Abstract
The objective of this study was to determine the protective effects of the mitochondrial division inhibitor 1 (Mdivi1) in striatal neurons that stably express mutant Htt (STHDhQ111/Q111) and wild-type (WT) Htt (STHDhQ7/Q7). Using gene expression analysis, biochemical methods, transmission electron microscopy (TEM) and confocal microscopy methods, we studied (i) mitochondrial and synaptic activities by measuring mRNA and the protein levels of mitochondrial and synaptic genes, (ii) mitochondrial function and (iii) ultra-structural changes in mutant Htt neurons relative to WT Htt neurons. We also studied these parameters in Mdivil-treated and untreated WT and mutant Htt neurons. Increased expressions of mitochondrial fission genes, decreased expression of fusion genes and synaptic genes were found in the mutant Htt neurons relative to the WT Htt neurons. Electron microscopy of the mutant Htt neurons revealed a significantly increased number of mitochondria, indicating that mutant Htt fragments mitochondria. Biochemical analysis revealed defective mitochondrial functioning. In the Mdivil-treated mutant Htt neurons, fission genes were down-regulated, and fusion genes were up-regulated, suggesting that Mdivil decreases fission activity. Synaptic genes were up-regulated, and mitochondrial function was normal in the Mdivi1-treated mutant Htt neurons. Immunoblotting findings of mitochondrial and synaptic proteins agreed with mRNA findings. The TEM studies revealed that increased numbers of structurally intact mitochondria were present in Mdivi1-treated mutant Htt neurons. Increased synaptic and mitochondrial fusion genes and decreased fission genes were found in the Mdivi1-treated WT Htt neurons, indicating that Mdivi1 beneficially affects healthy neurons. Taken together, these findings suggest that Mdivi1 is protective against mutant Htt-induced mitochondrial and synaptic damage in HD neurons and that Mdivi1 may be a promising molecule for the treatment of HD patients.
Introduction
Huntington's disease (HD) is an autosomal, dominantly inherited neurodegenerative disease, characterized by chorea, seizures, involuntary movements, dystonia, cognitive decline, intellectual impairment and emotional disturbances (1–3). HD is a midlife disease, with HD patients surviving for ∼10–20 years after disease onset. In HD patients, selective neuronal loss has been observed in the caudate and putamen of the striatum, cortex and hypothalamus, and to a lesser extent in hippocampal and subthalamus neurons (1,4–6). In addition, mutant huntingtin (Htt) protein aggregates have been found in pathological sites in HD postmortem brains (7). Currently, there are no drugs or agents available to treat or to prevent HD.
Htt is a 350 kDa protein, ubiquitously expressed in the brain and peripheral tissues of HD patients (8,9). Both wild-type (WT) and mutant Htt are reported to localize in the cytoplasm. Small numbers of mutant Htt also have also been found in subcellular organelles, including the nucleus, plasma membrane, mitochondria, lysosomes and endoplasmic reticulum. Mutant Htt has been found to impair the functions of subcellular organelles (10–15) and to interact with a large number of brain proteins, with the extent of this interaction dependent on the number of expanded polyQ repeats (16).
Several lines of evidence suggest that defective mitochondrial bioenergetics are involved in HD progression. (i) Biochemical studies of mitochondria in striatal neurons from late-stage HD patients and HD mouse models revealed reduced enzymatic activities of several components of oxidative phosphorylation, including complexes II, III and IV (17–19). (ii) Expanded polyQ repeats are associated with low levels of mitochondrial ATP and decreased mitochondrial adenosine diphosphate uptake (20). (iii) Calcium-induced mitochondrial permeability is a major factor in HD pathogenesis (21,22). (iv) Defective mitochondrial axonal transport in HD cortical and hippocampal neurons has been identified as another factor in HD pathogenesis (15,18,23–25). (v) Recent studies suggest that structural abnormalities in mitochondria are involved in HD and that these abnormalities are caused by an imbalance in highly conserved GTPase proteins that are essential for mitochondrial division (Drp1 and Fis1) and mitochondrial fusion (Mfn1, Mfn2 and Opa1) (20,23,26–28).
To determine whether the interaction between Drp1 and mHtt increases as HD progresses, Shirendeb et al. (23) performed co-immunoprecipitation analysis of Drp1 and mutant Htt, using a Drp1 antibody, and they conducted immunoblotting analysis, using the mHtt-specific antibody 1C2 and protein lysates of cortical tissues from harvested brains of control subjects and patients with Grade 3 and Grade 4 HD. They also used striatal protein lysates from 2-month-old BACHD mice. Immunoblotting analysis revealed an 80 kDa in the specimens from the Grade 3 and Grade 4 HD patients and in the 2-month-old BACHD mice. Further, the intensity of the 80 kDa band increased with HD progression, indicating that the interaction of Drp1 with mHtt may be involved in HD progression. Further, GTPase Drp1 enzymatic activity, which is critical for mitochondrial division, was elevated in the HD-affected neurons, in the HD postmortem bains, and in the brains from BACHD mice, indicating that Drp1 and mHtt interactions enhance mitochondrial fragmentation. These results suggest that Drp1 interacts with mHtt and participates in excessive mitochondrial fragmentation in HD neurons (23).
To identify mitochondrial fission inhibitors, several groups have independently screened chemical libraries and have found several fission inhibitors: Mdivi1 (29), Dynasore (30) and P110 (31). Among these, Mdivi1 has been extensively investigated with experimental rodent models for epilepsy and seizures (32,33), ischemia (34,35), oxygen glucose deprivation (36) and conditions such as aggregation of endosomes and vesicle fusion during exocytosis (37). In all of these diseased states or conditions, Mdivi1 was found to benefit affected tissues and cells by reducing excessive mitochondrial fission, maintaining the mitochondrial fission–fusion balance and maintaining the normal functioning of cells.
However, the efficacy of Mdivi1 has not been studied in HD. Mdivi1 is a cell-permeable, selective mitochondrial fission inhibitor with a molecular weight of 353.22. Mdivi1 is dissolved in DMSO. Mdivi1 inhibits GTPase Drp1 activity by blocking the self-assembly of Drp1, resulting in a reversible formation of elongated and tubular mitochondria in WT cells (29). The efficacy of Mdivi 1 in HD-affected neurons needs elucidation.
In the current study, using electron, confocal and fluorescence microscopy; gene expression analysis; and biochemical methods, we sought to determine (i) mitochondrial structure by measuring mRNA and the protein levels of Drp1, Fis1, Mfn1, Mfn2, Opa1, CypD, electron transport chain (ETC) and synaptic genes; (ii) mitochondrial function by measuring free radical production, lipid peroxidation, GTPase Drp1 enzymatic activity, mitochondrial ATP and cell viability; and (iii) ultra-structural changes of subcellular organelles in the cell, including mitochondria in mutant Htt and WT Htt neurons. We also sought to determine the above-mentioned parameters in Mdivi1-treated and untreated WT Htt and mutant Htt neurons.
Results
Gene expression differences between wild-type Htt and mutant Htt neurons
As shown in Table 1, the mRNA expression level of the fission gene Drp1 was increased by 1.8-fold (P = 0.03), and the mRNA expression level of the fission gene Fis1 was increased by 2.7 (P = 0.001). However, all fusion genes were decreased: Mfn1 was decreased by 2.0-fold (P = 0.001), Mfn2 by 1.3-fold (P = 0.07) and Opa1 by 2.1-fold (P = 0.002) in mutant striatal Htt (Q111/Q111) neurons relative to the WT striatal Htt (Q7/Q7) neurons, indicating the presence of abnormal mitochondrial dynamics in HD-affected neurons. mRNA expressions of ETC genes were unchanged. All synaptic genes in mutant Htt neurons were decreased: synaptophysin was decreased by 1.5-fold (P = 0.02), PSD95 by 1.4-fold (P = 0.01), synapsin 1 by 1.6-fold (P = 0.01), synapsin 2 by 1.3-fold (P = 0.09), neurogranin by 1.4-fold (P = 0.03), GAP43 by 1.6-fold (P = 0.01) and synaptopodin by 1.3-fold (P = 0.09). Taken together, all of these findings suggest that synaptic activity is low in mutant Htt neurons relative to WT Htt neurons.
Table 1.
mRNA fold changes of mitochondrial, ETC, and synaptic genes in mutant Htt neurons relative to WT Htt neurons
| Gene | mRNA fold changes |
|---|---|
| Mitochondrial dynamics genes | |
| DRP1 | 1.8* |
| FIS1 | 2.7** |
| Mfn1 | −2.0** |
| Mfn2 | −1.3 |
| Cyclophilin D | 1.3 |
| OPA1 | 2.1** |
| VDAC | 1.3 |
| Mitochondria-encoded electron transport chain genes | |
| ND1—Complex I | 1.2 |
| ND3—Complex I | 1.0 |
| ND6—Complex I | 1.0 |
| Cyt B—Complex III | 1.3 |
| COX1—Complex IV | 1.3 |
| COX2—Complex IV | 1.2 |
| COX3—Complex IV | 1.1 |
| ATP6—Complex V | 1.5* |
| Synaptic genes | |
| Synaptophysin | −1.5* |
| PSD-95 | −1.4* |
| Synapsin 1 | −1.6* |
| Synapsin 2 | −1.3 |
| Synaptobrevin 1 | 1.1 |
| Synaptobrevin 2 | 1.2 |
| Neurogranin | −1.4* |
| GAP43 | −1.6* |
| Synaptopodin | −1.3 |
Gene expression differences between Mdivi1-treated and untreated wild-type Htt neurons
To determine the effects of Mdivi1 on mitochondrial structure and on synaptic proteins, we independently treated WT Htt and mutant Htt neurons with Mdivil at concentrations of 25 and 50 μm, and we used real-time RT-PCR to measure mRNA levels before and after treatment.
mRNA expression levels of Drp1 and Fis1 were significantly decreased in neurons treated with Mdivi1 at 25 and 50 μm concentrations (Table 2). In contrast, the levels of mRNA expression of the mitochondrial fusion genes Mfn1, Mfn2 and Opa1 increased after treatment with Mdivil. These findings indicate that Mdivi1 enhances fusion activity in WT Htt neurons. Significantly reduced mRNA levels were found in CypD in Mdivi1-treated WT Htt neurons. Reduced mRNA expression levels were found in all ETC genes in WT Htt cells treated with Mdivi1 at 50 μm, and mRNA expression levels were unchanged in WT Htt neurons treated at 25 μm (Table 2).
Table 2.
mRNA fold changes of mitochondrial, ETC and synaptic genes in WT Htt neurons treated with Mdivi1
| Gene | mRNA fold changes |
|
|---|---|---|
| 25 µM Mdivi | 50 µM Mdivi | |
| Mean | Mean | |
| Mitochondrial dynamics genes | ||
| DRP1 | −1.2 | −1.4* |
| FIS1 | −1.2 | 1.0 |
| Mfn1 | 3.7** | 4.6** |
| Mfn2 | 2.1** | 1.4* |
| Cyclophilin D | −1.2 | −1.4* |
| OPA1 | 1.9* | 2.2** |
| VDAC | 1.1 | 1.4* |
| Mitochondria-encoded electron transport chain genes | ||
| ND1—Complex I | −1.1 | −1.8* |
| ND3—Complex I | 1.0 | −1.3 |
| ND6—Complex I | 1.1 | −1.3 |
| Cyt B—Complex III | 1.0 | −1.3 |
| COX1—Complex IV | 1.0 | −1.3 |
| COX2—Complex IV | 1.0 | −1.7* |
| COX3—Complex IV | 1.1 | −1.3 |
| ATP6—Complex V | 1.1 | −1.3 |
| Synaptic genes | ||
| Synaptophysin | 1.2 | 1.6* |
| PSD-95 | 1.1 | 1.3 |
| Synapsin 1 | 1.3 | 1.2 |
| Synapsin 2 | 1.3 | 1.8** |
| Synaptobrevin 1 | 1.5* | 2.0** |
| Synaptobrevin 2 | 1.3 | 1.3 |
| Neurogranin | 1.7** | 1.5* |
| GAP43 | 1.4* | 1.1 |
| Synaptopodin | 2.0** | 1.3 |
mRNA expression levels were increased in synaptic genes, including synaptophysin, PSD95, synapsin 1, synapsin 2, synaptobrevin 1, synaptobrevin 2, neurogranin, GAP43 and synaptopodin in WT Htt neurons treated with Mdivi1 at 25 and 50 μm concentrations relative to the untreated neurons (Table 2). These findings suggest that Mdivi1 enhances synaptic gene expressions in WT cells treated with Mdivi1 at 25 and 50 μm concentrations.
