Abstract
Null mutations in genes involved in V(D)J recombination cause a block in B- and T-cell development, clinically presenting as severe combined immunodeficiency (SCID). Hypomorphic mutations in the non-homologous end-joining gene DCLRE1C (encoding ARTEMIS) have been described to cause atypical SCID, Omenn syndrome, Hyper IgM syndrome and inflammatory bowel disease—all with severely impaired T-cell immunity. By whole-exome sequencing, we investigated the molecular defect in a consanguineous family with three children clinically diagnosed with antibody deficiency. We identified perfectly segregating homozygous variants in DCLRE1C in three index patients with recurrent respiratory tract infections, very low B-cell numbers and serum IgA levels. In patients, decreased colony survival after irradiation, impaired proliferative response and reduced counts of naïve T cells were observed in addition to a restricted T-cell receptor repertoire, increased palindromic nucleotides in the complementarity determining regions 3 and long stretches of microhomology at switch junctions. Defective V(D)J recombination was complemented by wild-type ARTEMIS protein in vitro. Subsequently, homozygous or compound heterozygous DCLRE1C mutations were identified in nine patients from the same geographic region. We demonstrate that DCLRE1C mutations can cause a phenotype presenting as only antibody deficiency. This novel association broadens the clinical spectrum associated with ARTEMIS mutations. Clinicians should consider the possibility that an immunodeficiency with a clinically mild initial presentation could be a combined immunodeficiency, so as to provide appropriate care for affected patients.
Introduction
Severe combined immunodeficiency (SCID) is a rare disorder presenting in infancy with life-threatening infections (bacterial, viral or fungal), failure to thrive and diarrhea (1). SCID can be caused by mutations in various genes, predominantly affecting T-cell immunity. In SCID, T-cell activation and function are impaired, or T-cell development is hampered causing low or absent peripheral T cells. Distinct genetic forms of SCID can be subdivided into T-B+, T-B− or T+B+ SCID, depending on the presence/absence of the respective cell line (2,3).
Among the genetic defects that cause T-B− SCID are biallelic mutations in DCLRE1C, initially identified in a subset of T-B− SCID patients with increased radiosensitivity (OMIM# 605988) (4,5). DCLRE1C encodes ARTEMIS, a nuclease with intrinsic 5′-3′ exonuclease activity on single-stranded DNA. After phosphorylation by and in complex with DNA-dependent protein kinase catalytic subunit, ARTEMIS acquires endonuclease activity on 5′ and 3′ overhangs, and hairpins. It is involved in non-homologous end-joining (NHEJ) and is essential for opening hairpins, which arise as intermediates during V(D)J recombination of the immunoglobulin and T-cell receptor genes in T- and B-cell development (6).
SCID with faulty V(D)J recombination can also be due to biallelic mutations in the recombination activating genes 1 and 2 (RAG1/2, OMIM# 179615/OMIM# 179616) (7). Hypomorphic mutations in either RAG1 or RAG2, which allow for residual recombination events, can be associated with clinical entities less severe than typical SCID, such as Omenn syndrome, atypical SCID or common variable immunodeficiency (CVID) (8,9).
Patients with hypomorphic mutations in DCLRE1C and a clinical diagnosis of atypical SCID, Omenn syndrome, Hyper IgM syndrome or inflammatory bowel disease have recently been described. Affected individuals presented with recurrent respiratory tract infections, candidiasis, immune dysregulation and malignancies in childhood, adolescence or even adulthood (10,11).
Patients with hypomorphic mutations in SCID genes present with less severe clinical courses and therefore are reminiscent of other primary immunodeficiencies (PID) such as antibody deficiencies (e.g. CVID). Antibody deficiencies are typically treated with immunoglobulin substitution, whereas SCID patients receive hematopoietic stem cell transplantation (HSCT). It is therefore essential, to validate or exclude the presence of hypomorphic mutations in SCID genes to consider appropriate treatment options upon disease progression. Here, we report on patients with DCLRE1C mutations who were diagnosed with an antibody deficiency.
Results
Autosomal-recessive inheritance of an antibody deficiency in a Turkish family
We first analyzed the genetic cause of an antibody deficiency in a family from Turkey. Patients 1 (P1), P2 and P3 were the index patients (Family A, Fig. 1A). Onset of disease was after their second year of life with recurrent respiratory tract infections, low B-cell counts and normal T-cell counts. At their initial immunological evaluation, all had reduced IgA levels and P3 also had low IgG (Table 1 and Supplementary Material, Table S1). Therefore, P1 and P2 were diagnosed with possible CVID and P3 with probable CVID at their initial presentation.
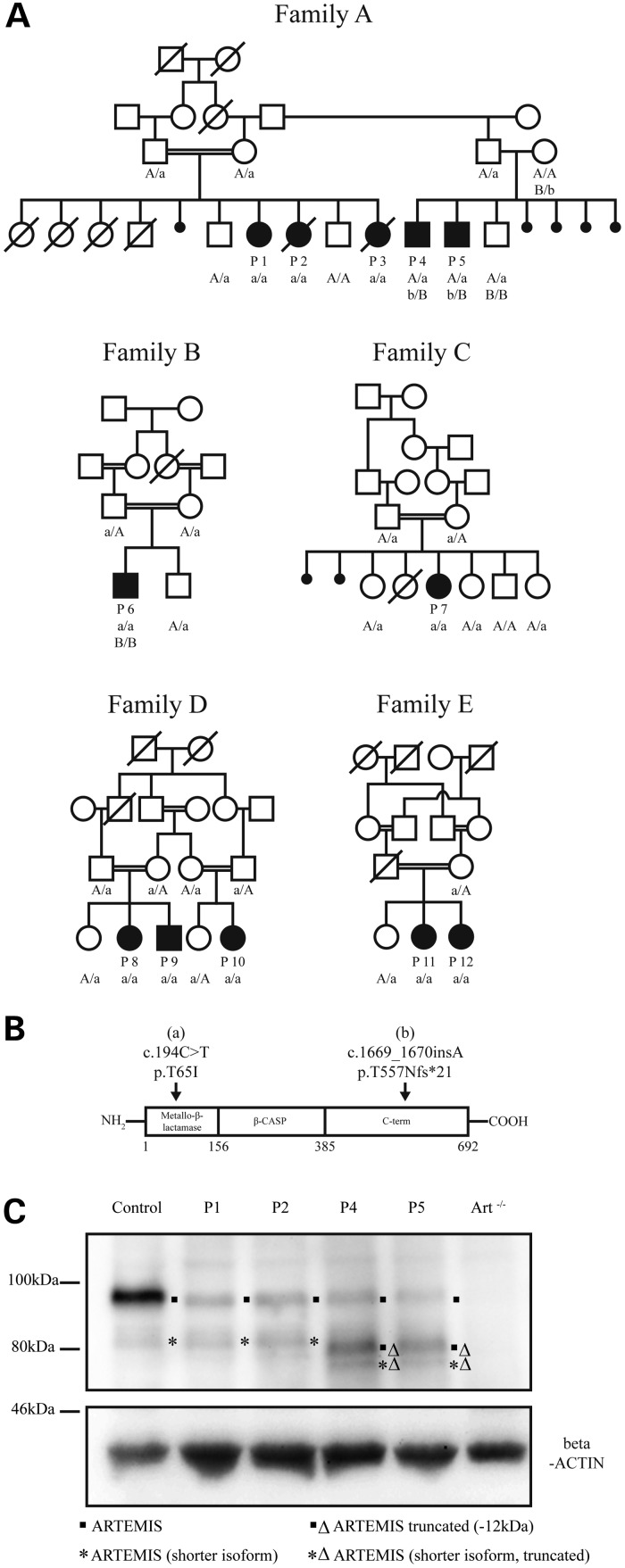
Figure 1.
Autosomal-recessive DCLRE1C variants in families with antibody deficiency cause reduced ARTEMIS expression. (A) Segregation of DCLRE1C variants with the phenotype. Circles, female; squares, male; open symbols, unaffected; filled symbols, affected; slashes, deceased; double horizontal lines, consanguineous marriage; P1–P12, patients; genotypes for the variants are indicated (uncapitalized letters, mutated alleles; capitalized letters, wild-type alleles). (B) Variants a (c.194C>T) and b (c.1669_1670insA) localize to distinct domains of ARTEMIS. (C) Fibroblasts from P1 and P2 express reduced amounts of ARTEMIS; P4 and P5 express full-length and the truncated protein at reduced levels; the band corresponding to the truncated ARTEMIS protein runs slightly lower than a shorter isoform of ARTEMIS also visible in the cells from the control, from P1, and from P2; an additional band only faintly visible in P4 and P5 most likely corresponds to a truncated form of the shorter isoform; fibroblasts from a healthy control and ARTEMIS negative cells (Art−/−) were used as controls; the beta-ACTIN control confirms equal loading.
