Abstract
Plectin is a highly versatile cytoskeletal protein that acts as a mechanical linker between intermediate filament (IF) networks and various cellular structures. The protein is crucial for myofiber integrity. Its deficiency leads to severe pathological changes in skeletal muscle fibers of patients suffering from epidermolysis bullosa simplex with muscular dystrophy (EBS-MD). Skeletal muscle fibers express four major isoforms of plectin which are distinguished solely by alternative, relatively short, first exon-encoded N-terminal sequences. Each one of these isoforms is localized to a different subcellular compartment and plays a specific role in maintaining integrity and proper function(s) of myofibers. The unique role of individual isoforms is supported by distinct phenotypes of isoform-specific knockout mice and recently discovered mutations in first coding exons of plectin that lead to distinct, tissue-specific, pathological abnormalities in humans. In this study, we demonstrate that the lack of plectin isoform 1 (P1) in myofibers of mice leads to alterations of nuclear morphology, similar to those observed in various forms of MD. We show that P1-mediated targeting of desmin IFs to myonuclei is essential for maintenance of their typically spheroidal architecture as well as their proper positioning and movement along the myofiber. Furthermore, we show that P1 deficiency affects chromatin modifications and the expression of genes involved in various cellular functions, including signaling pathways mediating mechanotransduction. Mechanistically, P1 is shown to specifically interact with the myonuclear membrane-associated (BAR domain-containing) protein endophilin B. Our results open a new perspective on cytoskeleton-nuclear crosstalk via specific cytolinker proteins.
Introduction
The mechanical coupling of the nucleus to the cytoskeleton as well as the proper organization of the nuclear structure and composition appear critical for the development and normal functioning of the cell. The bridge formed across the nuclear envelope is based on interactions between the inner nuclear membrane-resident SUN proteins with the outer nuclear membrane-resident KASH proteins (such as nesprins) within the perinuclear membrane. This assembly of proteins, commonly referred to as linkers of the nucleoskeleton and cytoskeleton (LINC) complex, is essential for coupling cytoplasmic elements to the nucleus, determining the spacing between the inner and outer nuclear membranes, controlling nuclear size and architecture, positioning and anchoring of nuclei, prevention of nuclear pore complex clustering and organization of the perinuclear cytoskeleton (1–4).
The mechanisms regulating the specific functions and interactions of LINC complexes, including those involved in connecting the nucleus to the cytoskeleton, are incompletely understood. Whereas the linkages between the nucleus and the actin and microtubule networks have been assessed in various systems, studies on the mechanism of intermediate filament (IF) network anchorage at the nucleus and its impact on cellular functions remain scarce. Early on, IFs have been suggested to play a role in mechanotransduction involving the nucleus (5) and they were implicated and proposed as a general mechanism for the proper positioning of nuclei in immobile cells in general (6,7). Furthermore, it has been proposed that desmin regulates the positioning of junctional nuclei and the spacing between extrajunctional nuclei in multinucleated muscle cells (8). For a number of reasons, skeletal muscle fibers would seem to be an optimal system for studying interactions and crosstalk between nuclei and cytoskeletal elements. First, their multinucleated state and large size facilitate the analysis of complex interaction patterns, and second, mechanotransduction is an immanent feature of their function.
With plectin, an exceptionally versatile IF-based cytolinker protein (9), there is a strong candidate for mediating IF network interactions with the nucleus. In fact, it has been shown that plectin binds to nesprin-3 and simultaneously recruits IFs to the nuclear membrane (10,11). However, the physiological relevance of this interaction, its regulation and the question whether nesprin-3 is the only outer nuclear membrane protein that specifically binds to plectin are issues that need to be clarified.
It has been shown that skeletal muscle fibers express predominantly four isoforms of plectin, namely plectin 1 (P1), P1b, P1d and P1f, each of which gets recruited to a specific cellular structure or compartment where it acts as a docking site for IF networks (12–14). The phenotypic analysis of myofibers from conditional (muscle-restricted) plectin knockout (MCK-Cre/cKO) mice revealed plectin to be crucial for IF (desmin) network organization, optimal functioning of mitochondria and the structural integrity of the contractile apparatus and of the myofiber as a whole (15). These studies also revealed that one of plectin's isoforms, P1, was specifically associated with desmin IF networks at the outer nuclear/sarcoplasmic membrane system, suggesting that it might have a specific function in organizing and regulating IF networks in perinuclear areas.
The questions we wanted to address in this study were the following: does plectin deficiency has an impact on IF network organization at the nuclear periphery, and in the case it does, is this phenomenon isoform-dependent? What role does plectin (or a certain isoform) play in maintaining the structural integrity and architecture of myonuclei? And finally, does the lack of plectin affect nuclear functions, positioning and dynamics, and if so, what are the underlying mechanisms? To tackle these issues, we performed comparative studies, where we analyzed two isoform-specific knockout mouse lines, one lacking P1 (P1-KO) (16), the other P1b (P1b-KO) (17), a plectin conditional knockout (MCK-Cre/cKO) mouse line, and desmin-null mice (18). Here we report that plectin not only is involved in desmin IF anchorage at the nuclear periphery of myofibers, but also plays a crucial role in signaling pathways mediating mechanotransduction in muscle.
Results
Desmin IF network integrity in perinuclear regions is plectin isoform P1 dependent
Previous studies on keratinocytes and myotubes suggested that isoform P1 is targeted to the outer nuclear/endoplasmic reticulum (ER) membrane system (14,19). To establish that the isoform-specific N-terminal domain of P1 was indeed responsible for such a targeting in myocytes, we transfected plectin-deficient (P-null) myoblasts with cDNA expression plasmids encoding EGFP-tagged versions of either an N-terminal fragment of P1 (P1-8; comprising the isoform-specific, exon 1-encoded sequence succeeded by the exons 2–8-encoded ABD; see below), or full-length P1. Both overexpressed fusion proteins were targeted toward the nucleus, accumulating in a ring-shaped pattern around the nucleus and diffusely in the cytoplasm (unpublished data). Fusion proteins using corresponding expression plasmids of mitochondrion-associated plectin isoform P1b did not reveal perinuclear localization (17,20; and unpublished data), consistent with the notion that targeting to the nucleus was isoform (P1)-specific.
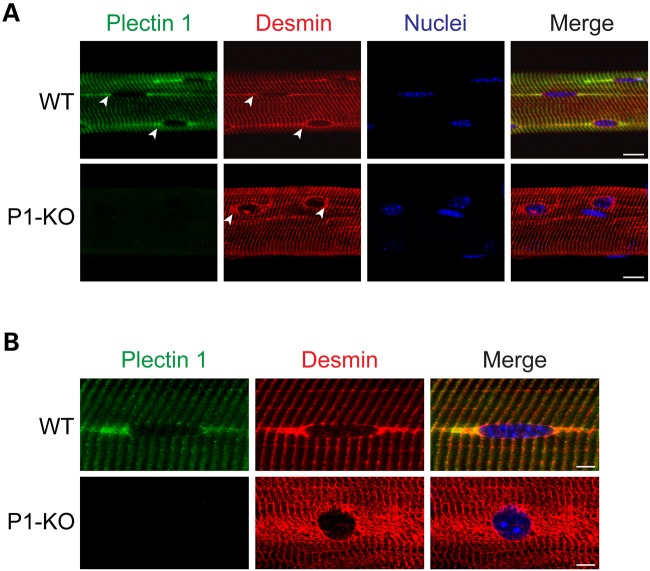
To investigate the effects of P1 deficiency on perinuclear IF network organization, teased myofibers of wild-type and isoform P1-KO mice were subjected to co-immunostaining using isoform-specific anti-P1 and anti-desmin antibodies. In wild-type cells, P1 showed an accumulation at tail-like appendages associated with the equatorial regions of typically spheroidal and quite flat myonuclei, consistent with previous studies (15,19). Longitudinally extending toward the neighboring nuclei linearly arranged along the myofiber, these structures most probably represented the sarcoplasmic reticulum (Fig. 1A and B, upper rows; and Fig. 2A). The desmin-specific perinuclear staining pattern was very similar to that of P1, except that in this case the nuclear surface seemed to be continuously outlined, whereas this hardly applied for P1. These observations were consistent with the notion that both proteins were associated with the outer nuclear membrane/ER membrane system and that IFs formed cage-like structure around the nucleus.
Figure 1.
Partial colocalization of P1 and desmin at the outer nuclear membrane and disarray of perinuclear IF networks upon P1 deficiency. Wild-type and P1-deficient myofibers were subjected to double immunolabeling using antibodies to P1 and desmin; nuclei were visualized by staining with Hoechst dye. (A) Confocal and (B) deconvolved IFM images. Note partial colocalization of P1 and desmin at tail-like structures longitudinally extending from the nuclear equator of wild-type myofibers (arrowheads in A, upper row) and along Z-disk striations. Note disarray of desmin-positive structures in the vicinity of nuclei (arrowheads, lower row) as well as more spherical shapes of nuclei in P1-deficient compared with wild-type myofibers. Scale bars, 10 µm.
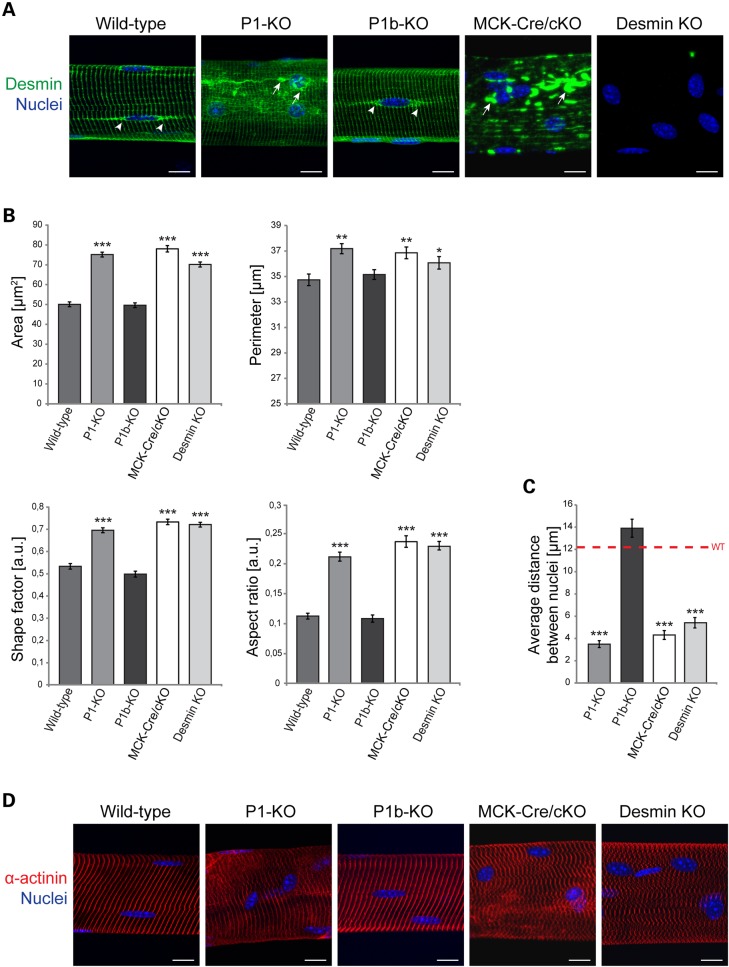
Figure 2.
