tanji K., Yamada T., Shimojo S., Koori T., Mora M., Riggs J.E., Oh S.J., Koga Y. et al. (2000) Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature, 406, 906–910. [DOI] [PubMed] [Google Scholar]
- 58.Charron P., Villard E., Sebillon P., Laforet P., Maisonobe T., Duboscq-Bidot L., Romero N., Drouin-Garraud V., Frebourg T., Richard P. et al. (2004) Danon's disease as a cause of hypertrophic cardiomyopathy: a systematic survey. Heart, 90, 842–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Selcen D., Muntoni F., Burton B.K., Pegoraro E., Sewry C., Bite A.V., Engel A.G. (2009) Mutation in BAG3 causes severe dominant childhood muscular dystrophy. Ann. Neurol., 65, 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee H.C., Cherk S.W., Chan S.K., Wong S., Tong T.W., Ho W.S., Chan A.Y., Lee K.C., Mak C.M. (2012) BAG3-related myofibrillar myopathy in a Chinese family. Clin. Genet., 81, 394–398. [DOI] [PubMed] [Google Scholar]
- 61.Arimura T., Ishikawa T., Nunoda S., Kawai S., Kimura A. (2011) Dilated cardiomyopathy-associated BAG3 mutations impair Z-disc assembly and enhance sensitivity to apoptosis in cardiomyocytes. Hum. Mutat., 32, 1481–1491. [DOI] [PubMed] [Google Scholar]
- 62.Norton N., Li D., Rieder M.J., Siegfried J.D., Rampersaud E., Zuchner S., Mangos S., Gonzalez-Quintana J., Wang L., McGee S. et al. (2011) Genome-wide studies of copy number variation and exome sequencing identify rare variants in BAG3 as a cause of dilated cardiomyopathy. Am. J. Hum. Genet., 88, 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Villard E., Perret C., Gary F., Proust C., Dilanian G., Hengstenberg C., Ruppert V., Arbustini E., Wichter T., Germain M. et al. (2011) A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur. Heart J., 32, 1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Towbin J.A. (2010) Left ventricular noncompaction: a new form of heart failure. Heart Fail. Clin., 6, 453–469, viii. [DOI] [PubMed] [Google Scholar]
- 65.Chen H., Zhang W., Li D., Cordes T.M., Mark Payne R., Shou W. (2009) Analysis of ventricular hypertrabeculation and noncompaction using genetically engineered mouse models. Pediatr. Cardiol., 30, 626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luxan G., Casanova J.C., Martinez-Poveda B., Prados B., D'Amato G., MacGrogan D., Gonzalez-Rajal A., Dobarro D., Torroja C., Martinez F. et al. (2013) Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat. Med., 19, 193–201. [DOI] [PubMed] [Google Scholar]
- 67.Finsterer J. (2009) Cardiogenetics, neurogenetics, and pathogenetics of left ventricular hypertrabeculation/noncompaction. Pediatr. Cardiol., 30, 659–681. [DOI] [PubMed] [Google Scholar]
- 68.Hussein A., Karimianpour A., Collier P., Krasuski R.A. (2015) Isolated noncompaction of the left ventricle in adults. J. Am. Coll. Cardiol., 66, 578–585. [DOI] [PubMed] [Google Scholar]
- 69.Diaz-Araya G., Vivar R., Humeres C., Boza P., Bolivar S., Munoz C. (2015) Cardiac fibroblasts as sentinel cells in cardiac tissue: receptors, signaling pathways and cellular functions. Pharmacol. Res., doi:10.1016/j.phrs.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 70.Belaid A., Ndiaye P.D., Klionsky D.J., Hofman P., Mograbi B. (2013) Signalphagy: scheduled signal termination by macroautophagy. Autophagy, 9, 1629–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schiller N.B., Shah P.M., Crawford M., DeMaria A., Devereux R., Feigenbaum H., Gutgesell H., Reichek N., Sahn D., Schnittger I. et al. (1989) Recommendations for quantitation of the left ventricle by two-dimensional echocardiography. American Society of Echocardiography Committee on Standards, Subcommittee on Quantitation of Two-Dimensional Echocardiograms. J. Am. Soc. Echocardiogr., 2, 358–367. [DOI] [PubMed] [Google Scholar]
- 72.Henry W.L., Gardin J.M., Ware J.H. (1980) Echocardiographic measurements in normal subjects from infancy to old age. Circulation, 62, 1054–1061. [DOI] [PubMed] [Google Scholar]
- 73.Richardson P., McKenna W., Bristow M., Maisch B., Mautner B., O'Connell J., Olsen E., Thiene G., Goodwin J., Gyarfas I. et al. (1996) Report of the 1995 World Health Organization/International Society and Federation of Cardiology Task Force on the Definition and Classification of cardiomyopathies. Circulation, 93, 841–842. [DOI] [PubMed] [Google Scholar]
- 74.Jenni R., Oechslin E., Schneider J., Attenhofer Jost C., Kaufmann P.A. (2001) Echocardiographic and pathoanatomical characteristics of isolated left ventricular non-compaction: a step towards classification as a distinct cardiomyopathy. Heart, 86, 666–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Muhammad E., Reish O., Ohno Y., Scheetz T., Deluca A., Searby C., Regev M., Benyamini L., Fellig Y., Kihara A. et al. (2013) Congenital myopathy is caused by mutation of HACD1. Hum. Mol. Genet., 22, 5229–5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.