Gene expression differences between Mdivi1-treated and untreated mutant Htt neurons
As shown in Tables 3 and 4, mRNA expression levels decreased in Drp1 and Fis1 in Mdivi1-treated mutant Htt neurons relative to Mdivi1-untreated mutant Htt neurons. Significantly increased mRNA expression levels were found in Mfn1, Mfn2 and Opa1. These results indicate that Mdivi1 at concentrations of 25 and 50 μm enhances fusion activity in mutant Htt neurons.
Table 3.
mRNA fold changes of mitochondrial, ETC and synaptic genes in mutant Htt neurons treated with Mdivi1
| Gene | mRNA fold changes |
|
|---|---|---|
| 25 µM Mdivi | 50 µM Mdivi | |
| Mean | Mean | |
| Mitochondrial dynamics genes | ||
| DRP1 | −1.6* | −1.4* |
| FIS1 | −1.6* | −1.1 |
| Mfn1 | 3.2** | 1.8** |
| Mfn2 | 2.2** | 1.9** |
| Cyclophilin D | −1.6* | −1.4* |
| OPA1 | 1.7* | 2.6** |
| VDAC | −1.4* | −1.1 |
| Mitochondria-encoded electron transport chain genes | ||
| ND1—Complex I | −1.1 | 1.7* |
| ND3—Complex I | −1.2 | 1.7* |
| ND6—Complex I | −1.2 | 1.5* |
| Cyt B—Complex III | −1.2 | 1.3 |
| COX1—Complex IV | −1.2 | 1.6* |
| COX2—Complex IV | −1.2 | 1.6* |
| COX3—Complex IV | −1.3 | 1.4* |
| ATP6—Complex V | −1.4* | 1.3 |
| Synaptic genes | ||
| Synaptophysin | 1.5* | 2.1** |
| PSD-95 | 1.2 | 1.3 |
| Synapsin 1 | 1.4* | 1.6* |
| Synapsin 2 | 3.3** | 1.8** |
| Synaptobrevin 1 | 2.6** | 2.2** |
| Synaptobrevin 2 | 1.2 | 1.2 |
| Neurogranin | 1.2 | 2.2** |
| GAP43 | 1.7* | 3.2** |
| Synaptopodin | 1.2 | 2.1* |
Table 4.
Summary of real-time RT-PCR oligonucleotide primers used in measuring mRNA expression in mitochondrial dynamics genes, ETC genes and synaptic genes in WT and mutant Htt neurons treated with Mdivi1
| Gene | DNA sequence (5′-3′) | PCR product size |
|---|---|---|
| Mitochondrial structural genes | ||
| Drp1 | Forward primer ATGCCAGCAAGTCCACAGAA | 86 |
| Reverse primer TGTTCTCGGGCAGACAGTTT | ||
| Fis1 | Forward primer CAAAGAGGAACAGCGGGACT | 95 |
| Reverse primer ACAGCCCTCGCACATACTTT | ||
| MFN1 | Forward primer GCAGACAGCACATGGAGAGA | 83 |
| Reverse primer GATCCGATTCCGAGCTTCCG | ||
| MFN2 | Forward primer TGCACCGCCATATAGAGGAAG | 78 |
| Reverse primer TCTGCAGTGAACTGGCAATG | ||
| Cyclophilin D | Forward primer AGATGTCAAATTGGCAGGGGG | 91 |
| Reverse primer TGCGCTTTTCGGTATAGTGCT | ||
| Opa1 | Forward primer ACCTTGCCAGTTTAGCTCCC | 82 |
| Reverse primer TTGGGACCTGCAGTGAAGAA | ||
| VDCA1 | Forward primer CTCCCACATACGCCGATCTT | 58 |
| Reverse primer GCCGTAGCCCTTGGTGAAG | ||
| Mitochondria-encoded electron transport chain genes | ||
| ND1—CI | Forward primer CGGGCCCCCTTCGAC | 72 |
| Reverse primer GGCCGGCTGCGTATTCT | ||
| ND3—CI | Forward primer TGTACTCAGAAAAAGCAAATCCATATG | 73 |
| Reverse primer AATAATAGAAATGTAATTGCTACCAAGAAAAA | ||
| ND6—CI | Forward primer CCGCAAACAAAGATCACCCAG | 79 |
| Reverse primer GAAGGAGGGATTGGGGTAGC | ||
| CYT B—CIII | Forward primer TTATCGCGGCCCTAGCAA | 75 |
| Reverse primer TAATCCTGTTGGGTTGTTTGATCC | ||
| COX 1—CIV | Forward primer ATCACTACCAGTGCTAGCCG | 84 |
| Reverse primer CCTCCAGCGGGATCAAAGAA | ||
| COX2—CIV | Forward primer CATCCCAGGCCGACTAAATC | 74 |
| Reverse primer TTTCAGAGCATTGGCCATAGAA | ||
| COX3—CIV | Forward primer CAGGATTCTTCTGAGCGTTCTATCA | 72 |
| Reverse primer AATTCCTGTTGGAGGTCAGCA | ||
| ATP6—CV | Forward primer TCCCAATCGTTGTAGCCATCA | 76 |
| Reverse primer AGACGGTTGTTGATTAGGCGT | ||
| Synaptic genes | ||
| Synaptophysin | Forward primer CTGCGTTAAAGGGGGCACTA | 81 |
| Reverse primer ACAGCCACGGTGACAAAGAA | ||
| PSD95 | Forward primer CTTCATCCTTGCTGGGGGTC | 90 |
| Reverse primer TTGCGGAGGTCAACACCATT | ||
| Synapsin 1 | Forward primer TGAGGACATCAGTGTCGGGTAA | 64 |
| Reverse primer GGCAATCTGCTCAAGCATAGC | ||
| Synapsin 2 | Forward primer TCCCACTCATTGAGCAGACATACT | 63 |
| Reverse primer GGGAACGTAGGAAGCGTAAGC | ||
| Synaptobrevin 1 | Forward primer TGCTGCCAAGCTAAAAAGGAA | 68 |
| Reverse primer CAGATAGCTCCCAGCATGATCA | ||
| Synaptobrevin 2 | Forward primer GGGACCAGAAGTTGTCGGAG | 89 |
| Reverse primer CTTGAGCTTGGCTGCACTTG | ||
| Neurogranin | Forward primer CTCCAAGCCAGACGACGATA | 83 |
| Reverse primer AACTCGCCTGGATTTTGGCT | ||
| GAP43 | Forward primer GCTGCGACCAAAATTCAGGC | 83 |
| Reverse primer GCTGGTGCATCACCCTTCT | ||
| Synaptopodin | Forward primer TCCTGCGCCCTGAACCTA | 70 |
| Reverse primer GACGGGCGACAGAGCATAGA | ||
| Housekeeping genes | ||
| β-Actin | Forward primer AGAAGCTGTGCTATGTTGCTCTA | 91 |
| Reverse primer TCAGGCAGCTCATAGCTCTTC | ||
| GAPDH | Forward primer TTCCCGTTCAGCTCTGGG | 59 |
| Reverse primer CCCTGCATCCACTGGTGC | ||
In mutant Htt neurons treated with Mdivi1, mRNA expression levels of matrix genes were significantly reduced in CypD. Reduced mRNA expressions were found in all ETC genes in mutant Htt neurons treated with Mdivi1 at a 25 μm concentration (Table 3). In contrast, mRNA levels of ETC genes increased in all ETC genes when mutant Htt neurons were treated with Mdivil at a 50 μm concentration (Table 3).
mRNA expression levels were increased in synaptic genes, including synaptophysin, PSD95, synapsin1, synapsin synaptobrevin1, synaptobrevin 2, GAP43 and synaptopodin, in Mdivi1-treated mutant Htt neurons relative to the untreated neurons. These findings suggest that Mdivi1 enhances synaptic gene expression in Mdivi1-treated mutant Htt neurons.
Immunoblotting analysis
Protein levels between WT Htt and mutant Htt neurons
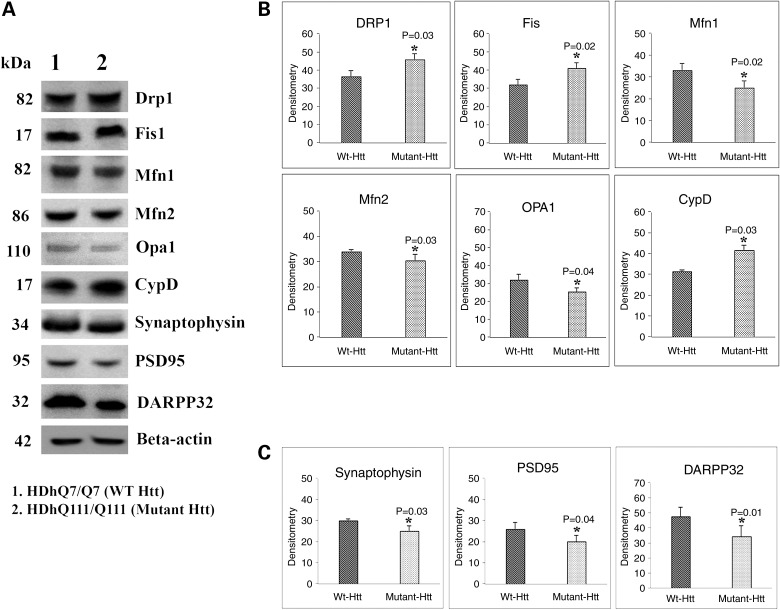
To understand the effects of mutant Htt on mitochondrial and synaptic proteins, we performed immunoblotting analysis using proteins of WT Htt (HDhQ7/Q7) cells and mutant Htt (HDhQ111/Q111) cells. Our quantitative densitometry analysis revealed that significantly increased levels of fission proteins Drp1 and Fis1 were found in mutant Htt neurons relative to WT Htt neurons (Fig. 1A and B). In contrast, decreased fusion proteins Mfn1, Mfn2 and Opa1 were found in mutant Htt neurons relative to the WT Htt neurons (Fig. 1A and B). Levels of the matrix protein CypD were significantly increased in the mutant Htt neurons. Expression levels of synaptic proteins, synaptophysin and PSD95 significantly decreased in mutant Htt neurons relative to WT Htt neurons (Fig. 1A and 2C). Expression levels of the medium-spiny neuronal protein marker DARPP32 were also significantly decreased in mutant Htt neurons (Fig. 1A and C).
Figure 1.