P1 deficiency in myofibers leads to alterations in nuclear parameters. (A) IFM of teased EDL muscle fibers, isolated from wild-type, P1-KO, P1b-KO, MCK-Cre/cKO and desmin KO mice, were subjected to IFM using antibodies to desmin; nuclei were visualized with Hoechst dye. Arrowheads, desmin structures extending from nuclei of wild-type and P1b-KO myofibers. Arrows, desmin-positive structures aggregating in the vicinity of nuclei (P1-KO and MCK-Cre/cKO) as well as throughout the sarcoplasm (MCK-Cre/cKO). Note altered nuclear morphology in P1-KO, MCK-Cre/cKO and desmin-null, but not P1b-KO myofibers. Scale bars, 20 µm. (B) Bar graphs representing statistical analyses of nuclear areas, perimeters, shape factors and aspect ratios. Note similarity of alterations observed for P1-KO and MCK-Cre/cKO myofibers. Note also that the perimeter chart x-axis starts at 25 µm. Error bars ± SEM, three experiments, n = 150 nuclei examined in each experiment. *P < 0.05; **P < 0.005; ***P < 0.001. (C) Statistical analysis of distances between nuclei (closest neighbors) determined in specimens as shown in (A). Error bars ± SEM, three experiments, n = 70 total measurements each. *P < 0.05; **P < 0.005; ***P < 0.001. (D) Like (A), using anti-α-actinin instead of anti-desmin antibodies. Note misalignment and disorganization of α-actinin-positive Z-disks in P1−/−, MCK-Cre/cKO and desmin-null muscle fibers. Scale bars, 20 µm.
In stark contrast, in P1-KO muscle fibers the desmin networks appeared as having collapsed, forming aggregates in perinuclear areas (Fig. 1A and B, lower rows). Overall, the desmin network of P1-deficient muscle appeared much more dispersed and irregularly distributed in the vicinity of nuclei, compared with the regular staining patterns observed in wild-type muscle. In addition, DNA-specific Hoechst staining revealed clearly evident alterations of nuclear morphology (see below). These observations suggested that P1 plays an important role in perinuclear IF network organization, with possible consequences for myonuclear functions.
Lack of P1 leads to alterations in size, shape and positioning of nuclei in skeletal muscle fibers
To investigate whether P1-mediated nuclear anchoring of IF networks and phenotypic alterations of myonuclei were interdependent, we subjected teased extensor digitorum longus (EDL) muscle fibers isolated from wild-type, P1-KO, P1b-KO, MCK-Cre/cKO and desmin-null mice to comparative analyses of nuclear and IF network parameters. P1b-KO and MCK-Cre/cKO specimens were included in this analysis to establish isoform-specificity of P1-KO phenotypes, whereas desmin-null muscle fibers served for assessing whether the lack of IF anchorage was equally effective in altering the nuclear morphology as was the deficiency of IFs per se. In this analysis, we excluded nuclei that were in the immediate vicinity of neuromuscular synapses, because they often show clustering, impeding the distinction of individual features; moreover, synaptic nuclei are functionally specialized to synthesize constituent proteins of the postsynaptic apparatus, whereas this does not apply for more distant myonuclei (21–24).
Our analysis revealed that P1-KO, MCK-Cre/cKO, as well as desmin-null myonuclei were bigger and more rounded compared with their wild-type and P1b-KO counterparts, which typically were elongated and of ellipsoidal shape (Fig. 2A). Furthermore, in contrast to wild-type and P1b-KO myofibers, in P1-KO, MCK-Cre/cKO and desmin KO tissues, nuclei were no longer linearly arranged and oriented such that their long axis was parallel to that of the muscle fiber. In these cases, the nuclei were irregularly spaced and frequently clustered (Fig. 2A). In P1-KO as well as in MCK-Cre/cKO muscle fibers, nuclear areas and perimeters were found to be significantly increased (50 and 20%, respectively) compared with wild-type specimens (Fig. 2B). Furthermore, shape factor and aspect ratio measurements clearly showed that P1-KO nuclei were more rounded and displayed smoother surfaces compared with the typically flat and elongated shape of control nuclei (Fig. 2B). The corresponding data obtained for P1b-KO muscle correlated with those of wild-type specimens, suggesting that P1 affects nuclei in an isoform-specific manner. Interestingly, the analysis of desmin-null myonuclei revealed phenotypic characteristics that were very similar to those of P1-deficient mice (Fig. 2A and B). This suggested that the proper docking of desmin IFs at the nuclear membrane via P1 was as important for nuclear morphology as filament network formation itself.
Examining the positioning of nuclei along myofibers, we found that in P1-deficient cells nuclei were no longer distributed in a regular pattern all along the fibers, but often showed clustering. In fact, the average distance between neighboring nuclei in P1-KO fibers was strikingly smaller (>3-fold) than that in wild-type or P1b-KO muscle specimens (Fig. 2C). These observations demonstrated that for performing their previously proposed functions in positioning and physically linking nuclei to sarcomeric structures (8,25), desmin IFs inevitably needed to be anchored at the nuclear membrane via plectin.
Albeit the nuclear parameters were affected to a similar extent in myofibers lacking just P1 (P1-KO) or all plectin isoforms combined (MCK-Cre/cKO), the magnitude of desmin IF network disorganization differed in both cell types. While in P1-KO muscle fibers desmin-positive aggregates were restricted to perinuclear areas, in MCK-Cre/cKO cells, massive aggregation was observed all over the sarcoplasm (Fig. 2A). Thus, the disruption of its nuclear docking alone affected IF network integrity as a whole to a much lesser extent than that of all plectin isoform-mediated IF linkages. Similarly, the α-actinin-positive Z-disk pattern of P1-KO myofibers deviated only slightly and in a spatially restricted manner (perinuclear) from the highly regular pattern typical of wild-type (and P1b-KO) specimens, whereas the corresponding phenotype manifestation in MCK-Cre/cKO myofibers (and desmin-null muscle fibers) was much stronger, affecting myofibrils throughout the whole sarcoplasm (Fig. 2D).
P1-deficient myoblasts undergo differentiation ex vivo forming myotubes with giant nuclei
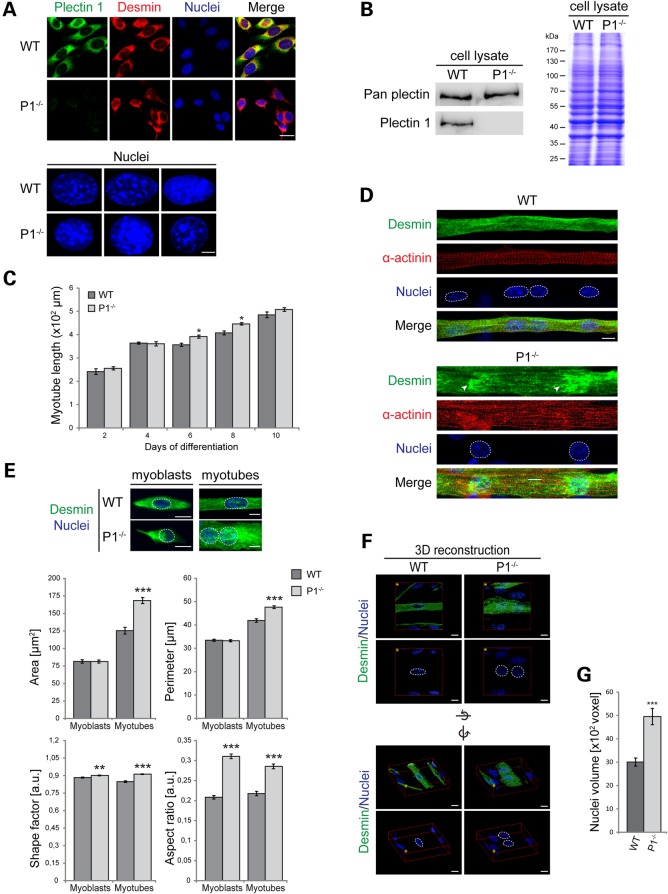
To study the mechanisms leading to the nuclear phenotype of isoform P1-deficient myofibers on molecular and cellular levels, we generated a P1 isoform-deficient immortalized myoblast cell line, using a previously established protocol that is based on the isolation of myoblast cultures from mutant mouse lines crossed into a p53−/− background (20). As shown by immunostaining and immunoblotting (IB) using isoform-specific and anti-pan plectin antibodies, cultures of immortalized myoblasts isolated from P1−/−/p53−/− double knockout mice (from now on referred to as P1−/− myoblasts) were devoid of P1, while other isoforms of plectin seemed to be expressed normally (Fig. 3A and B). Immunofluorescence microscopy (IFM) of P1+/+/p53−/− myoblasts (referred to as wild-type) using isoform P1 and desmin-specific antibodies revealed partial colocalization of both proteins predominantly in perinuclear areas (Fig. 3A). Moreover, a trend toward a more spherical shape of P−/− compared with wild-type nuclei became apparent at higher magnifications (Fig. 3A, lower panels; see also Fig. 3E).
Figure 3.
Nuclear alterations of plectin-deficient myofibers are mimicked by myotubes differentiated ex vivo from P1-deficient myoblasts. (A) Upper panels, double IFM of wild-type and P1−/− myoblasts using anti-P1 and anti-desmin antibodies, and visualization of nuclei with Hoechst dye; lower panels, higher magnification of stained nuclei from three different cells. Scale bars, 5 µm (upper panels), 10 µm (lower panels). (B) IB of total cell lysates prepared from wild-type or P1–/– myotubes using anti-pan plectin and anti-P1 antibodies. (C) Statistical analysis of the length of wild-type and P1−/− myotubes differentiated for the time indicated. Error bars ± SEM. *P < 0.05. Myotubes analyzed: WT, n = 76, P1–/–, n = 79 (day 2); WT, n = 285, P1–/–, n = 127 (day 4); WT, n = 335, P1–/–, n = 270 (day 6); WT, n = 539, P1–/–, n = 399 (day 8); WT, n = 363, P1–/–, n = 260 (day 10). (D) Double IFM of wild-type and P1−/− myotubes using antibodies to desmin and α-actinin; nuclei visualized with Hoechst dye are highlighted by a dashed line. Arrowheads indicate aggregates of desmin IFs at the nuclear periphery in P1−/− cells. Scale bars, 10 µm. (E) Upper panel: IFM of wild-type and P1−/− myotubes (day 7) using antibodies to desmin; nuclei were visualized with Hoechst dye. Lower panel: bar graphs representing statistical analyses of areas, perimeters, shape factors and aspect ratios of nuclei derived from undifferentiated myoblasts and multinucleated myotubes of wild-type and P1−/− cell lines. Error bars ± SEM, three experiments, n = 60 nuclei analyzed from undifferentiated and differentiated cells. **P < 0.005; ***P < 0.001. (F) Wild-type and P1−/− myotubes were immunolabeled using antibodies to desmin, and nuclei (highlighted by dashed lines) were visualized with Hoechst dye. 3D reconstructions of single WT and P1−/−cells were prepared from confocal Z-stack images using Huygens software. Images are shown from two perspectives: above the cells (upper panels), and sideways (lower panels). Image orientations (yellow asterisk) and rotation axes are indicated. Scale bars, 10 µm. (G) Statistical evaluation of the volume of nuclei present in wild-type and P1−/− myotubes. Error bars ± SEM, three experiments. Nuclei analyzed in each experiment: WT, n = 97; P1−/−, n = 101. ***P < 0.001.