Table 1.
Immunological data and clinical phenotype
| Patient | Age of onset | ALC (cells/µl) | B cells (cells/µl) | T cells (cells/µl) | Naive CD4 T cellsa | Naive CD8 T cellsb | Serum Ig (mg/dl) | Clinical phenotype |
|---|---|---|---|---|---|---|---|---|
| P1 | 4 years | 3700 | ↓ 118 | 1689 | ↓ 0.1 | ↓ 0.9 | IgG: 1110 IgM: 97 IgA: ↓ 6.6 |
Respiratory infections, gastroenteritis, otitis media |
| P2 | 4 years | 6000 | 480 | 2940 | ↓ 6.2 | ↓ 1.2 | IgG: 1710 IgM: 159 IgA: ↓ 6.6 |
Respiratory infections |
| P3 | 2 years 10 months | 2200 | ↓ 102 | 1562 | n.e. | n.e. | IgG: ↓ 4 IgM: 88 IgA: ↓ 1 |
Respiratory infections, granulomatous skin lesions, Hashimoto's thyroiditis, juvenile idiopathic arthritis |
| P4 | 3 years | 2170 | 238 | 1388 | ↓ 12.4 | ↓ 11.9 | IgG: 1340 IgM: 157 IgA: ↓ 6.6 |
Respiratory infections, gastroenteritis, diffuse varicella infection, vitiligo |
| P5 | 6 years | ↓ 791 | ↓ 31 | ↓ 490 | ↓ 4.3 | ↓ 5.6 | IgG: ↓ 489 IgM: 132 IgA: ↓ 64.3 |
Respiratory infections, granulomatous skin lesions, vitiligo |
| P6 | 2 years | ↓ 800 | ↓ 22 | ↓ 547 | ↓ 1.4 | ↓ 1.9 | IgG: ↓ 240 IgM: ↓ 35 IgA: ↓ 6.6 |
Verruca vulgaris, mycobacterial skin infection |
| P7 | 2 years | 1400 | ↓ 193 | ↓ 532 | ↓ 6.1 | ↓ 2.4 | IgG: ↓ 363 IgM: 149 IgA: ↓ 32.6 |
Respiratory infections, otitis media, gastroenteritis |
| P8 | 4 years | 2440 | ↓ 24 | 2025 | ↓ 2.2 | ↓ 2.3 | IgG: 1140 IgM: 86.5 IgA: ↓ 6.6 |
Respiratory infections, severe varicella infection, brucellosis, verruca vulgaris |
| P9 | 4 years | ↓ 900 | ↓ 36 | ↓ 558 | ↓21.7 | ↓ 5.6 | IgG: ↓ 560 IgM: 54 IgA: ↓ 19 |
Verruca vulgaris |
| P10 | 10 years | 1220 | ↓ 24 | ↓ 683 | ↓ 1.9 | ↓ 1.8 | IgG: 1190 IgM: ↓ 86.9 IgA: ↓ 4.5 |
Verruca vulgaris, respiratory infections |
| P11 | 5 years | 1240 | ↓ 13 | ↓ 603 | ↓ 6.3 | n.e. | IgG: 1040 IgM: ↓ 20.9 IgA: ↓ 24 |
Respiratory infections |
| P12 | 2 years | 2000 | ↓ 16 | ↓ 540 | ↓ 4.9 | n.e. | IgG: ↓ 135 IgM: ↓ 15 IgA: ↓ 25 |
Respiratory infections, aphtous stomatitis, undefined skin lesions |
Bold numbers indicate values outside of the reference range; arrows indicate an increase or reduction compared to the reference range. Ig, Immunoglobulins; n.e., not evaluated.
a(% of CD3+CD4+ cells).
b(% of CD3+CD8+ cells).
Identification of the candidate variant in DCLRE1C (ARTEMIS)
To identify the disease-causing gene in Family A, we employed whole-exome sequencing of P1, P3 and two healthy siblings. In DCLRE1C, encoding ARTEMIS, we detected a sequence variant (c.194C>T) that met our filtering criteria and predicted a damaging single amino acid change (p.T65I). Mutations in ARTEMIS cause T-B-NK+ SCID in individuals with two null mutations or combined immunodeficiency (CID) in individuals with at least one hypomorphic mutation (5,11).
Because of the consanguinity of the parents in Family A, we also employed homozygosity mapping. Multiple markers on both sides of the identified variant in DCLRE1C were heterozygous in at least one of the genotyped affected individuals; it appears that the c.194C>T variant arose twice on different founder haplotypes in Turkey. No other plausible candidate variants were identified using homozygosity mapping.
Severely reduced expression of ARTEMIS in patients with DCLRE1C variants
We used Sanger sequencing to test whether the variant segregates perfectly with the disease status in Family A (Fig. 1A). All affected individuals carried the ARTEMIS variant (c.194C>T; p.T65I). Whereas P1, P2 and P3 were homozygous for the variant, P4 and P5 were heterozygous. We therefore analyzed all coding exons of DCLRE1C in P4 and identified a frameshift variant in exon 14 (c.1669_1670insA; p.T577Nfs*21). This heterozygous variant was also detected in P5 (Fig. 1A).
We next screened other patients with low B cells, low IgA and not severely reduced T-cell counts from the same center in Turkey. Seven additional immunodeficient patients from four unrelated families were identified who had the homozygous c.194C>T variant. Clinical and immunological findings are given in Table 1; more detailed information (Supplementary Material, Table S1) and case reports are available in this article's supplement. None of their unaffected relatives was homozygous for the c.194C>T variant (Fig. 1A).
Western blot analyses demonstrated that ARTEMIS expression in fibroblasts from P1 and P2 was severely reduced (Fig. 1C). P4 and P5 fibroblasts also showed a reduced expression of ARTEMIS protein. However, an additional band was visible, likely corresponding to the truncated protein expressed from the allele carrying the frameshift mutation. The frameshift leads to a premature stop codon, predicted to reduce the molecular mass by ∼12 kDa. Western blot analyses of HEK293T cells transfected with plasmids encoding either the missense or the frameshift variant also yielded reduced expression of those ARTEMIS variants (Supplementary Material, Fig. S1). These observations suggest that the disease-causing mechanism is associated with ARTEMIS insufficiency.
Patient-derived fibroblasts show increased sensitivity to ionizing radiation
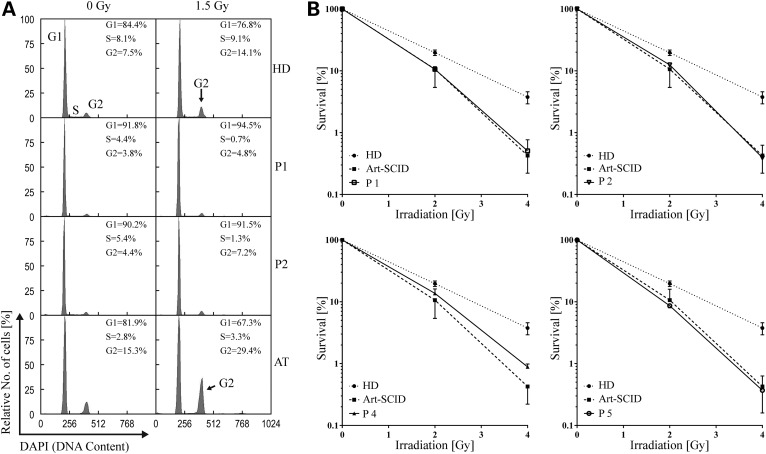
ARTEMIS was first described to be mutated in radiosensitive SCID patients (5). We therefore tested whether our patients have increased sensitivity to irradiation. Irradiation causes DNA double-strand breaks (DSBs) and stops progression through the cell cycle until DSBs are repaired. Cells from patients with defects in DNA DSB repair (e.g. ataxia telangiectasia) accumulate in G2/M phase after irradiation. In fibroblasts from P1 and P2, cell-cycle progression was not significantly altered 48 h after irradiation with 1.5 Gy (Fig. 2A). This observation is consistent with previous reports demonstrating that ARTEMIS is not required for cell-cycle checkpoint arrest mediated by ATM (Ataxia-telangiectasia-mutated, OMIM# 607585) (12).