Immunoblotting analysis of proteins in WT Htt (HDhQ7/Q7) and mutant Htt (HDhQ111/Q111) neurons. (A) Representative immunoblotting analysis of WT and mutant Htt neurons. (B) Quantitative densitometry analysis of mitochondrial dynamics and matrix proteins, Drp1, Fis1, Mfn1, Mfn2, Opa1 and CypD of WT Htt neurons relative to mutant Htt neurons. (C) Quantitative densitometry analysis of synaptic proteins synaptophysin, PSD95 and DARPP32 of mutant Htt neurons relative to WT Htt neurons. Significantly increased levels of fission proteins (Drp1 and Fis1) and significantly decreased levels of fusion and matrix proteins (Mfn1, Mfn2, Opa1 and CypD) in mutant Htt neurons relative to WT Htt neurons. Expression levels of synaptic protein levels synaptophysin and PSD95 and medium-spiny neuronal marker DARPP32 were significantly reduced in mutant Htt neurons.
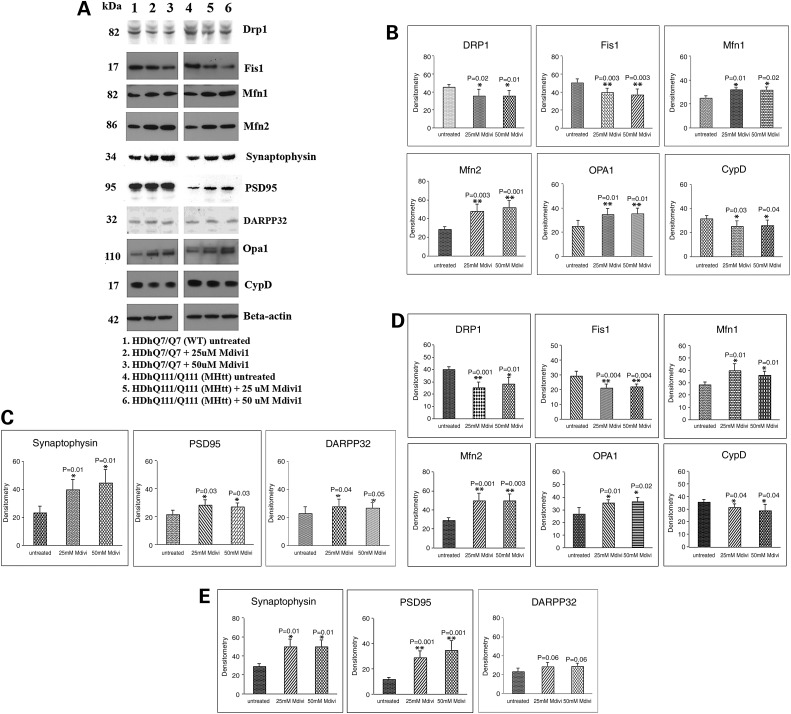
Figure 2.
Immunoblotting analysis of proteins in WT Htt and mutant Htt neurons treated with Mdivi1 and untreated. (A) Representative immunoblotting analysis of WT and mutant Htt neurons. (B) Quantitative densitometry analysis of mitochondrial dynamics and matrix proteins, Drp1, Fis1, Mfn1, Mfn2, Opa1 and CypD of WT Htt neurons treated with Mdivi1 relative to Mdivi1 untreated. (C) Quantitative densitometry analysis of synaptic proteins synaptophysin, PSD and DARPP32 of WT Htt neurons treated with Mdivi1 relative to untreated. (D) Quantitative immunoblotting analysis of mitochondrial dynamics and matrix proteins Drp1, Fis1, Mfn1, Mfn2 and Opa1 and CypD proteins of mutant Htt neurons treated with Mdivi1 relative to untreated. (E) Quantitative densitometry analysis of synaptic proteins, synaptophysin, PSD and DARPP32 of mutant Htt neurons treated with Mdivi1 relative to untreated. The fission proteins Drp1 and Fis1 were significantly decreased, and the fusion proteins, Mfn1, Mfn2 and Opa1 and the synaptic, synaptophysin and PSD95 and DARPP32 proteins were significantly increased in Mdivil at 25 and 50 μm concentrations, indicating that Mdivi1 reduces fission activity and enhances fusion and synaptic activity.
Protein levels between Mdivi1-treated and untreated WT Htt neurons
To determine the effects of Mdivi1 on mitochondrial structure and on synaptic proteins, we independently treated WT Htt and mutant Htt neurons with Mdivil at concentrations of 25 and 50 μm, and we used immunoblotting and quantitative densitometry to measure protein levels before and after treatment.
In Mdivi1-treated WT Htt neurons compared with untreated WT Htt neurons, significantly decreased levels of expression were found in Drp1 and Fis1 (Fig. 2A and B). In contrast, increased fusion protein levels were found in Mfn1, Mfn2 and Opa1 in WT Htt neurons treated with Mdivi1 compared with the untreated neurons (Fig. 2A and B). Levels of the matrix protein CypD were significantly decreased in the Mdivil-treated WT Htt neurons, but significantly increased levels of synaptophysin and PSD95 were present in the Mdivi1-treated WT Htt neurons (Fig. 2A and C). Expression levels of the medium-spiny neuronal protein marker DARRP32 were also significantly increased in the Mdivil-treated WT Htt neurons (Fig. 2A and C and Fig. 3).
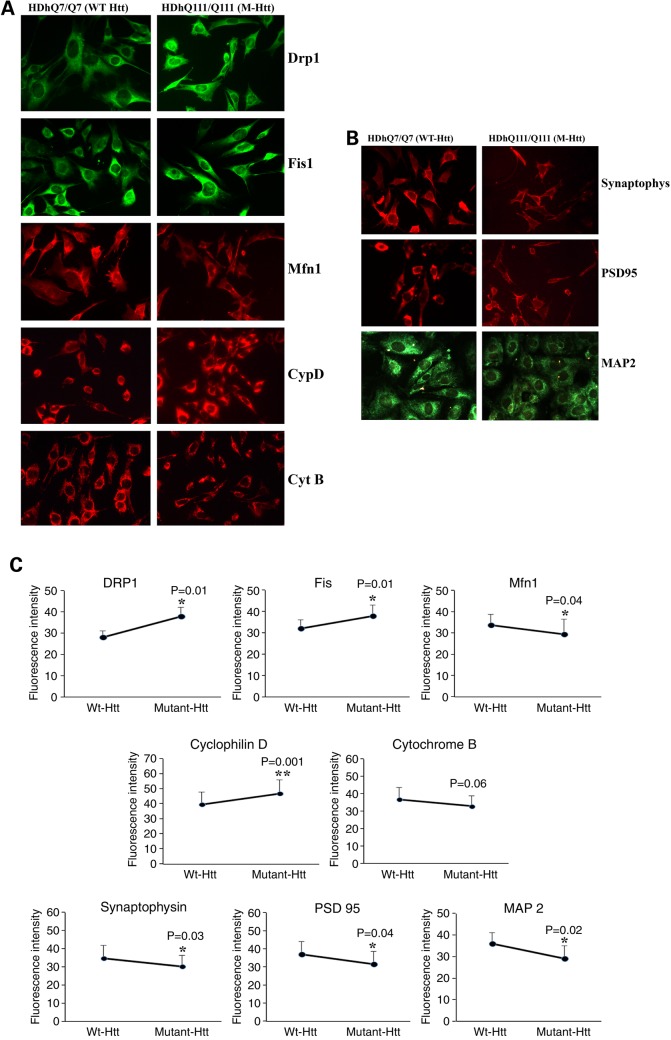
Figure 3.
Immunofluorescence analysis of proteins from WT Htt neurons and mutant Htt neurons. (A) Representative images of WT Htt and mutant Htt neurons from mitochondrial dynamics and matrix proteins. (B) Representative images of WT Htt and mutant Htt neurons from synaptic proteins. (C) Quantitative immunofluorescence analysis of mitochondrial dynamics and matrix and synaptic proteins, Drp1, Fis1, Mfn1, Mfn2, Opa1, CypD and synaptophysin, PSD95 and MAP2 of WT Htt and mutant Htt neurons.
Protein levels between Mdivi1-treated and untreated mutant Htt neurons
In Mdivi1-treated mutant Htt neurons, significantly decreased protein levels were found for Drp1 and Fis1, in contrast to the protein levels for Mfn1, Mfn2 and Opa1, which were increased in the Mdivil-treated mutant Htt neurons (Fig. 2A and D). These results suggest that Mdivi1 reduces fission activity. Decreased protein levels were found for CypD in the Mdivi1-treated mutant Htt neurons relative to the untreated neurons (Fig. 2A and D). Significantly increased protein levels were found for synaptophysin, PSD95 and DARPP32 in the Mdivi1-treated mutant Htt neurons relative to the untreated mutant Htt neurons, suggesting that Mdivi1 enhances synaptic activity (Fig. 2A and E).
Immunofluorescence analysis
Immunofluorescence levels between WT Htt and mutant Htt neurons
To determine the effect of mutant Htt on protein levels and localizations, we performed immunofluorescence analysis of WT Htt neurons and mutant Htt neurons on following proteins: Drp1 and Fis1, Mfn1, CypD, Cyt B (mitochondrial), synaptic (synaptophysin, PSD95) and dendritic (MAP2). We found significantly increased levels of Drp1 (P = 0.01), Fis1 (P = 0.01) and CypD (P = 0.001) and significantly reduced levels of Mfn1 (P = 0.04) in mutant Htt neurons relative to WT Htt neurons, indicating the presence of abnormal mitochondrial dynamics in mutant Htt neurons.
Immunofluorescence analysis of synaptic and dendritic proteins revealed that significantly decreased levels of synaptophysin (P = 0.001), PSD95 (P = 0.04) and MAP2 (P = 0.02) in mutant Htt neurons relative to WT neurons, suggesting that mutant Htt reduces synaptic and dendritic proteins.
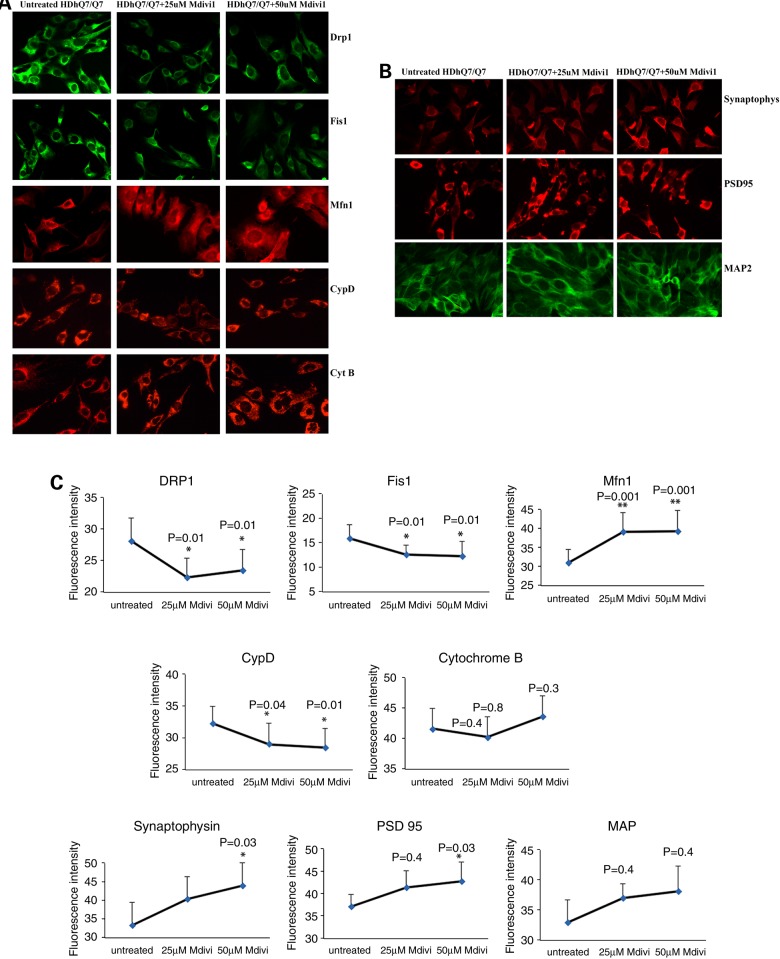
Immunofluorescence levels between Mdivi-treated and untreated WT Htt neurons
We treated WT Htt and mutant Htt neurons with Mdivil at 25 and 50 μm concentrations. We performed immunofluorescence analysis on mitochondrial, synaptic and dendritic proteins. As shown in Figure 4A and C, following Mdivil treatment of WT Htt neurons, we found significantly decreased levels of the following proteins: Drp1 at 25 and 50 μm concentrations (both P = 0.01), Fis1 at 25 and 50 μm (both P = 0.01), CypD at 25 μm (P = 0.04) and at 50 μm (P = 0.01). However, we found increased levels of Mfn1 at 25 and at 50 μm (both P = 0.001). Taken together, these results indicate that Mdivi1 at both concentrations reduces mitochondrial fission activity and enhances fusion activity. Our immunofluorescence findings agree with the immunoblotting results.