P1−/− myoblasts were able to fuse with each other and differentiate into multinucleated myotubes similar to their wild-type counterparts (Fig. 3C and D). However, while in wild-type cells, the desmin IF network, aside from its association with Z-disks, was rather uniformly distributed over the entire fiber, in P1−/− cells it showed an accumulation in aggregate-like structures surrounding the nuclei (Fig. 3D). Moreover, similar to P1-deficient muscle fibers, the nuclei in P1−/− myotubes showed distinct morphological features compared with wild-type specimens. In particular, an increase in the size of nuclei and a change in nuclear shape from ellipsoidal into more spherical could be noticed (Fig. 3D). Apart from altered nuclear morphology, P1-deficient myotubes appeared to be thicker, i.e. more expanded along their short axis, and the α-actinin-positive Z-disk striation pattern appeared less ordered compared with wild-type cells (Fig. 3D).
When we compared and statistically analyzed nuclear parameter alterations in proliferating (mononucleated) myoblasts versus differentiated (multinucleated) myotubes, no differences in nuclear areas or perimeters of wild-type and P1−/− nuclei were found at the myoblast stage, whereas in P1−/− myotubes these parameters were increased by near 30% (area) and 14% (perimeter) compared with the wild-type cells (Fig. 3E). However, similar to their differentiated counterparts, undifferentiated P1−/− myoblasts were distinguishable from wild-type myoblasts by the shape of their nuclei, manifesting as increased shape factors and aspect ratios. Similar analyses of primary p53+/+/P1−/− (single knockout) myoblasts revealed changes in all of the four nuclear parameters tested (unpublished data). Using three-dimensional (3D) deconvolution analysis, the total volume of nuclei in differentiated P1-deficient cells was found to be increased by ∼65% compared with wild-type nuclei (Fig. 3F and G). Thus, we concluded that nuclei in P1-deficient myoblasts and myotubes in general were more spheroidal and displayed smoother surfaces compared with their wild-type counterparts (Fig. 3E). In all, these results indicated that the decoupling of myonuclei from desmin IFs leads to an increase in their size, a more spherical shape and a larger volume.
Plectin isoform-specific rescue of nuclear phenotypes in P1−/− myocytes
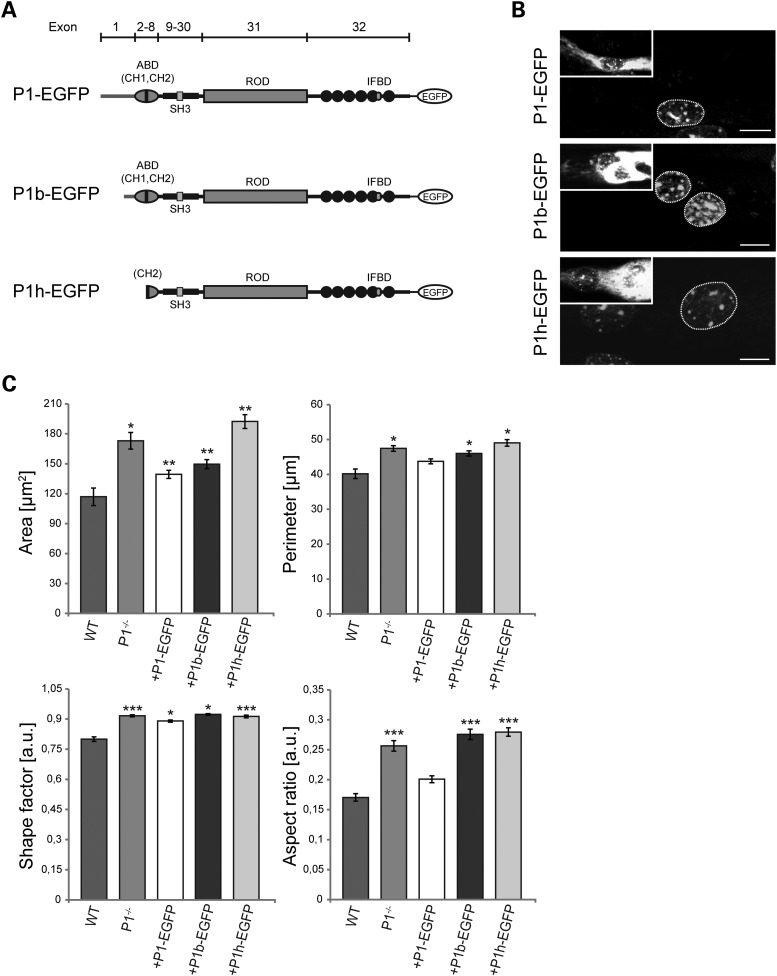
To assess the potential of distinct plectin isoforms to restore normal nuclear morphology, plasmids encoding EGFP-tagged versions of full-length P1, P1b or P1h were transfected into P1−/− myoblasts that subsequently were differentiated into myotubes. All of the plectin expressed isoforms contained the C-terminal IF-binding domain, but differed at their N termini (Fig. 4A). P1 and P1b started with the short sequences encoded by their alternative first exons, whereas P1h (having its start codon in exon 6) represented an isoform without isoform-specific N terminal sequences and without a functional ABD (14) (Fig. 4A). As shown in Figure 4B and C, the forced expression of P1 and P1b, both resulted in a reduction of the size and the perimeters of nuclei, albeit P1 was significantly more effective than P1b. Statistically, the differences between wild-type and P1−/− myotubes in size and perimeters of nuclei were reduced upon expression of P1 by ∼64 and 50%, respectively. Remarkably, only full-length P1, but not P1b, was effecting a rescue of the elliptical shape typical of wild-type nuclei (Fig. 4C, aspect ratio). In contrast, overexpressed P1h showed no effect on any of the nuclear parameters measured, and thus was lacking any rescue potential (Fig. 4C).
Figure 4.
P1-specific rescue of nuclear morphology in differentiated myotubes. (A) Schematic representation of EGFP-tagged plectin isoform cDNA expression constructs. Exon organization (bar on top) and important molecular domains are indicated. ABD, actin binding domain; CH1/2, calponin homology domains; SH3, Src homology 3 domain; IFBD, intermediate filament binding domain. (B) P1−/− myoblasts transfected with EGFP-tagged plectin isoforms (P1-EGFP, P1b-EGFP or P1h-EGFP) were subjected (7 days) to differentiation into multinucleated myotubes and nuclei visualized with Hoechst dye; nuclei of non-transfected myotubes served as controls. Scale bars, 10 µm. (C) Statistical analysis of area, perimeter, shape factor and aspect ratio of nuclei in myotubes transfected with different plectin cDNA construct. Note rescue of nuclear morphology upon P1-EGFP expression and partial rescue upon P1b-EGFP expression. Error bars ± SEM, three experiments, n = 33 nuclei analyzed. *P < 0.05; **P < 0.005; ***P < 0.001.
Reduced mobility and translocation of nuclei in P1-deficient myotubes
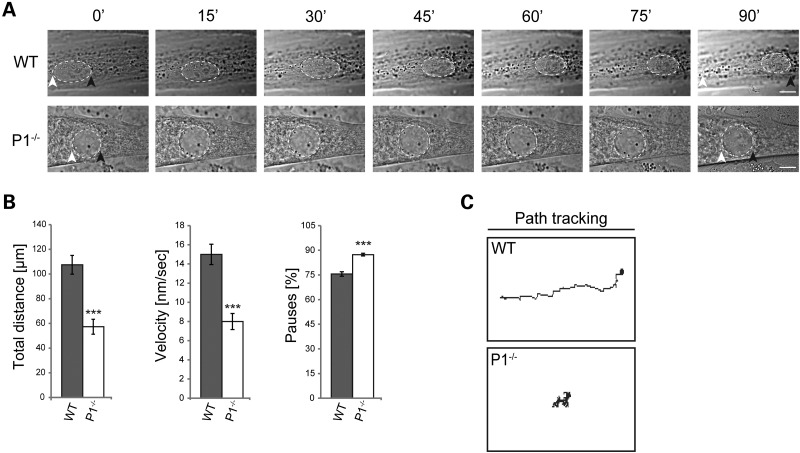
It has been shown that myonuclei translocate along the long axis of the myofibers in order to minimize transport distances and ensure sufficient transcriptional capacity on-site (26–28). To assess whether P1 deficiency affected the movement of nuclei along the myotube, fully differentiated P1−/− and wild-type myotubes were subjected to live cell video microscopy. The parameters measured, total distance, velocity and pause frequency revealed that the nuclear dynamics in P1 cells were drastically decreased compared with wild-type. While control nuclei could easily translocate within the myotube, nuclei from P1-deficient cells migrate to a lesser extent or sometimes seem not to move at all (Fig. 5A). During a 2-h observation period, wild-type nuclei covered distances of up to 100 µm with an average velocity of 15 nm/s, while the corresponding parameters of P1−/− nuclei were reduced to ∼50% (Fig. 5B). Moreover, we noticed that P1−/− nuclei paused more frequently compared with their wild-type counterparts (Fig. 5B). Trajectory tracking of nuclei clearly demonstrated that within the same timeframe, wild-type nuclei migrated over longer distances than P1−/− nuclei (Fig. 5C). The latter also showed slower movements and, instead of migrating directionally, they often oscillated or were rotating in one place (Fig. 6A and C). The observed decrease in nuclear dynamics in P1−/− myotubes would be consistent with a potential role of P1 in force transmission from the cytoskeleton to the nuclear envelope.
Figure 5.
P1 deficiency affects dynamic properties of nuclei. (A) Phase contrast images were taken from time-lapse recordings of selected wild-type and P1−/− myotube nuclei (dashed lines). Note slower movement of P1−/− compared with wild-type nuclei over time. White and black arrowheads, starting and endpoints of movement, respectively. Scale bars, 10 µm. (B) Statistical quantification of nuclear dynamics in wild-type and P1−/− myotubes. Error bars ± SEM, three experiments, n = 33 nuclei analyzed (each for WT and P1−/− cells). ***P < 0.001. (C) Migration traces (manual path tracking) of nuclei shown in (A).
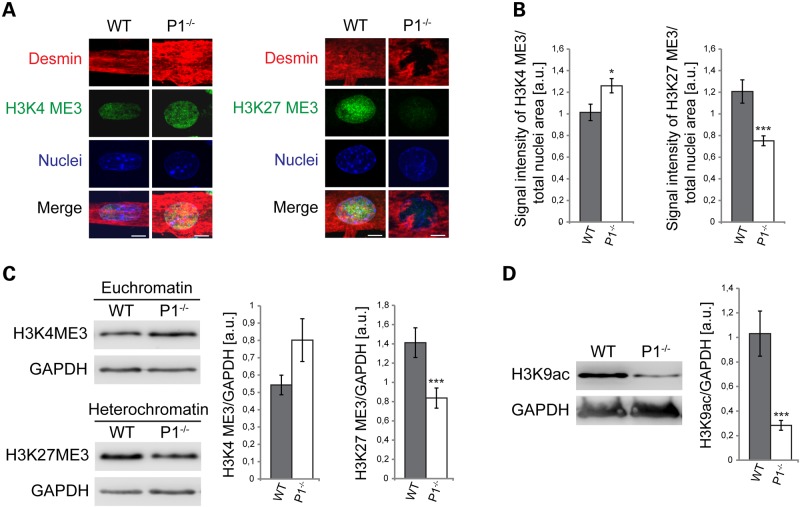
Figure 6.