Figure 2.
Increased radiosensitivity in fibroblasts from patients with DCLRE1C mutations. (A) Fibroblasts from P1 and P2 show normal cell-cycle progression after irradiation. HD, healthy donor fibroblasts; AT, fibroblasts from a patient with ataxia telangiectasia as a disease (positive) control; percentages of cells in S-Phase, G1-Phase or G2-Phase are indicated; DAPI, 4′,6-diamidino-2-phenylindole. (B) Decreased colony survival in fibroblasts from both homozygous (P1, P2) and compound heterozygous (P4, P5) DCLRE1C mutation carriers after irradiation, comparable to fibroblasts from a patient with ARTEMIS-SCID (Art-SCID); Error bars indicate standard error of the mean.
However, radiosensitivity in fibroblasts from patients with ARTEMIS variants was markedly increased, when analyzed independently from cell-cycle progression in colony survival assays after γ-irradiation (Fig. 2B). The degree of sensitivity was comparable to fibroblasts from a patient with T-B− SCID owing to a known homozygous mutation in DCLRE1C.
Patients’ T cells show a terminally differentiated phenotype and reduced proliferative capacity
Low B-cell numbers and increased radiosensitivity are features of faulty DSB repair and therefore suggested an NHEJ defect. However, patients had normal or near normal T-cell numbers. To exclude gene conversion to wild-type in T cells, or the maternal origin of T cells, DCLRE1C was sequenced from sorted CD3+CD8+ and CD3+CD4+ cells from P1. Both had the mutant germline configuration (data not shown). We therefore assessed, by flow cytometry, whether T-cell differentiation is affected in patients with ARTEMIS variants. Frequencies of regulatory T cells were normal (P1) or elevated (P4 and P5). Expression of marker cytokines (Interferon-γ, Interleukin-4 and Interleukin-17) for T-helper-cell differentiation, which was measured after stimulation, indicated normal capability to differentiate into T-helper-cell subsets in all assessed patients (Supplementary Material, Fig. S2A). In contrast, in peripheral lymphocytes from patients, a severe reduction of naïve T cells was observed whereas terminally differentiated T cells were increased (Table 1 and Supplementary Material, Table S1). Furthermore, CD4+ and CD8+ T cells from all tested patients showed reduced proliferation in vitro (Supplementary Material, Fig. S2B). As reduced T-cell proliferation is a criterion for T-cell deficiency [new clinical diagnostic criteria of the European Society for Immunodeficiencies (13)], these findings indicate that patients with ARTEMIS variants have a subclinical combined immunodeficiency, without clinical signs of T-cell dysfunction.
Restricted T-cell receptor repertoires and elongated palindromes at TCRβ coding joints in patients with ARTEMIS variants
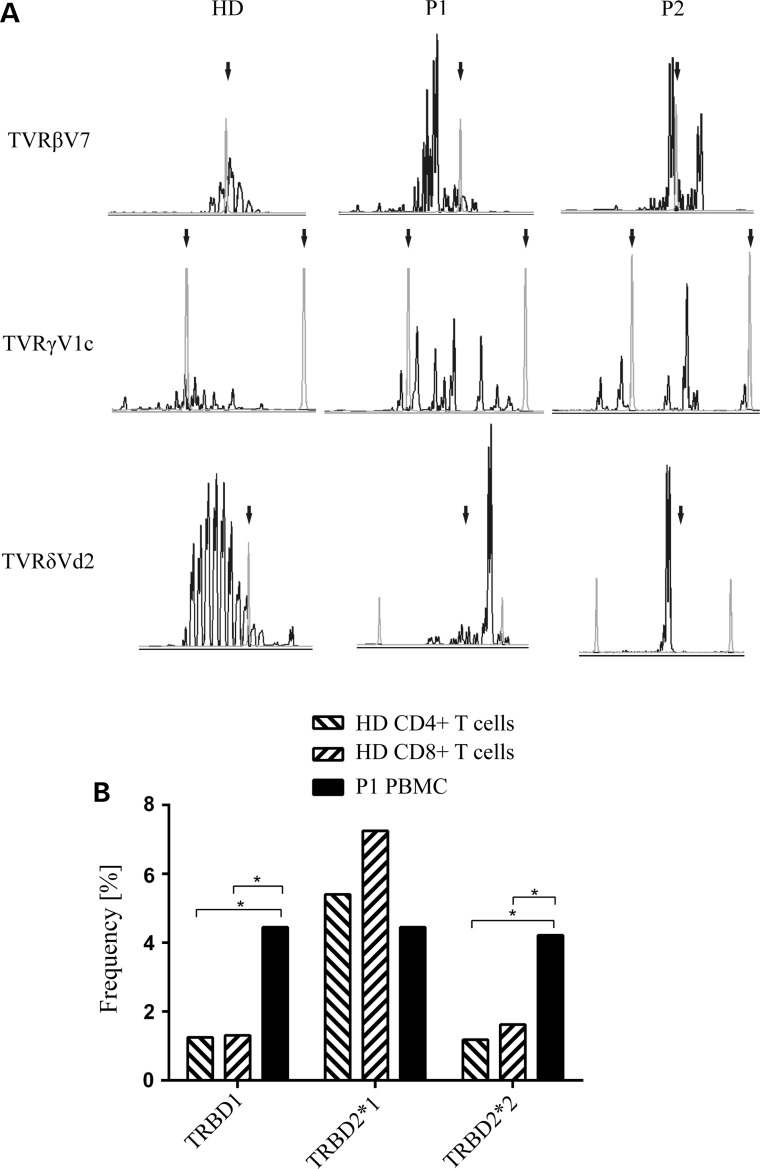
Based on the reduction of naïve T cells and limited T-cell proliferation, we hypothesized that in patients with ARTEMIS variants, T-cell numbers are maintained by few cells that complete maturation in the thymus and subsequently proliferate excessively in the periphery. We therefore assessed, by spectratyping on cDNA, whether there is a skewed or restricted T-cell receptor (TCR) repertoire. Indeed, P1 and P2 had oligoclonally expanded TCRβ sequences with dominant recombination events within a polyclonal background. TCRγ length distribution was even more restricted, and TCRδ rearrangements were also oligoclonal (Fig. 3A).
Figure 3.
Abnormal in vivo V(D)J recombination of TCR genes in patients carrying homozygous DCLRE1C mutations. (A) TCR spectratyping demonstrates the expression of a restricted TCR repertoire in P1 and P2. Length distribution of CDR3 of TCRβ, -γ and -δ families are shown; arrows, length standards. (B) Palindromic sequences adjacent to two of the three Dβ segments are more frequent in peripheral blood mononuclear cells (PBMC) from P1; (HD) healthy donor; *P < 0.005 (Fisher's exact test).
ARTEMIS nuclease opens DNA hairpins, which occur as intermediates during V(D)J recombination, at the tip or next to it (6). When ARTEMIS is defective, yet unknown mechanisms open these hairpins, however with slower kinetics and further away from the tip. Thus, ARTEMIS insufficiency results in elongated palindromic sequences at coding joints (14). In P1, we found significantly higher frequencies of TRBD1 and TRBD2*2 (two of the three possible TCRβ D segments) with adjacent palindromes than in controls (Fig. 3B). In summary, our analyses of TCR V(D)J recombination in vivo suggest that the recombination defect is caused by the variant c.194C>T in DCLRE1C.
Increased use of alternative end-joining during class switching to IgA
DNA DSBs occur physiologically in early B- and T-cell development when antigen receptors are re-arranged during V(D)J recombination. Additionally, mature B cells rearrange the constant part of the Ig heavy chain during class-switch recombination (CSR). CSR mainly depends on NHEJ (15), but alternative end-joining, which sometimes depends on sequence homology between two DNA ends, is preferred when classical NHEJ is dysfunctional (16).