Figure 4.
Immunofluorescence analysis of proteins from Mdivil-treated and untreated WT Htt neurons. (A) Representative images of Mdivi1-treated and untreated WT Htt neurons from mitochondrial dynamics and matrix proteins. (B) Representative images of Mdivi1-treated and untreated WT Htt neurons from synaptic proteins. (C) Quantitative immunofluorescence analysis of mitochondrial dynamics and matrix and synaptic proteins, Drp1, Fis1, Mfn1, Mfn2, Opa1, CypD and synaptophysin, PSD95 and MAP2 of WT Htt neurons treated with Mdivi1 and untreated. In the WT Htt neurons, the levels of both fission and the matrix proteins were significantly decreased, and the fusion protein and the ETC protein were significantly increased when treated with Mdivil at both 25 and 50 μm concentrations.
Immunofluorescence analysis showed significantly increased levels of synaptophysin and PSD95 when they were treated with a 50 μm concentration of Mdivil (P = 0.03) (Fig. 4B and C). Statistical significance was not observed for synaptophysin and PSD95 when they were treated with Mdivil at a 25 μm concentration.
Immunofluorescence levels between Mdivi-treated and untreated mutant Htt neurons
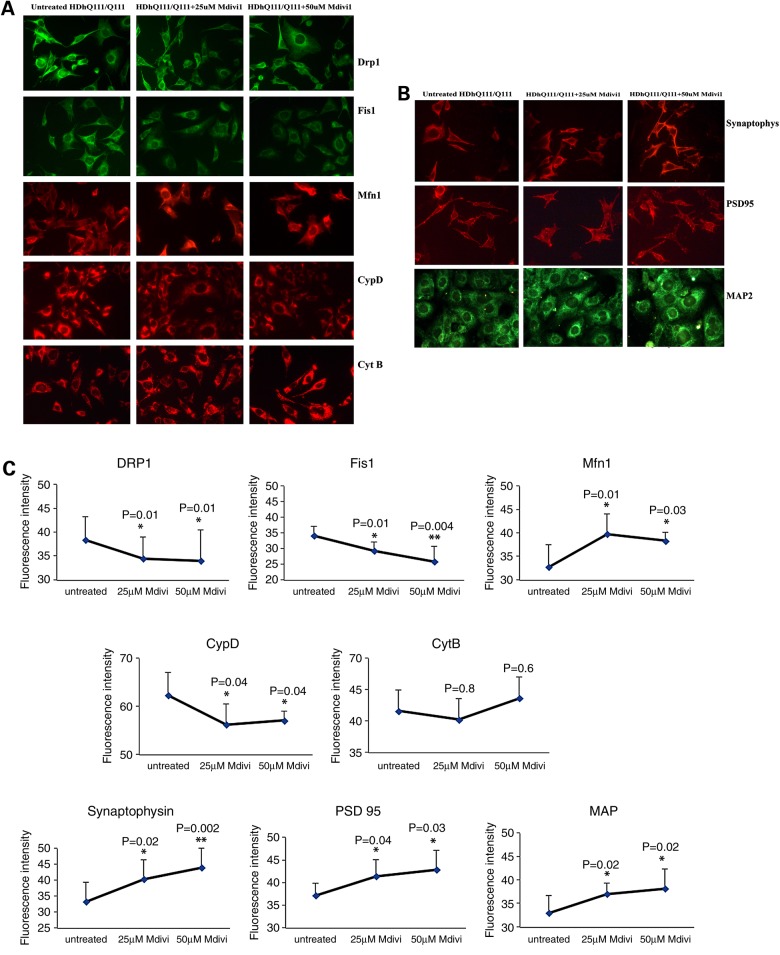
Following Mdivil treatment of mutant Htt neurons, we found significantly decreased levels of Drp1 at 25 and 50 μm concentrations of Mdivil (both P = 0.01), decreased levels of Fis1 at 25 μm (P = 0.01) and at 50 μm (P = 0.004), and decreased levels of CypD at 25 and 50 μm (both P = 0.04), as shown in Figure 5A and C. However, as shown in Figure 5A, Mfn1 increased when treated with Mdivil at 25 μm (P = 0.01) and 50 μm concentration (P = 0.03), as did synaptophysin at Mdivil concentrations of 25 μm (P = 0.02) and of 50 μm (P = 0.002), PSD95 at 25 μm (P = 0.04) and at 50 μm (P = 0.03) and MAP2 at 25 μm (P = 0.002) and 50 μm (P = 0.02) (Fig. 5B and C).
Figure 5.
Immunofluorescence analysis of proteins in Mdivil-treated and untreated mutant Htt neurons. (A) Representative images of Mdivi1-treated and untreated mutant Htt neurons from mitochondrial dynamics and matrix proteins. (B) Representative images of Mdivi1-treated and untreated mutant Htt neurons from synaptic proteins. (C) Quantitative immunofluorescence analysis of mitochondrial dynamics and matrix and synaptic proteins, Drp1, Fis1, Mfn1, Mfn2, Opa1, CypD and synaptophysin, PSD95 and MAP2 of mutant Htt (HDhQ111/Q111) neurons treated with Mdivi1 and untreated. The fission and matrix proteins were significantly decreased, and the fusion and ETC proteins were significantly increased upon treatment with Mdivil at 25 and 50 μm concentrations, indicating that Mdivi1 reduces mitochondrial fission activity and enhances fusion activity.
Transmission electron microscopy
Mitochondrial number between WT Htt and mutant Htt neurons
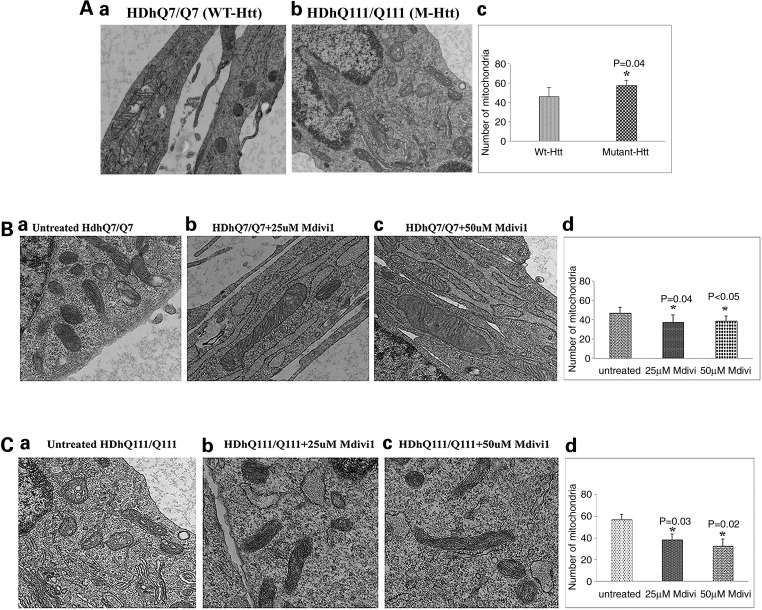
To determine the effect of mutant Htt on mitochondrial number, we performed transmission electron microscopy (TEM) using WT Htt neurons and mutant Htt neurons. As shown in Figure 6A, we found significantly increased number of mitochondria in mutant Htt neurons relative to WT Htt neurons (P = 0.04), suggesting that mutant Htt enhances mitochondrial fragmentation.
Figure 6.
Transmission electron microscopy of studies. (A) Mitochondrial number between WT Htt and mutant Htt neurons. (a) Healthy, intact mitochondria in the WT Htt neurons; (b) fragmented mitochondria in mutant Htt neurons. (c) Results from quantitative analysis of mitochondria. Significantly increased numbers of mitochondria were found in the mutant Htt neurons (P = 0.04) relative to WT Htt neurons. (B) Transmission electron microscopy of Mdivil-treated and untreated WT Htt neurons. (a) Healthy, intact mitochondria in the WT Htt neurons; (b) healthy, intact and elongated mitochondria in the 25 μm Mdivi1-treated WT Htt neurons. (c) Healthy, intact mitochondria in the 50 μm Mdivi1-treated WT Htt neurons. (d) Results from quantitative analysis of mitochondria. Significantly decreased numbers of mitochondria were found in the WT neurons treated with Mdivil at 25 μm (P = 0.04) and 50 μm (P = 0.04) concentrations compared with the mitochondria in mutant Htt neurons untreated with Mdivi1. (C) Transmission electron microscopy of Mdivil-treated and untreated mutant Htt neurons. (a) Fragmented and structurally damaged mitochondria in the mutant Htt neurons; (b) intact mitochondria in the 25 μm Mdivi1-treated WT Htt neurons. (c) Intact mitochondria in the 50 μm Mdivi1-treated mutant Htt neurons. (d) Results from quantitative analysis of mitochondria. Significantly decreased numbers of mitochondria were found in the mutant neurons treated with Mdivil at 25 μm (P = 0.03) and 50 μm (P = 0.02) concentrations compared with the mitochondria in mutant Htt neurons untreated with Mdivi1.
Mitochondrial number between Mdivi1-treated and untreated WT Htt neurons
To determine the effects of Mdivi1 on mitochondrial number, WT Htt neurons were treated with Mdivi1 at concentrations of 25 and 50 μm, and the number of mitochondria was assessed using TEM. The number of mitochondria significantly decreased following Mdivil treatment at 25 μm (P = 0.04) and at 50μm (P = 0.04) in the WT Htt neurons (Fig. 6B).
Mitochondrial number between Mdivi1-treated and untreated mutant Htt neurons
Similar to WT Htt neurons, we also treated mutant Htt neurons with Midivi1 and assessed mitochondrial number. The number of mitochondria significantly decreased in mutant Htt neurons treated with Mdivi1 at a concentration of 25 μm (P = 0.03) and of 50 μm (P = 0.02) (Fig. 6C).
Mitochondrial function
To determine differences in mitochondrial function between WT Htt and mutant Htt neurons, mitochondrial function as assessed in WT and mutant Htt neurons. The parameters included H2O2 production, lipid peroxidation, ATP production, cell viability and GTPase Drp1 enzymatic activity.