P1 deficiency leads to conformational changes of chromatin. (A) Triple labeling of nuclei and their cytoplasmic environment using Hoechst dye (nuclei) and antibodies to desmin and either euchromatin (H3K4ME3, left panels) or heterochromatin (H3K27ME3, right panels). Scale bars, 10 µm. (B) Bar graphs representing statistical analyses of euchromatin and heterochromatin content of nuclei shown in (A). Fluorescent signals of H3K4ME3 or H3K27ME3 were normalized to total nuclear areas. Error bars ± SEM, three experiments, n = 81 nuclei measured for each experiment. *P < 0.05; ***P < 0.001. (C) Quantitative IB analysis of euchromatin and heterochromatin in cell lysates derived from wild-type and P1−/− myotubes. Signals were normalized to GAPDH (see also Supplementary Material, Fig. S1). (D) Quantitation (as in C) of total amount of H3K9ac in wild-type and P1−/− myotube cell lysates. Note significant decrease in overall gene expression level in P1-deficient compared with wild-type cells.
P1-deficiency affects chromatin conformation and gene expression
Mechanotransduction, the mechanisms involving the transmission of mechanical signals across the nuclear envelope to the nuclear lamina and transcription machinery, requires the proper physical coupling of the nucleus to the cytoskeleton (29). To investigate whether the observed alterations in nuclear morphology and migration were paralleled by chromatin reorganization, nuclei from wild-type and P1-deficient (desmin-positive) myotubes were labeled for DNA (Hoechst dye) and either the euchromatin marker histone 3 tri methyl K4 (H3K4me3), or the heterochromatin marker H3K27me3, using site-specific antibodies (Fig. 6A). The quantitation of the chromatin marker signals revealed a 25% increased level of euchromatin with a simultaneous >30% downregulation of heterochromatin compared with the chromatin content of wild-type counterparts (Fig. 6B). These findings were confirmed by quantitative IB analysis of cell lysates (Fig. 6C; and Supplementary Material, Fig. S1). Thus P1−/− nuclei showed an upshift in total euchromatin levels and a significant decrease in heterochromatin content compared with wild-type specimens.
On the basis of the increased proportion of euchromatin in P1-deficient myotubes, one may have expected to find a general transcriptional activation in these cells. To verify this, we measured the total levels of histone 3 acetylated at lysine 9 (H3K9ac), a marker generally linked to gene activation, in P1−/− and wild-type myotube lysates by IB. Intriguingly, we found that the level of total H3K9ac was not elevated but decreased (to ∼30%) in P1−/− compared with wild-type cells, suggesting that although the euchromatin state was elevated in P1-deficient myotubes, transcription in general was not (Fig. 6D). These data suggested that due to the lack of P1 and the ensuing alterations in nuclear architecture and chromatin conformation, genes involved in a variety of cellular functions may show abnormalities in expression.
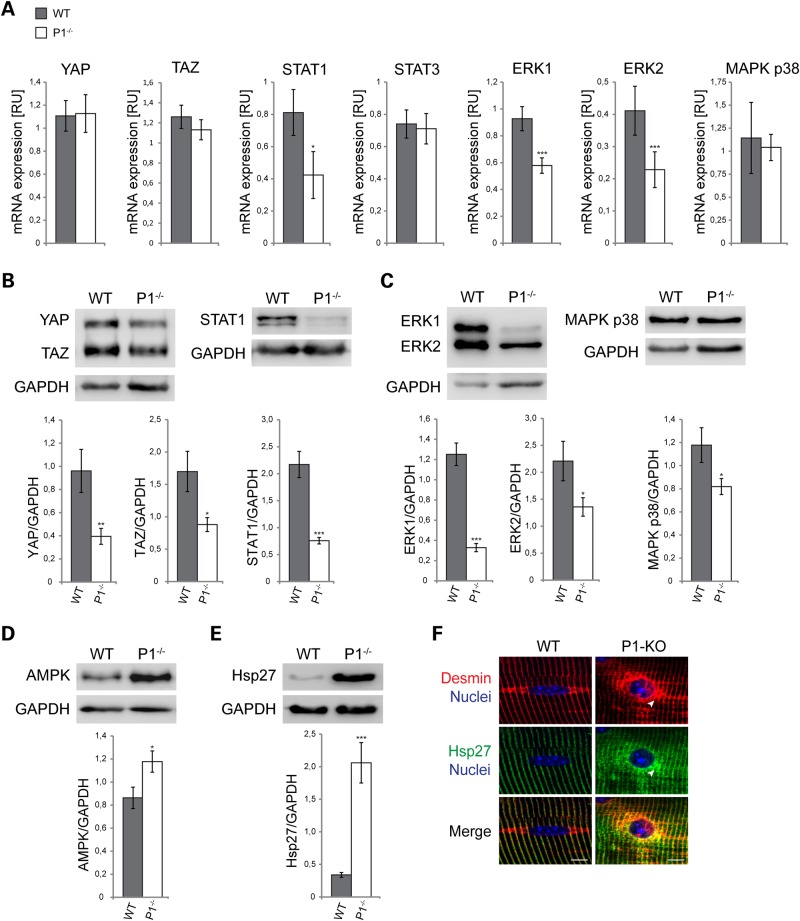
To assess whether proteins involved in mechanotransduction were affected, we measured mRNA and protein expression levels of the mechanosignaling-linked transcription factors YAP, TAZ/WWTR1, STAT1 and STAT3 (30–32), and of the MAP kinase pathway constituents ERK1/2, and MAPK p38. Using quantitative RT-PCR (qRT-PCR), we found mRNA levels of STAT1 as well as ERK1 and ERK2 to be significantly reduced in P1−/− compared with wild-type cells (by 48, 38, and 45%, respectively), whereas no significant differences were found for the remainder of the transcripts tested (Fig. 7A; and Supplementary Material, Fig. S2). Interestingly, however, on the protein level, not only STAT1 and ERK1/2, but all of these proteins tested, including YAP, TAZ and MAPK p38, were found to be significantly reduced, with STAT1 (70% reduction) and MAP kinase ERK1 (75%) showing the most dramatic effects (Fig. 7A and B). On the other hand, AMP-activated protein kinase (AMPK), a protein responsible for energy homeostasis that is recruited to Z-disks by plectin during myogenesis (33), was found to be upregulated in P1-deficient versus wild-type cells by 40% (Fig. 7C). Similarly, a striking (5-fold compared with wild-type) upregulation of heat shock protein hsp27 was observed for P1−/− myotube lysates (Fig. 7D), indicative of a massive chaperone activation in response to P1 deficiency-inflicted IF network collapse. A comparably strong upregulation of hsp27 had previously been observed in plectin-null myotubes (20). As expected, double IFM of teased P1−/− myofibers using antibodies to hsp27 and desmin revealed colocalization of hsp27 with desmin-positive perinuclear aggregates (Fig. 7E). In all, these results supported the notion that P1 not only is important for specific morphological features, translocation and compartmentalization of myonuclei, but also affects functional properties of myonuclei, including the expression of some genes involved in mechanotransduction, metabolism and stress response.
Figure 7.
Altered mRNA expression and protein homeostasis in P1-deficient muscle cells. (A) Quantitative analysis of mechanotransducer transcripts in wild-type and P1-deficient myotubes. Graphs show the relative expression levels of YAP, TAZ, STAT1/3, ERK1/2 and MAPK p38 in wild-type versus P1-deficient myotubes. RNA was isolated from at least three independent samples of (each) wild-type and P1-deficient cell cultures. RT-PCR analyses were performed in duplicates and measurements made using 96-well optical plates as described in the text. The housekeeping gene HPRT was used as a reference. Note significant downregulation of mRNA expression of STAT1, ERK1 and ERK2 in P1-deficient cells compared with wild-type controls. Error bars, ±SEM. *P < 0.05; ***P < 0.001. (B–E) Equivalent amounts of cell lysates derived from wild-type and P1–/– muscles were subjected to IB analysis using antibodies to the signal transducers and activators of transcriptions YAP/TAZ and STAT1 (B), the protein kinases ERK1/2 and MAPK p38 (C), AMP-activated protein kinase (AMPK) (D) and heat shock protein (hsp) 27 (E). GAPDH was used as loading control. Error bars ± SEM. *P < 0.05; **P < 0.005; ***P < 0.001. Note striking (5-fold) upregulation of hsp27 in P1-deficient cells (D). (F) Double IFM of the nuclear area of wild-type and P1-KO teased EDL muscle fibers using antibodies to desmin and hsp27; nuclei were visualized with Hoechst dye. Note accumulation and colocalization of desmin and hsp27 in the perinuclear area of the P1-KO fiber (arrowheads). Scale bars, 10 µm.
Isoform-specific interaction of P1 with endophilin B in skeletal muscle
To better understand the mechanism of P1-nuclear envelope docking, we assessed whether protein(s) forming the LINC complex were involved in this process. One potential P1-specific binding partner could have been the ubiquitously expressed outer nuclear membrane protein nesprin-3, which interacts with plectin's ABD via the first of its eight spectrin repeats (10,11), and has been proposed to stabilize the cytoskeletal anchorage and maintain the structural integrity and shape of the nucleus (34–36). However, the interaction between nesprin-3 and plectin so far has not been shown to be specific for any of the plectin isoforms. Furthermore, preliminary studies from our laboratory (in collaboration with Colin Stewart, Institute of Medical Biology, Singapore) indicated that nesprin-3-deficient skeletal muscle tissues did not exhibit the nuclear phenotype characteristic of P1-deficient tissues (unpublished data). These observations suggested that another or additional binding partner of P1 is mediating its targeting and desmin network recruitment to nuclei.