We assessed whether CSR in our patients is accomplished by alternative end-joining instead of NHEJ. For this purpose, switch junctions from P1 and P2 were compared with published sequences from adult healthy controls (17). In P1 and P2, we found that 23% of the Sµ–Sα junctions (indicating a switch from IgM to IgA) had a significantly increased microhomology of >9 bp compared with 3% in controls (Table 2). This contrasts with a previous report in which atypical SCID patients with hypomorphic DCLRE1C splice-site mutations did not show increased usage of long microhomology (18). However, in typical SCID patients and a progressive CID patient caused by DCLRE1C gene deletions or mutations, 39% of Sµ–Sα junctions showed microhomology of >9 bp (17).
Table 2.
Characterization of Sµ–Sα and Sµ–Sγ junctions
| Patients | Perfectly matched short homology |
Total no. of S fragments | |||||
|---|---|---|---|---|---|---|---|
| 0 bp |
1–3 bp | 4–6 bp | 7–9 bp | ≥10 bp | |||
| Direct end-joining | Small insertions | ||||||
| Sµ–Sα | |||||||
| P1+2 | 2 (7%) | 7 (23%) | 7 (23%) | 2 (7%) | 5 (17%) | 7 (23%)***↑ | 30 |
| ARTEMIS atypical SCID | 9 (22%) | 8 (20%) | 11 (27%) | 9 (22%) | 3 (7%) | 2 (5%) | 41 |
| ARTEMIS−/− | 0 (0%)**↓ | 6 (11%) | 10 (19%) | 8 (15%) | 9 (17%) | 21 (39%)***↑ | 54 |
| Controls (adult) | 28 (18%) | 39 (25%) | 56 (36%) | 15 (10%) | 11 (7%) | 5 (3%) | 154 |
| Controls (1–13 years) | 31 (17%) | 42 (23%) | 36 (20%) | 29 (16%) | 19 (10%) | 26 (14%) | 183 |
| Sµ–Sγ | |||||||
| P1+2 | 6 (32%) | 5 (26%) | 5 (26%)**↓ | 3 (16%) | 0 (0%) | 0 (0%) | 19 |
| Controls (adult) | 12 (20%) | 7 (12%) | 37 (63%) | 3 (5%) | 0 (0%) | 0 (0%) | 59 |
| ARTEMIS−/− | 5 (21%) | 4 (17%) | 14 (58%) | 1 (4%) | 0 (0%) | 0 (0%) | 24 |
| Controls (1–6 years) | 13 (22%) | 9 (16%) | 26 (45%) | 10 (17%) | 0 (0%) | 0 (0%) | 58 |
Sµ–Sα, switch junctions owing to CSR from IgM to IgA; Sµ–Sγ, switch junctions owing to CSR from IgM to IgG; ARTEMIS atypical SCID, published switch junctions from a patient with hypomorphic ARTEMIS mutations (18); ARTEMIS−/−, published switch junctions from patients with ARTEMIS-SCID (n = 3) and progressive CID (n = 1). The Sµ–Sγ junctions were derived from the patient with progressive CID, as no Sµ–Sγ junctions could be amplified from the cells from the ARTEMIS-SCID patients (17); Statistical analysis was performed using χ2 tests, and significant differences are indicated in bold.
**P < 0.01, ***P < 0.001; Patients 1+2 and atypical ARTEMIS were compared with controls (adult), whereas ARTEMIS−/− were compared with controls (1–13 years). Arrows indicate a significant increase or reduction compared with controls.
Comparing Sµ–Sγ junctions from P1 and P2, the use of 1–3 bp microhomology was decreased, whereas the use of >3 bp was marginally increased. Furthermore, no sequential switching was detected (i.e. switching from IgM through one IgG subclass to a different IgG subclass located downstream in the IgH locus) in Sµ–Sγ junctions from P1 and P2. An excess of such junctions has been reported in a patient with progressive CID owing to mutated ARTEMIS (17).
Taken together, in contrast to previously described patients with hypomorphic ARTEMIS mutations, class-switch to IgA was dependent on microhomology in P1 and P2, but not to the same degree as in ARTEMIS-SCID patients.
Complementation of defective V(D)J recombination by ARTEMIS
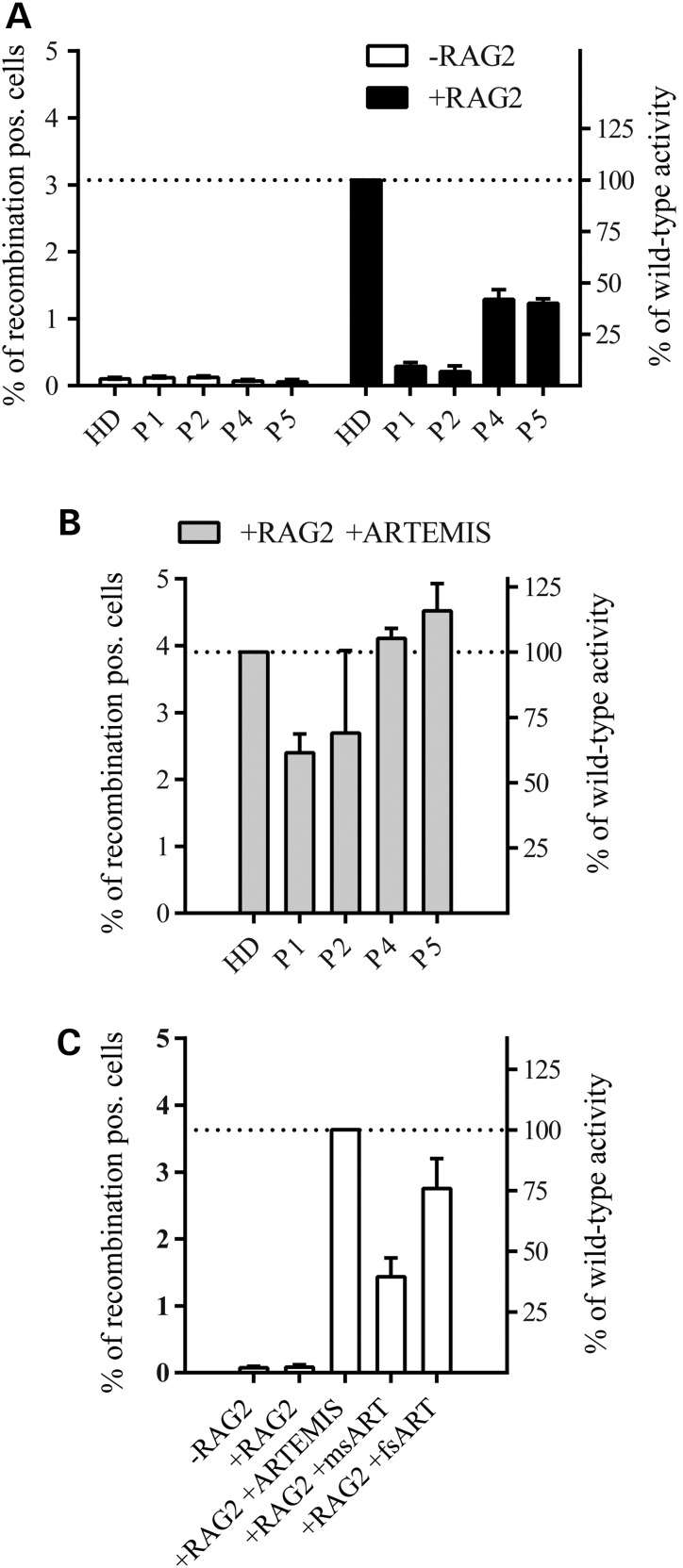
Our data indicated an NHEJ defect with residual V(D)J recombination. For confirmation, fibroblasts from patients were subjected to a plasmid-based reporter assay for inversional V(D)J recombination. After co-transfection of RAG1 and RAG2 expression plasmids and the recombination substrate, recombination efficiency in cells from P1 and P2 increased only slightly above background (cells not co-transfected with RAG2). However, in ARTEMIS compound heterozygous cells from P4 and P5, residual recombination of ∼40% of control levels was retained (Fig. 4A).
Figure 4.