Mitochondria function between WT Htt and mutant Htt neurons
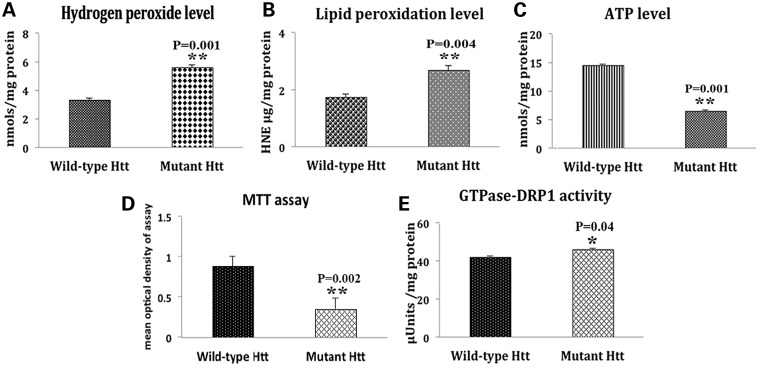
Significantly increased levels of H2O2 (P = 0.001) and 4-hydroxy-2-nonenol (P = 0.004) were found in mutant Htt neurons relative to WT Htt neurons. In contrast, significantly decreased ATP production (at P = 0.001) and cell viability (P = 0.002) were found in mutant Htt neurons relative to WT Htt neurons. The level of GTPase Drp1 activity was increased (P = 0.04) in mutant Htt neurons. These findings suggest that mitochondrial function is defective in mutant Htt neurons (Fig. 7).
Figure 7.
Mitochondrial functional parameters in WT Htt and mutant Htt neurons. Mitochondrial function was assessed in WT Htt (HDhQ7/HDhQ7) and mutant Htt (HDhQ111/Q111) neurons by measuring: (A) H2O2 production, (B) lipid peroxidation, (C) ATP levels, (D) cell viability and (E) GTPase Drp1 activity. Significantly increased levels of H2O2 (P = 0.001) and 4-hydroxy-2-nonenol (P = 0.004), and significantly decreased ATP production (P = 0.001) and cell viability (P = 0.002) were found in the mutant Htt neurons relative to the WT Htt neurons. The level of GTPase Drp1 activity was significantly increased (P = 0.04) in mutant Htt neurons.
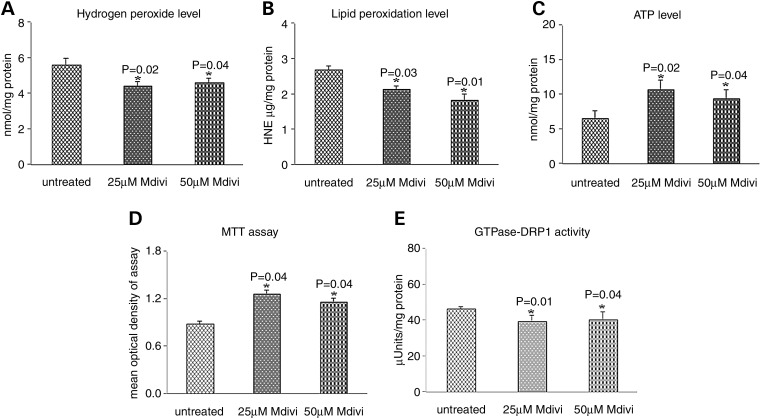
Mitochondria function between Mdivi1-treated and untreated WT Htt neurons
H2O2 production
Significantly decreased levels of H2O2 were found in mitochondria from WT Htt neurons that were treated with Mdivi 1 at concentrations of 25 μm (P = 0.02) and 50 μm (P = 0.04) relative to untreated WT Htt neurons (Fig. 8).
Figure 8.
Mitochondrial functional parameters in Mdivil-treated and untreated WT Htt neurons. Mitochondrial function was assessed in WT Htt neurons by measuring: (A) H2O2 production, (B) lipid peroxidation, (C) ATP levels, (D) cell viability and (E) GTPase Drp1 activity. Significantly decreased levels of the following mitochondrial functional parameters were found in the WT Htt neurons upon treatment with Mdivil: H2O2 with Mdivil at 25 μm (P = 0.02) and 50 μm (P = 0.04) concentrations; 4-hydroxy-2-nonenol with Mdivil at 25 μm (P = 0.01) and 50 μm (P = 0.02) concentrations; and GTPase Drp1 activity with Mdivil at 25 μm (P = 0.01) and 50 μm (P = 0.01) concentrations. In contrast, significantly increased mitochondrial functional parameters were found in the WT Htt neurons upon treatment with Mdivil: ATP at 25 μm (P = 0.02) and 50 μm (P = 0.04) concentrations, and cell viability at 25 μm (P = 0.01) and 50 μm (P = 0.01) concentrations.
Lipid peroxidation
To determine whether Mdivi1 reduces lipid peroxidation in WT Htt neurons treated with Mdivi1, we measured 4-hydroxy-2-nonenol, an indicator of lipid peroxidation. We found significantly decreased levels of lipid peroxidation in the WT Htt neurons treated with Mdivi1 at 25 μm concentration (P = 0.01) and at 50 μm (P = 0.02) relative to the untreated WT Htt neurons (Fig. 8).
ATP production
Significantly increased levels of ATP were found in WT Htt neurons treated with Mdivil at 25 μm concentration (P = 0.02) and at 50 μm (P = 0.04) relative to the untreated WT Htt neurons (Fig. 8).
Cell viability
Cell viability was significantly increased in WT Htt neurons treated with Mdivil at 25 and 50 μm concentrations (both P = 0.01) compared with the untreated WT Htt neurons (Fig. 8).
GTPase Drp1 activity
Significantly decreased levels of GTPase activity were found in WT Htt neurons treated with Mdivil at concentrations of 25 μm (P = 0.01) and 50 μm (P = 0.04) relative to untreated WT Htt neurons (Fig. 8), indicating that Mdivi1 decreases GTPase Drp1 activity in the treated WT Htt neurons.
Mitochondria function between Mdivi1-treated and untreated mutant Htt neurons
H2O2 production
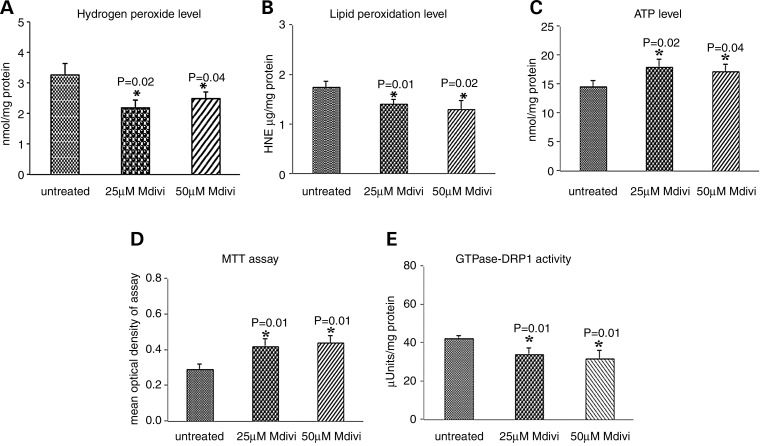
As shown in Figure 9, significantly decreased levels of H2O2 were found in mitochondria from mutant Htt neurons treated with Mdivi1 at concentrations of 25 μm (P = 0.02) and at 50 μm (P = 0.01) relative to untreated mutant Htt neurons, indicating that Mdivi1 decreases H2O2 levels.
Figure 9.
Mitochondrial functional parameters in Mdivil-treated and untreated mutant Htt neurons. Mitochondrial function was assessed by measuring: (A) H2O2 production, (B) lipid peroxidation, (C) ATP levels, (D) cell viability and (E) GTPase Drp1 activity. Significantly decreased levels were found in the following parameters in mutant Htt neurons upon Mdivil treatment: H2O2 with Mdivil at 25 μm (P = 0.02) and 50 μm (P = 0.01) concentrations, and 4-hydroxy-2-nonenol with Mdivil at 25 μm (P = 0.03) and 50 μm (P = 0.01) concentrations. In contrast, significantly increased levels were found in the following parameters upon Mdivil treatment: ATP production at 25 μm (P = 0.02) and 50 μm (P = 0.04) concentrations, cell viability at 25 μm (P = 0.04) and 50 μm (P = 0.04) concentrations and GTPase Drp1 activity at 25 μm (P = 0.01) and 50 μm (P = 0.04) concentrations.
Lipid peroxidation
Significantly decreased levels of 4-hydroxy-nonenol lipid were found in mutant Htt neurons treated with Mdivi1 at concentrations of 25 μm (P = 0.03) and 50 μm (P = 0.01) relative to the untreated mutant neurons, indicating that Mdivi1 reduces lipid peroxidation in the treated mutant Htt neurons (Fig. 9).
ATP production
Significantly increased levels of ATP were found in mutant Htt neurons treated with Mdivil at concentrations of 25 μm (P = 0.02) and at 50 μm (P = 0.04) relative to untreated Htt neurons, indicating that Mdivi1 enhances ATP levels in mutant Htt neurons (Fig. 9).
Cell viability
Cell viability was significantly increased in mutant Htt neurons treated with Mdivil at 25 and 50 μm concentrations (both P = 0.01) compared with untreated mutant Htt neurons, indicating that Mdivi1 enhances cell viability (Fig. 9).
GTPase Drp1 activity
Significantly decreased levels of GTPase activity were found in mutant Htt neurons treated with Mdivil at 25 μm (P = 0.01) and at 50 μm concentrations (P = 0.04) relative to untreated mutant Htt neurons, indicating that Mdivi1 decreases GTPase Drp1 activity in mutant Htt neurons. These findings suggest that Mdivi 1 reduces fission-linked GTPase activity (Fig. 9).
Discussion
Using gene expression analysis, biochemical methods, TEM and confocal microscopy methods, we studied (i) mitochondrial structure by measuring mRNA and the protein levels of mitochondrial and synaptic genes, (ii) mitochondrial function and (iii) ultra-structural changes in mutant Htt neurons relative to WT Htt neurons. We also studied the above-mentioned parameters in Mdivi1-treated and untreated WT Htt and mutant Htt neurons. In mutant Htt neurons, fission genes were up-regulated and fusion genes were down-regulated, suggesting that mutant Htt impairs mitochondrial dynamics in HD neurons. Synaptic genes were reduced, and mitochondrial function was defective and numbers of mitochondria were increased in mutant Htt neurons relative to WT Htt neurons. On the contrary, in mutant Htt neurons treated with Mdivi1, fission genes were down-regulated and fusion genes were up-regulated, suggesting that Mdivi1 may have a role in decreasing fission activity in mutant Htt neurons. In Mdivi1-treated mutant Htt neurons, we also found that synaptic genes were up-regulated and mitochondrial function was normal. The TEM studies revealed that reduced numbers of mitochondria were present in Mdivi1-treated mutant Htt neurons compared with the untreated Htt neurons. Increased synaptic and mitochondrial fusion genes and decreased fission genes were found even in WT neurons treated with Mdivi1, indicating that Mdivi1 may also beneficially affect healthy Htt neurons. These findings suggest that Mdivi1 is protective against mutant Htt-induced mitochondrial and synaptic damage in HD neurons.
mRNA levels
Excessive mitochondrial fragmentation and reduced fusion, defective mitochondrial function, impaired axonal transport and synaptic damage have been extensively reported in HD neurons (23,24,26,38,39). In the current study, our results (Table 1) further confirmed the presence of abnormal mitochondrial dynamics, reduced synaptic activity, increased mRNA levels of fission genes and reduced mRNA levels of fusion genes and synaptic genes in mutant Htt neurons relative to WT Htt neurons.
The current study examined Mdivi1 to determine any beneficial effects, particularly reduced mitochondrial fission, in striatal neurons that express expanded polyQ (HDhQ111/111) repeats. Using real-time RT-PCR analysis, we measured mRNA levels of mitochondrial fission, fusion, ETC and synaptic genes in WT Htt and mutant Htt neurons that we treated with Mdivil at concentrations of 25 and 50 μm concentrations (Tables 2 and 3). As expected, we found increased mRNA levels of fission genes and decreased levels of fusion genes in the Mdivil-treated mutant Htt neurons, strongly suggesting that Mdivi1 reduces fission activity and enhances fusion activity. Interestingly, similar results were found in WT Htt neurons (Table 2), indicating that Mdivi1 reduces fission activity in neurons with normal polyQ repeats.
mRNA levels in ETC genes were decreased in mutant Htt neurons treated with Mdivil at the higher 50 μm concentration, indicating that 25 μm may reduce ETC activity. However, mRNA levels in ETC genes were unchanged when WT Htt neurons were treated with Mdivil at a concentration of 25 μm. Overall, Mdivil at a 25 μm concentration decreased ETC activity in both WT and mutant Htt neurons and reduced OXPHOS activity, which may be beneficial, particularly for mutant Htt neurons.