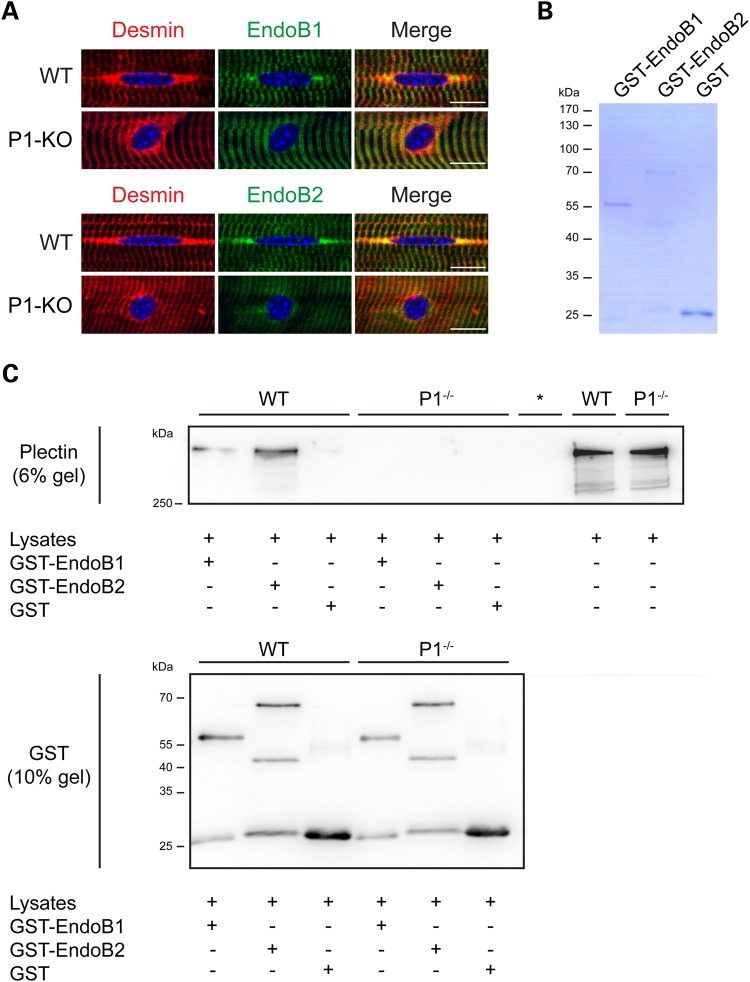
Recently, it has been shown that endophilin B2 (endoB2), a BAR domain-containing protein, binds specifically to P1 in HeLa cells (37). Because proteins with BAR domains are essential in controlling the shape and dynamics of intracellular membranes, we assessed whether this type of interaction occurs in myofibers. Analyzing the localization of endophilins in teased wild-type myofibers by IFM, using antibodies to endophilin B1 (endoB1) and endoB2 as well as desmin, we observed colocalization of both endophilins with desmin in characteristic tail-like structures at the nuclear periphery and partially also at Z-disks, whereby the signal for endoB2 at the nuclear edges was much stronger than that of endoB1 (Fig. 8A, upper panels). Intriguingly, in P1-deficient myofibers, neither one of these endophilins was found accumulated at the nuclear periphery any more. Their staining patterns were rather diffuse in perinuclear regions and more restricted to Z-disks, contrasting the perinuclear accumulation of desmin-positive aggregates (Fig. 8A, lower panels). These observations supported the notion that P1 indeed is an interaction partner of endophilins at the nuclear membrane of myofibers.
Figure 8.
Endophilin B2 colocalizes and interacts with P1 at the periphery of myofibers. (A) Teased EDL muscle fibers derived from wild-type and P1-KO mice were immunolabeled using antibodies to desmin and either endoB1 (upper panels) or endoB2 (lower panels). Note colocalization of both endophilins with desmin-positive tail-like structures at the nuclear periphery in wild-type fibers, in contrast to the diffuse distribution of endophilins in the vicinity of P1-KO myonuclei. Scale bars, 10 µm. (B) SDS-PAGE (Coomassie-staining) of recombinant proteins (GST-tagged endoB1 and endoB2, and GST protein) used in pull-down assays. (C) Myotubes differentiated from wild-type and P1–/– myoblasts were subjected to GST pull-down assays; GST protein served as negative control. Proteins pulled-down and total cell lysates (input) were analyzed by IB and probed with either anti-pan plectin or anti-GST antibodies using two separate (6 and 10% polyacrylamide) gels. Note positive plectin signals in GST-endoB1 as well as GST-endoB2 pull-down samples from wild-type lysates. *, empty lane.
To test plectin–endophilin binding on the molecular level, we performed GST pull-down assays, where cell lysates derived from P1−/− or wild-type myotubes were incubated with recombinant (GST-tagged) full-length endoB1 and endoB2 proteins coupled to glutathione Sepharose beads (Fig. 8B and C). The analysis of endophilin-bound plectin by IB using anti-pan plectin antibodies showed that P1 present in wild-type lysates bound to either one of the recombinant endophilin versions (Fig. 8C, WT). Although pull downs are limited in providing quantitative information, a clear trend for endoB2 pulling-down more plectin than endoB1 was systematically observed (five independent pull downs). The analysis of the material pulled-down from P1−/− cell lysates revealed no plectin-positive signals (Fig. 8C, P1−/−). As no other isoforms of plectin (albeit present in the lysates) were pulled-down, and consequently were able to substitute for P1, we concluded that the interaction of both endophilin versions with plectin was isoform P1-specific.
Discussion
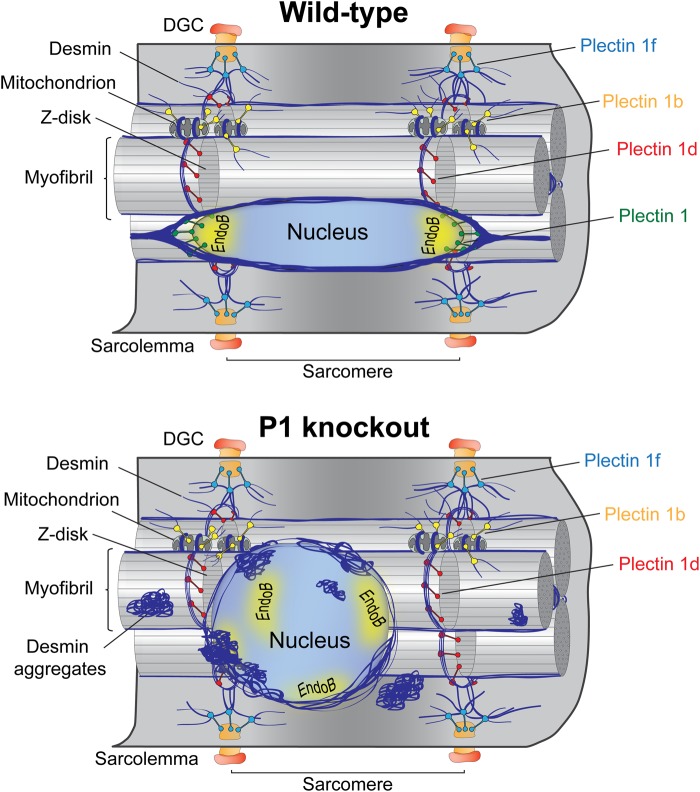
This study shows that P1, an ubiquitously expressed isoform of plectin, targets and anchors the desmin IF network to the outer membrane system of myonuclei. By phenotypically analyzing myofibers and myotubes derived from four genetically different mouse lines, including two isoform-specific knockout lines (P1-KO and P1b-KO), a muscle-restricted conditional (MCK/Cre) plectin knockout line and desmin-null mice, we could show that P1 is essential for maintaining the typically spheroidal morphology and the regular distribution of myonuclei along the myofiber. As depicted in the schematic model presented in Figure 9, our data suggest that P1-mediated anchorage and integration into the desmin IF network provides myonuclei with a physical connection to the contractile apparatus (via P1d), the sarcoplasmic membrane (via P1f) and mitochondria (via P1b). As previously shown for IFs expressed in other cell types, such as fibroblasts, keratinocytes and endothelial cells (38–42), desmin IF networks form a cage-like structure around myonuclei and mechanically connect and integrate the nucleus with the cytoskeleton, other organelles as well as plasma membrane-associated structures. In this way, these elements are hold under isometric tension (prestress) and provide mechanical stiffness for the whole cell (43). The lack of P1 and the ensuing decoupling of nuclei from the IF network (formation of perinuclear desmin-positive protein aggregates) lead to an expansion in the volume of nuclei and a shape transition from ellipsoidal, as is typical of their pre-stressed (wild-type) state, to more spherical. It has been shown that disruption, mutations or deletions of IF proteins, including lamins, result in decreased mechanical stiffness of the cell but also in cell injury in response to mechanical stress and reduced mechanical activation of gene transcription (44–46). Because we found that the decoupling of IFs from the nuclear membrane as a consequence of P1 deficiency phenotypically manifests with nuclear aberrations similar to those observed in desmin-null muscle, we conclude that nuclear docking of the IF network via plectin is indispensable for its role as mechanotransducer.
Figure 9.
Schematic model of cell organization and nuclear morphology typical of wild-type and P1-KO myofibers. (A) Each one of four major plectin isoforms expressed in myofibers is targeted to a different cellular compartment: P1 to the nuclear/ER membrane system, P1b to mitochondria, P1d to Z-disks and P1f to the sarcolemma. All of these isoforms recruit desmin IFs via their C-terminal IF-binding domain to these respective sites, thereby controlling IF network architecture and reinforcing the structural integrity of myofibers. P1 is predominantly localized at tail-like structures longitudinally extending from the nuclear equator. Desmin IFs recruited by P1 form a cage-like structure around nuclei, which holds nuclei under isometric tension, enabling their immediate response to mechanical stimuli. Ellipsoidally shaped, elongated nuclei are spatially distributed all along the myofiber. P1 targeting and nuclear docking occurs via nucleus-associated proteins, such as nesprin-3 and endophilin B. (B) In P1-deficient myofibers, desmin IFs lose their docking site at the nuclear membrane, leading to their collapse and aggregation in the vicinity and onto the surface of nuclei. The disconnection of nuclei from the IF network promotes alterations of nuclear morphology, dislocation of nuclei, decrease in their mobility and impaired mechanotransduction. Moreover, IF-uncoupling effects a structural reorganization of the nuclear interior, presenting as altered chromatin conformation and decreased gene expression. Note, for simplicity the IF network is depicted only in part. Yellow areas (A and B), perinuclear location of endophilin B.
Isoform P1's perinuclear localization, as revealed using isoform-specific antibodies, its ability to restore (contrary to other isoforms) the normal (ellipsoidal) shape of nuclei upon forced expression in P1-deficient myocytes and the observation that other isoform-specific knockout mouse lines, such as P1b-KO mice, do not show a similar phenotype, strongly suggest that P1 affects myonuclei in an isoform-specific manner. Albeit some parameters, such as nuclear area and perimeter, could be rescued to some extent also by reconstitution with full-length P1b (but not P1h), the crucial element responsible for P1-targeting to the nuclear/ER membrane system is clearly the unique N-terminal region of P1. P1 is characterized by the longest isoform-specific first exon-encoded sequence (180 amino acid residues) and it contains several binding motifs for interaction with distinct molecules. These include ubiquitin ligase Siah (Seven in absentia homolog), which binds to a RPVAxVxPxxR motif within residues 95–117 in exon 1 (47); endoB2 which interacts with P1 via a PETP motif within residues 150–153 (37) and β-synemin (48). Moreover, exon 1 contains numerous positively charged amino acids (especially arginines which mediate hydrogen bonding and amino-aromatic interactions) making P1 an attractive binding partner for negatively charged molecules. Interestingly, methylation of arginine disrupts hydrogen bonding and induces steric hindrance which could modulate intra- or inter-molecular interactions of target proteins or even disrupt protein folding (49–52). Additionally, exon 1, unlike other plectin isoform-specific exons, comprises a KKDRR sequence, which constitutes a classical monopartite [K(K/R)X(K/R)] nuclear localization signal (NLS) (53). The question still remains whether the predicted NLS within exon 1 is functional. Future studies addressing this issue may help to better understand the nuclear targeting mechanism of the protein or possibly reveal new function(s) of this isoform.
Regarding the question how P1, once targeted to the nucleus, is kept at the nuclear envelope, we hypothesize that the isoform-specific N terminal protein domain of P1 affects the interaction potential of other regions, including the contiguous multifunctional ABD and the more downstream SH3 domain, which are common to all isoforms. This could happen through triggering conformational changes within the molecule, or simply by positioning these domains in spatial proximity to their interaction partners at the nuclear membrane, creating a sort of plectin-dependent microenvironment. In this way, the N terminus of P1 could not only be responsible for its targeting toward the nuclear membrane, but also preserve and reinforce IF network integration of nuclei by undergoing multiple interactions, a feature expected to foster mechanotransduction. We speculate that other isoforms act in a similar way at other cellular locations. Isoform-specific targeting has been reported also for the cytolinker protein BPAG1, in which case an N-terminal fusion protein of BPAG1a was shown to colocalize with actin stress fibers and nuclei in C2C12 cells (54).