Reversion of defective recombination by ARTEMIS. In vitro V(D)J recombination in fibroblasts transfected with the V(D)J recombination substrate and an RAG1 expression vector (−RAG2, negative control). Cells were co-transfected with RAG2 (+RAG2) or both RAG2 and ARTEMIS (+RAG+ARTEMIS) expression vectors; error bars indicate standard deviation from three independent experiments. (A) Dramatically reduced recombination in patients with homozygous (P1, P2) and heterozygous (P4, P5) DCLRE1C mutations; HD, healthy donor; +RAG2, transfection of the substrate plus RAG1 and RAG2 expression vectors. (B) The recombination defect in patients with homozygous DCLRE1C mutations is considerably attenuated (P1; P2) and completely rescued in patients with heterozygous DCLRE1C mutations (P4; P5) by ectopic expression of ARTEMIS. (C) Expression of mutated ARTEMIS only partially rescues the phenotype in ARTEMIS-negative fibroblasts. msART, plasmid expressing the missense mutation (c.194C>T); fsART, plasmid expressing the frameshift mutation (c.1669_1670insA).
To assess whether the V(D)J recombination defect could be rescued by wild-type ARTEMIS protein, fibroblasts were co-transfected with a wild-type ARTEMIS expression vector and RAG1/2 plasmids (Fig. 4B). Wild-type ARTEMIS restored the recombination efficiency to different degrees compared with control cells (P1: 61% of control cell level; P2: 69%; P4: 105%; P5: 116%, Fig. 4B).
We sought to investigate the individual activity of each ARTEMIS mutation by subjecting ARTEMIS negative fibroblasts to the V(D)J recombination assay after transfecting them with either the missense (c.194C>T) or the frameshift (c.1669_1670insA) mutation. The missense mutation yielded a recombination efficiency of 40% of wild-type ARTEMIS whereas the frameshift mutation showed a recombination efficiency of 76%. This supports the hypothesis that the frameshift mutation retains more enzymatic activity on the reporter plasmid than the missense mutation.
These results demonstrate that both homozygous and compound heterozygous ARTEMIS mutations cause a defect of V(D)J recombination in patients of this study and show that the missense mutation causes a more severe phenotype than the frameshift mutation.
Discussion
This report identifies homozygous and compound heterozygous mutations in DCLRE1C as genetic causes for PID ranging from antibody deficiency to atypical SCID. Whole-exome sequencing identified homozygous variants in DCLRE1C segregating with the phenotype in P1, P2 and P3 in the index family (Family A). The disease-causing gene DCLRE1C localizes in an interval where many markers are heterozygous, demonstrating a pitfall in homozygosity mapping in a consanguineous background.
The index patients (P1, P2 and P3) presented at ages of 2 to 8 years with recurrent respiratory infections. All developed B-cell lymphopenia, whereas T-cell numbers were normal throughout their clinical courses. Reduced B-cell numbers have also been reported in patients with atypical SCID owing to mutations in ARTEMIS (11). However, none of these patients had sustained normal T-cell counts.
Reduced naïve T cells in our patients with ARTEMIS mutations indicated that T-cell development was affected, although total counts of T cells were normal. The two-part hypothesis that a hypomorphic mutation allows for residual V(D)J recombination in T cells, which proliferate at compensatory levels in the periphery, is confirmed by the restricted TCR repertoire in patients' T cells. Although T-cell numbers were normal, we observed an overall reduction of T-cell proliferation, presumably due to the terminally differentiated phenotype of T cells, which have a limited proliferative capacity. Despite a biased composition of T-cell subpopulations, we did not find evidence of T-cell dysfunction.
ARTEMIS is proposed to play similar roles in V(D)J recombination for T and B cells. The observation of a stronger reduction of B-cell than T-cell counts in our patients suggests however, that the central compartment of B-cell progenitors might be more prone to exhaustion than the T-cell compartment. It is also possible that the V(D)J recombination defect is not (or not only) due to faulty enzymatic activity but also due to insufficient ARTEMIS levels. Accordingly, ARTEMIS expression in fibroblasts from our patients was only weakly detectable (Fig. 1D), as has been reported for other patients with hypomorphic ARTEMIS mutations (19). A previous study of mutant ARTEMIS transcripts revealed differential expression among several cell types (18). Thus, tissue-specific expression of mutant ARTEMIS might make B cells more vulnerable than T cells.
Null mutations in ARTEMIS have been shown to affect CSR (17,20). In contrast to previously reported patients with hypomorphic ARTEMIS mutations, we observed a significant increase in long microhomologies at Sµ–Sα junctions from P1 and P2. However, those junctions were less frequent than those in ARTEMIS-SCID patients, arguing that the homozygous c.194C>T mutation is hypomorphic.
In colony survival assays, patients with homozygous (P1 and P2) and compound heterozygous ARTEMIS mutations all showed increased radiosensitivity, comparable to a patient with ARTEMIS-SCID (Fig. 2B). These results suggest that the numerous DSBs that occur after γ-irradiation in vitro cannot be repaired. In vivo, single DSBs during V(D)J recombination in one cell might be repaired by a mutated NHEJ factor with residual enzymatic activity.
The two hypomorphic ARTEMIS mutations identified in this study have distinct effects on pathogenesis, as confirmed in a V(D)J recombination assay. The c.194C>T mutation affects the metallo-beta-lactamase domain of ARTEMIS, which is essential for V(D)J recombination (21). Fibroblasts from P1 and P2 showed only marginal recombination ability. In contrast, compound heterozygous fibroblast from Patients P4 and P5 retained 40% of wild-type recombination efficiency. As the paternal allele carries the c.194C>T mutation, the defect of that ARTEMIS mutant is identical as in patients P1 and P2. In contrast, the frameshift mutation (c.1669_1670insA) on the maternal allele leads to a truncated ARTEMIS that leaves the metallo-beta-lactamase and beta-CASP domains unaffected and thus retains significant enzymatic activity as shown by the expression of this variant in an ARTEMIS-negative cell line (Fig. 3B).
The identification of seven additional patients from four unrelated families (P6 through P12) carrying the same homozygous c.194C>T mutation enabled us to study the phenotypic variability associated with this mutation. The surviving patients were 6 to 21 years old when this report was prepared. Clinical signs of impaired T-cell immunity were milder than those in atypical SCID patients with hypomorphic ARTEMIS mutations who had severe opportunistic infections. Signs of T-cell dysfunction were limited to generalized varicella infections in P4 and P8. P6, P8, P9 and P10 had verruca vulgaris and P12 aphtous stomatitis. Gastroenteritis is common in combined immunodeficiencies, however was present only in P1, P4 and P7. Autoimmune cytopenia was observed in patients carrying hypomorphic ARTEMIS mutations (11), but not in our cohort. P4 and P5 showed vitiligo, which is regarded as an autoimmune phenomenon. P3 was the only patient with widespread immune dysregulation, manifesting as Hashimoto's thyroiditis and juvenile idiopathic arthritis. She had formation of granulomatous skin lesions, which were described in a subset of RAG-deficient patients with hypomorphic mutations and were proposed to serve as a criterion for the differential diagnosis of a combined immunodeficiency in patients with a CVID diagnosis (8,22). P5, P6, P12 and a previously published patient with a hypomorphic ARTEMIS mutation (18) also had granulomatous skin lesions, suggesting that granuloma formation could also be a common feature among patients with hypomorphic ARTEMIS mutations. However, in P6 these were attributed to a probable mycobacterial skin infection. Different types of malignancies are described for hypomorphic ARTEMIS patients (11), but none of our patients developed solid tumors or hematological neoplasias to date. This difference in the clinical course may be due to specific characteristics of the c.194C>T mutation.
While this manuscript was in preparation, a patient suffering from T-B-NK+ atypical SCID with the same missense mutation (c.194C>T) was published (23). However, the relationship between genotype and phenotype cannot be evaluated, because the missense mutation was only found on one allele (the mutation status of the other allele was stated as being not available).
Our findings emphasize that normal T-cell counts do not necessarily imply intact T-cell immunity and vice versa. P9 was considered healthy before we identified the same mutation in his DNA as in his sister (P8) who was found to be immunodeficient 4 years earlier despite having normal T-cell numbers. Upon his first immunological evaluation, he presented with B- and T-cell lymphopenia and had low IgG and IgA levels. Thorough laboratory and genetic evaluations of apparently unaffected family members are required, particularly when evaluating possible donors for HSCT (24). The variable phenotype of patients with identical mutations may be attributed to genetic (epigenetics, modifying genes) and environmental (antigenic diversity, nutrition) factors.