Excessive mitochondrial fragmentation, reduced mitochondrial fusion, defective axonal mitochondrial transport, defective mitochondrial function and defective biogenesis have been extensively reported in such neurodegenerative diseases such as Alzheimer's (40–43), Huntington's (23,24), Parkinson's (44) and ALS (45). These mitochondrial abnormalities are precursors for synaptic degeneration and neuronal dysfunction that are known to occur in neurodegenerative diseases. In the current study, the reduced excessive fragmentation that we found in Mdivi1-treated HD neurons implicates Mdivi1 as a possible treatment in other neurodegenerative diseases, not only HD.
The significantly increased mRNA levels of synaptic genes that we observed in the Mdivi1-treated mutant Htt neurons indicates that reduced mitochondrial fission activity may also apply to synaptic gene expressions and synaptic activity in HD neurons, suggesting that Mdivi1 treatment may be beneficial in terms of reducing fission activity and enhancing fusion machinery or neurons that express normal polyQ repeats.
Protein levels
In the current study, Drp1 and Fis1 proteins were increased and Mfn1, Mfn2 and Opa1 proteins were decreased in mutant Htt neurons, confirming the presence of abnormal mitochondrial dynamics in mutant Htt neurons (Fig. 1A and B). On the contrary, fission proteins were decreased and fusion proteins were increased in mutant Htt neurons treated with Mdivil, suggesting that both mRNA and protein levels are altered via Mdivi1. In the present study, these changes were found to be dose dependent. Further, the matrix protein CypD was also decreased in a dose-dependent manner in the Mdivi1-treated mutant and WT Htt neurons, indicating that Mdivi1 reduces fission activity in Htt neurons, irrespective of the polyQ length. Our quantitative immunoblotting and immunofluorescence findings revealed that synaptic proteins increased not only in Mdivi1-treated mutant Htt neurons but also in WT Htt neurons, indicating that Mdivi1 alters synaptic proteins and enhances synaptic activity. However, the effect of Mdivi1 appears to be exceptionally high in mutant Htt neurons expressing an expanded polyQ (111) repeats than WT Htt neurons. These observations also suggest that Mdivi1 is a promising drug to treat HD neurons with increased numbers of polyQ repeats.
Transmission electron microscopy
Recent ultra-structural studies revealed increased mitochondrial fragmentation and structurally damaged mitochondria in neurons and lymphoblasts from HD patients, in primary neurons from HD mice (23,24,26–28), and now, in the current study, in striatal mutant Htt neurons from HD knockin mice. The increased numbers of mitochondria in mutant Htt neurons (Fig. 6A) in the present study were likely due to Drp1-mutant Htt-linked impaired balance. The changes in Drp1 levels may be responsible for synaptic damage in HD-affected neurons and for chorea and involuntary movements found in HD patients.
In the current study, Mdivi1 at both 25 and 50 μm concentrations decreased Drp1 in mutant Htt neurons, and mutant Htt was reported to promote excessive mitochondrial fragmentation. Further, the Mdivi1-treated mutant Htt neurons exhibited a more intact mitochondrial structure, suggesting that the Mdivi1 balances fission and fusion machinery. These findings agree with real-time RT-PCR and protein data of fission and fusion genes, mRNA findings of ETC genes and mitochondrial functional data (current study). In studies of TEM, elongated mitochondria were found in neuronal processes and terminals of Mdivi1-treated mutant Htt neurons relative to untreated mutant Htt neurons (data not shown), indicating that healthy mitochondria travel all along neuronal processes in mutant Htt neurons to supply ATP at nerve terminals. These observations suggest that Mdivi1 may be a promising molecule to study in terms of its efficacy in treating neuronal damage associated with HD in mice and HD patients.
Mitochondrial function
Mitochondrial function, mitochondrial bioenergetics and neuronal viability are key parameters in assessing neuronal function in patients with HD since they are all known to be defective in HD-affected neurons (17–20,23,24,26). In the current study, we also found mitochondrial function defective (Fig. 8) in mutant Htt neurons relative to WT Htt neurons.
In determining the effects of Mdivi1 on mitochondrial function on mutant and WT Htt neurons treated with Mdivi1 at 25 and 50 μm concentrations, we found that mutant Htt neurons exhibited less abnormal mitochondrial function than did untreated mutant Htt neurons, in terms of decreasing free radicals and lipid peroxidation and in terms of increasing ATP production and neuronal viability. Further, the levels of GTPase Drp1 enzymatic activity, an indicator of mitochondrial division (23), were significantly decreased in mutant Htt neurons treated with Mdivi1, strongly suggesting that Mdivi1 alters (i) these parameters, all of which are known to be critical for neuronal function in HD neurons (23) and (ii) mitochondrial dynamics (in particular, the fission–fusion balance), mitochondrial structure and function, neuronal viability and synaptic activity. Thus, all of our data in this study point to Mdivi1 as protecting HD neurons from mutant Htt-induced mitochondrial and synaptic toxicities.
This is the first study to report positive, potentially therapeutic effects of Mdivi1 on mutant Htt neurons. These findings warrant additional studies to investigate Mdivi1 as a potential drug capable of preventing or reducing the effects of HD on HD patients.
Materials and Methods
Cell cultures and Mdivi1 treatments
Immortalized striatal progenitor neurons expressing endogenous WT Htt (STHdhQ7/Q7) and the homozygous mutant Htt (STHdh Q111/Q111) were used in the current study. Cell lines were prepared from WT mice and homozygous HdhQ111/Q111 knock-in mice (46,47). Cells were grown at 37°C in Dulbecco's modified Eagle's medium (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum, 1% non-essential amino acids, 2 mm l-glutamine and 400 μg/ml G418 (Geneticin, Invitrogen, Carlsbad, CA, USA). They were treated with Mdivil at concentrations of 25 and 50 μm (Sigma-Aldrich, CA, USA) for 24 h and then were harvested. Pellets were prepared for RNA and protein studies.
Quantitative real-time RT-PCR
Using the reagent TRIzol (Invitrogen), total RNA was isolated from three independent Mdivi1 treatments of WT (n = 3) and mutant Htt neurons (n = 3) from control, untreated WT (n = 3) and mutant Htt (n = 3) neurons. Using primer express Software (Applied Biosystems, Carlsbad, CA, USA), we designed the oligonucleotide primers for the housekeeping genes β-actin, GAPDH, mitochondrial structural genes, fission genes (Drp1, Fis1), fusion genes (MFN1, MFN2, Opa1, VDAC1), the mitochondrial matrix protein CypD, mitochondrial-encoded ETC genes (Complex I—subunits 1, 3 and 6; Complex III—CytB; Complex IV—Cox1–3; and Complex V—ATPase), synaptic genes, synaptophysin, PSD95, synapsins1–2, synaptobrevins1–2, neurogranin, GPA43 and synaptopodin. The primer sequences and amplicon sizes are listed in Table 3. Using SYBR-Green chemistry-based quantitative real-time RT-PCR, we measured mRNA expression of the above-mentioned genes, as described by Manczak et al. (48).
The mRNA transcript level was normalized against β-actin and the GAPDH at each dilution. The standard curve was the normalized mRNA transcript level, plotted against the log-value of the input cDNA concentration at each dilution. To compare β-actin, GAPDH and neuroprotective markers, relative quantification was performed according to the CT method (Applied Biosystems). Briefly, the comparative CT method involved averaging triplicate samples, which were taken as the CT values for β-actin, GAPDH and neuroprotective markers. β-Actin normalization was used in the present study, because the β-actin CT values were similar for the Htt neurons treated with Mdivi1 and the untreated Htt neurons, for the mitochondrial ETC genes, mitochondrial structural genes and the synaptic genes. The ΔCT-value was obtained by subtracting the average β-actin CT value from the average CT-value of the synaptic mitochondrial ETC genes and the mitochondrial structural genes. The ΔCT of untreated Htt neurons was used as the calibrator. The fold change was calculated according to the formula 2−(Δ ΔCT), where ΔΔCT is the difference between ΔCT and the ΔCT calibrator value. To determine the statistical significance of mRNA expression, the CT value differences between the untreated WT and mutant Htt neurons, and the Mdivi1-treated WT and mutant Htt neurons were used in relation to β-actin normalization. Statistical significance was calculated using one-way ANOVA.
Immunoblotting analysis
To determine whether Mdivi1 alters the protein levels of mitochondrial structural, ETC and synaptic genes that showed altered mRNA expression, we performed immunoblotting analyses of protein lysates from Mdivil-treated and untreated WT and mutant Htt neurons as described in Manczak et al. (48). Twenty micrograms of protein lysates from the Mdivil-treated and untreated WT Htt and mutant Htt neurons was resolved on a 4–12% Nu-PAGE gel (Invitrogen). The resolved proteins were transferred to nylon membranes (Novax Inc., San Diego, CA, USA) and were then incubated for 1 h at room temperature with a blocking buffer (5% dry milk dissolved in a TBST buffer). The nylon membranes were incubated overnight with the primary antibodies shown in Table 5. The membranes were washed with a TBST buffer 3 times at 10-min intervals and were then incubated for 2 h with appropriate secondary antibodies, followed by three additional washes at 10-min intervals. Mitochondrial and synaptic proteins were detected with chemiluminescence reagents (Pierce Biotechnology, Rockford, IL, USA), and the bands from immunoblots were quantified on a Kodak Scanner (ID Image Analysis Software, Kodak Digital Science, Kennesaw, GA, USA). Briefly, image analysis was used to analyze gel images captured with a Kodak Digital Science CD camera. The lanes were marked to define the positions and specific regions of the bands. An ID fine-band command was used to locate and to scan the bands in each lane and to record the readings.
Table 5.