Our studies clearly demonstrated that within the same time frame P1−/− nuclei migrated over shorter distances compared with wild-type nuclei. However, they were not frozen in place, as they were still able to oscillate and/or rotate. Thus it is likely that some of the force providers known to be involved in positioning and translocation of nuclei, such as the actomyosin or microtubule based machineries, were still acting on them. This raises the question how the decoupling of nuclei from the desmin IF network mechanistically suppresses myonuclear translocation. One possible explanation could be that IFs provide a kind of stable rail or corridor system along which actomyosin or MT-based machineries become oriented and are able to develop directional forces. In such a scenario, the internal crosslinking and the anchorage of IFs at sarcolemmal costameres and Z-disks of the contractile apparatus via plectin molecules could provide the local mechanical constraint required for actomyosin or MT-driven directional force generation. However, in the absence of P1, this constraint would be missing as a result of the disorganization and hence dysfunction of the perinuclear IF network, leading to the observed non-directional and apparently uncontrolled movement of the nuclei. Another possible explanation would be that the desmin-positive protein aggregates present in close vicinity of P1-deficient myonuclei physically block nuclear translocation machineries. In any case, our observations suggest that a P1-mediated crosstalk between IFs and force-producing machineries is required for the positioning and translocation of myonuclei. It will be a challenging task for future studies to reveal the underlying, presumably quite complex, molecular mechanism in more detail.
On the basis of the increased euchromatin and substantially decreased heterochromatin levels found in P1-deficient myonuclei, one may have expected elevated transcriptional activation. However, we found gene activation markers, such as H3K9ac, to be low, indicating that in spite of euchromatin elevation, transcription was blocked and not proceeding like in wild-type controls. Thus, as a consequence of nuclear deformation, P1 deficiency led to changes in the conformational state of the chromatin, eventually causing alteration of transcription. This notion is supported by previous observations showing that altered nuclear morphology leads to changes in chromosome organization, which then can affect gene expression (55,56). Moreover, it has been shown that changes in the forces applied to the nucleus can evoke changes in nucleolar functions, chromatin folding or access of transcription factors to gene regulatory sites (43,57). In fact some of the mechanotransducer proteins tested in our study, including the transcription factor STAT1, and MAP kinases ERK1/2, showed a significant downregulation on the transcriptional level. As not only these, but all of the mechanotransducer proteins tested, showed significantly downregulated protein levels in P1-deficient cells, dysfunctions of intracellular signaling pathways are a likely consequence. In addition, the observed upregulation of AMPK, a protein responsible for energy balance regulation at the single-cell level, might affect the biosynthesis and contractility of the muscle. The striking increase in hsp27 protein levels, combined with the observed colocalization of hsp27 with perinuclear desmin aggregates, indicated a massive activation of chaperones in response to plectin deficiency, fully consistent with dysregulated protein homeostasis. Overall, these observations confirmed previous studies on plectin-null cells, which show massive desmin aggregation accompanied by a remarkable upregulation of hsp27 (20).
The nuclei in nesprin-3-deficient myofibers did not feature any of the morphological alterations found in P1-KO muscle fibers (unpublished data). Moreover, no alterations in protein levels or localization of nesprin-3 were observed, and a downregulation of the protein during myoblasts differentiation was typical for both, P1-deficient and wild-type myotubes (unpublished data). On these grounds, we concluded that if interactions between nesprin-3 and plectin were occurring in myofibers, they were unrelated to the pathway leading to the phenotypic alterations of P1-deficient myonuclei. Thus, P1-nesprin-3 interactions occurring at the outer nuclear membrane are likely to fulfill functions other than shaping the morphology or mediating the positioning of nuclei along the muscle fiber. Reinforcing and stabilizing cytoplasmic-nuclear coupling might be one of them (58,59), transmission of mechanical signals another (34). In all, our results are consistent with the idea that the BAR domain-containing protein endophilin B is directly involved in targeting IFs to myonuclei, albeit a role of nesprin-3 in nuclear targeting of P1 at early stages of myoblast differentiation cannot be ruled out. Vannier et al. (37), who were the first to show an interaction between endoB2 and P1, suggested that these proteins form a functional complex that acts as a membrane-anchoring device of vimentin in HeLa cells. We now confirm this interaction and extend it to myofibers, where we show that P1 and endophilins form a complex that recruits desmin IFs to myonuclei with consequences for nuclear morphology and gene expression.
According to our working model (Fig. 9), P1 binds via its unique N-terminal molecular domain to the nuclear/ER membrane system, where it interacts with membrane-associated proteins such as endophilin B and nesprin-3, and simultaneously recruits desmin IFs via its C-terminal IF-binding domain. We demonstrated that two isoforms of endophilin B (endoB1 and endoB2) colocalize with P1 and desmin along the characteristic tail-like structures at the nuclear periphery and partially also with peripheral, probably costameric, structures at the level of Z-disks (48). Thus, in wild-type myofibers, nucleus-associated endophilin B was found accumulated in the equatorial membrane regions showing the greatest curvature. To find endophilins particularly in these regions was in agreement with the notion that BAR domain proteins explicitly sense and sculpt curved cellular membranes (60). The diffuse distribution of endoB1 and endoB2 over the nuclear membrane without regional accentuation in P1-deficient muscle was likely due to its decoupling from the IF cytoskeleton. It has been shown that various arrangements of BAR domains on the membrane surface lead to distinct membrane curvatures and bending dynamics (60). Consequently, cytoskeleton-decoupling in P1-deficient myofibers might promote conformational rearrangements of endophilins, affecting their interaction potential and leading to their more dispersed localization and the more spherical shape of myonuclei. This could also explain the observation that P1-deficient myonuclei are characterized by smoother surfaces compared with wild-type cells. An endophilin B-P1-desmin IF complex might thus support the proper bending of the nuclear membrane and maintenance of the spheroidal shape of myonuclei. Because proteins with BAR domains are supposed to control the shape and dynamics of intracellular membranes, endophilin B could at least partially be responsible for the nuclear phenotype observed in P1-deficient myofibers. Future, more detailed studies are needed to confirm this hypothesis and unravel the underlying molecular mechanism.
Recently reported mutations of the plectin gene within exons 1f (61) and 1a (62), causing muscle-only limb girdle muscular dystrophy (MD) and skin-only epidermolysis bullosa simplex (EBS), respectively, as well as the analyses of diverse plectin-isoform-deficient mouse lines (63) clearly showed that the different isoforms of plectin are responsible for distinct functions. The fact that P1 is involved in proper nuclear architecture and function opens a new perspective on the role of cytolinkers in mechanotransduction. P1-deficient myofibers and myotubes offer good systems in which to study the mechanism(s) involved in the crosstalk between myonuclei and mechanical signal receptors via the IF cytoskeleton. Because the nucleus is the largest mechanosensitive organelle, it would also be interesting to examine alterations of nuclear mechanical properties and stiffness caused by P1 deficiency and their consequences for cellular stability and function. Moreover, it will be of interest to investigate the role of P1 and IFs in sculpting of the nuclear membrane and maintaining its curvature via interaction with BAR domain-containing proteins, analogous to studies which revealed the importance of different plectin isoforms in maintaining the integrity of focal adhesions (41), neuromuscular junctions (64), mitochondria (65) and endothelial adherens junctions (42).
Materials and Methods
Mice
Animal studies were approved by the Federal Ministry for Science, Research and Economy (BMWFW), Vienna, Austria. All experiments were performed using material derived from at least 3-month-old male mice. The striated muscle-restricted conditional plectin knockout (MCK-Cre/cKO) as well as the isoform P1 and the isoform P1b knockout mouse lines were described previously (15–17). The desmin knockout mouse line, kindly provided by D. Paulin (Université Paris), has been described by Li et al. (18). All mice were on a C57BL/6 background; a C57BL/6 mouse line served as wild-type control. Mice bearing floxed plectin alleles in the C57BL/6 background (MCK-Cre/cKO littermates negative for Cre recombinase) showed the same features as wild-type controls.
cDNA expression plasmids
Expression plasmids encoding full-length P1, P1b and P1h, all fused to C-terminal EGFP, were previously described (14). cDNA constructs of GST-tagged endoB1 and endoB2 used for bacterial expression were kindly provided by H. McMahon (MRC Laboratory of Molecular Biology, UK).
Antibodies
For IFM, rabbit anti-plectin antiserum #46 (66); mouse monoclonal Abs (mAbs) to desmin (clone D33; Dako, Glostrup, Denmark), and rabbit antibodies (Abs) to endoB1 and endoB2 (67) (kindly provided by P. De Camilli, Yale School of Medicine, USA; and H. McMahon, MRC Laboratory of Molecular Biology, UK) were used as primary Abs; donkey Rhodamine Red anti-mouse, goat Rhodamine Red anti-rabbit, donkey Alexa 488 anti-mouse and anti-rabbit and donkey Texas-Red anti-goat IgGs (all from Jackson Immunoresearch, West Grove, PA) as secondary Abs. Streptavidin conjugated with Cy5 (Jackson Immunoresearch) was used as an additional detection reagent for teased myofibers immunostained using Vector® M.O.M.™ Immunodetection Kit (Vector Laboratories). For IB, rabbit anti-plectin antiserum #9 (66), mouse mAbs to GST tags (Sigma-Aldrich), mouse Abs to ERK1/2 and rabbit Abs to STAT1, YAP/TAZ, p38 MAPK and AMPK (all from Cell Signaling) were used as primary antibodies; goat HRPO-conjugated anti-mouse (Jackson Immunoresearch) and anti-rabbit (Vector Laboratories, Burlingame, CA) IgGs as secondary immunoreagents. Affinity-purified rabbit Abs to P1 (16); mouse mAbs to α-actinin (Sigma-Aldrich); rabbit Abs to hsp27 (Abcam) and antibodies to H3K4me3 and H3K27me3 (Merck Millipore, Darmstadt, Germany) were used for both, IFM and IB. The latter, and antibodies to histone 3 (IB), were generously provided by C. Seiser (MFPL, Austria).
Cell culture and transfection of myoblasts
Primary WT and P1−/− myoblasts were isolated from neonatal skeletal limb muscles of WT and isoform P1 knockout mice by spreading cells onto collagen-coated culture dishes, and cultivation in 20% FCS and basic fibroblast growth factor-supplemented growth medium (20). Immortalized P1−/− and P0 myoblast cell lines were established in a similar way using muscle tissues from P1−/−/p53−/− or P0/p53−/− double knockout mice; in these cases p53−/− knockout mice (WT/p53−/−) served as controls (20). To induce myoblast fusion, the cell growth medium was switched to DMEM containing 5% horse serum (differentiation medium). For transient transfection of myoblasts with cDNA expression plasmids, Amaxa nucleofector kit for human dermal fibroblasts (Lonza, Basel, Switzerland) was used.