The fact that patients with mutations in SCID-causing genes might present with an antibody deficiency should raise the awareness of physicians working in the field of PID. Exposure to X rays has to be minimized during the clinical management in such patients (e.g. by performing magnetic resonance imaging instead of computed tomography when evaluation for lung involvement and avoiding routine X rays at dentist check-ups). Live vaccines should not be administered because on the one hand it is unlikely that patients achieve protective antibody titers; on the other hand, they might not be able to clear the attenuated virus. Furthermore, upon progression of disease, evaluation of HSCT should be initiated earlier, then it is currently done in other antibody deficiencies [e.g. severe cases of CVID (25)] and the patient's radiosensitive status has to be considered when choosing a chemotherapeutic regimen for conditioning to avoid additional adverse effects (26).
In this study, whole-exome sequencing—but not homozygosity mapping—led to the identification of disease-causing mutations in DCLRE1C in patients with antibody deficiency. Like in RAG deficiency, hypomorphic mutations in DCLRE1C may cause combinations of symptoms less severe than those observed in atypical SCID, with a high heterogeneity of laboratory and clinical findings, even among patients from the same family with identical disease-causing mutations.
Patients, Materials and Methods
Study participants
Informed consent/assent was obtained from all healthy individuals and patients according to local ethics committees' requirements. This study has been approved by the Medical Ethics Committee of the University of Freiburg (protocol number 295/13).
Homozygosity mapping
P1, P3, two unaffected siblings and both unaffected parents were genotyped using the Affymetrix Genome-wide Human SNP Array 6.0 as described (27). Homozygosity mapping was done using the in-house software findhomoz (28), searching for intervals in which markers were perfectly segregating with autosomal-recessive transmission of the disease and both genotyped, affected children are homozygous for the same haplotype. In light of sequencing results, markers in or near DCLRE1C on chromosome 10 were analyzed individually.
Whole-exome sequencing and data analysis
Genomic DNA from Patient 1, Patient 3 and two healthy siblings the DNA was analyzed by whole-exome sequencing at Beijing Genome Institute. The sequenced reads were mapped against the human reference genome build UCSC hg19 using Bowtie 2 v2.2.3 (29), reordered, sorted and converted to bam format, followed by the removal of PCR duplicates with Picard v1.115. Local realignment around InDels and base quality score recalibration as well as variant calling and variant quality score recalibration were performed with the GATK v3.1 (30) according to their best practice recommendations. Analysis of genetic variant data in the form of VCF files was conducted using the VCFtools program package (31).
For the annotation of variants with the IDs from the short genetic variants database dbSNP v138, we used SnpSift, which is part of the main distribution of the toolbox SnpEff v3.6 (32). Because Patients 1 and 3 were born to consanguineous parents, autosomal-recessive inheritance was assumed and variants not homozygous in the patients or homozygous in the unaffected siblings were excluded. The common variants [frequency >0.01 in dbsnp138 or ExAC(33)] were then eliminated. Genes were designated as related to the immune system as previously described (34). Candidate variants (Supplementary Material, Table S2) were prioritized according to frequency, a reported function in the immune system, and software predictions for a possible damaging effect of the variant (35,36).
Immunoblot analysis
Equal amounts of whole cell lysates, prepared from primary fibroblasts, were separated on 8% SDS–PAGE gels and blotted onto polyvinylidene difluoride membranes. Non-specific binding was blocked with 5% nonfat dry milk in TBS-T (137 mm sodium chloride, 20 mm Tris–HCL pH 8.0, 0.1% Tween 20). The following antibodies were used: rabbit anti-ARTEMIS (Cell Signaling, Frankfurt, Germany) and mouse anti-beta-ACTIN (Novus Biologicals). Signals were detected with horseradish peroxidase-coupled anti-rabbit IgG (#7074, Cell Signaling) and a chemiluminescent reagent (LumiGlo, Cell Signaling) using a Fusion-FX7 CCD camera (Peqlab, Erlangen, Germany).
Radiosensitivity assays
Cell-cycle analysis of irradiated fibroblasts was performed as previously described (37). For colony survival assays, primary skin fibroblasts were harvested during exponential growth. As a disease control, fibroblasts from an ARTEMIS-SCID patient (homozygous mutation c.110A>G, p.D37G) were used. Cells were irradiated and subsequently seeded in culture dishes coated with gelatin. After 14 days, dishes were washed, stained with 0.5% crystal violet, and colonies were counted. Plating efficiency for each dish was calculated (number of colonies divided by the number of seeded cells). Survival was determined by dividing plating efficiency of irradiated samples by plating efficiency of untreated samples. For each sample, three independent experiments were performed.
Sanger sequencing
Genomic DNA from family members was isolated using standard methods. Coding genomic regions including flanking intronic sequences of DCLRE1C were amplified from genomic DNA by standard PCR. PCR primers were used for Sanger sequencing according to standard techniques. Primer sequences are available on request.
Amplification and analysis of in vivo generated switch junctions
Genomic DNA was extracted from PBMCs or whole blood using a commercially available kit (QIAamp DNA mini kit, Qiagen, Hilden, Germany). The amplification of Sµ–Sα (38,39) and Sµ–Sγ (40) junctions and analysis (41) was performed as described.
T-cell receptor spectratyping and rearrangement studies
TCRβ, TCRγ and TCRδ spectratyping was performed from RNA isolated from PBMCs using a commercially available kit (RNeasy mini kit, Qiagen) following synthesis of oligo dT–primed cDNA as described previously (42). All fluorescent fragments were analyzed on an ABI 3130-XL capillary sequencer (Life Technologies).
V(D)J recombination assay in fibroblasts
V(D)J recombination assays were performed as described previously (43).
Cell culture
Peripheral blood mononuclear cells were isolated from whole blood by ficoll density gradient centrifugation, stored in the gas phase of liquid nitrogen and thawed for individual experiments. Primary skin fibroblasts were obtained from skin biopsies pretreated with collagenase type IV (Worthington, Lakewood, NJ, USA), dispase II (Roche, Mannheim, Germany) and subsequently cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum and 100 U/ml penicillin–streptomycin (all from Gibco, Life Technologies, Darmstadt, Germany). All cells were cultured at 37°C and 5% CO2 in a humidified atmosphere.
Flow cytometry
Flow cytometric analyses were performed on a Becton Dickinson Canto II (BD Biosciences, Heidelberg, Germany) and analyzed on FlowJo software version 10 (Treestar, Ashland, OR). Cells were stained with antibodies against:
TCRab (APC, IP26), CD4 (PerCp-Cy5.5, PRA-T4), CD21 (PE, HB5), CD27 (APC, 0323), CD127 (PE, eBioRDR5), FOXP3 (AF488, PCH101), IL-4 (APC, 8D4-8), IL-17 (PE, eBio64DEC17, all from eBioscience, Frankfurt, Germany), TCRgd (PE, IMMU510), CD16 (PE, 3G8), CD56 (PE, N901), all from Beckman Coulter, Krefeld, Germany), CD24 (BV510, ML5), CD38 (BV421, HIT2), CD45 (PerCP, HI30, all from BioLegend, London, UK), HLA-DR (FITC), CD3 (APC-H7, SK7), CD8 (PerCP-Cy5.5, SK1), CD19 (PE-Cy7, SJ25C1), CD25 (APC, 2A3), CD28 (PE, L293), CD45RA (PE-Cy7), CD45RO (PE-Cy7, UCHL-1), IFN-γ (FITC, B27, all from BD) and IgD (FITC, Life Technologies). Cell viability was analyzed with 7 AAD staining solution (BioLegend) and fixable viability dye (eFluor506, eBioscience). For the detection of regulatory T cells, commercially available reagents (eBioscience) were used, following the manufacturer's instructions.
Cytokine expression and proliferation of T cells
For cytokine measurements, 500 000 peripheral blood mononuclear cells were incubated overnight in Iscove's modified Dulbecco's medium supplemented with 10% fetal calf serum and 100 U/ml penicillin–streptomycin (all from Gibco). Cells were treated with brefeldin A (Sigma–Aldrich, Steinheim, Germany) and stimulated with 50 U/ml IL-2 (Novartis, Nürnberg, Germany), phorbol 12-myristate 13-acetate (PMA; 0.05 μg/ml, Sigma–Aldrich) and ionomycin (1 μg/ml, Sigma–Aldrich) for 4 h at 37°C. Cells were then stained with antibodies against CD3, CD4, CD45RO and fixable viability dye. After fixation and permeabilization with Cytofix/Cytoperm (BD), cells were stained with antibodies against IL-17, IL-4 and IFN-γ and analyzed by flow cytometry. Only CD3+, CD4+, CD45RO+ cells were analyzed for cytokine expression levels.