Summary of antibody dilutions and conditions used in immunoblotting analysis of mitochondrial dynamics and synaptic proteins in WT and mutant Htt neurons
| Marker | Primary antibody: species and dilution | Purchased from company, city and state | Secondary antibody, dilution | Purchased from company, city and state |
|---|---|---|---|---|
| Drp1 | Rabbit Polyclonal 1:500 | Novus Biological, Littleton, CO | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare, Amersham, Piscataway, NJ |
| Fis1 | Rabbit Polyclonal 1:500 | Protein Tech Group, Inc., Chicago, IL | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare, Amersham, Piscataway, NJ |
| Mfn1 | Rabbit Polyclonal 1:400 | Abcam, Cambridge, MA | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare, Amersham, Piscataway, NJ |
| Mfn2 | Rabbit Polyclonal 1:400 | Abcam, Cambridge, MA | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare, Amersham, Piscataway, NJ |
| Opa1 | Mouse Monoclonal 1:500 | BD Biosciences, San Jose, CA | Sheep anti-mouse HRP 1:10 000 | GE Healthcare, Amersham, Piscataway, NJ |
| CypD | Mouse Monoclonal 1:500 | EMD, Calobiochem Chemicals, Inc., Gibbstown, NJ | Sheep anti-mouse HRP 1:10 000 | GE Healthcare, Amersham, Piscataway, NJ |
| SYN | Rabbit Polyclonal 1:400 | Thermo Fisher Scientific, Inc., Rockford, IL | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare, Amersham, Piscataway, NJ |
| PSD95 | Rabbit Monoclonal 1:300 | Abcam, Cambridge, MA | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare, Amersham, Piscataway, NJ |
| DARPP-32 | Rabbit Polyclonal 1:200 | Santa Cruz Biotechnology, Inc., Dallas, TX | Donkey anti-rabbit HRP 1:10 000 | GE Healthcare, Amersham, Piscataway, NJ |
| B-actin | Mouse Monoclonal 1:500 | Sigma-Aldrich, St Luis, MO | Sheep anti-mouse HRP 1:10 000 | GE Healthcare, Amersham, Piscataway, NJ |
Immunofluorescence analysis and quantification
Immunofluorescence analysis was performed with the Mdivil-treated and untreated WT and mutant Htt neurons as described in Manczak et al (48). The Htt neurons were washed with warm PBS, fixed in freshly prepared 4% paraformaldehyde in PBS for 10 min and then washed with PBS and permeabilized with 0.1% Triton-X100 in PBS. They were blocked with a 1% blocking solution (Invitrogen) for 1 h at room temperature. All neurons were incubated overnight with primary antibodies (see Table 6). After incubation, the neurons were washed 3 times with PBS, for 10 min each. The neurons were incubated with a secondary antibody conjugated with Fluors 488 and 599 (Invitrogen) for 1 h at room temperature. The neurons were washed 3 times with PBS and mounted on slides. Photographs were taken with a multiphoton laser scanning microscope system (ZeissMeta LSM510). To quantify the immunoreactivity of mitochondrial and synaptic antibodies for each treatment, 10–15 photographs were taken at ×40 magnification. The signal intensity indicating the immunoreactivity of the cell body and neurite length was quantified for several randomly selected images, and statistical significance was assessed, using one-way ANOVA for mitochondrial and synaptic proteins.
Table 6.
Summary of antibody dilutions and conditions used in immunohistochemistry/immunofluorescence analysis of mitochondrial dynamics and synaptic proteins in WT and mutant Htt neurons
| Markers | Primary antibody: species and dilution | Purchased from company, city and state | Secondary antibody, dilution, Alexa dye | Purchased from company, city and state |
|---|---|---|---|---|
| Drp1 | Rabbit Polyclonal 1:300 | Novus Biological, Littleton, CO | Goat anti-rabbit Biotin 1:400, HRP-Streptavidin (1:200), TSA-Alexa488 |
KPL, Gaithersburg, MD VECTOR Laboratories, Inc. Burlingame, CA Molecular Probes, Grand Island, NY |
| Fis1 | Rabbit Polyclonal 1:300 | Protein Tech Group, Inc., Chicago, IL | Goat anti-rabbit Biotin 1:400, HRP-Streptavidin (1:200), TSA-Alexa488 |
KPL, Gaithersburg, MD VECTOR Laboratories, Inc. Burlingame, CA Molecular Probes, Grand Island, NY |
| Mfn1 | Rabbit Polyclonal 1:300 | Abcam, Cambridge, MA | Goat anti-rabbit Biotin 1:400, HRP-Streptavidin (1: 200), TSA-Alexa594 |
KPL, Gaithersburg, MD VECTOR Laboratories, Inc. Burlingame, CA Molecular Probes, Grand Island, NY |
| CypD | Mouse Monoclonal 1:500 | EMD, Calobiochem Chemicals, Inc., Gibbstown, NJ | Goat anti-mouse Biotin 1:400, HRP-Streptavidin (1:200), TSA-Alexa594 | KPL, Gaithersburg, MD VECTOR Laboratories, Inc., Burlingame, CA Molecular Probes, Grand Island, NY |
| Cyt B | Rabbit Polyclonal 1:500 | Santa Cruz Biotechnology, Inc., Dallas, TX | Goat anti-mouse Biotin 1:400, HRP-Streptavidin (1:200), TSA-Alexa594 | KPL, Gaithersburg, MD VECTOR Laboratories, Inc., Burlingame, CA Molecular Probe, Grand Island, NY |
| SYN | Rabbit Polyclonal 1:400 | Thermo Fisher Scientific, Inc., Rockford, IL | Goat anti-rabbit Biotin 1:400, HRP-Streptavidin (1:200), TSA-Alexa594 | KPL, Gaithersburg, MD VECTOR Laboratories, Inc., Burlingame, CA Molecular Probes, Grand Island, NY |
| PSD95 | Rabbit Monoclonal 1:300 | Abcam, Cambridge, MA | Goat anti-rabbit Biotin 1:400, HRP-Streptavidin (1:200), TSA-Alexa594 | KPL, Gaithersburg, MD VECTOR Laboratories, Inc., Burlingame, CA Molecular Probes, Grand Island, NY |
| MAP2 | Rabbit Polyclonal 1:600 | Santa Cruz Biotechnology, Inc., Dallas, TX | Goat anti-rabbit Biotin 1:400, HRP-Streptavidin (1:200), TSA-Alexa488 | KPL, Gaithersburg, MD VECTOR Laboratories, Inc., Burlingame, CA Molecular Probe, Grand Island, NY |
Transmission electron microscopy
To determine the effects of Mdivi1 on the numbers of mitochondria and any rescue effects of Mdivi1 on mitochondria in the mutant Htt neurons, we used TEM on WT Htt (n = 4) and mutant Htt (n = 4) treated and untreated with Mdivi1.
Mdivil-treated and untreated Htt neurons were fixed in 100 μm sodium cacodylate (pH 7.2), 2.5% glutaraldehyde, 1.6% paraformaldehyde, 0.064% picric acid and 0.1% ruthenium red. They were gently washed and post-fixed for 1 h in 1% osmium tetroxide plus 08% potassium ferricyanide, in 100 mm sodium cacodylate, pH 7.2. After a thorough rinsing in water, the Htt neurons were dehydrated, infiltrated overnight in 1:1 acetone:Epon 812 and infiltrated for 1 h with 100% Epon 812 resin. They were then embedded in the resin. After polymerization, 60 to 80 nm thin sections were cut on a Reichert ultramicrotome and stained for 5 min in lead citrate. They were rinsed and post-stained for 30 min in uranyl acetate and then were rinsed again and dried. Electron microscopy was performed at 60 kV on a Philips Morgagne TEM equipped with a CCD, and images were collected at magnifications of ×1000–37 000. The numbers of mitochondria were counted in the Mdivi1-treated WT and mutant Htt neurons and Mdivi-untreated WT and mutant Htt neurons, and statistical significance was determined, using one-way ANOVA.
Mitochondrial functional assays
H2O2 production
Using an Amplex® Red H2O2 Assay Kit (Molecular Probes, Eugene, OR, USA), the production of H2O2 was measured in independent experiments (n = 4) of Mdivil-treated and untreated mutant and WT Htt neurons, as described in Manczak et al. (48). Briefly, H2O2 production was measured in the mitochondria isolated from the Mdivil-treated and untreated mutant and WT Htt neurons. A BCA Protein Assay Kit (Pierce Biotechnology) was used to estimate protein concentration. The reaction mixture contained mitochondrial proteins (μg/μl), Amplex Red reagents (50 μm), horseradish peroxidase (0.1 U/ml) and a reaction buffer (1X). The mixture was incubated at room temperature for 30 min, followed by spectrophotometer readings of fluorescence (570 nm). Finally, H2O2 production was determined, using a standard curve equation expressed in nmol/μg mitochondrial protein.
Lipid peroxidation assay
Lipid peroxidates are unstable indicators of oxidative stress in neurons (49). The final product of lipid peroxidation is 4-hydroxy-2-nonenol (HNE), which was measured in the cell lysates from Mdivil-treated (n = 4) and untreated Htt (n = 4) WT and mutant Htt neurons, using an HNE-His ELISA Kit (Cell BioLabs, Inc., San Diego, CA, USA). Briefly, freshly prepared protein was added to a 96-well protein binding plate and incubated overnight at 4°C. It was then washed 3 times with a buffer. After the last wash, the anti-HNE-His antibody was added to the protein in the wells, which was then incubated for 2 h at room temperature and was washed again 3 times. Next, the samples were incubated with a secondary antibody conjugated with peroxidase for 2 h at room temperature, followed by incubation with an enzyme substrate. Optical density was measured (at 450 nm) to quantify the level of HNE.
ATP levels
ATP levels were measured in mitochondria isolated from Mdivi1-treated (n = 4) and untreated (n = 4) WT and mutant Htt neurons, using an ATP determination kit (Molecular Probes). The bioluminescence assay is based on the reaction of ATP with recombinant firefly luciferase and its substract luciferin. Luciferase catalyzes the formation of light from ATP and luciferin. It is the emitted light that is linearly related to the ATP concentration, which is measured with a luminometer. We measured ATP from mitochondrial pellets using a standard curve method.
Cell viability test (MTT assay). Mitochondrial respiration, an indicator of cell viability, was assessed in the Mdivil-treated (n = 4) and untreated (n = 4) WT and mutant Htt neurons, using the mitochondria-dependent reduction of 3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) to formazan. Briefly, in this reduction, the Htt neurons were seeded in 12-well plates at a density of ∼105 neurons per well. After treatment, MTT (5 mg/ml in PBS) was added to the plates, and the neurons (control and experimental) were incubated for 3 h. The medium was then replaced with an SDS/DMSO lysis buffer, and MTT absorption was measured at 570 nm. Results were expressed as the percentage of MTT reduction, assuming that the absorbance of untreated Htt neurons was 100%.
GTPase Drp1 enzymatic activity
Using a calorimetric kit (Novus Biologicals, Littleton, CO, USA), GTPase enzymatic activity was measured in Mdivil-treated (n = 4) and untreated (n = 4) mutant and WT Htt neurons, following GTPase assay methods described in Shirendeb et al.,23 based on GTP hydrolyzing to GDP and to inorganic Pi. GTPase activity was measured, based on the amount of Pi that the GTP produces. By adding the ColorLock Gold (orange) substrate to the Pi generated from GTP, we assessed GTP activity, based on the inorganic complex solution (green). Colorimetric measurements (green) were read in the wavelength range of 650 nm. GTPase activity in the Mdivil-treated and untreated WT and mutant Htt neurons was compared with untreated Htt neurons.
Funding
This research was supported by NIH grants AG042178 and AG047812, and the Garrison Family Foundation.
Acknowledgements
We thank Coriell cell repositories of the Coriell Institute for Medical Research for the HDhQ7/Q7 and HDhQ111/Q111 cell lines.
Conflict of Interest statement. None declared.