IFM and image analysis
Cells, fixed with 4% paraformaldehyde or methanol, were permeabilized with 0.1% Triton X-100, immunolabeled and mounted in Mowiol. Preparation and immunostaining of teased fibers of limb muscles have previously been described (15). Microscopy was performed using a fluorescence laser scanning microscope (LSM710; Carl Zeiss, Jena, Germany), equipped with Plan-Apochromat 40×/1.3NA and 63×/1.4NA objective lenses, and Zeiss ZEN (2010) software. All images were processed using ImageJ (version 1.43, NIH, USA) and Photoshop CS6 (Adobe, San Jose, CA) software packages. All measurements describing the morphology (area, perimeter, shape factor and aspect ratio) of nuclei were analyzed from maximum intensity projections of Z-stack images of teased muscle fibers or cells using ImageJ or Zeiss ZEN software. To measure eu- and heterochromatin content in cultured myotubes, confocal images were recorded below fluorescence intensity saturation levels and fluorescence emission intensities of distinct nuclei were measured using ImageJ; values were normalized to total size of nuclei. Deconvolution of Z-stack images was done using Huygens Professional software (version 4.4; SVI, Hilversum, the Netherlands) and blind deconvolution tool. The 3D reconstruction of perinuclear desmin filament networks was done after deconvolution of immunostained tissue fibers or myotubes and surface rendering using the Surface Renderer tool (Huygens Professional). To calculate the volume of nuclei in myotubes, voxel intensities of Hoechst-stained nuclei were measured after deconvolution of desmin immunostained cells using the Object Analyzer tool (Huygens Professional).
Time-lapse microscopy and tracing of nuclei
To monitor the dynamics of nuclei, differentiated myotubes adhering to laminin (Sigma-Aldrich)-coated dishes (Ibidi, Munich, Germany) were washed in PBS, switched to phenol-red free differentiation medium, and subjected to time-lapse microscopy under constant 5% CO2 flow, using a Zeiss LSM710 confocal microscope equipped with a heated stage (37°C) and a Plan-Apochromat 63×/1.4NA objective lens. Images were taken at 10 s intervals during 1–2 h recordings using the Time Series tool. To analyze mobility of the nuclei along myotubes, individual nuclei were traced using the Manual Tracking plug-in (ImageJ software).
Purification of recombinant proteins and IB
GST-tagged recombinant endoB1 and endoB2, and GST-protein were expressed in Escherichia coli BL21 (DE3) and purified on Glutathione Sepharose™ 4B (GS4B) beads as described in the manufacturer's instructions (GE Healthcare Life Sciences). Recombinant proteins and cell/tissue lysates (see below) were separated using standard SDS-(6 or 10%) PAGE. For IB, proteins were transferred to nitrocellulose membranes (blocked with 5% BSA in PBS supplemented with 0.1% Tween 20) prior to incubations with primary Abs, followed by HRP-conjugated secondary Abs. Chemiluminescence was detected with Fusion FX7 system (Peqlab, Erlangen, Germany) and bands were quantified with QuantiScan v1.5 software (Biosoft, Cambridge, UK).
Preparation of cell lysates and GST pull-down assay
The preparation of cell lysates from cultivated myocytes has previously been described (20). For GST pull-down assays, differentiated myotubes were washed briefly in ice-cold PBS and homogenized in 20 mm Tris–HCl/pH 8.0, 1 mm EDTA, 200 mm NaCl and 0.5% NP40, supplemented with complete mini protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail 1 and 3 (Sigma-Aldrich). Extracts were centrifuged at 14 000g for 10 min at 4°C, and supernatants were collected and used for GST pull-down assays. Beads coupled with GST-tagged recombinant proteins were incubated (rotation) with cell lysates for 2 h at 4°C. After washing three times with 20 mm Tris–HCl/pH 7.4, 0.1 mm EDTA, 300 mm NaCl, 0.5% Nonidet P-40 and 1% Triton X-100, the beads were heated in 5× SDS sample buffer for 5 min at 95°C, and the supernatants subjected to SDS-PAGE and IB. Bait and prey proteins were detected using anti-GST and anti-plectin antibodies, respectively.
RNA isolation and qRT-PCR
Total RNA was isolated from differentiated (7 days) wild-type/P1-deficient myotubes using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. One microgram of RNA was reversely transcribed with the iScript cDNA Synthesis Kit (Bio-Rad) and 1:20 dilutions of cDNA samples were analyzed by real-time PCR using the KAPA SYBR FAST qPCR kit (Peqlab) and the iCycler IQ system (Bio-Rad). Primer sequences are listed in Supplementary Material, Figure S2. Values were normalized to housekeeping gene hypoxanthine–guanine phosphoribosyltransferase (HPRT) transcript levels.
Statistical evaluation
Comparison between two groups or values of multiple groups was performed using unpaired, two-tailed Student's t-test (α=0.05) or one-way analyses of variance (ANOVA; α = 0.05), respectively. The significance between values of individual groups and controls was subsequently determined using Tukey's post hoc test. P values of less than 0.05 were considered statistically significant.
Supplementary Material
Funding
This work was supported by Austrian Science Research Fund (FWF) Grant I413-B09 (part of the Multilocation DFG-Research Unit 1228 ‘Molecular Pathogenesis of Myofibrillar Myopathies’) and doctoral program grant W1220. Funding to pay the Open Access publication charges for this article was provided by FWF.
Supplementary Material
Acknowledgements
We thank Colin Stewart (Institute of Medical Biology, Singapore) for generously providing samples of skeletal muscle derived from nesprin-3 knockout mice, Harvey McMahon (MRC Laboratory of Molecular Biology) for expression plasmids and antibodies and Pietro De Camilli (Yale School of Medicine) for antibodies. We also thank Christian Seiser and Anna Sawicka (both Max F. Perutz Laboratories, Vienna) for providing antibodies and for helpful discussions. Special thanks go to Lilli Winter and Maria J. Castañón for support and valuable feedback.
Conflict of Interest statement. None declared.
References
- 1.Kandert S., Luke Y., Kleinhenz T., Neumann S., Lu W., Jaeger V.M., Munck M., Wehnert M., Muller C.R., Zhou Z. et al. (2007) Nesprin-2 giant safeguards nuclear envelope architecture in LMNA S143F progeria cells. Hum. Mol. Genet., 16, 2944–2959. [DOI] [PubMed] [Google Scholar]
- 2.Crisp M., Liu Q., Roux K., Rattner J.B., Shanahan C., Burke B., Stahl P.D., Hodzic D. (2006) Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol., 172, 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Q., Pante N., Misteli T., Elsagga M., Crisp M., Hodzic D., Burke B., Roux K.J. (2007) Functional association of Sun1 with nuclear pore complexes. J. Cell Biol., 178, 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hale C.M., Shrestha A.L., Khatau S.B., Stewart-Hutchinson P.J., Hernandez L., Stewart C.L., Hodzic D., Wirtz D. (2008) Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models. Biophys. J., 95, 5462–5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang N., Butler J.P., Ingber D.E. (1993) Mechanotransduction across the cell surface and through the cytoskeleton. Science, 260, 1124–1127. [DOI] [PubMed] [Google Scholar]
- 6.Dupin I., Sakamoto Y., Etienne-Manneville S. (2011) Cytoplasmic intermediate filaments mediate actin-driven positioning of the nucleus. J. Cell Sci., 124, 865–872. [DOI] [PubMed] [Google Scholar]
- 7.Dupin I., Etienne-Manneville S. (2011) Nuclear positioning: mechanisms and functions. Int. J. Biochem. Cell Biol., 43, 1698–1707. [DOI] [PubMed] [Google Scholar]
- 8.Ralston E., Lu Z., Biscocho N., Soumaka E., Mavroidis M., Prats C., Lomo T., Capetanaki Y., Ploug T. (2006) Blood vessels and desmin control the positioning of nuclei in skeletal muscle fibers. J. Cell. Physiol., 209, 874–882. [DOI] [PubMed] [Google Scholar]
- 9.Wiche G., Osmanagic-Myers S., Castañón M.J. (2015) Networking and anchoring through plectin: a key to IF functionality and mechanotransduction. Curr. Opin. Cell Biol., 32, 21–29. [DOI] [PubMed] [Google Scholar]
- 10.Wilhelmsen K., Litjens S.H., Kuikman I., Tshimbalanga N., Janssen H., van den Bout I., Raymond K., Sonnenberg A. (2005) Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J. Cell Biol., 171, 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ketema M., Wilhelmsen K., Kuikman I., Janssen H., Hodzic D., Sonnenberg A. (2007) Requirements for the localization of nesprin-3 at the nuclear envelope and its interaction with plectin. J. Cell Sci., 120, 3384–3394. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs P., Zörer M., Rezniczek G.A., Spazierer D., Oehler S., Castañón M.J., Hauptmann R., Wiche G. (1999) Unusual 5′ transcript complexity of plectin isoforms: novel tissue-specific exons modulate actin binding activity. Hum. Mol. Genet., 8, 2461–2472. [DOI] [PubMed] [Google Scholar]
- 13.Elliott C.E., Becker B., Oehler S., Castañón M.J., Hauptmann R., Wiche G. (1997) Plectin transcript diversity: identification and tissue distribution of variants with distinct first coding exons and rodless isoforms. Genomics, 42, 115–125. [DOI] [PubMed] [Google Scholar]
- 14.Rezniczek G.A., Abrahamsberg C., Fuchs P., Spazierer D., Wiche G. (2003) Plectin 5′-transcript diversity: short alternative sequences determine stability of gene products, initiation of translation and subcellular localization of isoforms. Hum. Mol. Genet., 12, 3181–3194. [DOI] [PubMed] [Google Scholar]
- 15.Konieczny P., Fuchs P., Reipert S., Kunz W.S., Zeöld A., Fischer I., Paulin D., Schröder R., Wiche G. (2008) Myofiber integrity depends on desmin network targeting to Z-disks and costameres via distinct plectin isoforms. J. Cell Biol., 181, 667–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abrahamsberg C., Fuchs P., Osmanagic-Myers S., Fischer I., Propst F., Elbe-Burger A., Wiche G. (2005) Targeted ablation of plectin isoform 1 uncovers role of cytolinker proteins in leukocyte recruitment. Proc. Natl. Acad. Sci. USA, 102, 18449–18454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winter L., Abrahamsberg C., Wiche G. (2008) Plectin isoform 1b mediates mitochondrion-intermediate filament network linkage and controls organelle shape. J. Cell Biol., 181, 903–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z., Mericskay M., Agbulut O., Butler-Browne G., Carlsson L., Thornell L.E., Babinet C., Paulin D. (1997) Desmin is essential for the tensile strength and integrity of myofibrils but not for myogenic commitment, differentiation, and fusion of skeletal muscle. J. Cell Biol., 139, 129–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rezniczek G.A., Konieczny P., Nikolic B., Reipert S., Schneller D., Abrahamsberg C., Davies K.E., Winder S.J., Wiche G. (2007) Plectin 1f scaffolding at the sarcolemma of dystrophic (mdx) muscle fibers through multiple interactions with beta-dystroglycan. J. Cell Biol., 176, 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winter L., Staszewska I., Mihailovska E., Fischer I., Goldmann W.H., Schröder R., Wiche G. (2014) Chemical chaperone ameliorates pathological protein aggregation in plectin-deficient muscle. J. Clin. Invest., 124, 1144–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Englander L.L., Rubin L.L. (1987) Acetylcholine receptor clustering and nuclear movement in muscle fibers in culture. J. Cell Biol., 104, 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Couteaux R., Pecot-Dechavassine M. (1973) Ultrastructural and cytochemical data on the mechanism of acetylcholine release in synaptic transmission. Arch. Ital. Biol., 111, 231–262. [PubMed] [Google Scholar]