For measuring proliferation, peripheral blood mononuclear cells were labeled with the fluorescent dye carboxyfluorescein succinimidyl ester (CFSE, Molecular Probes, Life Technologies) for 15 min at 37°C with gentle agitation and washed with PBS. CFSE-labeled peripheral blood mononuclear cells (50 000 in 200 μl IMDM/10% FCS) were stimulated either with phytohemagglutinin (PHA; 10 µg/ml, Sigma–Aldrich) or with anti-CD3 and anti-CD28 coupled beads (Dynabeads, Life Technologies) at 37°C. No stimulants were added to control cells. After 5 days, cells were washed, surface-stained with anti-CD3, anti-CD4 and anti-CD8 and analyzed by flow cytometry.
High-throughput sequencing of TCRβ CDR3 region
The complementary-determining region 3 (CDR3) sequences of T-cell receptor beta (TCRβ) rearrangements were analyzed by a multiplex PCR assay combined with high-throughput sequencing as previously described (44). DNA was extracted according to standard protocols from sorted CD4+ and CD8+ T cells from a healthy donor and from PBMCs of P1 at one time point. Generated sequence data (9 545 772 reads on CD4+ T cells, 8 343 301 reads on CD8+ T cells and 648 715 reads on PBMC from P1) were analyzed by the open-source software MiTCR (45). Detection of palindromic nucleotides was performed by the alignment of 6-bp-long sequences spanning 5′ nucleotides of the D segments and palindromic nucleotides, respectively.
Western blot of transfected ARTEMIS variants
HEK293T cells were transfected using the Amaxa Cell Line Nucleofactor Kit V (Lonza, Cologne, Germany) with 5 μg empty pcDNA6/myc-His Version A plasmid, or plasmids coding for either wild-type or mutated versions of ARTEMIS fused to the myc-His tag. For the determination of transfection efficiencies, cells were co-transfected with 1 μg pmaxGFP plasmid (Lonza) encoding the green fluorescent protein. After 24 h, cells were harvested and lysed in the lysis buffer [50 mm Tris–HCl pH 8.0, 62.5 mm EDTA, 1% (w/v) NP-40, 0.4% (w/v) sodium deoxycholate]. Lysates were spun down in a microcentrifuge at 14 000×g to pellet-insoluble debris. Protein concentrations of the lysates were determined by the BioRad Dc Protein assay (BioRad, Hercules). A 30 μg portion of each lysate was run on an 8% SDS–polyacrylamide gel. The proteins were transferred to Immobilon-P membranes (Millipore, Billerica), and the blots were developed using monoclonal mouse anti-myc (Novex, Life Technologies, Darmstadt, Germany) and polyclonal rabbit anti-β-ACTIN (Abcam, Cambridge, UK) antibodies. Secondary antibodies used were goat anti-mouse IgG (H+L)-HRP-conjugate and goat anti-rabbit IgG (H+L)-HRP-conjugate (BioRad).
Supplementary Material
Funding
This study was supported by the German Federal Ministry of Education and Research (BMBF 01EO1303). The authors are responsible for the contents of this publication. This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Library of Medicine. T.V. received funding from the MOTIVATE program of the medical faculty Freiburg sponsored by the Else Kröner Fresenius Foundation. M.D.V. is a Helmholtz Young Investigator (Helmholtz Association, Germany). P.F. received funds from the Deutsche Jose Carreras Leukämie Stiftung (DJCLS R10/34f). Q.P.H, A.B. and L.H. received grants from the Swedish Research Council and the Swedish Cancer Society. D.S.'s group was supported by the Schroeder-Kurth fund.
Supplementary Material
Acknowledgements
We are grateful to all individuals participating in this study. We would like to thank C. Speckmann and G. Finkenzeller for providing cell lines, E.-M. Rump, M. Buchta, P. Mrovecova and F. Nussbaumer for excellent technical support and A. Durandy for reviewing the manuscript.
Conflict of Interest statement. None declared.
References
- 1.Dvorak C.C., Cowan M.J., Logan B.R., Notarangelo L.D., Griffith L.M., Puck J.M., Kohn D.B., Shearer W.T., O'Reilly R.J., Fleisher T.A. et al. (2013) The natural history of children with severe combined immunodeficiency: baseline features of the first fifty patients of the primary immune deficiency treatment consortium prospective study 6901. J. Clin. Immunol., 33, 1156–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Burg M., Gennery A.R. (2011) Educational paper. The expanding clinical and immunological spectrum of severe combined immunodeficiency. Eur. J. Pediatr., 170, 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pannicke U., Baumann B., Fuchs S., Henneke P., Rensing-Ehl A., Rizzi M., Janda A., Hese K., Schlesier M., Holzmann K. et al. (2013) Deficiency of innate and acquired immunity caused by an IKBKB mutation. N. Engl. J. Med., 369, 2504–2514. [DOI] [PubMed] [Google Scholar]
- 4.Li L., Moshous D., Zhou Y., Wang J., Xie G., Salido E., Hu D., de Villartay J.P., Cowan M.J. (2002) A founder mutation in Artemis, an SNM1-like protein, causes SCID in Athabascan-speaking Native Americans. J. Immunol., 168, 6323–6329. [DOI] [PubMed] [Google Scholar]
- 5.Moshous D., Callebaut I., de Chasseval R., Corneo B., Cavazzana-Calvo M., Le Deist F., Tezcan I., Sanal O., Bertrand Y., Philippe N. et al. (2001) Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell, 105, 177–186. [DOI] [PubMed] [Google Scholar]
- 6.Ma Y., Pannicke U., Schwarz K., Lieber M.R. (2002) Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell, 108, 781–794. [DOI] [PubMed] [Google Scholar]
- 7.Schwarz K., Gauss G.H., Ludwig L., Pannicke U., Li Z., Lindner D., Friedrich W., Seger R.A., Hansen-Hagge T.E., Desiderio S. et al. (1996) RAG mutations in human B cell-negative SCID. Science, 274, 97–99. [DOI] [PubMed] [Google Scholar]
- 8.Abolhassani H., Wang N., Aghamohammadi A., Rezaei N., Lee Y.N., Frugoni F., Notarangelo L.D., Pan-Hammarstrom Q., Hammarstrom L. (2014) A hypomorphic recombination-activating gene 1 (RAG1) mutation resulting in a phenotype resembling common variable immunodeficiency. J. Allergy Clin. Immunol., 134, 1375–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niehues T., Perez-Becker R., Schuetz C. (2010) More than just SCID--the phenotypic range of combined immunodeficiencies associated with mutations in the recombinase activating genes (RAG) 1 and 2. Clin. Immunol., 135, 183–192. [DOI] [PubMed] [Google Scholar]
- 10.Bajin I.Y., Ayvaz D.C., Unal S., Ozgur T.T., Cetin M., Gumruk F., Tezcan I., de Villartay J.P., Sanal O. (2013) Atypical combined immunodeficiency due to Artemis defect: a case presenting as hyperimmunoglobulin M syndrome and with LGLL. Mol. Immunol., 56, 354–357. [DOI] [PubMed] [Google Scholar]
- 11.Lee P.P., Woodbine L., Gilmour K.C., Bibi S., Cale C.M., Amrolia P.J., Veys P.A., Davies E.G., Jeggo P.A., Jones A. (2013) The many faces of Artemis-deficient combined immunodeficiency: two patients with DCLRE1C mutations and a systematic literature review of genotype-phenotype correlation. Clin. Immunol., 149, 464–474. [DOI] [PubMed] [Google Scholar]
- 12.Riballo E., Kuhne M., Rief N., Doherty A., Smith G.C., Recio M.J., Reis C., Dahm K., Fricke A., Krempler A. et al. (2004) A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol. Cell, 16, 715–724. [DOI] [PubMed] [Google Scholar]
- 13.Clinical diagnosis criteria for the ESID Registry. 2014. Available at: http://esid.org/Working-Parties/Registry/Diagnosis-criteria (4 February 2015, date last accessed).