References
- 1.Vonsattel J.P., Myers R.H., Stevens T.J., Ferrante R.J., Bird E.D., Richardson E.P. Jr (1985) Neuropathological classification of Huntington's disease. J. Neuropathol. Exp. Neurol., 44, 559–577. [DOI] [PubMed] [Google Scholar]
- 2.Folstein S.E. (1990) Huntington's Disease. Johns Hopkins University Press, Baltimore. [Google Scholar]
- 3.Bates G.P. (2005) History of genetic disease: the molecular genetics of Huntington disease - a history. Nat. Rev. Genet., 6, 766–773. [DOI] [PubMed] [Google Scholar]
- 4.Spargo E., Everall I.P., Lantos P.L. (1993) Neuronal loss in the hippocampus in Huntington's disease: a comparison with HIV infection. J. Neurol. Neurosurg. Psychiatry, 56, 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Politis M., Pavese N., Tai Y.F., Tabrizi S.J., Barker R.A., Piccini P. (2008) Hypothalamic involvement in Huntington's disease: an in vivo PET study. Brain, 131, 2860–2869. [DOI] [PubMed] [Google Scholar]
- 6.Soneson C., Fontes M., Zhou Y., Denisov V., Paulsen J.S., Kirik D., Petersén A; Huntington Study Group PREDICT-HD investigators (2010) Early changes in the hypothalamic region in prodromal Huntington disease revealed by MRI analysis. Neurobiol. Dis., 40, 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reddy P.H., Mao P., Manczak M. (2009) Mitochondrial structural and functional dynamics in Huntington's disease. Brain Res. Rev., 61, 33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reddy P.H., Williams M., Tagle D.A. (1999) Recent advances in understanding the pathogenesis of Huntington's disease. Trends Neurosci., 22, 248–255. [DOI] [PubMed] [Google Scholar]
- 9.Li S., Li X.J. (2006) Multiple pathways contribute to the pathogenesis of Huntington disease. Mol. Neurodegener., 1, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kegel K.B., Meloni A.R., Yi Y., Kim Y.J., Doyle E., Cuiffo B.G., Sapp E., Wang Y., Qin Z.H., Chen J.D. et al. (2002) Huntingtin is present in the nucleus, interacts with the transcriptional corepressor Cterminal binding protein, and represses transcription. J. Biol. Chem., 277, 7466–7476. [DOI] [PubMed] [Google Scholar]
- 11.Kegel K.B., Sapp E., Yoder J., Cuiffo B., Sobin L., Kim Y.J., Qin Z.H., Hayden M.R., Aronin N., Scott D.L. et al. (2005) Huntingtin associates with acidic phospholipids at the plasma membrane. J. Biol. Chem., 280, 36464–36473. [DOI] [PubMed] [Google Scholar]
- 12.Choo Y.S., Johnson G.V., MacDonald M., Detloff P.J., Lesort M. (2004) Mutant huntingtin directly increases susceptibility of mitochondria to the calcium-induced permeability transition and cytochrome c release. Hum. Mol. Genet., 13, 1407–1420. [DOI] [PubMed] [Google Scholar]
- 13.Truant R., Atwal R., Burtnik A. (2007) Hypothesis: Huntingtin may function in membrane association and vesicular trafficking. Biochem. Cell Biol., 84, 912–917. [DOI] [PubMed] [Google Scholar]
- 14.Atwal R.S., Xia J., Pinchev D., Taylor J., Epand R.M., Truant R. (2007) Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum. Mol. Genet., 16, 2600–2615. [DOI] [PubMed] [Google Scholar]
- 15.Orr A.L., Li S., Wang C.E., Li H., Wang J., Rong J., Xu X., Mastroberardino P.G., Greenamyre J.T., Li X.J. (2008) N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J. Neurosci., 28, 2783–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borrell-Pagès M., Zala D., Humbert S., Saudou F. (2006) Huntington's disease: from huntingtin function and dysfunction to therapeutic strategies. Cell Mol. Life Sci., 63, 2642–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Browne S.E., Bowling A.C., MacGarvey U., Baik M.J., Berger S.C., Muqit M.M., Bird E.D., Beal M.F. (1997) Oxidative damage and metabolic dysfunction in Huntington's disease: selective vulnerability of the basal ganglia. Ann. Neurol., 41, 646–653. [DOI] [PubMed] [Google Scholar]
- 18.Tabrizin S.J., Cleeter M.W., Xuereb J., Taanman J.W., Cooper J.M., Schapira A.H. (1999) Biochemical abnormalities and excitotoxicity in Huntington's disease brain. Ann. Neurol., 45, 25–32. [DOI] [PubMed] [Google Scholar]
- 19.Pandey M., Varghese M., Sindhu K.M., Sreetama S., Navneet A.K., Mohanakumar K.P., Usha R. (2008) Mitochondrial NAD+-linked State 3 respiration and complex-I activity are compromised in the cerebral cortex of 3- nitropropionic acid-induced rat model of Huntington's disease. J. Neurochem., 104, 420–434. [DOI] [PubMed] [Google Scholar]
- 20.Seong I.S., Ivanova E., Lee J.M., Choo Y.S., Fossale E., Anderson M., Gusella J.F., Laramie J.M., Myers R.H., Lesort M. et al. (2005) HD CAG repeat implicates a dominant property of huntingtin in mitochondrial energy metabolism. Hum. Mol. Genet., 14, 2871–2880. [DOI] [PubMed] [Google Scholar]
- 21.Panov A.V., Lund S., Greenamyre J.T. (2005) Ca2+-induced permeability transition in human lymphoblastoid cell mitochondria from normal and Huntington's disease individuals. Mol. Cell Biochem., 269, 143–152. [DOI] [PubMed] [Google Scholar]
- 22.Trushina E., Dyer R.B., Badger J.D. 2nd, Ure D., Eide L., Tran D.D., Vrieze B.T., Legendre-Guillemin V., McPherson P.S., Mandavilli B.S. et al. (2004) Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol. Cell Biol., 24, 8195–8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shirendeb U., Calkins M., Manczak M., Dufour B., McBride J., Mao P., Reddy P.H. (2012) Mutant huntingtin's association with mitochondria protein Drp1, and impaired axonal transport of mitochondria in Huntington's disease neurons. Hum. Mol. Genet., 21, 406–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song W., Chen J., Petrilli A., Liot G., Klinglmayr E., Zhou Y., Poquiz P., Tjong J., Pouladi M.A., Hayden M.R. et al. (2011) Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat. Med., 17, 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang D.T., Rintoul G.L., Pandipati S., Reynolds I.J. (2006) Mutant huntingtin aggregates impair mitochondrial movement and trafficking in cortical neurons. Neurobiol. Dis., 22, 388–400. [DOI] [PubMed] [Google Scholar]
- 26.Shirendeb U., Reddy A.P., Manczak M., Calkins M.J., Mao P., Tagle D.A., Reddy P.H. (2011) Abnormal mitochondrial dynamics, mitochondrial loss and mutant huntingtin oligomers in Huntington's disease: implications for selective neuronal damage. Hum. Mol. Genet., 20, 1438–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Costa V., Giacomello M., Hudec R., Lopreiato R., Ermak G., Lim D., Malorni W., Davies K.J., Carafoli E., Scorrano L. (2010) Mitochondrial fission and cristae disruption increase the response of cell models of Huntington's disease to apoptotic stimuli. EMBO Mol. Med., 2, 490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo X., Disatnik M.H., Monbureau M., Shamloo M., Mochly-Rosen D., Qi X. (2013) Inhibition of mitochondrial fragmentation diminishes Huntington's disease-associated neurodegeneration. J. Clin. Invest., 123, 5371–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cassidy-Stone A., Chipuk J.E., Ingerman E., Song C., Yoo C., Kuwana T., Kurth M.J., Shaw J.T., Hinshaw J.E., Green D.R. et al. (2008) Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev. Cell., 14, 193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macia E., Ehrlich M., Massol R., Boucrot E., Brunner C., Kirchhausen T. (2006) Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell., 10, 839–850. [DOI] [PubMed] [Google Scholar]
- 31.Qi X., Qvit N., Su Y.C., Mochly-Rosen D. (2013) A novel Drp1 inhibitor diminishes aberrant mitochondrial fission and neurotoxicity. J. Cell Sci., 126, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie N., Wang C., Lian Y., Zhang H., Wu C., Zhang Q. (2013) A selective inhibitor of Drp1, mdivi-1, protects against cell death of hippocampal neurons in pilocarpine-induced seizures in rats. Neurosci. Lett., 545, 64–68. [DOI] [PubMed] [Google Scholar]
- 33.Qiu X., Cao L., Yang X., Zhao X., Liu X., Han Y., Xue Y., Jiang H., Chi Z. (2013) Role of mitochondrial fission in neuronal injury in pilocarpine-induced epileptic rats. Neuroscience., 245, 157–165. [DOI] [PubMed] [Google Scholar]
- 34.Park S.W., Kim K.Y., Lindsey J.D., Dai Y., Heo H., Nguyen D.H., Ellisman M.H., Weinreb R.N., Ju W.K. (2011) A selective inhibitor of drp1, mdivi-1, increases retinal ganglion cell survival in acute ischemic mouse retina. Invest. Ophthalmol. Vis. Sci., 52, 2837–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X., Yan H., Yuan Y., Gao J., Shen Z., Cheng Y., Shen Y., Wang R.R., Wang X., Hu W.W., Wang G., Chen Z. (2013) Cerebral ischemia-reperfusion-induced autophagy protects against neuronal injury by mitochondrial clearance. Autophagy, 9, 1321–1333. [DOI] [PubMed] [Google Scholar]
- 36.Wappler E.A., Institoris A., Dutta S., Katakam P.V., Busija D.W. (2013) Mitochondrial dynamics associated with oxygen-glucose deprivation in rat primary neuronal cultures. PLoS One, 8, e63206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chlystun M., Campanella M., Law A.L., Duchen M.R., Fatimathas L., Levine T.P., Gerke V., Moss S.E. (2013) Regulation of mitochondrial morphogenesis by annexin A6. PLoS One, 8, e53774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J., Moody J.P., Edgerly C.K., Bordiuk O.L., Cormier K., Smith K., Beal M.F., Ferrante R.J. (2010) Mitochondrial loss, dysfunction and altered dynamics in Huntington's disease. Hum. Mol. Genet., 19, 3919–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H., Lim P.J., Karbowski M., Monteiro M.J. (2009) Effects of overexpression of huntingtin proteins on mitochondrial integrity. Hum. Mol. Genet., 18, 737–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manczak M., Calkins M.J., Reddy P.H. (2011) Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer's disease: implications for neuronal damage. Hum. Mol. Genet., 20, 2495–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calkins M.J., Manczak M., Mao P., Shirendeb U., Reddy P.H. (2011) Impaired mitochondrial biogenesis, defective axonal transport of mitochondria, abnormal mitochondrial dynamics and synaptic degeneration in a mouse model of Alzheimer's disease. Hum. Mol. Genet., 20, 4515–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calkins M.J., Reddy P.H. (2011) Amyloid beta impairs mitochondrial anterograde transport and degenerates synapses in Alzheimer's disease neurons. Biochim. Biophys. Acta, 1812, 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X., Su B., Lee H.G., Li X., Perry G., Smith M.A., Zhu X. (2009) Impaired balance of mitochondrial fission and fusion in Alzheimer's disease. J. Neurosci., 29, 9090–9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X., Yan M.H., Fujioka H., Liu J., Wilson-Delfosse A., Chen S.G., Perry G., Casadesus G., Zhu X. (2012) LRRK2 regulates mitochondrial dynamics and function through direct interaction with DLP1. Hum. Mol. Genet., 21, 1931–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magrané J., Cortez C., Gan W.B., Manfredi G. (2014) Abnormal mitochondrial transport and morphology are common pathological denominators in SOD1 and TDP43 ALS mouse models. Hum. Mol. Genet., 23, 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trettel F., Rigamonti D., Hilditch-Maguire P., Wheeler V.C., Sharp A.H., Persichetti F., Cattaneo E., MacDonald M.E. (2000) Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum. Mol. Genet., 9, 2799–2809. [DOI] [PubMed] [Google Scholar]
- 47.Wheeler V.C., White J.K., Gutekunst C.A., Vrbanac V., Weaver M., Li X.J., Li S.H., Yi H., Vonsattel J.P., Gusella J.F. et al. (2000) Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in HdhQ92 and HdhQ111 knock-in mice. Hum. Mol. Genet., 9, 503–513. [DOI] [PubMed] [Google Scholar]
- 48.Manczak M., Mao P., Calkins M.J., Cornea A., Reddy A.P., Murphy M.P., Szeto H.H., Park B., Reddy P.H. (2010) Mitochondria-targeted antioxidants protect against amyloid-beta toxicity in Alzheimer's diseaseneurons. J. Alzheimers Dis., 20(Suppl. 2), S609–S631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hall E.D., Bosken J.M. (2009) Measurement of oxygen radicals and lipid peroxidation in neural tissues. Curr. Protoc. Neurosci., Chapter 7:Unit 7.17.1-51. [DOI] [PubMed] [Google Scholar]