- 23.Morris N.R. (2000) Nuclear migration. From fungi to the mammalian brain. J. Cell Biol., 148, 1097–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaeffer L., de Kerchove d'Exaerde A., Changeux J.P. (2001) Targeting transcription to the neuromuscular synapse. Neuron, 31, 15–22. [DOI] [PubMed] [Google Scholar]
- 25.Shah S.B., Davis J., Weisleder N., Kostavassili I., McCulloch A.D., Ralston E., Capetanaki Y., Lieber R.L. (2004) Structural and functional roles of desmin in mouse skeletal muscle during passive deformation. Biophys. J., 86, 2993–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruusgaard J.C., Liestol K., Ekmark M., Kollstad K., Gundersen K. (2003) Number and spatial distribution of nuclei in the muscle fibres of normal mice studied in vivo. J. Physiol., 551, 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper W.G., Konigsberg I.R. (1961) Dynamics of myogenesis in vitro. Anat. Rec., 140, 195–205. [DOI] [PubMed] [Google Scholar]
- 28.Capers C.R. (1960) Multinucleation of skeletal muscle in vitro. J. Biophys. Biochem. Cytol., 7, 559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaalouk D.E., Lammerding J. (2009) Mechanotransduction gone awry. Nat. Rev. Mol. Cell Biol., 10, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan J., Fukuda K., Saito M., Matsuzaki J., Kodama H., Sano M., Takahashi T., Kato T., Ogawa S. (1999) Mechanical stretch activates the JAK/STAT pathway in rat cardiomyocytes. Circ. Res., 84, 1127–1136. [DOI] [PubMed] [Google Scholar]
- 31.Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S. et al. (2011) Role of YAP/TAZ in mechanotransduction. Nature, 474, 179–183. [DOI] [PubMed] [Google Scholar]
- 32.Low B.C., Pan C.Q., Shivashankar G.V., Bershadsky A., Sudol M., Sheetz M. (2014) YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Lett., 588, 2663–2670. [DOI] [PubMed] [Google Scholar]
- 33.Gregor M., Zeöld A., Oehler S., Andrä Marobela K., Fuchs P., Weigel G., Hardie D.G., Wiche G. (2006) Plectin scaffolds recruit energy-controlling AMP-activated protein kinase (AMPK) in differentiated myofibres. J. Cell Sci., 119, 1864–1875. [DOI] [PubMed] [Google Scholar]
- 34.Morgan J.T., Pfeiffer E.R., Thirkill T.L., Kumar P., Peng G., Fridolfsson H.N., Douglas G.C., Starr D.A., Barakat A.I. (2011) Nesprin-3 regulates endothelial cell morphology, perinuclear cytoskeletal architecture, and flow-induced polarization. Mol. Biol. Cell, 22, 4324–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taranum S., Sur I., Muller R., Lu W., Rashmi R.N., Munck M., Neumann S., Karakesisoglou I., Noegel A.A. (2012) Cytoskeletal interactions at the nuclear envelope mediated by nesprins. Int. J. Cell Biol., 2012, 736524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu W., Schneider M., Neumann S., Jaeger V.M., Taranum S., Munck M., Cartwright S., Richardson C., Carthew J., Noh K. et al. (2012) Nesprin interchain associations control nuclear size. Cell. Mol. Life Sci., 69, 3493–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vannier C., Pesty A., San-Roman M.J., Schmidt A.A. (2013) The Bin/amphiphysin/Rvs (BAR) domain protein endophilin B2 interacts with plectin and controls perinuclear cytoskeletal architecture. J. Biol. Chem., 288, 27619–27637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osmanagic-Myers S., Gregor M., Walko G., Burgstaller G., Reipert S., Wiche G. (2006) Plectin-controlled keratin cytoarchitecture affects MAP kinases involved in cellular stress response and migration. J. Cell Biol., 174, 557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burgstaller G., Gregor M., Winter L., Wiche G. (2010) Keeping the vimentin network under control: cell-matrix adhesion-associated plectin 1f affects cell shape and polarity of fibroblasts. Mol. Biol. Cell, 21, 3362–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiche G., Winter L. (2011) Plectin isoforms as organizers of intermediate filament cytoarchitecture. Bioarchitecture, 1, 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregor M., Osmanagic-Myers S., Burgstaller G., Wolfram M., Fischer I., Walko G., Resch G.P., Jörgl A., Herrmann H., Wiche G. (2014) Mechanosensing through focal adhesion-anchored intermediate filaments. FASEB J., 28, 715–729. [DOI] [PubMed] [Google Scholar]
- 42.Osmanagic-Myers S., Rus S., Wolfram M., Halak D., Goldmann W.H., Bonakdar N., Fischer I., Reipert S., Zuzuarregui A., Walko G., Wiche G. (2015) Plectin reinforces vascular integrity by mediating vimentin–actin network crosstalk. J. Cell Sci. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ingber D.E. (2006) Cellular mechanotransduction: putting all the pieces together again. FASEB J., 20, 811–827. [DOI] [PubMed] [Google Scholar]
- 44.Maniotis A.J., Chen C.S., Ingber D.E. (1997) Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. USA, 94, 849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eckes B., Dogic D., Colucci-Guyon E., Wang N., Maniotis A., Ingber D., Merckling A., Langa F., Aumailley M., Delouvee A. et al. (1998) Impaired mechanical stability, migration and contractile capacity in vimentin-deficient fibroblasts. J. Cell Sci., 111, 1897–1907. [DOI] [PubMed] [Google Scholar]
- 46.Lammerding J., Schulze P.C., Takahashi T., Kozlov S., Sullivan T., Kamm R.D., Stewart C.L., Lee R.T. (2004) Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest., 113, 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.House C.M., Frew I.J., Huang H.L., Wiche G., Traficante N., Nice E., Catimel B., Bowtell D.D. (2003) A binding motif for Siah ubiquitin ligase. Proc. Natl. Acad. Sci. USA, 100, 3101–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hijikata T., Nakamura A., Isokawa K., Imamura M., Yuasa K., Ishikawa R., Kohama K., Takeda S., Yorifuji H. (2008) Plectin 1 links intermediate filaments to costameric sarcolemma through beta-synemin, alpha-dystrobrevin and actin. J. Cell Sci., 121, 2062–2074. [DOI] [PubMed] [Google Scholar]
- 49.McBride A.E., Silver P.A. (2001) State of the arg: protein methylation at arginine comes of age. Cell, 106, 5–8. [DOI] [PubMed] [Google Scholar]
- 50.Wei Y., Horng J.C., Vendel A.C., Raleigh D.P., Lumb K.J. (2003) Contribution to stability and folding of a buried polar residue at the CARM1 methylation site of the KIX domain of CBP. Biochemistry, 42, 7044–7049. [DOI] [PubMed] [Google Scholar]
- 51.Bedford M.T., Richard S. (2005) Arginine methylation an emerging regulator of protein function. Mol. Cell, 18, 263–272. [DOI] [PubMed] [Google Scholar]
- 52.Gary J.D., Clarke S. (1998) RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol., 61, 65–131. [DOI] [PubMed] [Google Scholar]
- 53.Lange A., Mills R.E., Lange C.J., Stewart M., Devine S.E., Corbett A.H. (2007) Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem., 282, 5101–5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young K.G., Pool M., Kothary R. (2003) Bpag1 localization to actin filaments and to the nucleus is regulated by its N-terminus. J. Cell Sci., 116, 4543–4555. [DOI] [PubMed] [Google Scholar]
- 55.He S., Dunn K.L., Espino P.S., Drobic B., Li L., Yu J., Sun J.M., Chen H.Y., Pritchard S., Davie J.R. (2008) Chromatin organization and nuclear microenvironments in cancer cells. J. Cell. Biochem., 104, 2004–2015. [DOI] [PubMed] [Google Scholar]
- 56.Itano N., Okamoto S., Zhang D., Lipton S.A., Ruoslahti E. (2003) Cell spreading controls endoplasmic and nuclear calcium: a physical gene regulation pathway from the cell surface to the nucleus. Proc. Natl. Acad. Sci. USA, 100, 5181–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ingber D.E. (1997) Tensegrity: the architectural basis of cellular mechanotransduction. Annu. Rev. Physiol., 59, 575–599. [DOI] [PubMed] [Google Scholar]
- 58.Postel R., Ketema M., Kuikman I., de Pereda J.M., Sonnenberg A. (2011) Nesprin-3 augments peripheral nuclear localization of intermediate filaments in zebrafish. J. Cell Sci., 124, 755–764. [DOI] [PubMed] [Google Scholar]
- 59.Ketema M., Kreft M., Secades P., Janssen H., Sonnenberg A. (2013) Nesprin-3 connects plectin and vimentin to the nuclear envelope of Sertoli cells but is not required for Sertoli cell function in spermatogenesis. Mol. Biol. Cell, 24, 2454–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arkhipov A., Yin Y., Schulten K. (2008) Four-scale description of membrane sculpting by BAR domains. Biophys. J., 95, 2806–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gundesli H., Talim B., Korkusuz P., Balci-Hayta B., Cirak S., Akarsu N.A., Topaloglu H., Dincer P. (2010) Mutation in exon 1f of PLEC, leading to disruption of plectin isoform 1f, causes autosomal-recessive limb-girdle muscular dystrophy. Am. J. Hum. Genet., 87, 834–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gostynska K.B., Nijenhuis M., Lemmink H., Pas H.H., Pasmooij A.M., Lang K.K., Castañón M.J., Wiche G., Jonkman M.F. (2015) Mutation in exon 1a of PLEC, leading to disruption of plectin isoform 1a, causes autosomal-recessive skin-only epidermolysis bullosa simplex. Hum. Mol. Genet., 24, 3155–3162. [DOI] [PubMed] [Google Scholar]
- 63.Rezniczek G., Winter L., Walko G., Wiche G (2016). Methods in Enzymology. Elsevier, doi:10.1016/bs.mie.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 64.Mihailovska E., Raith M., Valencia R.G., Fischer I., Al Banchaabouchi M., Herbst R., Wiche G. (2014) Neuromuscular synapse integrity requires linkage of acetylcholine receptors to postsynaptic intermediate filament networks via rapsyn-plectin 1f complexes. Mol. Biol. Cell, 25, 4130–4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winter L., Kuznetsov A.V., Grimm M., Zeöld A., Fischer I., Wiche G. (2015) Plectin isoform P1b and P1d deficiencies differentially affect mitochondrial morphology and function in skeletal muscle. Hum. Mol. Genet., 24, 4530–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Andrä K., Kornacker I., Jörgl A., Zörer M., Spazierer D., Fuchs P., Fischer I., Wiche G. (2003) Plectin-isoform-specific rescue of hemidesmosomal defects in plectin (–/–) keratinocytes. J. Invest. Dermatol., 120, 189–197. [DOI] [PubMed] [Google Scholar]
- 67.Farsad K., Ringstad N., Takei K., Floyd S.R., Rose K., De Camilli P. (2001) Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J. Cell Biol., 155, 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.