- 14.van der Burg M., Verkaik N.S., den Dekker A.T., Barendregt B.H., Pico-Knijnenburg I., Tezcan I., vanDongen J.J., van Gent D.C. (2007) Defective artemis nuclease is characterized by coding joints with microhomology in long palindromic-nucleotide stretches. Eur. J. Immunol., 37, 3522–3528. [DOI] [PubMed] [Google Scholar]
- 15.Stavnezer J., Schrader C.E. (2014) IgH chain class switch recombination: mechanism and regulation. J. Immunol., 193, 5370–5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kotnis A., Du L., Liu C., Popov S.W., Pan-Hammarstrom Q. (2009) Non-homologous end joining in class switch recombination: the beginning of the end. Philos Trans R Soc Lond B Biol Sci, 364, 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du L., van der Burg M., Popov S.W., Kotnis A., van Dongen J.J., Gennery A.R., Pan-Hammarstrom Q. (2008) Involvement of Artemis in nonhomologous end-joining during immunoglobulin class switch recombination. J. Exp. Med., 205, 3031–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ijspeert H., Lankester A.C., van den Berg J.M., Wiegant W., van Zelm M.C., Weemaes C.M., Warris A., Pan-Hammarstrom Q., Pastink A., van Tol M.J. et al. (2011) Artemis splice defects cause atypical SCID and can be restored in vitro by an antisense oligonucleotide. Genes Immun., 12, 434–444. [DOI] [PubMed] [Google Scholar]
- 19.Woodbine L., Grigoriadou S., Goodarzi A.A., Riballo E., Tape C., Oliver A.W., van Zelm M.C., Buckland M.S., Davies E.G., Pearl L.H. et al. (2010) An Artemis polymorphic variant reduces Artemis activity and confers cellular radiosensitivity. DNA Repair, 9, 1003–1010. [DOI] [PubMed] [Google Scholar]
- 20.Rivera-Munoz P., Soulas-Sprauel P., Le Guyader G., Abramowski V., Bruneau S., Fischer A., Paques F., de Villartay J.P. (2009) Reduced immunoglobulin class switch recombination in the absence of Artemis. Blood, 114, 3601–3609. [DOI] [PubMed] [Google Scholar]
- 21.Poinsignon C., Moshous D., Callebaut I., de Chasseval R., Villey I., de Villartay J.P. (2004) The metallo-beta-lactamase/beta-CASP domain of Artemis constitutes the catalytic core for V(D)J recombination. J. Exp. Med., 199, 315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuetz C., Huck K., Gudowius S., Megahed M., Feyen O., Hubner B., Schneider D.T., Manfras B., Pannicke U., Willemze R. et al. (2008) An immunodeficiency disease with RAG mutations and granulomas. N. Engl. J. Med., 358, 2030–2038. [DOI] [PubMed] [Google Scholar]
- 23.Felgentreff K., Lee Y.N., Frugoni F., Du L., van der Burg M., Giliani S., Tezcan I., Reisli I., Mejstrikova E., de Villartay J.P. et al. (2015) Functional analysis of naturally occurring DCLRE1C mutations and correlation with the clinical phenotype of ARTEMIS deficiency. J. Allergy Clin. Immunol., 136, 140–150 e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuetz C., Pannicke U., Jacobsen E.M., Burggraf S., Albert M.H., Honig M., Niehues T., Feyen O., Ehl S., Debatin K.M. et al. (2014) Lesson from hypomorphic recombination-activating gene (RAG) mutations: Why asymptomatic siblings should also be tested. J. Allergy Clin. Immunol., 133, 1211–1215. [DOI] [PubMed] [Google Scholar]
- 25.Wehr C., Gennery A.R., Lindemans C., Schulz A., Hoenig M., Marks R., Recher M., Gruhn B., Holbro A., Heijnen I. et al. (2015) Multicenter experience in hematopoietic stem cell transplantation for serious complications of common variable immunodeficiency. J. Allergy Clin. Immunol., 135, 988–997 e986. [DOI] [PubMed] [Google Scholar]
- 26.Lobachevsky P., Woodbine L., Hsiao K.C., Choo S., Fraser C., Gray P., Smith J., Best N., Munforte L., Korneeva E. et al. (2015) Evaluation of severe combined immunodeficiency and combined immunodeficiency pediatric patients on the basis of cellular radiosensitivity. J. Mol. Diagn., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glocker E.O., Kotlarz D., Boztug K., Gertz E.M., Schaffer A.A., Noyan F., Perro M., Diestelhorst J., Allroth A., Murugan D. et al. (2009) Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med., 361, 2033–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glocker E.O., Hennigs A., Nabavi M., Schaffer A.A., Woellner C., Salzer U., Pfeifer D., Veelken H., Warnatz K., Tahami F. et al. (2009) A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N. Engl. J. Med., 361, 1727–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langmead B., Salzberg S.L. (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods, 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M. et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res., 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T. et al. (2011) The variant call format and VCFtools. Bioinformatics, 27, 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly, 6, 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Exome Aggregation Consortium (ExAC). Cambridge, MA. Available at: http://exac.broadinstitute.org. Accessed December 10, 2014).
- 34.Schubert D., Bode C., Kenefeck R., Hou T.Z., Wing J.B., Kennedy A., Bulashevska A., Petersen B.S., Schaffer A.A., Gruning B.A. et al. (2014) Autosomal dominant immune dysregulation syndrome in humans with CTLA4 mutations. Nat. Med., 20, 1410–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar P., Henikoff S., Ng P.C. (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc., 4, 1073–1081. [DOI] [PubMed] [Google Scholar]
- 36.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. (2010) A method and server for predicting damaging missense mutations. Nat Methods, 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schindler D., Hoehn H. (1999) In Wegner R.-D. (ed), Diagnostic Cytogenetics. Springer Berlin Heidelberg, pp. 269–281. [Google Scholar]
- 38.Pan Q., Petit-Frere C., Dai S., Huang P., Morton H.C., Brandtzaeg P., Hammarstrom L. (2001) Regulation of switching and production of IgA in human B cells in donors with duplicated alpha1 genes. Eur. J. Immunol., 31, 3622–3630. [DOI] [PubMed] [Google Scholar]
- 39.Pan Q., Petit-Frere C., Lahdesmaki A., Gregorek H., Chrzanowska K.H., Hammarstrom L. (2002) Alternative end joining during switch recombination in patients with ataxia-telangiectasia. Eur. J. Immunol., 32, 1300–1308. [DOI] [PubMed] [Google Scholar]
- 40.Pan-Hammarstrom Q., Jones A.M., Lahdesmaki A., Zhou W., Gatti R.A., Hammarstrom L., Gennery A.R., Ehrenstein M.R. (2005) Impact of DNA ligase IV on nonhomologous end joining pathways during class switch recombination in human cells. J. Exp. Med., 201, 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stavnezer J., Bjorkman A., Du L., Cagigi A., Pan-Hammarstrom Q. (2010) Mapping of switch recombination junctions, a tool for studying DNA repair pathways during immunoglobulin class switching. Adv. Immunol., 108, 45–109. [DOI] [PubMed] [Google Scholar]
- 42.Ehl S., Schwarz K., Enders A., Duffner U., Pannicke U., Kuhr J., Mascart F., Schmitt-Graeff A., Niemeyer C., Fisch P. (2005) A variant of SCID with specific immune responses and predominance of gamma delta T cells. J. Clin. Invest., 115, 3140–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pannicke U., Ma Y., Hopfner K.P., Niewolik D., Lieber M.R., Schwarz K. (2004) Functional and biochemical dissection of the structure-specific nuclease ARTEMIS. EMBO J., 23, 1987–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ritter J., Seitz V., Balzer H., Gary R., Lenze D., Moi S., Pasemann S., Seegbarth A., Wurdack M., Hennig S. et al. (2015) Donor CD4 T Cell Diversity Determines Virus Reactivation in Patients After HLA-Matched Allogeneic Stem Cell Transplantation. Am J Transplant, 15, 2170–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolotin D.A., Shugay M., Mamedov I.Z., Putintseva E.V., Turchaninova M.A., Zvyagin I.V., Britanova O.V., Chudakov D.M. (2013) MiTCR: software for T-cell receptor sequencing data analysis. Nat Methods, 10, 813–814. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.